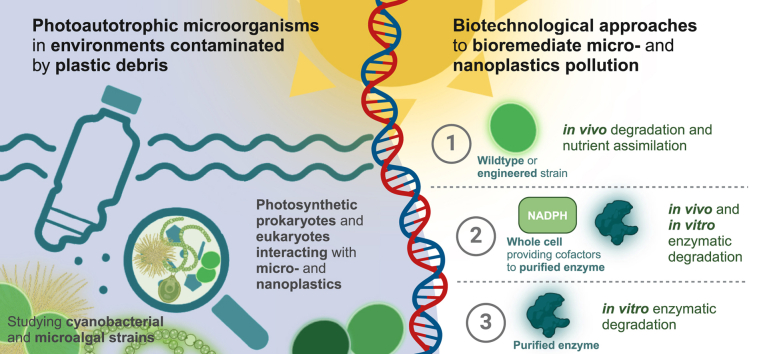
Abstract
Mismanaged plastics, upon entering the environment, undergo degradation through physicochemical and/or biological processes. This process often results in the formation of microplastics (MPs), the most prevalent form of plastic debris (<1 mm). MPs pose severe threats to aquatic and terrestrial ecosystems, necessitating innovative strategies for effective remediation. Some photosynthetic microorganisms can degrade MPs but there lacks a comprehensive review. Here we examine the specific role of photoautotrophic microorganisms in water and soil environments for the biodegradation of plastics, focussing on their unique ability to grow persistently on diverse polymers under sunlight. Notably, these cells utilise light and CO2 to produce valuable compounds such as carbohydrates, lipids, and proteins, showcasing their multifaceted environmental benefits. We address key scientific questions surrounding the utilisation of photosynthetic microorganisms for MPs and nanoplastics (NPs) bioremediation, discussing potential engineering strategies for enhanced efficacy. Our review highlights the significance of alternative biomaterials and the exploration of strains expressing enzymes, such as polyethylene terephthalate (PET) hydrolases, in conjunction with microalgal and/or cyanobacterial metabolisms. Furthermore, we delve into the promising potential of photo-biocatalytic approaches, emphasising the coupling of plastic debris degradation with sunlight exposure. The integration of microalgal-bacterial consortia is explored for biotechnological applications against MPs and NPs pollution, showcasing the synergistic effects in wastewater treatment through the absorption of nitrogen, heavy metals, phosphorous, and carbon. In conclusion, this review provides a comprehensive overview of the current state of research on the use of photoautotrophic cells for plastic bioremediation. It underscores the need for continued investigation into the engineering of these microorganisms and the development of innovative approaches to tackle the global issue of plastic pollution in aquatic and terrestrial ecosystems.
Keywords: Environmental biotechnology, Microplastic, Nanoplastic, Microalgae, Cyanobacteria
Graphical abstract
Highlights
-
•
Photosynthetic microorganisms for bioremediation of plastic debris were reviewed.
-
•
Microalgal-bacterial consortia offer attractive possibilities for plastic degradation.
-
•
More research is needed on plastic polymers other than polyethylene terephthalate.
1. Introduction
In today's society, the fundamental reliance on a wide variety of plastic materials has become indispensable in our daily lives. Their production and use have increased exponentially over the last 40 years. However, the large manufacture of these long-chain polymers in many different and diverse applications (e.g., food packaging, agriculture, clothes, and construction) has converted them into an environmental concern due to their dispersal in all ecosystems. This quickly became a significant ecological issue, especially once waste management technologies could not follow the same pace as the plastic production consumption rate [[1], [2], [3], [4]]. The dispersion mechanisms of polymers across diverse ecosystems are a complex topic that has been studied by several researchers. One study by Kane and Clare [5] comprehensively reviewed the dispersion, accumulation, and fate of microplastics (MPs) in deep-marine environments. The authors harnessed existing knowledge of seafloor microplastic distribution and integrated this with process-based sedimentological models of particle transport, providing new insights and identifying future research challenges [5]. The integration of process-based sedimentological and stratigraphic knowledge with insights from modern sedimentary systems, including biological activity within them, will provide essential constraints on MPs transfer to deep-marine environments, their distribution and fate, and the implications that these have for the wider ecosystem [5]. Prabhu et al. [6] conducted a literature survey on the presence of MPs in sediments, water, and biota samples across the globe. The survey found that MPs pervade the global seafloor, from abyssal plains to submarine canyons and deep-sea trenches.
Furthermore, the recent emergence of the COVID-19 crisis raised significant implications at multiple levels, including economic, health, and social affairs [7]. This has also caused environmental consequences connected to the increased use of safety items (e.g., protective masks, gloves, and COVID tests) based on different plastic polymers that will not be recycled soon [[7], [8], [9]]. Peng et al. [10] showed that since the beginning of the pandemic, an estimated 8.4 million tons of plastic waste has been generated from 193 countries. The same study also revealed that approximately 25,000 tons of plastic COVID waste, consisting of personal protective equipment (PPE) such as masks and gloves, has leaked into the ocean. Pandemic epicentres, in particular, have faced challenges in managing waste [11]. Hospital waste represents 73% of the bulk of global discharge, and 72% of the global discharge is from Asia, which indicates the need for better management of medical waste in developing countries [10].
Most of the daily objects are made from conventional plastic polymers (e.g., (i) low- and high-density polyethylene (LDPE and HDPE), (ii) polystyrene (PS), (iii) polypropylene (PP), (iv) polyethylene terephthalate (PET), and (v) polyamides (PA)), requiring several decades or centuries to be degraded in the environment [1,12]. The current procedures for global waste plastic management include (i) recycling (around 9% of globally produced plastics), (ii) incineration for energy production (12–19%), and (iii) landfilling and mismanagement (71–79%). Landfilling is the most used method because of the lower cost and the impossibility of recycling all plastics [1,[12], [13], [14]]. The issue of plastic pollution demands either a different approach to traditional mitigation measures or the development of new home-compostable polymers that are biodegradable and disintegrable in the environment [2,3,15,16]. The ecotoxicological implications of some ‘compostable’ bioplastics are not well studied in some cases or have been reported to have low environmental degradation under realistic conditions and similar toxicity to conventional plastics [15,17]. In addition, many of these may be blends or contain chemical additives, resulting in difficulty or impossibility of recycling [2,3,12]. Various countries are adopting various waste management plans, strategies, and methods [18]. However, various challenges are associated with waste management in different geographical contexts. Limited resources, including infrastructure and funding, intersect with sociocultural complexities like public awareness, diverse practices, and communication gaps, further exacerbated by geographical and technological constraints [18,19]. These challenges can be addressed by implementing effective strategies tailored to the specific needs. Furthermore, the lack of proper waste management infrastructure and recycling facilities during the COVID-19 pandemic emergency has increased pollution with more plastic [20]. Governments and other stakeholders should work together to develop and implement sustainable practices that are environmentally friendly and economically viable. In Europe, one of the major challenges in waste management is the lack of coordination between generators, collectors, and disposal facilities [21]. In developing countries, waste management is a critical issue due to rapid population growth, inadequate funding, and lack of awareness [22].
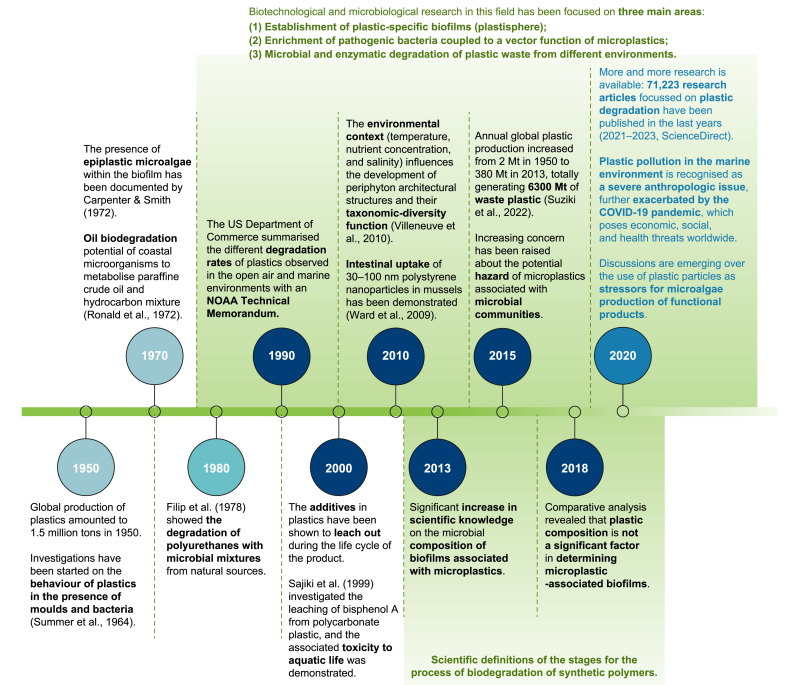
In the past few decades, research on plastic waste pollution has focused on identifying its sources, its impact on the environment and human health, and developing effective waste management strategies [23]. However, the current challenges of plastic waste pollution are more complex and require a more comprehensive approach. The main emerging information accompanying this scientific field, especially concerning biotechnology and microbiology, has been summarised in Fig. 1.
