Abstract
Background
Inflammatory bowel disease (IBD), which includes Crohn’s disease (CD) and ulcerative colitis (UC), has been associated with several cancer risks in observational studies, but the observed associations have been inconsistent and may face the bias of confounding and reverse causality. The potential causal relationships between IBD and the risk of cancers remain largely unclear.
Methods
We performed genome-wide linkage disequilibrium score regression (LDSC), standard two-sample Mendelian randomization (MR), and colocalization analyses using summary genome-wide association study (GWAS) data across East Asian and European populations to evaluate the causal relationships between IBD and cancers. Sensitivity analyses for the MR approach were additionally performed to explore the stability of the results.
Results
There were no significant genetic correlations between IBD, CD, or UC and cancers (all P values > 0.05) in East Asian or European populations. According to the main MR analysis, no significant causal relationship was observed between IBD and cancers in the East Asian population. There were significant associations between CD and ovarian cancer (odds ratio [OR] = 0.898, 95% CI = 0.844–0.955) and between UC and nonmelanoma skin cancer (OR = 1.002, 95% CI = 1.000–1.004, P = 0.019) in the European population. The multivariable MR analysis did not find any of the above significant associations. There was no shared causal variant to prove the associations of IBD, CD, or UC with cancers in East Asian or European populations using colocalization analysis.
Conclusions
We did not provide robust genetic evidence of causal associations between IBD and cancer risk. Exposure to IBD might not independently contribute to the risk of cancers, and the increased risk of cancers observed in observational studies might be attributed to factors accompanying the diagnosis of IBD.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-024-03352-9.
Keywords: Inflammatory bowel disease, Cancers, Mendelian randomization, Genetic correlation, Colocalization analysis
Background
Cancers, which are important causes of morbidity and mortality and carry an enormous disease burden, remain a formidable challenge [1]. An increasing number of studies have shown that the incidence of cancer in various organs diagnosed in adults younger than 50 years old has been increasing globally [2–4]. Therefore, identifying risk factors in young people is crucial for preventing early-stage cancer.
Inflammatory bowel disease (IBD), which includes Crohn’s disease (CD) and ulcerative colitis (UC) and is more common in young people, is a debilitating and progressive disorder of the gastrointestinal tract [5]. The symptoms of IBD patients, such as abdominal pain, diarrhea, bloody stool, and frequent bowel movements, seriously affect quality of life, affecting millions of people worldwide and increasing in prevalence [5–7]. IBD is a chronic disorder characterized by intestinal inflammation that can induce adverse outcomes [8].
Because IBD is difficult to treat, there is an increased risk of cancer without formal treatment [9]. The well-established connection between IBD and intestinal cancer, particularly colorectal cancer (CRC), is supported by numerous observational studies [10–12]. This heightened risk of intestinal cancer may be attributed to long-term chronic inflammation [13]. Observational studies have also identified associations between IBD and extraintestinal cancer, such as skin, hepatobiliary, and lung cancer [12, 14]. However, traditional observational studies have found it challenging to mitigate the biases caused by confounding and reverse causation. In addition, an umbrella review and reanalysis of meta-analyses of observational studies have indicated that associations between IBD and different cancers are inconsistent [15]. Despite several studies that have examined the potential causal associations between IBD and cancer risk by using the Mendelian randomization (MR) approach [16–20], which can overcome the limitations of traditional observational study designs by using Mendel’s law, most of the MR studies were based on European population, and only one MR study explored the association between IBD and hepatobiliary pancreatic cancer in the East Asian population [18]. However, whether lifelong exposure to IBD has a causal association with cancers is still largely unknown.
Summarized data derived from genome-wide association studies (GWAS) are widely reported to make two-sample MR studies easy and popular to implement [21, 22]. A recent GWAS with a larger sample size reported loci associated with IBD in East Asian populations [23], which allowed us to explore the associations between IBD and cancers in the East Asian population. Although the MR approach utilizes the random allocation of genotypes during meiosis to yield natural randomized controlled trials, effectively mitigating potential unmeasured confounding and reverse causation [24, 25], this approach still faces some challenges.
Recently, increasing attention has been given to combining genetic correlation with MR analyses [26, 27] or combining MR with colocalization analyses [28, 29] as analytical frameworks to illuminate causal associations. Linkage disequilibrium score regression (LDSC) enables the assessment of genetic correlation between two traits, offering a broad range of genetic insights without being affected by sample overlap; its principal limitation lies in its incapacity to infer causality [30]. MR analysis provides evidence similar to randomized controlled trial [24], but the drawback is the need to satisfy certain assumptions and susceptibility to pleiotropy and weak instrumental variable bias. Colocalization analysis can ascertain whether two or more traits or diseases share the same genetic variants as their potential causal variants, offering specific genetic variant information and assessing potential pleiotropy; however, this approach is limited by its statistical power [31].
Therefore, we integrated genetic correlation, MR, and colocalization approaches to investigate the causal relationship between IBD and cancer risk. This is the first study using such a comprehensive approach, balancing the genetic evidence from these three methods to provide a robust understanding of the causal association between IBD and cancer risk across East Asian and European populations.
Methods
Study design
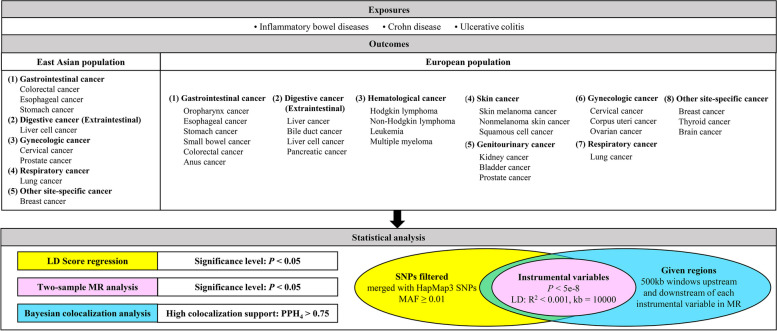
Our study was based on summary-level GWAS data available for East Asian and European populations to explore the potential causal associations between IBD (including UC and CD) and the risk of cancers. In the East Asian population, eight site-specific cancers were included in the analysis: colorectal, esophageal, stomach, liver cell, cervical, prostate, lung, and breast cancers. In the European population, 27 site-specific cancers were selected, including oropharynx, esophageal, stomach, small bowel, colorectal, anus, liver, bile duct, liver cell, pancreatic, Hodgkin lymphoma, non-Hodgkin lymphoma, leukemia, multiple myeloma, skin melanoma, nonmelanoma skin, squamous cell, kidney, bladder, prostate, cervical, corpus uteri, ovarian, lung, breast, thyroid, and brain cancers. Genome-wide LDSC was used to assess the genetic association between IBD and cancer. A standard two-sample MR analysis was performed to clarify the causal relationship between IBD and cancer. Colocalization analysis was used to investigate the local genetic structure shared between IBD and cancer and to assess whether the causal association was due to chance. Figure 1 provides an overview of the design and process of our analysis.
