Abstract
Cauliflower mosaic virus pregenomic 35S RNA begins with a long leader sequence containing an extensive secondary structure and up to nine short open reading frames (sORFs), 2 to 35 codons in length. To test whether any of these sORFs are required for virus viability, their start codons were mutated either individually or in various combinations. The resulting viral mutants were tested for infectivity on mechanically inoculated turnip plants. Viable mutants were passaged several times, and the stability of the introduced mutations was analyzed by PCR amplification and sequencing. Mutations at the 5′-proximal sORF A and in the center of the leader resulted in delayed symptom development and in the appearance of revertants. In the central leader region, the predicted secondary structure, rather than the sORF organization, was restored, while true reversions or second-site substitutions in response to mutations of sORF A restored this sORF. Involvement of sORF A and secondary structure of the leader in the virus replication cycle, and especially in translation of the 35S RNA via ribosome shunting, is discussed.
Cauliflower mosaic virus (CaMV) is the prototype member of the caulimoviruses, which together with the plant badnaviruses and the animal hepadnaviruses form the family of pararetroviruses (33, 35, 50). The replication of the circular, double-stranded viral DNA involves reverse transcription of a terminally redundant pregenomic RNA which is transcribed from extrachromosomal DNA copies of viral genomes in the nucleus of an infected cell. It is very probable that for all viruses of this family, reverse transcription occurs on RNA molecules that have been packaged into virion or virionlike particles (3, 43). In addition to their function as replicative intermediates, the pregenomic RNAs also serve as complex, polycistronic mRNAs and in some cases as substrates for RNA splicing (for reviews, see references 37 and 50).
For CaMV, the pregenomic RNA (35S RNA) has a length of about 8.2 kb and begins with a highly structured leader sequence of about 600 nucleotides (nt) which contains up to nine short open reading frames (sORFs) with coding potentials of 2 to 35 amino acids. The leader sequence is followed by seven longer ORFs which (with the exception of ORF VI, which is encoded by the subgenomic 19S RNA) are translated either from the 35S RNA or from its spliced derivatives, which also contain most parts of the leader sequence. Translation of these complex RNAs involves a ribosome shunt that allows the bypassing of inhibitory elements in the center of the leader (19, 22) and translation reinitiation that requires the activity of a CaMV-encoded translational transactivator (4, 20, 36, 54).
A number of important steps in the CaMV infection cycle are regulated or influenced by elements located within the leader sequence. The primer binding site (PBS) for reverse transcription is located at the 3′ end of the leader, and template switching during reverse transcription must occur within the terminal redundancy, i.e., the first 190 nt of the leader (3). A sequence near the 5′ end was shown to enhance expression from the 35S promoter at either the transcriptional or the posttranscriptional level (14, 16, 48). The leader greatly influences translation of ORFs downstream of it (2, 18), and sequence regions with an inhibitory effect on translation and regions which at least partially alleviate this inhibition have been recognized (19, 22). The leader also contains the viral polyadenylation signal, which has to be used inefficiently in the leader to allow synthesis of full-length 35S RNA (53). Also, an inefficient splice donor site is located toward the end of the leader (36). The computer-predicted hairpinlike structure of the leader (17) has been confirmed in vitro (30). A sequence located at the top of this hairpin is highly conserved among all caulimoviruses (17) and in rice tungro bacilliform virus (RTBV) (29); its function is unknown at present. Structural analysis of the leader revealed the potential for dimerization (30), which might be important for reverse transcription (cf. true retroviruses). Also, by analogy with related animal viruses, it is likely that at least parts of a packaging signal for encapsidation of the RNA for reverse transcription are located within the leader sequence.
The importance of some leader regions for isolated steps like translation (19, 22) and polyadenylation (51, 52) has been analyzed in transient expression systems, but these studies did not clarify whether structural features of the leader or the primary sequence were responsible for the observed effects. Such experiments also do not allow investigation of the correct interplay of the different functions in the viral infection cycle. An alternative approach to elucidate structure and sequence requirements is the in vivo analysis of mutant viruses. Even though the lack of a reliable single-cell infection system for caulimoviruses precludes the determination of the precise step influenced by a given mutation, the whole-plant system with several subsequent infection cycles allows the study of the evolution of the best-adapted sequences in a small-scale experiment. Evolution is enhanced by the error-prone process of replication by reverse transcription, which generates sequence variants as soon as even a limited amount of replication occurs (49). In fact, such analyses provided the first data indicating that translation of CaMV follows rules that are highly unusual for eukaryotic cells (9, 10, 55).
Here we describe an analysis of mutations of the sORFs in the leader. In a number of systems, fine regulation of gene expression is accomplished by sORFs (for reviews, see references 26, 27, and 31), and it also has been suggested that RNA usage for packaging or translation is regulated by sORFs (12, 13, 57). Testing mutations of all sORFs in the leader of the CaMV 35S RNA, either alone or in various combinations, for their effects on infectivity in repeated infection cycles revealed that the structure in the center of the leader and a translation event at sORF A are major determinants of viral competitiveness. These data provide new insight into the process of ribosome shunting, which appears to be a general mechanism utilized by different viruses, including RTBV (23), adenovirus (59), Sendai virus (7), and budgerigar fledgling disease virus (41).
MATERIALS AND METHODS
Virus and plants.
CaMV strain Ca540, which was used as a reference wild-type virus in this study, is not a natural isolate. Its genome consists of the BamHI (1927)-BstEII (127) portion of the strain S genome (which includes the leader and which was modified by the insertion of a 9-bp BamHI-SmaI linker [GATCCCGGG] behind the first ATG of ORF V [3634 to 3636]) and the complementing BstEII (127)-BamHI (1927) portion of the genome of strain CM4-184 (25, 34) bearing a natural deletion of 420 nt (spanning position 1387 to 1806) within the aphid transmission factor gene. (The numbering in this section is in accordance with that of the strain S sequence [15].) Neither the artificial insertion nor the natural deletion interferes with viral infectivity. However, the latter feature destroys aphid transmissibility. Therefore, the possibility of cross-contamination by insect-transmitted viruses during parallel analyses of different mutants could be excluded. In all mutant viruses, the S leader of Ca540 has been replaced with the strain CM4-184 leader containing the sORF mutations. In a control experiment, it was established that a wild-type Ca540 virus and its derivative with an unmodified CM4-184 leader had the same infection characteristics (i.e., latency period and severity of symptoms). The sequence differences between the leaders (see Fig. 1) allowed a clear differentiation between mutated or reverted viruses and the wild-type control virus.
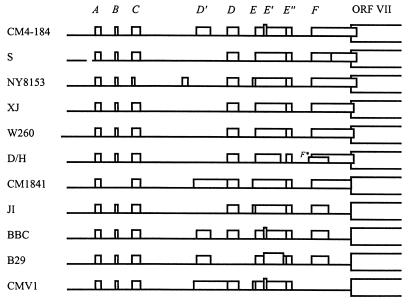
FIG. 1.
Comparison of sORFs in the 35S RNA leaders of different CaMV strains and isolates found in the GenEMBL database. The leader sequence preceding the first large ORF (ORF VII) is depicted as a thick line; the sORFs (A to F) are indicated by boxes. Vertical lines define positions of sORF start and stop codons.
Plasmid pCa540 contains the strain Ca540 DNA genome, linearized at a unique SalI site (4837) and cloned into SalI of pUC8V209, a derivative of pUC8 lacking the SmaI site (the SalI linker GGTCGACC was inserted between the SmaI and HindIII sites of pUC8).
The wild-type and mutant viruses were propagated in turnip (Brassica rapa-rapifera cv. Just Right) in a phytobox with illumination for 16 h/day at 20 or 26°C. To mechanically inoculate plants, viral DNA was amplified in Escherichia coli DH5α and excised from the plasmid vector with SalI. Then, the digested DNA was ethanol precipitated and dissolved in sterile water to a concentration of 1 μg/μl (unless otherwise stated). In the presence of a small amount of celite abrasive, 25 μl of inoculum was gently rubbed with gloved fingers onto the first and second true leaves of three-leaf-stage seedlings. Passages of infectious viruses to new plants were performed by homogenizing 0.2 g of infected tissue of young leaves in 0.5 ml of 10 mM Tris-HCl (pH 7.5)–1 mM EDTA in a sterile mortar, clarifying the extract by centrifugation, and applying 50 μl of the supernatant onto seedlings as described above. Special care was taken to avoid cross-contamination by different isolates during all procedures. Typically, symptoms appeared after 20 days for infections with cloned viral DNA and after 14 days for infections with sap from infected plants. Observed delays are expressed as a percentage of the time required for infection by the wild-type virus.
Construction of virus mutants.
