INTRODUCTION
Effective colorectal cancer (CRC) screening can reduce the incidence and mortality of this all-too-common disease, but nearly one-third of the screen-eligible population is not up to date with current recommendations (1). The US Preventive Services Task Force, American Cancer Society, Multisociety Task Force (MSTF), and other national organizations all endorse multiple options for average-risk CRC screening (2–4), including the multitarget stool DNA (mt-sDNA) assay, marketed as Cologuard (Exact Sciences, Madison, WI), introduced into clinical practice in August 2014. Here, key concepts and practical questions related to mt-sDNA screening are discussed to provide background for informed patient-provider discussions.
PRECLINICAL DEVELOPMENT
Compared with the normal colorectal epithelium, neoplastic colonocytes exhibit increased proliferation, decreased apoptosis, and altered intercellular adhesion, providing a strong biologic rationale for fecal sampling and molecular interrogation (Figure 1) (5). Through a series of laboratory discovery and early translational studies (6–9), a focused set of methylation (NDRG4 and BMP3) and mutation (KRAS) markers was identified for screening assay development, in combination with hemoglobin and ACTB (an indicator of total human DNA). Specimen processing and analysis are performed using a high-throughput automated assay system, with qualitative results (positive or negative) based on an integrated, multimarker logistic regression algorithm (Figure 2). Additional details regarding the mt-sDNA assay collection kit, analytical performance, quality controls, and technical specifications are described in the US FDA Premarket Approval application (P130017) (10).
Figure 1.
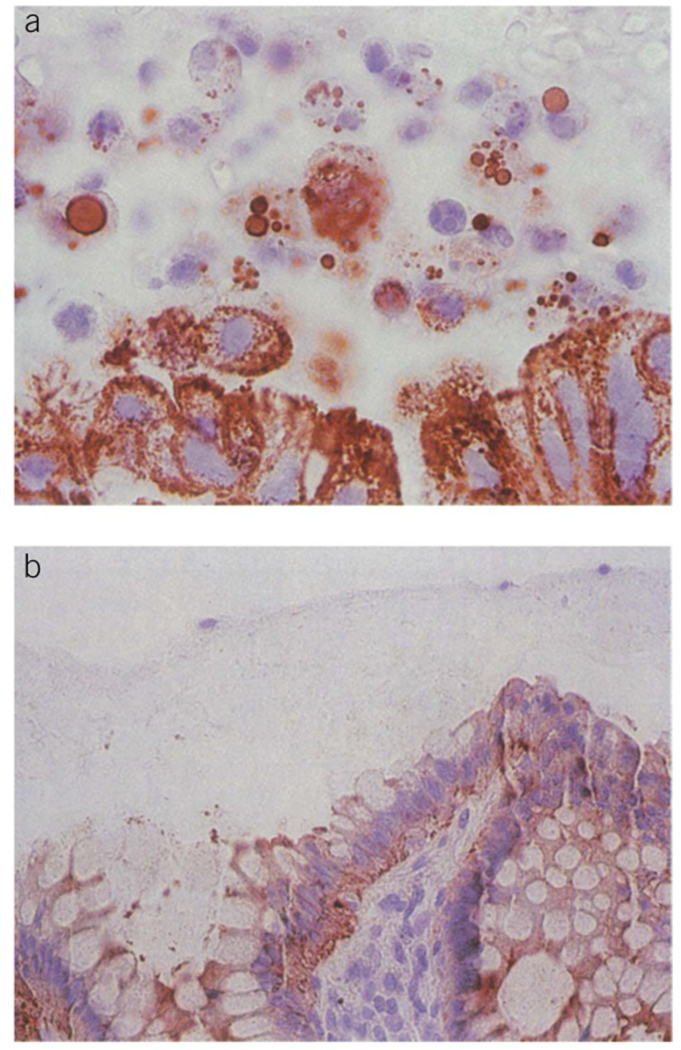
Exfoliated colonocytes. (a) Mucocellular layer above colorectal cancer. (b) Mucocellular layer above normal colonic mucosa. From Ahlquist DA, Harrington JJ, Burgart LJ,and Roche PC. Morphometric analysis of the “mucocellular layer” overlying colorectal cancer and normal mucosa: relevance to exfoliation and stool screening. Hum Pathol 2000;31(1):51–7. Used with permission from Elsevier.
Figure 2.
