Abstract
Nucleoside analog-resistant variants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) that displayed higher in vitro polymerase fidelity were previously identified via nucleotide insertion and mispair extension assays. To evaluate the contribution of increased nucleotide insertion and primer extension fidelities on the overall error rate of HIV-1 RT, we have measured the impact of two such mutations, E89G and M184V, on DNA copying fidelity in an M13 phage-based forward mutation assay. Using this assay, we observed mutation frequencies of 8.60 × 10−3, 6.26 × 10−3, 5.53 × 10−3, and 12.30 × 10−3 for wild-type, E89G, M184V, and double-mutant E89G/M184V HIV-1 RTs, respectively. Therefore, the overall polymerase fidelities of wild-type, E89G, M184V, and E89G/M184V HIV-1 RTs are similar (less than twofold differences) for DNA-dependent DNA synthesis. Thus, rather large increases in fidelity of deoxynucleoside triphosphate insertion and mispair extension observed previously appear not to influence the overall error rate of these mutants. However, a qualitative analysis of the mutations induced revealed significant differences in the mutational spectra between the wild-type and mutant enzymes.
One of the obstacles to successful antiretroviral treatment of human immunodeficiency virus (HIV) infections is the eventual emergence of drug-resistant viruses. A primary contributor to this process is the inherently high genetic variability of HIV, commonly encountered in virus populations in the infected individuals (9, 19, 25). This high level of variability is directly related to the viral mutation rate which, in turn, is thought to be significantly influenced by the polymerase fidelity of HIV reverse transcriptase (RT).
Mutants of HIV type 1 (HIV-1) RT resistant to nucleoside analog drugs that coincidentally display higher in vitro polymerase fidelity in gel-based nucleotide insertion and primer extension assays have been identified (6, 11, 20, 21, 24, 31). These include the multi-dideoxynucleoside triphosphate-resistant E89G and the 3TC-resistant M184V RT variants. The M184V mutation arises in patients in response to antiviral therapy with (−)2′,3′ dideoxy-3′-thiacytidine (3TC; lamivudine) (27) and confers up to a 1,000-fold increase in viral resistance to 3TC (7, 26, 30) as well as a low-level cross-resistance to ddC and ddI (8). The E89G mutation, initially isolated via a phenotypic bacterial screening assay (22), confers a broad cross-resistance to deoxynucleotide (dNTP) analogs including ddATP, ddCTP, ddGTP, ddTTP, 3TCTP, and phosphonoformic acid (foscarnet; foscavir). This mutation has also been observed in in vitro-selected, 3TC-resistant variants of HIV-1 (7). The influence of these mutations on drug sensitivity is thought to be due to changes in the geometry of the dNTP-binding pocket brought about by direct interaction of the altered residue with the incoming dNTP in the case of M184V (12, 21, 29) or via template repositioning in the case of E89G (4, 29).
Previous observations showing that E89G and M184V RTs display increased fidelity of dNTP insertion and mispair extension suggested that these enzymes may be less prone to polymerase errors that lead to base substitution mutations. However, polymerase errors that contribute to mutation rates are not limited to uncorrected polymerase misinsertions. Both base substitutions and frameshift errors can be generated through mechanisms not involving direct misinsertion but rather involving template-primer slippage or dislocation (16, 28). In fact, a large proportion of errors by HIV-1 RT have been shown to be of this type (2). Thus, fidelities of dNTP insertion and mispair extension cannot solely be used to determine an overall polymerase error rate.
In this communication, we report the overall polymerase error rates of the RT mutants E89G and M184V measured via an M13-based forward mutation assay (2, 3). The double mutant E89G/M184V was also included in the study to examine if the effects of the two mutations on the overall error rate would be cumulative. Our findings indicate that the overall levels of polymerase fidelity of wild-type, E89G, M184V, and E89G/M184V HIV-1 RTs are similar (less than twofold differences). However, the mutational spectra of the variant enzymes revealed altered error specificities. It is unclear what impact such changes will have on viral variation, and this remains to be experimentally examined.
MATERIALS AND METHODS
Phage DNA and bacterial strains.
Bacteriophage M13mp2, a strain that carries the lacZα gene of Escherichia coli, was used to prepare gapped duplex DNA substrate. E. coli NR9099 [Δ(pro-lac) thi ara recA56/F′ (proAB lacIq ZΔM15)] served as the bacterial host for the preparation of both single-stranded and replicative-form M13 DNA. Electrocompetent E. coli MC1061 [hsdR hsdM+ araD Δ(ara leu) Δ(lacIPOZY) galU galK strA] cells were used to produce phage from the products of the fill-in reactions. E. coli CSH50 [Δ(pro-lac) thi ara strA/F′ (proAB lacIq ZΔM15 traD36)] was the α-complementation strain used to score mutant phage.