All point mutations were introduced by the method of PCR-mediated site-directed mutagenesis described by Galvez and DeLumen (24). Briefly, with a pair of primers flanking the leader and a pair of complementary overlapping primers containing a mutation, two overlapping PCR products were produced. Subsequent PCR ligation of these products, in the presence of the flanking primers, yielded the whole leader sequence containing the desired mutation. The latter PCR product either was used as a template for further mutation or was cloned back into the original template plasmid, pV322. pV322 contains the fragment of the strain CM4-184 genome comprising the 35S RNA promoter, the leader, ORF VII, and the beginning of ORF I. (pV322 is described as pCAT1 in reference 4.) Flanking primers 1 and 2 corresponded, respectively, to positions 7331 to 7355, including the EcoRV site within the 35S RNA promoter, and positions 137 to 114 of the complementary strand, including the BstEII site within ORF VII. The EcoRV and BstEII sites, both unique in pV322, were used for the subcloning of PCR products. All mutations created as described above are shown in Table 1. Combinations of different mutations, used to produce the ATG-less leader mutant MALL and some other mutants shown in Table 2, were constructed by taking advantage of two restriction sites, BsmBI (7689) and AlwNI (7870), which divide the leader into three parts.
TABLE 1.
Individual sORF mutations
| sORF | Original start-codon contexta (strengthb) | Mutation
|
Restriction site created | |
|---|---|---|---|---|
| Designation | Positionc | |||
| 6172 | ||||
| | | | ||||
| A | ATA ATA ATG TGT GAG TAG (M) | MA(TTG) | A61T | SspI |
| MA(TTT) | ATG61TTT | SspI | ||
| MA(ACG) | T62C | AflIII | ||
| MA(ACA) | TG62CA | NspI | ||
| MA(gcT)d | TG64GC | |||
| MA(TGg)d | T66G | |||
| MA(stop)d | G72C | ScaI | ||
| B | TCG CTC ATG TGT (W) | MB(GAG) | AT114GA | XhoI |
| C | CTT AGT ATG TAT (M) | MC(ACT) | TG144CT | ScaI |
| D′ | GCC CAG ATG CCG (W) | MD′(CGG) | AT292CG | EagI |
| D | GAA AAG ATG CTA (M) | MD(GCG) | AT344GC | HaeII |
| E | AAT ATC ATG AAG (M) | ME(CTT) | ATG398CTT | AflII |
| E′ | GAA GAC ATG GAA (S) | ME′(AAG) | T428Ae | |
| E" | AAG ACG ATG GAA (S) | ME"(TCG) | AT467TC | AatII |
| F | AAG GGA ATG CTT (M) | MF(AAG) | T519A | HindIII |
Start codon in boldface.
Strength of context according to Kozak (38). W, weak; M, moderate; S, strong.
Numbering is from the 5′ end of the strain CM4-184 35S RNA (see Fig. 4); when a doublet or triplet is mutated, the number of the 5′-proximal nucleotide is given.
The sORF A ATG remains intact.
Occurs in strain S and in other CaMV strains lacking sORF E′ (Fig. 1).
TABLE 2.
Combinations of individual sORF mutationsa
| Mutant | Individual sORF mutations combined
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D′ | D | E | E′ | E" | F | |
| MAB | MA(TTG) | × | |||||||
| MAC | MA(TTG) | × | |||||||
| MABC | MA(TTG) | × | × | ||||||
| MDEb | × | × | × | ||||||
| MABCDEb | MA(TTG) | × | × | × | × | × | |||
| MD′DEb | × | × | × | × | |||||
| ME"F | × | × | |||||||
| MALL+D′ | MA(TTG) | × | × | × | × | × | × | × | |
| MALL | MA(TTG) | × | × | × | × | × | × | × | × |
| MALL(TTT) | MA(TTT) | × | × | × | × | × | × | × | × |
| MALL+A(ACA) | MA(ACA) | × | × | × | × | × | × | × | × |
| MALL+A(gcT) | MA(gcT)c | × | × | × | × | × | × | × | × |
| MALL+A(TGg) | MA(TGg)c | × | × | × | × | × | × | × | × |
| MALL+A(stop) | MA(stop)c | × | × | × | × | × | × | × | × |
Individual sORF mutations are described in Table 1.
Carries an in-frame ATG within sORF F (GTA545ATG), originating from strain S.
The sORF A ATG remains intact.
Mutant versions of the leader were transferred to the viral genome by replacing the EcoRV (7343)-BstEII (127) fragment of pCa540 with the corresponding one from the derivatives of pV322.
DNA preparation and PCR, cloning, and sequencing of viral progeny from infected plants.
Total DNA from infected plants was prepared by a fast method described by Padegimas et al. (45). Small pieces of young infected leaves were ground in liquid nitrogen, and about 50 to 100 mg of plant tissue powder was transferred to an Eppendorf tube containing 850 μl of TELT extraction buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 4 M LiCl, 4% [wt/vol] Triton X-100). Extraction was carried out by shaking the tube for 15 min at 37°C. Then RNA was precipitated by incubation on ice for 5 min followed by centrifugation in an Eppendorf centrifuge at 12,000 rpm for 5 min. To 700 μl of supernatant was added an equal volume of phenol-chloroform (1:1), and then this mixture was shaken for 15 min at 37°C. The centrifugation step was repeated, and 500 μl of the resulting upper phase was transferred to a new tube. The DNA was precipitated with 2 volumes of ethanol, washed with 1 ml of 70% ethanol, air dried, and dissolved in 100 μl of sterile bidistilled water.
A 0.4- to 2-μl volume of total DNA (about 20 to 100 ng) was taken for PCR analysis. PCR was carried out in a 25-μl reaction mixture (1× Vent polymerase buffer [New England Biolabs] supplemented with additional MgSO4 [to 6 mM], 200 μM each deoxynucleoside triphosphate, 30 pmol of each flanking primer [no. 1 and no. 2; see above], and 0.125 U of Vent polymerase [New England Biolabs]) for 35 cycles of 15 s at 95°C, 30 s at 65°C, and 30 s at 72°C in a DNA thermal cycler (Perkin-Elmer Cetus). The resulting PCR product of the expected size (834 nt) was extracted from low-melting-point agarose, trimmed with EcoRV and BstEII (the sites located within primers 1 and 2, respectively), purified by agarose gel electrophoresis, and cloned into the EcoRV and BstEII sites of pV322. For the introduction of the point mutations (see above), the same PCR conditions and cloning procedure were used. Sequencing of mutant versions of the leader showed that no additional substitutions occurred during these manipulations.
Plasmid DNA was sequenced either manually, using a Sequenase 2.0 kit (U.S. Biochemicals), or automatically, with AmpliTaq FS DNA polymerase and dye terminators, on an ABI 377 DNA sequencer (Perkin-Elmer Applied Biosystems). In the latter case, one run of the reaction with primer 1, annealing 80 nt upstream of the 35S RNA leader, allowed us to read reliably the whole leader sequence (612 nt in length).
Computer prediction of RNA secondary structure.
RNA secondary structure was predicted with the MFold program (Wisconsin Package, version 9.0; Genetics Computer Group, Madison, Wis.) based on the method of Zuker and Stiegler (61). The first 912 nt of the 35S RNA, comprising the leader and ORF VII, was folded at 20 or 26°C to fit our in planta experimental conditions (see the legend to Fig. 2 for more details).
FIG. 2.
Restoration of leader features on passage of sORF mutants in turnip plants.
RESULTS
Comparison of sORFs present in the 35S RNA leaders of CaMV isolates.
The 612-nt 35S RNA leader of strain CM4-184 that was mutated in this study harbors nine sORFs, designated A, B, C, D′, D, E, E′, E", and F (the start codons of sORFs E′ and E" are located within sORF E). Comparison of 11 different CaMV isolates clearly showed that most of these sORFs are well conserved (Fig. 1). Alignment of the leader sequences revealed nucleotide substitutions, short insertions, and deletions at 80 positions (13%); 36 of them fell in the sORFs. However, in only 10 cases did two or more strains have the same change at a given position within a sORF.
sORFs A and E", both three codons in length, are present in all strains and are not affected by any substitution. sORFs B, C, D, and E appeared to be less conserved, and some sORFs (including D′ and E′) are present in only some strains (Fig. 1).
sORF F is of particular interest since it is located immediately upstream of the first long ORF (VII) in a region required for ribosome shunting (22), close to the PBS. Translation of this sORF might influence the function of these latter elements and translation of ORF VII. However, sORF F turned out to be the most variable. It overlaps ORF VII by either 14 or 8 nt or not at all. In one strain (S), it contains an internal ATG codon, and in another strain (CMV1), it is missing altogether (Fig. 1). This variability suggests that sORF F is not very important for virus viability.
Strategy for studying the roles of the sORFs.
