Abstract
Degenerate PCR primers which amplify a conserved region of the DNA polymerase genes of the herpesvirus family were used to provide sequence evidence for a new bovine herpesvirus in bovine B-lymphoma cells and peripheral blood mononuclear cells (PBMC). The sequence of the resultant amplicon was found to be distinct from those of known herpesvirus isolates. Alignment of amino acid sequences demonstrated 70% identity with ovine herpesvirus 2, 69% with alcelaphine herpesvirus 1, 65% with bovine herpesvirus 4, and 42% with bovine herpesvirus 1. Phylogenetic analysis placed this putative virus within the tumorigenic Gammaherpesvirinae subfamily, and it is tentatively identified as bovine lymphotropic herpesvirus. This novel agent was expressed in vitro from infected PBMC, and cell-free supernatants were used to transfer infection to a bovine B-cell line, BL3. Analysis, with specific PCR primers, of DNA from bovine PBMC and lymphoma cells identified infection in blood of 91% of adult animals (n = 101), 63% of lymphomas (n = 32), and 38% of juveniles (n = 13). Of the adults, herpesvirus infection was present in 94% of animals that were seropositive for bovine leukemia virus (BLV) (n = 63) and in 87% of BLV-seronegative animals (n = 38). Of the seropositive group, 17 animals exhibited persistent lymphocytosis, and 100% of these were herpesvirus positive by PCR. A role for bovine lymphotropic herpesvirus as a cofactor in BLV pathogenesis is considered.
The ability of retroviruses and herpesviruses to act as cofactors in pathogenesis has been demonstrated in chickens coinfected with avian leukosis virus and Marek’s disease virus, which results in significant increases of lymphoid leukosis (2, 22). This synergism may result from herpesvirus transactivation of retrovirus expression, as has been demonstrated in a number of in vitro systems, and is attributed largely to the action of immediate-early herpesvirus products (for reviews, see references 10 and 16).
Known herpesviruses of cattle include bovine herpesvirus 1 (BHV1), which causes infectious bovine rhinotracheitis, BHV2, the cause of bovine mammalitis, BHV4, isolated from peripheral blood mononuclear cells (PBMC) and associated with a wide range of clinical signs, and BHV5, a neurovirulent strain of BHV1. Cattle are also subject to zoonotic infection with two agents of malignant catarrhal fever (MCF): alcelaphine herpesvirus 1 (AHV1), from wildebeest; and ovine herpesvirus 2 (OHV2) (27).
Enzootic bovine leukosis (EBL) is a neoplastic lymphoproliferative disease of cattle associated with infection by bovine leukemia virus (BLV) (17). EBL occurs in less than 5% of BLV-infected animals after a prolonged latent period, but the BLV genome is invariably found to be monoclonally integrated into the DNA of tumor cells, and no preferred sites for integration have been identified (18). BLV is also associated with persistent B lymphocytosis (PL) in about 30% of infected cattle (11). Sheep can be experimentally infected with BLV and are subject to PL and lymphoma development (7, 8). The BLV-encoded protein, Tax, a trans-acting transcriptional activator, is presumed to be a mediator of tumorigenesis (17, 36). Paradoxically, there is a paucity of BLV transcription in vivo, and the sequence of events that lead to tumorigenesis remains to be elucidated.
A means of detecting herpesvirus DNA, devised by VanDevanter et al. (35), relies on highly conserved amino acid motifs contained in the DNA polymerase genes of herpesviruses as the basis for PCR amplification with degenerate primers. This method has been used to partially sequence the polymerase genes of several existing herpesvirus isolates and to characterize new herpesviruses (28). We used this method to address the possibility that a herpesvirus cofactor may be present in EBL. A novel herpesvirus sequence was identified and used to determine the prevalence of this agent, designated bovine lymphotropic herpesvirus (BLHV), in cattle and its association with BLV infection and BLV-associated pathogenesis.
MATERIALS AND METHODS
Animals.
Blood samples were obtained from commercial dairy herds in Colorado, New York, and New Jersey over the 12-year period 1985 to 1997. Bovine tumors were collected over the same time period from naturally infected animals at necropsy. Sheep were experimentally infected with BLV via injection of either PBMC from BLV-positive cattle with PL, cell-free BLV, or bovine embryonic spleen cells transfected with a molecular clone of BLV, pBLV913 (15, 29). Sera were screened for antibodies against BLV antigens by agar gel immunodiffusion (Leukassay B; Rhone Merieux, Inc., Athens, Ga.). Cattle were diagnosed with PL if their lymphocyte counts exceeded 9.5 × 103/μl on consecutive evaluations at ≥3-month intervals.
Cell isolation.
PBMC were isolated by density gradient centrifugation on Ficoll-Hypaque (Histopaque 1077; Sigma Chemical Co., St. Louis, Mo.) and washed four times with phosphate-buffered saline supplemented with 1% bovine serum albumin. Lymphoma cells were derived from the interior of the tumor mass. Fresh tissues were lacerated to release single cells, and these were banded on Ficoll-Hypaque as described above.
Viruses and cell lines.
