Abstract
Mycobacterium tuberculosis (Mtb) is the causative agent of tuberculosis (TB), an infectious disease that is a major killer worldwide. Due to selection pressure caused by the use of antibacterial drugs, Mtb is characterised by mutational events that have given rise to multi drug resistant (MDR) and extensively drug resistant (XDR) phenotypes. The rate at which mutations occur is an important factor in the study of molecular evolution, and it helps understand gene evolution. Within the same species, different protein-coding genes evolve at different rates. To estimate the rates of molecular evolution of protein-coding genes, a commonly used parameter is the ratio dN/dS, where dN is the rate of non-synonymous substitutions and dS is the rate of synonymous substitutions. Here, we determined the estimated rates of molecular evolution of select biological processes and molecular functions across 264 strains of Mtb. We also investigated the molecular evolutionary rates of core genes of Mtb by computing the dN/dS values, and estimated the pan genome of the 264 strains of Mtb. Our results show that the cellular amino acid metabolic process and the kinase activity function evolve at a significantly higher rate, while the carbohydrate metabolic process evolves at a significantly lower rate for M. tuberculosis. These high rates of evolution correlate well with Mtb physiology and pathogenicity. We further propose that the core genome of M. tuberculosis likely experiences varying rates of molecular evolution which may drive an interplay between core genome and accessory genome during M. tuberculosis evolution.
Keywords: Comparative genomics, dN/dS, molecular evolution, Mycobacterium tuberculosis
Introduction
Mycobacterium tuberculosis, a deadly pathogen
Mycobacterium tuberculosis (Mtb) is the causative agent of tuberculosis (TB), an infectious disease that is a major killer worldwide. In spite of a general decrease in the absolute number of TB deaths until 2019, TB remains a serious public health concern, exacerbated by the recent coronavirus disease 2019 (COVID-19) pandemic but also resistance of Mtb to first-line drugs. The proportion of people diagnosed with bacteriologically confirmed pulmonary TB who were resistant to rifampicin, one of the most effective first-line drugs, has been on the rise since 2018. 1 Globally in 2019, there were nearly 500 000 incident cases of rifampicin-resistant TB, of which 78% were multidrug resistant (MDR) TB. 2 MDR-TB is defined as TB that is resistant to the 2 most effective first-line drugs, isoniazid and rifampicin.
Evolutionary changes in Mtb: The case of resistance
Drug resistance often points to significant mutational events and hence evolution of Mtb. The emergence of extensively drug resistant (XDR) TB and MDR TB in KwaZulu-Natal, South Africa, was shown to have taken place via several independent evolutionary events which involved accumulation of stepwise resistance mutations over decades, for example, mutations in rpoB, pncA, rrs, gyrA. In particular, the evolution of the historical Tugela Ferry XDR isolate from initial isoniazid and streptomycin resistance to full-blown XDR spanned nearly 4 decades. Furthermore, MDR and XDR evolved de novo at least 56 and 9 independent times respectively, with concomitant appearance of novel compensatory mutations that may have restored bacterial fitness and facilitated transmission of drug-resistant strains. 3
Other studies have corroborated Mtb evolution to antibiotic resistance (AR). In an investigation of the M strain MDR-TB outbreak that occurred in Buenos Aires, Argentina, it was found that Mtb M strain evolved to XDR through accumulation of resistance mutations over the years. Many of these mutations were non-synonymous, hence suggested positive selection. The ancestor of the M strain qualified for MDR status as early as 1973. In a time frame of approximately 4 decades, this MDR Mtb strain successfully spread and evolved resistance to several additional drugs, thus gaining pre-XDR and ultimately XDR status. 4
Very recently, whole genome sequencing (WGS) was used to demonstrate the microevolution of a parental Mtb strain into clonal variants. The main driver of the microevolution was drug selective pressure, favoured by intermittent antibiotic therapy. Within 7 months, the most recent common ancestor of the Mtb strain evolved from MDR to pre-XDR status as a result of point mutations and insertions-deletions (indels) that were responsible for phenotypic resistance to a wide range of antibiotics. 5 Mutations in the form of indels and single nucleotide polymorphisms (SNPs) were further shown to contribute to Mtb genome evolution via enrichment of AR genes. 6 Finally, de novo evolution of resistance, particularly to fluoroquinolones, was suggested to have driven the development and spread of XDR-TB in Belarus, a country known to have a high incidence of MDR-TB. 7
Molecular evolutionary rates of Mtb
Mtb is characterised by extensive mutation and evolution, necessary for adaptation to its host or to selection pressure imposed by antibacterial drugs. A number of prior studies corroborate this fact. For example, comparative analysis of the genomes of drug susceptible and drug resistant Mtb isolates suggests rapid evolution of Mtb. 8 In addition, within-host microevolution of Mtb is characterised by elevated rates of nucleotide substitution which have implications for the appearance of AR. 5 On a larger scale, high levels of mutation resulting in significant genomic diversity and the rapid evolution of Mtb have consistently been identified in different regions of the world shedding light on the emergence of AR.3,4,8 -11 Among the numerous Mtb virulence factors, PE/PPE genes have been shown to evolve at high rates due to particularly high frequency of mutations.12,13 At the same time, varying rates of mutation or evolution of Mtb have been demonstrated to occur during human infection, with decreasing mutation rates through latent infection 14 and high mutation rates during active disease. 9
As outlined above, the majority of studies on Mtb evolutionary rates have focussed on genes involved in colonisation or infection dynamics, virulence, and drug resistance. However, there is paucity of information on the rates of molecular evolution of individual biological processes and molecular functions in Mtb, irrespective of the participation of these processes and functions in a specific pathogen-related mechanism. Here, we assess the evolutionary rates of select processes and functions in Mtb using the dN/dS method. dN/dS (symbolised ω) measures the ratio of the rates of non-synonymous substitutions to synonymous substitutions and is indicative of the level of natural selection acting on protein-coding genes.
Pan genome estimation of M. tuberculosis
We perform a large-scale molecular evolutionary analysis of the core genes of Mtb amongst 264 strains. Prior studies of Mtb evolutionary pangenomics have dealt with smaller numbers of strains. For example, in their core and pan genome analysis, Wan et al 15 used complete genomes of 21 Mtb strains to shed light on genetic diversity in and evolution of the Mtb Manila family. Other authors have investigated 36 strains to better understand how molecular evolution has shaped the primary (core) and secondary (accessory) genome of Mtb. 16 In another recent study, Zakham et al 17 probed the core genome and virulence determinants of 168 Mtb strains plus other human-adapted Mycobacterium species. A phylogenetic analysis based on the core genome revealed 3 different clades of Mtb strains. 17 Furthermore, although a previous study used 1595 sequenced strains, the approach and overall aim were different from what we present here since machine learning was employed to elucidate the complexity of AR evolution in the Mtb pan genome. 18
Yang et al 16 coined the term ‘super core genes’ which are defined as core genes with 2 or more copies in more than 90% of Mtb strains, and they suggested the action of evolutionary forces that changed the copy numbers. Copy number variations in turn might favour adaptation of Mtb to a human host and contribute to phenotypic differences between Mtb and M. bovis, in other words, individuality of the Mtb species. Another interesting finding is that copy number variations drive interconversion amongst super core genes, single-copy core genes, dispensable genes and strain-specific genes, testifying to a dynamic process of alteration in gene copy numbers during evolution. Importantly, the super core genes were found to code for proteins known to be involved in Mtb pathogenicity, for example, PE/PPE, virulence factors, antigens and transposases. They were also characterised by high dN/dS values, suggesting that they are likely undergoing adaptive evolution. 16 Moreover, contrary to common belief, Zakham et al 17 concluded that even the core genome contains several virulence-associated genes.
