Abstract
Adenovirus type 12 (Ad12) infection of human cells induces four chromosomal fragile sites corresponding to the U1 small nuclear RNA (snRNA) genes (the RNU1 locus), the U2 snRNA genes (RNU2), the U1 snRNA pseudogenes (PSU1), and the 5S rRNA genes (RN5S). Ad12-induced fragility of the RNU2 locus requires U2 snRNA transcriptional regulatory elements and viral early functions but not viral replication or integration, or chromosomal sequences flanking the RNU2 locus. We now show that Ad12 cannot induce the RNU1, RNU2, or PSU1 fragile sites in Saos-2 cells lacking the p53 and retinoblastoma (Rb) proteins but that viral induction of fragility is rescued in these cells when the expression of wild-type p53 or selected hot-spot mutants (i.e., V143A, R175H, R248W, and R273H) is restored by transient expression or stable retroviral transduction. We also observed weak constitutive fragility of the RNU1 and RNU2 loci in cells belonging to xeroderma pigmentosum complementation groups B and D (XPB and XPD) which are partially defective in the ERCC2 (XPD) and ERCC3 (XPB) helicase activities shared between the repairosome and the RNA polymerase II basal transcription factor TFIIH. We propose a model for Ad12-induced chromosome fragility in which interaction of p53 with the Ad12 E1B 55-kDa transforming protein (and possibly E4orf6) induces a p53 gain of function which ultimately perturbs the RNA polymerase II basal transcription apparatus. The p53 gain of function could interfere with chromatin condensation either by blocking mitotic shutdown of U1 and U2 snRNA transcription or by phenocopying global or local DNA damage. Specific fragilization of the RNU1, RNU2, and PSU1 loci could reflect the unusually high local concentration of strong transcription units or the specialized nature of the U1 and U2 snRNA transcription apparatus.
Fragile sites are nonstaining gaps in metaphase chromosomes caused by incompletely condensed chromatin or, more rarely, by frank chromosome breaks. Fragile sites frequently colocalize with recurrent cancer breakpoints (55), with preferential targets for viral integration (62, 89), with sites of sister chromatid exchange (19, 21; but see reference 86), and with sites of chromosome translocations in primate lineages (55). Although these observations support the view that fragile sites are recombinogenic, it is important to distinguish between physical fragility (as assayed cytologically) and genetic instability (as assayed by rearrangements on an individual or evolutionary time scale). Physical fragility might be directly responsible for the observed genetic fragility, for example, by rendering the DNA more accessible to the recombination apparatus, but it is also possible that both physical and genetic fragility reflect some other underlying phenomenon which facilitates recombination and disrupts chromatin packing.
It has been known for 30 years that infection of human primary embryonic kidney cells at a low multiplicity with adenovirus type 12 (Ad12), but not adenovirus 2 or 5, induces four sites of metaphase chromosome fragility (94); at a higher multiplicity both viruses induce generalized chromosome fragility (67, 94). The four Ad12 chromosome “modification” sites were subsequently found to colocalize with four tandemly repeated multigene families (6, 49, 50, 73): the human U1 small nuclear RNA (snRNA) genes at 1p36 (the RNU1 locus), the U2 snRNA genes at 17q21-22 (RNU2), an ancient family of U1 snRNA pseudogenes at 1q12-q22 (PSU1), and the 5S rRNA genes at 1q42-43 (RN5S). Ad12 efficiently induces fragility of the RNU1, RNU2, and PSU1 loci in several different cell lines (4, 16, 20, 42, 67); RN5S fragility has not been visualized except in primary human embryonic kidney cells (94) and may be weak, cell type specific, or abolished by immortalization. Colocalization of the four Ad12-inducible fragile sites with highly transcribed, tandemly repeated multigene families encoding small, abundant structural RNAs suggested that concentrated transcriptional activity might somehow interfere with local chromatin condensation (49); however, it was puzzling that none of the four modification sites appeared to be constitutively fragile in the absence of viral infection, and it was not known what viral or cellular functions were required to induce fragility.
Cytological colocalization of the four virally inducible fragile sites with four tandemly repeated multigene families strongly suggested that the small RNA genes themselves were necessary and sufficient for virally induced fragility. We (4) and others (20, 42) subsequently demonstrated that an artificial tandem array of active, but not inactive, U2 snRNA genes creates a new Ad12-inducible fragile site. Thus, Ad12-induced fragility of the RNU2 locus requires the minimal U2 snRNA transcription unit but not the chromosomal sequences flanking the RNU2 locus (44, 60) nor any other sequences from the 6.1-kb U2 repeat unit, including the CT microsatellite (43) and the solo long terminal repeat (45, 60).
The best-characterized fragile sites appear to be caused by defects in DNA replication which interfere, either directly or indirectly, with subsequent chromatin packing (reviewed in reference 79). Such chromatin-packing defects can be induced by a variety of drugs, including aphidicolin (which directly inhibits the DNA polymerases α, δ, and ɛ) and methotrexate (which affects replication by causing an imbalance in the deoxynucleotide triphosphate pools). Alternatively, packing defects can be caused by chromosomal alterations such as the CGG triplet repeat expansion, which locally retards DNA replication in the FRAXA form of fragile X syndrome (28) and potentiates CpG hypermethylation (25). In none of these instances is it clear why incomplete or delayed DNA replication, or hypermethylation, would interfere with chromatin condensation; the initiation of chromatin condensation might be coupled to the completion of DNA replication, or independent cell cycle controls might initiate chromatin condensation in late G2 phase regardless of whether DNA replication is truly complete. Here we examine a very different kind of chromosome fragility which may be caused by a defect in transcription rather than a defect in replication.
Stable expression of the E1 transforming region of Ad12 is sufficient to induce constitutive fragility of the RNU2 locus in a somatic-cell hybrid containing an intact human chromosome 17 in a mouse background (15). The E1 region of both Ad12 and Ad2/5 consists of two independent transcription units, although the Ad2/5 system is far better studied. Alternative splicing of the Ad2/5 E1A transcript generates 13S, 12S, and 9S mRNAs encoding related multifunctional proteins; E1B encodes 19- and 55-kDa proteins in overlapping reading frames. The E1A proteins bind pRB- and p300/CBP-family proteins (10). In addition, both the 12S- and 13S-encoded E1A proteins can inhibit the p53 transactivation (75), and the 13S-encoded E1A protein can relieve p53 repression (32). E1A-induced, p53-dependent apoptosis of infected cells (88) is prevented by the E1B 55-kDa protein, which inhibits p53 transactivation by binding to the N terminus of p53 (33, 92; reviewed in reference 7), and by the E1B 19-kDa protein, which shares structural and functional homology with the cellular antiapoptotic protein Bcl-2 (27, 63). Although encoded outside the E1 region, the E4orf6 34-kDa protein may modulate transformation by inhibiting p53 transactivation (13) and repression (58) or by relocalizing cytoplasmic E1B 55-kDa protein to the nucleus (22).
Mutations in the Ad12 E1B 55-kDa protein but not in the 19-kDa protein significantly reduced Ad12-induced fragility (67). Since the Ad12 E1B 55-kDa protein also directly represses the p53 transactivation domain (7, 33, 92), we wondered whether Ad12-induced fragility of the RNU1 and RNU2 loci might reflect viral inactivation of p53 functions. We found, however, that the RNU1 and RNU2 loci are not constitutively fragile in cells lacking p53 function and that Ad12 cannot induce fragility of either locus in the absence of wild-type or mutant p53 function. Since steady-state levels of U1 and U2 are within the normal range in cells lacking p53, we conclude that fragility of the RNU1 and RNU2 loci reflects an Ad12-induced p53 gain of function.
