Abstract
The vaccinia virus (VV) A33R gene encodes a highly conserved 23- to 28-kDa glycoprotein that is specifically incorporated into the viral outer envelope. The protein is expressed early and late after infection, consistent with putative early and late promoter sequences. To determine the role of the protein, two inducible A33R mutants were constructed, one with the late promoter and one with the early and late A33R promoter elements. Decreased A33R expression was associated with small plaques that formed comets in liquid medium. Using both an antibiotic resistance gene and a color marker, an A33R deletion mutant, vA33Δ, was isolated, indicating that the A33R gene is not essential for VV replication. The plaques formed by vA33Δ, however, were tiny, indicating that the A33R protein is necessary for efficient cell-to-cell spread. Rescue of the large-plaque phenotype was achieved by inserting a new copy of the A33R gene into the thymidine kinase locus, confirming the specific genetic basis of the phenotype. Although there was a reduction in intracellular virus formed in cells infected with vA33Δ, the amount of infectious virus in the medium was increased. The virus particles in the medium had the buoyant density of extracellular enveloped viruses (EEV). Additionally, amounts of vA33Δ cell-associated extracellular enveloped viruses (CEV) were found to be normal. Immunogold electron microscopy of cells infected with vA33Δ demonstrated the presence of the expected F13L and B5R proteins in wrapping membranes and EEV; however, fully wrapped vA33Δ intracellular enveloped viruses (IEV) were rare compared to partially wrapped particles. Specialized actin tails that propel IEV particles to the periphery and virus-tipped microvilli (both common in wild-type-infected cells) were absent in cells infected with vA33Δ. This is the first deletion mutant in a VV envelope gene that produces at least normal amounts of fully infectious EEV and CEV and yet has a small-plaque phenotype. These data support a new model for VV spread, emphasizing the importance of virus-tipped actin tails.
Vaccinia virus (VV), the most intensively studied member of the Orthopoxvirus genus of the Poxviridae, is closely related to variola and monkeypox viruses, the causative agents of pandemic smallpox and a smallpox-like disease in Zaire, respectively (17, 35). VV, previously used as a smallpox vaccine, is a vector in recombinant vaccine strategies to prevent numerous infectious diseases and to treat cancer (33, 34). The characteristic features of orthopoxviruses are the following: a large, nearly 200,000-bp genome; a cytoplasmic site of replication; regulated early, intermediate, and late gene expression; and a complex mode of assembly involving multiple viral membranes (33). The latter aspect of VV replication is the subject of this study.
The first viral structures, visible by electron microscopy of infected cells, are crescent-shaped membranes associated with dense granular material (8). These structures evolve into spherical particles containing nucleoids and then into brick-shaped intracellular mature virions (IMV). IMV have two closely apposed outer membranes (48), comprise the majority of intracellular virus particles, and are infectious upon disruption of infected cells. Some IMV are destined to form intracellular enveloped viruses (IEV) by acquisition of two additional membrane layers from the trans-Golgi cisternae (23, 27, 43). The four membrane IEV may be propelled by actin tails to the cell periphery to form specialized microvilli (7, 22, 24, 49). The outermost viral membrane of IEV fuses with the plasma membrane (1), and the externalized particles, with three remaining membranes, remain attached to the cell surface as cell-associated extracellular enveloped viruses (CEV) or are released as extracellular enveloped viruses (EEV) (3, 27, 32, 39, 43). The CEV have been thought to mediate local cell-to-cell spread, whereas the EEV may provide long-range transmission.
Since enveloped virus particles are responsible for the spread of infection (1, 2, 5, 38, 52), the formation and function of the IEV, CEV, and EEV are of considerable interest. The proteins incorporated into the outer envelope are encoded by at least six genes, namely, the A56R (40, 46), F13L (2, 25), B5R (14, 29), A36R (36), A34R (11, 31), and A33R (41) genes. Information regarding the roles of the individual envelope proteins has been obtained by deleting or repressing the expression of the cognate genes. Mutants with a deletion of the F13L, B5R, A36R, or A34R gene produce small plaques in vitro and where tested are severely attenuated in vivo (2, 11, 15, 28, 31, 36, 44, 54). Since these mutations have little effect on IMV formation, IMV must not mediate efficient cell-to-cell spread. The small-plaque phenotype, however, appears to have several different causes. Deletion of the B5R or F13L gene severely inhibits formation of IEV and extracellular particles because of a defect in wrapping (2, 15, 54). By contrast, EEV formation is increased when the A34R gene is deleted (31, 55) and moderately reduced when the A36R gene is deleted (36). The small-plaque phenotype of the A34R mutant has been attributed to an 80% decrease in the specific infectivity of mutant EEV (31) and to the absence of actin tails and specialized microvilli (55), although the relative contributions of the two effects are unclear.
The A33R gene was recently shown to encode a type II integral membrane protein found in EEV but not IMV (41). The A33R gene is highly conserved in all orthopoxviruses (41, 42), and a homolog is present in distantly related Molluscum contagiosum virus (45), suggesting that it has an important role. Despite its conservation in poxviruses, the A33R gene product shows little homology to other proteins, precluding any functional insights. Here, we describe the effects of mutations in the promoter or coding regions of the A33R gene on the VV life cycle. A severe reduction in cell-to-cell spread was attributed to a defect in the formation of actin tails and specialized, virus-tipped microvilli.
MATERIALS AND METHODS
Cells and antibodies.
VV stocks were prepared in HeLa cells as described previously (12); BS-C-1 cells served for plaque assays, immunoprecipitations, and Western blotting; and RK13 cells were used to propagate VV for CsCl purification. Cells were grown in Eagle minimal essential medium (EMEM) with 10% fetal bovine serum, and infections were carried out in EMEM with 2.5% fetal bovine serum. For titrations of VV and analysis of plaque size, monolayers were fixed and stained with 0.1% crystal violet in 20% ethanol. Mouse monoclonal antibody (MAb) 20 (B5R specific) and MAb 4 (A33R specific) have been described before (37, 41). The F13L peptide RLVETLPENMDFRSDHLTTFEC was injected into rabbits to prepare anti-37N antibody.
Construction of inducible VV A33R mutants.
DNA segments containing an intact A33R open reading frame (ORF), a modified promoter, the Escherichia coli xanthine guanine phosphoribosyl transferase (gpt) gene under the p7.5 promoter, and flanking sequences were constructed by linking together five pieces of DNA by recombinant PCR (53). The flanking sequence that included part of the adjacent A32L coding region was amplified by using the primer pair CCTAATATTGGTACGTGTCTA and GATTTATTATCAAATTAATTTAC. The gpt gene was amplified by using a first primer containing sequences of the A32L promoter region (underlined), namely, CTAAATTAATTTGATAATAAATCTTAGCGACCGGAGATTGG; the second primer, CGACCTTAGTTTTCCATATTTTCACTAATTCCAAACCCACC (used for the virus named 13E14 or vA33full) or GCCTTCTTTGTTCTCCTCCCACTAATTCCAAACCCACC (for the virus named 9M2B or vA33late) contained the A33R early or late promoter (sequence underlined), respectively. The promoter region of A33R was amplified with AAAATATGGAAAACTAAGGTCG or GGAGGAGAACAAAGAAGGC to either include or exclude the putative early promoter, respectively. The other end of the promoter region was amplified with GAATTGTGAGCGCTCACAATTCTATTTATGTCACGATGT, containing sequences of the E. coli lac operator (lacO; underlined). The A33R gene was amplified with primers GTGAGCGCTCACAATTCACATTTATTATCATGATG containing sequences of the lacO (underlined) and AAAATAAATATTAGTTCATTGTT. The six DNA segments (there were two alternate A33R promoter pieces used) were amplified by PCR individually, purified by using Promega PCR Preps, and then joined by recombinant PCR in a stepwise fashion. The final 1,963- and 1,934-bp PCR products were cloned in a TA vector (Invitrogen, San Diego, Calif.) and sequenced by using a Prism Dye Deoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, Calif.) in conjunction with a model 373 DNA sequencer (Applied Biosystems). The plasmids were mixed with N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl sulfate (DOTAP) (Boehringer Mannheim) and transfected into cells infected with the recombinant vaccinia virus vlacI (19), which contains the lac repressor (lacI) in the TK locus. Viruses were grown under semisolid agarose (GIBCO/BRL, Grand Island, N.Y.) containing mycophenolic acid (MPA; Sigma) for selection of recombinant virus (16) and 5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for induction of the A33R gene. Viruses were plaque purified three times and amplified.