To study the requirement for translational events at the sORFs in the leader and/or their potential products, we mutated their ATG start codons. To account for a possible redundancy of sORF functions, we not only mutated every sORF individually but removed them in different combinations from the 5′ region (A, B, and C), the central region (D′, D, E, and E′), and the 3′ region (E" and F) of the leader. Furthermore, an ATG-less leader lacking all sORFs was also constructed. All individual point mutations and combinations thereof are summarized in Tables 1 and 2, respectively. In most cases, the sORF mutations introduced new restriction sites (Table 1), allowing them to be followed at different steps. Our previous results had suggested that sORF A might play an important role in the ribosome shunt mechanism (22, 48a). The conservation of this sORF among all known CaMV strains supports this hypothesis. To evaluate critical parameters of a translational event at this sORF and to determine if its 3-amino-acid product is required, several additional mutations to the sORF A sequence were created (Table 1). Mutant leaders were introduced into a non-aphid-transmissible recombinant strain of CaMV.
Almost all of the mutant viruses, including the one with a sORF-less leader, were infectious to turnip plants, resulting eventually in the development of symptoms identical to those caused by wild-type CaMV. However, a delay in symptom appearance of between 10 and 200% (with respect to the time required for symptom appearance with the wild-type virus) was observed in some cases, suggesting a deleterious effect of the respective mutations. Because suboptimal viral genomes should be outcompeted by better-adapted genomes generated during error-prone transcription or reverse transcription, virus populations from plants infected with mutant viruses were propagated by serial passage to new plants. Since the mutations might also influence structural features of the leader and thus be affected by temperature, infections were performed at both 20 and 26°C.
Routinely, on every passage, a mixture of saps from two plants infected with the same mutant virus was applied onto two new plants. The same mixture was used for analysis of the viral progeny by PCR amplification, cloning, and sequencing of the leader. In most cases, any originally observed delay of symptom appearance vanished within a few passages, suggesting that more-efficient revertants had outcompeted the original mutants. The appearance of revertants was confirmed by sequencing of individual clones from the progeny of mutant viruses. The results of infectivity studies and sequencing analyses are summarized in Fig. 2 and 3. In the following sections, we describe in detail how the sORF mutations affected the viability of the virus and which types of reversions (if any) evolved in response to the individual and combined mutations in different regions of the leader.
FIG. 3.
Restoration of leader features on passage of other sORF mutants in turnip plants.
Analysis of an ATG-free leader.
Removal of all nine sORFs from the leader (mutant MALL) resulted in a delay of symptom appearance of 50% in the primary infection cycle. The leader sequences of 14 clones isolated from the infected plants were determined, but a sequence change (from G to C) was observed in only one case at one position (nt 63). The latter mutation was not found in any later generation obtained by further inoculation of plants with this virus pool and therefore seems to be rather accidental and not to confer a selective advantage. This result indicates that CaMV without any ATG codons in the leader sequence of the 35S RNA is viable. All cis-acting signals required for replication must still be functional in this mutant. It is less certain that all signals required for proper gene expression are also still functional, since it is possible that a small proportion of reverted genomes (<7%) might have contributed protein products in trans.
To study further the eventual evolution of more-efficient viral genomes, new plants were inoculated with the sap from the primary-infected plants at either 20 or 26°C. After the first passage, there was still a significant delay in symptom appearance of 50% (20°C) or 71% (26°C). Upon further passaging, the delay time gradually decreased; after the fourth passage at 20°C, no difference from that of the wild-type virus was discernible. Already after the second passage, all recovered viral genomes contained sequence differences compared to the original inoculum (Fig. 2). In all recovered mutants, the originally altered base of the start codon of sORF D′ had been changed (from a C to an A or a T at position 292 [c292A or c292T]). In no case was a new start codon generated at this position, suggesting that either a primary sequence motif or a structural determinant was affected by the original mutation. This reversion was in almost all cases accompanied by one or two additional mutations which created one or two upstream sORFs. One of these mutations (G249A) created a sORF of nine codons that does not exist in any CaMV strain. It also slightly increases the stability of the predicted secondary structure by changing a GU base pair to AU within stem section 3 (Fig. 4) and thus may partially compensate for the reduced stability of this stem due to the original mutation in the start codon of sORF E (Fig. 4). The other mutation (t61A) restored the wild-type sORF A.
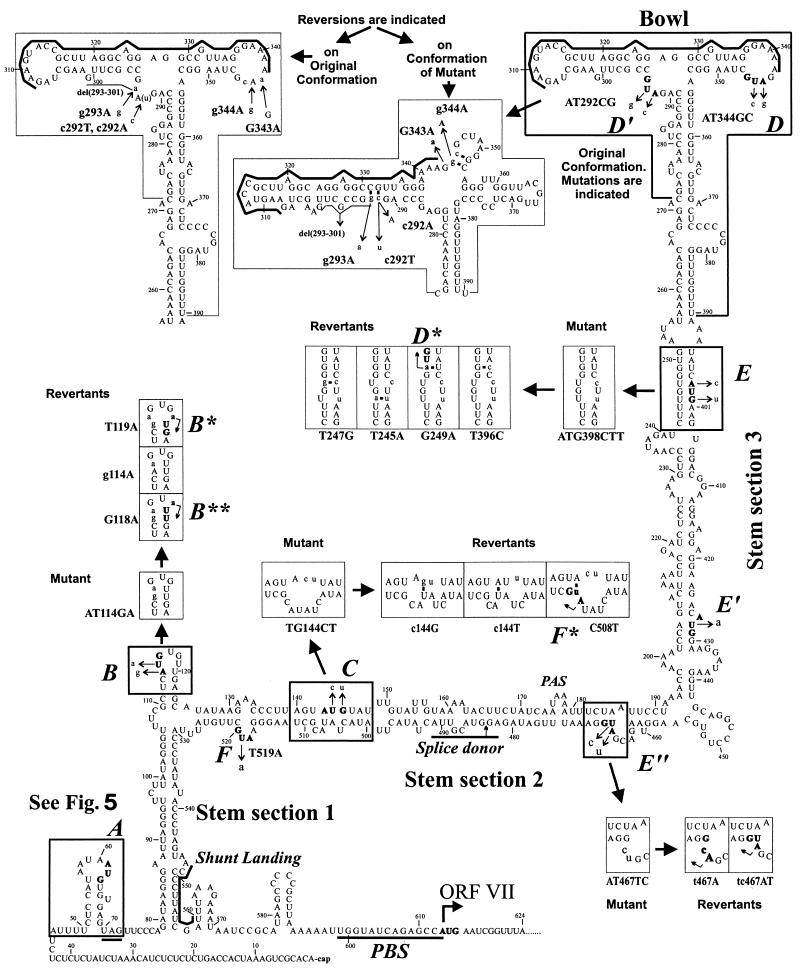
FIG. 4.
Reversions of the 35S RNA leader secondary structure and sORFs generated in planta. The MFold-predicted (20°C) hairpinlike secondary structure of the strain CM4-184 leader is shown. Here we refer to three parts of the main stem, which are divided by the side branches, as stem sections 1, 2, and 3 in ascending order and to the top of the structure as the bowl. Elements of this structure, which are influenced by the sORF mutations and the ensuing reversions, are boxed. The start codons of the sORFs are indicated in boldface and also by uppercase italic letters (A to F) along the sequence. Mutations are indicated by thin arrows. Mutant and revertant conformations of the structural elements are drawn in boxes nearby and indicated by thick arrows. Bent thin arrows within boxes show novel start codons created by the reversions in some cases (B*, B**, D*, F*). Nucleotides that do not occur in the wild-type sequence are in lowercase letters. Thick lines define the conserved purine-rich sequence of the bowl, the shunt landing sequence, the sORF A stop codon, the PBS, and the splice donor. PAS, polyadenylation signal.
While the G249A mutation was present in only a subset of the viral genomes, a reverted sORF A was present in all viral genomes recovered after further propagation. Besides the true reversion to the wild-type sORF A sequence, a second-site substitution, generating a novel ATG upstream of the original sORF A start codon, was frequently observed (A60G in Fig. 2 and 5). After the third and fourth passages, additional mutations in stem section 3 (T247G), increasing the stability of the stem (Fig. 4), and the mutated start codon of sORF D (g344A, not restoring a start codon) became predominant. A tendency for the occurrence of reversions in the region of the start codon for sORF E" existed (Fig. 2 and 4), but all other original mutations appeared to be stable. Very few mutations in other regions of the CaMV 35S RNA leader were observed in the sequences of 68 independent clones (Fig. 2), suggesting either that the replication mechanism is not so error prone after all or that selection for an active leader is tight and allows very few changes.
FIG. 5.
