Abstract
Background
The 2022 global outbreak of Monkeypox virus (MPXV) highlighted challenges with polymerase chain reaction detection as divergent strains emerged and atypical presentations limited the applicability of swab sampling. Recommended testing in the United States requires a swab of lesions, which arise late in infection and may be unrecognized. We present MPXV detections using plasma microbial cell-free DNA (mcfDNA) sequencing.
Methods
Fifteen plasma samples from 12 case-patients were characterized through mcfDNA sequencing. Assay performance was confirmed through in silico inclusivity and exclusivity assessments. MPXV isolates were genotyped using mcfDNA, and phylodynamic information was imputed using publicly available sequences.
Results
MPXV mcfDNA was detected in 12 case-patients. Mpox was not suspected in 5, with 1 having documented resolution of mpox >6 months previously. Six had moderate to severe mpox, supported by high MPXV mcfDNA concentrations; 4 died. In 7 case-patients, mcfDNA sequencing detected coinfections. Genotyping by mcfDNA sequencing identified 22 MPXV mutations at 10 genomic loci in 9 case-patients. Consistent with variation observed in the 2022 outbreak, 21 of 22 variants were G > A/C > T. Phylogenetic analyses imputed isolates to sublineages arising at different time points and from different geographic locations.
Conclusions
We demonstrate the potential of plasma mcfDNA sequencing to detect, quantify, and, for acute infections with high sequencing coverage, subtype MPXV using a single noninvasive test. Sequencing plasma mcfDNA may augment existing mpox testing in vulnerable patient populations or in patients with atypical symptoms or unrecognized mpox. Strain type information may supplement disease surveillance and facilitate tracking emerging pathogens.
Keywords: metagenomic sequencing, microbial cell-free DNA, outbreak surveillance, mpox, HIV
In 2022, mpox (formerly known as monkeypox), caused by the zoonotic double-stranded DNA Monkeypox virus (MPXV) from the Orthopoxvirus genus in the family Poxviridae, evaded surveillance and established a global chain of human-to-human transmission [1]. By the end of the year, >80 000 mpox cases were attributed to the outbreak [2]. Past outbreaks originated from zoonotic transmission in endemic areas in Central and West Africa and caused illness comprising a brief febrile prodromal period followed by a rash that evolved over the course of several weeks [3]. In contrast, the epidemiology and clinical presentation of the 2022 outbreak have tended to be quite different. Most outbreak cases and clusters have been associated with transmission through close, intimate contact, especially through sexual activity and in nonendemic countries. This has resulted in considerable variation in clinical course and rash development [3, 4]. Further complicating efforts to recognize and control ongoing MPXV transmission have been findings that some infected persons may spread the virus during the prodromal period before rash development [5–7].
The mpox diagnostic test recommended by the Centers for Disease Control and Prevention (CDC) guidance is a non-MPXV-specific pan-Orthopoxvirus real-time polymerase chain reaction (PCR) test [8]. Positive samples are subsequently processed directly by CDC with a confirmatory MPXV-specific PCR test. However, PCR-based MPXV tests exhibit several limitations. First, such assays have only been approved by the US Food and Drug Administration (FDA) for detection of MPXV DNA from lesion swab samples [9, 10]. The FDA has further advised against using other sample types to prevent false results [11]. The guidance restricts testing to patients with suspected MPXV lesions and may therefore miss those in the prodromal yet potentially infectious phase, patients with atypical presentations (as seen in the current outbreak), and patients with unrecognized lesions. In other countries, PCR testing samples from other nonlesion body sites or fluids in symptomatic individuals (eg, upper respiratory, rectal, or seminal fluid) have been reported to have reasonable sensitivity (69%–100%), and a small proportion of samples (2%–6.5%) from presymptomatic individuals (eg, pharyngeal, anorectal) have yielded MPXV [12]. However, the World Health Organization recommends, similar to the CDC, that the best diagnostic specimens are taken directly from the mpox rash, and the recommended test is detection of viral DNA by PCR [13]. Second, PCR-based assays have the potential for false-negative results related to genomic divergence in outbreak strains at primer-targeted sites, observed during the 2022 outbreak caused by a deletion of the tumor necrosis factor receptor gene [14]. The large genomic changes observed in MPXV strains could impede primer site recognition [15], implying a need for ongoing assay optimization [14]. Finally, PCR tests provide data only on the pathogens tested and do not detect concurrent infections. Given the heightened risk among immunocompromised individuals during an mpox outbreak as well as increasing reports of clinically important coinfections (including human immunodeficiency virus [HIV and sexually transmitted infections 10, 16, 17]) in addition to well-recognized HIV-related opportunistic infections, the potential for coinfection is a substantial consideration.
Microbial cell-free DNA (mcfDNA) metagenomic sequencing from plasma has been used for pathogen-agnostic infectious disease diagnostics [18] and has been demonstrated to detect diagnostically challenging clinical infections [19–21]. The pathogen-agnostic nature of mcfDNA testing may support detecting emerging infections. For example, it has been used to detect Mycobacterium chimaera infections associated with contamination of heater-cooler devices for cardiac surgery [22] as well as pathogens of porcine origin in the first patient with a genetically modified porcine-to-human cardiac xenotransplantation [23]. Detection and quantification of microbes through mcfDNA sequencing relies on the presence of their DNA sequences or markers within a database. Provided a database augmented with the novel sequence, a method-based analytical validation can be used to characterize the robustness of the assay to the emerging concern with no changes to the underlying chemistry [24]. We describe the detections and quantification of MPXV in 12 patients via mcfDNA sequencing and explore subtyping directly from mcfDNA, which may potentially augment public health surveillance.
MATERIALS AND METHODS
Sample Material and Case-Patient Details
Peripheral blood samples for plasma mcfDNA sequencing ordered by clinicians from across the United States (US) as part of their diagnostic management of patients were eligible for this study. The subset of samples yielding a MPXV detection was then included. Samples had been collected from July 2022 through April 2023. Each clinician for the respective patient was contacted and provided relevant demographic, clinical, and orthogonal testing information for their patient. They also completed for their respective patients an mpox severity scoring system (MPOX-SSS) tool developed by a group of academic and CDC collaborators [25, 26]. This study was determined to be exempt from institutional review board (IRB) oversight by the Advarra Institutional Review Board based on Department of Health and Human regulation 45 Code of Federal Regulations 46.104(d)(4).