Cells infected with human herpesvirus 1 (HHV1; herpes simplex virus type 1 [HSV1] strain F), and stocks of BHV1 (Colorado strain), BHV2 (field isolate), BHV4 (strain DN 599), and AHV1 (strain WC-11) were used for DNA isolation. Bovine and alcelaphine viruses were generously provided by E. Dubovi (Diagnostic Laboratory, New York State College of Veterinary Medicine, Cornell University, Ithaca, N.Y.). DNA positive for OHV2 was kindly provided by H. W. Reid (Moredun Research Institute, Edinburgh, United Kingdom) (3). BLV-positive cell lines FLK-BLV (34) and NBC-13 (12) were cultured in Dulbecco’s minimum essential medium supplemented with 10% fetal calf serum and l-glutamine. NBC-13 cells were provided by J. F. Ferrer (New Bolton Center, University of Pennsylvania, Kennett Square, Pa.). BL3 cells (ATTC CRL 8037) were cultured in Leibovitz-15 medium supplemented with 10% fetal calf serum and 2 μM β-mercaptoethanol. Bovine PBMC and tumor cells were grown in RPMI 1640 medium supplemented with 20% fetal calf serum, 2 μM β-mercaptoethanol, 10−8 M phorbol myristate acetate, and 5 μg of dexamethasone per ml. Supernatants were clarified by centrifugation at 10,000 × g for 15 min followed by 2 h at 100,000 × g.
DNA isolation.
Cellular DNA was isolated by sodium dodecyl sulfate (SDS)-proteinase K treatment and phenol and chloroform extraction (4), or total cell lysates were prepared as described by Higuchi (14). Viral DNA was purified by addition of SDS and proteinase K to 1 ml of cell supernatant followed by phenol and chloroform extraction, ethanol precipitation, and resuspension in 50 μl of deionized water. Pellets from ultracentrifugation of cell supernatants were suspended in 100 μl of SDS-proteinase K lysis buffer, extracted, precipitated, and resuspended in 50 μl of water.
Consensus sequence PCR amplification.
Amplification of a portion of herpesvirus DNA polymerase was performed, with modifications, as previously described (35), with nested, degenerate primers targeted to highly conserved amino acid motifs. Primary amplification of 1 μg of cellular DNA or 5 μl of viral template DNA (10 to 50 ng) was performed in a 50-μl reaction mix with two upstream primers, DFA and ILK, and one downstream primer, KG1 (Table 1). Secondary amplification of 1 μl of the first reaction mix was performed with one upstream primer, TGV, and one downstream primer, IYG. Reaction mixtures contained 1 μM each primer, 200 μM each deoxynucleoside triphosphate, 2 mM MgCl2, 2.5% dimethyl sulfoxide, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, and 2.5 U of Taq polymerase (Gibco, Gaithersburg, Md.). One half of the reaction volume, including buffer, deoxynucleoside triphosphates, and primers, was overlaid with paraffin (Ameriffin; Baxter, McGaw Park, Ill.). Polymerase and sample DNA in buffer were then added, and samples were heated to 94°C for 3 min, 60°C for 2 min, and 72°C for 1 min to complete the first cycle. An additional 44 cycles were performed at 94°C for 30 s, 46° for 1 min, and 72°C for 1 min in an Omnigene thermal cycler (Hybaid Limited, Middlesex, United Kingdom). Twenty microliters from the secondary reaction was electrophoresed in 3% Nusieve–1% GTG agarose in Tris-acetate buffer. Products of the appropriate length (200 to 250 bp) were excised from the gel, purified with a Qiaex II gel extraction kit (Qiagen, Inc., Chatsworth, Calif.), and suspended in 30 μl of water. An aliquot was then directly sequenced with primers TGV and IYG with fluorescent-dye terminators and Taq polymerase on an ABI 373A automated sequencer (Applied Biosystems, Inc., Foster City, Calif.) at the Biotechnology Resource Center, Cornell University. The same material was cloned into pBluescript (SK−) (Stratagene, La Jolla, Calif.), which was prepared as previously described for the cloning of unmodified PCR products (19), and this clone was identified as pBLHV.
TABLE 1.
Sequences and positions of oligonucleotide primers
| Primer | Sequence (5′-3′) | Nucleotide position |
|---|---|---|
| DFA | GAYTTYGCNAGYYTNTAYCC | 2149–2168a |
| ILK | TCCTGGACAAGCAGCARNYSGCNMTNAA | 2405–2434a |
| KG1 | GTCTTGCTCACCAGNTCNACNCCYTT | 2857–2883a |
| TGV | TGTAACTCGGTGTAYGGNTTYACNGGNGT | 2440–2469a |
| IYG | CACAGAGTCCGTRTCNCCRTADAT | 2647–2671a |
| 5′BLHV | GTCCGGTTTGCTTCCTTGCT | 3–22b |
| 3′BLHV | CACGCGCAAGCTAGGATCGGC | 154–174b |
| 5′BLHVP | GAAAATTGCAGAAACGGTCA | 24–43b |
| 3′BLHVP | CGCCCCTCTGCGACTGCAAT | 133–152b |
| 5′BHV4 | TTCAGGAATTCTCCCATGCA | 3–22b |
| 3′BHV4 | TACTCTAAATGAGGCATCTGGGT | 142–165b |
| 3′OHV2 | CACTGTGAATCTCGGGGTCGGGT | 154–177b |
| 5′ACT | GAGAAGATCTGGCACCACAC | 19–40c |
| 3′ACT | AGCCATCTCCTGCTCGAAGT | 437–457c |
| 5′LTR | GAGCTCTCTTGCTCCCGAGAC | 256–275d |
| 3′LTR | CGGAGATAGGGCTCGCGATGGTCTCAGCCG | 337–366d |
| 556 | AGTCTGGGTATATGAATCCAGATGGCTCTC | 38–68e |
| 755 | AAGATAAGCACCAGTTATGCATCTGATAAA | 431–460e |
Nucleotide position in the DNA polymerase sequence of HHV1 (35).