Here, we present an analysis of the molecular evolutionary rates of 815 core Mtb protein-coding genes with the aim of uncovering the nature of the genes with high rates of evolution and the biological pathways in which they are involved.
Methods
Mtb data collation using TAGOPSIN
Data retrieval and collation were performed using TAGOPSIN. 19 In brief, TAGOPSIN is a Java command line programme for rapid and systematic retrieval of select data from 7 public biological databases relevant to comparative genomics and protein structure studies. The programme allows a user to retrieve organism-centred data and assemble them in a single local database in PostgreSQL (https://www.postgresql.org). This local database constitutes a useful resource for running specific queries easily. Table 1 gives the statistics of the dataset built by TAGOPSIN for Mtb.
Table 1.
The statistics of the dataset built by TAGOPSIN for Mtb.
| No. of organisms | From Taxonomy | 2859 |
|---|---|---|
| From Nucleotide | 123 | |
| No. of curated genomes | 265 | |
| No. of CDSs | 1 122 747 | |
| No. of proteins | 2595 | |
| No. of GO terms | 1932 | |
| No. of protein domain families | 1420 | |
| No. of protein 3D structures | 1706 | |
| Approx. runtime (hours) | 28 | |
Identification of proteins by Gene Ontology term
Gene Ontology 20 (GO, version 2021-07) terms of the ‘biological process’ (BP) and ‘molecular function’ (MF) namespace that describe vital processes and functions in the cell were manually chosen using the ontology editor, Protégé. 21 The number of representative UniProtKB/Swiss-Prot proteins for each term and all its corresponding children terms were obtained from the local database. This task was done programmatically for all available strains of Mtb. Nine terms (Table 2) were selected for evaluation of the evolutionary rates of their associated genes. They are mainly terms that have at least 50 representative proteins (ie, Swiss-Prot entries) and/or that describe essential processes and functions in the cell.
Table 2.
GO terms and corresponding number of unique RefSeq gene products used in the calculation of dN/dS values.
| GO term | No. of gene products |
|---|---|
| Carbohydrate metabolic process (GO:0005975) | 38 |
| Cell wall organization or biogenesis (GO:0071554) | 25 |
| Cellular amino acid metabolic process (GO:0006520) | 58 |
| Active transmembrane transporter activity (GO:0022804) | 12 |
| Efflux transmembrane transporter activity (GO:0015562) | 3 |
| Inorganic molecular entity transmembrane transporter activity (GO:0015318) | 17 |
| Kinase activity (GO:0016301) | 28 |
| Passive transmembrane transporter activity (GO:0022803) | 9 |
| Peptidase activity (GO:0008233) | 15 |
In addition, to identify proteins with low evolutionary rates that may be of use as drug target candidates, GO terms that describe functions associated with membrane proteins were chosen since the latter are amongst the first contacts a small molecule drug would have with the cell. Hence, children terms of ‘transmembrane transporter activity (GO:0022857)’ were chosen to cover membrane proteins or enzymes (Table 2), and this term incorporates roughly 100 proteins of all available Mtb strains. Mechanisms that commonly occur in bacteria like active and passive transport, but also efflux transport in drug-resistant bacteria, were considered here. Examples of gene products in these groups include adenosine triphosphate (ATP) synthase, protein translocase, and ATP binding cassette (ABC) transporter permease. Supplemental Table S1 lists the details of these terms, in particular their GO IDs and children or parent terms.
For each one of the 9 terms and its corresponding children terms, the representative protein entries from Swiss-Prot as well as RefSeq were identified. In this step, RefSeq entries were included so as to cover a maximum number of strains of Mtb for phylogenetic analysis. The numbers of unique RefSeq gene products identified for each term and used in downstream evolutionary analyses are given in Table 2. If gene products were common or overlapping among different GO terms, this was disregarded and they were included in their respective GO terms.
Estimation of the rates of molecular evolution for select GO processes and functions
Next, maximum likelihood phylogenetic analysis was performed. In brief, for every GO term listed in Table 2, coding sequences (CDSs) were retrieved for all gene products of the 264 strains of Mtb from the local database. Because the genomic features are annotated differently, the reference genome for Mtb (genome AC ‘NC_000962’) was excluded from data selection so as to maintain consistency in the dataset and in all subsequent analyses. However, to cater for this, the genome AC ‘NC_018143’ was used for the reference laboratory strain Mtb H37Rv. Codon-based multiple sequence alignment (MSA) was then performed using PRANK 22 (version 170427). The alignment file was used to estimate a phylogenetic tree with IQ-TREE 23 (version 1.6.12). DNA models of nucleotide substitution as inferred by IQ-TREE were applied. dN/dS was computed using the null model one-ratio (‘site model M0’) as implemented in the CODEML programme of PAML 24 (version 4.9). This model does a maximum likelihood estimation of ω by averaging over all sites (codons) in the sequence and across all branches in the tree. Therefore, only a single ω value was generated and was calculated by evaluation of the tree topology specified in the tree structure file. Supplemental Figure S2 illustrates the steps in the calculation of dN/dS for each gene product.
For comparative analysis, genes known to evolve or hypothesised as evolving at high rate based on known dN/dS values or high proportion of SNPs respectively were identified from literature (Table 3). Moreover, genes that determine species-specific traits like virulence and drug resistance were hypothesised as evolving at a high rate. On the other hand, genes known to evolve or hypothesised as evolving at low rate based on known dN/dS values or participation of the gene in housekeeping biological pathways like DNA replication and cell division were also identified from literature (Table 4). These 2 sets of genes constitute our ‘reference genes’. The evolutionary rate of a query GO BP or GO MF was estimated by comparison with that of ‘reference’ genes submitted to the same phylogenetic analysis.
Table 3.
Fast-evolving reference genes of Mtb used for calculation of dN/dS values.
| Gene product (as per NCBI GenBank annotation) | Reference |
|---|---|
| Known antibiotic resistance genes | |
| DNA-directed RNA polymerase subunit beta | Cohen et al 3 , Farhat et al 25 |
| 30S ribosomal protein S12 | |
| FAD-containing monooxygenase EthA | |
| Enhanced intracellular survival protein Eis | |
| Virulence genes | |
| Type VII secretion system ESX-1 subunit EccD1 | Mikheecheva et al 26 |
| Type VII secretion system ESX-1 target EspA | Folkvardsen et al 27 , Mikheecheva et al 26 |
| Ribonuclease VapC37 | Mikheecheva et al 26 |
| Ribonuclease VapC47 | Folkvardsen et al 27 , Mikheecheva et al 26 |
| Iron import ATP-binding/permease IrtA | Mikheecheva et al 26 |
| Cell wall homoeostasis | |
| UDP-N-acetylmuramoyl-l-alanine–d-glutamate ligase | Farhat et al 25 |
| Phthiocerol type I polyketide synthase PpsA | |
| DNA replication and repair | |
| DNA-directed RNA polymerase subunit beta’ | Farhat et al 25 |
| Transcription-repair coupling factor | Folkvardsen et al 27 , Dos Vultos et al 28 |
| PE/PPE genes | |
| PE family protein PE3, PE27 | Farhat et al 25 |
| PPE family protein PPE38 | McEvoy et al 13 |
| Intermediary metabolism and respiration (includes nucleotide metabolism) | |
| Adenine phosphoribosyltransferase | Folkvardsen et al 27 , Farhat et al 25 |
| NADH-quinone oxidoreductase subunit G | Folkvardsen et al 27 , Mikheecheva et al 26 , Farhat et al 25 |
| UTP–glucose-1-phosphate uridylyltransferase | Folkvardsen et al 27 |
| Adenylate cyclase | Folkvardsen et al 27 , Farhat et al 25 |
Table 4.