MATERIALS AND METHODS
Cells, FISH, and recombinant DNA.
Saos-2, SBN, and SEN osteosarcoma cells were obtained from W.-H. Lee and propagated as described previously in Dulbecco’s modified Eagle medium with 10% fetal bovine serum at 37°C under 5% CO2 (9). Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines GM5927 (normal), GM01855 (XPB), GM02285 (XPB), and GM02455 (XPD) were purchased from the Coriell Institute and propagated in RPMI 1640 medium with 20% fetal bovine serum. The fluorescent in situ hybridization (FISH) protocol and U2 gene probe were as described previously (4); U1 probes were as described by Lindgren et al. (50). p53 expression constructs (5, 35) were the kind gift of B. Vogelstein.
Complementation assay for p53 function.
In preliminary experiments with the pCH110 β-galactosidase expression vector (Pharmacia) as described by Lim and Chae (46), we tested Saos-2 transfection efficiencies over a range of electroporation voltages from 0.15 to 0.55 kV; 0.25 kV was optimal, with decreased efficiency at 0.15 kV and decreased survival at 0.55 kV. For the complementation assay, Saos-2 cells in 150-mm-diameter plates were grown overnight in fresh medium to 30 to 50% confluence and then infected with Ad12 for 1 h at a multiplicity of 10. Infected cells were collected by trypsinization and resuspended in cold phosphate-buffered saline (PBS). Electroporation was performed at 0.25 kV and 960 μF, using 0.4-cm-diameter cuvettes and a Bio-Rad Gene Pulser with a capacitance extender. For each electroporation, 107 cells were suspended in 0.6 ml of PBS containing 1.25% dimethyl sulfoxide, and 20 μg of total DNA (18 μg of the p53 vector and 2 μg of the pCH110 β-galactosidase vector) was added to the cell suspension. After electroporation, the cuvette was immediately chilled in ice, and the cell suspension was divided among four 100-mm-diameter plates containing 10 ml of Dulbecco’s modified Eagle medium. After 6 h the medium was replaced, and 16 h after transfection the cells were treated with Colcemid (0.1 μg/ml) for 3 h. Cells from three plates were then collected by trypsinization, and metaphase spreads were prepared by the standard techniques of hypotonic shock and methanol-acetic acid fixation (4). The fourth plate was stained for β-galactosidase as described previously (46).
RESULTS
p53 function is required for Ad12-induced chromosome fragility.
Analysis of metaphase chromosomes from a cell line entirely lacking functional p53, the Saos-2 osteosarcoma (9, 82), revealed that the RNU2 locus was not constitutively fragile in these cells nor could fragility be induced by Ad12 infection (Fig. 1 and Table 1). Thus, although loss of p53 often results in genomic and chromosomal instability, the induction of fragility by Ad12 cannot simply reflect viral inactivation of p53 functions. Nor can these negative results be explained trivially by the failure of Ad12 to infect p53-deficient cells; Saos-2 cells can be efficiently infected with Ad12 (77), and most cells in our infected cultures were dead within several days.
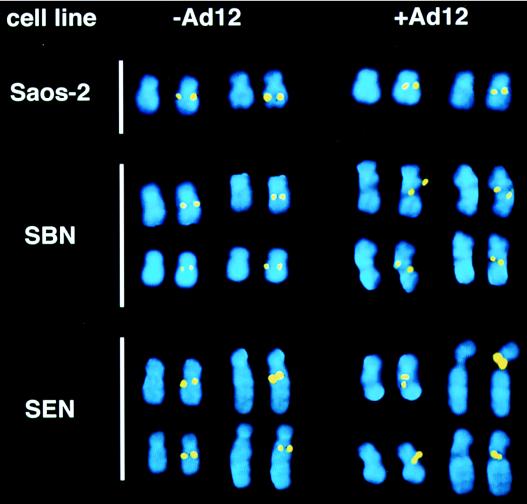
FIG. 1.
Ad12-induced fragility of the human RNU2 locus requires p53 function. Representative metaphase chromosomes are shown from the indicated cell lines with and without Ad12 infection. Each RNU2-containing chromosome is represented by a pair of images: the total DNA visualized by DAPI (4′,6-diamidino-2-phenylindole) staining is on the left, and the U2 arrays visualized by FISH and superimposed on the DAPI image are on the right. One of the two RNU2 loci in Saos-2 cells is located on a der(17) chromosome resulting from an unidentified translocation distal to 17q22. (See Table 1 for quantitation.) Note that Ad12-induced fragility covers a broad range of chromosome morphologies, including (i) frank separation or thinning of one or both chromatids; (ii) dislocation of the chromosome axis; and (iii) splitting, smearing, obvious intensification, or displacement of the RNU2 signal from the body of the chromosome as imaged by the DAPI stain (4). As seen in the upper panel (+Ad12), the fragile phenotype can be subtle: the RNU2 signal may coincide with thinning of one (upper- and lower-right SBN chromosomes) or both chromatids (upper- and lower-right SBN chromosomes); it may appear immediately adjacent to the site of chromatid thinning (upper- and lower-right SBN chromosomes); it may fail to coincide with the DAPI stain (upper- SBN and lower-left SEN chromosomes); or the two RNU2 signals may be displaced from normal lateral register (lower- SBN and upper-left SEN chromosomes). These morphologies are almost never seen in the absence of infection.
TABLE 1.
p53 dependence in Ad12-induced RNU2 fragility
| Cell line | p53 status |
RNU2 fragilitya as shown by:
|
|
|---|---|---|---|
| % of cells | % of RNU2 loci | ||
| HT1080 | Wild typeb | 92 | 75 |
| Saos-2 | Null | 6 | 3 |
| SEN8 | R273H | 67 | 44 |
| SEN9 | R273H | 72 | 52 |
| SBN40 | Wild type | 90 | 64 |
| SBN45 | Wild type | 87 | 61 |
At least 50 metaphases were examined for each datum. SEN, SBN, and Saos-2 cells (9) were generously provided by Wen-Hwa Lee. An RNU2 locus was considered fragile if there was obvious fragmentation of the FISH signal or if the signal was far more intense than most others in the field. An MOI of 10 was used for all cell lines.
We next investigated, using SBN and SEN cells (9), whether stable reintroduction into Saos-2 cells of the gene for either wild-type p53 (SBN cells) or the p53 mutant R273H (SEN cells) on a retroviral vector would restore Ad12-induced fragility (Fig. 1 and Table 1). Remarkably, virally induced fragility was restored nearly as well by the R273H mutant as by wild-type p53, although the transcriptional transactivation function of R273H is moderately to severely impaired (18, 59, 85, 93). These results exclude an essential role for Rb protein in virally induced fragility because Saos-2 cells also lack functional Rb protein (82). Most importantly, the absence of p53 in Saos-2 cells does not cause constitutive fragility of the RNU1 or RNU2 loci (Fig. 1) nor does it affect the steady-state levels of U1 and U2 snRNA (3a). Thus, the Ad12 E1B 55-kDa protein, perhaps working in concert with other viral early proteins (67, 74) such as E4orf6 (13, 22, 58), must induce a p53 gain of function that differs from the p53 functions normally responsible for regulating cell cycle progression and apoptosis.
Complementation assay for p53 function.