Plasma mcfDNA Sequencing
Plasma mcfDNA sequencing was performed as previously described [27] in the Karius, Inc, clinical laboratory (Redwood City, California), certified under the Clinical Laboratory Improvement Amendments of 1988 and accredited by the College of American Pathologists. Sequencing data were analyzed using the validated Karius Test bioinformatic pipeline version DC-3.13, which was designed to detect mcfDNA from >1500 microbes [27], including MPXV, and reports the absolute plasma concentration of mcfDNA in molecules per microliter (MPM) for each microbe detected. In line with FDA guidance for the development of MPXV diagnostic tests [28], we further characterized the performance of mcfDNA sequencing in the context of MPXV, its diversity, and related Orthopoxvirus species (see Supplementary Material, Supplementary Tables 1 and 2).
Variant Calling
Isolates were genotyped directly from mcfDNA fragments attributed to MPXV. The mcfDNA sequencing reads were aligned to the human MPXV B.1 clade genome reference (National Center for Biotechnology Information accession number ON563414.1) using bowtie2 (-very-sensitive-local mode) [29]. Duplicate reads were removed using Picard MarkDuplicates [30]. Joint variant calling was performed on the case-patient read alignments (BAM files) as input and using bcftools [31] mpileup (-q 20) piped to bcftools call with the following arguments (-m–ploidy 1-q 30-p 0.0000000001). Variants exhibiting potential heterozygosity in any sample, as evidenced by non-zero phred-scaled likelihood scores for both alleles, were discarded. In cases of extensively high coverage, mcfDNA sequencing additionally permits de novo genome assembly and thus analysis of structural variation (see Supplementary Material, Supplementary Figure 1).
Phylogeny of Case-Patient MPXV Strains
The phylogenetic context for the MPXV isolate from each case-patient was analyzed within the wider context of the 2022 outbreak using the Nextstrain toolkit [32]. The genome sequence for each isolate was derived from the MPXV B.1 clade reference (accession number ON563414.1), modified by the MPXV single-nucleotide polymorphisms genotyped for each case-patient. Conversely, genomic regions with insufficient sequencing coverage for variant calling were retained as ambiguous nucleotides. The reconstructed isolate genomes were used as direct input to Nextstrain, alongside the reference MPXV dataset provided by the Nextstrain team. The default hMPXV (human MPXV) configuration was utilized for downstream analysis, with the additional filtering rule to force inclusion of all Karius-derived sequences (-include-where ‘abbr_authors = Kariusdx’).
Statistical Analysis
The Pearson correlation coefficient (R) was used to assess the relationship between log10(MPM) and MPOX-SSS (R version 4.3.1) [33]. The α value was .05.
RESULTS
We present findings for 12 case-patients (Table 1), all of whom were male. The mode of suspected transmission was sexual (male having sex with males) for 7 and was unknown for the others. For 7 case-patients, the respective clinicians either suspected or had confirmed mpox infection before ordering mcfDNA sequencing to determine what other pathogen(s) might be contributing to the case-patient's clinical condition. However, for the remaining 5 case-patients, mpox was not suspected or a current concern, and 3 of these case-patients (2, 5, and 12) presented with findings not expected for mpox. Other clinically relevant coinfections were detected in 7 case-patients. Six case-patients had progressive or prolonged mpox, as supported by high MPXV mcfDNA concentration (median MPM, 175 366 [range, 192–1 752 438]) and MPOX-SSS >15 of maximum possible score 23 (median MPOX-SSS, 19 [range 0–23) 25, 26]. There was a statistically significant positive correlation between log(MPM) and MPOX-SSS (R = 0.926, P < .001) (Supplementary Figure 2).
Table 1.
Characteristics at the Time of Plasma Sample Collection From Patients in Whom Microbial Cell-Free DNA Sequencing Detected Monkeypox virus, July 2022–April 2023
| CP No. | Age, y; Sex | HIV Status (CD4 Count. cells/μl) | Mode of Mpox Spread | Description of Lesions if Present at Time of mcfDNA Sequencing | mcfDNA Sequencing Detections (MPM)a | Mpox Suspected or Diagnosed | Orthopoxvirus PCR Result | Mpox Severity Score, 1–23 [25, 26] | Additional Commentsb |
|---|---|---|---|---|---|---|---|---|---|
| 1c | 30–39; male | HIV/AIDS (<35) | Sexual (MSM) | Perirectal and perioral lesions, progressed to disseminated (pustular and necrotic) | HSV-2 (538) CMV (12 900) HHV-8 (316) MPXV (746 852d) PJP (1838) |
Yes | Positive | 20 | Received 3 courses of TPOXX with 2nd course PO at 1st sampling and 3rd course IV at 2nd sampling. Required hospitalization in ICU. Diagnoses included mpox, PJP, CMV colitis, HSV-2, Kaposi sarcoma. Was discharged to hospice care. |
| HHV-8 (176) MPXV (1 752 437d) PJP (1007) Proteus mirabilis (3313) Pseudomonas aeruginosa (44 638) |
|||||||||
| 2 | 20–29; male | Negative | Unknown | No reported lesions | MPXV (192) | Not suspected | Not done | 2 | Presented with fever, difficulty breathing, chest pain, requiring brief hospitalization. No TPOXX. Diagnosed with acute myocarditis. |
| 3 | 30–39; male | HIV/AIDS (<1) | Unknown | Extensive distribution of lesions, especially on face and inner right thigh | CMV HSV-2 HHV-8 MPXV Primate tetraparvovirus Fusobacterium necrophorum Lactobacillus jensenii Prevotella bivia Staphylococcus aureus Streptococcus anginosus |
Yes | Positive | 23 | Received IV TPOXX. Hospitalized in ICU. Diagnosed with mpox, right thigh myositis, pneumonia, disseminated CMV. Subsequently suffered overwhelming septic shock and died. |