True and second-site reversions of sORF A generated in planta. Different types of sORF A mutations (designated on the left) and the corresponding reversions are grouped in panels. The coding sequences of the wild-type sORF A (in the uppermost panel) and the reverted sORF As are boxed. Lowercase letters indicate nucleotides that do not occur in the wild-type sequence. The mini-sORF created in place of sORF A by some mutations is also boxed (a dashed box shows the same mini-sORF which might be opened by the TTG codon). In mutant MA(stop) (in the bottom panel), sORF A extends for an additional 23 codons.
Our analysis indicated that besides sORF A, no other sORF is required for optimal leader function. Other revertants seemed to restore structural features but not sORFs. At 20°C, reversion of sORF A, strengthening of stem section 3, and some changes in the sORF D′ and D ATG regions were sufficient to produce a fully competent viral leader sequence which no longer caused a delay in symptom production. After the fourth passage at 20°C, the virus pool appeared to be quite uniform; all clones analyzed contained a wild-type sORF A, the T247G mutation in stem section 3, and the c292A and g344A mutations in the start-codon regions of sORFs D′ and D, respectively. A true reversion of the sORF E" ATG codon was evident in some clones (tc467AT [Fig. 2 and 5]). Strikingly, the sORF-producing mutation G249A was missing in this population, although it was present in the inoculum (Table 2). The pool recovered at 26°C was more variable and contained individuals with the additional sORF; in this case, no E" reversion was detected. It is unclear whether these differences represent different selection criteria at the two temperatures and whether they cause the slight delay (14%) in symptom appearance at 26°C.
Mutations at the 3′ end of the leader.
As a first step in investigating which mutations contribute to the 50% delay in symptom appearance associated with MALL, the start codons of sORFs E" and F were mutated individually or in combination (yielding ME", MF, and ME"F). These sORFs are particularly interesting because sORF E" is absolutely conserved in all CaMV strains, the start codon of E" is in a perfect sequence context for translation initiation (Table 1), and sORF F overlaps the first longer ORF downstream of the leader (Fig. 1). Both features might therefore influence downstream translation. However, all mutant viruses infected plants with the same characteristics as the wild-type virus (with the exception of ME", with which symptom appearance was slightly delayed on passage at 20°C [Fig. 3]), indicating that these sORFs do not play an important role in virus fitness. In later passages, a first-site mutation in sORF F (a519C) was observed in one clone of ME"F; however, this mutation did not restore the ATG codon and thus might instead produce a more favorable primary- or secondary-structure element.
Mutations in the center of the leader.
The start codons of sORFs D′, D, and E were mutated together in mutant MD′DE. Symptom appearance with this mutant virus was delayed by 25%, but already after two passages, reversion to wild-type appearance of infection occurred (Fig. 3). At 20°C, only those substitutions at positions 292 and 247 described above were detected. At 26°C, most clones contained the g344A reversion and two new mutations, g293A (instead of the c292A/T mutation) and T245A. The last mutation is equivalent to the T247G mutation described above in increasing the stability of stem section 3 (Fig. 4). A double mutation in the sORF E ATG region was observed in one clone (Tt399CA). This mutation would also lead to increased stability of stem section 3 if the original leader sequence were present at positions 245 and 246. In this particular case, however, the increased stability was somewhat compromised by the T245A mutation present in the same clone. In general, substitutions that increase the stability of stem section 3 are more frequently observed at 26°C than at 20°C with this mutant (Fig. 3).
The G249A mutation, which was so predominant in early revertants obtained after infection with the MALL mutant, was not found with the MD′DE mutant.
These data indicate that stem section 3 has considerable importance as a virulence determinant. However, mutation of the sORF E start codon, which destabilizes this stem, either alone or in combination with mutation of sORF D did not cause any delay in symptom appearance (ME and MDE, Fig. 3). Also, in mutants in which all ATGs besides that of D′ have been mutated (MALL+D′) or in which the ATGs of sORFs A, B, C, D, and E have been mutated (MABCDE), the only reversion after the third passage was in the start codon of sORF A (Fig. 3). However, without restoration of stem section 3 stability, these revertants are still delayed in symptom appearance by about 70% and therefore clearly have not yet evolved to an optimal configuration.
We noticed that whenever sORF D′ was present, no reversions of the mutation in the D start codon occurred.
From these data, it appears that certain structures other than sORFs have to be conserved in the center of the leader. For mutations within stem section 3, the importance of structure is obvious because of the restoration of base pairing. For mutations in sORF D′ and D, this is less apparent, since these mutations reside in single-stranded loops. However, computer predictions for the structure of this region suggest that an optimal conformation is indeed altered in the mutant RNAs, and such a mutant conformation is weakened when the observed reversions or second-site substitutions are incorporated (Fig. 4). Our computer predictions favor the structure previously termed a bowl (17). Chemical and enzymatic structural analyses have led to the suggestion of an alternative crosslike structure (30) that shares with the bowl the two stem-loops but has a different organization of the intervening sequence. One revertant, represented by two of nine clones obtained from the MD′DE mutant (Fig. 3), exhibited a deletion of 9 nt that removed a sequence that in the cross structure is largely unpaired but in the bowl structure is paired. However, structure prediction for this revertant suggests a third conformation different from all of the others (data not shown). This deletion is always accompanied by another mutation increasing the stability of stem section 3 (T396C [Fig. 4]).
Structural comparisons revealed that 4 of 11 CaMV isolates (NY8153, CM1841, JI, and CMV1) carry the base pair inversion (C397-G248 to G397-C248) maintaining the structure of stem section 3 but at the same time introducing an in-frame ATG just upstream of the start codon of sORF E (Fig. 1; note that CM1841 lacks the latter ATG because of an additional substitution [A400G]). Since sORF E itself appears to be dispensable for infectivity, the latter finding again supports the importance of the secondary structure in this region.
Mutations at the beginning of the leader.
A recurrent feature of the mutants MALL, MABCDE, MALL+D′ is the reappearance of sORF A after two passages. For these mutants, we never observed reversions in the sORF C region and only occasionally found a reversion in sORF B (g114A, in only two clones of the MALL offspring [Fig. 2]). Also, when sORF A was mutated in combination with either sORF B or C, only substitutions restoring sORF A were found (mutants MAB and MAC [Fig. 3]). In contrast, in one particular case of mutant MABC, besides restoration of sORF A, changes in the sORF B and C regions or in the complementary regions of the stem-loop structure were frequently observed (Fig. 2 and 4). The most frequent substitution in sORF B did not change the GAG codon to which the original start codon had been altered but introduced a new start codon by a T119A transversion. The new sORF was not in frame with the original sORF B. This mutation might also restore structural features at the top of the sORF B side stem (Fig. 4). The second-most-frequent change was a G118A transition, which did not alter the predicted structure (Fig. 4). This transition might create a new sORF with a non-ATG start codon (ATT in a good sequence context for translation initiation), but since such non-ATG codons are only weak initiators unless they are supported by specific structural features, an unknown structural effect is as likely as a translational effect. The third substitution restores one of the mutated bases (g114A) and might again improve the structure (Fig. 4). These changes were usually accompanied by a C508T transition, which improves the base pairing in the sORF C region and, in addition, creates a new sORF which overlaps with sORF F (Fig. 4, F*; also occurs naturally in strain D/H [Fig. 1]). In addition, we found subpopulations in the MABC offspring containing other first-site substitutions in the sORF C region (c144T or c144G, not restoring an ATG) which might improve either the base pairing (Fig. 4) or some primary sequence motif, while the sORF B region remained unchanged (Fig. 2).
A translatable sORF A is an important determinant of infectivity.
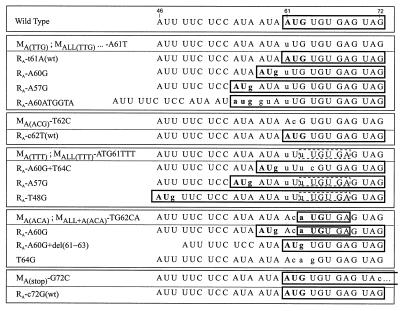
In the mutants MAB, MAC, MABC, MABCDE, MALL+D′, and MALL, the sORF A start codon has been altered from ATG to TTG. In planta, in all these mutants a sORF A was recreated by point mutations that introduced an ATG at either the original position (t61A) or one of two immediately upstream triplets (A60G and A57G). When this mutation was tested individually in mutant MA(TTG), the same substitutions occurred as well as an additional one creating an ATG by an insertion of 5 nt (Fig. 2 and 5).