Fig. 1.
Significant achievements in the scientific field of plastics and their degradation, especially with enzymes and/or microorganisms. The chronological succession of the related events in approximately 70 years is shown. Especially since 1980, an increasing number of theories and evidence regarding marine basins polluted by plastics have started to circulate. Several scientific definitions of processional stages for the biodegradation of synthetic polymers were formulated between 2008 and 2018. Studies focussing on the interactions between microplastics (MPs), nanoplastics (NPs), and microorganisms have increased in number during the last decade. NOAA: National Oceanic and Atmospheric Administration.
Over the past decade, biotechnology and microbiology research has shifted significantly towards three key areas: (i) identification of plastic-specific biofilm/plastisphere [[24], [25], [26], [27], [28]]; (ii) enrichment of pathogenic bacteria, especially the members of the genus Vibrio, linked to a vector function of MPs [29,30]; (iii) microbial and enzymatic degradation of plastic debris from terrestrial and aquatic environments [[31], [32], [33], [34], [35], [36], [37]]. Once in the environment, plastic materials generally undergo degradation when exposed to physicochemical and/or biological factors (e.g., ultraviolet (UV) radiation, mechanical abrasion, temperature, and microbial degradation), as schematically summarised in Fig. 2. The environmental factors generate abrasion, shear (compressive, tensile, and flexural), and torsional impact [[38], [39], [40], [41]]. Several manuscripts have reviewed the biotic and abiotic treatment of MPs [[41], [42], [43], [44], [45], [46], [47]]. Chen et al. [48] summarised the current technologies used to eliminate MPs from the environment, highlighting two key aspects towards this goal: (i) catalytic degradation of MPs into carbon dioxide (CO2) and water and (ii) catalytic recycling and upcycling plastic wastes into fuels and valorised chemicals.
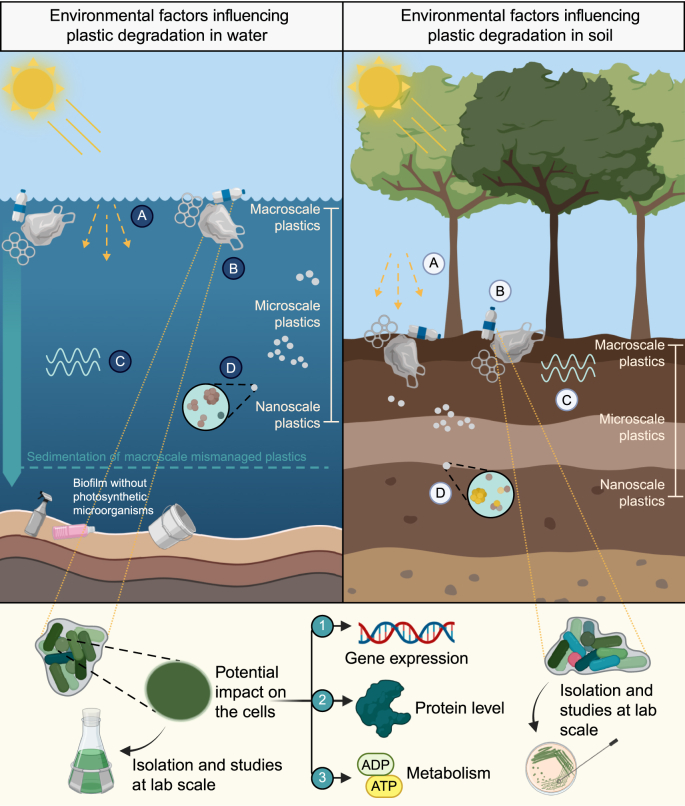
Fig. 2.
Schematic overview of the biological and abiotic degradation of MPs in marine and freshwater environments (left) and in soil (right). The abiotic methods usually rely on mechanical disruption, advanced oxidation, and photo-degradation. The biofilms on the surface of different plastics are characterised by the presence of microalgae and/or cyanobacteria only if light is available. The interactions with plastics and photosynthetic microorganisms are schematised, showing the potential impacts on the cellular level for gene expression, protein level, and metabolism. A: photodegradation. B: biofilm with photosynthetic microorganisms. C: mechanical degradation. D: microbial degradation.
The abiotic methods rely on mechanical disruption, advanced oxidation, and photo-degradation [41,49,50]. Biodegradation is mainly accomplished by the activities of microorganisms, resulting in biodeterioration, depolymerization, digestion, and mineralisation [51]. Photodegradation is the most significant process that starts plastic decomposition in aquatic and terrestrial environments in direct sunlight; this method is abiotic [52]. Plastic degradation usually results in tiny particles of irregular shapes and sizes (<1 mm), defined as MPs, which are the main form of plastic debris found in the environment impacting aquatic and terrestrial ecosystems [53,54]. These can easily be disrupted and dispersed by wind and water erosion, consequently entering the food chain, which increases the potential to affect several ecosystem functions at several levels. Different factors influencing the degradation have been identified, especially in the aquatic environments [41,55,56]. It is worth noting that both abiotic and biotic processes have their advantages and disadvantages. The choice of method depends on the type of plastic waste, the environmental setting, and the desired outcome [52]. Regarding applicability, abiotic methods are more suitable for large-scale plastic waste management, while biotic methods are generally more suitable for small-scale [52,57]. Also, abiotic ones are more appropriate for degrading hard plastics [52].
The environmental conditions and polymer chemistry significantly influence plastic degradation processes [12,13,53,58,59]. In principle, due to the release of toxic additives and metallic and organic compounds from their surfaces, MPs threaten the lives of several creatures: their harmful effects on marine organisms, especially the primary producers of the food chains, can influence food web consumers such as fish, aquatic birds, and even humans [60,61]. Microalgae and cyanobacteria in aquatic ecosystems could suffer from this contamination, but at the current environmental concentrations, this seems to have limited effects on parameters such as chlorophyll content, photosynthesis activity, and reactive oxygen species (ROS) [62]. The antibiotic adsorption on MPs and, therefore, the enrichment of potentially pathogenic plus antibiotic-resistant bacteria and antibiotic-resistance genes through horizontal gene transfer are other related concerns [61]. Because of all these factors, the bioremediation of related MPs-contaminated habitats is of fundamental importance. Differently, new (bio-)degradable plastics are usually made with natural raw materials which, after the action of different living microorganisms (e.g., heterotrophic and photoautotrophic bacteria, algae, microalgae, and fungi), could be metabolised and subsequently converted into biomass and/or secondary metabolites. Hydrobiodegradable polymers, including polyesters and thermoplastic starch, undergo degradation through abiotic and biotic hydrolysis processes [63]. In contrast, polyolefins, characterised by their hydrophobic nature as hydrocarbon polymers, exhibit resistance to hydrolysis, rendering them non-hydrobiodegradable.
The various cross-linked chemicals or the disparate hydrophobicity of the plastic debris can limit the potential of living cells to both colonise and degrade the material [41]. The degradation of plastics is challenging due to their highly stable chemical structure, which makes them resistant to most types of natural degradation [64,65]. Additives (e.g., catalysts, agents for vulcanisation, curing, blowing, pigments, dyes, plasticiser, flame retardants, heat stabilisers, photostabilisers, antioxidants, and biocide) are utilised to produce the polymer for the attainment of certain physical properties of the product, such as molecular weight, molar volume, density, degree of polymerisation, the crystallinity of material, etc. [66,67]. Nonrenewable petrochemical plastics are generally highly engineered materials with precise physical properties: the properties that make plastics so versatile for humans have also caused their degradation to be challenging [64].
Plastic microbial degradation often involves using waste material as a carbon source. These metabolic processes result in an overall beneficial impact on some ecosystem's functions [12,15,17,68,69]. Such a metabolic option is an environmentally friendly and sustainable solution [9,13,58,68,[70], [71], [72]]. Regarding bioreactors for the bioremediation of water bodies using microorganisms and to reach some estimation for big-scale applications, the airlift bioreactor is often utilised due to its advantages over bubble column and stirred bioreactors (e.g., reduction in cell damage and greater mass transfer capacity) [[73], [74], [75], [76]].
An increasing number of studies are showing wildtype [26,[77], [78], [79]] or engineered [[80], [81], [82], [83], [84]] strains able to degrade plastic fragments, also elucidating the related enzymatic mechanisms [[85], [86], [87], [88], [89], [90]]. Since some of the prokaryotes and eukaryotes from the environment could be unculturable in the laboratory, culture-independent methods are often utilised to select plastic-decomposing microorganisms: the metagenome analysis and in silico mining led to a deeper investigation towards the finding of efficient enzymes for MPs bioremediation [61]. The main wildtype and engineered microorganism able to degrade plastic debris, reported in literature, are shown in Table 1. Further, applying photosynthetic bacteria and unicellular eukaryotic algae, especially with engineered strains, may represent a possible option for plastic degradation in terrestrial and aquatic ecosystems [72,[90], [91], [92]]. Independently from the growth in open ponds or enclosed bioreactors, their main requirements are the availability of light, water, CO2, and elementary nutrients. Numerous species of green algae are Generally Recognized as Safe (GRAS) for consumption by humans and animals [93]. This group offers high metabolic diversity, besides more than 8000 species, and some strains can grow in darkness with a reduced carbon source − heterotrophically [94]. Furthermore, microalgae possess the most-used type of alternative biomass in current wastewater treatment applications: these can grow and even flourish in different conditions, including a wide variety of wastewater [95,96]. The wide diversity of functional groups in their cell wall allows the binding of pollutants to the membrane surface through the biosorption phenomenon [97,98]. Other chemicals are taken up and biodegraded by living cells [96].