Recombinant viral genomes were analyzed by PCR. Primers were chosen so that they amplified from within the A32L gene (ATCTGGGTTATAAACGGGTG) to within the A33R gene (ACCAATCACGCGTTTGCGTT), such that DNA from wild-type (WT) or inducible viruses would yield PCR products of approximately 400 and 1,250 bp, respectively. PCR (25 cycles) was carried out by standard procedures using Taq polymerase (Boehringer) with a 50°C annealing temperature.
Immunoprecipitation and Western blotting.
Immunoprecipitates and Western blots were made essentially as described previously (41).
Deletion of the A33R gene.
To create an A33R deletion mutant, the A33R flanks were amplified by PCR and cloned into the pZippy neo/GUS vector (47), containing the neomycin resistance gene (neo) (18) as well as the color marker β-glucuronidase (GUS) (6). The A32L flanking region was amplified with the primer pair GCGAAGCTTCCTAATATTGGTACGTGTCTA (HindIII restriction site underlined) and GCGGTCGACGATTTATTATCAAATTAATTTAG (SalI restriction site underlined). The A34R flanking region was amplified with the primer pair GCGGAGCTCTATCACAAGAAGTTAGAAAGT (SacI site underlined) and GCGAGATCTCATTTTTTGTTGTCACTTGTA (BglII site underlined). The PCR products and vector were digested, purified, ligated (Rapid Ligation Kit; Boehringer Mannheim), and used to transform E. coli (One Shot; Invitrogen), which was selected on ampicillin plates. The correct sequence of VV-derived DNA was confirmed, and the plasmids were transfected into cells infected with vA33late and incubated under a geneticin (GIBCO)-containing semisolid agarose overlay. After 2 days, infected monolayers were overlaid with medium containing 0.2 mg of 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Glu; Clontech Laboratories, Palo Alto, Calif.) per ml. Two days later, blue plaques were picked. After three rounds of plaque purification, the virus was amplified in six-well plates and viral DNA was analyzed by PCR. Primers ATCTGGGTTATAAACGGGTG and AAAATAAATATTAGTTCATTGTT were chosen to amplify from inside the A32L ORF to the junction of the A33R ORF and A34R promoter, such that WT DNA would yield a PCR product of 870 bp, the vA33late DNA with the inserted gpt gene would yield a product of 1,670 bp, and the knockout vA33Δ DNA with the neo/GUS genes would yield a product of 3,270 bp.
Construction of the A33R rescue virus.
The A33R gene was amplified by PCR using primers with restriction sites for cloning into the modified pSC11 vector containing TK flanking regions and the lacZ gene (13). The A33R gene was inserted after the P7.5 promoter of the vector so that both an early and a late promoter would regulate transcription. The upstream primer was CGCGCGTCGACATAAATAACATTTATTATC (SalI site underlined); the other primer was CTGAGCGGCCGCATTAGTTCATTGTTTTAACACA (NotI site underlined). The purified PCR product and vector plasmid were digested, repurified, and ligated together. Bacterial colonies were grown on agar plates containing ampicillin. Plasmids were prepared by using Promega Mini Preps and transfected into cells infected with vA33Δ. The infected cells were overlaid with semisolid plaque medium containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Promega) for detection of recombinant virus containing the lacZ gene. Blue plaques were picked three times in succession, and the purified virus was amplified. The viral DNA was analyzed by PCR for the presence of pSC11 flanking sequences contiguous with the A33R sequence and also for the absence of WT A33R in the A33R locus. The primer with the A33R promoter sequence was GGAGGAGAACAAAGAAGGC; the other was AAAATAAATATTAGTTCATTGTT. By using these primers, WT DNA yielded the predicted 615-bp product. To further confirm that the viruses were derived from vA33Δ, infected monolayers were stained with X-Glu to detect the GUS gene used to replace the A33R gene.
Electron microscopy.
For transmission electron microscopy, RK13 cells in 60-mm-diameter dishes were infected with VV at a multiplicity of 10 for 24 h, fixed in 2% glutaraldehyde, and embedded in Embed-812 (Electron Microscopy Sciences, Fort Washington, Pa.) or fixed with increasing concentrations of paraformaldehyde and prepared for immunoelectron microscopy as previously described (55). Thawed cryosections were incubated with either rabbit antipeptide antibody to F13L or MAb 20 recognizing the B5R protein (37). Samples were washed, incubated with gold particles conjugated to protein A (Department of Cell Biology, Utrecht University School of Medicine, Utrecht, The Netherlands), and viewed with a Philips CM 100 electron microscope.
For scanning electron microscopy, HeLa cells were grown on coverslips, infected with VV at a multiplicity of 10, and fixed and prepared for analysis on an Amray 1820D microscope at an accelerating voltage of 15 kV as described previously (55).
One-step growth curves.
Confluent BS-C-1 or RK13 cell monolayers in six-well plates were infected with VV at a multiplicity of 10 for 2 h. The inocula were removed, the cells were washed, and fresh medium was added. At intervals, the medium from an individual well was harvested and centrifuged at 1,800 × g for 5 min to pellet detached cells. The resulting cell pellet was combined with infected cells that had been scraped from the plate into 1 ml of fresh medium. Cells were frozen and thawed three times and sonicated. Viruses from the media and cells were titrated in duplicate on BS-C-1 cell monolayers.
Virus purification.
Wrapped and unwrapped virus particles were purified on the basis of their buoyant densities in CsCl as described previously (41).
Syncytium formation.
Confluent BS-C-1 cell monolayers were infected at a multiplicity of 10 for 2 h, washed, and incubated in medium for an additional 10 h as described before (2, 54). Cells were washed and treated with fusion buffer [phosphate-buffered saline with 10 mM 2-(N-morpholino)ethanesulfonic acid and 10 mM HEPES] at pH 5.5 or 7.4 for 2 min at 37°C. Afterwards, fusion buffer was replaced with medium and the cells were incubated at 37°C and then observed by phase-contrast microscopy.
Immunofluorescence microscopy.
Fluorescence microscopy was performed as described previously (55). Infected HeLa cell monolayers on coverslips were washed with phosphate-buffered saline, fixed in 3% paraformaldehyde, and permeabilized with 0.05% saponin (Calbiochem, San Diego, Calif.). Actin filaments were visualized with fluorescein isothiocyanate (FITC)-conjugated phalloidin (Molecular Probes, Eugene, Oreg.). Enveloped virus particles were stained by incubating them first with a rabbit polyclonal antiserum that recognizes the B5R and F13L gene products (21) and then with rhodamine-conjugated swine antirabbit antibody (Dako Corporation, Carpinteria, Calif.). Samples were viewed and images were collected with an MRC 1024 confocal microscope.
RESULTS
Construction of inducible A33R mutant viruses.
After unsuccessful attempts to isolate an A33R deletion mutant (cited in reference 41), construction of a virus containing an inducible A33R gene was undertaken. The strategy of inserting a 22-bp lacO sequence just downstream of a late-promoter TAAAT element in a virus (vlacI) that contains a constitutively expressed E. coli lacI has been used to characterize numerous essential and nonessential genes expressed under late VV promoters (11, 57, 58). However, for several reasons, the system has not been used successfully with early promoters.