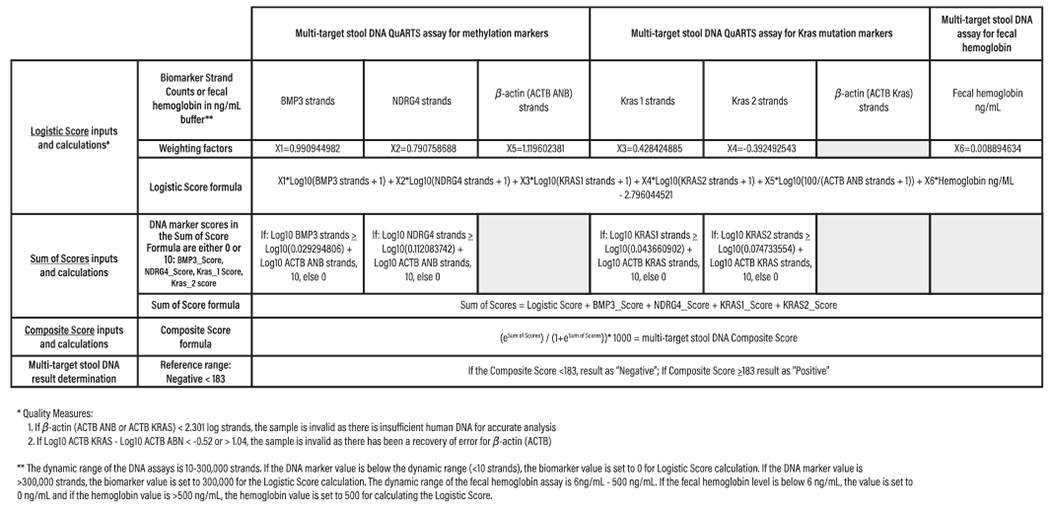
Integrated algorithm for determining commercially available multitarget stool DNA assay (Cologuard) results. The integrated algorithm has 3 main components: (i) logistic score, calculated from a logistic regression formula that uses group-level marker data to discriminate between patient populations with vs without screen-relevant colorectal neoplasia; (ii) sum of scores, which combines the logistic score with individual marker scores, to insure that if any DNA marker (but not hemoglobin) value exceeds the 99.5 percentile of normal,the assay will yield a positive result; and (iii) composite score, generated by subjecting the sum of scores to an exponential equation that generates an overall assay result ranging from 0 to 1,000, with positive (≥183)and negative (<183) thresholds determined through extensive preclinical testing. Using the integrated regression algorithm, higher sensitivities were observed for both colorectal cancers and advanced precancerous lesions (APLs), as compared to the individual marker approach (8). From Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370(14):1287–97. Used with permission from the Massachusetts Medical Society.
CLINICAL VALIDATION
Screening-setting performance of mt-sDNA was estimated and compared with the fecal immunochemical test (FIT) in a large, cross-sectional study of 9,989 asymptomatic subjects aged 50–84 years (ClinicalTrials.gov Identifier: NCT01397747) (11). Using colonoscopy as the reference standard, 65 subjects with CRC and 757 subjects with at least 1 advanced precancerous lesion (APL; defined as adenomas with high-grade dysplasia, ≥ 25% villous features, or ≥ 1 cm in diameter; or sessile serrated polyps ≥ 1 cm in diameter) were identified. Sensitivity of the mt-sDNA assay was statistically significantly higher than FIT for all comparisons (Figure 3), including any CRC (92.3% vs 73.8%, P = 0.002), early-stage (I-III) CRC (93.3% vs 73.3%, P = 0.002), any APL (42.4% vs 23.8%, P < 0.001), APLs with high-grade dysplasia (69.2% vs 49.2%, P = 0.004), and sessile serrated polyps ≥ 1 cm in diameter (42.4% vs 5.1%, P < 0.001). Conversely, specificity was higher for FIT than mt-sDNA, based on the colonoscopic absence of CRC or APLs (94.9% vs 86.6%, P < 0.001) or any neoplasia (96.4% vs 89.8%, P < 0.001). Mt-sDNA has also been studied vs FIT in Alaska Native (12) and African American (13) populations, respectively, with similar results.
Figure 3.