Nucleotide position in the DNA polymerase sequence nested between primers TGV and ILK.
Nucleotide position in the bovine actin sequence of Degen et al. (5).
Nucleotide position in the BLV sequence of Sagata et al. (31).
Nucleotide position in the OHV2 clone Bp4a1 of Baxter et al. (3).
Additional sequences were obtained by a primary amplification with consensus primers DFA and KG1 as described above, but with a total of 35 cycles. Secondary amplification of the first reaction mix was performed with DFA upstream and specific downstream primers derived from sequence data. This reaction was performed as described above, but with a total of 35 cycles and an annealing temperature of 60°C rather than 46°C. Specific primers included 3′BLHV, 3′OHV2, and 3′BHV4 as appropriate (Table 1).
Specific PCR amplification.
Specific herpesvirus primers, 5′BLHV and 3′BLHV (Table 1), were used alone or with bovine β-actin primers or BLV primers at 1 μmol/liter in a multiplex format. Actin primers 5′ACT and 3′ACT were used at 100 nmol/liter and BLV long terminal repeat (LTR)-specific primers 5′LTR and 3′LTR were used at 50 nmol/liter. Nested herpesvirus primers 5′BLHVP and 3′BLHVP, BHV4-specific primers 5′BHV4 and 3′BHV4, and OHV2-specific primers 556 and 755 were used at 2 μmol/liter. Conditions were as described above except for an increase of the dimethyl sulfoxide concentration to 5%, an initial cycle of 94°C for 3 min, 65°C for 2 min, and 72°C for 1 min, and parameters of 94°C for 30 s, 63°C for 30 s, and 72°C for 15 s, with a total of 35 cycles. Samples included either 1 μg of purified DNA or 15 μl of cell lysate (9 × 104 mononuclear cells). Products, separated by electrophoresis as described above, were blotted onto nylon membranes and hybridized with a DNA probe generated by PCR amplification of 10 pg of the herpesvirus pBluescript (SK−) clone pBLHV with 5′BLHVP and 3′BLHVP. Product of this amplification was gel purified and random prime labeled with [α-32P]CTP as instructed by the manufacturer (Boehringer Mannheim, Indianapolis, Ind.). The sensitivity of PCR with primers 5′BLHV and 3′BLHV was tested on serial 10-fold dilutions of pBLHV in 1 μg of control fetal bovine DNA.
Alignments and phylogenetic analysis.
DNA and corresponding amino acid sequences of amplified products, excluding the primed regions, were analyzed with MegAlign software (DNAStar, Inc., Madison, Wis.). Alignments were performed with the PAM250 residue weight table, and identities were determined from the PAM250 alignments and are reported as the percentage of identical amino acids. Additional sequences were obtained from GenBank. Phylogenetic analysis of DNA and amino acid alignments were performed with the PHYLIP package (University of Washington, Seattle) based on distance matrices obtained by the maximum likelihood approach and unweighted pair group method by arithmetic averaging analyses with bootstrap evaluation of 100 data sets.
Nucleotide sequence accession numbers.
The DNA polymerase sequences determined herein have been deposited in the National Center for Biotechnology Information database, and GenBank accession numbers are as follows: BLHV, AF031808; AHV1, AF031809; BHV2, AF031810; BHV4, AF031811; OHV2, AF031812.
RESULTS
Amplification of herpesvirus DNA polymerase gene sequences.
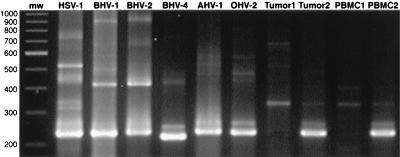
A nested PCR assay, based on conserved amino acid sequences of herpesvirus DNA polymerase genes (35), was used to amplify DNA purified from tumor cells of a BLV-positive, lymphosarcomatous cow. For comparison, viral DNAs from BHV1, BHV2, BHV4, and AHV1 and cellular DNA containing HSV1 and OHV2 genomes were amplified in an identical manner. In addition, DNAs from (i) a BLV-negative, congenital malignant lymphoma, (ii) PBMC of one BLV-positive and one BLV-negative calf, and (iii) FLK-BLV, BL3, and NBC-13 cell lines were amplified. Analysis of the second round of PCR by gel electrophoresis and ethidium bromide staining revealed one predominant band of approximately 232 bp from the BLV-positive bovine lymphosarcoma, BLV-positive calf PBMC, and OHV2 DNAs (Fig. 1). This product migrated in the size range predicted of herpesvirus DNA polymerase genes (200 to 250 bp). BHV4 DNA yielded a predominant band at 218 bp. HSV1, BHV1, and BHV2 yielded amplicons of 226, 229, and 232 bp, respectively. The BLV-negative tumor and calf PBMC DNAs had no discernible bands of the appropriate size. No amplification products were visible from cell line DNAs and buffer controls (not shown).