The first step in designing an inducible A33R mutant was to predict the type and location of the promoter so that lacO could be correctly positioned. Synthesis of the A33R protein was reported to occur predominantly late in infection (37), and the TAAAT element of a late promoter is located from −18 to −14 relative to the initiation codon of the ORF. Invariably, late RNA synthesis starts within the TAAAT; therefore, we decided to place the lacO sequence just downstream of TAAAT for optimal repression (19, 57). Further scrutiny, however, revealed the sequence AAAATATGGAAAACTA, which resembles the consensus core sequence of early promoters (9), between nucleotides −77 and −62. The finding of two copies of the early gene transcription termination sequence, TTTTTNT, starting 22 bp before and 7 bp after the A33R stop codon, was also consistent with early expression (41, 56). The presence of the second promoter presented a dilemma: if early expression is important, the full promoter region might be required; on the other hand, repression might be leaky if transcription started from the early promoter, due to both the timing of expression and distant location of the promoter relative to lacO. Therefore to evaluate the importance of the putative early promoter, we made two recombinant viruses, one (vA33full) containing the full promoter region and the other (vA33late) containing a truncated DNA segment without the predicted early promoter. In each case, lacO was positioned adjacent to the late-promoter TAAAT element. A recombinant PCR protocol was used to construct the two different promoter- and lacO-modified A33R genes with the adjacent gpt selectable marker and flanking sequences. After cloning, the sequences of the relevant parts of the plasmid were confirmed and the plasmid was transfected into cells infected with vlacI for recombination to occur. Virus was plaque purified in medium that contained MPA, to select for gpt-expressing recombinant viruses, and IPTG, to allow expression of the lacO-controlled A33R gene. After three plaque purifications, small virus stocks were prepared and analyzed by PCR. The sizes of the PCR products from 11% of the viruses were consistent with the insertion of the predicted recombinant DNA and deletion of WT DNA. The other virus isolates still retained the unmodified WT A33R gene, indicating the presence of single-crossover recombinants or WT virus. The relevant portions of the genomes of WT-A33 (vlacI), vA33full, and vA33late are depicted in Fig. 1.
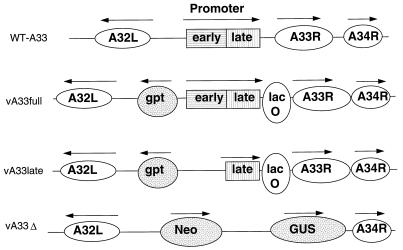
FIG. 1.
Representation of the genomes of A33R inducible and deletion mutants. In the top row, relevant features of the genome of WT-A33 (vlacI) are shown. The A32L, A33R, and A34R ORFs are represented by ovals, and the tandem early and late promoter elements of the A33R gene are shown boxed. Arrows indicate the directions of transcription. In the second row, the genome of vA33full is represented. The locations of the E. coli gpt gene and lacO are indicated. The vA33late mutant, depicted in the third row, is similar to vA33full except that the A33R early promoter has been deleted. The last row represents part of the genome of vA33Δ with the neo/GUS genes replacing A33R.
Plaque phenotypes of inducible mutant viruses.
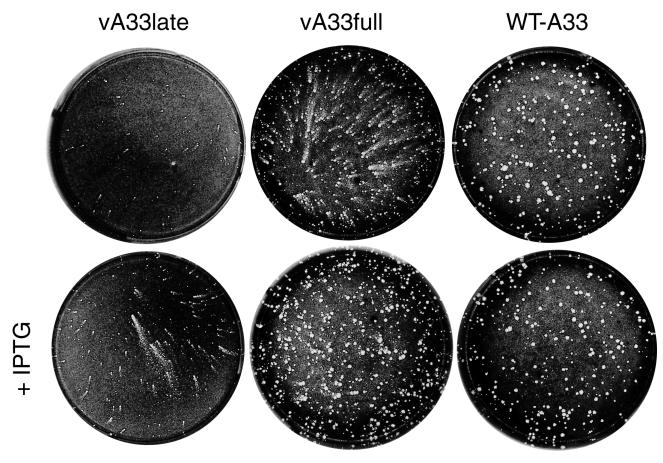
The size and appearance of the plaques formed by the mutant viruses, in the presence or absence of IPTG, were compared to those of the parental vlacI. The latter, like vA33full and vA33late, contains the lac repressor in the TK locus but is designated WT-A33 because the A33R gene was unmodified. After 29 h, the monolayers were fixed and stained with crystal violet. The virus retaining only the late promoter (vA33late) produced small plaques in the absence of IPTG and slightly larger plaques in the presence of inducer (Fig. 2). Surprisingly, the plaques were elongated and had the appearance of comets, a phenotype previously associated with a specific mutation in the A34R gene (4). The virus containing both the early and late promoters (vA33full) produced medium-size comet-shaped plaques without IPTG and larger, normal-size, round plaques with IPTG (Fig. 2).
FIG. 2.
Appearance of plaques formed by A33R inducible mutants. BS-C-1 cell monolayers were infected with vA33late, vA33full, or WT-A33 virus in the absence (top row) or presence (bottom row) of IPTG. After 29 h, the medium was removed and the monolayers were fixed and stained with 0.1% crystal violet in 20% ethanol.
The results demonstrated the importance of the A33R protein in cell-to-cell spread of VV. The inability of even high concentrations of IPTG to induce vA33late to produce large plaques, while the virus with both the early and late promoters produced WT-sized plaques with IPTG, indicated that the early promoter was important.
Expression of the A33R protein.
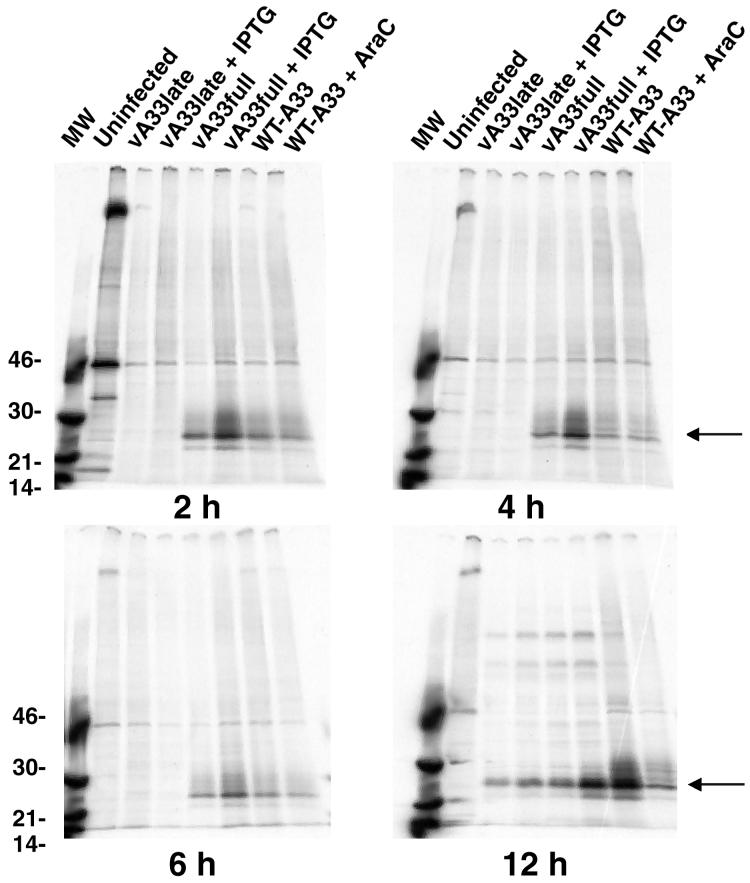
The temporal expression of the A33R gene by WT-A33 and mutant viruses was determined by metabolically labeling infected cells with [35S]methionine, immunoprecipitating the A33R protein from the lysates, and analyzing the protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography (Fig. 3). In cells infected with WT-A33, synthesis of the A33R protein was detected at 2 h, suggesting that the putative early promoter was indeed functional. This is in contrast to a previous report describing only late A33R expression (37). The large increase in A33R protein in WT-A33 at 12 h was consistent with the presence of the late promoter consensus element. AraC, an inhibitor of DNA replication and therefore late protein synthesis, decreased A33R levels at 12 h, as expected. The mutant vA33full synthesized A33R protein at early times, even in the absence of IPTG, indicating that the early promoter was not stringently repressed. Nevertheless, there was an increase in A33R protein expression in the presence of IPTG. In two experiments, the expression of A33R protein appeared somewhat greater in vA33full than in WT-A33 at early times, which may be due to an A/T-rich region in the adjacent gpt sequence in the mutant virus. In cells infected with the mutant vA33late, no A33R protein was detected at early times, indicating that the functional early promoter sequence had been deleted. At 12 h after infection, the A33R protein was detected, even in the absence of IPTG. Although the amount of protein is only slightly elevated in the presence of IPTG, this difference is biologically important, as indicated by the plaque phenotype changes (Fig. 2). At 12 h postinfection, both mutant viruses made less A33R protein than WT-A33R did, even in the presence of IPTG, probably due to negative effects of the lacO sequence adjacent to the late promoter (58).