| 4 | 30–39; male | HIV/AIDS (278) | Unknown | Disseminated lesions including face, hands, feet, and perianal region | MPXV (205 051) | Yes | Positive | 16 | Received IV TPOXX. History of recent treatment for syphilis. Required hospitalization. Diagnosed with mpox. |
| 5 | 30–39; male | HIV (545) | Sexual (MSM) | Few dry, healing skin lesions on fingers bilaterally | MPXV (336) Streptococcus pneumoniae (606) |
Not suspected for current presentation | Not done | 0 | History of recent treatment for syphilis. Presented with acute left-sided neck mass and history of acute signs/symptoms consistent with mpox but unconfirmed and resolved ∼2 wk previously; also history of exposure to reportedly mpox-infected contact. No TPOXX. Required hospitalization. Diagnosed with left neck mass/lymphadenitis (S pneumoniae, also yielded from needle aspiration). |
| 6 | 30–39; male | HIV/AIDS (80) | Unknown | Disseminated lesions over entire body including face | HHV-8 (1376) HSV-1 (140) MPXV (145 681) |
Yes | Positive | 17 | History of Kaposi sarcoma, anal warts (suspected anal condyloma). Received PO TPOXX. Required hospitalization. Diagnosed with mpox. |
| 7c | 20–29; male | HIV/AIDS (2.14) | Suspected sexual (MSM) | Initial perirectal lesions and general skin rash progressed to diffusely spread lesions | Adenovirus B (611) HHV-8 (56) MPXV (288 440) Primate tetraparvovirus 1 (315) |
Yes | Positive | 19 | Received TPOXX × 2 (PO, then IV). Required hospitalization. Diagnosed with mpox. |
| EBV (333) Primate tetraparvovirus 1 (444) MPXV (519 762d) |
|||||||||
| 8 | 20–29; male | HIV (190) | Sexual (MSM) | Very confluent rash spread, >40 round and crusted spots on body | MPXV (15 468) | Not suspected | Not done | 11 | Endemic mycoses (eg, sporotrichosis, paracoccidioidomycosis) suspected and received empiric antifungal therapy. Subsequent to mcfDNA sequencing detection of MPXV, history of mpox exposure elicited. Received IV TPOXX following mcfDNA sequencing detection of MPXV with rapid resolution of lesions. Required hospitalization for treatment. Diagnosed with mpox. |
| 9 | 50–59; male | HIV/AIDS (1.8) | Suspected sexual (MSM) | Lesions diffusely present over perianal region, inguinal region, face, forearm, fingers, toes | MPXV (83 199) | Yes | Positive | 20 | Received IV TPOXX. Required hospitalization. Diagnoses included ocular syphilis, bacterial osteomyelitis of thumb, mpox at time of mcfDNA sequencing, on treatment for all; subsequently, also Mycobacterium avium infection yielded from (pulmonary nodule culture). |
| 10c | 40–49; male | HIV/AIDS (44) | Sexual (MSM) | Extensive, progressive perianal/perirectal lesions with resulting rectal stenosis and intestinal obstruction | Alphapapillomavirus 11 (81) HSV-2 (1676) EBV (297) CMV (18 181) HHV-8 (559) MPXV (834 588d) Enterocytozoon bieneusi (7608) |
Yes | Positive | 21 | Received prolonged IV TPOXX. Required hospitalization in ICU. Diagnosed with mpox with complications of GI obstruction, MRSA (PICC line–associated). Subsequently developed septic shock and rapidly died. |
| HHV-8 (103) MPXV (571 364d) Enterocytozoon bieneusi (7608) Escherichia coli (417) Staphylococcus aureus (1526) |
|||||||||
| 11 | 30–39; male | HIV/AIDS (29) | Unknown | One single lesion on elbow | MPXV (18 052) | Not suspected | Not done (after mcfDNA sequencing detection of MPXV, skin biopsy sample tested by PCR and yielded positive) | 7 or 8, if pulmonary nodules secondary to mpox (being investigated) | Suspected fungal etiology; lesion biopsied with inconclusive dermatological assessment; chest CT demonstrated pulmonary nodules. History of confirmed mpox >6 mo previously with documented resolution of lesions. Diagnosed with mpox and PO TPOXX subsequent to mcfDNA sequencing detection of MPXV. |
| 12 | 40–49; male | HIV/AIDSe | Suspected sexual (MSM) | No reported lesions, although 1 rectal ulcer of unclear etiology | HSV-2 (203) CMV (578) HHV-8 (7376) MPXV (6610) Histoplasma capsulatum (62) Pneumocystis jirovecii (62) |
Not suspected | Not done (after mcfDNA sequencing detection of MPXV, PCR from blind rectal swab performed and yielded negative result) | 0 | Presented with severe pulmonary disease with hemorrhaging; conventional testing positive for Histoplasma. Required hospitalization in ICU. Complete diagnosis unclear but included histoplasmosis, Kaposi sarcoma, PJP, and presumptive mpox (latter 3 subsequent to mcfDNA sequencing detections). Subsequently died from pulmonary complications not thought to be mpox related. |
Abbreviations: CMV, cytomegalovirus; CP, case-patient; CT, computed tomography; EBV, Epstein-Barr virus; GI, gastrointestinal; HHV-8, human herpesvirus 8; HIV, human immunodeficiency virus; HSV-1, herpes simplex virus type 1; HSV-2, herpes simplex virus type 2; ICU, intensive care unit; IV, intravenous; mcfDNA, microbial cell-free DNA; MPM, molecules per microliter; MPXV, Monkeypox virus; MRSA, methicillin-resistant Staphylococcus aureus; MSM, men who have sex with men; PCR, polymerase chain reaction; PICC, peripherally inserted central catheter; PJP, Pneumocystis jirovecii pneumonia; PO, per os (oral); TPOXX, tecovirimat.
aDetections are provided in MPMs except when a specimen did not meet the minimum sequencing coverage requirements and therefore the reported organism(s) could not be quantified as was the case for case-patient 3.
bAdditional relevant clinical and treatment data are provided when available.
cCase-patients 1, 7, and 10 had repeat mcfDNA sequencing on plasma samples obtained approximately 3–4 weeks after the first respective sample.
dConcentration estimate above validated assay range; reported as >316 000.
eCD4 count unavailable for this patient.