Cells lacking p53 function usually undergo apoptosis when full or partial p53 function is restored; accordingly, SBN cells were more difficult to isolate, grew more slowly, and displayed decreased viability relative to SEN cells (9). To explore the role of additional p53 mutants in Ad12-induced RNU2 fragility, we therefore sought a more convenient transient-expression assay. The basic approach was to complement the defect in Saos-2 cells by transfection with p53 expression vectors; the key experimental decision was whether to transfect Saos-2 with p53 before Ad12 infection or to infect Saos-2 with Ad12 before p53 transfection. In addition, we also required that p53 transfection be very efficient, because only 3 to 5% of the cells normally produce scorable metaphases; the 20% transfection efficiency typically achieved by calcium phosphate precipitation would generate only 1% scorable metaphases, an unworkably low number. Since Saos-2 cells grow as an adherent monolayer, our initial strategy was to perform the entire assay without detaching the cells. We infected the monolayer by overlaying with Ad12 virus for 1 h, transfected with the wild-type p53 expression vector in the presence of a cationic lipid such as Lipofectin or Lipofectamine, and then washed out the excess cationic lipid before adding fresh medium. Unfortunately, all available cationic lipids caused high levels of speckled background in the FISH assay (42a); presumably this was due to nonspecific adsorption of biotinylated probe DNA on cellular components bearing residual cationic detergent. Use of higher concentrations of sonicated salmon sperm carrier DNA during hybridization failed to reduce this background, and we therefore resorted to electroporation.
Adherent Saos-2 cells were infected with Ad12 for 1 h, detached by trypsinization, electroporated, and replated (see Materials and Methods for details). Optimal electroporation conditions for Saos-2 (0.25 kV, 960 μF, PBS buffer containing 1.25% dimethyl sulfoxide) were established by a β-galactosidase color assay (46), and transfection efficiencies of 30 to 40% were routinely obtained. Most cells survived the combined infection-transfection protocol, as evidenced by reattachment within 6 h of replating. Cells were arrested with Colcemid 18 h after replating and harvested for FISH 3 h later. To ensure consistency and an adequate number of metaphases, 2 μg of the β-galactosidase expression vector pCH110 was always cotransfected with 18 μg of the p53 expression vector, and one of the four resulting plates was stained for β-galactosidase.
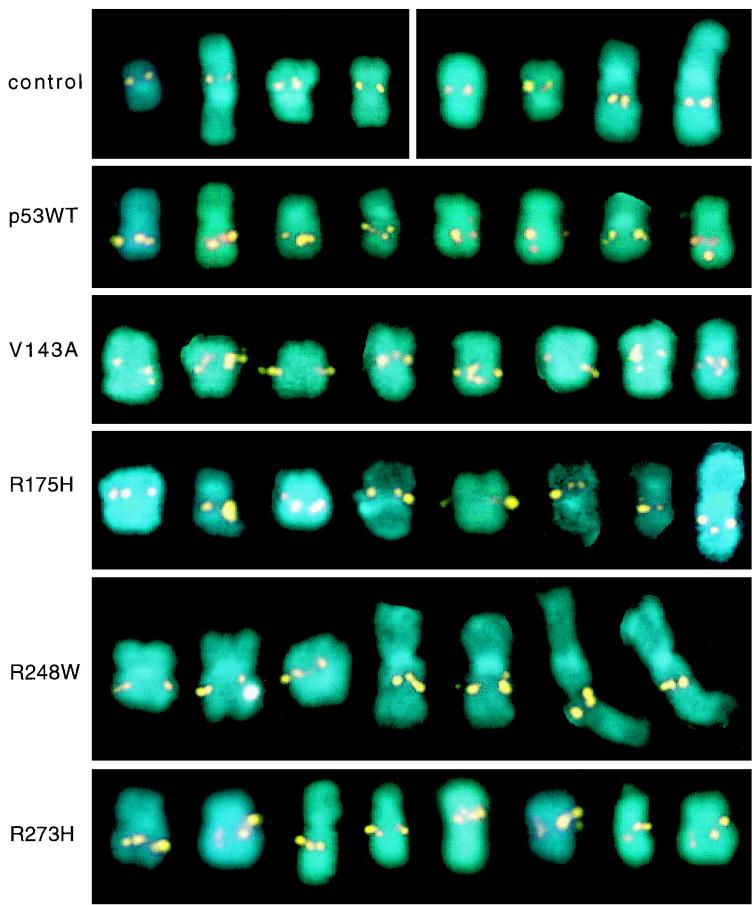
In this transient assay, Saos-2 cells were infected with Ad12 at low multiplicities (multiplicity of infection [MOI], 1 to 10) and then transfected with expression vectors encoding wild-type p53, one of four most common p53 hot-spot mutants (V143A, R175H, R248W, or R273H), or the pCH110 β-galactosidase expression vector alone. The p53 cDNA expression vectors were driven by the cytomegalovirus promoter-enhancer; rabbit β-globin splicing and polyadenylation signals ensured efficient expression (5, 35). As shown in Fig. 2 and summarized in Table 2, wild-type p53 and all four hot-spot mutants complemented the Saos-2 defect, restoring RNU2 fragility to near-normal levels resembling those observed upon infection of HT1080 cells expressing resident p53. Although >70% of all chromosomes 17 in transfected cells exhibit some RNU2 fragility (either fragmentation or clear intensification of the RNU2 signal), occasionally only one sister chromatid exhibits fragility; thus, it is not unexpected that a small number of chromosomes 17 (usually <20%) apparently exhibit no RNU2 fragility at all.
FIG. 2.
Complementation assay for the role of p53 in Ad12-induced RNU2 fragility. Saos-2 cells were infected with Ad12 in a monolayer for 1 h, detached by trypsinization, transfected by electroporation with the indicated p53 expression vectors (wild type [WT], V143A, R175H, R248W, or R273H), and replated. The cells were then grown for 17 h, arrested with Colcemid for 3 h, and harvested for FISH as described previously (4). A gallery of eight metaphase chromosomes 17 is shown for each transfection; extended chromosomes 17 are from early metaphase. The top gallery represents two controls: Saos-2 without infection or transfection (left four chromosomes) and Saos-2 infected with Ad12 and transfected with 20 μg of the pCH110 β-galactosidase expression vector (right four chromosomes).
TABLE 2.
A complementation assay for p53 function in Ad12-induced RNU2 fragility
| p53 expression | MOI |
RNU2 fragilitya as shown by:
|
|
|---|---|---|---|
| % of cells | % of RNU2 loci | ||
| Wild type | 10 | 32 | 23 |
| V143A | 10 | 29 | 20 |
| R175H | 10 | 22 | 16 |
| R248W | 10 | 26 | 19 |
| R273H | 10 | 28 | 21 |
| Control | 0 | 9 | 5 |
At least 100 metaphases were examined for each datum. The p53 expression vectors have been described previously (5, 35). The complementation assay with Saos-2 cells was performed as detailed in Materials and Methods. All data were normalized to transfection efficiency as judged by the β-galactosidase color assay (46). Control cells were transfected with 20 μg of the pCH110 β-galactosidase expression vector. Note that the values for percent fragility refer to total number of metaphases examined; when we assumed a 100% infection and corrected for transfection efficiency (30 to 40%), RNU2 fragility was usually observed in >70% of the cells transfected with wild-type or mutant p53.
Defects in ERCC2 or ERCC3 function cause weak constitutive RNU2 fragility.
U1 and U2 snRNA are RNA polymerase II transcripts, and U2 transcriptional regulatory elements are required for Ad12-induced fragility of an artificial U2 array (4, 20). Although originally identified as essential components of the DNA “repairosome,” the excision repair cross-complementing ERCC2 and ERCC3 helicase proteins in fact belong to a common core of five subunits shared between the repairosome and the basal RNA polymerase II transcription factor TFIIH (14, 48, 53, 80). The ability of p53 to interact with three different subunits of TFIIH, including ERCC2, ERCC3, and p62 (40, 83), led us to ask whether the p53 gain of function induced by Ad12 might cause p53 to interact aberrantly with TFIIH.