Enzymes.
Recombinant wild-type heterodimeric RT derived from HIV-1Hxb2 and its variants with E89G, M184V, and E89G/M184V substitutions were bacterially expressed from derivatives of plasmid pL6H-PROT (14). The construction of these derivatives is described elsewhere (10). The recombinant enzymes were purified by Ni2+-nitrilotriacetic acid-hexahistidine chromatography followed by DEAE- and S-Sepharose chromatography as described earlier (15). The purified wild-type, E89G, M184V, and E89G/M184V RTs had specific activities of 220, 680, 280, and 89 U/mg, respectively, and were found to be nuclease free (data not shown). One unit is defined as the amount of enzyme required to incorporate 1 nmol of dTMP into DNA on a poly(rA)-oligo(dT) template-primer at 37°C in 10 min.
Forward mutation assay.
Gapped duplex M13mp2 DNA was prepared as described previously (3) as the template for DNA fill-in synthesis reactions. This template contained a single-stranded region of 361 nucleotides including the upstream regulatory sequences and the first 107 coding nucleotides of the lacZα gene. Gap-filling synthesis reactions were performed by combining purified RT (0.07 to 0.45 U) with gapped duplex DNA (75 ng) in a reaction buffer containing 75 mM Tris-Cl (pH 8.0), 80 mM KCl, 6 mM MgCl2, 10 mM dithiothreitol, and 500 μM each dATP, dCTP, dGTP, and dTTP (Boehringer Mannheim, Indianapolis, Ind.) in a total volume of 25 μl, and incubation for 1 h at 37°C. Complete gap closure was confirmed via agarose gel electrophoresis.
Polymerization products were electroporated into E. coli MC1061 host cells. After a brief (10-min) recovery period, transformants were plated on a bacterial indicator lawn (CSH50) in the presence of 0.195 mM 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Labscientific Inc., Livingston, N.J.) to generate plaques from released phages. Following incubation for approximately 15 h at 37°C, plaques were scored for α-complementation phenotype. Plaques produced from phage with unmutated lacZα gene display wild-type α complementation, efficiently hydrolyzing the chromogenic X-Gal substrate to appear dark blue. Mutant phage containing lacZα target mutations generate plaques whose phenotypes range from nearly wild-type dark blue to colorless. Plaques of phage designated as mutant were removed from initial screening plates and placed in 0.9% NaCl (1 ml). Mutant phenotypes were confirmed by plating equal inputs of wild-type and putative mutant phage (from diffused 1 ml of stock) on an indicator lawn as described above for initial screening. Once confirmed, mutant plaques were picked and stored at 4°C in 0.9% NaCl (1 ml) until needed for sequence analysis.
Mutational specificity.
Single-stranded phage DNA was prepared as the template for sequence analysis as previously described (3). DNA sequencing reactions were performed with a Sequenase 2.0 DNA sequencing kit (Amersham Life Sciences, Arlington Heights, Ill.) with the oligonucleotide 5′GCGCAGCTGTTGGGAAGGGCG3′ as primer. Sequencing reaction products were resolved on 6% denaturing polyacrylamide gels by using a GENOMYXLR DNA sequencer.
Calculating error frequencies and error rates.
Error frequencies were calculated as the ratio of confirmed mutant plaques to the total plaques screened. Spontaneous background mutation frequency was determined by electroporating uncopied gapped DNA and scoring for mutants as with DNA synthesized by RT. Corrected frequencies were then obtained by subtracting the spontaneous background from the frequency observed for a given RT (3).
Specific error rates were derived by multiplying the corrected overall error frequency with the percentage, in the total mutations, of the particular class of errors being examined (e.g., frameshifts). This value was then divided by 0.6 (the likelihood of expression of the newly synthesized phage minus strand in E. coli [17]) and then divided by the total number of sites that are susceptible to the given class of errors (3). Comparative statistical analysis of hotspot error rates was performed by using Fisher’s exact test.
RESULTS
Overall mutation frequencies.