Table 1.
Exemplary wildtype and engineered micro (organisms) reported in the literature are able to degrade plastic debris.
| Organism | Polymer | Experimental conditions | Outcome | Reference |
|---|---|---|---|---|
| Wildtype | ||||
| Ideonella sakaiensis | PET | Isolation from field samples | Degraded amorphous PET at ambient temperature and assimilation of its degradation monomers due to PET hydrolase (IsPETase) | [77] |
| Moraxella sp. Strain and Oleispira antarctica | PET | Isolation from the Antarctica environment | PET degradation at ambient temperatures (25 °C) | [99] |
| Pseudomonas and Bacillus consortium | PET | Isolated from petroleum-polluted soils | PET degradation | [100] |
| Klebsiella sp. | PVC | Larva's gut microbiota from Spodoptera frugiperda | Depolymerization and utilisation of PVC as sole energy source | [78] |
| Enterobacter | PHB | Mesophilic conditions | Methane production from PHB | [79] |
| Cupriavidus | Thermophilic conditions | |||
| Moorella | PHB, PLA | Methane production from PLA | ||
| Tepidimicrobium | ||||
| Clostridium | TPS | Mesophilic conditions | Methane production from TPS | |
| Rhodanobacter sp. Rs, Bacillus aryabhattai | PE | Inoculation on soil suspensions | Under co-culture, the ability for polyethylene mulching film degradation | [26] |
| Pseudomonas pseudoalcaligenes | PBAT | Incubation | Proteomic screening allowed the identify a new esterase, PpEst, that is involved in PBAT degradation. | [101] |
| Engineered | ||||
| Phaeodactylum tricornutum | PET | Microalgae transformation | Expression and secretion of PETase in the algal system under (mesophilic marine) growth conditions | [80] |
| Chlamydomonas reinhardtii | PET | Microalgae transformation | PET hydrolyzation. TPA, a fully degraded form of PET, was detected. | [81] |
| Ideonella sakaiensis | PET | Gene disruption system | PETase and MHETase are essential enzymes for PET digestion. | [83] |
| Improvement of the thermostability of leaf-branch compost cutinase | Improved PETase can reach up to 90% of PET degradation. | [86] | ||
| Pseudomonas putida | PBAT, PET | Modified M9 minimal medium. Electrocompetent cells were prepared by a modified standard protocol | Plastic biodegradation assays with the best PET hydrolase expression constructs are genomically integrated into our monomer metabolism. This resulted in various degrees of plastic depolymerization. The surface display of the PET hydrolase and the secretion were successfully achieved. | [84] |
PBAT: Polybutylene adipate co-terephthalate. PE: Polyethylene. PET: Polyethylene terephthalate. PHB: Polyhydroxybutyrate. PLA: Poly(lactic acid). PVC: Polyvinyl chloride. TPA: Terephthalic acid. TPS: thermoplastic starch.
In this review, we summarise various photosynthetic microorganisms that have been proven capable of colonising the surfaces of different types of plastic. Using the available literature, the answers to the following scientific questions were sought: what is the specific role of photosynthetic microorganisms in water and soil for plastic biodegradation? How can they be utilised for the MPs and NPs bioremediation? How can these be engineered for this purpose? Several of these still need to be answered with further laboratory experiments, and it is not yet possible to have valid explanations. We outline various hydrolases and metabolic pathways reported in the literature, likely exploitable with engineered microalgal and/or cyanobacterial strains. Thanks to developments in the engineering of these cells in recent decades, the tools are available for expressing heterologous enzymes with hydrolysis activity against fragments of PET and other types. Through this review, the aim is also to encourage further research towards obtaining more engineered photosynthetic strains and the discovery of those natively able to potentially degrade plastic debris in polluted ecosystems. The potential of photo-biocatalytic approaches related to the degradation of plastic debris is highlighted. Furthermore, the role of microalgal-bacterial consortia for biotechnological applications against MP pollution is also included.
2. Candidates of microalgae and cyanobacteria for plastic biodegradation in terrestrial or aquatic ecosystems
Once plastic debris reaches terrestrial and aquatic ecosystems, different processes start to interact with them [64,102]. Typically, abiotic degradation pathways (UV and thermal degradation, hydrolysis, and oxidation processes) are the first to appear and are often chronologically connected with microbial degradation. The structure of the polymer influences the material's degradation, and the conventional polymers (polyethylene (PE), PP, PS, polyvinyl chloride (PVC), PET and polyurethane (PU)) can be divided according to their chemistry and degradation pathways [64,102,103]. Briefly, polymers with a carbon-carbon backbone in their main chain (PE, PP, PS, and PVC) have high molecular weight and lack functional groups susceptible to degradation (oxygen, hydroxyl, and carboxylic acid). Therefore, abiotic processes have a primary role in their degradation since they are needed to initiate the insertion of favourable functional groups. For instance, photolysis and photo-oxidation lead to oxygen group insertion within the material backbone. The carbon-rich backbone is reduced to smaller and lower molecular weight fragments, which are more susceptible to biotic degradation [102].
Sometimes, different objects made of plastic have additives like antioxidants and stabilisers to inhibit or reduce polymer degradation. In other cases, pro-oxidants (such as oxygen and starch) increase their biodegradability by microbial enzymes [102]. The richness in oxygen groups characterising bio-based hydrophobic polymers is the main driver towards their degradation in natural environments and industrial composting facilities. In this respect, polymers with heteroatom in their main chains can be vulnerable to hydrolysis, photo- and biodegradation by microorganisms [64,102,104]. PS is the most resistant polymer in the environment and less susceptible to biodegradation due to its very high weight total fraction of carbon–carbon (C–C) backbone, while PU is instead the most prone to biodegradation due to their complex structure plus a relatively high abundance of O and N in the main chain [102,105]. Abiotic and biotic processes can also work in tandem, with an abiotic factor producing smaller particles which might be easily degradable by microorganisms via the mineralisation of oligomers. These processes may take a long time, several decades or centuries, even with pro-oxidants as catalysers. The existence of the bacteria on plastic surfaces is not always indicative that degradation of the specific polymer occurs. Solano et al. [106] observed that Actinobacteria and Proteobacteria were the most abundant on the surface of plastics with different degrees of biodegradability (white PE, oxo-degradable plastic, and polylactic acid-based plastic (PLA)). Even after 31 weeks in a composting test, none of the plastics showed any signs of degradation, like weight loss or colour changes [106].
Despite these observations, some bacteria, fungi, and cyanobacteria showed the ability to degrade plastics in terrestrial ecosystems. Sangale et al. [107] and Kumari et al. [108] reviewed the capability of plastics biodegradation from bacterial and fungal isolates of agricultural soils or anthropised areas (e.g., dumping and recycling sites) [107]. Wide interest in microalgae and cyanobacteria as degraders of plastics arose from evidence of their ability in bioremediation under environmentally friendly and low-cost approaches [31,103,108,109]. Within an aqueous environment, depending upon the type of plastic fragment, the debris can either float at the surface or sink to the bottom. Either way, these fragments represent a new habitat for microbes, so much so that a diverse community of heterotrophs, autotrophs, symbionts, and predators usually start growing on their surfaces [110,111]. In most cases, floating plastic communities are dominated by microalgal cells, as these photosynthetic organisms are known to exude polymers, which allow for their attachment and rapid growth. Most other buoyant plastics tend to sink to the sediment after the accumulation of microorganisms colonising the surface, resulting in the biofouling process [112,113]. Although a good amount of data is available for communities developing on plastic fragments localised at the top of aqueous bodies (mainly the cyanobacteria Phormidium and Rivularia), only limited information exists on the composition of items with microbial communities sampled from the seafloor [110,[114], [115], [116]]. When the subsurface plastisphere was investigated, commonly observed taxa include Bacteroidetes (Flavobacteriaceae) and Proteobacteria (Rhodobacteraceae and Alcanivoracaceae) [110,115,116].