FIG. 1.
Gel analysis of products of degenerate herpesvirus primers. BHV1, BHV2, BHV4, and AHV1 were amplified from viral DNA preparations. HSV1-, OHV2-, and BLV-negative (Tumor1) and -positive (Tumor2) bovine tumors and BLV-negative (PBMC1) and -positive (PBMC2) PBMC were amplified from 1 μg of cellular DNA. Position of molecular weight markers (mw) are indicated in base pairs.
Sequence analysis of consensus PCR products.
The above-specified amplicons were excised and directly sequenced with TGV and IYG primers (Table 1). Figure 2A shows the alignment of sequences derived from the bovine lymphoma DNA and known herpesvirus agents of cattle. The DNA sequence from AHV1 was identical to that subsequently published for this region (9). The sequences derived from the BLV-positive lymphoma and calf PBMC were identical and were predicted to encode an amino acid sequence with 53% identity to AHV1, 51% to BHV4, 50% to OHV2, and 33 and 37% to BHV1 and BHV2, respectively. The genome giving rise to this amplicon is proposed to be from a previously unidentified herpesvirus and is designated BLHV. Phylogenetic analysis of homologous DNA polymerase fragments of 32 different herpesviruses was in close agreement with the known clustering of the herpesviruses into alpha, beta, and gamma subfamilies and indicated that this virus was most closely related to members of the subfamily Gammaherpesvirinae (not shown).
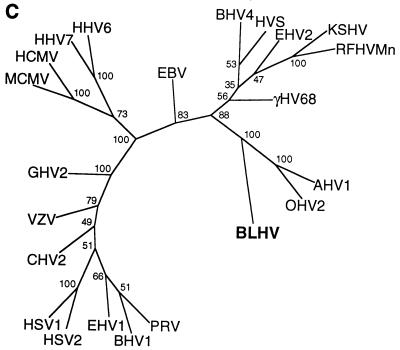
FIG. 2.
Analysis of DNA polymerase sequences of bovine herpesviruses. (A) Alignment of DNA sequences excluding TGV- and IYG-primed regions. The sequence obtained from bovine tumor is designated BLHV. Sequences for AHV1, BHV2, BHV4, OHV2, and BLHV were derived from the amplicons shown in Fig. 1. The sequence for BHV1 was obtained from GenBank (accession no. emb Z78205) (B) Alignment of amino acid sequences of gammaherpesviruses of cattle. Sequence corresponds to 478-bp of DNA internal to the DFA- and IYG-primed regions. Additional BLHV, BHV4, and OHV2 upstream sequences were obtained from PCR amplicons. Upstream AHV1 sequence was obtained from GenBank (accession no. AF005370) (C) Phylogenetic tree resulting from analysis of sequences in panel B and additional herpesvirus sequences from GenBank: CHV1 (canine herpesvirus 1; accession no. emb X89500), EBV (HHV4; accession no. V01555), EHV1 (equine herpesvirus 1; accession no. M86664), EHV2 (accession no. U20824), γHV68 (murine gammaherpesvirus 68; accession no. U97553), HCMV (human cytomegalovirus, HHV5; accession no. M14709), HHV6 (accession no. emb X83413), HHV7 (accession no. U43400), HSV1 (accession no. emb X04771), HSV2 (HHV2; accession no. M16321), HVS (saimiriine herpesvirus 2; accession no. M31122), KSHV (accession no. U93872), GHV2 (Marek’s disease virus, gallid herpesvirus 2; accession no. L40431), MCMV (murine cytomegalovirus; accession no. U68299), PRV (pseudorabies virus; accession no. L24487), RFHVMn (retroperitoneal fibromatosis herpesvirus of Macaca nemestrina; accession no. AF005478), and VZV (varicella-zoster virus, HHV3; accession no. emb X04370).
Additional upstream sequences for BLHV, OHV2, and BHV4 were obtained by PCR with degenerate primers, DFA and KG1 (Table 1), followed by seminested PCR with DFA and specific downstream primers derived from the sequences presented in Fig. 2A. A total sequence of 478 bp between degenerate regions DFA and IYG (Table 1) was used for further analysis. Figure 2B shows the amino acid alignment of BLHV with the gammaherpesviruses which infect cattle. Figure 2C shows the results of phylogenetic analysis of an homologous region of known herpesviruses. Bootstrap evaluation indicated that BLHV clusters with the MCF agents, AHV1 and OHV2.
Detection of herpesvirus DNA polymerase gene consensus sequence in ovine cells.
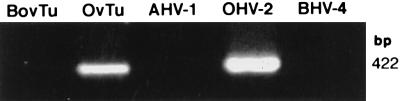
DNAs from five experimentally induced BLV-positive ovine lymphomas were also assayed for the presence of herpesvirus sequences with the degenerate PCR primers. Two sheep had been infected by BLV-positive bovine PBMC, one had been infected with cell-free BLV, and two had been infected with the BLV full-length clone, pBLV913, via transfected cells (15, 29). Two of the five tumor DNAs yielded a band at 232 bp (not shown). One of these tumors was from an animal that had been infected with BLV-positive bovine PBMC, and one was from a sheep infected with pBLV913. The sequence derived from the 232-bp amplicon was found to be identical to the sequence derived from OHV2 DNA. The identity of OHV2 in the sheep lymphomas was confirmed with primers 556 and 755 (Table 1), which amplify a 422-bp region of an OHV2 tegument protein gene (3) (Fig. 3). DNA preparations that yielded BLHV pol amplicons were also amplified with primers 556 and 755 and were negative, confirming that OHV2 DNA did not give rise to the amplicons in these preparations.