FIG. 3.
Synthesis of A33R protein. BS-C-1 cells were infected with vA33late, vA33full, or WT-A33 virus with IPTG or AraC as indicated. After 2, 4, 6, or 12 h, the medium was removed, the cells were incubated for 1 h with [35S]methionine, and the cells were lysed. The A33R protein was immunoprecipitated with MAb 4, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and autoradiographed. The molecular weights (MW; 103) of markers are shown on the left. The arrows on the right point to the A33R protein.
Although we could not obtain stringent repression of this complex promoter, the experiments demonstrated the presence of a functional early promoter regulating expression of the A33R gene. Moreover, a comparison of Fig. 2 and 3 suggested a correlation between the amount of A33R produced and plaque size and comet formation. Expression was the least and the plaques were the smallest when the early promoter was deleted and no inducer was added. We could not, however, distinguish between the importance of temporal and quantitative effects of the early promoter on gene expression.
Construction and characterization of an A33R deletion mutant.
The very small plaques that formed under leaky repressive conditions suggested that plaques formed in the absence of A33R gene expression would be even smaller and hence deletion mutants might be exceptionally difficult to isolate from a large-plaque-forming parental virus. Our strategy, therefore, was to delete the A33R coding region from vA33late, which makes very small plaques. In addition, we used both antibiotic selection to enrich recombinants and a color marker to identify the plaques.
DNA corresponding to the A32L and the A34R ORFs, flanking the A33R ORF, were cloned into a plasmid on either side of the genes coding for neomycin resistance and the GUS color marker. Upon recombination, these genes would replace the A33R promoter and coding region (as well as the lacO and the gpt gene) of vA33late (Fig. 1). The DNA encoding the last 14 amino acids of the A33R ORF was retained so as not to interfere with the nearby A34R promoter. This DNA was transfected into cells infected with vA33late in the absence of IPTG. Virus was grown in geneticin, and tiny plaques staining blue in the presence of X-Glu were picked and plaque purified three times. Analysis of these viruses by PCR indicated that 3 of 18 viruses (17%) were A33R deletion mutants; the others were single-crossover recombinants or mixtures (data not shown). One of the deletion mutants was chosen for further study and named vA33Δ. Immunoprecipitation of lysates from metabolically labeled cells confirmed the absence of A33R protein from cells infected with vA33Δ, while F13L protein levels appeared normal (data not shown). The successful isolation of vA33Δ indicated that the A33R gene is not essential.
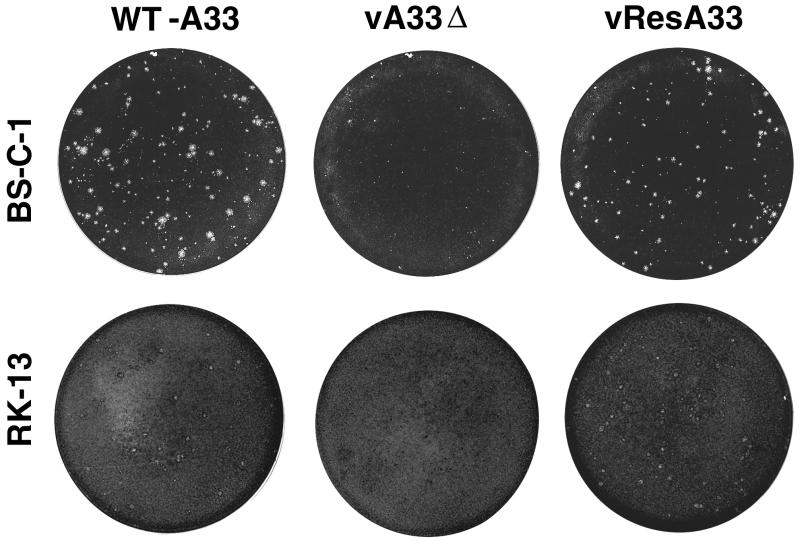
The pinpoint plaques produced by vA33Δ on BS-C-1 cells (shown at 48 h in Fig. 4) were even smaller than the uninduced vA33late plaques (shown at 29 h postinfection in Fig. 2). The plaques exhibited a comet shape (data not shown) similar to that of vA33late (Fig. 2). The plaque size of vA33Δ was also severely reduced on RK13 cells, yielding foci of infection visible as darkly staining points rather than plaques at 48 h (Fig. 4) and 72 h (data not shown). These results suggested that the phenotype is not host dependent.
FIG. 4.
Appearance of plaques formed by the A33R deletion mutant. BS-C-1 or RK13 monolayers were infected with WT-A33, vA33Δ, or vResA33. After 48 h, the medium was removed and the monolayers were fixed and stained with 0.1% crystal violet in 20% ethanol. At least 30 infectious foci or plaques are present in each well shown.
Rescue of the A33R deletion mutant.
When deletion mutants are made by insertion of a marker gene, the original gene can be restored to rule out the possibility that the phenotype is due to mutations in other parts of the genome. However, when the gene is reinserted into its original location, the procedure does not eliminate the possibility that either the original deletion itself or the expression of an inserted marker gene caused the effect via neighboring genes. In the present situation, this was particularly important because the neighboring gene, A34R, is required for efficient plaque formation (11, 31, 55). To avoid such ambiguity, we inserted a new A33R gene into the distal TK locus of vA33Δ (replacing the lacI gene).
The A33R gene was PCR cloned into pSC11 (under the early/late P7.5 promoter) containing the TK flanking regions and the lacZ gene. Plaques of the rescued virus, vResA33, were identified by their blue color in the presence of X-Gal. The vResA33 plaques were also blue in the presence of X-Glu, confirming that the virus was derived from vA33Δ. vResA33 was analyzed by PCR for the presence of neo/GUS genes in the A33R locus and A33R in the TK locus. Insertion of a new A33R gene in a distant locus resulted in plaques nearly the size of WT-A33 (Fig. 4), even though the neo/GUS genes were not removed from their position between the A32L and A34R ORFs. In addition to vResA33 producing large plaques, it should be noted that the vResA33 did not produce comets, indicating that the comet phenotype of vA33Δ is due to the lack of A33R protein and not an independent mutation elsewhere in the VV genome.
Formation of infectious intracellular and extracellular virus.
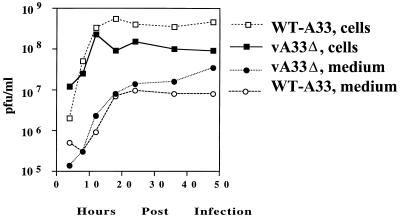
Deletion of other genes encoding EEV-specific proteins either increased or decreased EEV formation (2, 31, 36, 54). To quantitate the infectious virus produced by vA33Δ, a one-step growth analysis was performed. Replicate wells containing BS-C-1 cells were inoculated with WT-A33 or vA33Δ and washed after 2 h. At intervals, the medium and cells were harvested separately and the yields of infectious virus were determined by plaque assay. In three separate experiments, the titers of fresh extracellular vA33Δ virus were two- to fourfold higher than those of WT-A33 (Fig. 5). Freeze-thawing was shown to increase the infectivity of A34 deletion mutant EEV (31). However, disruption of the unpurified vA33Δ EEV outer membrane in this manner did not increase the infectivity, suggesting that the EEV particles are fully infectious. The vA33Δ mutant produced one-half to one-third of the cell-associated virus that WT-A33 did; this decrease in production was similar to the small reduction in IMV that occurs in other EEV protein deletion mutants (31, 36, 54). The total amount of virus produced by vA33Δ was 35 to 55% of that of WT-A33, since the majority of virus was cell associated. Similar results were obtained with BS-C-1 cells, although less vA33Δ and less WT-A33 were released into the medium than with RK13 cells.
FIG. 5.
Yields of extracellular and cell-associated infectious virus. RK13 cells were infected with WT-A33 or vA33Δ at a multiplicity of 10. The inocula were removed after 2 h and replaced with fresh medium. At the indicated times, the medium was removed and centrifuged to pellet detached cells. Adherent cells were scraped into 1 ml of fresh medium and combined with pelleted cells, frozen, and thawed three times, and sonicated for 30 s. The titers of the virus from the medium and the cells were determined in duplicate on BS-C-1 cell monolayers.