Reversion to the wild-type sequence also occurred when the sORF A start codon was changed to ACG instead of TTG [mutant MA(ACG)]. After the second passage, the c62T transition was predominant in the entire progeny at both temperatures (Fig. 3). The presence of all of these revertants suggests that the restoration of a translatable sORF is important. However, symptom development with mutants MA(TTG) and MA(ACG) was delayed by only 10%, and no reversions were found in the progeny of the primary infection cycle. Thus, even though the presence of a sORF A apparently increases the competitiveness of a viral genome so much that all of the progeny from a number of different sORF A mutants have acquired it, the mutant virus is almost as infectious as a virus with a wild-type sORF A. We had altered the ATG codon in only one position and created codons that might function as non-ATG start codons (see Discussion). We therefore produced an additional mutant with a TTT triplet instead of the ATG. Symptom development with this mutant [MA(TTT)] was delayed by 20 to 45% in the primary-infected plants. This delay was only slightly reduced in the first passage. In the second passage, no delay was observed and all the sequenced progeny clones had regained a sORF A via a second-site substitution upstream of the original site (A57G or T48G [Fig. 3 and 5]). The TTT mutation in the context of a sORF free leader [mutant MALL(TTT)] showed a delay of up to 100%. Second-passage progeny showed (besides the substitutions in the center of the leader described above) a sORF A created by an A60G transition. This reversion was always accompanied by a T64C transition (Fig. 5). It is noteworthy that in the TTT mutants, second-site substitutions restoring an in-frame ATG by a single base change at each of three possible positions upstream of the original start codon were generated, whereas the three other positions at which an ATG would be out of frame were never used (Fig. 5). Such an out-of-frame start codon was artificially generated by construction of the mutant MA(ACA). In this mutant, the sORF A sequence ATG.TGT.GAG has been converted to ACA.TGT.GAG (an ATG start codon is underlined), creating the shortest possible sORF, with a start codon followed immediately by a stop codon. Initial attempts to infect plants with this mutant failed. After a fivefold increase in the DNA concentration of the inoculum and after a delay of 200%, one of four inoculated plants developed symptoms. In half of the analyzed clones, a normal sORF A was recreated by an A60G transition; in one clone, the mini sORF was destroyed by a T64G transversion (Fig. 5). The remaining five clones showed no alterations, suggesting that complementation between the reverted and the nonreverted clones can occur. However, these sequences were lost upon further passaging, as was the delay in symptom development (Fig. 3). The progeny consisted only of clones with the A60G transition, either alone or together with a further modification that removed the ACA codon, a deletion of nt 61 to 63 [del(61–63)]. Consequently, the latter revertants had a sORF A with the wild-type sequence (Fig. 5). The same ACA mutation in the context of the ATG-free leader [MALL+A(ACA)] also resulted in a long delay (175%) before symptom development. The progeny invariably contained the sORF A reversion (A60G) together with some of the typical substitutions in stem section 3 and the sORF D′-D region (Fig. 3).
These results may indicate that the nature of the sORF is of some importance. We therefore changed the second codon of sORF A from TGT to either GCT or TGG. In the GCT mutant [MA(gcT)], the codon change also improved the context of the start codon (Table 1). Even though these mutants showed a slight delay in some infection experiments, the majority of the offspring still contained the original mutations and no characteristic revertants accumulated (Fig. 3). The delay was more pronounced with the TGG mutant [MA(TGg)]. This was particularly evident when the mutants were tested in the context of the ATG-free leader, wherein the TGG mutant [MALL+A(TGg)] is still delayed for symptom development by about 50% in the second passage. In the offspring of the GCT mutant [MALL+A(gcT)], the typical reversions in stem section 3 and the sORF D′-D region are represented (Fig. 3). With the MALL+A(TGg) mutant, only the reversions in the sORF D′-D region are found.
In mutants MA(stop) and MALL+A(stop), the stop codon of sORF A was changed from TAG to TAC, which extended the sORF by an additional 23 codons. When a mutant with this elongated sORF in an otherwise ATG-free leader [MALL+A(stop)] was tested, it proved to be noninfectious. After infection with a fivefold-concentrated inoculum of a mutant with an otherwise normal leader [MA(stop)], one plant out of four developed symptoms after a 200% delay. In all of the offspring clones, the original mutant had reverted to the wild-type sequence (Fig. 3 and 5).
Microvariations in mutant progeny.
In addition to the reversions described above, we found in the offspring of mutant viruses a few occasional substitutions, deletions, and insertions, most of which occurred only once in one clone (Fig. 2, 3, and 6). Such microvariations were found in 21% of the 349 sequenced clones. Most of these were simple base changes probably caused by misincorporation during transcription or reverse transcription. The overall ratio of transitions to transversions was about 1.5. It was almost the same for both occasional and dominant substitutions. Among occasional substitutions the most frequent were the C-to-T transition and the T-to-A transversion. Overall, the pattern of nucleotide sequence variation reported here is strikingly similar to that found among CaMV isolates by Chenault and Melcher (6). The only exception is an increased frequency of A-to-G transitions in our case. However, most of them represent the dominant reversions of sORF A, for which a strong selective pressure apparently exists. Among the occasional substitutions at other sites, the A-to-G transition occurred sixfold less frequently.
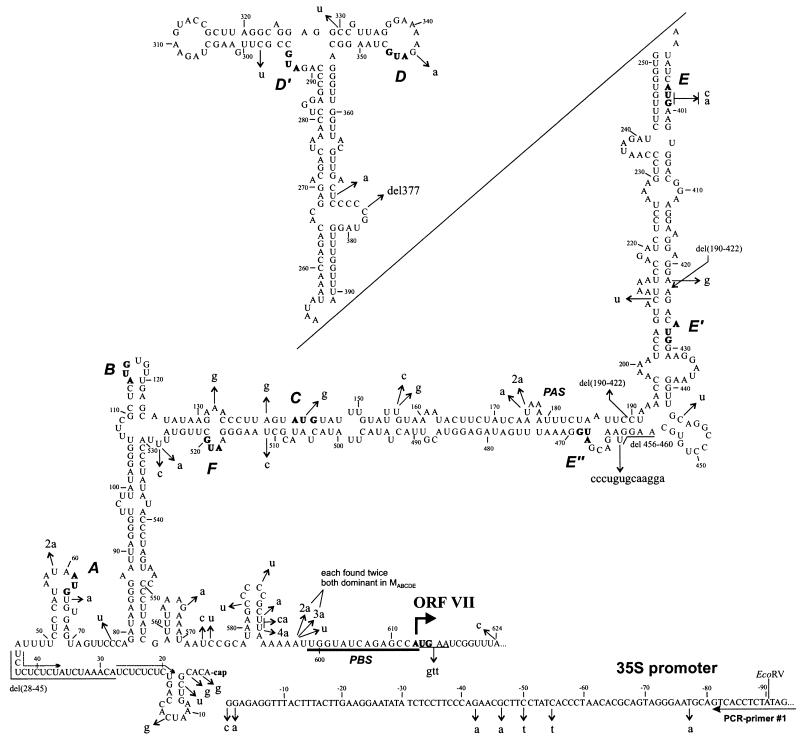
FIG. 6.
Microvariants of the 35S RNA leader found in the offspring of different sORF mutants. Nucleotide substitutions, deletions, and insertions are in lowercase letters and are indicated by thin arrows on the hairpinlike structure (disrupted in stem section 3 for the sake of compactness). A bent thick arrow shows the position of the ORF VII start codon; the CaMV PBS is underlined. The contiguous sequence of the CaMV 35S RNA promoter is shown at the bottom; the annealing site of PCR primer 1 is indicated. The start codons of sORFs are indicated in boldface and also by uppercase italic letters (A to F). Dotted arrows show a 10-nt CT repeat. PAS, polyadenylation signal.
These additional variations were rather scattered along the leader, although two hot spots of mutation appeared to exist. The first is immediately upstream of the PBS (nt 599 to 613) and the second is around the transcription start site (Fig. 6), respectively suggesting that the frequency of errors made by the reverse transcriptase is higher near the critical regions of binding and template switching (see reference 3 for a review). In particular, insertions of additional adenosines in the oligo(A) stretch at nt 592 to 597 were frequently observed (Fig. 6).
Most of the occasional substitutions are not disruptive for the hairpinlike secondary structure because they either fall into single-stranded regions or, in many cases, produce G-U base pairs (Fig. 6). Again, this suggests an importance of the secondary structure in the CaMV leader.
The most extreme mutation was found in one clone of the MALL progeny. A deletion spanning nt 190 to 422 removed half of the leader sequence (Fig. 2 and 6). It most probably occurred via premature template switching of the reverse transcriptase from position 422 to the 3′ end of the 35S RNA 3′ untranslated region, which has been mapped to position 190 or 192 (51). This deletion is probably lethal in the absence of a helper virus. Another deletion which can be explained by premature template switching spans positions 28 to 45 in one clone of the MABC progeny. It could have occurred via template switching between two consecutive 10-nt CT repeats (Fig. 6).
In two clones of the MABC progeny, after the third passage the T at position 175 within the polyadenylation signal (PAS), AATAAA, was replaced with either A or AA (Fig. 2 and 6). It has been shown that in plant protoplasts the CaMV PAS can tolerate various mutations, including the substitution of A for T (51).