As widely reported, the interaction between MPs and photosynthetic microorganisms likely affects aquatic environments at the ecosystem level. Indeed, due to the high surface area, availability of sunlight, and abundance of nitrogen and phosphorus within the aquatic media, plastic debris provides a superb growth matrix for microalgae. The optimal temperature for their growth varies depending on the species, but generally, this corresponds to values between 20 and 30 °C [117,118]. Temperatures exceeding the optimal range negatively impact the growth rate [117]. Regarding pH, these cells typically function optimally within the pH range of 6–9, facilitating their specific metabolic activities [119]. Resulting from their growth, eutrophication may detrimentally influence the aquatic ecosystems [111]. Cyanobacteria have been reported as the most prevalent organisms for plastic Interactions in agricultural fields and highly anthropised areas [109,120]. Microorganisms in a consortium can potentially increase plastic degradation, more than a single photoautotrophic or heterotrophic strain, due to metabolic cross-feeding for co-metabolic degradation [109,121]. In this respect, Jacksonvillea sp. ISTCYN1 was isolated from rice fields: the biofilm formation was reported in the presence of LDPE, HDPE, and PP as carbon sources, plus the ability to produce extracellular polymeric substances (EPS) and enzymes (e.g., laccase, esterase, lipase, and peroxidase) being involved in these processes [109]. These cells were grown in BG-11 medium adjusted to pH 7.4, incubated for 15 days at 25 ± 1 °C and light 12:12 h with an intensity of 3500 L × [109]. The growth conditions utilised for Jacksonvillea sp. ISTCYN1 are frequently employed for various other strains of cyanobacteria. This strain could be an effective option for plastic degradation under natural growth conditions. Generally, unless they are extremophiles, the temperature range to maintain alive photoautotrophic microorganisms on the surface of different types of plastic waste could assume values between 15 and 32 °C (pH 7–9 in aquatic environments). This is a wide range, which also contains suboptimal temperatures. While Mishra et al. [109] observed biofilm formation of Jacksonvillea sp. ISTCYN1 on plastics, the expression of the enzymes is not yet experimentally validated, even if these are predicted from its genome. MacLean et al. [122] found cyanobacteria (Tychonema CCAP 1459-11B and Nostoc sp.) to dominate plastic debris as a growth substrate. They suggested that cyanobacteria promote biocrust formation, increasing soil fertility by contributing to positive net production. In addition, these authors showed microbes growing in the cracks after UV-weathering and then suggested that microbes may be degrading their samples of plastics dumping sites in Germany. Finally, some phototrophic bacteria from the phyla Actinobacteria (e.g., genera Actinomycetospora, Arthrobacter, Rhodococcus, Rubrobacter, and Cellulosimicobium) and Proteobacteria have been indicated with the ability to grow on plastic surface collected from microbial communities in field dumping sites [122].
Samples dominated by these cells contained a mix of plastic debris with a dark organic matrix, with Nostocales being the most abundant. Other species from Arthrospira, Calothrix, Hydrocoleum, Lyngbya, Phormidium, Oscillatoria, or Spirulina genera have been identified for potential plastic degradation [109,120,122,123]. This suggests that cyanobacteria may be vital in ecological succession as colonisers. Similar results highlight that microalgae can colonise, grow, and degrade PE through ligninolytic and exopolysaccharide enzymes in aquatic and terrestrial ecosystems [103,108,124]. To facilitate their attachment to the plastic surface, microalgae and cyanobacteria generally produce exopolysaccharides around and outside their cells or filaments; this could provide an appropriate environment for polymer degradation [125]. Besides, soil-water diatoms (Navicula dicephala, Navicula minuta, Nitzschia intermedia, Synedra tabulate) have been identified from plastic bags from dumping sites with the ability to grow on LDPE surface [120], although more research is needed. Additionally, when biodegradable PLA and conventional PE film were incubated under different temperatures, these microorganisms were the most abundant on plastic surfaces [126].
When the potential of microalgae to colonise MPs of LDPE, HDPE, PET, or a mix of these was investigated at different sites, the cells successfully colonised this debris regardless of the condition examined. Sarmah and Rout indicated that some species (Chlorella sp., Scenedesmus quadricauda, or Stigeoclonium tenue) can grow on rubbish bags isolated from domestic dumping sites [120,123]. In studies by Sarmah and Rout [127] focused on the colonisation of Oscillatoria on submerged polyethene in domestic sewage water of Silchar town in India, the temperature of domestic sewage water (pH varying from 5.8 ± 0.10 to 8.1 ± 0.23) ranged between 28 and 34 °C. Nava et al. [111] assessed the factors affecting the distribution of different photosynthetic microorganisms, identifying the cyanobacterium Aphanocapsa incerta. Besides this species, other frequent microalgae included Cocconeis placentula Ehr., Peridiniopsis elpatiewskyi (Ostenf.) Bourrelly, Achnanthidium minutissimum (Kütz.) Czarnecki, Cocconeis pediculus Ehr., and Planktolyngbya limnetica [111].
In addition to the abovementioned reports on the growth of microorganisms on the plastic surface and plastic degradation, some authors [13,[128], [129], [130], [131]] indicated that PVC, LDPE or PLA weight loss due to potential degradation could also be related to degraded plasticisers by photosynthetic microorganisms since they could use these as a source of carbon and energy. This concept has been highlighted by Ru et al. [13] for PVC since materials based on this polymer have the highest proportion of plasticisers (up to 50%), and they are more susceptible to microbial attack. According to these authors, plastic weight loss is more related to plasticised PVC-degrading microorganisms than those capable of degrading PVC. Therefore, more analysis is needed to verify the potential plastic degradation since weight loss analysis, CO2 evolution, and gas chromatography cannot accurately assess this circumstance. Wright et al. [132] performed a detailed proteogenomic and metabolomic analysis on two marines selected strains from plastic debris (Halomonas sp. ATBC28 and Mycobacterium sp. DBP42), allowing the identification of the enzymes and the pathways involved in the biodegradation of three plasticisers (dibutyl phthalate (DBP), bis(2-ethylhexyl)phthalate (DEHP), and acetyltributylcitrate (ATBC)): this integrated multi-OMIC study also revealed the mechanisms used for ester side-chain removal (esterases and enzymes involved in the β-oxidation pathway) as well as the molecular response to deal with toxic intermediates like phthalate. Furthermore, this study demonstrated the metabolic potential in the biofilms colonising plastics to effectively biodegrade related additives and flag the importance of microbes in reducing environmental plastic toxicity [132].
3. Hydrolases with activities against PET for photosynthetic microorganisms
Microalgae show promise in biodegradation processes, possessing the capability to produce a variety of enzymes, including hydrolases. The degradation of plastics by these cells offers a potential eco-friendly and sustainable solution to plastic contamination [133]. One of the main issues with plastics is the wide variety in use and how they are an integral part of our everyday lives. PET is a material denser than water and a major ocean polluter; therefore, microbial remediation of PET polymers has been traditionally focused on these polymers, as highlighted in Table 1. According to a systematic review published in 2022, the most prevalent wild-type PET-degrading microorganisms were bacteria (56.3%, 36 genera), fungi (32.4%, 30 genera), microalgae (1.4%, the genus of Spirulina sp.), and invertebrate associated microbiota (2.8%) [134]. Among fungi and bacteria, the most prevalent genera were Aspergillus sp. And Bacillus sp. Among the publications analysed by Benavides Fernández et al. [134], 51.8% (71 articles) applied wild microorganisms, and 32.8% (45 articles) exploited genetically modified microorganisms. Regarding the percentage of PET degradation, Ideonella sakaiensis still corresponds to the most efficient wild-type microorganism to degrade PET [134].