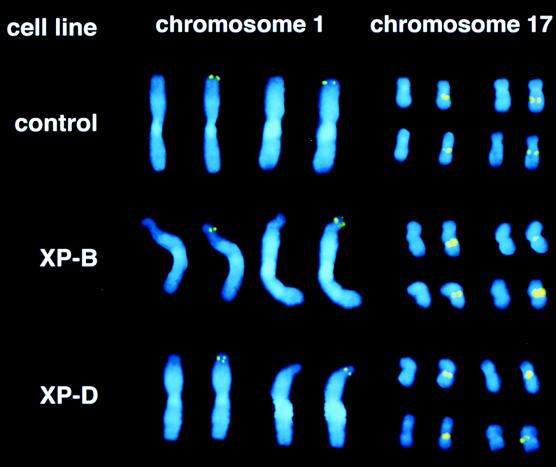
ERCC3 is mutated in xeroderma pigmentosum complementation group B (XPB) (65) and ERCC2 in complementation group D (XPD) (78). We therefore examined RNU1 and RNU2 fragility in EBV-transformed XPB (ERCC3) and XPD (ERCC2) lymphoblastoid cell lines (Fig. 3 and Table 3). Although the weak phenotype was somewhat obscured in highly condensed late-metaphase chromosomes, a clear phenotype could be observed in the more extended chromosomes of early metaphase. The RNU1 and RNU2 loci were constitutively fragile in both XPB and XPD cell lines, compared with the low level of fragility observed for normal EBV-transformed lymphoblastoid control cells (Fig. 3 and Table 3). As discussed below, the weak phenotype may reflect the fact that the primary defects in xeroderma pigmentosum are in the repair pathway, and these defects may only mildly (or fortuitously) affect the transcriptional functions of ERCC2 and ERCC3.
FIG. 3.
ERCC2 and ERCC3 mutations cause weak constitutive fragility of the human RNU1 and RNU2 loci. Representative metaphase chromosomes of the EBV-transformed lymphoblastoid cell lines XPB (GM01855) and XPD (GM02485), which are defective in ERCC3 and ERCC2 function, respectively. Compared to wild-type cells, XPB and XPD cells often exhibited a dislocation of the chromosome axis or a constriction of the chromatin near 1pter, and the RNU1 signal derived from a single chromatid occasionally split (see second chromosome 1 in XPB and first chromosome 1 in XPD). The XPB and XPD phenotypes are more apparent at RNU1 than RNU2. Other details are as described for Fig. 1. See Table 3 for quantitation.
TABLE 3.
Constitutive fragility of RNU1 and RNU2 loci in XPB and XPD cells
| Cell line | Mutant TFIIH subunit | % of cells exhibiting fragilitya
|
|
|---|---|---|---|
| RNU1 locus | RNU2 locus | ||
| GM5927 | None | <5 | 21 |
| GM01855 (XPB) | ERCC3 | 22 | 62 |
| GM02285 (XPB) | ERCC3 | 18 | 59 |
| GM02455 (XPD) | ERCC2 | 25 | 61 |
At least 100 metaphases were examined for each datum. Chromosomes were examined at comparable stages of condensation lest overcompaction obscure weak fragility. Cells in the GM series are EBV-transformed human lymphoblastoid lines obtained from the Coriell Institute. The low level of constitutive (i.e., nonadenoviral) fragility exhibited by the wild-type GM5927 line could reflect interaction of viral proteins such as EBNA-5 or BZLF1(Z) with p53 (1).
DISCUSSION
We have shown that p53 function is required for Ad12-induced fragility of the human RNU2 locus. In one set of experiments (Fig. 1 and Table 1), Saos-2 cells lacking detectable p53 expression were stably transduced with retroviral constructs expressing wild-type p53 or the R273H mutant. In the other set of experiments (Fig. 2 and Table 2), Saos-2 cells were transfected with vectors expressing wild-type p53 or one of four selected mutants (V143A, R175H, R248W, and R273H). In both sets of experiments, Ad12-induced metaphase fragility of the RNU2 locus was substantially restored by the expression of wild-type p53 and each of the four mutants tested. Each of these four mutations falls within one of the four highly conserved regions of the core DNA-binding region of p53 designated II (V143A), III (R175H), IV (R248W), and V (R273H). The R175H, R248W, and R273H mutants are also hot-spot mutations at codons which account for >20% of all mutations thus far reported (26); V143 is not a hot spot, but it was included to provide a disabling mutation in region II. While all four mutants are partially defective in DNA binding, the degree of the defect would almost certainly depend on the particular cell, assay, and temperature used (18, 36). For example, transcriptional transactivation of the WAF1 promoter by R273H appears to be only slightly impaired in primary human fibroblasts (85) but is nearly abolished in Saos-2 cells (59). In contrast, transactivation by R273H is nearly normal when assayed on an hsp70 promoter driven by a p53 consensus binding site in K562 cells (93). The observation that all four p53 hot-spot mutants were as effective as wild-type p53 implies that DNA binding is not required or that weak binding is sufficient to rescue fragility.
Our data show that Ad12 induces a p53 gain of function. Steady-state levels of U1 and U2 snRNA are within the normal range in Saos-2 cells lacking p53 function (3a), and the same is presumably true in countless tumor cell lines that are partially or completely lacking p53 function. Thus, p53 cannot be required for U1 or U2 transcription, and Ad12 proteins (probably E1B 55-kDa protein, perhaps working together with the E1A and/or E4orf6 products as discussed below) must cause a p53 gain of function that specifically interferes with metaphase chromatin packing of the RNU1, RNU2, and PSU1 loci. Other p53 gains of function have been characterized (12, 47; reviewed in reference 41). In most cases, these p53 gain-of-function mutants have lost tumor suppressor functions but also contribute to tumor progression. Some of these mutants may simply be dominant negatives (8), while others represent a genuine gain of function (12, 47, 56).
Ad12-induced fragility is unlikely to involve the p53-dependent apoptotic pathway (87). First, fragilization of the RNU1, RNU2, and PSU1 loci is highly specific at low MOIs; generalized fragility or chromosome “pulverization” reminiscent of apoptosis is observed only at high multiplicities (67). Second, the human group C adenovirus dl1520 (a double mutant expressing no E1B 55K protein whatsoever) grows well on C33A cervical carcinoma cells expressing the R273H mutant, but not on U20S osteocarcinoma cells expressing wild-type p53 (7). Thus, R273H fully restores RNU2 fragility in the Saos-2 osteosarcoma cells, although it cannot support E1A-induced apoptosis in the C33A cervical carcinoma cells. Third, three of the p53 mutants that support Ad12-induced RNU2 fragility in our assay (V143A, R175H, and R248W) were completely unable to induce apoptosis when transiently expressed in primary human fibroblasts; the fourth mutant (R273H) induced apoptosis only weakly (85). Thus, mutants in the core DNA binding regions of p53 that affect transactivation (18, 59, 85) can still support Ad12-induced fragility but not p53-dependent apoptosis.
At least three Ad2/5 proteins are known to interact, physically or functionally, with p53: the E1A proteins encoded by the 12S and 13S alternatively spliced mRNAs, the E1B 55-kDa protein, and the E4orf6 34-kDa protein. The 12S- and 13S-encoded E1A proteins inhibit the p53 transactivation function, possibly by inducing oligomerization of the N-terminal region of the protein (75), whereas the 13S-encoded E1A protein can relieve p53 repression, apparently by dissociating the complex between the C-terminal region of p53 and the TATA-binding protein (32, 52). The Ad2/5 and Ad12 E1B 55-kDa proteins both repress transactivation by binding to the N-terminal region of p53 (33, 91, 92; reviewed in reference 36). The Ad2/5 E4orf6 34-kDa protein can block p53 transactivation by interfering with contact between the N-terminal region of p53 and TAFII31, a component of TFIID (13), and can also block p53 repression, most probably by binding to the C-terminal repression region of p53 (58).