It has been recently suggested that many of the sequence variations observed in virus populations are artifacts of the analysis, which generally involves many PCR amplification steps (56). By using a PCR polymerase with a proofreading activity and optimized PCR conditions (see Materials and Methods), we could minimize this problem. In fact, in our mutagenesis experiments we never found any accidental misincorporations, deletions, or insertions within about 800 nt of the amplified product, even after as many as 140 amplification cycles in total. We therefore conclude that the observed sequence variations have been generated in planta.
DISCUSSION
Many viruses use the 5′ ends of their genomic or pregenomic RNAs in processes coordinating gene expression and replication. This requires the presence of regulatory sequences or structures which may influence each other and may also vary in response to viral or cellular factors such that identical viral RNAs can perform different functions in the course of the infection cycle. Paradigms for such complex leaders can be found, e.g., in the animal picornaviruses and in some complex retroviruses. All plant pararetroviruses possess complicated leader sequences containing a number of sORFs and having extensive secondary structure. An elegant way to study sequence requirements in these elaborate and condensed systems is to allow nature to choose the optimal virus genome by forcing evolution from suboptimal genomes. This method is particularly applicable for RNA viruses and retroviruses which employ in their replication enzymes lacking proofreading capability and therefore are supposed to evolve quickly. For the pararetroviruses, these enzymes are the cellular RNA polymerase II and the virus-encoded reverse transcriptase, which is known to produce errors not only by misincorporation but also by template switching within one or between different RNA molecules (for a review, see reference 32).
We have used this approach of studying mutations in the leader sequence of CaMV, and the ensuing reversions, to address the question of whether any of the sORFs are important for viral infectivity. In the first cycle, plants were infected with in vitro-generated, uniform mutant DNA. For subsequent passages, sap from infected plants, which contained a pool of viral genomes packaged into particles, was used. Different coinfecting CaMV genotypes can be propagated together, and since recombinants between such genotypes can be generated, they must exist together in one cell. Cross-protection has been reported for CaMV; 2 days’ advantage is sufficient for an otherwise inferior virus to preclude infection by an otherwise dominant variant (60). It is likely that competition between genomes will occur mainly during the very early phases of infection, when only very low copy numbers of the given genomes exist in the nucleus. In newly developing leaves that show a characteristic mosaic pattern, all virus genomes are probably derived from the pool of virus genomes that occupies the infected area near the growing tip (1, 40). If selection occurs before this pool is built up, there will be only a small number of competitive events. Alternatively, such events may be numerous if this pool contains a large variety of clones. We very rarely observed reversions in the sequence pool analyzed after the primary inoculation, even though the considerable delays in infection rates showed that the mutant viruses were less efficient than the wild-type virus. However, this pool certainly contained a low percentage such revertants, since they invariably dominated in subsequent passages. This suggests that only a few selective infections occur during one plant infection cycle. More-effective genomes would probably be present only in some of the many cells infected in parallel and could exert their advantage only in later passages.
We have altered start codons of sORFs in the leader and, thus, certainly changed translational features. These changes and the direct effect of the sequence alterations also alter the structure of the leader, and this in turn may directly or indirectly influence reverse transcription, e.g., by affecting the packaging of RNA. CaMV genomes with an ATG-free leader proved to be infectious, although symptom appearance was delayed. Most reversions involved stem section 3 and the sequence at the top of the leader hairpin. Computer-aided predictions of structure suggest that the previously described (17) bowl conformation of this region is the most stable one (Fig. 4); however, a number of suboptimal structures with only slightly higher energy are conceivable. One of those is a crosslike structure that has recently been suggested on the basis of in vitro probing (30). The formation of either structure is disturbed by the mutations in the start codons of sORFs D′ and D (Fig. 4) and, according to the predictions obtained with the MFold program, also by mutations in stem section 3 (e.g., sORF E’s start codon). All of the ensuing reversions and second-site mutations generated in planta lead to at least partial restoration of the structure (Fig. 4). The D′ region seems to be of special importance, since it invariably reverted very quickly while reversions in the D region were detected only occasionally, later in the passaging, and never when the D′ region was intact. Also, mutations in stem section 3 were tolerated when the D′ region was left intact (MALL+D′ and MABCDE). Strikingly, reversions in response to mutations in D′ and D occurred in almost all cases directly at the mutated nucleotides and not—as for almost all other mutants—at secondary sites, which is consistent with a requirement for not only the secondary structure but also a conserved primary sequence in the region. In fact, the top of the hairpin harbors the most conserved sequence in the caulimoviral leaders (Fig. 4) (17). A similar purine-rich sequence is also found in the badnavirus RTBV (29).
The bowl region has been shown to be dispensable for the unusual translation mechanisms of CaMV (17, 19, 22) and is not known to contain sequences affecting any other step of gene expression. The infectivity of the respective mutants and the ease with which revertants are generated argue for its importance late in the infection cycle, e.g., in the production of stable virions or in (long-distance) transport.
For mutations affecting secondary structure, temperature-dependent effects are conceivable. In our experiments, we observed a slight increase in stem section 3 restoration at 26°C compared to that at 20°C, but the effect was not drastic (Fig. 2 and 3).
One of the frequently occurring mutations (G249A) found in early-passage progeny of the MALL mutant only slightly improved the predicted stability of stem section 3 by altering a G-U base pair to A-U. This mutation, however, also created a new sORF of nine codons. This particular mutation was dominant only in a pool of viral sequences that contained only a few members with an intact sORF A or with a reversion in the D region. It was outcompeted later by other revertants containing a sORF A, reversions in the D region, and another stem section 3-restoring mutation (T247G [Fig. 2]). It remains to be investigated whether the novel sORF had a function in altering the ribosome flow on this otherwise ATG-free leader or whether the effect of the mutation was only structural.
In other cases, very few sORFs were created by reversions. Since most start codons had been mutated at two positions, direct reversions would have been difficult, but usually nearby codons could have been used to create an ATG codon by single-site mutations. It therefore appears that none of the sORFs is absolutely required for virus infectivity. However, all mutants tested acquired a sORF A after at most two additional passages. ATG-creating mutations occurred at different sites at or upstream of the original sORF A ATG (Fig. 5), suggesting that the presence of a sORF per se, rather than a structural feature, provides for the selective advantage while the length and the N-terminal coding potential of the sORF is not important. Sequence changes within the wild-type sORF A were also tolerated [MA(TGg), MALL+A(TGg), MA(gcT), and MALL+A(gcT)]. The TGG mutant, however, showed a slightly increased delay period length and therefore may be impaired in infectivity. In the different mutants that lack sORF A, ATG reversions occurred at every one of the three nearby codons at which an ATG in frame with the original one could be generated by a single-site mutation. Significantly, none of the three nearby out-of-frame possibilities was used (Fig. 5). The introduction of an out-of-frame ATG immediately followed by a stop codon [MA(ACA) and MALL+A(ACA)] resulted in a dramatic decrease in fitness. Mutant MA(ACA) evolved either by a knockout of the out-of-frame ATG by point mutation to AGG or, in a dominant subpopulation, by creation of an in-frame ATG at an upstream position. Furthermore, the artificial ACA codon was deleted from some genomes of the latter progeny. This is an indication that either the additional start codon was detrimental even when located within the sORF A or the mutated sORF A coding sequence was suboptimal. It is noteworthy that about half of the progeny of the primary infection with MA(ACA) retained the mutant sequence, suggesting that these genomes could be propagated together with the revertant ones although they were severely compromised when present alone. This, together with the severely reduced infectivity and the very early occurrence (and selection) of revertants, led us to conclude that the mutation affected an aspect of gene expression and that these mutants contain all of the cis-acting sequences necessary for replication and transport.
The infection characteristics of a mutant in which the stop codon of sORF A was altered [MA(stop)] were similar to those of MA(ACA). This mutation causes an elongation of sORF A from 4 to 27 codons and resulted in only occasional infection, and only after a 200% delay period. In all sequenced clones, a true reversion with recreation of the stop codon had occurred. Interestingly, a reversion was detected only at the original position and not at various other nearby codons at which a stop codon could have been generated by a single-site mutation. This is in contrast to the different second-site mutations restoring a sORF A start codon and may indicate that termination of sORF A translation at the original site is critical, either positionally or mechanistically. All functional sORFs A terminate in GAGTAG (Fig. 5). It remains to be investigated whether the type of termination signal, the last codon, or the position of the signal with respect to other leader regions (e.g., stem section 1; see below) is important. In other systems, unknown features of sORF coding potential or termination have been shown to influence the control of translation by such sORFs. In particular, reinitiation capacity could be influenced (see reference 31 for a review). We have shown previously that the sORF A region is important for CaMV ORF VII translation by the ribosome shunt mechanism in artificial reporter gene constructs (22) and also that sORFs can be important for the reinitiation mechanism activated by the CaMV transactivator protein on polycistronic mRNAs (20, 21). The effects of the different mutations and reversions on these translation processes are currently being investigated.