Genome coverage from mcfDNA sequencing ranged from 0.1 times to 101 times and was uniform across the MPXV genome in all cases (Supplementary Figure 3A). The sequencing coverage was sufficient to enable subtyping for most case-patients (Supplementary Figure 3B, Table 2). Therefore, genetic variation was assessed relative to the MPXV lineage responsible for the 2022 outbreak (hMPXV1 B.1 lineage [34]). We found mutations at 10 loci in the MPXV genome among the 12 case-patients studied (Table 2, Supplementary Table 3). Of the 22 variants we detected, 21 were G > A or C > T, consistent with a major known source of MPXV variation in the current outbreak (apolipoprotein B mRNA editing catalytic polypeptide-like 3 enzyme–mediated mutation) [35]. None of the detected mutations were found to be within the PCR primer targets.
Table 2.
Summary of the Detected Monkeypox virus Variants
| Locus | Allelea | Case-Patient Genotypeb | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref | Alt | 1 | 1b | 2 | 3 | 4 | 5 | 6 | 7 | 7b | 8 | 9 | 10 | 10b | 11 | 12 | |
| 20173 | A | G | . | . | / | . | . | / | . | . | . | . | G | . | . | . | . |
| 45798 | G | A | . | . | / | . | A | / | . | . | . | . | . | . | . | / | / |
| 63125 | A | T | . | . | / | . | . | / | . | T | T | . | . | . | . | . | . |
| 130095 | C | T | . | . | / | . | . | . | T | . | . | . | . | . | . | . | . |
| 149410 | G | A | . | . | / | A | . | . | . | . | . | . | . | . | . | . | . |
| 149823 | C | T | . | . | / | . | T | / | . | . | . | . | . | . | . | . | / |
| 155757 | C | T | . | . | T | . | . | / | . | . | . | . | . | . | . | . | T |
| 167669 | C | T | . | . | / | . | . | / | . | T | T | . | . | . | . | . | . |
| 186144 | G | A | . | . | / | A | A | / | . | A | A | / | / | . | . | A | / |
| 188370 | C | T | . | . | / | T | T | / | . | T | T | T | / | . | . | T | / |
Genomic coordinates are shown relative to the human MPXV1 B.1 reference strain (clade B1; ON563414.1). Genomic coordinates relative to the reference strain used by Nextstrain (clade IIb; NC_063383.1) are shown in Supplementary Table 3.
Abbreviation: MPXV, Monkeypox virus.
aThe B.1 reference nucleotide and alternate nucleotide for each locus are shown.
bLoci that match the reference genome are indicated with a dot. Loci that could not be genotyped are indicated with “/”.
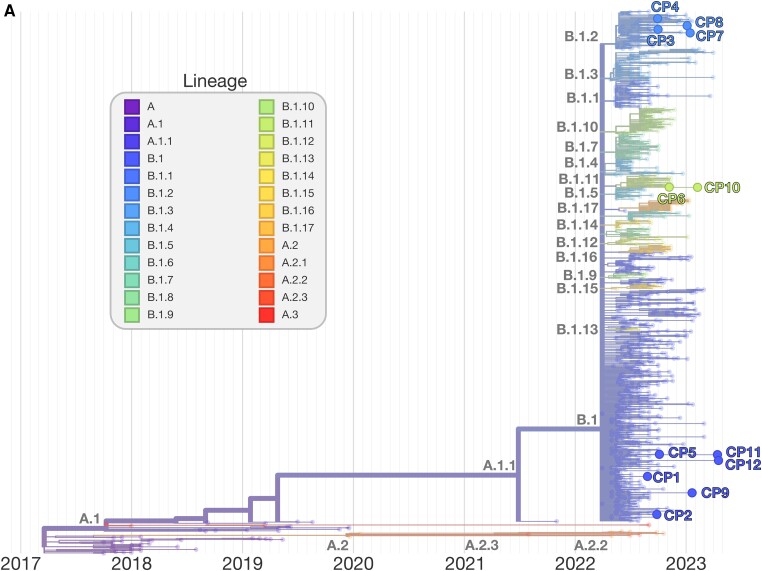
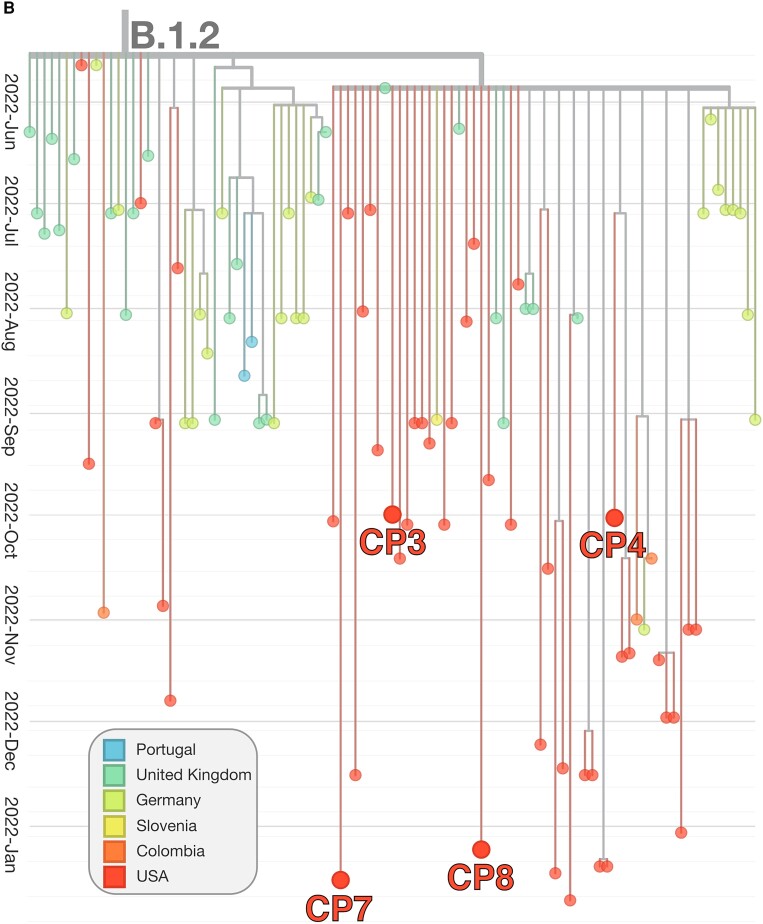
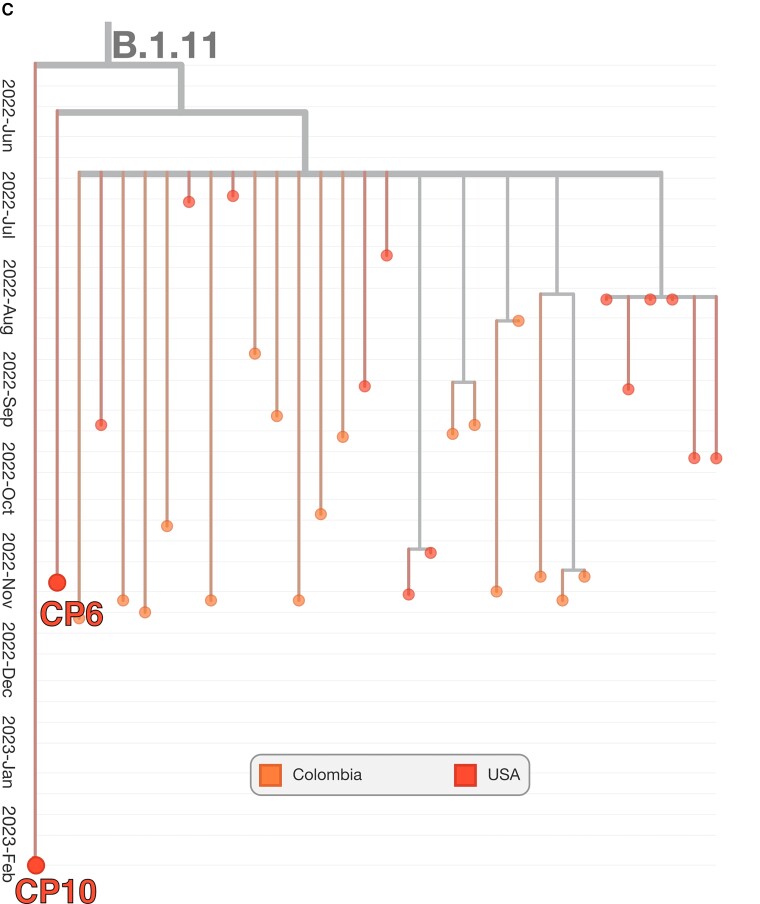
The genotype for each case-patient facilitated the phylogenetic placement of each isolate, relative to MPXV isolates from the 2022 outbreak. MPXV from case-patients 6 and 10 form a small phylogenetic clade with several other genomes isolated at similar times in the US and Colombia (sublineage B1.11), consistent with transmission in the US in late May 2022 [34]. MPXV from case-patients 3, 4, 7, and 8 derived from the B.1.2 sublineage, defined by the G- > A transition