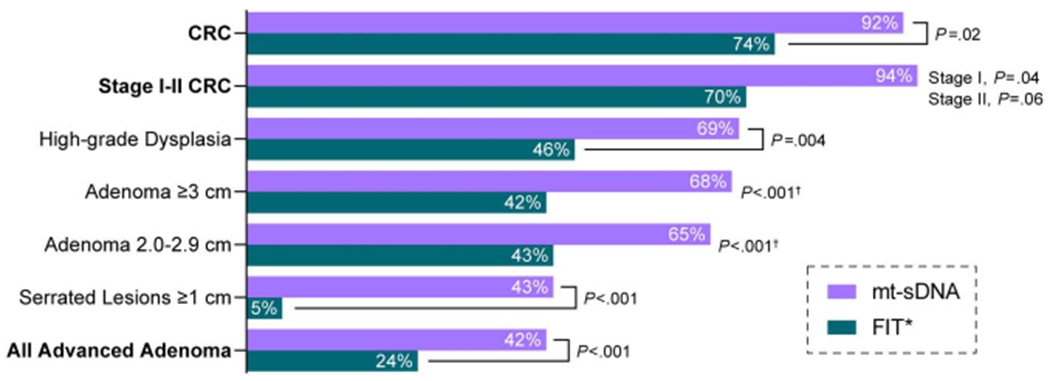
Sensitivity of the mt-sDNA and FIT assays for screen-detecting clinically relevant neoplasia in asymptomatic adults, aged 50–84 years. Data from (11) and Cologuard Physician Brochure. Exact Sciences Corporation. Madison, WI. CRC, colorectal cancer; FIT, fecal immunochemical test; mt-sDNA, multitarget stool DNA assay. *One time FIT. Pvalue is for the trend.
CLINICAL APPLICATION
Mt-sDNA is US FDA-approved for average-risk CRC screening in asymptomatic adults, aged 45 years and older, by prescription only. It should not be used in patients with ongoing melena or hematochezia; personal history of CRC, colorectal adenoma(s), or inflammatory bowel disease; family history of CRC in a first-degree relative diagnosed at age ≤ 60 years; 2 or more first-degree relatives diagnosed at any age; or heritable cancer syndromes.
For patients who remain at average - risk for CRC, mt-sDNA screening is recommended once every 3 years (2–4). Patients and providers who select mt-sDNA screening should commit to completing a follow-up colonoscopy if the stool test result is positive. Importantly, the mt-sDNA test should not be positioned as a replacement for diagnostic or surveillance colonoscopy in any clinical setting.
NAVIGATION SUPPORT
Mt-sDNA screening with Cologuard by Exact Sciences Laboratories includes comprehensive patient navigation support by mail, telephone, email, or SMS, based on patient preferences; this includes live patient and provider support, 24 h/d × 365 d/yr, with translation services available in more than 240 languages. With this navigation-assisted approach, the mt-sDNA assay completion rate within 12 months of test order is 68% overall, with slightly higher adherence (71%) reported for Medicare patients (14).
PATIENT ADOPTION
In a retrospective review of Medicare patients in a multispecialty group practice, Prince et al. (15) assessed the impact of mt-sDNA assay availability on cross-sectional CRC screening adherence, along with follow-up colonoscopy for those with a positive mt-sDNA assay result. Among 393 patients, 347 (88.3%) completed the mt-sDNA test, 51/347 (14.7%) had a positive result, and 49/51 (96.1%) went on to follow-up colonoscopy. Positive predictive values were 4/49 (8%) for CRC, 21/49 (51%) for advanced adenoma, and 40/49 (82%) for any neoplasia, respectively. Finney Rutten et al. (16) analyzed patterns and predictors of mt-sDNA use among screen-eligible residents of Olmsted County, MN, aged 50–75 years, from November 2014 through November 2015. Mt-sDNA test use increased over time (P = 0.01), while the screening colonoscopy rate concomitantly declined (P < 0.001). The highest rates of mt-sDNA adoption were in patients who were aged 50–54 years, female, white, or with a history of previous CRC screening.
DIAGNOSTIC YIELD AT FOLLOW-UP COLONOSCOPY
Johnson et al. (17) retrospectively compared colorectal neoplasia detection rates in a “blinded group” (n = 72) of DeeP-C study (NCT01397747) participants whose follow-up colonoscopies were performed by endoscopists unaware of the positive mt-sDNA results vs an “unblinded group” (n = 172) of clinically identified patients whose follow-up colonoscopies were performed by endoscopists aware of the positive mt-sDNA results. Any adenomatous or sessile serrated polyps (70% vs 53%, P = 0.01) and APLs (28% vs 21%, P = 0.27) were more commonly detected in the unblinded vs the blinded group, with a higher number of polyps detected per patient as well (median [inter-quartile range] of 2 [1–4] vs 1 [0–2]; P = 0.0007). Similarly, Eckmann et al. conducted a large, retrospective, single-center study of average-risk patients with positive mt-sDNA results, reporting that among 1,558 (87%) patients who completed the follow-up procedure (median of 44 [inter-quartile range 28–72] days from mt-sDNA result), any colorectal neoplasia was identified in 1w%). Among the subset of patients who reported previous (median 10.3 years) screening colonoscopy, 580 were found to have colorectal neoplasia (PPV = 63%) (18).