Ad12-induced fragility of the RNU1 and RNU2 loci is significantly inhibited by mutations in the E1B 55-kDa protein but not in the E1B 19-kDa protein (67). Therefore, the simplest hypothesis to explain Ad12-induced metaphase fragility is that the Ad12 E1B 55-kDa protein binds p53 and induces a gain of function; residual fragility induced by the Ad12 E1B 55-kDa mutant virus might reflect inhibition of p53 transactivation and/or repression by E1A, E4orf6, or other adenovirus gene products. The Ad2/5 and Ad12 E1B 55-kDa proteins both interact with and stabilize p53 (24, 74), and both block p53 transactivation functions (91); however, the Ad12 55-kDa protein, unlike the Ad2/5 protein, does not form easily detected immunoprecipitable complexes with p53 and fails to sequester p53 in the cytoplasm (24, 74). On the other hand, the Ad5 E4orf6 34-kDa protein causes a dramatic reduction in steady-state p53 levels (13), and interaction with the E4orf6 protein can relocalize the predominantly cytoplasmic Ad5 E1B 55-kDa protein to the nucleus (22). These differences in the interaction of the Ad2/5 and Ad12 E1B 55-kDa proteins with p53 may account for the observation that Ad12 induces four specific sites of chromosome fragility, whereas Ad2/5 only induces generalized fragility at higher multiplicities (67, 94).
The XPB (ERCC3) and XPD (ERCC2) helicases belong to a common core of five subunits shared between the repairosome and the basal RNA polymerase II transcription factor TFIIH (14, 48, 53, 66, 80). p53 can interact with at least three subunits of TFIIH, including XPB (ERCC3), XPD (ERCC2), and p62; the N-terminal activation domain of p53 interacts with p62 (40, 90), and the C-terminal domain interacts with XPB and XPD (40, 83–85). Binding of the C-terminal domain of p53 (residues 319 to 393) to XPB (ERCC3) and/or XPD (ERCC2) is functionally significant since it can induce p53-dependent apoptosis in primary human fibroblasts (85). The ability of mutations in the XPB (ERCC3) and XPD (ERCC2) helicases to induce weak constitutive fragility of the RNU1 and RNU2 loci (Fig. 3 and Table 3) therefore supports a model (Fig. 4) in which the Ad12-induced p53 gain of function acts directly on the XPB (ERCC3) and/or XPD (ERCC2) helicases either as part of TFIIH or of the repairosome. The relatively weak constitutive phenotype of these particular xeroderma pigmentosum mutants may indicate that they are preferentially defective in repair; although the XPB and XPD helicases serve a dual role in repair and transcription, only a minority of XPD mutations cause the severe developmental defects that characterize Cockayne syndrome or trichothiodystrophy, and that may be indicative of defects in transcription (11, 38).
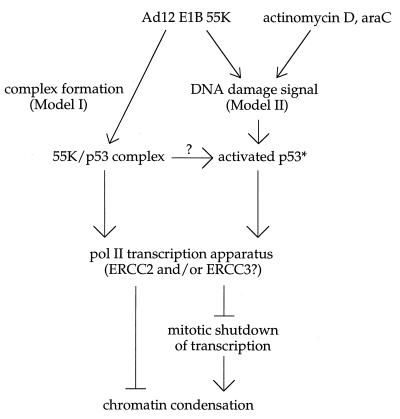
FIG. 4.
Models for p53- and transcription-dependent Ad12-induced fragility of the human RNU1 and RNU2 loci. In model I, the Ad12 E1B 55-kDa transforming protein (55K), perhaps assisted by the E4orf6 product, interacts with p53 to induce a p53 gain of function. (We postulate a p53 gain of function because RNU1 and RNU2 are not fragile in cells lacking p53 and steady-state levels of U1 and U2 snRNA are within the normal range.) In model II, the 55K protein generates or phenocopies a DNA damage signal which activates p53 (p53*). The 55K/p53 complex or activated p53* would then interfere with local chromatin condensation by binding directly to the transcription apparatus, by blocking mitotic shutdown of transcription, or by sequestering essential transcription factors involved in transcription-coupled repair (42a). In either model, specific fragilization of the RNU1, RNU2, and PSU1 loci could reflect the unusually high local concentration of independent transcription units (which might normally hinder chromatin condensation) or the specialized nature of the U1 and U2 snRNA transcription apparatus (which might interact uniquely with 55K/p53, p53*, or downstream factors). araC, 1-β-d-arabinylfuranosylcytosine.
We can imagine two very different models to explain Ad12-induced, locus-specific, metaphase chromosome fragility (Fig. 4). In the first model, the Ad12-induced p53 gain of function perturbs cell cycle regulation of U1, U2, and perhaps 5S transcription, causing transcription to persist into metaphase and to interfere directly with chromatin packing. Although it is often assumed that metaphase chromatin packing is dominant over transcription, there is good reason to believe that transcription must cease before proper metaphase chromatin packing can take place. Otherwise, it is difficult to explain why TATA binding protein (39, 69), RNA polymerase II transcription factors such as Oct-1 (68) and Sp1 (54), and TFIIIB (23, 88) are inactivated by phosphorylation as metaphase approaches (for a review, see reference 76) and nascent transcripts are aborted (72). The effect of p53 on the RNU1 and RNU2 transcription units might be mediated by interactions with the ERCC2 (XPD), ERCC3 (XPB), or p62 components of TFIIH as described above and/or with other components of the transcription machinery (36). In either case, it is tantalizing that TFIIH also contains a cdk7-cyclin H couple, but there is no direct evidence that TFIIH plays a role in the cell cycle regulation of RNA polymerase II transcription (70).
In the second model, the Ad12-induced p53 gain of function causes p53 to behave as though it had directly sensed (37) or had been alerted to DNA damage (reviewed in references 34 and 36). Although most DNA damage causes a global block in cell cycle progression, certain kinds of DNA damage could conceivably block local chromatin packing just as damage locally blocks transcription. For example, the Ad12-induced p53 gain of function might block chromatin packing by consolidating a transient or fortuitous complex between p53 and TFIIH or other transcription factors bound to the specialized U1 and U2 snRNA transcription apparatus (see below). In fact, new data strongly suggest that DNA damage, or the perception of such damage, may be the key to understanding Ad12-induced metaphase fragility. Thus, DNA-damaging reagents such as 1-β-d-arabinylfuranosylcytosine (52a) and actinomycin D that either cause or phenocopy DNA damage (92a) can efficiently induce p53- and transcription-dependent fragility of the RNU2 locus. The connection between transcription, repair, and fragility is further supported by the observation that defects in the CSB protein (defective in Cockayne syndrome complementation group B), which is involved in transcription-coupled repair, can cause constitutive unpacking of the RNU1, RNU2, and RN5S loci (42a). The fact that transcription-coupled repair is normal in primary Li-Fraumeni fibroblasts with homozygous p53 mutations but global DNA repair is deficient (17) reinforces the idea that a p53 gain of function and a CSB protein defect might be alternative means of phenocopying DNA damage. We therefore propose that Ad12 infection, CSB protein mutations, 1-β-d-arabinofuranosylcytosine, and low concentrations of actinomycin D all generate a similar or identical damage arrest signal (perhaps a posttranscriptional modification, altered conformation, or relocalization of p53) to which the actively transcribed RNU1 and RNU2 loci are especially or uniquely sensitive.