Fig. 1.
An overview of the study design and process. LD, linkage disequilibrium; MAF, minor allele frequency; MR, Mendelian randomization
Our study followed the STROBE-MR guidelines [22]. The STROBE-MR checklist is available in Additional file 1: Table S1.
Data sources
GWAS data for IBD
The GWAS summary-level data for IBD/UC/CD patients based on East Asian and European populations were released in the published study [23, 32]. The meta-GWAS for the East Asian population included 14,393 patients with IBD and 15,456 controls, 7372 patients with CD and 14,946 controls, and 6862 patients with UC and 15,456 controls [23]. The meta-GWAS for the European population included 25,042 patients with IBD and 34,915 healthy controls, 12,194 with CD and 28,072 healthy controls, and 12,366 with UC and 33,609 healthy controls [32]. More detailed information can be found in Additional file 1: Table S2.
GWAS data for cancers
GWAS summary statistics of eight cancers for the East Asian population were obtained from a large-scale GWAS conducted by Kazuyoshi Ishigaki et al. [33] from the Biobank Japan. The samples were collected from 12 medical institutions across Japan and included approximately 200,000 participants. The sample size of the GWAS data for cancers ranged from 90,336 to 212,453, and the number of cases ranged from 605 to 7062. More details of the GWAS data for cancers are listed in Additional file 1: Table S2.
With respect to the European population, we used data from the latest available GWAS data, which had the largest sample size or the largest sample size of patients for the outcome under investigation. The GWAS summary statistics for 27 cancers were obtained mainly from the following sources: (i) Rashkin SR et al. conducted GWAS across 18 types of cancer within two population-based cohorts: the UK Biobank and the Kaiser Permanente Genetic Epidemiology Research on Adult Health and Aging cohorts [34]; (ii) Jiang L et al. utilized fastGWA-GLMM to the UK Biobank data and subsequently procured full summary statistics [35]; (iii) A study conducted by Kimberley Burrows et al. presented detailed information on the GWAS focusing on pan-cancer and site-specific cancers among participants from the UK Biobank [36]; (iv) Meta-analysis with Transdisciplinary Research of Cancer in Lung of the International Lung Cancer Consortium and Lung Cancer Cohort Consortium, performed by McKay JD et al. [37]; and (v) Seviiri M et al. executed a multitrait genetic analysis of more than 300,000 participants from Europe, Australia, and the USA [38]. The sample size of the GWAS data ranged from 85,716 to 456,348, and the number of cases ranged from 104 to 29,266. More details of the GWAS data for cancers are listed in Additional file 1: Table S2.
Data analysis
Linkage disequilibrium score regression
Genome-wide LDSC [30] was used to assess the genetic association between IBD and cancer (https://github.com/bulik/ldsc). The LDSC calculates genetic correlation by considering the impact of all single nucleotide polymorphisms (SNPs), even those that do not achieve genome-wide significance. We removed SNPs that did not merge with HapMap3 SNPs and those with a minor allele frequency less than 0.01. The findings are presented as genetic correlation (rg) with standard error (SE). The results of LDSC analysis could not be available if either one or both traits exhibited too low heritability [39, 40].
P values less than 0.05 were considered suggestive of evidence for a potential genetic correlation. Statistical analysis was performed using LDSC v1.0.1.
Mendelian randomization analysis
MR analysis is an instrumental variable analysis that uses genetic variants as instrumental variables (IVs) to study causality. Additional file 2: Fig. S1 provides an overview of our MR design. MR analysis is based on three main assumptions: (i) genetic instruments are associated with exposure, (ii) genetic instruments are independent of any confounder, and (iii) genetic instruments affect outcome only through exposure.
Conditionally uncorrelated variants strongly (P < 5 × 10−8) and independently (linkage disequilibrium [LD] r2 < 0.001, window size = 10,000 kb) associated with IBD/UC/CD were extracted as IVs. The LD proxies were defined using 1000 genomes from East Asian and European samples. We calculated the overall R2 and F-statistics by summing the estimated R2 [R2 = 2 × EAF × (1-EAF) × beta2] and F-statistics [F = beta2/se2] for each SNP. The F-statistics for all traits under consideration exceeded 10 [41], indicating no potential weak instrument bias. In addition, the mRnd website tool (https://shiny.cnsgenomics.com/mRnd/) was used to calculate the statistical power of the MR analysis.
We selected the random-effects inverse-variance weighted (IVW) method as the primary analysis, and sensitivity analyses, including the weighted median (WM), penalized weighted median (PWM), MR-Egger, MR pleiotropy residual sum and outlier (PRESSO), and MR-robust adjusted profile score (RAPS) analyses, were performed to further explore the stability of the results. In addition, the intercepts of the MR-Egger analysis and MR-PRESSO global test were calculated to evaluate pleiotropy. When the global test P values in the MR-PRESSO analysis were less than 0.05, the MR-PRESSO estimates were the results after outlier removal. We conducted a Steiger directionality test to rule out potential reverse causality.
In addition, to further avoid potential pleiotropy, we scanned PhenoScanner (on February 6th, 2024; http://www.phenoscanner.medschl.cam.ac.uk) for identifying traits associated with instrumental variables (R2 ≥ 0.8, P values ≤ 5 × 10−8), and performed MR after removing SNPs associated with confounding factors (body mass index, waist circumference, hip circumference, waist-hip ratio, percentage of body fat, smoking, alcohol consumption, insomnia, depression, and physical activity). For significant MR findings, we also conducted multivariable MR (MVMR) analysis to obtain estimates independent of these confounding factors.
Pleiotropy poses a challenge to interpreting MR results. Therefore, we reported the primary IVW results, combined with methods for detecting and correcting for pleiotropy, to fully account for the bias from pleiotropy.
P values less than 0.05 were considered suggestive of evidence for a potential causal association. The IVW method, sensitivity analyses (excluding MR-RAPS), Steiger directionality test, and MVMR were implemented using the “TwoSampleMR” (version 0.5.7) package in R version 4.3.1. The MR-RAPS analysis was performed using the “mr.raps” (version 0.2) package in R version 4.3.1.
Bayesian colocalization analysis
We used this method to assess whether two associated traits were consistent with shared causal variant(s) according to the included IVs. Five mutually exclusive hypotheses were tested: (1) there is no causal genetic variant for either trait (H0); (2) there is one causal genetic variant for trait 1 only (H1); (3) there is one causal genetic variant for trait 2 only (H2); (4) there are two distinct causal genetic variants, one for each trait (H3); and (5) there is a causal genetic variant for both traits (H4). The posterior probability (PP) is used to quantify the support of each hypothesis and is expressed as PPH0, PPH1, PPH2, PPH3, and PPH4 [31]. We selected regions that had 500 kb windows upstream and downstream of each instrumental variable in MR for analysis, and the average value of PPH4 across all regions was taken as the final colocalization result.