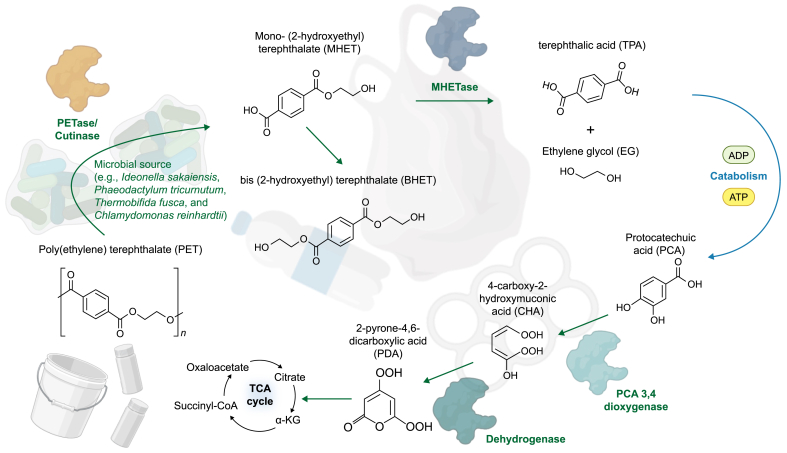
Many current techniques for dealing with these major plastic polluters (PE and PET) require large amounts of energy. For this reason, and others outlined by the United Nation's Sustainable Development Goals (SDGs), additional, less intensive, and economical processing methods are in desperate need. An important factor to consider when looking for appropriate enzymatic methods for plastic breakdown is the conditions in which they operate; otherwise, the same energy input issues will be encountered. Hydrolases are a wide group of enzymes that catalyse bond cleavages by reaction with water. This class of enzymes catalyses biochemical reactions between different functional groups, including peptide or carbon-halide bonds, thioesters, and ureas. The action of these enzymes results in the addition of –H or –OH groups (from H2O) to the plastic chains on the substrate's surface [135]. From the late 1970s, commercial lipases and even an esterase have been reported to hydrolyze polyester materials effectively: the discovery of specific PET hydrolases supported the interest in PET degradation by microorganisms. It stimulated the characterisation, enhancement, and identification of further analogous enzymes [136]. Most microbes able to secrete PET hydrolases are not ideal for industrial processes due to their complex genetic backgrounds and genetic modifications. If they are applied to industrial environments, ethical implications and regulatory frameworks would need to be addressed. Unfortunately, native microorganisms exhibit low expression rates of PET hydrolases, limiting their efficiency in plastic degradation. Therefore, enhancing expression levels is a crucial avenue to explore, particularly if microalgae are employed as a viable platform for plastic breakdown [137]. Work conducted in that area on Escherichia coli (E. coli) expression rates was limited by temperature stability, and this would be the next obstacle for microalgal species. Thus, genetic hosts such as Clostridium thermocellum, a thermophilic microorganism, could be used as they have proved to be a promising platform for lignocellulosic processes [138]. Recently, there has been significant progress in specific PET hydrolases, such as lipases, cutinases, and esterases derived from biological sources [139]. A great milestone was the isolation of Ideonella sakaiensis 201-F6 from a waste PET recycling station, which is capable of degrading both PET and the reaction intermediate mono (2-hydroxyethyl) terephthalate (MHET), using a PETase and an MHETase [77]. The discovery and modification of PETase and MHETase have provided an important basis for the degradation of PET at ambient temperature conditions. Since the knowledge of the protein structure is fundamental for its engineering, shortly after, several manuscripts focussing on the structure of Ideonella sakaiensis PETase (IsPETase, EC 3.1.1.101) were published. This hydrolase possesses a strictly conserved active site based on the Ser-His-Asp catalytic triad, containing an optimal substrate binding site to hold four MHET moieties of PET. Usually, wildtype PETase exhibits an optimum pH range of 7–9 and stability around pH 6–10 [140]. When the purified PETase was applied to PET film, pH 9.0 was identified as optimal and 30 °C as the optimum temperature [141]. Furthermore, in contrast to cutinase, this enzyme demonstrates lower activity on p-nitrophenol-linked aliphatic esters and exhibits 5.5- to 120-fold higher activity towards PET [[141], [142], [143]]. TfCa, a versatile carboxylesterase derived from Thermobifida fusca, has exhibited an intriguing ability to hydrolyze intermediates involved in the degradation of PET; furthermore, the enzymatic structure of TfCa was recently determined, shedding light on its functional characteristics [90]. Once more, artificial intelligence (AI) designing strategies have been used to optimise PET hydrolytic activity. With recent advances in AI, microalgal candidate hosts could be found for plastic processing, such as thermophilic species from the genera Cyanidioschyzon, Galdieria, and Thermochromatium. These have been studied for their ability to perform photosynthesis and produce biomass at elevated temperatures if required. They already possess heat-resistant enzymes and heat shock proteins that help them cope with thermal stress [144]. DuraPETase and FAST-PETase can significantly enhance PET degradation efficiency at mild-temperature environments, opening potentials for enzymatic plastic biodegradation at industrial-scale applications [145,146]. The schematic representation of the breakdown mechanism of PET is summarised in Fig. 3, where the polymer is proposed to degrade into CO2 and water via several intermediates by using different microorganisms (including microalgae) as sources of hydrolases [147,148]. PET is converted into TPA, MHET, and bis (2-hydroxyethyl) terephthalate (BHET). Then, MHET is hydrolysed by MHETase to TPA and ethylene glycol.
Fig. 3.
Breakdown mechanism of PET using an active form of PETase derived from microalgae. Microorganisms (Ideonella sakaiensis, Phaeodactylum tricurnutum, Thermobifida fusca, and Chlamydomonas Reinhardtii) were used as sources of hydrolases, which were able to catalyse the bond cleavages by reaction with H2O. BHET: bis (2-hydroxyethyl) terephthalate. CHA: 4-carboxy-2-hydroxymuconic acid. EG: ethylene glycol. MHET: mono-(2-hydroxyethyil) terephthalate. PCA: protocatechuic acid. PDA: 2-pyrone-4,6-dicarboxylic acid. PET: poly (ethylene) terephthalate. TCA cycle: tricarboxylic acid cycle. TPA: terephthalic acid.
TPA may then be metabolised to protocatechuic acid and 2-pyrone-4,6-dicarboxylic acid before entering the Krebs or Citric Acid Cycle. The degradation monomers can easily be recycled, although, at this point, the monomers may become pyruvate, oxalo-acetate, or eventually water and carbon dioxide molecules. The green microalgal species Chlamydomonas reinhardtii (C. reinhardtii) was modified to synthesise PETase. When comparing two strains of Chlamydomonas (C. reinhardtii CC-124 and CC-503), it was observed that CC-124 expressed more enzymes at 30 °C for up to four weeks [81]. After incubation, TPA was detected by high-performance liquid chromatography, and via microscopy, holes and dents were observed on the surface of the PET films. Interestingly, C. reinhardtii is generally recognized as safe (GRAS) and, therefore, suitable for biotechnological applications in natural habitats [81].
Due to the molecular structure of PET, in vitro, enzymatic degradation would be a logical step even if the purification plus preparation process of PET hydrolases is still expensive and time-consuming [137]. PET and similar plastics have high molecular weight and robust crystalline polymers. For these reasons, various thermal/chemical/mechanical pre-treatments of the plastics may be employed. The successful isolation of PETase from I. sakaiensis has opened the door for more studies into microbially-based research for plastic degradation. Further work should be conducted on other major plastics like PE, PP, or PS with or without pre-treatments. The marine diatom Phaeodactylum tricornutum with heterologous expression of PETase has been studied [80]. However, that strain did not grow as well as the green algae Chlamydomonas and Chlorella [149,150]. So far, C. reinhardtii is a potential leader microorganism for further research into PETase expression, being a freshwater microalga cultivable on a large scale. The rapid advances in cyanobacteria and prokaryotic cells for use as green cell factories for directly producing chemicals from CO2 and solar energy make them the most interesting hosts as biocatalysts for the degradation and use of waste plastics [91,151].
It is also important to mention the industrial status regarding the biodegradation and biorecycling of plastic waste. Direct use of cells expressing enzymes capable of degrading polymers, especially when coupled with the ability to utilise energy from light via photosynthesis as in photoautotrophic microorganisms, is at a state closer to the laboratory than the industrial scale. The successful implementation of this technology based on alive microorganisms needs detailed knowledge of the polluted site of metabolism, growth, and function of native microbial population: this could be overcome with the utilisation of cell immobilisation, nano bioremediation, biosurfactant, metagenomics, chemotaxis, and proteomics [152,153]. Especially regarding PET, its recycling with biocatalytic in vitro reactions based on engineered enzymes has notably advanced in recent years. Scaling up bioremediation enzymes for industrial use is a complex process that requires careful consideration of various factors such as the type of enzyme, the nature of the contaminant or, more specifically, the type of polymer in case of plastic waste degradation, the scale of the project, and the economic feasibility [[154], [155], [156]]. Considering large-scale industrial applications, the four best-performing enzymes for PET degradation were recently compared [156]. Interestingly, the next generation of LCCICCG enzyme will be utilised in the world's first PET biorecycling plant commissioned in 2025 [157].
4. Photolysis, photocatalytic and photobiocatalytic transformations as tools to remediate plastic debris pollution
Pyrolysis is a conventional technology used to mitigate MPs pollution, even if this approach has shown some disadvantages: high energy consumption, generation of secondary pollution, and difficulty controlling [41,47,158]. Photolysis is another abiotic approach to degrade MPs, which is based on the presence of light. Most MPs are recorded on land or floating on the ocean surface, being exposed to sunlight for extended periods [159,160]. Different manuscripts focus on some aspects of this wide topic or MPs photolysis [[161], [162], [163]]. Some MPs, such as polycarbonate, are responsive to light due to the presence of photoactive groups in their structures, namely chromophores [164,165]. Alkylphenol, aromatic amines, and thiopropionate esters are frequently brought into plastic manufacturing to tolerate oxidation, which has a low effective photolysis [166,167]. Therefore, light plays a crucial role in polymer degradation. Sunlight is known to be the natural energy source that degrades plastic waste at a very slow rate. Mimicking the role of sunlight, the photocatalytic degradation process could significantly accelerate the rate thanks to the photocatalyst that drastically facilitates the photochemical reactions involved in the degradation process [168].