FIG. 3.
Amplification of cellular and viral DNAs with OHV2-specific primers. BHV4 and AHV1 viral DNA preparations and 1 μg of DNA from BLHV-positive bovine tumor (BovTu), OHV2-positive ovine tumor (OvTu), and OHV2-positive control (OHV-2) were subjected to PCR with OHV2 tegument protein gene-specific primers 556 and 755, which yield a predicted 422-bp amplicon.
Specific amplification of BLHV.
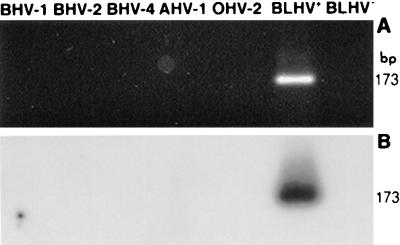
The BLHV-specific primers 5′BLHV and 3′BLHV (Table 1) were derived from the sequence data presented in Fig. 2A. These primers allowed the detection of femtogram quantities of cloned target DNA, pBLHV, diluted in 1 μg of control cellular DNA (data not shown). PCR with primers 5′BLHV and 3′BLHV yielded a predicted 173-bp product from the DNAs previously identified as positive for the BLHV sequence and not from viral DNAs from BHV1, BHV2, BHV4, AHV2, and OHV2 (Fig. 4A). Nested primers 5′BLHVP and 3′BLHVP were used to generate an internal probe to confirm the sequence of PCR products by Southern blot hybridization (Fig. 4B).
FIG. 4.
Amplification of viral and cellular DNAs with BLHV-specific primers. (A) Viral DNA preparations of BHV1, BHV2, BHV4, and AHV1 and cellular DNA from OHV2-positive cells and BLHV-positive (BLHV+) and -negative (BLHV−) tumors were subjected to PCR with primers 5′BLHV and 3′BLHV, which yield a predicted 173-bp amplicon. (B) Southern blot of the gel in panel A hybridized with a nested, BLHV-specific probe.
Transfer of infection with cell-free supernatants.
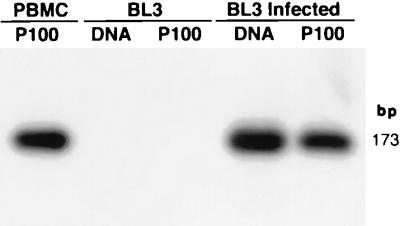
Primers 5′BLHV and 3′BLHV were used in a PCR assay to monitor BLHV expression in PBMC and tumor cells cultured for 8 days in the presence of phorbol myristate acetate and dexamethasone. Media from these cultures was clarified by centrifugation and ultracentrifuged at 100,000 × g. DNAs prepared from the resulting pellets were found to contain BLHV DNA by PCR, and the sequence was confirmed by Southern hybridization with an internal probe (Fig. 5). Clarified supernatant from cultured BLHV-positive cells was then applied to the bovine B-lymphocyte cell line, BL3. After 16 h, the cells were rinsed once with phosphate-buffered saline and resuspended in fresh medium. After an additional 72 h, cells and media were harvested separately. The cells were lysed for DNA isolation, the medium was clarified, and material was pelleted at 100,000 × g. The resultant cellular DNA, as well as DNA from the pellet, was positive for BLHV. DNA from untreated cells and material pelleted from their media were negative (Fig. 5).
FIG. 5.
Southern blot analysis of transfer of BLHV infection. DNA purified from material pelleted at 100,000 × g (P100) as well as cellular DNA (DNA) was amplified with BLHV-specific primers 5′BLHV and 3′BLHV, blotted, and hybridized with a nested, BLHV-specific probe. The first lane shows the result of amplification of DNA from material pelleted from BLHV-positive PBMC culture medium. The remaining lanes show the products of amplification of either cellular DNAs or pellets from medium of uninfected or infected BL3 cells.
Survey of bovine PBMC and lymphomas.