A PPH4 level greater than 75% was considered suggestive of evidence for a causal genetic variant for both traits. These PPs were calculated using the “coloc (version 5.2.3)” package in R version 4.3.1.
Possible results and explanations
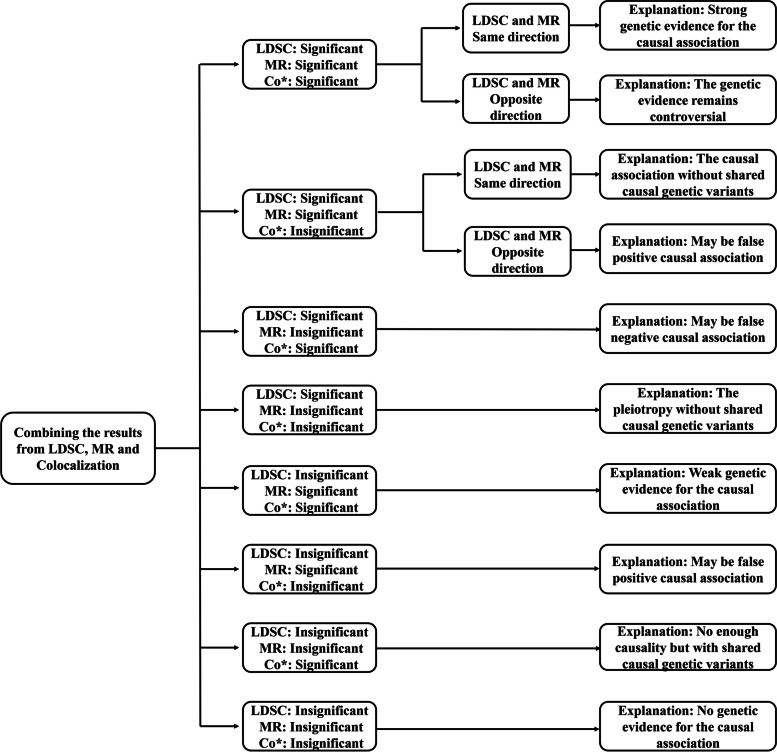
As shown in Fig. 2, we summarized ten possible results and nine explanations combining the results from genetic correlation, MR, and colocalization analyses based on the effects and levels of statistical significance/direction. The results from genetic correlation and MR analyses performed both statistically significant and direct, while colocalization analysis had only statistically significant. The results of MR comprehensively considered the primary analysis of the IVW method and excluded the potential bias of pleiotropy.
Fig. 2.
Summarize possible results and explanations. P values less than 0.05 in the LDSC and MR analyses were considered suggestive of evidence for a potential association, and a PPH4 level greater than 75% was considered suggestive of evidence for a causal genetic variant for both traits. Co*, colocalization; LDSC, linkage disequilibrium score regression; MR, Mendelian randomization
The specific results and explanations used were as follows: (Explanation i) when all three results were significant and in the same direction, it was interpreted as strong genetic evidence for the causal association; (Explanation ii) when all three results were significant and in the opposite direction, it was interpreted that the genetic evidence remains controversial; (Explanation iii) when the results from genetic correlation and MR analyses were significant and in the same direction, it was interpreted as the causal association without shared causal genetic variants; (Explanation iv) when the results from genetic correlation and MR analyses were significant and in the opposite direction or when only the result from MR analysis was significant, it might be a false positive causal association; (Explanation v) when the results from genetic correlation and colocalization analyses were significant, it might be a false negative causal association; (Explanation vi) when only the result from LDSC analysis was significant, it was interpreted as pleiotropy without shared causal genetic variants; (Explanation vii) when the results from MR and colocalization analyses were significant, it was interpreted as weak genetic evidence for causal association; (Explanation viii) when only the result from colocalization analysis was significant, it was interpreted as no enough causality but with shared causal genetic variants; (Explanation ix) when all three results were insignificant, it was interpreted as no genetic evidence for the causal association.
Risk of bias assessment
To assess the quality of the MR studies, we considered 8 potential biases: (1) weak instrument bias, (2) pleiotropy bias, (3) bias from sample overlap, (4) bias from crowd stratification, (5) bias from inconsistency with sensitivity analyses, (6) bias from lack of repeatability, (7) bias from inconsistency with other study design evidence, and (8) reporting bias. Each domain was judged as having a low, moderate (no information was classified as moderate bias), or high risk of bias. The detailed risk bias assessment criteria used in the Mendelian randomization studies can be found in Additional file 1: Table S3.
Results
Genetic correlation between IBD and cancers
As shown in Table 1, the genetic correlations of IBD, UC, and CD with cancer in the East Asian population ranged from 0.009 to 0.202. There were no significant genetic correlations between IBD, CD, or UC and cancer (all P values > 0.05).
Table 1.
The genetic correlations between IBD and cancers caused by LDSC in the East Asian population
| Outcomes | IBD | CD | UC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| rg | se | P | rg | se | P | rg | se | P | |
| Colorectal cancer | − 0.082 | 0.080 | 0.305 | − 0.151 | 0.078 | 0.052 | − 0.022 | 0.099 | 0.820 |
| Esophageal cancer | − 0.040 | 0.069 | 0.565 | − 0.099 | 0.071 | 0.161 | − 0.013 | 0.084 | 0.879 |
| Stomach cancer | 0.073 | 0.075 | 0.334 | 0.069 | 0.073 | 0.340 | 0.025 | 0.089 | 0.778 |
| Liver cell cancer | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Cervical cancer | − 0.140 | 0.112 | 0.214 | − 0.202 | 0.122 | 0.096 | − 0.069 | 0.126 | 0.586 |
| Prostate cancer | − 0.062 | 0.072 | 0.388 | − 0.084 | 0.078 | 0.283 | − 0.022 | 0.087 | 0.804 |
| Lung cancer | − 0.009 | 0.120 | 0.941 | − 0.051 | 0.125 | 0.683 | − 0.025 | 0.146 | 0.864 |
| Breast cancer | − 0.049 | 0.066 | 0.452 | − 0.088 | 0.068 | 0.194 | − 0.022 | 0.078 | 0.776 |
CD Crohn’s disease, IBD inflammatory bowel disease, LDSC linkage disequilibrium score regression, NA not available, UC ulcerative colitis
As shown in Table 2, the genetic correlations of IBD, UC, and CD with cancer ranged from 0.0004 to 0.133 in the European population. There were no significant genetic correlations between IBD, CD, or UC and cancer (all P values > 0.05).
Table 2.