CsCl gradient centrifugation of virus particles.
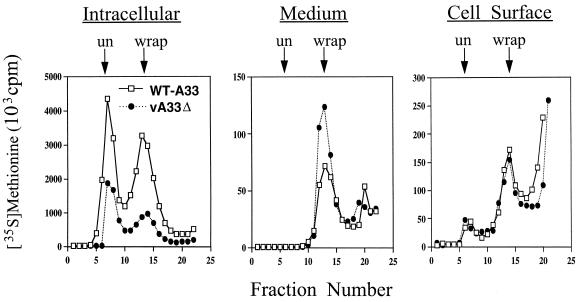
It was important to determine whether the extracellular virions produced by vA33Δ were enveloped, since IMV could be released by lysis of some cells. Enveloped virions, either IEV or EEV, can be distinguished from IMV by their lower buoyant density due to one or two additional membranes. Infected cells were metabolically labeled with [35S]methionine, and the virus from the medium and cell lysates was purified by CsCl density centrifugation. The two peaks of intracellular particles, corresponding to IMV and IEV from cells infected with vA33Δ, were lower than those from cells infected with WT-A33 (Fig. 6). Analysis of the medium revealed only a single peak, corresponding to EEV (97%), for both WT-A33 and vA33Δ, indicating that IMV contamination in media is negligible. There was, however, about threefold more EEV produced by vA33Δ than by WT-A33 (or vResA33 [data not shown]) and a corresponding increase in infectivity. Amounts of EEV greater than those of WT-A33 were also produced by vA33late without inducer (data not shown). Thus, deletion of the A33R gene increased the amount of infectious EEV released.
FIG. 6.
Purification of wrapped and unwrapped virus particles by buoyant density centrifugation. [35S]methionine-labeled WT-A33 or vA33Δ virus was harvested from the medium, the lysed cells (intracellular), or intact, washed cells that were trypsin treated to release virus attached to the cell surface. Virus samples were centrifuged in CsCl gradients, and the positions of wrapped (wrap) and unwrapped (un) particles are indicated with arrows.
Since CEV rather than EEV may be responsible for virus spread to adjacent cells during plaque formation, we measured the amount of [35S]methionine-labeled virus that could be released from washed cells by trypsin (3). The amounts of labeled material that sedimented as enveloped virus were similar for vA33Δ (Fig. 6), WT-A33 (Fig. 6), and vResA33 (data not shown). The trypsin also released some labeled material that remained at the top of the gradient and a small amount of material that sedimented as IMV.
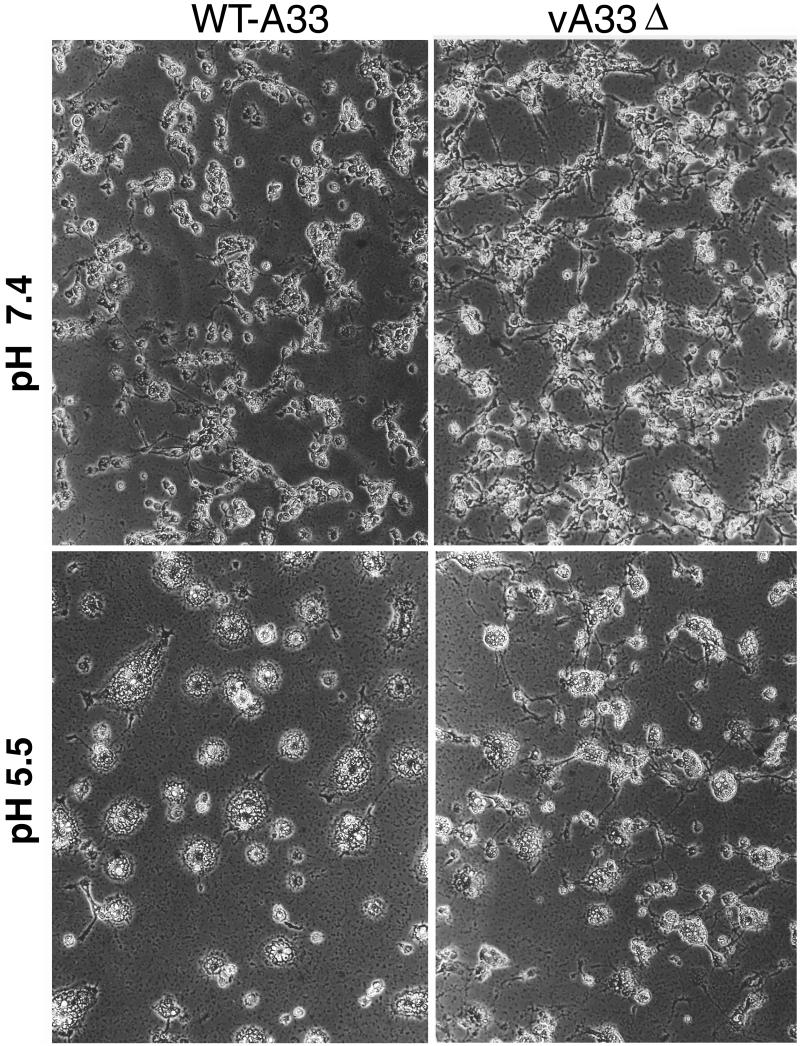
Fusion of vA33Δ-infected cells.
Cells infected with VV can be induced to fuse by brief treatment with low-pH buffer (10, 20). Formation of wrapped virus is necessary, but not always sufficient, for acid-induced polykaryon formation (2, 10, 54). We evaluated the ability of vA33Δ to mediate the formation of acid-induced syncytia. Cells were infected for 12 h with either WT-A33 or vA33Δ, incubated for 2 min in buffer at pH 5.5, and examined microscopically after 2 and 5 h. As controls, WT-A33 virus-infected cells treated with pH 7.4 buffer and uninfected cells treated with pH 5.5 buffer showed no fusion. However, by 2 h, cells infected with either vA33Δ or WT-A33 virus showed polykaryon formation, although those of vA33Δ appeared slightly smaller. Photographs taken at 5 h are shown in Fig. 7. The A34R and F13L deletion mutant viruses were also included as controls and induced no cell fusion, as previously reported (2, 55).
FIG. 7.
Induction of syncytia by WT-A33 or vA33Δ. BS-C-1 cells were infected for 12 h, treated for 2 min with buffer at pH 5.5 or 7.4, and then returned to the growth medium for 5 h and examined by phase-contrast microscopy.
Electron microscopic examination of cells infected with vA33Δ.
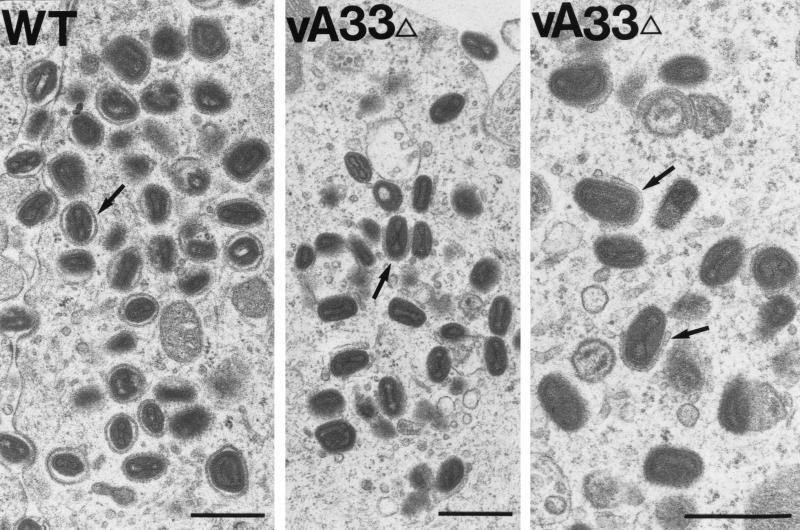
The biochemical studies demonstrated that deletion of the A33R gene reduced the amount of intracellular virus that could be recovered from cell lysates but increased the amount of EEV. Electron microscopy was used to further analyze the effect of the mutation on morphogenesis. In cells infected with vA33Δ, immature virions and IMV appeared to form normally. There also seemed to be an increase in membrane vesicles. Many of the vA33Δ IMV were associated with intracellular membranes, and many appeared to be partially wrapped by a double membrane (Fig. 8). Fully wrapped IEV, however, were rare or absent at both 24 and 48 h. In contrast, completely wrapped IEV were numerous in cells infected with WT-A33 (Fig. 8) or vResA33 (data not shown). This partial-wrapping phenotype occurred in both RK13 and BS-C-1 cells infected with vA33Δ, as well as with vA33late in the absence of inducer (data not shown).