Slow-evolving reference genes of Mtb used for calculation of dN/dS values.
| Gene product (as per NCBI GenBank annotation) | Rv number a | Reference |
|---|---|---|
| DNA replication, recombination and repair | ||
| Holliday junction resolvase RuvX | Rv2554c | Comas et al 29 |
| DNA gyrase subunit B | Rv0005 | |
| Uracil-DNA glycosylase | Rv2976c | |
| DNA polymerase III subunit gamma/tau | Rv3721c | |
| Transcription | ||
| Transcription termination factor Rho | Rv1297 | Comas et al 29 |
| Transcription termination/antitermination protein NusA | Rv2841c | |
| RNA polymerase sigma factor SigA | Rv2703 | |
| Translation, ribosomal structure and biogenesis | ||
| Elongation factor Tu | Rv0685 | Comas et al 29 |
| 30S ribosomal protein S3 | Rv0707 | |
| 30S ribosomal protein S4 | Rv3458c | |
| 50S ribosomal protein L22 | Rv0706 | |
| 50S ribosomal protein L23 | Rv0703 | |
| Alanine–tRNA ligase | Rv2555c | |
| Glycine–tRNA ligase | Rv2357c | |
| Metabolism and respiration | ||
| Cytochrome c oxidase subunit 3 | Rv2193 | Comas et al 29 |
| Electron transfer flavoprotein subunit beta | Rv3029c | |
| Prokaryotic ubiquitin-like protein Pup | Rv2111c | |
| Dihydroorotase | Rv1381 | |
| Cell division and/or chromosome segregation | ||
| Cell division protein FtsZ | Rv2150c | Comas et al 29 |
| Cell division protein FtsQ | Rv2151c | |
Median dN/dS values of query GO BPs and GO MFs were compared with those of the sets of reference genes. To avoid bias towards infinite or out-of-range values, dN/dS values greater than 1 were considered equal to 1. A Kruskal-Wallis test was carried out to assess the statistical significance of the observed differences in dN/dS. This was followed by pairwise comparisons using Wilcoxon’s test to identify which pairs of groups were significantly different. All statistical analyses were performed in R (version 4.1.1) and P-values <.05 were deemed statistically significant.
Estimation of the rates of molecular evolution for core genes in Mtb
The core genome is defined here as the set of genes common to all 264 strains of Mtb including the reference laboratory strain Mtb H37Rv (genome AC ‘NC_000962’). This set of genes comprises a total of 815 gene products after preprocessing. For each gene product, codon-based MSA was performed in PRANK 22 (version 170427), and the alignment file was used to estimate a phylogenetic tree in IQ-TREE 30 (version 2.1.3). dN/dS was computed using the null model one-ratio (‘site model M0’) as implemented in the CODEML programme of PAML 24 (version 4.4).
GO BP terms were assigned programmatically to each one of the 815 gene products along with the dN/dS values from the calculations described above. Where such a GO assignment was not available, for example in the case of gene products that map onto TrEMBL entries which are not covered by TAGOPSIN, GO terms were assigned based on commonness of function of gene products. Alternatively, GO BP terms were assigned manually by querying the UniProtKB (https://www.uniprot.org) and QuickGO web-based resources (https://www.ebi.ac.uk/QuickGO/) using the TrEMBL AC or RefSeq gene product name.
All gene products belonging to the same generic biological pathway were manually clustered based on the GO BP terms assigned in the previous step. GO terms that were ambiguous as well as clusters that contained less than 3 gene products were excluded from further analysis. A total of 338 gene products could thus be unambiguously classified into clusters. For each cluster of gene products, the distribution of dN/dS values was represented in a box plot. To avoid bias towards out-of-range or infinite values, dN/dS values greater than 1 were considered equal to 1. All statistical analyses were performed in R (version 4.0.4).
Identification of the pan genome
In addition to core genes, dispensable and strain-specific genes were identified so as to estimate the Mtb pan genome of 264 strains. Dispensable genes were defined as those genes that are present in at least 2 strains and at most 263 strains, while strain- specific genes were defined as those genes that are present in only 1 strain. The pan genome was estimated based on the total number of CDSs in each category and for each strain. These numbers were obtained by querying the local database by the gene product names.
Results
Mtb processes evolve at different rates
Maximum likelihood estimates of dN/dS revealed significant differences among groups of genes for all 3 Mtb BPs (Kruskal-Wallis P < .05). In all cases, there is a significant difference in dN/dS between the fast-evolving and slow-evolving reference genes, as confirmed by Wilcoxon’s test (P = .001, significance level α = .05, Figure 1). This means that the 2 sets of 20 Mtb reference genes listed in Tables 3 and 4 gave the results that concur with the initial grouping. Median dN/dS values of all groups of genes that were analysed are listed in Table 5.
Figure 1.
Box plots showing the distribution of dN/dS values for fast-evolving reference genes, slow-evolving reference genes, and genes of 3 biological processes of Mtb species, namely Cellular Amino Acid Metabolic Process (GO:0006520), Carbohydrate Metabolic Process (GO:0005975) and Cell Wall Organisation or Biogenesis (GO:0071554).
Table 5.
Median dN/dS values for all groups of genes analysed.
| GO term | Median dN/dS |
|---|---|
| Carbohydrate metabolic process | 0.248 |
| Cell wall organization or biogenesis | 0.124 |
| Cellular amino acid metabolic process | 0.354 |
| Active transmembrane transporter activity | 0.379 |
| Efflux transmembrane transporter activity | 0.052 |
| Inorganic molecular entity transmembrane transporter activity | 0.00 |
| Kinase activity | 0.353 |
| Passive transmembrane transporter activity | 0.00 |
| Peptidase activity | 0.263 |
| Reference I – Genes evolving at high rate | 0.666 |
| Reference II – Genes evolving at low rate | 0.155 |
Genes participating in the cellular amino acids (AA) metabolic process have a median ω higher than that of slow-evolving reference genes but smaller than that of fast-evolving reference genes (Kruskal-Wallis P = .00063, significance level α = .05). Pairwise comparisons showed the difference between slow-evolving genes and genes of the cellular AA metabolic process to be significant (Wilcoxon’s P = .008, α = .05, Figure 1). The difference between fast-evolving reference genes and genes of the cellular AA metabolic process was however not significant (Wilcoxon’s P = .28, α = .05).
Similarly, median ω of the carbohydrate metabolic process was larger than that of slow-evolving reference genes but smaller than that of fast-evolving reference genes (Kruskal-Wallis P = .001, α = .05). The median dN/dS value of the query genes was significantly smaller than that of fast-evolving reference genes, as revealed by Wilcoxon’s test (P = .026, α = .05, Figure 1). However, the difference between query genes and slow-evolving reference genes was not significant (Wilcoxon’s P = .303, significance level α = .05).