The genetic correlations between IBD and cancers caused by LDSC in the European population
| Outcomes | IBD | CD | UC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| rg | se | P | rg | se | P | rg | se | P | |
| Oropharynx cancer | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Esophageal cancer | 0.057 | 0.036 | 0.110 | 0.042 | 0.034 | 0.217 | 0.054 | 0.043 | 0.214 |
| Stomach cancer | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Small bowel cancer | − 0.066 | 0.138 | 0.636 | − 0.092 | 0.166 | 0.579 | 0.022 | 0.119 | 0.855 |
| Colorectal cancer | − 0.032 | 0.045 | 0.476 | − 0.017 | 0.043 | 0.685 | − 0.030 | 0.053 | 0.566 |
| Anus cancer | 0.011 | 0.035 | 0.748 | 0.043 | 0.037 | 0.244 | − 0.003 | 0.043 | 0.954 |
| Liver cancer | 0.035 | 0.100 | 0.727 | − 0.008 | 0.089 | 0.925 | 0.133 | 0.316 | 0.673 |
| Bile duct cancer | 0.011 | 0.052 | 0.830 | 0.042 | 0.063 | 0.510 | 0.026 | 0.069 | 0.702 |
| Liver cell cancer | − 0.026 | 0.031 | 0.397 | − 0.031 | 0.030 | 0.310 | − 0.029 | 0.038 | 0.452 |
| Pancreatic cancer | 0.038 | 0.045 | 0.395 | 0.002 | 0.044 | 0.974 | 0.052 | 0.050 | 0.299 |
| Hodgkin lymphoma | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Non-Hodgkin lymphoma | − 0.026 | 0.033 | 0.417 | − 0.017 | 0.033 | 0.612 | − 0.044 | 0.038 | 0.250 |
| Leukemia | − 0.056 | 0.046 | 0.225 | − 0.040 | 0.043 | 0.353 | − 0.020 | 0.053 | 0.714 |
| Multiple myeloma | 0.004 | 0.030 | 0.898 | − 0.024 | 0.030 | 0.434 | 0.037 | 0.039 | 0.338 |
| Skin melanoma cancer | 0.002 | 0.020 | 0.939 | 0.006 | 0.019 | 0.772 | 0.003 | 0.026 | 0.909 |
| Nonmelanoma skin cancer | − 0.001 | 0.012 | 0.935 | 0.006 | 0.013 | 0.657 | − 0.004 | 0.016 | 0.790 |
| Squamous cell cancer | 0.005 | 0.009 | 0.527 | 0.0004 | 0.009 | 0.961 | 0.014 | 0.011 | 0.204 |
| Kidney cancer | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Bladder cancer | 0.060 | 0.036 | 0.092 | 0.047 | 0.035 | 0.175 | 0.055 | 0.040 | 0.163 |
| Prostate cancer | − 0.024 | 0.016 | 0.135 | − 0.012 | 0.015 | 0.429 | − 0.030 | 0.019 | 0.126 |
| Cervical cancer | 0.002 | 0.029 | 0.944 | − 0.005 | 0.028 | 0.846 | 0.015 | 0.032 | 0.645 |
| Corpus uteri cancer | − 0.064 | 0.062 | 0.306 | − 0.038 | 0.056 | 0.504 | − 0.088 | 0.080 | 0.270 |
| Ovarian cancer | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Lung cancer | − 0.006 | 0.015 | 0.676 | − 0.002 | 0.014 | 0.877 | − 0.001 | 0.019 | 0.966 |
| Breast cancer | 0.003 | 0.016 | 0.825 | − 0.006 | 0.016 | 0.698 | 0.010 | 0.018 | 0.586 |
| Thyroid cancer | 0.010 | 0.035 | 0.779 | 0.037 | 0.037 | 0.321 | − 0.004 | 0.044 | 0.932 |
| Brain cancer | − 0.011 | 0.044 | 0.808 | − 0.017 | 0.041 | 0.682 | − 0.010 | 0.056 | 0.858 |
CD Crohn’s disease, IBD inflammatory bowel disease, LDSC linkage disequilibrium score regression, NA not available, UC ulcerative colitis
The causal association between IBD and cancers according to Mendelian randomization analysis
In the East Asian population, the number of IVs for all considered traits ranged from 28 to 56, the R2 varied from 55.06 to 122.39%, the F-statistics varied from 1860 to 4217, and the statistical power varied from 6 to 53% (Additional file 1: Table S4). In the European population, the IVs ranged from 44 to 116, the R2 varied from 41.04 to 85.37%, the F-statistics for all traits under consideration varied between 3226 and 8196, and the statistical power varied from 5 to 91% (Additional file 1: Table S5). All F-statistics were greater than 10, suggesting no potential weak instrument bias.
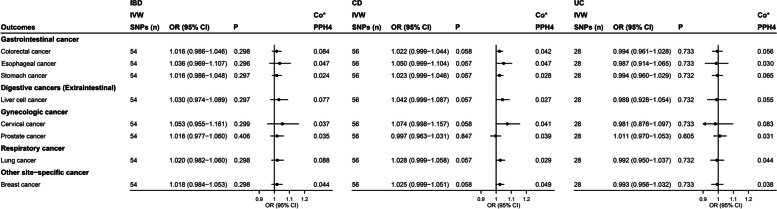
According to our primary analysis of IVW data, IBD, including CD and UC, had no significant associations with any cancer in the East Asian population (all P values > 0.05; Fig. 3). There were some suggestive associations between CD and CRC (odds ratio [OR] = 1.036, 95% confidence interval [CI] = 1.000–1.074, P = 0.048) according to the WM method; between IBD and stomach cancer (OR = 1.050, 95% CI = 1.001–1.101, P = 0.044), CD and liver cell cancer (OR = 1.071, 95% CI = 1.001–1.145, P = 0.048), and between CD and lung cancer (OR = 1.048, 95% CI = 1.000–1.097, P = 0.049; Additional file 1: Table S6) according to the PWM method. The MR-Egger intercept and MR-PRESSO test did not reveal evidence of horizontal pleiotropy (all P values > 0.05; Additional file 1: Table S6). Steiger directionality test results indicated that all causal directions were correct (Additional file 1: Table S6). There were pleiotropic instrumental variables for IBD and CD, but no pleiotropic instrumental variables were identified for UC (Additional file 1: Table S7). After removing pleiotropic instrumental variables, no significant association between IBD and cancer was found using any of the MR methods (all P values > 0.05; Additional file 1: Table S8).
Fig. 3.