FIG. 8.
Electron micrographs of Epon-embedded infected cells. RK13 cells were infected with WT-A33 or vA33Δ. After 24 h, the cells were fixed in glutaraldehyde and embedded in Epon. In WT-A33-infected cells (left panel), the arrow indicates an example of a fully wrapped IEV particle. In vA33Δ-infected cells (middle and right panels) arrows indicate partially wrapped IMV (incomplete IEV). It is important to note the numerous membrane vesicles with associated virus particles in the middle panel. Bars, 0.5 μm.
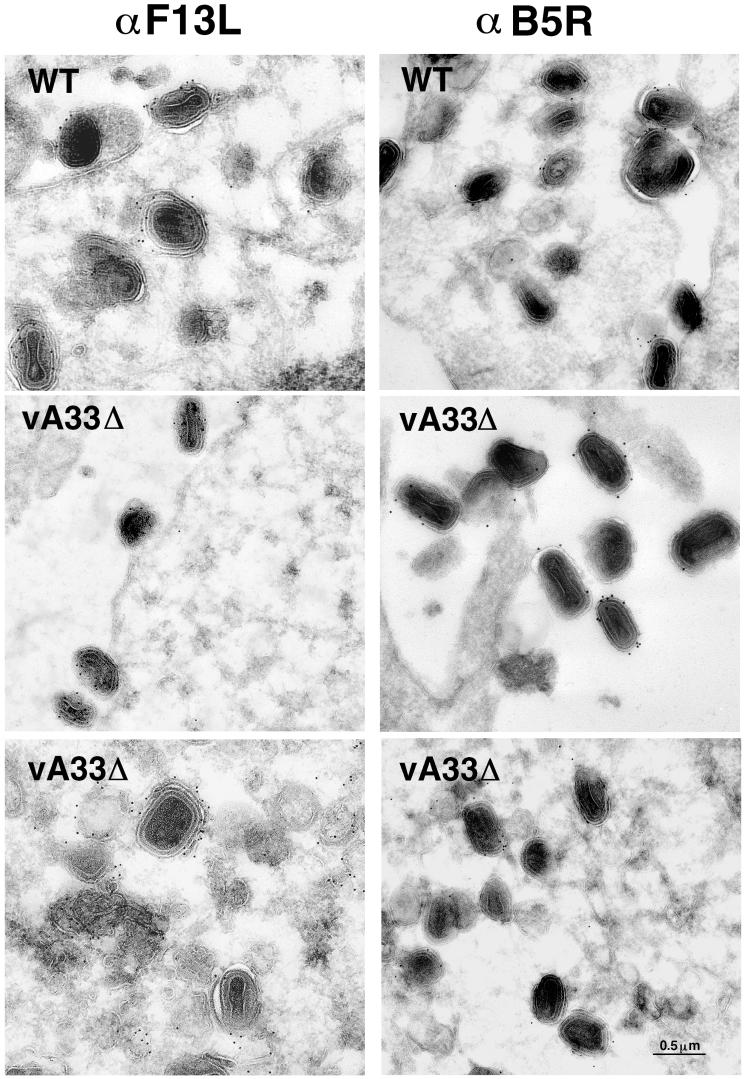
The increase in vA33Δ EEV and the apparent lack of fully wrapped IEV (the presumed precursor) were puzzling. To gain further insight, we investigated the membranes seen wrapping vA33Δ IMV to determine whether the membranes contained other proteins known to localize in the wrapping membranes derived from the trans-Golgi compartment (43). We labeled the EEV proteins encoded by the B5R and F13L genes by using specific antibodies and protein A-gold. We found that both proteins were present in the partially wrapped IEV and the membranes of extracellular particles released by cells infected with vA33Δ (Fig. 9). These proteins were also detected by Western blotting of CsCl-purified vA33Δ EEV (data not shown).
FIG. 9.
Immunogold labeling of viral membranes. RK13 cells were infected with WT-A33 or vA33Δ for 24 h, fixed in paraformaldehyde, cryosectioned, and incubated with antibodies to the F13L or B5R EEV-specific proteins and then protein A-gold.
Detection of actin filaments by immunofluorescence.
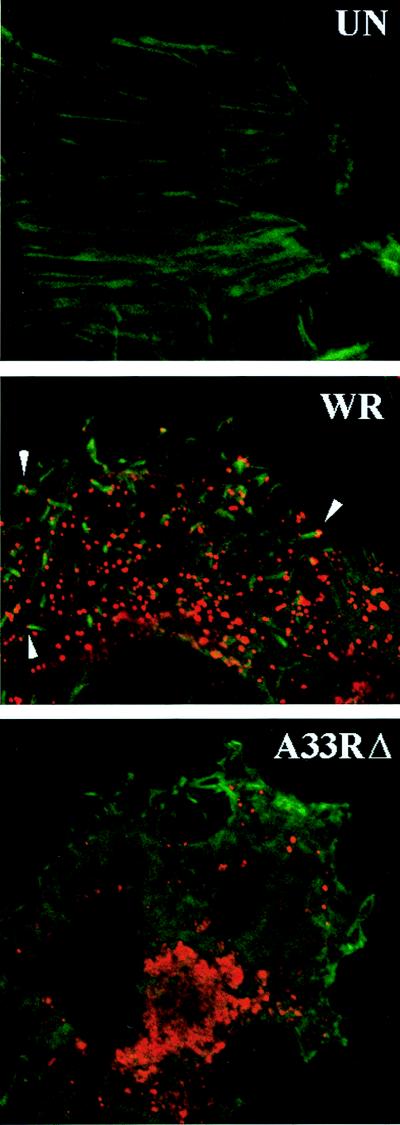
Since the small-plaque phenotype of the vA33Δ could not be explained by the failure to form IMV, CEV, or EEV, we investigated VV-induced formation of specialized actin tails. Infected cells were fixed and stained with FITC-phalloidin to detect actin filaments and incubated with antibody to the B5R and F13L membrane proteins to visualize fully or partially wrapped virus particles (stained red with rhodamine). In uninfected cells, thin actin stress fibers were visible throughout the cell (Fig. 10). The numerous virus-tipped, thick actin tails seen in cells infected with WT VV (strain WR) virus were not present in cells infected with vA33Δ. Consistent with biochemical data, fewer virus particles were present in the vA33Δ-infected cells. Occasionally, we saw vA33Δ virus particles adjacent to short, slender actin filaments but could not determine whether these were true tails. Also, the perinuclear area staining with VV Golgi-localized proteins was enlarged in the vA33Δ-infected cells.
FIG. 10.
Detection of actin filaments in VV-infected cells. HeLa cells were left uninfected (UN) or were infected with WT VV (strain WR) or vA33Δ. At 16 h, the cells were fixed, permeabilized, and incubated with FITC-conjugated phalloidin and a rabbit polyclonal serum that recognizes the B5R and F13L proteins, followed by rhodamine-conjugated antirabbit antibody. The images were examined by confocal microscopy.
Scanning electron microscopy.
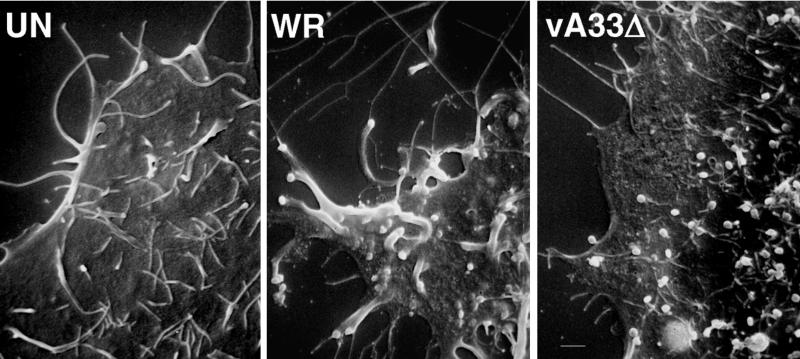
Virus-tipped, thick, actin-containing microvilli project from the surfaces of VV-infected cells (7, 22, 24, 30, 49). These virus-tipped, specialized microvilli were clearly visible by scanning electron microscopy on the surfaces of cells infected with WT VV (strain WR) (Fig. 11). In cells infected with vA33Δ, there were numerous virus particles on the cell surface but they were not associated with specialized microvilli. Thus, synthesis of the A33R protein is required for the formation of actin-containing specialized microvilli and virus spread but not for the transport of virus particles to the cell surface or the release of EEV.