at locus 186165. All viruses additionally fall within a common subclade of B.1.2 defined by C- > T transition at site 188391, common of US B.1.2 isolates (Figure 1B ). In addition to the 2 alleles described, MPXV from case-patients 3, 4, 7, and 8 were determined to have 1, 2, 2, and 0 unique mutations, respectively (see Table 2 for specific loci), not found in other public MPXV strains and consistent with the continued evolution of the B.1.2 sublineage. Conversely, case-patients 6 and 10 derived from the B.1.11 sublineage, a minor subvariant predominately geographically isolated to North America, in particular the US, and Colombia (Figure 1C ).
Figure 1.
Phylogenetic tree of Monkeypox virus (MPXV) outbreak, including case-patient derived isolates. Phylogenetic tree of the outbreak human MPXV lineage B.1. MPXV detections by microbial cell-free DNA in the respective case-patients are shown within their phylogenetic context (A). The tree was constructed and visualized using the Nextstrain toolkit [32]. The MPXV detected in case-patients 1, 2, 5, 9, 11, and 12, respectively, were observed to descend directly from the ancestral B.1 strain. For case-patients 2, 5, and 12, this is likely related to insufficient coverage for exhaustive genotyping. The MPXV from case-patients 3, 4, 7, and 8, respectively, were genotyped to be derived from lineage B.1.2 (B). Two main clades within the sublineage are observed. All 4 isolates were imputed to originate from the predominantly North American subclade. The MPXV detected in case-patients 6 and 10, respectively, were found to be from sublineage B.1.11, which likely arose in late May 2022 and has been detected only within the United States (USA) and Colombia (C).
DISCUSSION
Plasma mcfDNA sequencing holds the promise to both facilitate clinical management and enhance situational awareness for public health agencies and scientists tracking emerging pathogens. Effective outbreak control strategies require extensive and immediate testing availability [36, 37]. The validated assay utilized in this study [27] can readily quantify mcfDNA in a pathogen-agnostic manner from a blood sample. Initial observations, although conducted on a limited cohort, suggest a possible correlation between pathogen abundance and the severity of illness. Conversely, PCR-based diagnostics require considerable lead time to reach the requisite capacity needed to reliably test a population; in the best case, as in mpox, the assay exists but must be further optimized and scaled. The ability to detect MPXV directly from blood positions mcfDNA sequencing as a quantitative alternative to PCR testing. Finally and perhaps most critical to public health needs, mcfDNA sequencing has the possibility of detecting MPXV in atypical presentations in which lesions may be absent, resolving, or unclear in etiology as demonstrated in our small cohort (case-patients 2, 5, and 12).