Furthermore, although Kruskal-Wallis test indicates a significant difference in median ω among groups of genes for the cell wall organisation or biogenesis process (P = .0038, α = .05), the difference between either set of reference genes and the query genes was not significant, as revealed by Wilcoxon’s test (P > .05, Figure 1). The only significant difference was between the 2 sets of reference genes, that is, fast-evolving genes and slow-evolving genes (Wilcoxon’s P = .001, α = .05, Figure 1). Altogether, these results indicate a significantly high rate of molecular evolution for the cellular AA metabolic process and a significantly low rate of evolution of carbohydrate metabolic process in the Mtb species.
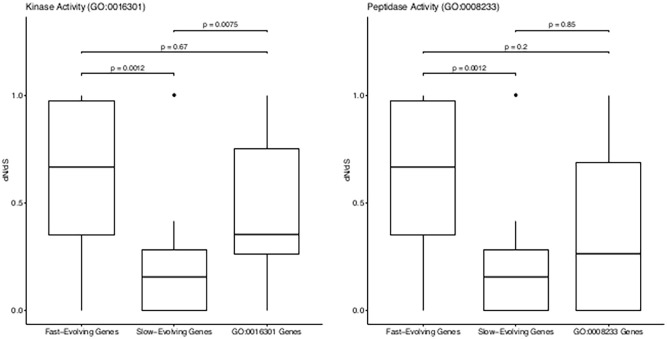
Mtb kinase function evolves at significantly high rate
There was a statistically significant difference in median dN/dS among groups of genes for the ‘kinase activity’ MF (Kruskal-Wallis P = .00053, α = .05). The median ω is greater than that of reference genes evolving at low rate and smaller than that of reference genes evolving at high rate (Figure 2). Pairwise comparisons showed a significant difference between genes of kinase activity function and slow-evolving reference genes (Wilcoxon’s P = .008, α = .05). The difference between kinase activity genes and fast-evolving reference genes was however not significant (Wilcoxon’s P = .666, α = .05, Figure 2). It can thus be deduced that genes coding for proteins functioning as kinases in the Mtb species evolve at significantly high rate, with a median dN/dS value of 0.353 (Table 5).
Figure 2.
Box plots showing distribution of dN/dS for fast-evolving reference genes, slow-evolving reference genes and genes of kinase activity (GO:0016301) and peptidase activity (GO:0008233) functions in the Mtb species.
Evolutionary rate of Mtb peptidase function
Median dN/dS was also significantly different among groups of genes of the ‘peptidase activity’ MF (Kruskal-Wallis P = .0022, α = .05). Nevertheless, only the 2 sets of reference genes were significantly different (Wilcoxon’s P = .001, α = .05). There was no significant difference between the query genes and either set of reference genes (P > .05, Figure 2).
Evolutionary rates of Mtb transmembrane transporter functions
Maximum likelihood estimates of dN/dS revealed significant differences among slow-evolving, fast-evolving and transmembrane transporter function groups of genes for the Mtb species (Kruskal-Wallis P < .05). The passive transmembrane transporter activity has a median ω lower than that of either set of reference genes (Kruskal-Wallis P = .00017, significance level α = .05), and is in fact equal to zero (Figure 3). There was a significant difference between the query genes and fast-evolving reference genes (Wilcoxon’s P = .006, α = .05), but not between the query genes and slow-evolving reference genes (Wilcoxon’s P = .381, α = .05, Figure 3).
Figure 3.
Box plots showing distribution of dN/dS for fast-evolving reference genes, slow-evolving reference genes and genes of transmembrane transporter functions (namely passive GO:0022803, active GO:0022804, inorganic molecular entity GO:0015318 and efflux GO:0015562) in the Mtb species.
Similarly, the inorganic molecular entity transmembrane transporter activity has a median ω of zero, lower than that of either set of reference genes (Kruskal-Wallis P = .00011, α = .05, Figures 3 and 4). Median dN/dS was significantly different between the query genes and fast-evolving reference genes (Wilcoxon’s P = .001, α = .05). However, the difference between query genes and slow-evolving reference genes was not significant (Wilcoxon’s P = .711, α = .05, Figure 3).
Figure 4.
(A) Distribution of dN/dS values for different metabolic pathways in Mtb and (B) distribution of dN/dS values for different metabolic pathways of Mtb.
Moreover, there was a statistically significant difference among groups of genes for the MFs ‘active transmembrane transporter activity’ (Kruskal-Wallis P = .0015, α = .05) and ‘efflux transmembrane transporter activity’ (Kruskal-Wallis P = .0016, α = .05). Nevertheless, for these MFs, the difference between the query genes and either set of reference genes was not significant (P > .05, Figure 3). Only the 2 sets of reference genes were significantly different from each other (Wilcoxon’s P = .001, α = .05, Figure 3).
Overall, these results suggest a significantly low rate of evolution of the inorganic molecular entity transmembrane transporter and passive transmembrane transporter activities. With median ω values of zero, these 2 functions possibly experience purifying selection. It is very likely that the median ω of the ‘efflux transmembrane transporter activity’ function is biased towards sample size. Indeed, the Wilcoxon test has little power with small samples. In fact, if a sample has 5 or fewer values, the Wilcoxon test will almost always give a P-value greater than .05, which is the case here for the efflux transmembrane transporter function (Table 2 and Figure 3).
Rates of evolution of Mtb core genes by pathway
A total of 33 different clusters were obtained following manual clustering of gene products along with their dN/dS values and based on the associated GO BP terms. The distribution of dN/dS for each cluster is represented in Figure 4A and B.
In general, pathways that are associated with low dN/dS values include DNA replication, carbohydrate/monosaccharide metabolic process, growth, and protein folding. For the purpose of an evolutionary analysis however, more emphasis is placed here on those pathways with high dN/dS since core genes under the action of high selection pressure have been previously identified. Examples of pathways that are characterised by high dN/dS values include nucleotide/nucleoside or base salvage, AA biosynthetic process, AA metabolic process, proteolysis and catabolic process (Figure 4A and B).
We define a benchmark for interpretation, as used by Verma et al, 31 gene products with a dN/dS between 0.35 and 0.70 are categorised as ‘moderately diversifying’ and genes with a dN/dS > 0.70 are categorised as ‘diversifying’. In addition, dN/dS values ⩾ 1.00 are indicative of positive diversification while genes with a dN/dS < 0.35 are deemed to be stabilised. 31 Within the nucleotide/ nucleoside biosynthetic process, the products ‘nicotinate-nucleotide– dimethylbenzimidazole phosphoribosyltransferase’, ‘formyltetrahydrofolate deformylase’ and ‘phosphoribosylaminoimidazolesuccinocarboxamide synthase’ have particularly high ω values. From our GO assignment, the former participates in the nucleoside biosynthetic process and undergoes positive diversification, while the latter 2 participate in the de novo inosine monophosphate (IMP) biosynthetic process. Phosphoribosylaminoimidazolesuccinocarboxamide synthase is moderately diversifying whereas formyltetrahydrofolate deformylase undergoes positive diversification. Nonetheless, our results show that the generic pathway is stabilised, with a median ω < 0.35 (Figure 4A). Furthermore, an interesting finding is that core genes involved in salvage of nucleotide, ribonucleoside and nitrogenous base likely evolve at a high rate, with all genes in this cluster being diversified (ω > 0.70). The gene coding for adenylate kinase in particular potentially undergoes positive diversification (ω > 1).