Forest plot of the results of IVW analysis and colocalization analysis for the association of IBD with cancers in the East Asian population. Co*, colocalization analysis; the average value of PPH4 across all regions was taken as the final colocalization result. CD, Crohn’s disease; CI, confidence interval; IBD, inflammatory bowel disease; IVW, inverse-variance-weighted; OR, odds ratio; SNPs, single nucleotide polymorphisms; UC, ulcerative colitis
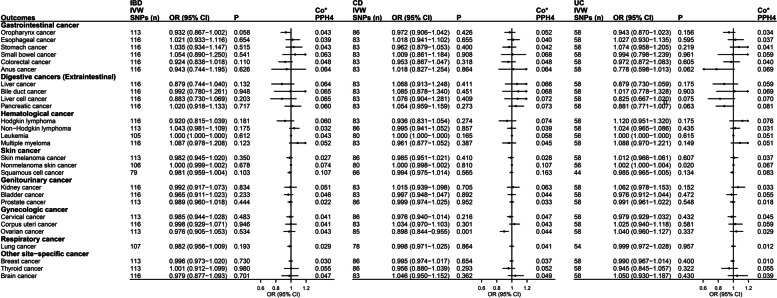
The primary analysis of the IVW data revealed significant associations between CD and ovarian cancer (OR = 0.898, 95% CI = 0.844–0.955, P = 0.0007) and between UC and nonmelanoma skin cancer (OR = 1.002, 95% CI = 1.000–1.004, P = 0.019; Fig. 4) in the European population. These findings were also found in several sensitivity analyses (Additional file 1: Table S9). The MR-Egger intercept test did not reveal evidence of horizontal pleiotropy between CD and ovarian cancer (P = 0.739) or between UC and nonmelanoma skin cancer (P = 0.115). The global test P values from the MR-PRESSO analysis did not indicate potential pleiotropy between CD and ovarian cancer (P = 0.812); however, it revealed evidence of potential pleiotropy between UC and nonmelanoma skin cancer (P < 0.001). After removing outliers, the MR-PRESSO results still suggested a significant association between UC and nonmelanoma skin cancer (OR = 1.001, 95% CI = 1.000–1.003, P = 0.037). Additionally, the global test P values of the MR-PRESSO analysis for IBD and squamous cell cancer, as well as for UC and squamous cell cancer, were less than 0.001. After outlier exclusion, the MR-PRESSO results revealed a suggestive association between IBD and squamous cell cancer (OR = 0.970, 95% CI = 0.955–0.984, P = 0.0002) and between UC and squamous cell cancer (OR = 0.972, 95% CI = 0.959–0.986, P = 0.0003; Additional file 1: Table S9). The results of the Steiger directionality test indicated that all causal directions were correct (Additional file 1: Table S9). After removing pleiotropic instrumental variables (Additional file 1: Table S10), the MR results revealed a significant association between CD and ovarian cancer (OR = 0.890, 95% CI = 0.835–0.949, P = 0.0003), whereas there was no significant association between UC and nonmelanoma skin cancer (OR = 1.001, 95% CI = 0.999–1.002, P = 0.424; Additional file 1: Table S11). The MVMR analysis was further performed to show a lack of significant association between CD and ovarian cancer (OR = 0.940, 95% CI = 0.856–1.033, P = 0.201) or between UC and nonmelanoma skin cancer (OR = 0.983, 95% CI = 0.903–1.070, P = 0.693; Additional file 1: Table S12–13) after adjusting for confounding factors.
Fig. 4.
Forest plot of results from IVW analysis for the association of IBD with cancers in the European population. Co*, colocalization analysis; the average value of PPH4 across all regions was taken as the final colocalization result. CD, Crohn’s disease; CI, confidence interval; IBD, inflammatory bowel disease; IVW, inverse variance weighted; OR, odds ratio; SNP, single nucleotide polymorphism; UC, ulcerative colitis
Colocalization analysis for IBD and cancers
There was no shared causal variant to prove the associations of IBD, CD, or UC with cancers in East Asian or European populations using colocalization analysis based on the average value of PPH4 across all regions (PPH4 < 75%; Figs. 3 and 4). In addition, the values of PPH4 across each region are presented in Additional file 1: Tables S14–15. The colocalization results suggested that there may be no common biological mechanism between IBD and cancer.
Evaluation of evidence from the MR approach
Overall, the potential biases were rated high due to pleiotropy bias, inconsistency with the sensitivity analyses, and lack of repeatability (Additional file 1: Table S3). However, we performed analyses to avoid potential pleiotropy with the use of PhenoScanner tools and the MVMR approach. In addition, we comprehensively considered the results of genetic correlation and MR and colocalization analyses and different populations.
Discussion
After fully balancing the genetic evidence from genetic correlation, MR and colocalization analyses, we used the latest and largest datasets to investigate the causal associations of genetically determined IBD with the risk of cancers across East Asian and European populations. In the East Asian population, neither of the three approaches provided genetic evidence for the associations of IBD with eight common cancers. In the European population, we found evidence for associations between CD and ovarian cancer and between UC and nonmelanoma skin cancer via the MR method. However, these findings were not supported by the results of LDSC and colocalization analyses. Genetic backgrounds vary between different ethnicities, which might lead to differences in the results of MR analysis. However, we did not provide robust genetic evidence showing the causal associations between IBD and cancer risk in either East Asian or European populations.
Our findings were somewhat inconsistent with previous MR study results. Previous MR studies have focused on examining the association of genetically predicted IBD with intestinal cancers, including CRC, oral cancer, and pharyngeal cancer; hepatobiliary pancreatic cancer [16, 18–20]; and only one of the extraintestinal cancers, prostate cancer [17]. In addition, a recent comprehensive MR analysis exploring the associations between IBD and 32 cancers in the European population showed causal associations of predicted IBD with the oral cavity and breast cancer [42]. The different findings in these studies may be due to the use of different populations and different summary-level GWAS data for exposure and outcome. Compared to other studies, we utilized the latest and largest GWAS summary data across East Asian and European populations. In addition, we combined the genetic evidence from different methods.
To the best of our knowledge, no previous studies have systematically and comprehensively investigated the genetic association between IBD and various cancer sites, balancing genetic evidence from genetic correlation, MR, and colocalization analyses. Our study presented one result and explanation: MR analysis was significant, whereas LDSC and colocalization analyses were not, which suggested that the findings in MR may be prone to false positive bias. When the proportion of heritability explained by genome-wide significant SNPs, as determined by MR approaches, is low, the accuracy of MR results may be inferior to LDSC [30]. MR analysis is more liberal than colocalization analysis, and significant MR findings without evidence of colocalization may suggest distinct causal variants for exposure and outcome that are in linkage disequilibrium, indicating that the assumption of MR may have been violated and that the results are unreliable [29]. In addition, we evaluated and corrected pleiotropy using several methods, including MR-Egger analysis, the MR-PRESSO global test, the removal of instrumental variables associated with confounding factors, and MVMR analysis, to comprehensively assess the MR results. Overall, our findings suggest that the associations between CD and ovarian cancer and between UC and nonmelanoma skin cancer according to MR analysis may be false. Our study summarized ten possible results and explained them, filling the gap in this area by combining the evidence from genetic correlation, MR, and colocalization analyses.