FIG. 11.
Scanning electron microscopy of cells infected with VV. HeLa cells were left uninfected (UN) or were infected at a multiplicity of 10 with WT VV (strain WR) or vA33Δ. After 17 h, the cells were fixed with glutaraldehyde and the samples were coated with gold-palladium alloy and viewed with an Amray 1820D microscope.
DISCUSSION
We combined genetic, biochemical, and microscopic approaches to investigate the role of the A33R glycoprotein, the most recently discovered EEV membrane component (41). Mutants in which the A33R gene promoter was altered or the coding sequence was deleted were used to demonstrate parallel reductions in A33R glycoprotein synthesis and plaque size. The presence of an early promoter sequence was required for normal plaque formation, although it is not known if early expression is needed or simply contributes quantitatively to the A33R protein effect. The tiny plaques formed by vA33Δ were even smaller than those produced by a B5R deletion mutant (54), approximately the same size as those produced by the A34R and A36R (31, 36) deletion mutants, but slightly larger than those of an F13L deletion mutant (3) when compared directly (data not shown). Small-plaque phenotypes have been associated with reductions in the quantity (3, 36, 54) or infectivity (31) of extracellular virus. Neither factor is relevant for the A33R mutant, since the amounts of infectious EEV were larger than those for WT-A33 controls. In addition, similar amounts of CEV were released by trypsin treatment of WT-A33- and mutant-infected cells.
Another mechanism of small-plaque formation was recently suggested by Wolffe et al. (55), who attributed the plaque size of an A34R deletion mutant to a defect in formation of actin tails and specialized virus-tipped microvilli. Nevertheless, the reported lower infectivity of A34R mutant EEV (31) may also have contributed to impaired cell-to-cell spread. vA33Δ is the first small-plaque mutant virus that produces normal levels of fully infectious EEV and CEV. vA33Δ has defects in actin tail formation and specialized microvilli, providing strong support for a model in which efficient cell-to-cell spread is mediated by virus particles at the tips of specialized microvilli.
The inability to detect actin tails by microscopy in cells infected with the A33R deletion mutant is apparently due to their failure to form, although the transient existence of tails cannot be ruled out. The A33R proteins might have direct or indirect roles in the actin nucleation or polymerization steps. In cells infected with A33R or A34R (11, 55) deletion mutants, there appeared to be a paucity of fully wrapped cytoplasmic particles to which actin tails could attach. Although there are many partially wrapped IEV in cells infected with the A33R deletion mutant, actin tails are not seen. Thus, either the A33R protein or fully closed IEV are required for attachment of actin tails. Therefore, the absence of actin tails and specialized microvilli may be secondary to a reduction in fully wrapped IEV.
Some strains of VV, such as IHD, form plaques under liquid medium that have an elongated comet shape (for an example, see Fig. 2). Comets are associated with increased amounts of EEV and with a single amino acid substitution in the A34R ORF (3, 4). In the absence of WT levels of A33R protein, the small plaques of A33R mutants had comet tails. Tiny comets were detected with vA33Δ, but larger comets were found with the leaky inducible mutants. vA33full, however, produced round plaques without comets in the presence of IPTG, indicating that comet formation was due to reduced A33R protein levels rather than a coincidental mutation elsewhere in the genome. Nevertheless, the proximity of the A34R gene, which also encodes an EEV membrane glycoprotein associated with comet formation, made it necessary to consider indirect effects caused by the genetic alterations at the A33R locus. To evaluate this potential problem, we inserted a new A33R gene into a distal site within the genome of vA33Δ. The rescued virus formed large plaques without comets, despite retention of the alterations at the site of the original A33R gene. Therefore, we could confidently rule out significant neighboring gene effects on the mutant phenotype. Since a risk of local disturbances at deletion or insertion sites occurs frequently, distal gene insertion may be a generally useful alternative to constructing revertants. Comets appear to form when the amounts of infectious EEV are increased. It is also possible that the A33R and A34R proteins might be involved in other ways, e.g., in the binding and release of the virus particles from the cell surface.
Cells infected with VV can be induced to fuse by brief low-pH treatment (10, 20), suggesting that one mechanism of virus entry is endosomal (26). The assembly inhibitor rifampin or deletion of the F13L or B5R gene inhibits fusion, presumably due to the absence of CEV on the cell surface (2, 10, 54). The failure of A34R deletion mutants to induce fusion suggests that actin-containing microvilli might be required (55). However, microvilli are not required since vA33Δ induced fusion without specialized microvilli. It is likely, therefore, that in the case of the A34R deletion mutant, either CEV are not present on the surface of infected cells or the particles are not competent to fuse.
Fully wrapped IEV were rare or absent in vA33Δ-infected cells, whereas partially enveloped IMV were numerous. This seemed paradoxical since large amounts of EEV were produced. The partially wrapped IMV could represent (i) the form that fuses with the membrane to release EEV, (ii) intermediate forms that would become completely wrapped, or (iii) dead-end structures resulting from defective wrapping machinery. If partially enveloped IMV fuse with the plasma membrane (hypothesis i), this could raise topological problems and lead to incomplete or missing EEV membranes. If the incomplete IMV do eventually become fully wrapped (hypothesis ii), then the fully wrapped A33R-deficient IEV must exit the cells very rapidly so they are not captured by electron microscopy. In that scenario, one role of the A33R protein may be to retard the transit of IEV in order to permit actin tails to form. If the incompletely wrapped IMV are dead-end structures (hypothesis iii), this would suggest that the A33R protein is needed for complete wrapping and that there must be alternative pathways of EEV formation, such as budding through Golgi or plasma membranes, both of which contain EEV-specific proteins (43). Although budding particles are not commonly seen, virus particles are sometimes found inside of membrane cisternae, and Tsutsui et al. (50, 51) described plasma membrane budding of the IHD-W strain of VV in FL cells. Budding as well as wrapping and fusion mechanisms would result in EEV that have one additional membrane relative to those of IMV.
In conclusion, the A33R protein is required for normal plaque formation, actin tails, and specialized virus-tipped microvilli. These data indicate that the formation of EEV and CEV is not sufficient for plaque formation and suggest that the microvilli are responsible for efficient cell-to-cell spread.
ACKNOWLEDGMENTS
We thank T. Shors for the pZippy neo/GUS vector, N. Cooper for cells, and J. Sisler for sequencing of plasmids.