The fidelity of DNA synthesis by wild-type, E89G, M184V, and E89G/M184V HIV-1 RTs was measured in an M13 phage gap-filling assay. Copying errors resulting in altered function of the phage lacZα reporter gene were phenotypically scored. Gap-filling DNA synthesis reactions (two independent syntheses per enzyme) were performed with purified wild-type, E89G, M184V, and E89G/M184V HIV-1 RTs, and completion of the reaction was confirmed via agarose gel electrophoresis (data not shown). M13 DNA was electroporated into host E. coli, and plaques were generated by plating the transformants on a bacterial indicator lawn. Mutation frequencies for each enzyme were compiled from ratios of total mutant to total wild-type plaques obtained from one to three electroporations per gap-filling reaction per enzyme. The observed mutation frequencies for the wild-type, E89G, M184V, and E89G/M184V HIV-1 RTs were 8.60 × 10−3, 6.26 × 10−3, 5.53 × 10−3, and 12.30 × 10−3, respectively (Table 1). These represent background-corrected values, adjusted by using the spontaneous mutation frequency of uncopied gapped DNA of 1.5 × 10−3. These results indicated that in terms of overall polymerase fidelity, the wild-type, E89G, M184V, and E89G/M184V HIV-1 RTs exhibit similar levels of fidelity (less than twofold differences). Of these, the E89G and M184V RTs were slightly less error prone than wild-type RT, while the E89G/M184V double mutant was slightly more error prone than wild-type RT.
TABLE 1.
Overall mutation frequencies
| RT | No. of plaques screened | No. of mutants | Mutation frequencya (10−3) |
|---|---|---|---|
| Wild type | 13,662 | 138 | 8.60 |
| E89G | 13,916 | 108 | 6.26 |
| M184V | 14,804 | 104 | 5.53 |
| E89G/M184V | 7,173 | 99 | 12.30 |
Corrected for background by subtracting the spontaneous background frequency of 1.50 × 10−3 from the frequency observed for a given RT. Spontaneous background was measured by using uncopied gapped M13mp2 DNA.
Analysis of mutation spectra of the variant RTs.
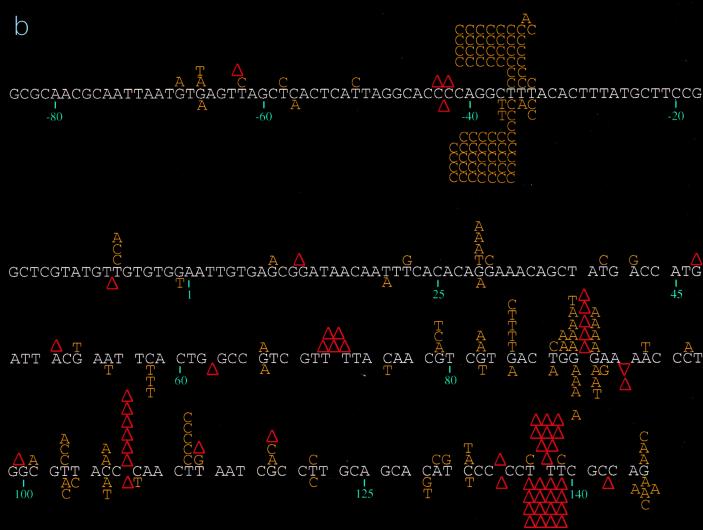
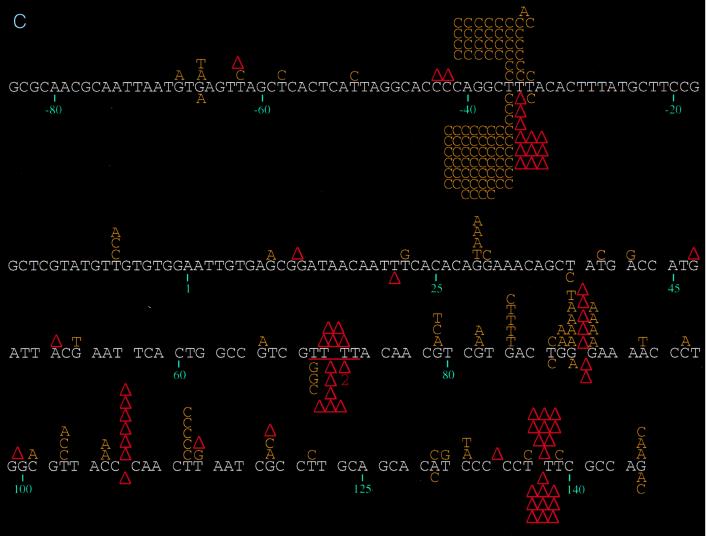
DNA sequence analysis of the phage mutants generated by the individual RTs revealed significant differences in the distribution and specificity of the mutations induced (Fig. 1). Mutations induced by wild-type, E89G, and M184V RTs were fairly well distributed throughout the target region, with some clustering at hotspots (sites with at least five mutations), while those produced by E89G/M184V RT were almost exclusively clustered at hotspots (see below). Close inspection of hotspot distribution shows that certain hotspots are shared by all RTs (Table 2). For example, the base substitution hotspot at position −36 is well represented in the spectra of all of the RTs studied here, as is the frameshift hotspot at positions 137 to 139 (Fig. 1; Table 2). In contrast, the base substitution hotspot observed in the mutation spectrum of wild-type RT at position 112 and the deletion hotspot at positions 106 to 108 (Fig. 1) are conspicuously absent from the spectra of the mutant RTs. In fact, with the mutant RTs, these sites are nearly devoid of mutations (Fig. 1). Similarly, the frameshift hotspot found at positions −34 to −36 in the spectra of the E89G and E89G/M184V RTs is completely absent in the spectra of wild-type and M184V RTs (P < 0.0001 for both E89G [versus wild-type and M184V RTs] and E89G/M184V [versus wild-type and M184V RTs]) (Fig. 1). Furthermore, a greater percentage of total mutations induced by E89G/M184V (83.8%) are within hotspots compared to mutations induced by the other RTs (wild type, 53.6% [P < 0.0001]; E89G, 60.2% [P = 0.0002]; M184V, 60.6% [P = 0.0003]). For the E89G/M184V variant, four sites (positions −36, −34 to −36, 70 to 73, and 137 to 139) account for almost 84% (83/99) of all mutations, with the −34 to −36 positions alone accounting for 68% (67/99) of total mutations observed (Fig. 1c; Table 2).