It is noteworthy that the generic pathways of AA biosynthetic process and AA metabolic process are associated with particularly high dN/dS values (Figure 4A and B). Here, dN/ dS estimates further show that genes involved in the biosynthesis of histidine tend to be the most diversified (ω > 0.35). By contrast, genes of the leucine biosynthetic process are the most stabilised with dN/dS < 0.35.
Perhaps unsurprisingly, the general pathway of lipid biosynthetic/metabolic process has high variability around the median, as indicated by the large interquartile range (Figure 4B). It is conceivable since mycobacteria possess a distinctive mycolic acid cell wall. Many genes participating in this generic pathway show signs of positive diversifying selection (ω > 1), for example, genes coding for lipoyl synthase, CDP- diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase, glycosyltransferase family 1 protein, rhamnosyl O-methyltransferase and (3R)-hydroxyacyl-ACP dehydratase subunit HadB (data not shown). The gene encoding the enzyme acyl-ACP thioesterase potentially experiences diversifying selection (ω > 0.70).
Classified as participating in ‘negative regulation of growth’, the ribonuclease VapC toxins VapC32, VapC39 and VapC46 are identified as Mtb core genes and are possibly undergoing positive diversifying selection with dN/dS values greater than 1 for all 3 gene products (Figure 4A). Again, this is plausible since VapC toxins have been reported to determine Mtb virulence. Moreover, similar to the general pathway of nucleoside/nucleotide salvage, the BP ‘proteolysis’ potentially evolves at a high rate since most gene products grouped in this cluster have ω values above 0.7 with 3 of them having an ω above 1 (median ω > 0.70, Figure 4B). Table 6 shows fast-evolving genes related to pathogenicity and resistance.
Table 6.
Fast-evolving genes related to pathogenicity and resistance.
| Fast evolving genes | Associated phenotype |
|---|---|
| VapC, VapC32, VapC39, VapC46 | Pathogenicity-related |
| Adenylate kinase | |
| Protein kinases | |
| PPE genes | |
| Genes of amino acid biosynthetic pathway | |
| Genes of carbohydrate metabolic pathway | |
| Transposases | Resistance-related |
| IS elements |
Other generic pathways that demonstrate high variability around the median include ‘catabolic process’, ‘post-translational modifications’, ‘cell cycle’, ’cofactor/coenzyme biosynthetic process’, ‘translation’ and ‘DNA-templated transcription’. Their large interquartile range shows that dN/dS spreads over high and low values, suggesting variable rates of molecular evolution. However, all these pathways have a median ω between 0.35 and 0.70 which indicates that they are moderately diversifying. Taken together, our results point to high rates of evolution with diversifying selection even among core genes that participate in housekeeping biological processes. Within the generic pathway ‘translation’ for example, many 30S and 50S ribosomal proteins were associated with high dN/dS values.
Pan genome estimation of 264 strains of Mtb
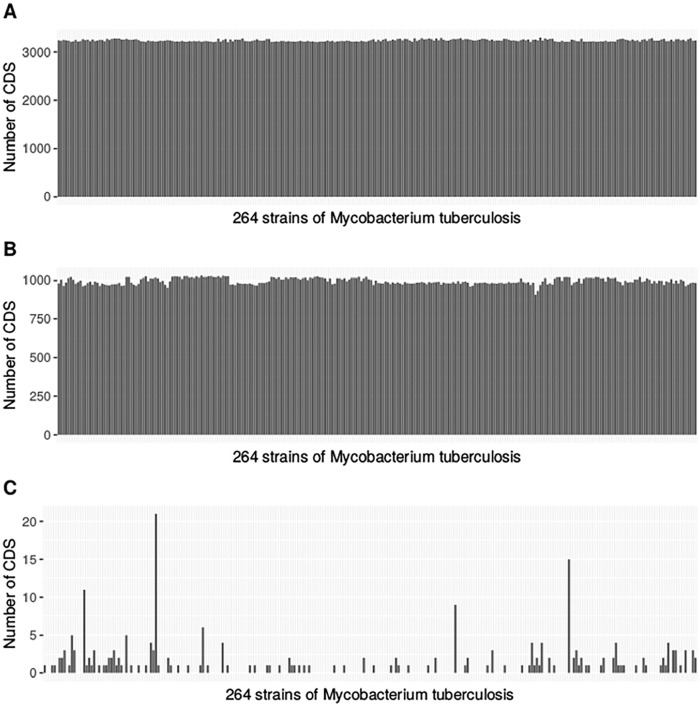
Figure 5 shows the distribution of core, dispensable and strain-specific genes across 264 strains of Mtb. Considering multiple copies of a gene, the average number of core genes is 3241 which gives a mean percentage of core genes of 76.5%. When disregarding multiple copies however, the average core gene count is 1113 and this is in fact constant throughout our Mtb population. The mean percentage of core genes in this case is 54.0%.
Figure 5.
The distribution of core (A), dispensable (B) and strain-specific (C) genes across 264 strains of Mtb.
Our analysis also shows that dispensable genes occur in multiple copies. We obtain an average of 996 dispensable genes taking into account multi-copy genes. It represents a mean percentage of 23.5% of the pangenome. Excluding multiple copies of a gene, the average dispensable gene count is 948 which is 46.0% of the pangenome.
Our preliminary analysis indicates that there are 318 genes on average which have at least 2 copies in more than 90% of strains. Actually, we find that super core genes are present in all 264 strains utilised here, and they represent a mean percentage of 28.6% of the total single-copy core gene count, that is, 1113.
Co-occurrence of dispensable genes
In order to get better insight into the evolution of the Mtb species, we carried out a preliminary analysis of the dispensable genes since they can play important roles in phenotypic variation and genome evolution. The dispensable genes identified in the previous step were analysed by groups of strains belonging to the same geographical location. In particular, the KZN strains of Mtb are known to have originated from KwaZulu-Natal, South Africa, while the Beijing/Beijing-like strains originated from the Beijing area, China. Gene products common to the KZN strains for example were obtained from the local database. A total of 955 gene products of the dispensable genome co-occurred in Mtb KZN strains.
It is noteworthy that on one hand, CDSs of some of these gene products exist in multiple copies, and on the other hand, some gene products do not appear in all KZN strains. For example, the gene product ‘beta-ketoacyl-ACP synthase’ appears in 4 out of 5 KZN strains while the product ‘beta-ketoacyl-[acyl- carrier-protein] synthase II’ appears in only 1 out of 5 strains. Importantly, we note that among the genes that exist in multiple copies are those annotated as transposases, suggesting a potential role in the pathogenicity of Mtb KZN strains.
Certainly, the function of transposases and IS elements in the evolution of antibiotic resistance and virulence is well-established. There is for instance an average of 12 copies of the gene coding for ‘IS3-like element IS987 family transposase’ per KZN strain. Moreover, a prior study revealed a high rate of molecular evolution of the PPE38 genomic region and suggested that this region is antigenic and hypervariable. 13 Our results show that the CDS of PPE38 exists in multiple copies in Mtb KZN strains. Of all PPE family proteins, PPE38 is the only one to have a multi-copy CDS, a finding that could confirm its role as Mtb virulence factor. Among other virulence factors, ‘VapC toxin family PIN domain ribonuclease’ exists in 3 KZN strains only (out of 5), pointing to a possible strain specificity of this toxin in giving a pathogenic phenotype. The ESAT-6-like proteins EsxI and EsxK belong to the ESAT-6 family of virulence factors. Here, EsxI is present in 2 copies per KZN strain while EsxK is present in only 1 strain, suggesting high rate of evolution and strain specificity respectively. Unsurprisingly, there are 2 copies of the gene product ‘multidrug-efflux transporter’ per Mtb KZN strain, which likely explains an AR phenotype.