In both models, the preferential effect of Ad12 on metaphase condensation of the RNU1, RNU2, and PSU1 loci (49, 94) could be explained either by the unusually high local concentration of short, independent transcription units at each affected locus, by the specialized nature of the U1 and U2 snRNA promoter (3, 29, 64) and termination factors (30, 31, 57), or by the highly structured nature of the nascent U1, U2, and 5S RNA transcripts (42a). A high local concentration of strong transcription units could render chromatin packing especially sensitive to persistent transcription or to a global damage arrest signal. Alternatively, specialized U1 and U2 snRNA transcription factors could be required to interact with the p53 gain of function or to sense the global arrest signal. A third possibility is that specialized factors might be required to release polymerases whose progress has been blocked by secondary structure in the nascent RNA transcript (42a).
ACKNOWLEDGMENTS
We thank David C. Ward, June Menninger, and Patricia Bray-Ward for cheerful instruction in FISH and for generous access to superb image capture and processing equipment in the early stages of this work; Wen-Hwa Li for graciously providing the SBN and SEN cell lines; Bert Vogelstein for wild-type and mutant p53 expression vectors; the Coriell Institute for prompt provision of GM cell lines; Silvia Bacchetti for communicating results in advance of publication; and Daiqing Liao and Arnold D. Bailey for critical comments on the manuscript.
This work was supported by NIH awards GM31073 and GM41624.
REFERENCES
- 1.Allday M J, Sinclair A, Parker G, Crawford D H, Farrell P J. Epstein-Barr virus efficiently immortalizes human B cells without neutralizing the function of p53. EMBO J. 1995;14:1382–1391. doi: 10.1002/j.1460-2075.1995.tb07124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson M J, Casey G, Fasching C L, Stanbridge E J. Evidence that wild-type TP53, and not genes on either chromosome 1 or 11, controls the tumorigenic phenotype of the human fibrosarcoma HT1080. Genes Chromosomes Cancer. 1994;9:266–281. doi: 10.1002/gcc.2870090407. [DOI] [PubMed] [Google Scholar]
- 3.Bai L, Wang Z, Yoon J B, Roeder R G. Cloning and characterization of the beta subunit of human proximal sequence element-binding transcription factor and its involvement in transcription of small nuclear RNA genes by RNA polymerases II and III. Mol Cell Biol. 1996;16:5419–5426. doi: 10.1128/mcb.16.10.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Bailey, A. D., and A. M. Weiner. Unpublished data.
- 4.Bailey A D, Li Z, Pavelitz T, Weiner A M. Adenovirus 12-induced fragility of the human RNU2 locus requires U2 snRNA transcriptional regulatory elements. Mol Cell Biol. 1995;15:6246–6255. doi: 10.1128/mcb.15.11.6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker J S, Markowitz S, Fearon R E, Willson V K J, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1992;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein L B, Manser T, Weiner A M. Human U1 small nuclear RNA genes: evidence for extensive conservation of flanking sequences suggests cycles of gene amplification and transposition. Mol Cell Biol. 1985;5:2159–2171. doi: 10.1128/mcb.5.9.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bischoff J R, Kim D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann R K, Vidal M, Boeke J D. Dominant-negative p53 mutations selected in yeast hit cancer hot spots. Proc Natl Acad Sci USA. 1996;93:4091–4095. doi: 10.1073/pnas.93.9.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P L, Chen Y M, Bookstein R, Lee W H. Genetic mechanisms of tumor suppression by the human p53 gene. Science. 1990;250:1576–1580. doi: 10.1126/science.2274789. [DOI] [PubMed] [Google Scholar]
- 10.Chiou S K, White E. p300 binding by E1A cosegregates with p53 induction but is dispensable for apoptosis. J Virol. 1997;71:3515–3525. doi: 10.1128/jvi.71.5.3515-3525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu G, Mayne L. Xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy: do the genes explain the diseases? Trends Genet. 1996;12:187–192. doi: 10.1016/0168-9525(96)10021-4. [DOI] [PubMed] [Google Scholar]
- 12.Dittmer D, Pati S, Zambetti G, Chu S, Teresky A K, Moore M, Finlay C, Levine A J. Gain of function mutations in p53. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 13.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 14.Drapkin R, Le Roy G, Cho H, Akoulitchev S, Reinberg D. Human cyclin-dependent kinase-activating kinase exists in three distinct complexes. Proc Natl Acad Sci USA. 1996;93:6488–6493. doi: 10.1073/pnas.93.13.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durnam D M, Smith P P, Menninger J C, McDougall J K. The E1 region of human adenovirus type 12 determines the sites of virally induced chromosomal damage. In: Botchan M, et al., editors. Cancer cells 4: DNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. pp. 349–354. [Google Scholar]
- 16.Durnam D M, Menninger J C, Chandler S H, Smith P P, McDougall J K. A fragile site in the human U2 small nuclear RNA gene cluster is revealed by adenovirus type 12 infection. Mol Cell Biol. 1988;8:1863–1867. doi: 10.1128/mcb.8.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ford J M, Hanawalt P C. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedlander P, Legros Y, Soussi T, Prives C. Regulation of mutant p53 temperature-sensitive DNA binding. J Biol Chem. 1996;271:25468–25478. doi: 10.1074/jbc.271.41.25468. [DOI] [PubMed] [Google Scholar]
- 19.Gaddini L, Pelliccia F, Limongi M Z, Rocchi A. Study of the relationships between common fragile sites, chromosome breakages and sister chromatid exchanges. Mutagenesis. 1995;10:257–260. doi: 10.1093/mutage/10.3.257. [DOI] [PubMed] [Google Scholar]
- 20.Gargano S, Wang P, Rusanganwa E, Bacchetti S. The transcriptionally competent U2 gene is necessary and sufficient for adenovirus type 12 induction of the fragile site at 17q21-22. Mol Cell Biol. 1995;15:6256–6261. doi: 10.1128/mcb.15.11.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glover T W, Stein C K. Induction of sister chromatid exchanges at common fragile sites. Am J Hum Genet. 1987;41:882–890. [PMC free article] [PubMed] [Google Scholar]
- 22.Goodrum F D, Shenk T, Ornelles D A. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J Virol. 1996;70:6323–6335. doi: 10.1128/jvi.70.9.6323-6335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottesfeld J M, Wolf V J, Dang T, Forbes D J, Hartl P. Mitotic repression of RNA polymerase III transcription in vitro mediated by phosphorylation of a TFIIIB component. Science. 1994;263:81–84. doi: 10.1126/science.8272869. [DOI] [PubMed] [Google Scholar]
- 24.Grand R J, Grant M L, Gallimore P H. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology. 1994;203:229–240. doi: 10.1006/viro.1994.1480. [DOI] [PubMed] [Google Scholar]
- 25.Hagerman R J, Hull C E, Safanda J F, Carpenter I, Staley L W, O’Connor R A, Seydel C, Mazzocco M M, Snow K. K, Thibodeau S N, et al. High functioning fragile X males: demonstration of an unmethylated fully expanded FMR-1 mutation associated with protein expression. Am J Med Genet. 1994;51:298–308. doi: 10.1002/ajmg.1320510404. [DOI] [PubMed] [Google Scholar]