FIG. 1.
(a) Spectrum of mutations induced by wild-type HIV-1 RT and its E89G variant. Comparison of mutations generated in the lacZα target by wild-type HIV-1Hxb2 and E89G HIV-1 RTs is shown. Mutations induced by wild-type HIV-1Hxb2 RT are represented above the template sequence (white), and mutations induced by E89G HIV-1 RT are shown below the template sequence. Substitutions are indicated by the letter corresponding to the new base (in yellow) above or below the base in the template sequence it is replacing. Nucleotide deletions are indicated by an upright (red) triangle over or under the corresponding run of template bases; nucleotide additions are indicated by an inverted (red) triangle over or under the corresponding run of template bases. Triangles with numerals (2 or 3) underneath them indicate deletions involving more than one base (the bases deleted or are thought to be involved in the deletion are underlined in red). A two-base deletion with the asterisk indicates where it was not possible to determine whether bases 140 and 141 or 141 and 142 were deleted. Nucleotide positions are indicated below the template sequence. (b) Spectrum of mutations induced by wild-type and the M184V mutant HIV-1 RTs. Comparison of mutations generated in the lacZα target by wild-type HIV-1Hxb2 and M184V HIV-1 RTs is shown. Mutations induced by wild-type HIV-1Hxb2 RT are represented above the template sequence (white), and mutations induced by M184V HIV-1 RT are shown below the template sequence. (c) Spectrum of mutations induced by wild-type and E89G/M184V mutant RTs. Comparison of mutations generated in the lacZα target by wild-type HIV-1Hxb2 and E89G/M184V HIV-1 RTs is shown. Mutations induced by wild-type HIV-1Hxb2 RT are represented above the template sequence (white), and mutations induced by E89G/M184V HIV-1 RT are shown below the template sequence.
TABLE 2.
Error rates at selected mutational hotspots
| Site | Wild type
|
E89G
|
M184V
|
E89G/M184V
|
||||
|---|---|---|---|---|---|---|---|---|
| No. of errors | Error rate | No. of errors | Error rate | No. of errors | Error rate | No. of errors | Error rate | |
| Base substitutions | ||||||||
| −36 | 26 | 1/370 | 35 | 1/296 | 37 | 1/305 | 55 | 1/87.5 |
| −35 | 8 | 1/1,203 | 0 | CDd | 1 | 1/11,284 | 0 | CD |
| 84 | 5 | 1/1,926 | 0 | CD | 1 | 1/11,284 | 0 | CD |
| 89 | 5 | 1/1,926 | 4 | 1/2,588 | 5 | 1/2,257 | 1 | 1/4,829 |
| 112 | 5 | 1/1,926 | 0a | CD | 0a | CD | 0a | CD |
| 145 | 3 | 1/3,209 | 1 | 1/10,351 | 5 | 1/2,257 | 2 | 1/2,415 |
| Frameshifts | ||||||||
| −34 to −36 | 0b | CD | 23b | 1/1,350 | 0b | CD | 12b | 1/1,207 |
| 70 to 73 | 5 | 1/7,702 | 2 | 1/20,702 | 0 | CD | 6 | 1/3,220 |
| 88 to 90 | 5 | 1/5,777 | 1 | 1/31,054 | 0 | CD | 2 | 1/7,244 |
| 106 to 108 | 6 | 1/4,814 | 1c | 1/31,054 | 1c | 1/33,852 | 1c | 1/14,488 |
| 137 to 139 | 9 | 1/3,209 | 7 | 1/4,436 | 16 | 1/2,116 | 10 | 1/1,449 |
| Overall | 138 | 1/17,651 | 108 | 1/24,249 | 104 | 1/27,450 | 99 | 1/12,341 |
In comparison of each mutant RT to wild type, P values ranged from 0.069 to 0.076.