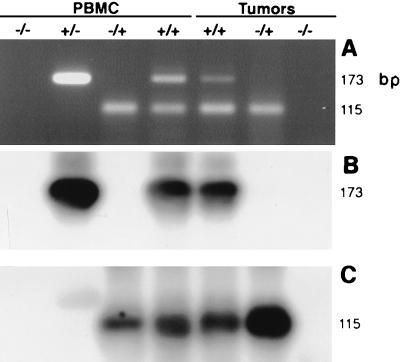
The BLHV-specific primers 5′BLHV and 3′BLHV were also used to analyze additional bovine DNAs. Dual amplification of BLV and BLHV was performed in a multiplex assay in which BLV LTR primers yield a 115-bp product in addition to the 173-bp BLHV product. Representative assays are shown in Fig. 6, and complete results for 147 samples are presented in Table 2. The BLHV sequence was detected in the DNA of PBMC from 91% of adult animals (n = 101) and 38% of juveniles (n = 13) and in the DNA of tumor cells of 63% of lymphomas (n = 32). Of the adults, the occurrence of herpesvirus infection was 94% in animals that were seropositive for BLV (n = 63) and 87% in BLV-seronegative animals (n = 38). In the BLV-seropositive group, 17 animals exhibited PL, and all were BLHV positive. Within the BLV-seronegative group, nine animals were positive for BLV LTR sequence by PCR, and all were BLHV positive. DNA from BL3 cells was negative for both BLV and BLHV, and DNA from FLK-BLV and NBC-13 cells were positive for BLV and negative for BLHV (data not shown). Dual-negative samples were reanalyzed by multiplex PCR with primers for BLHV and bovine actin, and all were positive for amplification of actin DNA target while remaining BLHV negative. DNAs from all BLHV-negative tumors were subjected to further PCR analysis with both the degenerate herpesvirus primers and the BHV4-specific primers 5′BHV4 and 3′BHV4 (Table 1). Amplification with degenerate primers was negative. The BHV4-specific primers, derived from sequence data in Fig. 2A, yielded the predicted 163-bp product when used to amplify BHV4 viral DNA and did not amplify any product from BLHV-positive DNAs or from BLHV-negative tumor DNAs (not shown).
FIG. 6.
Dual amplification of bovine PBMC and tumor DNAs with BLHV- and BLV-specific primers. (A) Gel analysis of samples which represent the variety of results for all 147 samples tested (BLHV and BLV). Samples were amplified from 1 μg of DNA or 15 μl of cell lysate (9 × 104 mononuclear cells) with primer pairs 5′BLHV-3′BLHV and 5′LTR-3′LTR, which yield 173-bp BLHV and 115-bp BLV amplicons, respectively. (B) Southern blot of the gel in panel A hybridized with a nested, BLHV-specific probe. (C) Southern blot as in panel B stripped and reprobed with a BLV-specific probe.
TABLE 2.
Occurrence of BLV and BLHV infections in cattle
| Determination | No. (%)
|
|||
|---|---|---|---|---|
| Total | BLV+ | BLHV+ | BLV+ BLHV+ | |
| Congenital tumor | 1 | 0 | 0 | 0 |
| Calves | 13 | 3 (23) | 5 (38) | 2 (15) |
| BLV seronegative | 38 | 9 (24) | 33 (87) | 9 (24) |
| BLV seropositive | ||||
| Asymptomatic | 46 | 46 (100) | 42 (91) | 42 (91) |
| Lymphocytotic | 17 | 17 (100) | 17 (100) | 17 (100) |
| Lymphosarcoma | 32 | 32 (100) | 20 (63) | 20 (63) |
| Total | 147 | 107 (73) | 117 (80) | 80 (54) |
DISCUSSION
The sequence data indicate the existence of a previously unclassified bovine herpesvirus and suggest that this virus is distributed ubiquitously in cattle. This sequence was PCR amplified from B-lymphoma cells associated with BLV pathogenesis as well as from normal bovine PBMC. Phylogenetic analysis indicated that BLHV is a member of the gammaherpesvirus subfamily, and its sequence is most similar to those of the MCF agents of cattle, AHV1 (69%) and OHV2 (70%), which are classified in the genus Rhadinovirus. No defined pathology has been identified in association with infection.
Analysis of five BLV-positive B lymphomas of sheep with degenerate and specific PCR primers detected the presence of a distinct gammaherpesvirus, OHV2, in two of five animals. Zoonotic OHV2 infection of cattle and other ruminants results in T-lymphocyte proliferation and transformation (3). OHV2 tropism in sheep and its pathogenic potential have not been defined. The association of OHV2 with BLV-positive malignant B lymphoma, like that of BLHV, not only indicates a B-cell tropism but suggests a possible role in the progression of BLV pathogenesis in sheep.
The BLHV-specific primers 5′BLHV and 3′BLHV were derived from sequence data and used to monitor the expression of BLHV in PBMC and tumor cell cultures. The PCR signal obtained from 100,000 × g pellets of clarified supernatants could possibly represent cellular DNA contamination from fragmented cells present in these cultures. Therefore, cell supernatants were used to transfer infection to a bovine B-cell line, BL3. It is unlikely that the transmission to and reisolation from in vitro target cells represents residual contamination, and this forms a base from which to pursue conditions for virus isolation and characterization. However, the infection of BL3 cells established here was transient and not associated with overt cytopathic effects. Efforts are ongoing to establish a cell culture system for the propagation and characterization of this virus. Poor growth in vitro is not uncharacteristic of the gammaherpesviruses: AHV1 and BHV4 grow slowly and generate low viral titers (13), whereas OHV2 has not been isolated and remains identified only as a cell-associated DNA genome (3). A herpesvirus previously isolated from bovine lymphosarcoma, Pennsylvania 47 strain, required prolonged incubations for detection and isolation, was distinguished from known bovine herpesviruses (21, 33), and may be similar to BLHV. Likewise, Kaposi’s sarcoma herpesvirus (KSHV) has not been transmitted to uninfected cells in vitro (23), and Epstein-Barr virus (EBV), isolated by virtue of its transforming capacity in vitro, maintains a predominantly latent infection both in vivo and in vitro (26).