Photocatalysis has long been considered for approaches regarding sustainability, such as micropollutant degradation, CO2 reduction, and H2 evolution reaction [[169], [170], [171]]. The main difference between MPs photocatalytic degradation and photolysis concerns the input of photocatalysts. For the photocatalytic degradation process, the addition of photocatalysts enriches the amount and type of ROS, stimulating depolymerization, degradation, and mineralisation in the aqueous environment [41]. Light is used as a source of energy to degrade contaminants, mostly exploiting the formation of ROS successfully [171,172]. The increasing number of published studies has highlighted the method of photocatalytic degradation as a tool to diminish MPs pollution in marine and freshwater environments [161,173]. Ouyang et al. [174] and Wang et al. [175] suggest that the use of photocatalytic technology represents a highly promising approach to transforming plastic wastes into value-added products through green and mild methods. The photocatalytic plastic waste upcycling reaction routes can be modulated by regulating experimental conditions (e.g., solvents and atmosphere), thus resulting in controllable product selectivity [175]. There are relatively few studies on photocatalytic degradation of plastic fragments. In the typical experiments, TiO2|Pt, CdS/CdOx and CNx|Ni2P photocatalysts are exploited [174]. However, TiO2|Pt can only absorb UV light (approximately 4% of the total energy of sunlight) and are relatively expensive, and the CdS/CdOx quantum dots are toxic [174]. To know the details of case studies or real-world examples and investigated types of polymers, showing the practical applications of these photocatalysts, the review published by Ouyang et al. [174] contains specific information. For example, using CdS/CdOx against 50 mg mL−1 PLA, 25 mg mL−1 PET, and 25 mg mL−1 polyurethane (PUR), the following experimental conditions were applied: in a sealed photoreactor having the solution volume of 2 mL for 10 M sodium hydroxide (NaOH), 0.5 mM quantum dots were irradiated for 4 h at 25 °C with simulated solar light (AM 1.5G, 100 mW cm−2) [174,176]. Aiming for sustainability, it is necessary to design non-toxic photocatalysts driven by visible light to transform plastics into H2 fuel and various organic chemicals. Still, the progress in the synthesis and modification methods of photocatalysts, coupled with a more in-depth understanding of photodegradation mechanisms, will enable the achievement of higher activities than those currently obtained [174,175,177].
Applying enzymes from several microorganisms and the parallel exploitation of the energy from light can correspond to a more effective degradation [178]. MPs and NPs are potentially decomposable with photocatalysis, headed for smaller molecules or additionally mineralised to CO2. Valuable compounds may be formed due to photocatalytic CO2 reduction [41,179], and those with smaller molecular weight could be assimilated into the anabolic pathways of different microorganisms and give rise to complete carbon cycles [180].
Photosynthesis is a biological process that utilises light energy to convert inorganic carbon into organic matter: photoautotrophic microorganisms generally have high photosynthetic and metabolic capacities and can produce a variety of valuable metabolites, such as lipids, carbohydrates, pigments, and proteins [[181], [182], [183]]. By exploiting the coupling with the energy obtained using available light via photosynthesis, the capability to fix CO2, and the ability of selected enzymes to produce desired molecules, microalgae and cyanobacteria may prove to be more advantageous and sustainable as hosts for the photobiocatalytic reactions by removing the need for the external addition of co-factors. Generally, pH and temperature for these approaches have values for the optimal activity of the heterologous enzyme (e.g., pH 7–9 and 30 °C for PETase) and for the optimal function of specific intracellular metabolic pathways (e.g., pH 7.0–7.6 and generally between 20 and 30 °C for model cyanobacteria and microalgae) of the used photosynthetic microorganisms (e.g., energy in form of adenosine triphosphate (ATP) and nicotinamide adenine dinucleotide phosphate (NADPH) from photosynthesis). The methods available for their engineering allow the expression of different heterologous enzymes: the gene expressions can be tuned by either exploiting self-replicative plasmids or integrative vectors under different synthetic and native promoters of varying strength [[184], [185], [186], [187], [188]]. The ability of cyanobacteria to recycle NADPH, using light-driven water oxidation to supply electrons, makes them attractive as photobiocatalysts for different uses [[189], [190], [191], [192], [193], [194], [195], [196], [197]]. Particularly, their capacity to utilise light for the biotransformation of specific substrates into the desired products is a unique advantage compared to heterotrophic bacteria. Some studies also found the reductive ability of in vivo biotransformations using terpenes, hydrocortisone, chalcones, and oxophosphonates [198,199]. Studies of photobiocatalysis were also performed using microalgae like C. reinhardtii [200]. The developments in recombinant cyanobacterial and microalgal applications may facilitate the remediation of plastic pollution, expressing selected enzymes able to degrade these polymers and exploiting light. Especially when employing engineered strains by genetic modification or evolved adaptation, NPs might be considered as substrates for photobiocatalytic reactions: the potential application of light-driven biotransformations, converting nanoscopic plastics particles into valuable products with photosynthetic microorganisms, could lead to an innovative scientific topic of applied biotechnology.
5. Beneficial interactions between heterotrophic bacteria and photosynthetic microorganisms in mixed-species biofilms
Most microorganisms naturally occur in mixed-species communities. This allows them to interact like mutual biofilm development [201,202]. The already mentioned plastispheres, which globally develop on plastics, rely on various interaction mechanisms involving bacteria and microalgae: this was confirmed in plastispheres of the Mediterranean, where such interactions were shown to frequently occur [203]. Sun et al. [202] recently published a detailed review focussing on biofilm for the bioremediation of MPs in contaminated freshwater environments. In the conclusion of their work, these authors mentioned that the method of degrading MPs using biofilms is feasible, even if their degradation is currently not sufficiently significant: previous studies have found that the highest degradation that can occur is approximately 20% [202]. The main reasons are (i) the structural stability of MPs, (ii) the biodegradation process of MPs is carried out in multiple steps which are not simultaneously feasible, and (iii) the fact that it is a slow process which requires a long time to be entirely completed. Future research on biofilm applied towards the degradation of MPs should consider (i) improving the speed and quality of its formation, (ii) better efficiency of its utilisation, and (iii) screening and effectively isolating different functional strains plus culturing them [202].
Future bioremediation approaches using either wild-type or recombinant microalgae and/or cyanobacteria will likely benefit from introducing beneficial bacteria, fungi or archaea in mixed-species biofilms. In this regard, various heterotrophic bacteria can substantially improve the growth and fitness of photosynthetic microorganisms’ populations by providing vitamin B12, phytohormones, minerals, and other important micro-compounds [[204], [205], [206]]. Such beneficial interactions were verified in laboratory conditions, where substantial increases in biomass production were observed when certain microalgae were co-cultivated with specific bacteria [207]. For the biotechnologically relevant algae Scenedesmus vacuolatus and Haematococcus lacustris, up to 14-fold increases in biomass production were observed [207].
Methylobacteria that elicited the growth promotion were subjected to genomic analyses and found to harbour genes involved in the synthesis of vitamins, siderophores, and plant hormones [207]. Although the response process to the widespread phytohormone indole-3-acetic acid (IAA) differs between algae and land plants, this can lead to increased growth in both [208]. Implementing these observations for plastic degradation will require targeted approaches in the next years, evaluating viable applications outside of controlled bioreactors, where inoculation can be precisely regulated to achieve and maintain specific characteristics like certain cell density. Although the exploitable interactions between fungi and microalgae have a relation to plastics bioremediation, the approaches have not yet been extensively investigated. However, recent studies have highlighted the industrial potential of their co-applications for other purposes [[209], [210], [211]]. Indications for species-specific co-occurrence of microalgae and fungi were also found in environmental studies [211], although most have remained un-verified.
Applying microbial communities, Niu et al. [212] and Narciso-Ortiz et al. [76] showed promising achievements towards degraded plastic debris. In detail, Niu et al. [212] recently provided new insights on the natural degradation potential of MPs by microbial communities in rivers. Potential PE/PP degrading bacterial communities were enriched and screened by outdoor and indoor culture experiments for 1150 days in this study. Furthermore, Narciso-Ortiz et al. [76] showed results from the bioprospection of a bacterial consortium composed of four bacteria (Xanthomonas, Acinetobacter bouvetii, Shewanella, and Aquamicrobium lusatiense) for hydrocarbon biodegradation in an airlift bioreactor: testing for ten days, this consortium was able to successfully degrade the maximum diesel concentration at 20 g L−1. This consortium began consuming the substrate from the initial time point, accelerating its consumption before the biomass lag phase finished (0–4 days). Hexadecane was removed by the pure cultures of Acinetobacter bouvetii (72 ± 4%), Xanthomonas sp. (46 ± 4%) and Defluvibacter. Lusatiensis (40 ± 6%) [213]. Mixed cultures showed an enhanced hexadecane removal, particularly with the whole consortium (79 ± 3%) and the combination of Xanthomonas sp. And Acinetobacter bouvetii (74 ± 7%). The role of A. bouveti coincided with that of one of the biosurfactant producers [213]. Furthermore, different strains were isolated from the soil samples, and three of the five isolated colonies could degrade PET: Bacillus muralis, Brevibacterium, and Serratia proteamaculans. PET debris was incubated with each strain for three days. These authors also reported the first visual evidence of PET degradation by an isolated forest-native bacterial strain, showing that Bacillus muralis is the most efficient degrader [76]. Changes in PET surfaces were observed by scanning electron microscopy (SEM), and the treated and control samples of PET were examined using Fourier transform infrared (FTIR) spectroscopy: a difference in transmittance percentage in the carboxylic group was observed between the samples, probably because this corresponds to the site of action for hydrocarbon-degrading enzymes [76]. No sufficient specific enzymatic or other activity values have been reported to compare it with other microorganisms capable of degrading PET. According to the systematic review by Benavides Fernández et al. [134] Bacillus cereus and Bacillus gottheilii are two species belonging to the genus Bacillus that can degrade PET more efficiently than other microorganisms. Still within this genus, A recent study published in 2023 reports the biodegradation of PET by Bacillus safensis YX8 [214]. A review published in 2021 reports the biodegradation of PET by PETase produced by genetically modified microorganisms such as E. coli [143]. Comparing their activities between each other, taking into account the difficulty in doing so considering that the applied conditions are different between these studies, the modified strains expressing engineered PETase by site-directed mutagenesis (often introducing modifications to the amino acid chain to enhance thermal stability, helping in maintaining activity for a longer time) show increased enzymatic activity compared to the wildtype form [142,143].