In comparison of E89G to wild type or M184V as well as in comparison of E89G/M184V to wild type or M184V, P values were <0.0001 each.
In comparison of each mutant RT to wild type, P values ranged from 0.139 to 0.244.
CD, cannot determine.
Sequence analysis also revealed that wild-type and M184V RTs generated predominantly base substitution mutations, roughly 75% (103/138) and 77% (80/104), respectively, of all mutations (Table 3). On the other hand, E89G RT generated substitution and frameshift mutations (mainly single-base deletions) at almost equal proportions (57:51) (Table 3). Interestingly, the E89G/M184V double mutant had a phenotype intermediate to that of single mutants with respect to base substitution/frameshift ratio, with approximately 67% (66/99) of RT-induced mutations being of the substitution class (Table 3).
TABLE 3.
Summary of error rates for wild-type, E89G, M184V, and E89G/M184V RTs by class
| Mutation type | Wild type
|
E89G
|
M184V
|
E89G/M184V
|
||||
|---|---|---|---|---|---|---|---|---|
| No. of errors | Error rate | No. of errors | Error rate | No. of errors | Error rate | No. of errors | Error rate | |
| All classes | 138 | 1/17,651 | 108 | 1/24,249 | 104 | 1/27,450 | 99 | 1/12,341 |
| Base substitutions | 103 | 1/11,876 | 57 | 1/23,041 | 80 | 1/17,921 | 66 | 1/9,294 |
| Frameshifts | 35a | 1/40,650 | 51 | 1/30,030 | 24a | 1/69,444 | 33a | 1/21,645 |
| At runs | 32 | 1/23,753 | 43 | 1/19,011 | 24 | 1/37,037 | 33 | 1/11,567 |
| At non-runs | 3 | 1/222,222 | 8 | 1/89,286 | 0 | CDb | 0 | CD |
In comparison of E89G to wild type, M184V, and E89G/M184V, P values were 0.0004, 0.0003, and 0.048, respectively.
CD, cannot determine.
Most of the frameshift mutations detected involved the gain (+1) or loss (−1) of one base. Frameshift mutations involving the deletion of two bases were seen only with E89G (four occurrences out of 51 frameshift mutations; Fig. 1a) and E89G/M184V RTs (one occurrence out of 33 frameshifts; Fig. 1c). Frameshift mutations generated by E89G also included one deletion involving three bases (Fig. 1a). Major deletions (>50 bp) were also induced; however, they were uncommon and seen only with wild-type (2 of 138 mutants scored) and E89G (3 of 108 mutants scored) RTs.
DISCUSSION
Viral mutation rates are the products of the combined effects of overall polymerase (copying) error rates, replication rates, and biological selection (5). Presumably, increases or decreases in any of these factors could result in corresponding changes in mutation rate. This notion has previously not been tested for any variant of HIV-1 RT. The variants of HIV-1 RT that were recently reported to possess increased fidelities of dNTP insertion and primer extension in in vitro assays (6, 11, 20, 21, 24, 31) appeared suitable to examining the contributions of polymerase fidelity to overall mutation rate in vivo. Thus, we have determined the overall copying fidelity of the E89G, M184V, and E89G/M184V RT variants. We had earlier reported increases of three- to sixfold in average nucleotide insertion fidelity for the M184V and E89G RTs (6, 31). When changes in the fidelity of formation of specific mispairs are considered, the increases of 2- to 45-fold (6, 21, 24, 31) have been seen. With respect to terminal mismatch extension efficiency, increases in fidelity of primer extension, ranging from 3- to 66-fold greater (for extension from specific mispairs) than for wild-type RT, have been reported for E89G and M184V RTs (11, 20, 24). In contrast to these large increases in fidelities of dNTP insertion and primer extension previously observed for specific types of errors, the M184V and E89G RTs in the current study showed only slightly decreased overall polymerase error rates whereas the E89G/M184V variant rate was slightly increased (less than twofold differences when compared to the wild type) (Table 1). This discrepancy could be a result of the influence of certain features of the M13 forward mutation assay, such as host repair and the ability to detect slippage-mediated errors in addition to nucleotide misinsertion and mispair extension. In fact, a substantial proportion of the total mutations observed occurred within the homonucleotide runs in the template sequence which are primary sites for slippage-mediated mutagenesis. Thus, in light of the modest differences observed in overall error rates, it appears that any increases in fidelity of these mutants that may be afforded by the increased dNTP insertion/primer extension fidelity are muted by the contributions of other mutagenic mechanisms.