A survey of DNAs from the PBMC of cattle with specific primers indicated that BLHV is ubiquitous. Infection likely occurs at a young age, because at 2 weeks of age 38% of calves were positive for BLHV DNA. A large portion of the samples tested were preselected for BLV infection and therefore do not represent a random population. However, there was a high rate of infection, 87%, among adult, BLV-negative animals. The rate of detection of BLHV infection in tumors was lower than expected considering the high rate of infection in PBMC from adult animals. DNAs from the peripheral blood of animals with tumor were not assayed for BLHV in this study; BLHV may have been present in PBMC of these animals and excluded from the tumor.
The combined amplification of BLHV and BLV by multiplex PCR provides direct detection of dual infection with these viruses. Previous studies have demonstrated high rates of BLV infection in U.S. dairy herds (4), and so it is not surprising that a significant number of animals were dually infected with BLV and BLHV. The question remains as to whether there is any significant pathogenic potential as a result of this coinfection. There is precedent for the induction of lymphocytosis by members of the gammaherpesvirus subfamily. In particular, several members, saimiriine herpesvirus 2, ateline herpesvirus 2, EBV, KSHV, AHV1, and BHV4, carry bcl-2 homologs (1, 9, 20, 30), and high Bcl-2 levels and protection from apoptosis have been implicated in the maintenance of PL (6, 25, 32). Another plausible role for BLHV in PL is transactivation of the BLV promoter, as has been demonstrated in herpesvirus/retrovirus systems (10, 16). PBMC of lymphocytotic animals carry unintegrated BLV DNA as a result of reinfection (24), which indicates induction of BLV replication at this stage of disease.
Assay of all BLHV-negative tumors for other herpesviruses, with degenerate and specific PCR primers, yielded negative results, thereby precluding a general requirement for herpesvirus infection in the maintenance of this particular set of tumors, yet the concurrence of BLHV and BLV infection in the majority of tested tumors and the dual infection of all PL animals indicates that a role for BLHV as a cofactor in BLV-associated tumorigenesis should be considered.
ACKNOWLEDGMENTS
This work was supported in part by USDA grant 93-37204-9213. J.R. was supported by USDA National Needs graduate fellowship 92-38420-7366.
We thank Gary L. Cockerell and Thomas J. Divers for generous provision of bovine and ovine samples and Allan Eaglesham for critical review of the manuscript.
REFERENCES
- 1.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittman S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacon L D, Witter R L, Fadly A M. Augmentation of retrovirus-induced lymphoid leukosis by Marek’s disease herpesvirus in white leghorn chickens. J Virol. 1989;63:504–512. doi: 10.1128/jvi.63.2.504-512.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxter S I F, Pow I, Bridgen A, Reid H W. PCR detection of the sheep-associated agent of malignant catarrhal fever. Arch Virol. 1993;132:145–159. doi: 10.1007/BF01309849. [DOI] [PubMed] [Google Scholar]
- 4.Cockerell G L, Rovnak J. The correlation between the direct and indirect detection of bovine leukemia virus infection in cattle. Leukocyte Res. 1988;12:465–469. doi: 10.1016/0145-2126(88)90112-9. [DOI] [PubMed] [Google Scholar]
- 5.Degen J L, Neubauer M G, Degen S J, Seyfried C E, Morris D R. Regulation of protein synthesis in mitogen-activated bovine lymphocytes. Analysis of actin-specific and total mRNA accumulation and utilization. J Biol Chem. 1983;258:12153–12162. [PubMed] [Google Scholar]
- 6.Dequiedt F, Hanon E, Kerkhofs P, Pastoret P-P, Portetelle D, Burny A, Kettman R, Willems L. Both wild-type and strongly attenuated bovine leukemia viruses protect peripheral blood mononuclear cells from apoptosis. J Virol. 1997;71:630–639. doi: 10.1128/jvi.71.1.630-639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimmock C K, Rogers R J, Chung Y S, Mackenzie A R, Waugh P D. Differences in the lymphoproliferative response of cattle and sheep to bovine leucosis virus infection. Vet Immunol Immunopathol. 1986;11:325–331. doi: 10.1016/0165-2427(86)90035-8. [DOI] [PubMed] [Google Scholar]
- 8.Djilali S, Parodi A L, Levy D, Cockerell G L. Development of leukemia and lymphosarcoma induced by bovine leukemia virus in sheep: a hematopathological study. Leukemia. 1987;1:777–781. [PubMed] [Google Scholar]
- 9.Ensser A, Pflanz R, Fleckenstein B. Primary structure of the alcelaphine herpesvirus 1 genome. J Virol. 1997;71:6517–6525. doi: 10.1128/jvi.71.9.6517-6525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evermann J F, Derse D, Dorn P L. Interactions between herpesviruses and retroviruses: implications in the initiation of disease. Microb Pathog. 1991;10:1–9. doi: 10.1016/0882-4010(91)90060-n. [DOI] [PubMed] [Google Scholar]