Considering the pathogenesis of chronic systemic inflammation and immune dysregulation, the disorders associated with IBD extend beyond intestinal diseases, leading to an increased risk of extraintestinal manifestations in IBD patients [43]. Our results are mostly contrary to the hypothesis of pathogenesis and the findings of umbrella reviews based on observational studies [15]. The previously reported observational associations between IBD and cancer risk might be affected by detection bias, unobserved confounding factors, and pleiotropy. First, the excess diagnosis due to endoscopic screening and surveillance in patients with IBD might contribute to the rising risk of common cancers in observational studies. A Scandinavian register-based cohort study from 1969 to 2017 recently showed that the incidence of CRC increased during the first year after IBD diagnosis but significantly decreased after the first year of follow-up [44]. A greater extent of medical examinations, particularly a higher frequency of colonoscopy/sigmoidoscopy, in IBD patients than in the control population induces detection bias [45] and results in a false association between IBD and CRC. During long-term follow-up after IBD diagnosis, it is possible that the decrease in CRC incidence could be related to changes in lifestyle or medical management during the treatment of IBD. In addition, treatment following the diagnosis of IBD might contribute to the increased risk of common cancers in observational studies. A systematic review summarized the protective and risk factors for CRC in IBD patients and showed that primary sclerosing cholangitis, postinflammatory polyps, and colon segment resection were risk factors for the incidence of CRC in IBD patients [46]. Because genetically predicted IBD was also associated with a higher probability of taking anti-IBD medication, it could be that exposure to such drugs, rather than having IBD, was associated with the risk of cancer. Increasing evidence has shown that common treatments for IBD patients, such as glucocorticoids, thiopurines, and immunomodulatory agents, promote the development of cancer [47–51]. Moreover, our MR findings suggest an association between CD and a reduced risk of ovarian cancer, which could be attributed to pleiotropy. Our MVMR analysis showed an insignificant causal association between CD and ovarian cancer.
Overall, our findings suggest that IBD might not be a trigger for developing cancers, and the observed association between IBD and cancer needs to be interpreted very cautiously in clinical practice. Future studies are warranted to investigate the common risk factors or confounding factors underlying the association between IBD and cancer risk. In addition, the increased risk of cancer in observational studies might be attributed to factors accompanying the diagnosis of IBD, especially treatments for IBD patients. A full understanding of cancer as an adverse consequence of IBD is necessary to ensure appropriate vigilance against cancer. These findings should be confirmed by better-designed epidemiologic studies.
Strengths and limitations
Our study has notable strengths. The genetic correlation, MR, and colocalization analyses have specific strengths and limitations that can complement each other to some extent to mitigate some false negative and false positive results. Our work is a landmark study in that it provides guidance on how to integrate evidence from genetics-driven studies accumulated to date to enable a more reliable interpretation of the epidemiological relationship between IBD and cancer. In addition, we used data from the latest available GWAS data with the largest sample size or the largest sample size of patients for exposure and outcome under investigation, and we repeated our findings using different methods and different ancestries. The objective estimates of the association between IBD and cancer may be more precise than previous estimates and are less prone to bias.
We acknowledge the limitations of this study. First, because either one or both traits had too low heritability, some results from LDSC analysis were not available. Second, a key assumption of MR is that SNPs are not associated with any confounders of the exposure or the outcome. Even if we considered the pleiotropic bias, any MR study could not completely rule out pleiotropic bias. Third, we investigated three exposed phenotypes and used different sets of IVs, but the power of our analyses with the different instruments varied, which could have induced weak instrumental variable bias. Furthermore, the colocalization analysis of genes and proteins is widely used but is still in the development stage for determining the optimal dichotomy. We included all IVs in the colocalization analysis and chose a cut-off value of 0.75 for PPH4. Besides, we used multiple data sources to increase the statistical power, but the relatively small number of cancer patients reduced the power of the MR analyses. In addition, despite leveraging data from genetic studies with very large sample sizes, our study was insufficient to detect very small effects. Moreover, we could not further adjust for potential confounders due to a lack of individual-level data. Finally, due to the limited cancer GWAS data on Asians, the results for more than half of the cancers could not be validated in the Asian population. We should carefully utilize our findings in racially and ethnically diverse populations.
Conclusions
In summary, according to comprehensive genetic correlation, MR, and colocalization analyses, inherited lifetime exposure to IBD was not strongly associated with the risk of cancer. Exposure to IBD might not independently contribute to the risk of cancers, underlying that the increased risk of cancers observed in observational studies might be attributed to factors accompanying the diagnosis of IBD, especially treatments for IBD patients.
Supplementary Information
Additional file 1: Table S1. STROBE Statement-Checklist of items in reports of Mendelian randomization. Table S2. Characteristics of the summary GWAS data. Table S3. Assessment criteria for risk of bias assessment via the MR approach. Table S4. Characteristics of the instrumental variables in the East Asian population. Table S5. Characteristics of the instrumental variables in the European population. Table S6. MR estimates of the causal associations between IBD and cancers in the East Asian population. Table S7. Related traits of genetic variants used for Mendelian randomization analysis in the East Asian population. Table S8. MR estimates of the causal associations between IBD and cancers after removing instrumental variables associated with confounding factors in the East Asian population. Table S9. MR estimates of the causal associations between IBD and cancers in the European population. Table S10. Related traits of genetic variants used for Mendelian randomization analysis in the European population. Table S11. MR estimates of the causal associations between IBD and cancers after removing instrumental variables associated with confounding factors in the European population. Table S12. MVMR estimates of the causal associations between CD and ovarian cancer after adjusting for confounding factors. Table S13. MVMR estimates of the causal associations between UC and nonmelanoma skin cancer after adjusting for confounding factors. Table S14. Colocalization analysis for the associations between IBD and cancers in the East Asian population. Table S15. Colocalization analysis for the associations between IBD and cancers in the European population.
Additional file 2: Fig. S1. Mendelian randomization model. Solid arrows = causal effects; dashed arrows = causal effects prohibited by MR assumptions II and III. Assumption I: Genetic instruments are associated with exposures; Assumption II: Genetic instruments are independent of confounding factors; Assumption III: Genetic instruments affect outcomes only through exposures. IVW: inverse-variance-weighted.
Acknowledgements
The authors thank all the investigators and participants who contributed to the summary GWAS data.
Abbreviations
- CD
Crohn’s disease
- CI
Confidence interval
- CRC
Colorectal cancer
- GWAS
Genome-wide association study
- IBD
Inflammatory bowel disease
- IVs
Instrumental variables
- IVW
Inverse-variance weighted
- LD
Linkage disequilibrium
- LDSC
Linkage disequilibrium score regression
- MR
Mendelian randomization
- MVMR
Multivariable Mendelian randomization
- OR
Odds ratio
- PP
Posterior probability
- PRESSO
Pleiotropy residual sum and outlier
- PWM
Penalized weighted median
- RAPS
Robust adjusted profile score
- rg
Genetic correlation
- SE
Standard error
- SNPs
Single nucleotide polymorphisms
- UC
Ulcerative colitis
- WM
Weighted median
Authors’ contributions
Study conception: DL, YL, and YXW; study design: DL, YXW, and MLC; data extraction and analyses: DL, MLC, HTW, and CGZ; results presentation and interpretation: DL, MLC, HTW, WJC, and YXW; manuscript drafting and revising: DL, MLC, and HTW. The work reported in the paper has been performed by the authors, unless clearly specified in the text. All authors read and approved the final manuscript.