With the entire genome of an organism serving as a target, we demonstrated that pathogen-agnostic mcfDNA sequencing may offer intrinsic robustness toward emerging DNA pathogens and their subtypes. Through a detailed in silico study (see Supplementary Material), we showed that the existing Karius genomic reference database is sufficient to detect and quantify novel MPXV strains, including the outbreak strain, in an already validated assay. Additionally, the database can be easily updated without modifying underlying chemistry to extend the assay to cover additional variants and better represent the underlying biological diversity. For instance, expansion of a genomic reference database to include additional strains of the cowpox virus clades may further reduce the risk of cross-reactivity with MPXV.
In addition to mcfDNA detections contributing to individual diagnoses, we demonstrated the ability to accurately determine the phylogenetic lineage from which the given analyte derives in cases of sufficient coverage. When such assays are combined with spatiotemporal tracking at scale, findings across multiple sites and geographies can be leveraged to provide rapid phylodynamic analysis from the onset of a public health emergency or disease outbreak, similar to state-of-the-art techniques [32]. Furthermore, subtyping directly from mcfDNA sequencing samples, in combination with patient response to treatment, may be used to support efforts required to computationally associate mutations to viral phenotypes [38], such as antiviral resistance [39].
Mpox detection in certain case-patients highlighted various scenarios of clinical importance and further consideration. In case-patient 1, a follow-up sample collected >4 weeks after the first exhibited a >2-fold increase in MPM, which is consistent with the case-patient's severe progressive mpox disease despite 3 courses of tecovirimat, the last given intravenously. This scenario suggests the potential application for individual clinical monitoring and therapy as has been suggested for other types of infections [40, 41]. Case-patient 3 was in critical condition and died shortly after the sample collection for mcfDNA sequencing. The high content of human DNA in this sample precluded accurate quantification, likely reflecting the patient's critical condition [42]. Case-patient 2 presented with signs and symptoms consistent with acute myocarditis without any lesions or other manifestations typical for mpox. Whether this case-patient had previously resolved lesions is uncertain. However, acute myocarditis approximately 1 week after onset of mpox symptoms has been reported recently in another patient [43]. Case-patient 5, who had evidence of resolving lesions and a history of unconfirmed illness preceded by reported exposure to a person with mpox, also presented with findings other than expected for mpox. This case-patient had a left-sided neck mass assessed as probable lymphadenitis, likely secondary to Streptococcus pneumoniae, which was identified both by mcfDNA sequencing and cultivation of the needle aspiration fluid. Whether the mpox detected in these 2 case-patients represented active or declining virus activity, they both highlight concerns regarding possible serious complications of mpox infection as well as the need for clinicians to maintain a high suspicion for mpox, especially with the current persistent low-level circulation of MPXV globally. Case-patient 11's scenario suggests mpox reinfection potential despite past history of confirmed and clinically resolved mpox disease, as has been reported by others [44, 45] versus MPXV persistence, especially in people with AIDS. Finally, the case-patients with AIDS and polymicrobial detections (1, 3, 6, 7, 10, 12) serve as reminders of the high likelihood of clinically complicating opportunistic coinfections in these patients.
This study had several limitations. First, our study relied on a convenience sample, which introduces bias, potentially misses patients positive for MPXV by PCR but negative by mcfDNA sequencing (ie, false negatives), and limits the generalizability of our findings. A controlled, prospective study could address this limitation. However, notably, mcfDNA sequencing detections were correlated with clinical and orthogonal data for each case-patient, including 1 patient who was demonstrated to be positive by PCR subsequent to mcfDNA detection, thereby strengthening our findings. Second, while MPXV-derived mcfDNA was detected in both suspected and nonsuspected infected individuals in this study, the underlying temporal relationship between the status of MPXV infection and the presence of plasma mcfDNA is unclear and under active research. Similarly, the viral kinetics of MPXV in different sample types in general is still being investigated with MPXV DNA level in blood at any given time point observed by PCR to be low [46]. Prospective serial monitoring of MPXV in multiple patients could help elucidate these dynamics. Third, genotyping of the MPXV from mcfDNA was constrained to samples with an adequate concentration of the corresponding mcfDNA. Future studies are needed to precisely determine the frequency and requisite conditions for successful genotyping of outbreak strains from mcfDNA. In fact, the relatively high amounts of DNA detected in study samples would seem to be characteristic to at least the mpox detections in our cohort, especially patients with moderate to severe disease, compared with mcfDNA sequencing detections of other microbes in general. Whether this characteristic is unique to the current mpox infections or to patients with severe mpox disease or whether it reflects mpox viremia or copious viral shedding from other infected sites, or both, remains to be elucidated. Finally, an extensive investigation of each case-patient's epidemiological history and exposures was not undertaken and may have been informative in conjunction with the presented phylogenetic analyses.
In conclusion, we present the first detections of MPXV using plasma mcfDNA sequencing. The study demonstrates the potential of mcfDNA sequencing to detect, quantify, and subtype infecting pathogens using a single noninvasive test. Microbial cell-free DNA sequencing may offer key advantages to support the currently recommended PCR assay for mpox diagnosis and clinical monitoring, including potentially for genetic markers of emerging antiviral resistance, as well as concurrent detection of opportunistic infections in critically ill immunocompromised patients. During this period of persistent, low-level global MPXV circulation, mcfDNA sequencing may further support identifying those presenting with atypical, unsuspected mpox disease. The potential information from MPXV detections and subtyping may contribute to understanding and monitoring the evolving mpox epidemiology.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Sarah Y Park, Medical Affairs, Karius, Inc, Redwood City, California.
Martin S Lindner, Analytics, Karius, Inc., Redwood City, California.
Kevin Brick, Analytics, Karius, Inc., Redwood City, California.
Nicholas Noll, Analytics, Karius, Inc., Redwood City, California.
Rachid Ounit, Analytics, Karius, Inc., Redwood City, California.
Luis J Noa, Infectious Disease Section, AdventHealth Orlando, Florida.
Rabeeya Sabzwari, Infectious Diseases, Edward Hines Jr Veterans Affairs Hospital, Hines, Illinois.
Ronald Trible, Georgia Infectious Diseases, PC, Atlanta.
Jason C Sniffen, Infectious Disease Section, AdventHealth Orlando, Florida.