The lack of increase in fidelity for M184V RT seen in this study would appear to be concordant with the biological data regarding viral evolution of M184V mutant viruses. Drug-resistant variants of such viruses were found to emerge in cell culture at rates equal to those of wild-type viruses (1, 13), suggesting that mutation rates were unaffected by the RT mutation. It is likely that compensatory factors similar to those operating in the M13 forward assay (as described above) were modulating the effect of the viral 184V substitution.
Although both E89G and M184V mutations increase nucleotide insertion/primer extension fidelity, it seems likely that they exert their effects via distinct mechanisms. The residues affected by these mutations lie within the enzyme active site in or near the dNTP-binding pocket (12, 29). Structural data place these residues at opposing sides of the active site, with the E89 residue contacting the phosphate-deoxyribose backbone of the template strand and the M184 residue interacting with the sugar moiety of the primer terminus (12, 29). Consequently, as mutations at these residues alter different surfaces of the dNTP-binding pocket, it might be anticipated that the effects of these modifications on the dNTP-binding pocket conformation would differ. Such differences are reflected in the divergence seen in the distribution and specificity of mutations induced by the variants. For instance, the E89G mutation had an enhancing effect on frameshift mutagenesis (both run and non-run associated). In contrast, the M184V mutation had a minimal effect on this class of errors (Table 3). It is generally thought that most frameshift mutations (insertions and deletions) involving few (one to three) nucleotides arise from transiently misaligned or slipped template-primer intermediates (28). The E89 residue is located within the β5a strand of the palm region of RT, where it is thought to help the polymerase grip the template (12). It is conceivable that the glycine substitution at this residue, by virtue of glycine’s smaller side chain, results in a looser grip, which might enable the primed template to more easily form misaligned intermediates while still bound to the enzyme. Alternatively, a looser template grip stemming from the 89G mutation might allow the RT to more readily dissociate from the template, providing greater opportunity for the formation of misaligned template-primers. A determination of dissociation rates using purified RTs and model template-primers may help differentiate between the two possibilities. The M184 residue participates in dNTP binding (21, 32) rather than template-primer grip. Thus, it is possible that substitutions at this position affect utilization of frameshift intermediates rather than the enzyme’s ability to promote their formation.
To date, the E89G substitution has been found only in viral isolates bearing the M184V mutation as well. Therefore, the E89G/M184V double mutant was studied in order to understand the cumulative effect of these two mutations on error rate. Curiously, when present independently, the individual mutations increased polymerase fidelity slightly while a combination of the two led to a slight decrease in the overall polymerase fidelity (Table 1). More unexpected was the highly clustered distribution of the mutations induced by the E89G/M184V RT, considering that both E89G and M184V RTs generated mutations that were fairly well distributed. Yet the influence of both RT mutations appears to be seen in the mutational specificity of the double mutant. Hotspots common to both single variants (positions −36 and 137 to 139) are reproduced by the E89G/M184V variant. The strong representation of the deletion hotspot at the −34 to −36 site seems to suggest some dominance of the 89G mutation; however, the lack of non-run-associated frameshifts for the double mutant appears to indicate a significant contribution by the 184V residue to frameshift specificity. Thus, the error specificities of each single mutation are preserved to a degree in the double mutant while the effects of each individual substitution on polymerase fidelity are somewhat modulated by the other. Since these two residues occupy opposite sides of the active site, it may be possible that steric constraints imposed by these substitutions are additive, thus further restricting the range of stable premutational intermediates. If this is so, it may offer an explanation for the limited mutation distribution exhibited by E89G/M184V RT.