Similarly, a total of 984 gene products co-occurring in Mtb Beijing and Beijing-like strains were obtained. In particular, the gene coding for ABC transporter ATP-binding protein/permease is present in 2 copies on average. This transporter is reported to be involved in Mtb virulence. We also note the presence of the following products in only 1 Beijing/Beijing-like strain (out of a total of 7 strains): ‘3-hydroxyacyl-thioester dehydratase HtdY’, ‘4Fe-4S dicluster domain-containing protein’, ‘AbrB/MazE/SpoVT family DNA-binding domain-containing protein’, ‘DNA or RNA helicase of superfamily II’, ‘DNA topoisomerase (ATP-hydrolysing) subunit A’, ‘IS110-like element IS1547 family transposase’, ‘PPE family protein PPE66’, ‘PPE family protein PPE67’, ‘antitoxin VapB47’, ‘multidrug transporter’, ‘ribonuclease VapC38’, among others. The majority of these products are present in 1 and the same strain, M. tuberculosis Beijing (genome AC ‘NZ_CP011510’). As with Mtb KZN, many transposases exist in multiple copies in the Beijing and Beijing-like strains, indicating a crucial role in the development of AR and virulence in Mtb Beijing. For example, each Mtb Beijing strain has on average 16 copies of the gene coding for IS3 family transposase. In addition, ESAT-6-like proteins EsxI and EsxK are present in 2 copies per Beijing or Beijing-like strain. In order to better understand which gene products confer specificity on KZN and Beijing strains, the products were distinctively identified.
Discussion
These findings point to the continuous process of evolution in genes that are part of essential pathways such as transcription and translation, amino acid and lipid metabolism. This study investigated the evolutionary rates of sets of genes involved in different pathways as per the GO grouping. Genes for the salvage nucleotides synthesis pathway, the amino acids biosynthesis and those for kinases were found to have high dN/dS values. Nucleotides are constitutively synthesised for energy and biosynthesis of nucleic acids. They are usually produced through the normal biosynthetic (de novo) processes from small precursor molecules. The nucleotide salvage pathway can also produce nucleotides from purines and pyrimidines and ensures that the organism can survive if the normal pathways are compromised. The salvage pathway is therefore an alternative way for producing vital nucleotides and can be subjected to high mutation rates without threatening the survival of the organism. Adenylate kinase is a phosphotransferase which interconverts adenosine phosphates: 2 ADP ↔ AMP + ATP. 32 This nucleotide exchange is important in energy metabolism and signalling. Several isoforms of the enzymes have been described. Adenylate kinase phosphorylates to adenine to form adenosine as part of the salvage pathway.
High rates of evolution linked to M. tuberculosis survival and pathogenicity
Here, we show that the cellular AA metabolic process and the kinase activity function likely evolve at a significantly high rate in the Mtb species. These findings correlate well with Mtb pathogenicity and survival. Indeed, several studies have highlighted the essentiality of amino acid biosynthesis and protein kinase activity during chronic infection and pathogenesis.33 -38
Mycobacteria can synthesise all 20 amino acids. 39 It can evade the host defencive mechanism of histidine starvation, by making its own through the de novo pathway. Amino acids are essential precursors of protein synthesis and for many other metabolic intermediates. Glutamate and glutamine are the main nitrogen source for other molecules. Amino acid biosynthetic pathways and protein kinases are crucial for pathogenicity. 33 There could also be a potential link with the synthesis of virulence factors, such as PE/PPE proteins of Mtb, 40 which have a conserved N-terminal domain that incorporates Pro-Glu (PE) or Pro-Pro-Glu (PPE) residues. The glutamine family amino acid metabolic process was included in evolutionary rate estimation. This family of AAs comprises arginine, glutamate, glutamine and proline. It is possible that the cellular AA metabolic process evolves at a significantly high rate in Mtb for the synthesis of diverse forms of PE/PPE proteins. Phylogenetic analysis of several amino acid biosynthesis genes has shown that gene duplication and horizontal transfer contributed to the genomes of Corynebacteria. 41 The paralogues glnA (glutamine synthase I) and ocd (ornithine cyclodeaminase) of C. efficiens were acquired by horizontal gene transfer and not by gene duplication. Gene transfer is likely to be important in the evolution of the amino acid biosynthesis enzymes. It is reasonable to say that our result of a significantly high rate of evolution of the cellular AA metabolic process correlates with Mtb physiology and pathogenicity by favouring efficient synthesis and/or biological activity of diverse virulence factors.
There is convincing evidence that kinases also play an important role in Mtb physiology and pathogenicity. Kinases are essential for protein activation through phosphorylation and are particularly important in signal transduction pathways. They have been classified into His-, Tyr- and Ser/Thr-kinases as per the amino acid they phosphorylate. There are 11 Ser/Thr kinases in Mycobacteria : Pkn-A, B, D, E, F, G, H, I, J, K and L which are differentially distributed among different species. They are organised into sub-domains and have conserved amino acids motifs. The activation loop of Ser/Thr kinases have been shown to be highly variable indicating that they interact with different molecules. The loop is phosphorylated for enzyme activity. 42 The protein tyrosine kinase PtkA is essential for growth of Mtb in macrophages, the preferred niche of the pathogen. Likewise, PknB is a serine/threonine protein kinase that is necessary for survival of Mtb both in vitro and in vivo, and for host immune evasion. It is also required to establish an infection and cause disease. 37
Low rate of molecular evolution revealed
Our findings also reveal a significantly low rate of molecular evolution for the carbohydrate metabolic process, to what is previously reported about Mtb carbohydrate metabolism in vivo and in vitro. Mtb has been classified as an obligate aerobe whereby respiration is a vital component of its physiology and is subject to change in different host micro-environments. 43 Moreover, many enzymes involved in gluconeogenesis were considered in our evolutionary analysis, and during infection carbohydrate synthesis in Mtb occurs primarily through conversion of lipid and/or AA intermediates by gluconeogenesis. 44 A mechanism of carbon co-catabolism that enables flow of carbon via both glycolysis and gluconeogenesis has also been revealed. 45 We thus expect a high rate of molecular evolution of the carbohydrate metabolic process in Mtb. Nonetheless, we hypothesise that our result of a low evolutionary rate might be relevant when the pathogen is in a state of latency.