Funding
This work was supported by the China Postdoctoral Science Foundation (grant number 2021M703366) and the National Key R&D Program of China (grant number 2017YFE0118800). The funders had no role in the study design, data analysis, interpretation of the data, or preparation of the manuscript.
Availability of data and materials
The data underlying this article are available in the article and in its online supplementary material.
Declarations
Ethics approval and consent to participate
The analyses for this study were based on publicly available summary datasets, and no additional ethics approval or consent was needed.
Consent for publication
This manuscript does not include details, images, or videos relating to an individual person; therefore, consent for publication is not needed beyond the informed consent provided by all study participants as described above.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Di Liu and Meiling Cao contributed equally to this work.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Ugai T, Sasamoto N, Lee HY, Ando M, Song M, Tamimi RM, et al. Is early-onset cancer an emerging global epidemic? Current evidence and future implications. Nat Rev Clin Oncol. 2022;19(10):656–673. doi: 10.1038/s41571-022-00672-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.GBD 2019 Adolescent Young Adult Cancer Collaborators The global burden of adolescent and young adult cancer in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Oncol. 2022;23(1):27–52. doi: 10.1016/S1470-2045(21)00581-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jianhui Z, Liying X, Jing S, Mingyang S, Lijuan W, Shuai Y, et al. Global trends in incidence, death, burden and risk factors of early-onset cancer from 1990 to 2019. BMJ Oncology. 2023;2(1):e000049. doi: 10.1136/bmjonc-2023-000049. [DOI] [Google Scholar]
- 5.GBD 2017 Inflammatory Bowel Disease Collaborators The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol Hepatol. 2020;5(1):17–30. doi: 10.1016/S2468-1253(19)30333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390(10114):2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan GG, Windsor JW. The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2021;18(1):56–66. doi: 10.1038/s41575-020-00360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodson R. Inflammatory bowel disease. Nature. 2016;540(7634):S97. doi: 10.1038/540S97a. [DOI] [PubMed] [Google Scholar]
- 9.Parigi TL, D'Amico F, Abreu MT, Dignass A, Dotan I, Magro F, et al. Difficult-to-treat inflammatory bowel disease: results from an international consensus meeting. Lancet Gastroenterol Hepatol. 2023;8(9):853–859. doi: 10.1016/S2468-1253(23)00154-1. [DOI] [PubMed] [Google Scholar]
- 10.Jess T, Gamborg M, Matzen P, Munkholm P, Sørensen TIA. Increased risk of intestinal cancer in Crohn's disease: a meta-analysis of population-based cohort studies. Off J Am College Gastroenterol ACG. 2005;100(12):2724–9. doi: 10.1111/j.1572-0241.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 11.Castaño-Milla C, Chaparro M, Gisbert JP. Systematic review with meta-analysis: the declining risk of colorectal cancer in ulcerative colitis. Aliment Pharmacol Ther. 2014;39(7):645–659. doi: 10.1111/apt.12651. [DOI] [PubMed] [Google Scholar]
- 12.Wu S, Xie S, Yuan C, Yang Z, Liu S, Zhang Q, et al. Inflammatory bowel disease and long-term risk of cancer: a prospective cohort study among half a million adults in UK biobank. Inflamm Bowel Dis. 2023;29(3):384–395. doi: 10.1093/ibd/izac096. [DOI] [PubMed] [Google Scholar]
- 13.Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140(6):1807–1816. doi: 10.1053/j.gastro.2011.01.057. [DOI] [PubMed] [Google Scholar]
- 14.Lo B, Zhao M, Vind I, Burisch J. The risk of extraintestinal cancer in inflammatory bowel disease: a systematic review and meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol. 2021;19(6):1117–38.e19. doi: 10.1016/j.cgh.2020.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Piovani D, Hassan C, Repici A, Rimassa L, Carlo-Stella C, Nikolopoulos GK, et al. Risk of cancer in inflammatory bowel diseases: umbrella review and reanalysis of meta-analyses. Gastroenterology. 2022;163(3):671–684. doi: 10.1053/j.gastro.2022.05.038. [DOI] [PubMed] [Google Scholar]
- 16.Chen G, Xie J, Liang T, Wang Y, Liao W, Song L, et al. Inflammatory bowel disease promote oral cancer and pharyngeal cancer: new evidence of Mendelian randomization. Acta Otolaryngol. 2022;142(2):191–196. doi: 10.1080/00016489.2022.2035431. [DOI] [PubMed] [Google Scholar]
- 17.Cheng W, Liao Y, Mou R, Xiao X, Jia Y. Inflammatory bowel disease and prostate cancer risk: a two-sample Mendelian randomization analysis. Front Immunol. 2023;14:1157313. doi: 10.3389/fimmu.2023.1157313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Li X, Hong J, Huang L, Jiang Q, Guo S, et al. Inflammatory bowel disease increases the risk of hepatobiliary pancreatic cancer: A two-sample Mendelian randomization analysis of European and East Asian populations. Cancer Med. 2023;12(12):13599–13609. doi: 10.1002/cam4.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F, Liu Y, Wang Z, Zhao Q, Li Y, Tang T. A mendelian randomization study with populations of European ancestry rules out a causal relationship between inflammatory bowel disease and colorectal cancer. Front Genet. 2022;13:949325. doi: 10.3389/fgene.2022.949325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min Y, Wei X, Peng X. Letter: Mendelian randomization-New perspective in inflammatory bowel disease and pancreatic cancer? Aliment Pharmacol Ther. 2023;58(1):143–144. doi: 10.1111/apt.17537. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Meng X, Tian Q, Cao W, Fan X, Wu L, et al. Vitamin D and multiple health outcomes: an umbrella review of observational studies, randomized controlled trials, and Mendelian randomization studies. Adv Nutr. 2022;13(4):1044–1062. doi: 10.1093/advances/nmab142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skrivankova VW, Richmond RC, Woolf BAR, Davies NM, Swanson SA, VanderWeele TJ, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375:n2233. doi: 10.1136/bmj.n2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Liu R, Gao H, Jung S, Gao X, Sun R, et al. Genetic architecture of the inflammatory bowel diseases across East Asian and European ancestries. Nat Genet. 2023;55(5):796–806. doi: 10.1038/s41588-023-01384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies NM, Holmes MV, Davey SG. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Austin-Zimmerman I, Levey DF, Giannakopoulou O, Deak JD, Galimberti M, Adhikari K, et al. Genome-wide association studies and cross-population meta-analyses investigating short and long sleep duration. Nat Commun. 