Since the influence of these mutations on overall RT fidelity appears minimal, could differences in error specificity have a significant biological effect on viruses harboring these mutations? It is difficult to predict any specific effects of 184V alteration since the change observed in overall fidelity was small. In the case of E89G variant viruses, perhaps the enhancement in frameshift mutagenesis associated with the 89G substitution when expressed in vivo results in a higher proportion of virions with single-base insertions and deletions which are far more likely to be lethal than base substitutions. This may partially account for the failure of this mutation to be seen in a viral isolate independent of the 184V substitution. The ability of the M184V mutation to reduce the frameshift mutagenicity of E89G could permit a variant HIV with both mutations to replicate and to be recovered via in vitro selection protocols (7).
Finally, it is known that sequence context contributes substantially to the mutagenic potential of a given nucleotide site (2, 18, 23), and this likely results in the site bias that produces hotspots. The studies reported here were conducted in the context of the lacZα gene, which may not be representative of viral sequences. Therefore, these RT mutations could impart to replicating viral genomes overall fidelities higher or lower than those observed here. In this context, the double mutant E89G/M184V RT in particular needs to be tested for its effect on viral mutation rate since it appears to display few mutations outside the hotspots which are so characteristic of lacZα gene. It should be possible through single-cycle infection studies with HIVs containing these mutations to establish the in vivo significance of our findings.
ACKNOWLEDGMENTS
We thank Clyde A. Hutchison III (University of North Carolina, Chapel Hill) for providing phasmids of the M184V mutant RT cDNA, B. D. Preston for providing the M13mp2 phage, T. A. Kunkel (National Institute for Environmental Health Sciences) for providing the bacterial strains for the M13 forward mutation assay, Kenneth Curr, Clark Choi, and Jayanthi Manne for technical assistance, Ganjam V. Kalpana, William Franklin, and Lisa Rezende for critically reading the manuscript, and the Oligonucleotide Synthesis Facility of the Albert Einstein College of Medicine’s Cancer Center for DNA oligonucleotides.
This work was supported by Public Health Service grants AI-30861 and AI-40375 (to V.R.P.). W.C.D. acknowledges support from institutional training grant T32-AI07501.
REFERENCES
- 1.Balzarini J, Pelemans H, Karlsson A, De Clercq E, Kleim J P. Concomitant combination therapy for HIV infection preferable over sequential therapy with 3TC and non-nucleoside reverse transcriptase inhibitors. Proc Natl Acad Sci USA. 1996;93:13152–13157. doi: 10.1073/pnas.93.23.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bebenek K, Abbotts J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 3.Bebenek K, Kunkel T A. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 4.Boyer P L, Tantillo C, Jacobo-Molina A, Nanni R G, Ding J, Arnold E, Hughes S H. Sensitivity of wild-type human immunodeficiency virus type 1 reverse transcriptase to dideoxynucleotides depends on template length; the sensitivity of drug-resistant mutants does not. Proc Natl Acad Sci USA. 1994;91:4882–4886. doi: 10.1073/pnas.91.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin J. HIV viral dynamics. AIDS. 1996;10:S75–S84. [PubMed] [Google Scholar]
- 6.Drosopoulos W C, Prasad V R. Increased polymerase fidelity of E89G, a nucleoside analog-resistant variant of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1996;70:4834–4838. doi: 10.1128/jvi.70.7.4834-4838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao Q, Gu Z, Parniak M A, Cameron J, Cammack N, Boucher C, Wainberg M A. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1993;37:1390–1392. doi: 10.1128/aac.37.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu Z, Gao Q, Li X, Parniak M A, Wainberg M A. Novel mutation in the human immunodeficiency virus type 1 reverse transcriptase gene that encodes cross-resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine. J Virol. 1992;66:7128–7135. doi: 10.1128/jvi.66.12.7128-7135.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn B H, Shaw G M, Taylor M E, Redfield R R, Markham P D, Salahuddin S Z, Wong-Staal F, Gallo R C, Parks E S, Parks W P. Genetic variation in HTLV III/LAV over time in patients with AIDS or at risk for AIDS. Science. 1986;232:1548–1553. doi: 10.1126/science.3012778. [DOI] [PubMed] [Google Scholar]