Pangenome of Mtb
In general, our numbers of core and dispensable genes are lower than those found by other authors. This is because the addition of more strains shrinks the core gene count in favour of an increase in dispensable and strain-specific genes. However, the numbers of strain-specific genes obtained in this analysis are low compared with other studies. Here, the maximum number of strain-specific genes is 21 for the strain Mtb EAI5/NITR206. Conversely, Yang et al 16 for example estimated 97 strain-specific genes for that same strain. On the whole, they obtained higher numbers of strain-specific genes and lower numbers of dispensable genes for any given strain because they analysed much fewer strains than in our study. Notably, it can be hypothesised that genes identified as strain-specific by Yang et al have in fact spread throughout a larger population, possibly by HGT, and become dispensable genes. Certainly, other authors have demonstrated that HGT is a key mechanism that has shaped the evolution of the species via chromosomal DNA transfer. 46
A second original aspect described here is the identification of genes that potentially determine the respective specificities of Mtb KZN and Beijing/Beijing-like strains. A total of 104 and 133 gene products specific to Mtb KZN and Beijing respectively were thus identified. The PPE family proteins have been established as important Mtb virulence factors. Our analysis shows that PPE1, PPE17, PPE22, PPE32, PPE38, PPE44 and PPE50 are specific to the KZN strains and not to the Beijing or Beijing-like strains. Conversely, PPE66 and PPE67 possibly confer specificity on the Beijing/Beijing-like strains and contribute to their virulence. Moreover, among the ESAT-6 family of virulence factors, ESAT-6-like proteins EsxO, EsxP and EsxW are specific to Mtb KZN while EsxL is specific to Mtb Beijing/Beijing-like. These findings suggest that different selection pressures prevailing in different geographical regions could have caused varying levels of mutation and rates of evolution which in turn could have led to the appearance of distinct virulence factors of the PPE and ESAT-6 families in Mtb KZN and Beijing strains. It has been previously reported that PPE38 for instance is encoded by a genomic region that is characterised by rapid molecular evolution. 13 The emergence of strain-specific virulence factors is in addition to the numerous PPE and ESAT-6 proteins common to both groups of strains. A similar conclusion can be drawn for the toxins MazF3 and MazF8 which seem to be specific to Mtb KZN. Serine-threonine kinases have been shown to play important roles in Mtb pathogenicity.37,47,48 Here, a comparison of dispensable genes between Mtb KZN and Beijing/Beijing-like strains suggests that ‘Stk1 family PASTA domain-containing Ser/Thr kinase’ is specific to Mtb Beijing/Beijing-like.
Interplay between core genome and accessory genome
Our results indicate a high proportion of the dispensable genome (46.0% excluding multiple copies of a gene) relative to the core genome (54.0% excluding multiple copies). Other authors have identified a generally higher share of the core genome, albeit using fewer strains. We do obtain a high percentage of the core genome, that is, 76.5%, but only when considering multiple copies of a gene. In that case, there is a concomitant decrease in the percentage of dispensable genes (from 46.0% to 23.5% of the pangenome). It is thus conceivable that there is indeed interconversion amongst core genes and dispensable genes through copy number variations as proposed by Yang et al 16 Interconversion may also happen via changes in the number of copies of the same gene within the same strain. Besides the relative share of core and dispensable genome with respect to the pangenome, our discovery of high rates of molecular evolution amongst core genes further support this concept. It is possible that strain-specific genes also participate in such a dynamic process. Hence, during evolution of Mtb, we cannot rule out an interplay between the core genome and the accessory genome (dispensable + strain-specific) that potentially facilitates the emergence of pathogenic strains and the development of traits related to virulence. Some genes might evolve at high rate in some strains to enable the pathogen to adapt to and survive in its host, but also to cause disease. The core genome is involved probably because it encodes functions required for housekeeping cellular processes and organismal survival. One hypothesis is that genes determining pathogenicity (which were formerly part of the accessory genome) and genes necessary for pathogen survival are co-located and are co-transcribed in the Mtb core genome. The region harbouring those genes could thus be undergoing rapid evolution.
Conclusion
Here, we presented the results of an investigation of the rates of molecular evolution of select biological processes and molecular functions in Mtb. We have shown that the cellular AA metabolic process and the kinase activity function evolve at significantly high rate while the carbohydrate metabolic process evolves at significantly low rate in the Mtb species. We have supported our findings with evidence reporting that the high rates of evolution correlate well with Mtb physiology and pathogenicity.
We also corroborated the findings of previous authors by showing that, even genes of the Mtb core genome evolve at high rate and encode virulence-associated traits, and, that there is indeed an interplay between the core genome and the accessory genome that drives the evolution of Mtb. We have also shown that core genes participating in AA biosynthetic/metabolic process evolve at a high rate, and pathways like post-translational modifications, translation and DNA-templated transcription experience variable rates of molecular evolution. This study is however limited by the fact that groups of genes were analysed together under a GO term. Understanding the evolution at the single gene level would probably point to those that are mutating at differential rates under selection pressures. This can also be extended to an understanding of why some (or parts of) proteins are more likely to bear structural changes than others. Furthermore, our investigation relied on existing GO annotations and therefore newly characterised genes from recent studies have not been included here. Future work should consider such transcribed products which are yet to be annotated. 49
Supplemental Material
Supplemental material, sj-docx-1-evb-10.1177_11769343241239463 for Large-scale Pan Genomic Analysis of Mycobacterium tuberculosis Reveals Key Insights Into Molecular Evolutionary Rate of Specific Processes and Functions by Eshan Bundhoo, Anisah W Ghoorah and Yasmina Jaufeerally-Fakim in Evolutionary Bioinformatics
Acknowledgments
We acknowledge support from the University of Mauritius and the Human Heredity and Health in Africa Bioinformatics Network (H3ABioNet).
Footnotes
Author Contributions: YJF conceived the study. EB and AWG wrote scripts for data analysis and visualisation. EB designed and performed the analysis. EB wrote the original draft. AWG and YJF edited the manuscript. All authors read and approved the final manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the Higher Education Commission of Mauritius [scholarship to EB].
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplemental Material: Supplemental material for this article is available online.