- 26.Hainaut P, Soussi T, Shomer B, Hollstein M, Greenblatt M, Hovig E, Harris C C, Montesano R. Database of p53 gene somatic mutations in human tumors and cell lines: updated compilation and future prospects. Nucleic Acids Res. 1997;25:151–157. doi: 10.1093/nar/25.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J, Sabbatini P, Perez D, Rao L, Modha D, White E. The E1B 19K protein blocks apoptosis by interacting with and inhibiting the p53-inducible and death-promoting Bax protein. Genes Dev. 1996;10:461–477. doi: 10.1101/gad.10.4.461. [DOI] [PubMed] [Google Scholar]
- 28.Hansen R S, Canfield T K, Fjeld A D, Mumm S, Laird C D, Gartler S M. A variable domain of delayed replication in FRAXA fragile X chromosomes: X inactivation-like spread of late replication. Proc Natl Acad Sci USA. 1997;94:4587–4592. doi: 10.1073/pnas.94.9.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry R W, Ma B, Sadowski C L, Kobayashi R, Hernandez N. Cloning and characterization of SNAP50, a subunit of the snRNA-activating protein complex SNAPc. EMBO J. 1996;15:7129–7136. [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez N, Weiner A M. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986;47:249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez N, Lucito R. Elements required for transcription initiation of the human U2 snRNA gene coincide with elements required for snRNA 3′ end formation. EMBO J. 1988;7:3125–3134. doi: 10.1002/j.1460-2075.1988.tb03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horikoshi N, Usheva A, Chen J, Levine A J, Weinmann R, Shenk T. Two domains of p53 interact with the TATA-binding protein, and the adenovirus 13S E1A protein disrupts the association, relieving p53-mediated transcriptional repression. Mol Cell Biol. 1995;15:227–234. doi: 10.1128/mcb.15.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kao C C, Yew P R, Berk A J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 34.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 35.Kern E S, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler W K, Vogelstein B. Oncogenic forms of p53 inhibit p53 regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 36.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 37.Lee S, Elenbaas B, Levine A, Griffith J. p53 and its 14 kDa C-terminal domain recognize primary DNA damage in the form of insertion/deletion mismatches. Cell. 1995;81:1013–1020. doi: 10.1016/s0092-8674(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 38.Lehmann A R. Nucleotide excision repair and the link with transcription. Trends Biochem Sci. 1995;20:402–405. doi: 10.1016/s0968-0004(00)89088-x. [DOI] [PubMed] [Google Scholar]
- 39.Leresche A, Wolf V J, Gottesfeld J M. Repression of RNA polymerase II and III transcription during M phase of the cell cycle. Exp Cell Res. 1996;229:282–288. doi: 10.1006/excr.1996.0373. [DOI] [PubMed] [Google Scholar]
- 40.Leveillard T, Andera L, Bissonnette N, Schaeffer L, Bracco L, Egly J M, Wasylyk B. Functional interactions between p53 and the TFIIH complex are affected by tumour-associated mutations. EMBO J. 1996;15:1615–1624. [PMC free article] [PubMed] [Google Scholar]
- 41.Levine A J, Wu M C, Chang A, Silver A, Attiyeh E F, Lin J, Epstein C B. The spectrum of mutations at the p53 locus. Evidence for tissue-specific mutagenesis, selection of mutant alleles, and a “gain of function” phenotype. Ann N Y Acad Sci. 1995;768:111–128. doi: 10.1111/j.1749-6632.1995.tb12115.x. [DOI] [PubMed] [Google Scholar]
- 42.Li Y P, Tomanin R, Smiley J R, Bacchetti S. Generation of a new adenovirus type 12-inducible fragile site by insertion of an artificial U2 locus in the human genome. Mol Cell Biol. 1993;13:6064–6070. doi: 10.1128/mcb.13.10.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Liao, D., and A. M. Weiner. Unpublished data.
- 43.Liao D, Weiner A M. Concerted evolution of the tandemly repeated genes encoding primate U2 small nuclear RNA (the RNU2 locus) does not prevent rapid diversification of the (CT)n · (GA)n microsatellite embedded within the U2 repeat unit. Genomics. 1995;30:583–593. doi: 10.1006/geno.1995.1280. [DOI] [PubMed] [Google Scholar]
- 44.Liao D, Pavelitz T, Kidd J, Kidd K, Weiner A M. Concerted evolution of the tandemly repeated genes encoding human U2 snRNA (the RNU2 locus) involves rapid intrachromosomal homogenization and rare interchromosomal gene conversion. EMBO J. 1997;16:588–598. doi: 10.1093/emboj/16.3.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao, D., T. Pavelitz, and A. M. Weiner. Characterization of a novel class of interspersed LTR elements in primate genomes: structure, genomic distribution, and evolution. J. Mol. Evol., in press. [DOI] [PubMed]
- 46.Lim K, Chae C B. A simple assay for DNA transfection by incubation of the cells in culture dishes with substrates for β-galactosidase. BioTechniques. 1989;7:576–579. [PubMed] [Google Scholar]
- 47.Lin J, Teresky A K, Levine A J. Two critical hydrophobic amino acids in the N-terminal domain of the p53 protein are required for the gain of function phenotypes of human p53 mutants. Oncogene. 1995;10:2387–2390. [PubMed] [Google Scholar]
- 48.Lindahl T, Karran P, Wood R D. DNA excision repair pathways. Curr Opin Genet Dev. 1997;7:158–169. doi: 10.1016/s0959-437x(97)80124-4. [DOI] [PubMed] [Google Scholar]
- 49.Lindgren V, Ares M, Weiner A M, Francke U. Human genes for U2 small nuclear RNA map to a major adenovirus 12 modification site on chromosome 17. Nature. 1985;314:115–116. doi: 10.1038/314115a0. [DOI] [PubMed] [Google Scholar]
- 50.Lindgren V L, Bernstein L B, Weiner A M, Francke U. Human U1 small nuclear RNA pseudogenes do not map to the site of the U1 genes in 1p36 but are clustered in 1q12-q22. Mol Cell Biol. 1985;5:2172–2180. doi: 10.1128/mcb.5.9.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linke S P, Clarkin K C, Di Leonardo A, Tsou A, Wahl G M. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 52.Liu X, Miller C W, Koeffler P H, Berk A J. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol Cell Biol. 1995;13:3291–3300. doi: 10.1128/mcb.13.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.MacArthur, H. L., M. L. Agarwal, and S. Bacchetti. Induction of fragility at the human RNU2 locus by cytosine arabinoside is dependent upon a transcriptionally competent U2 small nuclear RNA gene and the expression of p53. Somat. Cell Mol. Genet., in press. [DOI] [PubMed]
- 53.Marinoni J C, Roy R, Vermeulen W, Miniou P, Lutz Y, Weeda G, Seroz T, Gomez D M, Hoeijmakers J H, Egly J M. Cloning and characterization of p52, the fifth subunit of the core of the transcription/DNA repair factor TFIIH. EMBO J. 1997;16:1093–1102. doi: 10.1093/emboj/16.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez-Balbas M A, Dey A, Rabindran S K, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 55.Miro R, Clemente I C, Fuster C, Egozcue J. Fragile sites, chromosome evolution, and human neoplasia. Hum Genet. 1987;75:345–349. doi: 10.1007/BF00284105. [DOI] [PubMed] [Google Scholar]
- 56.Muller B F, Paulsen D, Deppert W. Specific binding of MAR/SAR DNA-elements by mutant p53. Oncogene. 1996;1:1941–1952. [PubMed] [Google Scholar]
- 57.Neuman de Vegvar H E, Lund E, Dahlberg J E. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986;47:259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]