PRACTICAL QUESTIONS
The following section includes a series of practical questions related to mt-sDNA screening that may be commonly encountered, along with pragmatic responses to support accurate discussion and decision-making.
Why are not the mt-sDNA molecular marker results reported separately?
The commercially available mt-sDNA assay (Cologuard) result is derived from a previously reported, multiparameter mathematical algorithm (6,8) (Figure 2), which was developed and locked before the data analyses from 2 screen-setting clinical trials (11,12). The algorithm-based approach was found to be more sensitive than a single marker cutoff approach in premarket analyses and was therefore included as a specification of the assay’s US FDA approval as a qualitative test. Since the individual marker data are less accurate and have not been separately validated, the mt-sDNA marker panel results are reported in aggregate, as positive or negative.
How often is the mt-sDNA assay result reported as positive?
The expected mt-sDNA positivity rate in average-risk CRC screening patients is approximately 16% (11). All positive mt-sDNA results should prompt a timely follow-up colonoscopy because data from FIT-based screening programs demonstrate increased CRC mortality risk if colonoscopy is delayed or neglected (19,20).
Are false-positive results more common with the mt-sDNA assay or the FIT assay?
In the pivotal DeeP-C study (NCT01397747), the cross-sectional point estimate for mt-sDNA specificity was lower than FIT (86.6% vs 94.9%, respectively), translating to false-positive rates of 13.4% and 5.1% for 1 cycle of CRC screening. However, consideration of longitudinal screening provides a different perspective. For example, over a 3-year screening period, the cumulative specificity for annual FIT screening is simply estimated as 94.9% × 94.9% × 94.9% = 85.5% (14.5% false-positive rate), whereas the cumulative specificity of triennial mt-sDNA screening stays at 86.6% (13.4% false-positive rate). Comparability in longitudinally derived false-positive rates for the mt-sDNA and FIT assays has been appropriately acknowledged in the 2017 MSTF guidelines (4).
If the mt-sDNA assay result is positive and the follow-up colonoscopy is negative, is further evaluation needed?
Expert opinion and existing data indicate that high-quality colonoscopy, even if negative, is sufficient follow-up for a positive mt-sDNA assay result, as recommended in the 2017 MSTF guidelines (4). Supporting this recommendation, Berger et al. (21) recently reported outcomes for patients with discordant (positive Cologuard, negative colonoscopy; n = 205) or concordant (negative Cologuard, negative colonoscopy; n = 1,011) findings at baseline.
After a median follow-up period of 5.4 years, the rate of aerodigestive cancers identified in the former and latter groups was not statistically significant (n = 5 and n = 11, respectively; adjusted risk ratio of 2.2; 95% confidence interval, 0.8–6.2; P = 0.15). Moreover, the incidence of observed aerodigestive cancers in the discordant group did not differ from the expected rate (n = 6), based on Surveillance, Epidemiology, and End Results data (risk ratio 0.8; 95% confidence interval, 0.3–1.9; P = 0.599).
SUMMARY
Increased patient and provider engagement in effective, longitudinal CRC screening has tremendous potential to positively improve public health. The COVID-19 pandemic has raises several new challenges for timely CRC screening, which can be best overcome through thoughtful consideration and informed discussion with patients about all available, guideline-endorsed options, along with close communication between primary and specialty care providers to ensure appropriate follow-up for positive screening test results. Compelling scientific data and growing practical experience support home-based mt-sDNA screening as an effective, acceptable, and accessible option for average-risk patients. Expanded collaborations between clinicians, health system administrators, industry collaborators, advocacy partners, and other interested stakeholders are required to ensure that population-based screening can have an even greater impact on CRC incidence and mortality rates moving forward.
ACKNOWLEDGEMENTS
Confirmation of accurate descriptions related to the Cologuard assay was provided by Exact Sciences staff. Data visualization assistance was provided by David K Edwards V, PhD (Exact Sciences, Madison, WI).