- 11.Ferrer J F, Marshak R R, Abt D A, Kenyon S J. Relationship between lymphosarcoma and persistent lymphocytosis in cattle: a review. J Am Vet Med Assoc. 1979;175:705–708. [PubMed] [Google Scholar]
- 12.Ferrer J F, Stock N D, Lin P. Detection of replicating C-type viruses in continuous cell cultures established from cows with leukemia: effect of the culture medium. J Natl Cancer Inst. 1971;47:613–621. [PubMed] [Google Scholar]
- 13.Henry B E, Ota R, Evermann J F. Genetic relatedness of disease-associated field isolates of bovid herpesvirus type 4. Am J Vet Res. 1986;47:2242–2246. [PubMed] [Google Scholar]
- 14.Higuchi R. Rapid, efficient DNA extraction for PCR from cells or blood. Perkin-Elmer Cetus Amplif Forum PCR Users. 1989;2:1–3. [Google Scholar]
- 15.Jensen W A, Rovnak J, Cockerell G L. In vivo transcription of the bovine leukemia virus tax/rex region in normal and neoplastic lymphocytes of cattle and sheep. J Virol. 1991;65:2484–2490. doi: 10.1128/jvi.65.5.2484-2490.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaguchi Y, Mikami T. Molecular interactions between retroviruses and herpesviruses. J Vet Med Sci. 1995;57:801–811. [PubMed] [Google Scholar]
- 17.Kettmann R, Burny A, Callebaut I, Droogmans L, Mammericks M, Willems L, Portetelle D. Bovine leukemia virus. In: Levy J A, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. p. 39. [Google Scholar]
- 18.Kettmann R, Cleuter Y, Mammerickx M, Meunier-Rotival M, Bernardi G, Burny A, Chantreene H. Genomic integration of bovine leukemia virus: comparison of persistent lymphocytosis with lymph node tumor form of enzootic bovine leukosis. Proc Natl Acad Sci USA. 1980;77:2577–2581. doi: 10.1073/pnas.77.5.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchuk D, Drumm M, Saulino A, Collins F S. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osorio F A, Reed D E, Van der Maaten M J, Metz C A. Comparison of the herpesviruses of cattle by DNA restriction endonuclease analysis and serologic analysis. Am J Vet Res. 1985;46:2104–2109. [PubMed] [Google Scholar]
- 22.Peters W P, Kufe D, Schlom J, Frankel J W, Prickett C O, Groupe V, Spiegelman S. Biological and biochemical evidence for an interaction between Marek’s disease herpesvirus and avian leukosis virus in vivo. Proc Natl Acad Sci USA. 1973;70:3175–3178. doi: 10.1073/pnas.70.11.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 24.Reyes R A, Cockerell G L. Unintegrated bovine leukemia virus DNA: association with viral expression. J Virol. 1996;70:4961–4965. doi: 10.1128/jvi.70.8.4961-4965.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reyes, R. A., and G. L. Cockerell. Bovine leukemia virus associated-leukemogenesis is correlated with suppression of programmed cell death and increased expression of bcl-2. Submitted for publication.
- 26.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott; 1996. pp. 2397–2446. [Google Scholar]
- 27.Roizman B, DesRosiers R C, Fleckenstein B, Lopez C, Minson A C, Studdert M J. The family Herpesviridae: an update. Arch Virol. 1992;123:425–449. doi: 10.1007/BF01317276. [DOI] [PubMed] [Google Scholar]
- 28.Rose T M, Strand K B, Schultz E R, Schaefer G, Rankin G W, Thouless M E, Tsai C, Bosch M L. Identification of two homologs of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rovnak J, Boyd A L, Casey J W, Gonda M A, Jensen W A, Cockerell G L. Pathogenicity of molecularly cloned bovine leukemia virus. J Virol. 1993;67:7096–7105. doi: 10.1128/jvi.67.12.7096-7105.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russo J J, Bohenzky R A, Chien M-C, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sagata N, Yasunaga T, Tsuzuku-Kawamura J, Ohishi K, Ogawa Y, Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci USA. 1985;82:677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz-Cornil I, Chevallier N, Belloc C, Le Rhun D, Laine V, Berethelemy M, Mateo A, Levy D. Bovine leukemia virus-induced lymphocytosis in sheep is associated with reduction of spontaneous B cell apoptosis. J Gen Virol. 1997;78:153–162. doi: 10.1099/0022-1317-78-1-153. [DOI] [PubMed] [Google Scholar]
- 33.Van der Maaten M J, Boothe A D. Isolation of a herpes-like virus from lymphosarcomatous cattle. Arch Gesamte Virusforsch. 1972;37:85–96. doi: 10.1007/BF01241154. [DOI] [PubMed] [Google Scholar]
- 34.Van Der Maaten M J, Miller J M. Replication of bovine leukemia virus in monolayer cell cultures. Bibl Haematol. 1976;43:360–362. doi: 10.1159/000399166. [DOI] [PubMed] [Google Scholar]
- 35.VanDevanter D R, Warrener P, Bennett L, Schultz E R, Coulter S, Garber R L, Rose T M. Detection and analysis of diverse herpesviral species by consensus primer PCR. J Clin Microbiol. 1996;34:1666–1671. doi: 10.1128/jcm.34.7.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willems L, Heremans H, Chen G, Portetelle D, Billiau A, Burny A, Kettmann R. Cooperation between bovine leukemia virus transactivator protein and Ha-ras oncogene in cellular transformation. EMBO J. 1990;9:1577–1581. doi: 10.1002/j.1460-2075.1990.tb08277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]