- 58.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ory K, Legros Y, Auguin C, Soussi T. Analysis of the most representative tumour-derived p53 mutants reveals that changes in protein conformation are not correlated with loss of transactivation or inhibition of cell proliferation. EMBO J. 1994;13:3496–3504. doi: 10.1002/j.1460-2075.1994.tb06656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pavelitz T, Rusché L, Matera A G, Scharf J M, Weiner A M. Concerted evolution of the tandem array encoding primate U2 snRNA occurs in situ, without changing the cytological context of the RNU2 locus. EMBO J. 1995;14:169–177. doi: 10.1002/j.1460-2075.1995.tb06987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pellegata N S, Antoniono R J, Redpath J L, Stanbridge E L. DNA damage and p53-mediated cell cycle arrest: a reevaluation. Proc Natl Acad Sci USA. 1996;93:15209–15214. doi: 10.1073/pnas.93.26.15209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Popescu N C, Zimonjic D, DiPaolo J A. Viral integration, fragile sites, and proto-oncogenes in human neoplasia. Hum Genet. 1990;84:383–386. doi: 10.1007/BF00195804. [DOI] [PubMed] [Google Scholar]
- 63.Rao L, Debbas M, Sabbatini P, Hockenbery D, Korsmeyer S, White E. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc Natl Acad Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sadowski C L, Henry R W, Kobayashi R, Hernandez N. The SNAP45 subunit of the small nuclear RNA (snRNA) activating protein complex is required for RNA polymerase II and III snRNA gene transcription and interacts with the TATA box binding protein. Proc Natl Acad Sci USA. 1996;93:4289–4293. doi: 10.1073/pnas.93.9.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers J H, Chambon P, Egly J M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 66.Schaeffer L, Moncollin V, Roy R, Staub A, Mezzina M, Sarasin A, Weeda G, Hoeijmakers J H, Egly J M. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J. 1994;13:2388–2392. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schramayr S, Caporossi D, Mak I, Jelinek T, Bacchetti S. Chromosomal damage induced by human adenovirus type 12 requires expression of the E1B 55-kilodalton viral protein. J Virol. 1990;64:2090–2095. doi: 10.1128/jvi.64.5.2090-2095.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Segil N, Roberts S B, Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- 69.Segil N, Guermah M, Hoffmann A, Roeder R G, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 70.Seroz T, Hwang J R, Moncollin V, Egly J M. TFIIH: a link between transcription, DNA repair and cell cycle regulation. Curr Opin Genet Dev. 1995;5:217–221. doi: 10.1016/0959-437x(95)80011-5. [DOI] [PubMed] [Google Scholar]
- 71.Sharma S, Schwarte-Waldhoff I, Oberhuber H, Schafer R. Functional interaction of wild-type and mutant p53 transfected into human tumor cell lines carrying activated ras genes. Cell Growth Differ. 1993;4:861–869. [PubMed] [Google Scholar]
- 72.Shermoen A W, O’Farrell P H. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sorensen P D, Lomholt B, Frederiksen S, Tommerup N. Fine mapping of human 5S rRNA genes to chromosome 1q42.11-q42.13. Cytogenet Cell Genet. 1991;57:26–29. doi: 10.1159/000133107. [DOI] [PubMed] [Google Scholar]
- 74.Steegenga W T, Shvarts A, van Laar T, van der Eb A J, Jochemsen A G. Altered phosphorylation and oligomerization of p53 in adenovirus type 12-transformed cells. Oncogene. 1995;11:49–57. [PubMed] [Google Scholar]
- 75.Steegenga W T, van Laar T, Riteco N, Mandarino A, Shvarts A, van der Eb A J, Jochemsen A G. Adenovirus E1A proteins inhibit activation of transcription by p53. Mol Cell Biol. 1996;16:2101–2109. doi: 10.1128/mcb.16.5.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stukenberg P T, Lustig K D, McGarry T J, King R W, Kuang J, Kirschner M W. Systematic identification of mitotic phosphoproteins. Curr Biol. 1997;7:338–348. doi: 10.1016/s0960-9822(06)00157-6. [DOI] [PubMed] [Google Scholar]
- 77.Subramanian T, Tarodi B, Chinnadurai G. p53-independent apoptotic and necrotic cell deaths induced by adenovirus infection: suppression by E1B 19K and Bcl-2 proteins. Cell Growth Differ. 1995;6:131–137. [PubMed] [Google Scholar]
- 78.Sung P, Bailly V, Weber C, Thompson L H, Prakash L, Prakash S. Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature. 1993;365:852–855. doi: 10.1038/365852a0. [DOI] [PubMed] [Google Scholar]
- 79.Sutherland G R, Richards R I. The molecular basis of fragile sites in human chromosomes. Curr Opin Genet Dev. 1995;5:323–327. doi: 10.1016/0959-437x(95)80046-8. [DOI] [PubMed] [Google Scholar]
- 80.Svejstrup J Q, Vichi P, Egly J M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 81.Thut C J, Chen J L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 82.Uzvolgyi E, Classon M, Henriksson M, Huang M, Szekely L, Lee W H, Klein G, Sumegi J. Reintroduction of a normal retinoblastoma gene into retinoblastoma and osteosarcoma cells inhibits the replication associated function of SV40 large T antigen. Cell Growth Differ. 1991;2:297–303. [PubMed] [Google Scholar]
- 82a.Wahl, G. M. Personal communication.
- 83.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J R, Harris C C. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Freidberg E C, Evans M K, Taffe B G, et al. p53 modulation of TFIIH-associated nucleotide excision repair activity. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 85.Wang X W, Vermeulen W, Coursen J D, Gibson M, Lupold S E, Forrester K, Xu G, Elmore L, Yeh H, Hoeijmakers J H, Harris C C. The XPB and XPD DNA helicases are components of the p53-mediated apoptosis pathway. Genes Dev. 1996;10:1219–1232. doi: 10.1101/gad.10.10.1219. [DOI] [PubMed] [Google Scholar]
- 86.Wenger S L. Chemical induction of sister chromatid exchange at fragile sites. Cancer Genet Cytogenet. 1995;85:72–74. doi: 10.1016/0165-4608(95)00137-9. [DOI] [PubMed] [Google Scholar]
- 87.White E. Regulation of p53-dependent apoptosis by E1A and E1B. Curr Top Microbiol Immunol. 1995;199:34–58. [PubMed] [Google Scholar]
- 88.White R J, Gottlieb T M, Downes C S, Jackson S P. Mitotic regulation of a TATA-binding-protein-containing complex. Mol Cell Biol. 1995;15:1983–1992. doi: 10.1128/mcb.15.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilke C M, Hall B K, Hoge A, Paradee W, Smith D I, Glover T W. FRA3B extends over a broad region and contains a spontaneous HPV16 integration site: direct evidence for the coincidence of viral integration sites and fragile sites. Hum Mol Genet. 1996;5:187–195. doi: 10.1093/hmg/5.2.187. [DOI] [PubMed] [Google Scholar]
- 90.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, et al. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 92.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 92a.Yu, A., A. D. Bailey, and A. M. Weiner. Metaphase fragility of the human RNU1 and RNU2 loci is induced by actinomycin D through a p53-dependent pathway. Hum. Mol. Genet., in press. [DOI] [PubMed]
- 93.Zhang W, Funk W D, Wright W E, Shay J W, Deisseroth A B. Novel DNA binding of p53 mutants and their role in transcriptional activation. Oncogene. 1993;8:2555–2559. [PubMed] [Google Scholar]
- 94.zur Hausen H. Induction of specific chromosomal aberrations by adenovirus type 12 in human embryonic kidney cells. J Virol. 1967;1:1174–1185. doi: 10.1128/jvi.1.6.1174-1185.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]