6. Conclusions
In conclusion, photosynthetic prokaryotic and eukaryotic microorganisms emerge as promising candidates for addressing plastic pollution in terrestrial and aquatic ecosystems. Their inherent advantages, including low cost, scalability, and amenability to genetic modification through mutagenesis, breeding, or transformation, position them as versatile tools for sustainable environmental remediation. Microalgae and cyanobacteria, in particular, exhibit proficiency in reducing nitrogen, phosphate, and other hazardous compounds in wastewater and serve as crucial biomanufacturing platforms for producing several biochemicals. Leveraging sunlight as their primary energy source, these microorganisms present an eco-friendly alternative to traditional systems, significantly reducing the carbon footprint associated with the overall process.
The diverse functional groups within the cell walls of these cells enable effective biosorption of pollutants, showcasing their potential in mitigating plastic pollution. Notably, the process of binding pollutants to their surface can occur independently of their viability, while other aggregates may be actively taken up and biodegraded by living cells. Exploring microorganisms inhabiting plastic-polluted areas worldwide holds great promise for identifying strains capable of facilitating plastic interactions and promoting biodegradation. Towards future scientific studies, targeted research is imperative to unravel the full potential of genetically modified microorganisms, such as cyanobacteria and microalgae, engineered to produce enzymes like PETase and MHETase. urethanase, and alkane hydroxylase. These enzymes could serve as the foundation for transforming MPs and NPs into valuable biomass or specific bioactive compounds, thus closing the loop on plastic waste.
The use of genetically modified organisms (GMOs) in environmental applications has been debated for decades. While GMOs can potentially provide significant benefits, ethical and regulatory concerns must be considered. From an ethical standpoint, the population is generally worried about the potential risks of releasing GMOs into the environment, creating new species that could harm different ecosystems. Additionally, there is a concern that GMOs could have unintended consequences, such as creating new allergens or toxins. From a regulatory standpoint, several agencies oversee the use of GMOs in environmental applications (e.g., in the United States, the Environmental Protection Agency (EPA); in the European Union, the European Food Safety Authority (EFSA)). The direct use of genetically modified cyanobacteria and/or microalgae in natural environments contaminated by MPs and NPs does not satisfy the ethical and regulatory points.
Despite recent strides, the field of microorganism-mediated plastic remediation remains in its infancy, presenting different possibilities for expansion and exciting discoveries. While their efficacy in treating plastic pollutants in terrestrial ecosystems, marine environments, and freshwater basins is not yet fully understood, their demonstrated ability to interact as consortia or in isolation on various plastic waste surfaces underscores the need for broader scientific exploration. Regardless of the promising role of photosynthetic prokaryotic and eukaryotic microorganisms in addressing plastic pollution, some limitations and potential biases need to be considered. For instance, the effectiveness of these microorganisms in breaking down plastics depends on various factors such as the type of plastic, the environmental conditions, and the microbial community. Additionally, using these microorganisms may not be a complete solution to plastic pollution but rather a complementary approach that can be used in conjunction with other strategies. While using GMOs in environmental applications can provide significant benefits, it is important to carefully consider their ethical and regulatory implications. In this direction, the biotechnologies based on modified cyanobacteria and microalgae (e.g., in vivo bioremediation, photobiocatalysis, and in combination with in vitro approaches based on photocatalysts) should be ensured to be utilised safely and responsibly (e.g., bioremediation via bioreactors, where GMOs can degrade and assimilate plastic waste taken from contaminated environments, avoiding environmental issues due to the introduction of new microorganisms in the natural ecosystems).
In the years ahead, widening the scientific niche for the environmental treatment of plastic waste through comprehensive studies on the engineering, interactions, and potential applications of photosynthetic microorganisms promises to unlock novel solutions to the global plastic pollution crisis.
CRediT authorship contribution statement
Giovanni Davide Barone: Conceptualization, Data Curation, Formal Analysis, Investigation, Methodology, Supervision, Validation, Funding Acquisition, Visualization, Writing - Original Draft, Writing - Review & Editing. Andrés RodrÍguez-Seijo, Mattia Parati, Brian Johnston: Data Curation, Formal Analysis, Investigation, Visualization, Writing - Original Draft, Writing - Review & Editing. Elif Erdem, Tomislav Cernava, Zhi Zhu, Xufeng Liu: Writing - Original Draft, Writing - Review & Editing. Ilka Maria Axmann, Peter Lindblad: Supervision, Writing - Original Draft, Writing - Review & Editing. Iza Radecka: Conceptualization, Data Curation, Supervision, Validation, Funding Acquisition, Writing - Original Draft, Writing - Review & Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge the financial support by the University of Graz (Open Access Publishing Agreement). ARS would like to acknowledge the support given through ED431C2021/46-GRC attributed to Universidade de Vigo by Xunta de Galicia and IJC2020-044197-I through the Universidade de Vigo, MCIN/AEI/10.13039/501100011033 and the European Union through “NextGenerationEU/PRTR”. This article/publication is based upon work from COST Action CA20101 Plastics monitoRIng detectiOn RemedIaTion recoverY - PRIORITY, supported by COST (European Cooperation in Science and Technology), www.cost.eu. This work was partially supported the University of Wolverhampton Research Investment Fund (RIF4). The figures were created with BioRender.com.
Contributor Information
Giovanni Davide Barone, Email: giovanni.barone@uni-graz.at.
Iza Radecka, Email: I.Radecka@wlv.ac.uk.