- 10.Hamburgh, M. E., W. C. Drosopoulos, and V. R. Prasad. The influence of 3TC-resistance mutations M184V and E89G in the human immunodeficiency virus reverse transcriptase on mispair extension efficiency. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 11.Hsu M, Inouye P, Rezende L, Richard N, Li Z, Prasad V R, Wainberg M A. Higher fidelity of RNA-dependent DNA mispair extension by M184V drug-resistant than wild type reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res. 1997;25:4532–4536. doi: 10.1093/nar/25.22.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobo-Molina A, Ding J, Nanni R G, Clark A D J, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S, Arnold E. Crystal structure of human immunodeficiency virus reverse transcriptase complexed with double stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keulen W, Nijhuis M, Schuurman R, Berkhout B, Boucher C. Reverse transcriptase and HIV variation. Science. 1997;275:229. . (Comment.) [PubMed] [Google Scholar]
- 14.Kew, Y., L. Olsen, A. Japour, and V. R. Prasad. Insertions into the b3-b4 hairpin loop of HIV-1 reverse transcriptase reveal a role for fingers subdomain in processive polymerization. J. Biol. Chem., in press. [DOI] [PubMed]
- 15.Kew Y, Song Q, Prasad V. Subunit selective mutagenesis of Glu89 residue in human immunodeficiency virus reverse transcriptase. J Biol Chem. 1994;269:15331–15336. [PubMed] [Google Scholar]
- 16.Kunkel T A, Alexander P S. The base substitution fidelity of eucaryotic DNA polymerases. Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J Biol Chem. 1986;261:160–166. [PubMed] [Google Scholar]
- 17.Kunkel T A, Soni A. Exonucleolytic proofreading enhances the fidelity of DNA synthesis by chick embryo DNA polymerase-gamma. J Biol Chem. 1988;263:4450–4459. [PubMed] [Google Scholar]
- 18.Mendelman L V, Boosalis M S, Petruska J, Goodman M F. Nearest neighbor influences on DNA polymerase insertion fidelity. J Biol Chem. 1989;264:14415–14423. [PubMed] [Google Scholar]
- 19.Meyerhans A, Cheynier R, Albert J, Seth M, Kwok S, Sninsky J, Morfeldt-Manson L, Asjo B, Wain-Hobson S. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell. 1989;58:901–910. doi: 10.1016/0092-8674(89)90942-2. [DOI] [PubMed] [Google Scholar]
- 20.Oude Essink B B, Back N K T, Berkhout B. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 1997;25:3212. doi: 10.1093/nar/25.16.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey V N, Kaushik N, Rege N, Sarafianos S G, Yadav P N S, Modak M J. Role of methionine 184 in human immunodeficiency virus type 1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 22.Prasad V R, Lowy I, De Los Santos T, Chiang L, Goff S P. Isolation and characterization of a dideoxyguanosine triphosphate-resistant HIV-1 reverse transcriptase expressed in bacteria. Proc Natl Acad Sci USA. 1991;88:11363–11367. doi: 10.1073/pnas.88.24.11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ricchetti M, Buc H. Reverse transcriptases and genomic variability: the accuracy of DNA replication is enzyme specific and sequence dependent. EMBO J. 1990;9:1583–1593. doi: 10.1002/j.1460-2075.1990.tb08278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubinek T, Bakhanashvili M, Hizi A. The fidelity of 3′ misinsertion and mispair extension during DNA synthesis exhibited by two drug-resistant mutants of the reverse transcriptase of human immunodeficiency virus type 1 with Leu74Val and Glu89Gly. Eur J Biochem. 1997;247:238. doi: 10.1111/j.1432-1033.1997.00238.x. [DOI] [PubMed] [Google Scholar]
- 25.Saag M S, Hahn B H, Gibbons J, Li Y, Parks E S, Parks W P, Shaw G M. Extensive variation of human immunodeficiency virus type-1 in vivo. Nature. 1988;334:440–444. doi: 10.1038/334440a0. [DOI] [PubMed] [Google Scholar]
- 26.Schinazi R F, Lloyd R M, Jr, Nguyen M, Cannon D L, McMillan A, Ilksoy N, Chu C K, Liotta D C, Bazmi H Z, Mellors J W. Characterization of human immunodeficiency viruses resistant to Oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 28.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 30.Tisdale M, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wainberg M A, Drosopoulos W C, Salomon H, Hsu M, Borkow G, Parniak M, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad V R. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 32.Wilson J E, Aulabaugh A, Caligan B, McPherson S, Wakefield J K, Jablonski S, Morrow C D, Reardon J E, Furman P A. Human immunodeficiency virus type-1 reverse transcriptase. Contribution of Met-184 to binding of nucleoside 5′-triphosphate. J Biol Chem. 1996;271:13656–13662. doi: 10.1074/jbc.271.23.13656. [DOI] [PubMed] [Google Scholar]