The apparent advantage of sORF A revertants raises the questions of why most of the sORF A mutants are infectious with only a slight delay and why revertants are selected only in later passages. We assume that the important feature in sORF A may still function partially even in most of the mutants. The strongly structured stem section 1, 15 nt downstream of the sORF A start codon (Fig. 4), is suitably located to enhance translation initiation at codons in an unfavorable sequence context or at non-ATG codons (39). Therefore, a certain level of translation initiation should occur at the ATG codons that have been changed at only one position (TTG or ACG [28]) or at the naturally occurring ATA codon immediately upstream of sORF A (the latter codon might be used when the ATG is replaced with noninitiating triplet ACA or TTT). This low level of initiation could be sufficient for the sORF to exert its function if a mechanism like ribosome stalling is involved. Stalling seems to be the mechanism by which a sORF in the leader of the human cytomegalovirus gpUL4 RNA controls downstream translation (5) and the Neurospora crassa arg-2 gene sORF attenuates translation in the presence of arginine (58). Such a mechanism might be also responsible for the effects of other sORFs (see, e.g., references 8, 42 and 44). Stalling depends on sequence features but is not significantly affected by the initiation frequency. In the case of CaMV, stalling might be caused by the sequence around the termination codon, which was unchanged in all functional sORFs A. It can be further proposed that stalling of the translating or terminating ribosome near the sORF A stop codon is an initial step of the ribosome shunt mechanism. Strikingly, the predicted secondary structure brings sORF A into the close spatial vicinity of the shunt landing sequence (22) that is involved in the formation of the base of stem section 1 (Fig. 4). Stalling of the ribosome before dissociation, which might be further prompted by the close proximity of stem section 1, might allow a physical interaction of the 40S ribosomal subunit and the landing sequence. In this model, precise positioning of the sORF A stop codon 6 nt upstream of the base of the stem should be critical, which is consistent with our in planta findings. Structural comparison reveals strikingly similar positioning of a 5′-proximal sORF near the base of large hairpin structures in the pregenomic RNA leaders of different plant pararetroviruses (48a). Significantly, in RTBV RNA (which has been shown to utilize a ribosome shunt mechanism [3]), a precisely defined shunt landing sequence is partially involved in the formation of the hairpin base (on the descending strand) and a 5′-proximal sORF 1 terminates in GAGTAG 7 nt upstream of the base of the hairpin. This similarity to CaMV strongly supports the proposed model of ribosome shunting.
Besides sORF A, the only other sORF absolutely conserved in all CaMV strains is sORF E". Its removal had no apparent detrimental effect, but interestingly, true revertants were obtained in one experiment (MALL after four passages) even though two mutations are required for the sORF E" restoration. This sORF is present in a region to which a cellular protein of unknown function binds (11), and its importance remains to be investigated.
Mutations in the leader sequence, which could not be connected to reversion events due to the incorporated mutations, were observed at a surprisingly low frequency even after four passages. This suggests either that the selection for functional genomes is tight and few alterations are tolerated or that the polymerase is more precise than has been assumed. The notion that reverse transcriptases are error prone is mainly derived from studies of retroviral enzymes. However, even for retroviruses, the error frequency of the reverse transcriptase is different for different viruses and seems to depend at least in part on the nature of amino acid X in the conserved YXDD center (47). In mammalian viruses, a certain error frequency may be an evolutionary advantage because it allows the generation of new variants which may escape immune surveillance. In plants, virus defense mechanisms are less well studied. They may also include the recognition of a specific protein (46), but this seems not to be a general mechanism, and therefore rapid alteration of sequence information may not be as advantageous.
ACKNOWLEDGMENTS
We are grateful to Matthias Müller, Sandra Corsten, and Diana Lewetag for excellent technical assistance. Special thanks go to Herbert Angliker for expert sequencing work. We thank Witold Filipowicz and Helen Rothnie for critical reading of the manuscript and Diana Dominguez, Lyubov Ryabova, and Etienne Herzog for helpful discussions. M.M.P. thanks his wife, Natalia Kolman, for understanding and moral support and Konstantin Skryabin for his steady help and interest.
This work was partially supported by an EMBO East-European postdoctoral fellowship to M.M.P.
REFERENCES
- 1.Al-Kaff N S, Covey S N. Unusual accumulations of cauliflower mosaic virus in local lesions, dark green leaf tissue, and roots of infected plants. Mol Plant-Microbe Interact. 1996;9:357–363. [Google Scholar]
- 2.Baughman G, Howell S H. Cauliflower mosaic virus 35S RNA leader region inhibits translation of downstream genes. Virology. 1988;167:125–135. doi: 10.1016/0042-6822(88)90061-x. [DOI] [PubMed] [Google Scholar]
- 3.Bonneville J-M, Hohn T. A reverse transcriptase for cauliflower mosaic virus: state of the art, 1992. In: Skalka N, Goff S, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 357–390. [Google Scholar]
- 4.Bonneville J-M, Sanfaçon H, Fütterer J, Hohn T. Posttranscriptional trans-activation in cauliflower mosaic virus. Cell. 1989;59:1135–1143. doi: 10.1016/0092-8674(89)90769-1. [DOI] [PubMed] [Google Scholar]
- 5.Cao J, Geballe A P. Inhibition of nascent-peptide release at translation termination. Mol Cell Biol. 1996;16:7109–7114. doi: 10.1128/mcb.16.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chenault K D, Melcher U. Patterns of nucleotide sequence variation among cauliflower mosaic virus isolates. Biochimie. 1994;76:3–8. doi: 10.1016/0300-9084(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 7.Curran J, Kolakofsky D. Scanning independent ribosomal initiation of the Sendai virus X protein. EMBO J. 1988;7:2869–2874. doi: 10.1002/j.1460-2075.1988.tb03143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damiani R D, Jr, Wessler S R. An upstream open reading frame represses expression of Lc, a member of the R/B family of maize transcriptional activators. Proc Natl Acad Sci USA. 1993;90:8244–8248. doi: 10.1073/pnas.90.17.8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon L K, Hohn T. Initiation of translation of the cauliflower mosaic virus genome from a polycistronic mRNA: evidence from deletion mutagenesis. EMBO J. 1984;3:2731–2736. doi: 10.1002/j.1460-2075.1984.tb02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon L K, Jiricny J, Hohn T. Oligonucleotide directed mutagenesis of the cauliflower mosaic virus genome DNA using a repair-resistant nucleoside analogue: identification of an agnogene initiation codon. Gene. 1986;41:225–231. doi: 10.1016/0378-1119(86)90102-2. [DOI] [PubMed] [Google Scholar]
- 11.Dominguez D I, Hohn T, Schmidt-Puchta W. Cellular proteins bind to multiple sites of the leader region of cauliflower mosaic virus 35S RNA. Virology. 1996;226:374–383. doi: 10.1006/viro.1996.0665. [DOI] [PubMed] [Google Scholar]
- 12.Donzé O, Damay P, Spahr P-F. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donzé O, Spahr P-F. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowson-Day M J, Ashurst J L, Mathias S F, Watts J W. Plant viral leaders influence expression of a reporter gene in tobacco. Plant Mol Biol. 1993;23:97–109. doi: 10.1007/BF00021423. [DOI] [PubMed] [Google Scholar]
- 15.Franck A, Guilley H, Jonard G, Richards K, Hirth L. Nucleotide sequence of cauliflower mosaic virus DNA. Cell. 1980;21:285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- 16.Fütterer J, Bonneville J-M, Gordon K, DeTapia M, Karlsson S, Hohn T. Expression from polycistronic cauliflower mosaic virus pregenomic RNA. In: McCarthy J E G, Tuite M F, editors. Post-transcriptional control of gene expression. NATO ASI series. H49. Berlin, Germany: Springer-Verlag; 1990. pp. 347–357. [Google Scholar]
- 17.Fütterer J, Gordon K, Bonneville J-M, Sanfaçon H, Pisan B, Penswick J, Hohn T. The leading sequence of caulimovirus large RNA can be folded into a large stem-loop structure. Nucleic Acids Res. 1988;16:8377–8390. doi: 10.1093/nar/16.17.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fütterer J, Gordon K, Pfeifer P, Sanfaçon H, Pisan B, Bonneville J-M, Hohn T. Differential inhibition of downstream gene expression by the cauliflower mosaic virus 35S RNA leader. Virus Genes. 1989;3:45–55. doi: 10.1007/BF00301986. [DOI] [PubMed] [Google Scholar]