Prerana Roth, Infectious Diseases, Prisma Health–Upstate, Greenville, South Carolina.
Amir Khan, Infectious Diseases, Carle Foundation Hospital, Urbana, Illinois.
Anamaria Rodriguez, Infectious Diseases, Orlando Health, Florida.
Syeda Sahra, Department of Infectious Diseases, Oklahoma University Medical Center, Oklahoma City.
Michael J Davis, Department of Infectious Diseases and International Medicine, University of Minnesota, Minneapolis, MN.
Inderjeet S Brar, Infectious Diseases, Baptist Memorial Health Care, Memphis, Tennessee.
Gayathri Balasundaram, Medical Affairs, Karius, Inc, Redwood City, California.
Frederick S Nolte, Medical Affairs, Karius, Inc, Redwood City, California.
Timothy A Blauwkamp, Executive Team, Karius, Inc., Redwood City, California.
Bradley A Perkins, Executive Team, Karius, Inc., Redwood City, California.
Sivan Bercovici, Executive Team, Karius, Inc., Redwood City, California.
Notes
Author contributions. Study concept and design: S. Y. P., M. S. L., K. B., N. N., S. B. Acquisition of data: S. Y. P., M. S. L., N. N., K. B., R. O., L. J. N., R. S., R. T., J. C. S., P. R., A. K., A. R., S. S., M. J. D., I. S. B. Analysis of the data: S. Y. P., M. S. L., N. N., K. B., R. O. Visualization: K. B., N. N. Interpretation of the data: S. Y. P., M. S. L., N. N., K. B., S. B. Drafting of the manuscript: M. S. L., K. B., N. N., S. Y. P., S. B. Review and revision of the manuscript: All authors.
Acknowledgments. The authors thank Alec Ford for his thoughtful comments and inspiring the communication of these results, and Karius, Inc, for supporting the investigation. We also thank Asim A. Ahmed, MD, formerly with Karius, Inc, for his early contributions related to the work. We appreciate the advice of Dr Jason Zucker, Columbia University, regarding application of the mpox severity scoring system and thank Caitlin Ryan, PhD, Karius, Inc, for providing statistical analysis support. Finally, we acknowledge the patients and their families and the contribution of their data toward hopefully benefitting many others.
Financial support. This work was supported by Karius, Inc, via paid employment of M. S. L., K. B., N. N., S. B., S. Y. P., R. O., G. B., T. A. B., and F. S. N. Funding to pay the Open Access publication charges for this article was provided by Karius, Inc.
Supplement sponsorship. This article appears as part of the supplement “Mpox: Challenges and Opportunities Following the Global 2022 Outbreak,” sponsored by the Centers for Disease Control and Prevention (Atlanta, GA).
References
- 1. Alakunle EF, Okeke MI. Monkeypox virus: a neglected zoonotic pathogen spreads globally. Nat Rev Microbiol 2022; 20:507–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization . Multi-country outbreak of mpox, External situation report #11—1 December 2022. https://www.who.int/publications/m/item/multi-country-outbreak-of-mpox--external-situation-report--11---1-december-2022. Accessed 12 December 2022.
- 3. Titanji BK, Tegomoh B, Nematollahi S, Konomos M, Kulkarni PA. Monkeypox: a contemporary review for healthcare professionals. Open Forum Infect Dis 2022; 9:ofac310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thornhill JP, Barkati S, Walmsley S, et al. Monkeypox virus infection in humans across 16 countries—April–June 2022. N Engl J Med 2022; 387:679–91. [DOI] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention . Signs and Symptoms [Internet]. https://www.cdc.gov/poxvirus/mpox/symptoms/index.html. Accessed 19 October 2023.
- 6. Ward T, Christie R, Paton RS, Cumming F, Overton CE. Transmission dynamics of monkeypox in the United Kingdom: contact tracing study. BMJ 2022; 379:e073153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Madewell ZJ, Charniga K, Masters NB, et al. Serial interval and incubation period estimates of monkeypox virus infection in 12 jurisdictions, United States, May–August 2022. Emerg Infect Dis 2023; 29:818–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Centers for Disease Control and Prevention . Guidelines for collecting and handling specimens for mpox testing. 2023. . https://www.cdc.gov/poxvirus/monkeypox/clinicians/prep-collection-specimens.html. Accessed 14 June 2023.
- 9. US Food and Drug Administration . Monkeypox (mpox) and medical devices. 2023. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/monkeypox-mpox-and-medical-devices. Accessed 6 July 2023.
- 10. Zucker J. CROI 2023: epidemiology, diagnosis, and management of mpox. Top Antivir Med 2023; 31:510–9. [PMC free article] [PubMed] [Google Scholar]
- 11. US Food and Drug Administration . For monkeypox testing, use lesion swab samples to avoid false results: FDA safety communication. 2022. https://www.fda.gov/medical-devices/safety-communications/monkeypox-testing-use-lesion-swab-samples-avoid-false-results-fda-safety-communication. Accessed 14 June 2023.
- 12. Lim CK, Roberts J, Moso M, et al. Mpox diagnostics: Review of current and emerging technologies. J Med Virol 2023; 95:e28429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peterson H, Adler BL, Ochoa MT. Mpox (monkeypox) clinical pearls. Cutis 2023; 111:170–1. [DOI] [PubMed] [Google Scholar]
- 14. Centers for Disease Control and Prevention . Lab alert: MPXV TNF receptor gene deletion may lead to false negative results with some MPXV specific LDTs. 2022. https://www.cdc.gov/locs/2022/09-02-2022-lab-alert-MPXV_TNF_Receptor_Gene_Deletion_May_Lead_False_Negative_Results_Some_MPXV_Specific_LDTs.html. Accessed 15 June 2023.
- 15. Gigante CM, Plumb M, Ruprecht A, et al. Genomic deletions and rearrangements in monkeypox virus from the 2022 outbreak, USA. bioRxiv [Preprint]. Posted online 17 September 2022. https://www.biorxiv.org/content/10.1101/2022.09.16.508251. Accessed 13 June 2023.