References
- 1. World Health Organization. Global Tuberculosis Report 2021. World Health Organization; 2021. [Google Scholar]
- 2. World Health Organization. Global Tuberculosis Report 2020. World Health Organization; 2020. [Google Scholar]
- 3. Cohen KA, Abeel T, Manson McGuire A, et al. Evolution of extensively drug-resistant tuberculosis over four decades: whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLoS Med. 2015;12(9):e1001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eldholm V, Monteserin J, Rieux A, et al. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat Commun. 2015;6:7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fernandez Do, Porto DA, Monteserin J, Campos J, et al. Five-year microevolution of a multidrug-resistant Mycobacterium tuberculosis strain within a patient with inadequate compliance to treatment. BMC Infect Dis. 2021;21:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Godfroid M, Dagan T, Merker M, et al. Insertion and deletion evolution reflects antibiotics selection pressure in a Mycobacterium tuberculosis outbreak. PLoS Pathog. 2020;16(9):e1008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wollenberg KR, Desjardins CA, Zalutskaya A, et al. Whole-genome sequencing of Mycobacterium tuberculosis provides insight into the evolution and genetic composition of drug-resistant tuberculosis in Belarus. J Clin Microbiol. 2017;55:457-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang WF, Lu MJ, Cheng TR, et al. Genomic analysis of Mycobacterium tuberculosis isolates and construction of a Beijing lineage reference genome. Genome Biol Evol. 2020;12:3890-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hakamata M, Takihara H, Iwamoto T, et al. Higher genome mutation rates of Beijing lineage of Mycobacterium tuberculosis during human infection. Sci Rep. 2020;10:17997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Isakova J, Sovkhozova N, Vinnikov D, et al. Mutations of rpoB, katG, inhA and ahp genes in rifampicin and isoniazid-resistant Mycobacterium tuberculosis in Kyrgyz Republic. BMC Microbiol. 2018;18:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ilina EN, Shitikov EA, Ikryannikova LN, et al. Comparative genomic analysis of Mycobacterium tuberculosis drug resistant strains from Russia. PLoS One. 2013;8(2):e56577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Phelan JE, Coll F, Bergval I, et al. Recombination in pe/ppe genes contributes to genetic variation in Mycobacterium tuberculosis lineages. BMC Genomics. 2016;17:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McEvoy CR, van Helden PD, Warren RM, Gey van Pittius NC. Evidence for a rapid rate of molecular evolution at the hypervariable and immunogenic Mycobacterium tuberculosis PPE38 gene region. BMC Evol Biol. 2009;9:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colangeli R, Gupta A, Vinhas SA, et al. Mycobacterium tuberculosis progresses through two phases of latent infection in humans. Nat Commun. 2020;11:4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wan X, Koster K, Qian L, et al. Genomic analyses of the ancestral Manila family of Mycobacterium tuberculosis. PLoS One. 2017;12(4):e0175330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang T, Zhong J, Zhang J, et al. Pan-genomic study of Mycobacterium tuberculosis reflecting the primary/secondary genes, generality/individuality, and the interconversion through copy number variations. Front Microbiol. 2018;9:1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zakham F, Sironen T, Vapalahti O, Kant R. Pan and core genome analysis of 183 Mycobacterium tuberculosis strains revealed a high inter-species diversity among the human adapted strains. Antibiotics. 2021;10:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kavvas ES, Catoiu E, Mih N, et al. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat Commun. 2018;9:4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bundhoo E, Ghoorah AW, Jaufeerally-Fakim Y. TAGOPSIN: collating taxa-specific gene and protein functional and structural information. BMC Bioinformatics. 2021;22:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gene Ontology Consortium. The gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325-D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Musen MA. The protégé project: a look back and a look forward. AI Matters. 2015;1:4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Löytynoja A. Phylogeny-aware alignment with PRANK and PAGAN. Methods Mol Biol. 2021;2231:17-37. [DOI] [PubMed] [Google Scholar]
- 23. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586-1591. [DOI] [PubMed] [Google Scholar]
- 25. Farhat MR, Shapiro BJ, Kieser KJ, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet. 2013;45(10):1183-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mikheecheva NE, Zaychikova MV, Melerzanov AV, Danilenko VN. A nonsynonymous SNP catalog of Mycobacterium tuberculosis virulence genes and its use for detecting new potentially virulent sublineages. Genome Biol Evol. 2017;9(4):887-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Folkvardsen DB, Norman A, Andersen AB, et al. A major Mycobacterium tuberculosis outbreak caused by one specific genotype in a low-incidence country: exploring gene profile virulence explanations. Sci Rep. 2018;8(1):11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dos Vultos T, Mestre O, Rauzier J, et al. Evolution and diversity of clonal bacteria: the paradigm of Mycobacterium tuberculosis. PLoS One. 2008;3(2):e1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Comas I, Chakravartti J, Small PM, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42(6):498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Minh B, Schmidt H, Chernomor O, et al. IQ-TREE 2: New Models and efficient methods for phylogenetic inference in the genomic era [published correction appears in Mol Biol Evol. 2020 Aug 1;37(8):2461]. Mol Biol Evol. 2020;37:1530-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Verma H, Nagar S, Vohra S, et al. Genome analyses of 174 strains of Mycobacterium tuberculosis provide insight into the evolution of drug resistance and reveal potential drug targets. Microb Genomics. 2021;7:mgen000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dzeja P, Terzic A. Adenylate kinase and AMP signaling networks: metabolic monitoring, signal communication and body energy sensing. Int J Mol Sci. 2009;10:1729-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hasenoehrl EJ, Rae Sajorda D, Berney-Meyer L, et al. Derailing the aspartate pathway of Mycobacterium tuberculosis to eradicate persistent infection. Nat Commun. 2019;10:4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tiwari S, van Tonder AJ, Vilchèze C, et al. Arginine-deprivation-induced oxidative damage sterilizes Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2018;115:9779-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wong D, Li W, Chao JD, et al. Protein tyrosine kinase, PtkA, is required for Mycobacterium tuberculosis growth in macrophages. Sci Rep. 2018;8:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berney M, Berney-Meyer L, Wong KW, et al. Essential roles of methionine and S-adenosylmethionine in the autarkic lifestyle of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2015;112:10008-10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chawla Y, Upadhyay S, Khan S, et al. Protein kinase B (PknB) of Mycobacterium tuberculosis is essential for growth of the pathogen in vitro as well as for survival within the host. J Biol Chem. 2014;289:13858-13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Awasthy D, Gaonkar S, Shandil RK, et al. Inactivation of the ilvB1 gene in Mycobacterium tuberculosis leads to branched-chain amino acid auxotrophy and attenuation of virulence in mice. Microbiology. 2009;155:2978-2987. [DOI] [PubMed] [Google Scholar]
- 39. Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence [published correction appears in Nature 1998 Nov 12;396(6707):190]. Nature. 1998;393:537-544. [DOI] [PubMed] [Google Scholar]
- 40. Ly A, Liu J. Mycobacterial virulence factors: surface-exposed lipids and secreted proteins. Int J Mol Sci. 2020;21:3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nishio Y, Nakamura Y, Usuda Y, et al. Evolutionary process of amino acid biosynthesis in Corynebacterium at the whole genome level. Mol Biol Evol. 2004;21:1683-1691. [DOI] [PubMed] [Google Scholar]
- 42. Janczarek M, Vinardell JM, Lipa P, Karaś M. Hanks-type serine/threonine protein kinases and phosphatases in bacteria: roles in signaling and adaptation to various environments. Int J Mol Sci. 2018;19:2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baer CE, Rubin EJ, Sassetti CM. New insights into TB physiology suggest untapped therapeutic opportunities. Immunol Rev. 2015;264:327-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fieweger RA, Wilburn KM, VanderVen BC. Comparing the metabolic capabilities of bacteria in the Mycobacterium tuberculosis complex. Microorganisms. 2019;7:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ehrt S, Schnappinger D, Rhee KY. Metabolic principles of persistence and pathogenicity in Mycobacterium tuberculosis. Nat Rev Microbiol. 2018;16:496-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boritsch EC, Khanna V, Pawlik A, et al. Key experimental evidence of chromosomal DNA transfer among selected tuberculosis-causing mycobacteria. Proc Natl Acad Sci USA. 2016;113:9876-9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cowley S, Ko M, Pick N, et al. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol. 2004;52:1691-1702. [DOI] [PubMed] [Google Scholar]
- 48. Walburger A, Koul A, Ferrari G, et al. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. 2004;304:1800-1804. [DOI] [PubMed] [Google Scholar]
- 49. Li J, Singh U, Arendsee Z, Wurtele ES. Landscape of the dark transcriptome revealed through Re-mining massive RNA-seq data. Front Genet. 2021;12 :722981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-evb-10.1177_11769343241239463 for Large-scale Pan Genomic Analysis of Mycobacterium tuberculosis Reveals Key Insights Into Molecular Evolutionary Rate of Specific Processes and Functions by Eshan Bundhoo, Anisah W Ghoorah and Yasmina Jaufeerally-Fakim in Evolutionary Bioinformatics