Footnotes
Potential competing interests: Mayo Clinic has an intellectual property agreement with Exact Sciences under which J.B.K. is listed as an inventor and has received royalties paid to Mayo Clinic. P.J.L. serves as Chief Medical Officer for Screening at Exact Sciences through a contracted services agreement with Mayo Clinic. P.J.L. has received royalties paid to Mayo Clinic.
REFERENCES
- 1.Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70(3):145–64. [DOI] [PubMed] [Google Scholar]
- 2.Bibbins-Domingo K, Grossman DC, Curry SJ, et al. Screening for colorectal cancer: US Preventive Services Task Force recommendation statement. JAMA 2016;315(23):2564–75. [DOI] [PubMed] [Google Scholar]
- 3.Wolf AMD, Fontham ETH, Church TR, et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin 2018;68(4):250–81. [DOI] [PubMed] [Google Scholar]
- 4.Rex DK, Boland CR, Dominitz JA, et al. Colorectal cancer screening: Recommendations for physicians and patients from the U.S. Multi-society Task Force on colorectal cancer. Am J Gastroenterol 2017;112(7):1016–30. [DOI] [PubMed] [Google Scholar]
- 5.Ahlquist DA, Harrington JJ, Burgart LJ, et al. Morphometric analysis of the “mucocellular layer” overlying colorectal cancer and normal mucosa: Relevance to exfoliation and stool screening. Hum Pathol 2000;31(1):51–7. [DOI] [PubMed] [Google Scholar]
- 6.Zou H, Allawi H, Cao X, et al. Quantification of methylated markers with a multiplex methylation-specific technology. Clin Chem 2012;58(2):375–83. [DOI] [PubMed] [Google Scholar]
- 7.Ahlquist DA, Zou H, Domanico M, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology 2012;142(2):248–56; quiz e25-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lidgard GP, Domanico MJ, Bruinsma JJ, et al. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin Gastroenterol Hepatol 2013;11(10):1313–8. [DOI] [PubMed] [Google Scholar]
- 9.Ahlquist DA, Sargent DJ, Loprinzi CL, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med 2008;149(7):441–50, w81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Exact Sciences Corporation. Summary of Effectiveness and Safety Data. Premarket Approval Application (PMA) Number: P130017. U.S. Food & Drug Administration: Silver Spring, MD, 2014. [Google Scholar]
- 11.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370(14):1287–97. [DOI] [PubMed] [Google Scholar]
- 12.Redwood DG, Asay ED, Blake ID, et al. Stool DNA testing for screening detection of colorectal neoplasia in Alaska native people. Mayo Clin Proc 2016;91(1):61–70. [DOI] [PubMed] [Google Scholar]
- 13.Cooper GS, Markowitz SD, Chen Z, et al. Performance of multitarget stool DNA testing in African American patients. Cancer 2018;124(19):3876–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swartz R, Weiser E, Parks P, et al. Su1660—colorectal cancer screening: Compliance with multitarget stool Dna testing among Medicare beneficiaries. Gastroenterology 2019;156(6):S–601. [Google Scholar]
- 15.Prince M, Lester L, Chiniwala R, et al. Multitarget stool DNA tests increases colorectal cancer screening among previously noncompliant Medicare patients. World J Gastroenterol 2017;23(3):464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finney Rutten LJ, Jacobson RM, Wilson PM, et al. Early adoption of a multitarget stool DNA test for colorectal cancer screening. Mayo Clin Proc 2017;92(5:726–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson DH, Kisiel JB, Burger KN, et al. Multitarget stool DNA test: Clinical performance and impact on yield and quality of colonoscopy for colorectal cancer screening. Gastrointest Endosc 2017;85(3):657–65.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckmann JD, Ebner DW, Bering J, et al. Multitarget stool DNA screening in clinical practice: High positive predictive value for colorectal neoplasia regardless of exposure to previous colonoscopy. Am J Gastroenterol 2020;115(4):608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doubeni CA, Fedewa SA, Levin TR, et al. Modifiable failures in the colorectal cancer screening process and their association with risk of death. Gastroenterology 2019;156(1):63–74.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meester RG, Zauber AG, Doubeni CA, et al. Consequences of increasing time to colonoscopy examination after positive result from fecal colorectal cancer screening test. Clin Gastroenterol Hepatol 2016;14(10):1445–51.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger BM, Kisiel JB, Imperiale TF, et al. Low incidence of aerodigestive cancers in patients with negative results from colonoscopies, regardless of findings from multitarget stool DNA tests. Clin Gastroenterol Hepatol 2020;18:864–71. [DOI] [PMC free article] [PubMed] [Google Scholar]