- 16. Ortiz-Saavedra B, Montes-Madariaga ES, Cabanillas-Ramirez C, et al. Epidemiologic situation of HIV and monkeypox coinfection: a systematic review. Vaccines (Basel) 2023; 11:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Curran KG, Eberly K, Russell OO, et al. HIV and sexually transmitted infections among persons with monkeypox—eight U.S. jurisdictions, May 17–July 22, 2022. MMWR Morb Mortal Wkly Rep 2022; 71:1141–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park SY, Chang EJ, Ledeboer N, et al. Plasma microbial cell-free DNA sequencing from over 15,000 patients identified a broad spectrum of pathogens. J Clin Microbiol 2023; 9:e0185522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Foong KS, Mabayoje M, AlMajali A. Clinical impact of noninvasive plasma microbial cell-free deoxyribonucleic acid sequencing for the diagnosis and management of Pneumocystis jirovecii pneumonia: a single-center retrospective study. Open Forum Infect Dis 2022; 9:ofac652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zou RH, Nguyen VDB, Zigrossi PA, et al. Non-invasive diagnosis of pulmonary Kaposi sarcoma: a case report. J Infect Dis Epidemiol 2020; 6:161. [Google Scholar]
- 21. Roy M, Siddique N, Bathina B, Ahmad S. Rare presentation of toxoplasma pneumonitis in the absence of neurological symptoms in an AIDS patient and use of next-generation sequencing for diagnosis. Eur J Case Rep Intern Med 2020; 7:001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tan NY, Tarabochia AD, DeSimone DC, et al. Updated experience of Mycobacterium chimaera infection: diagnosis and management in a tertiary care center. Open Forum Infect Dis 2021; 8:ofab348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Griffith BP, Goerlich CE, Singh AK, et al. Genetically modified porcine-to-human cardiac xenotransplantation. N Engl J Med 2022; 387:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. US Food and Drug Administration . Infectious disease next generation sequencing based diagnostic devices: microbial identification and detection of antimicrobial resistance and virulence markers; draft guidance for industry and Food and Drug Administration staff; availability. Fed Regist 2016:29869–70. https://www.federalregister.gov/documents/2016/05/13/2016-11237/infectious-disease-next-generation-sequencing-based-diagnostic-devices-microbial-identification-and. Accessed 19 October 2023 [Google Scholar]
- 25. He J. Development and pilot of an mpox severity scoring system (mpox-SSS). In: Conference on Retroviruses and Opportunistic Infections. 2023. https://www.croiconference.org/abstract/development-and-pilot-of-an-mpox-severity-scoring-system-mpox-sss/. Accessed 9 July 2023.
- 26. Zucker J. Monkeypox severity score development and validation. MPOX Severity Score System (MPOX-SSS). https://mpoxseverityscore.com/mpx_dev_val/. Accessed 14 July 2023.
- 27. Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 2019; 4:663–74. [DOI] [PubMed] [Google Scholar]
- 28. US Food and Drug Administration . Monkeypox (mpox) Emergency Use Authorizations for Medical Devices. https://www.fda.gov/medical-devices/emergency-use-authorizations-medical-devices/monkeypox-mpox-emergency-use-authorizations-medical-devices. Accessed 19 October 2023.
- 29. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Broad Institute . Picard tools. http://broadinstitute.github.io/picard/. Accessed 21 June 2023.
- 31. Danecek P, Bonfield JK, Liddle J, et al. Twelve years of SAMtools and BCFtools. Gigascience 2021; 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34:4121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ripley BD. The R project in statistical computing. MSOR Connect. Educational Development Unit, University of Greenwich. 2001. https://www.R-project.org/. Accessed 27 September 2023.
- 34. Aksamentov I, Roemer C, Hodcroft E, Neher R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw 2021; 6:3773. [Google Scholar]
- 35. Isidro J, Borges V, Pinto M, et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat Med 2022; 28:1569–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu W-K, Liou J-M, Hsu C-C, Lin Y-H, Wu M-S. Pandemic preparedness in Taiwan. Nat Biotechnol 2020; 38:932–3. [DOI] [PubMed] [Google Scholar]
- 37. Dighe A, Cattarino L, Cuomo-Dannenburg G, et al. Response to COVID-19 in South Korea and implications for lifting stringent interventions. BMC Med 2020; 18:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neher RA, Bedford T, Daniels RS, Russell CA, Shraiman BI. Prediction, dynamics, and visualization of antigenic phenotypes of seasonal influenza viruses. Proc Natl Acad Sci U S A 2016; 113:E1701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rabaan AA, Abas AH, Tallei TE, et al. Monkeypox outbreak 2022: what we know so far and its potential drug targets and management strategies. J Med Virol 2023; 95:e28306. [DOI] [PubMed] [Google Scholar]
- 40. Eichenberger EM, Degner N, Scott ER, et al. Microbial cell-free DNA identifies the causative pathogen in infective endocarditis and remains detectable longer than conventional blood culture in patients with prior antibiotic therapy. Clin Infect Dis 2023; 76:e1492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heldman MR, Ahmed AA, Liu W, et al. Serial quantitation of plasma microbial cell-free DNA before and after diagnosis of pulmonary invasive mold infections after hematopoietic cell transplant recipients. J Infect Dis 2024; 229:576–87. [DOI] [PubMed] [Google Scholar]
- 42. Kitsios GD, Bain W, Al-Yousif N, et al. Plasma microbial cell-free DNA load is associated with mortality in patients with COVID-19. Respir Res 2021; 22:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pinho AI, Braga M, Vasconcelos M, et al. Acute myocarditis: a new manifestation of monkeypox infection? JACC Case Rep 2022; 4:1424–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Golden J, Harryman L, Crofts M, et al. Case of apparent mpox reinfection. Sex Transm Infect 2023; 99:283–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Musumeci S, Najjar I, Amari EBE, et al. A case of mpox reinfection. Clin Infect Dis 2023; 77:135–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim H, Kwon R, Lee H, et al. Viral load dynamics and shedding kinetics of mpox infection: a systematic review and meta-analysis. J Travel Med 2023; 30:taad111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.