2023;14(1):6059. doi: 10.1038/s41467-023-41249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prince C, Sharp GC, Howe LD, Fraser A, Richmond RC. The relationships between women's reproductive factors: a Mendelian randomisation analysis. BMC Med. 2022;20(1):103. doi: 10.1186/s12916-022-02293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J, Lu J, Lu Q, Weng W, Guan Z, Wang Z. Mendelian randomization and colocalization analyses reveal an association between short sleep duration or morning chronotype and altered leukocyte telomere length. Commun Biol. 2023;6(1):1014. doi: 10.1038/s42003-023-05397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuber V, Grinberg NF, Gill D, Manipur I, Slob EAW, Patel A, et al. Combining evidence from Mendelian randomization and colocalization: Review and comparison of approaches. Am J Hum Genet. 2022;109(5):767–782. doi: 10.1016/j.ajhg.2022.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49(2):256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishigaki K, Akiyama M, Kanai M, Takahashi A, Kawakami E, Sugishita H, et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet. 2020;52(7):669–679. doi: 10.1038/s41588-020-0640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rashkin SR, Graff RE, Kachuri L, Thai KK, Alexeeff SE, Blatchins MA, et al. Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat Commun. 2020;11(1):4423. doi: 10.1038/s41467-020-18246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang L, Zheng Z, Fang H, Yang J. A generalized linear mixed model association tool for biobank-scale data. Nat Genet. 2021;53(11):1616–1621. doi: 10.1038/s41588-021-00954-4. [DOI] [PubMed] [Google Scholar]
- 36.Burrows K, Haycock P, Bull C, Dudding T, Gormley M, Robinson T, et al. Genome-wide association study of cancer risk in UK Biobank. University of Bristol. 202110.5523/bris.aed0u12w0ede20olb0m77p4b9
- 37.McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet. 2017;49(7):1126–1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seviiri M, Law MH, Ong JS, Gharahkhani P, Fontanillas P, Olsen CM, et al. A multi-phenotype analysis reveals 19 susceptibility loci for basal cell carcinoma and 15 for squamous cell carcinoma. Nat Commun. 2022;13(1):7650. doi: 10.1038/s41467-022-35345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 42.Gao H, Zheng S, Yuan X, Xie J, Xu L. Causal association between inflammatory bowel disease and 32 site-specific extracolonic cancers: a Mendelian randomization study. BMC Med. 2023;21(1):389. doi: 10.1186/s12916-023-03096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SH, Kwon JE, Cho ML. Immunological pathogenesis of inflammatory bowel disease. Intest Res. 2018;16(1):26–42. doi: 10.5217/ir.2018.16.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Everhov ÅH, Erichsen R, Järås J, Pedersen L, Halfvarson J, Askling J, et al. Colorectal cancer in elderly-onset inflammatory bowel disease: a 1969–2017 Scandinavian register-based cohort study. Aliment Pharmacol Ther. 2022;56(7):1168–1182. doi: 10.1111/apt.17175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun D, Zhou T, Heianza Y, Li X, Fan M, Fonseca VA, et al. Type 2 Diabetes and Hypertension. Circ Res. 2019;124(6):930–937. doi: 10.1161/CIRCRESAHA.118.314487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wijnands AM, de Jong ME, Lutgens M, Hoentjen F, Elias SG, Oldenburg B. Prognostic factors for advanced colorectal neoplasia in inflammatory bowel disease: systematic review and meta-analysis. Gastroenterology. 2021;160(5):1584–1598. doi: 10.1053/j.gastro.2020.12.036. [DOI] [PubMed] [Google Scholar]
- 47.Khan N, Abbas AM, Lichtenstein GR, Loftus EV, Jr, Bazzano LA. Risk of lymphoma in patients with ulcerative colitis treated with thiopurines: a nationwide retrospective cohort study. Gastroenterology. 2013;145(5):1007–15.e3. doi: 10.1053/j.gastro.2013.07.035. [DOI] [PubMed] [Google Scholar]
- 48.Kreijne JE, Goetgebuer RL, Erler NS, De Boer NK, Siebers AG, Dijkstra G, et al. Cumulative exposure to immunomodulators increases risk of cervical neoplasia in women with inflammatory bowel disease. Aliment Pharmacol Ther. 2023;58(2):207–217. doi: 10.1111/apt.17555. [DOI] [PubMed] [Google Scholar]
- 49.Obradović MMS, Hamelin B, Manevski N, Couto JP, Sethi A, Coissieux MM, et al. Glucocorticoids promote breast cancer metastasis. Nature. 2019;567(7749):540–544. doi: 10.1038/s41586-019-1019-4. [DOI] [PubMed] [Google Scholar]
- 50.Tang S, Zhang Z, Oakley RH, Li W, He W, Xu X, et al. Intestinal epithelial glucocorticoid receptor promotes chronic inflammation-associated colorectal cancer. JCI Insight. 2021;6(24):e151815. doi: 10.1172/jci.insight.151815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yadav S, Singh S, Harmsen WS, Edakkanambeth Varayil J, Tremaine WJ, Loftus EV., Jr Effect of medications on risk of cancer in patients with inflammatory bowel diseases: a population-based cohort study from Olmsted county. Minnesota Mayo Clin Proc. 2015;90(6):738–746. doi: 10.1016/j.mayocp.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. STROBE Statement-Checklist of items in reports of Mendelian randomization. Table S2. Characteristics of the summary GWAS data. Table S3. Assessment criteria for risk of bias assessment via the MR approach. Table S4. Characteristics of the instrumental variables in the East Asian population. Table S5. Characteristics of the instrumental variables in the European population. Table S6. MR estimates of the causal associations between IBD and cancers in the East Asian population. Table S7. Related traits of genetic variants used for Mendelian randomization analysis in the East Asian population. Table S8. MR estimates of the causal associations between IBD and cancers after removing instrumental variables associated with confounding factors in the East Asian population. Table S9. MR estimates of the causal associations between IBD and cancers in the European population. Table S10. Related traits of genetic variants used for Mendelian randomization analysis in the European population. Table S11. MR estimates of the causal associations between IBD and cancers after removing instrumental variables associated with confounding factors in the European population. Table S12. MVMR estimates of the causal associations between CD and ovarian cancer after adjusting for confounding factors. Table S13. MVMR estimates of the causal associations between UC and nonmelanoma skin cancer after adjusting for confounding factors. Table S14. Colocalization analysis for the associations between IBD and cancers in the East Asian population. Table S15. Colocalization analysis for the associations between IBD and cancers in the European population.
Additional file 2: Fig. S1. Mendelian randomization model. Solid arrows = causal effects; dashed arrows = causal effects prohibited by MR assumptions II and III. Assumption I: Genetic instruments are associated with exposures; Assumption II: Genetic instruments are independent of confounding factors; Assumption III: Genetic instruments affect outcomes only through exposures. IVW: inverse-variance-weighted.
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.