REFERENCES
- 1.Appleyard G, Hapel A J, Boulter E A. An antigenic difference between intracellular and extracellular rabbitpox virus. J Gen Virol. 1971;13:9–17. doi: 10.1099/0022-1317-13-1-9. [DOI] [PubMed] [Google Scholar]
- 2.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco R, Moss B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco R, Sisler J R, Moss B. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J Virol. 1993;67:3319–3325. doi: 10.1128/jvi.67.6.3319-3325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulter E A, Appleyard G. Differences between extracellular and intracellular forms of poxvirus and their implications. Prog Med Virol. 1973;16:86–108. [PubMed] [Google Scholar]
- 6.Carroll M W, Moss B. E. coli β-glucuronidase (GUS) as a marker for recombinant vaccinia viruses. BioTechniques. 1995;19:352–354. [PubMed] [Google Scholar]
- 7.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 8.Dales S, Siminovitch L. The development of vaccinia virus in Earles L strain cells as examined by electron microscopy. J Biophys Biochem Cytol. 1961;10:475–503. doi: 10.1083/jcb.10.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davison A J, Moss B. The structure of vaccinia virus early promoters. J Mol Biol. 1989;210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 10.Doms R W, Blumenthal R, Moss B. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J Virol. 1990;64:4884–4892. doi: 10.1128/jvi.64.10.4884-4892.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan S A, Smith G L. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J Virol. 1992;66:1610–1621. doi: 10.1128/jvi.66.3.1610-1621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earl P L, Cooper N, Moss B. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Wiley Interscience; 1991. pp. 16.16.1–16.16.7. [Google Scholar]
- 13.Eisenlohr L C, Bacik I, Bennink J R, Bernstein K, Yewdell J W. Expression of a membrane protease enhances presentation of endogenous antigens to MHC class I-restricted T lymphocytes. Cell. 1992;71:963–969. doi: 10.1016/0092-8674(92)90392-p. [DOI] [PubMed] [Google Scholar]
- 14.Engelstad M, Howard S T, Smith G L. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- 15.Engelstad M, Smith G L. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 16.Falkner F G, Moss B. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J Virol. 1988;62:1849–1854. doi: 10.1128/jvi.62.6.1849-1854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenner F, Henderson D A, Arita I, Jezek Z, Ladnyi I D. Smallpox and its eradication. 1st ed. Geneva, Switzerland: World Health Organization; 1988. [Google Scholar]
- 18.Franke C A, Rice C M, Strauss J H, Hruby D E. Neomycin resistance as a dominant selectable marker for selection and isolation of vaccinia virus recombinants. Mol Cell Biol. 1985;5:1918–1924. doi: 10.1128/mcb.5.8.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuerst T R, Fernandez M P, Moss B. Transfer of the inducible lac repressor/operator system from Escherichia coli to a vaccinia virus expression vector. Proc Natl Acad Sci USA. 1989;86:2549–2553. doi: 10.1073/pnas.86.8.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong S C, Lai C F, Esteban M. Vaccinia virus induces cell fusion at acid pH and this activity is mediated by the N-terminus of the 14-kDa virus envelope protein. Virology. 1990;178:81–91. doi: 10.1016/0042-6822(90)90381-z. [DOI] [PubMed] [Google Scholar]
- 21.Hiller G, Eibl H, Weber K. Characterization of intracellular and extracellular vaccinia virus variants: N1-isonicotinoyl-N2-3-methyl-4-chlorobenzoylhydrazine interferes with cytoplasmic virus dissemination and release. J Virol. 1981;39:903–913. doi: 10.1128/jvi.39.3.903-913.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiller G, Jungwirth C, Weber K. Fluorescence microscopical analysis of the life cycle of vaccinia virus in chick embryo fibroblasts. Virus-cytoskeleton interactions. Exp Cell Res. 1981;132:81–87. doi: 10.1016/0014-4827(81)90085-9. [DOI] [PubMed] [Google Scholar]
- 23.Hiller G, Weber K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol. 1985;55:651–659. doi: 10.1128/jvi.55.3.651-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology. 1979;98:142–153. doi: 10.1016/0042-6822(79)90533-6. [DOI] [PubMed] [Google Scholar]
- 25.Hirt P, Hiller G, Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichihashi Y, Dales S. Biogenesis of poxviruses: interrelationship between hemagglutinin production and polykaryocytosis. Virology. 1971;46:533–543. doi: 10.1016/0042-6822(71)90057-2. [DOI] [PubMed] [Google Scholar]
- 27.Ichihashi Y, Matsumoto S, Dales S. Biogenesis of poxviruses: role of A-type inclusions and host cell membranes in virus dissemination. Virology. 1971;46:507–532. doi: 10.1016/0042-6822(71)90056-0. [DOI] [PubMed] [Google Scholar]
- 28.Isaacs, S., R. Blasco, and B. Moss. Unpublished data.
- 29.Isaacs S N, Wolffe E J, Payne L, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krempien U, Schneider L, Hiller G, Weber K, Katz E, Jungwirth C. Conditions for pox virus-specific microvilli formation studied during synchronized virus assembly. Virology. 1981;113:556–564. doi: 10.1016/0042-6822(81)90183-5. [DOI] [PubMed] [Google Scholar]
- 31.McIntosh A G, Smith G L. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J Virol. 1996;70:272–281. doi: 10.1128/jvi.70.1.272-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan C. Vaccinia virus reexamined: development and release. Virology. 1976;73:43–58. doi: 10.1016/0042-6822(76)90059-3. [DOI] [PubMed] [Google Scholar]
- 33.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. New York, N.Y: Lippincott-Raven Press; 1996. [Google Scholar]
- 34.Moss B. Vaccinia virus: a tool for research and vaccine development. Science. 1991;252:1662–1667. doi: 10.1126/science.2047875. [DOI] [PubMed] [Google Scholar]
- 35.Mukinda V B, Mwema G, Kilundu M, Heymann D L, Khan A S, Esposito J J. Re-emergence of human monkeypox in Zaire in 1996. Lancet. 1997;17:1449–1450. doi: 10.1016/S0140-6736(05)63725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a M(r) 43–50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 37.Payne L G. Characterization of vaccinia virus glycoproteins by monoclonal antibody preparations. Virology. 1992;187:251–260. doi: 10.1016/0042-6822(92)90313-e. [DOI] [PubMed] [Google Scholar]
- 38.Payne L G. Significance of extracellular virus in the in vitro and in vivo dissemination of vaccinia virus. J Gen Virol. 1980;31:147–155. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- 39.Payne L G, Kristensson K. Mechanism of vaccinia virus release and its specific inhibition by N1-isonicatinoyl-N2-3-methyl-4-chlorobenzoylhydrazine. J Virol. 1979;32:614–622. doi: 10.1128/jvi.32.2.614-622.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Payne L G, Norrby E. Presence of haemagglutinin in the envelope of extracellular vaccinia virus particles. J Gen Virol. 1976;32:63–72. doi: 10.1099/0022-1317-32-1-63. [DOI] [PubMed] [Google Scholar]
- 41.Roper R L, Payne L G, Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmaljohn, A. Personal communication.
- 43.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seki M, Oie M, Ichihashi Y, Shida H. Hemadsorption and fusion inhibition activities of hemaggluttinin analyzed by vaccinia virus mutants. Virology. 1990;175:372–384. doi: 10.1016/0042-6822(90)90422-n. [DOI] [PubMed] [Google Scholar]
- 45.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 46.Shida H. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology. 1986;150:451–462. doi: 10.1016/0042-6822(86)90309-0. [DOI] [PubMed] [Google Scholar]
- 47.Shors, T., and B. Moss. Unpublished data.
- 48.Sodeik B, Doms R W, Ericcson M, Hiller G, Machamer C E, van’t Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stokes G V. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J Virol. 1976;18:636–643. doi: 10.1128/jvi.18.2.636-643.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsutsui K. Release of vaccinia virus from FL cells infected with the IHD-W strain. J Electron Microsc. 1983;32:125–140. [PubMed] [Google Scholar]
- 51.Tsutsui K, Uno F, Akatsuka K, Nii S. Electron microscopic study on vaccinia virus release. Arch Virol. 1983;75:213–218. doi: 10.1007/BF01315275. [DOI] [PubMed] [Google Scholar]
- 52.Turner G S, Squires E J. Inactivated smallpox vaccine: immunogenicity of inactivated intracellular and extracellular vaccinia virus. J Gen Virol. 1971;13:19–25. doi: 10.1099/0022-1317-13-1-19. [DOI] [PubMed] [Google Scholar]
- 53.Turner P C, Moyer R W. A PCR-based method for manipulation of the vaccinia virus genome that eliminates the need for cloning. BioTechniques. 1992;13:764–771. [PubMed] [Google Scholar]
- 54.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolffe E J, Katz E, Weisberg A, Moss B. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J Virol. 1997;71:3904–3915. doi: 10.1128/jvi.71.5.3904-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuen L, Moss B. Oligonucleotide sequence signalling transcriptional termination of vaccinia virus early genes. Proc Natl Acad Sci USA. 1987;84:6417–6421. doi: 10.1073/pnas.84.18.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Moss B. Inducer-dependent conditional-lethal mutant animal viruses. Proc Natl Acad Sci USA. 1991;88:1511–1515. doi: 10.1073/pnas.88.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Moss B. Vaccinia virus morphogenesis interrupted when expression of the gene encoding an 11-kilodalton phosphoprotein is prevented by the Escherichia coli lac repressor. J Virol. 1991;65:6101–6110. doi: 10.1128/jvi.65.11.6101-6110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]