- 19.Fütterer J, Gordon K, Sanfaçon H, Bonneville J-M, Hohn T. Positive and negative control of translation by the leader sequence of cauliflower mosaic virus pregenomic 35S RNA. EMBO J. 1990;9:1697–1707. doi: 10.1002/j.1460-2075.1990.tb08293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fütterer J, Hohn T. Translation of a polycistronic mRNA in the presence of the cauliflower mosaic virus transactivator protein. EMBO J. 1991;10:3887–3896. doi: 10.1002/j.1460-2075.1991.tb04958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fütterer J, Hohn T. Role of an upstream open reading frame in the translation of polycistronic mRNAs in plant cells. Nucleic Acids Res. 1992;20:3851–3857. doi: 10.1093/nar/20.15.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fütterer J, Kiss-László Z, Hohn T. Non-linear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- 23.Fütterer J, Potrykus I, Bao Y, Li L, Burns T M, Hull R, Hohn T. Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J Virol. 1996;70:2999–3010. doi: 10.1128/jvi.70.5.2999-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galvez A F, DeLumen B O. Generation of an Nco I restriction site for translation fusions using PCR-mediated, site-directed mutagenesis. Plant Mol Biol Rep. 1995;13:232–242. [Google Scholar]
- 25.Gardner R C, Howarth A J, Hahn P, Brown-Luedi M, Shepherd R J, Messing J. The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing. Nucleic Acids Res. 1981;9:2871–2888. doi: 10.1093/nar/9.12.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geballe A P. Translational control mediated by upstream AUG codons. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 173–197. [Google Scholar]
- 27.Geballe A P, Morris D R. Initiation codons within 5′-leaders of mRNA as regulators of translation. Trends Biochem Sci. 1994;19:159–164. doi: 10.1016/0968-0004(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 28.Gordon K, Fütterer J, Hohn T. Efficient initiation of translation at non-AUG triplets in plant cells. Plant J. 1992;2:809–813. [PubMed] [Google Scholar]
- 29.Hay J M, Jones M C, Blakebrough M L, Dasgupta I, Davies J W, Hull R. An analysis of the sequence of an infectious clone of rice tungro bacilliform virus, a plant pararetrovirus. Nucleic Acids Res. 1991;19:2615–2621. doi: 10.1093/nar/19.10.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hemmings-Mieszczak M, Steger G, Hohn T. Alternative structures of the cauliflower mosaic virus 35S RNA leader: implications for viral expression and replication. J Mol Biol. 1997;267:1075–1088. doi: 10.1006/jmbi.1997.0929. [DOI] [PubMed] [Google Scholar]
- 31.Hinnebusch A G. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 199–244. [Google Scholar]
- 32.Hohn T. Recombination of plant pararetroviruses. In: Paszkowski J, editor. Homologous recombination and gene silencing in plants. Dordrecht, The Netherlands: Kluwer Academic Press; 1994. pp. 25–38. [Google Scholar]
- 33.Hohn T, Fütterer J. The proteins and functions of plant pararetroviruses: knowns and unknowns. Crit Rev Plant Sci. 1997;16:133–161. [Google Scholar]
- 34.Howarth A J, Gardner R C, Messing J, Shepherd R J. Nucleotide sequence of naturally occurring deletion mutants of cauliflower mosaic virus. Virology. 1981;112:678–685. doi: 10.1016/0042-6822(81)90313-5. [DOI] [PubMed] [Google Scholar]
- 35.Hull R, Covey S N. Retroelements: propagation and adaptation. Virus Genes. 1995;11:105–118. doi: 10.1007/BF01728652. [DOI] [PubMed] [Google Scholar]
- 36.Kiss-László Z, Blanc S, Hohn T. Splicing of cauliflower mosaic virus 35S RNA is essential for viral infectivity. EMBO J. 1995;14:3552–3562. doi: 10.1002/j.1460-2075.1995.tb07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiss-László Z, Hohn T. Pararetro- and retrovirus RNA: splicing and the control of nuclear export. Trends Microbiol. 1996;4:480–485. doi: 10.1016/s0966-842x(97)82909-5. [DOI] [PubMed] [Google Scholar]
- 38.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leisner S M, Turgeon R, Howell S H. Long distance movement of cauliflower mosaic virus in infected turnip plants. Mol Plant-Microbe Interact. 1992;5:41–47. [Google Scholar]
- 41.Li J. Molecular analysis of late gene expression in budgerigar fledgling disease virus. Ph.D. thesis. Giessen, Germany: University of Giessen; 1996. [Google Scholar]
- 42.Lohmer S, Maddaloni M, Motto M, Salamini F, Thompson R D. Translation of the mRNA of the maize transcriptional activator Opaque-2 is inhibited by upstream open reading frames present in the leader sequence. Plant Cell. 1993;5:65–73. doi: 10.1105/tpc.5.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mesnard J M, Carriere C. Comparison of packaging strategy of retroviruses and pararetroviruses. Virology. 1995;213:1–6. doi: 10.1006/viro.1995.1540. [DOI] [PubMed] [Google Scholar]
- 44.Michelet B, Lukaszewicz M, Dupriez V, Boutry M. A plant plasma membrane proton-ATPase gene is regulated by development and environment and shows signs of a translational regulation. Plant Cell. 1994;6:1375–1389. doi: 10.1105/tpc.6.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Padegimas L, Shulga O A, Skryabin K G. Screening of transgenic plants with polymerase chain reaction. Engl Transl Mol Biol (Moscow) 1993;27:583–585. [PubMed] [Google Scholar]
- 46.Padgett H S, Beachy R N. Analysis of a tobacco mosaic strain capable of overcoming N gene-mediated resistance. Plant Cell. 1993;5:577–586. doi: 10.1105/tpc.5.5.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandey V N, Kaushik N, Rege N, Sarafianos S G, Yadav P N S, Modak M J. Role of methionine 184 of human immunodeficiency virus type 1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 48.Pierce D A, Mettler I J, Lachmansingh A R, Pomeroy L M, Weck E A, Mascarenhas D. Effect of 35S leader modifications on promoter activity. In: Key J L, McIntosh L, editors. Plant gene systems and their biology. New York, N.Y: Alan R. Liss, Inc.; 1987. pp. 301–310. [Google Scholar]
- 48a.Pooggin, M. M. Unpublished data.
- 49.Riederer M A, Grimsley N H, Hohn B, Jiricny J. The mode of cauliflower mosaic virus propagation in the plant allows rapid amplification of viable mutant strains. J Gen Virol. 1992;73:1449–1456. doi: 10.1099/0022-1317-73-6-1449. [DOI] [PubMed] [Google Scholar]
- 50.Rothnie H M, Chapdelaine Y, Hohn T. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv Virus Res. 1994;44:1–67. doi: 10.1016/s0065-3527(08)60327-9. [DOI] [PubMed] [Google Scholar]
- 51.Rothnie H M, Reid J, Hohn T. The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3′-end formation in plants. EMBO J. 1994;13:2200–2210. doi: 10.1002/j.1460-2075.1994.tb06497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanfaçon H, Brodmann P, Hohn T. A dissection of the cauliflower mosaic virus RNA polyadenylation signal. Genes Dev. 1991;5:141–149. doi: 10.1101/gad.5.1.141. [DOI] [PubMed] [Google Scholar]
- 53.Sanfaçon H, Hohn T. Proximity to the promoter inhibits recognition of cauliflower mosaic virus polyadenylation signal. Nature. 1990;346:81–84. doi: 10.1038/346081a0. [DOI] [PubMed] [Google Scholar]
- 54.Scholthof H B, Gowda S, Wu F C, Shepherd R J. The full-length transcript of a caulimovirus is a polycistronic mRNA whose genes are trans activated by the product of gene VI. J Virol. 1992;66:3131–3139. doi: 10.1128/jvi.66.5.3131-3139.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sieg K, Gronenborn B. Abstracts of the NATO Advanced Studies Institute/FEBS advanced course on structure and function of plant genomes, Porto Portese, Italy. 1982. Introduction and propagation of foreign DNA in plants using cauliflower mosaic virus as a vector; p. 154. [Google Scholar]
- 56.Smith D B, McAllister J, Casino C, Simmonds P. Virus ‘quasispecies’: making a mountain out of a molehill? J Gen Virol. 1997;78:1511–1519. doi: 10.1099/0022-1317-78-7-1511. [DOI] [PubMed] [Google Scholar]
- 57.Sonstegard T S, Hackett P B. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J Virol. 1996;70:6642–6652. [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Sachs M S. Ribosome stalling is responsible for arginine-specific translation attenuation in Neurospora crassa. Mol Cell Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yueh A, Schneider R J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 60.Zhang X S, Melcher U. Competition between isolates and variants of cauliflower mosaic virus in infected turnip plants. J Gen Virol. 1989;70:3427–3437. [Google Scholar]
- 61.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]