References
- 1.Geyer R., Jambeck J.R., Law K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017;3(7) doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrelle S.B., Ringma J., Law K.L., Monnahan C.C., Lebreton L., McGivern A., Murphy E., Jambeck J.R., Leonard G.H., Hilleary M.A., Eriksen M., Possingham H.P., De Frond H., Gerber L.R., Polidoro B.A., Tahir A., Bernard M., Mallos N.J., Barnes M.D., Rochman C.M. Predicted growth in plastic waste exceeds efforts to mitigate plastic pollution. Science. 2020;369(6510):1515–1518. doi: 10.1126/science.aba3656. [DOI] [PubMed] [Google Scholar]
- 3.Lau W.W., Shiran Y., Bailey R.M., Cook E., Stuchtey M.R., Koskella J., Velis C.A., Godfrey L., Boucher J., Murphy M.B., Thompson R.C., Jankowska E., Castillo Castillo A., Pilditch T.D., Dixon B., Koerselman L., Kosior E., Favoino E., Gutberlet J., Baulch S., Atreya M.E., Fischer D., He K.K., Petit M.M., Sumaila U.R., Neil E., Bernhofen M.V., Lawrence K., Palardy J.E. Evaluating scenarios toward zero plastic pollution. Science. 2020;369(6510):1455–1461. doi: 10.1126/science.aba9475. [DOI] [PubMed] [Google Scholar]
- 4.Larue C., Sarret G., Castillo-Michel H., Pradas Del Real A.E. A critical review on the impacts of nanoplastics and microplastics on aquatic and terrestrial photosynthetic organisms. Small. 2021 doi: 10.1002/smll.202005834. [DOI] [PubMed] [Google Scholar]
- 5.Kane I.A., Clare M.A. Dispersion, accumulation, and the Ultimate fate of microplastics in deep-marine environments: a review and future directions. Front. Earth Sci. 2019;7:80. doi: 10.3389/feart.2019.00080. [DOI] [Google Scholar]
- 6.Prabhu P.P., Pan K., Krishnan J.N. Microplastics: global occurrence, impact, characteristics and sorting. Front. Mar. Sci. 2022;9 doi: 10.3389/fmars.2022.893641. [DOI] [Google Scholar]
- 7.Sethi S., Manuelpillai N., Mandal A., Simpson O., Morrissey H., Ball P., Sharrod-Cole H., Ford C., Whittaker A.C., Drayson M., Race A., Bateman J., Basu S., Cotton J. COVID-19 seroprevalence after the first UK wave of the pandemic and its association with the physical and mental wellbeing of secondary care healthcare workers. Brain Behav. Immun. 2022;24 doi: 10.1016/j.bbih.2022.100492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ammendolia J., Saturno J., Brooks A.L., Jacobs S., Jambeck J.R. An emerging source of plastic pollution: environmental presence of plastic personal protective equipment (PPE) debris related to COVID-19 in a metropolitan city. Environ. Pollut. 2021;269 doi: 10.1016/j.envpol.2020.116160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patrício Silva A.L., Prata J.C., Duarte A.C., Barcelò D., Rocha-Santos T. An urgent call to think globally and act locally on landfill disposable plastics under and after covid-19 pandemic: pollution prevention and technological (Bio) remediation solutions. Chem. Eng. J. (Lausanne) 2021;426 doi: 10.1016/j.cej.2021.131201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng Y., Wu P., Schartup A.T., Zhang Y. Plastic waste release caused by COVID-19 and its fate in the global ocean. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2111530118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.United Nations Environment Programme . Institute for Global Environmental Strategies, United Nations Environment Programme; 2020. Waste Management during the COVID-19 Pandemic from Response to Recovery.https://wedocs.unep.org/20.500.11822/33416 [Google Scholar]
- 12.Hahladakis J.N., Velis C.A., Weber R., Iacovidou E., Purnell P. An overview of chemical additives present in plastics: migration, release, fate and environmental impact during their use, disposal and recycling. J. Hazard Mater. 2018;344:179–199. doi: 10.1016/j.jhazmat.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Ru J., Huo Y., Yang Y. Microbial degradation and valorization of plastic wastes. Front. Microbiol. 2020;11:442. doi: 10.3389/fmicb.2020.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.OECD . OECD Publishing; Paris: 2022. Global Plastics Outlook: Economic Drivers, Environmental Impacts and Policy Options. [DOI] [Google Scholar]
- 15.Napper I.E., Thompson R.C. Environmental deterioration of biodegradable, oxo-biodegradable, compostable, and conventional plastic carrier bags in the sea, soil, and open-air over a 3-year period. Environ. Sci. Technol. 2019;53(9):4775–4783. doi: 10.1021/acs.est.8b06984. [DOI] [PubMed] [Google Scholar]
- 16.Gu J.D. Biodegradability of plastics: the issues, recent advances, and future perspectives. Environ. Sci. Pollut. Res. 2021;28:1278–1282. doi: 10.1007/s11356-020-11501-9. [DOI] [PubMed] [Google Scholar]
- 17.Zimmermann L., Dombrowski A., Völker C., Wagner M. Are bioplastics and plant-based materials safer than conventional plastics? In vitro toxicity and chemical composition. Environ. Int. 2020;145 doi: 10.1016/j.envint.2020.106066. [DOI] [PubMed] [Google Scholar]
- 18.Badola N., Chauhan J.S. In: Bioremediation of Environmental Pollutants. Suyal D.C., Soni R., editors. Springer; Cham: 2022. Waste management: challenges and opportunities; pp. 1–23. [DOI] [Google Scholar]
- 19.Barati B., Zafar F.F., Wang S. In: Waste-to-Energy. Abomohra A.E.F., Wang Q., Huang J., editors. Springer; Cham: 2022. Different waste management methods, applications, and limitations; pp. 21–58. [DOI] [Google Scholar]
- 20.Ali S., Bukhari D.A., Rehman A. Call for biotechnological approach to degrade plastic in the era of COVID-19 pandemic. Saudi J. Biol. Sci. 2023;30:3. doi: 10.1016/j.sjbs.2023.103583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Debrah J.K., Teye G.K., Dinis M.A.P. Barriers and challenges to waste management hindering the circular economy in sub-saharan africa. Urban Sci. 2022;6:57. doi: 10.3390/urbansci6030057. [DOI] [Google Scholar]
- 22.Jagun Z.T., Daud D., Ajayi O.M., Samsudin S., Jagun Jubril A., Abdul Rahman M.S. Waste management practices in developing countries: a socio-economic perspective. Environ. Sci. Pollut. Res. 2023;30:116644–116655. doi: 10.1007/s11356-022-21990-5. [DOI] [PubMed] [Google Scholar]
- 23.Kibria M.G., Masuk N.I., Safayet R., Nguyen H.Q., Mourshed M. Plastic waste: challenges and opportunities to mitigate pollution and effective management. Int. J. Environ. Res. 2023;17:20. doi: 10.1007/s41742-023-00507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberbeckmann S., Bartosik D., Huang S., Werner J., Hirschfeld C., Wibberg D., Heiden S.E., Bunk B., Overmann J., Becher D., Kalinowski J., Schweder T., Labrenz M., Markert S. Genomic and proteomic profiles of biofilms on microplastics are decoupled from artificial surface properties. Environ. Microbiol. 2021;23:3099–3115. doi: 10.1111/1462-2920.15531. [DOI] [PubMed] [Google Scholar]
- 25.Rosato A., Barone M., Negroni A., Brigidi P., Fava F., Biagi E., Candela M., Zanaroli G. Bacterial colonization dynamics of different microplastic types in an anoxic salt marsh sediment and impact of adsorbed polychlorinated biphenyls on the plastisphere. Environ. Pollut. 2022;315 doi: 10.1016/j.envpol.2022.120411. [DOI] [PubMed] [Google Scholar]
- 26.Wang P., Liu J., Han S., Wang Y., Duan Y., Liu T., Hou L., Zhang Z., Li L., Lin Y. Polyethylene mulching film degrading bacteria within the plastisphere: Co-culture of plastic degrading strains screened by bacterial community succession. J. Hazard Mater. 2023;442 doi: 10.1016/j.jhazmat.2022.130045. [DOI] [PubMed] [Google Scholar]
- 27.Koh J., Bairoliya S., Salta M., Cho Z.T., Fong J., Neo M.L., Cragg S., Cao B. Sediment-driven plastisphere community assembly on plastic debris in tropical coastal and marine environments. Environ. Int. 2023;179 doi: 10.1016/j.envint.2023.108153. [DOI] [PubMed] [Google Scholar]
- 28.Miao L., Li W., Adyel T.M., Yao Y., Deng Y., Wu J., Zhou Y., Yu Y., Hou J. Spatio-temporal succession of microbial communities in plastisphere and their potentials for plastic degradation in freshwater ecosystems. Water Res. 2023;229 doi: 10.1016/j.watres.2022.119406. [DOI] [PubMed] [Google Scholar]
- 29.Kirstein I.V., Kirmizi S., Wichels A., Garin-Fernandez A., Erler R., Löder M., Gerdts G. Dangerous hitchhikers? Evidence for potentially pathogenic Vibrio spp. on microplastic particles. Mar. Environ. Res. 2016;120:1–8. doi: 10.1016/j.marenvres.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Tavelli R., Callens M., Grootaert C., Abdallah M.F., Rajkovic A. Foodborne pathogens in the plastisphere: can microplastics in the food chain threaten microbial food safety? Trends Food Sci. Technol. 2022;129:1–10. doi: 10.1016/j.tifs.2022.08.021. [DOI] [Google Scholar]
- 31.Abomohra A., Hanelt D. Recent advances in micro-/nanoplastic (MNPs) removal by microalgae and possible integrated routes of energy recovery. Microorganisms. 2022;10:12. doi: 10.3390/microorganisms10122400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khandare S.D., Agrawal D., Mehru N., Chaudhary D.R. Marine bacterial based enzymatic degradation of low-density polyethylene (LDPE) plastic. J. Environ. Chem. 2022;10:3. doi: 10.1016/j.jece.2022.107437. [DOI] [Google Scholar]
- 33.Skariyachan S., Taskeen N., Preethi Kishore A., Venkata Krishna B. Recent advances in plastic degradation – from microbial consortia-based methods to data sciences and computational biology driven approaches. J. Hazard Mater. 2022;426 doi: 10.1016/j.jhazmat.2021.128086. [DOI] [PubMed] [Google Scholar]
- 34.Branson Y., Söltl S., Buchmann C., Wei R., Schaffert L., Badenhorst C.P.S., Reisky L., Jäger G., Bornscheuer U.W. Urethanases for the enzymatic hydrolysis of low molecular weight carbamates and the recycling of polyurethanes. Angew. Chem. Int. Ed. 2023;62 doi: 10.1002/anie.202216220. [DOI] [PubMed] [Google Scholar]
- 35.Geppner L., Ramer G., Tomasetig D., Grundhöfer L., Küss J., Kaup M., Henjakovic M. A novel enzymatic method for isolation of plastic particles from human blood. Environ. Toxicol. Pharmacol. 2023;104 doi: 10.1016/j.etap.2023.104318. [DOI] [PubMed] [Google Scholar]
- 36.Nagendranatha Reddy C., Kallem P., Mounika K.V.S.S.N., Muqeet A., Raj J.C.J., Aishwarya C.V.S., Kumar Gupta R., Polisetti V., Mishra B., Yadavalli R., Kumar Mandal S., Hedenqvist M.S., Banat F. Review of microplastic degradation: understanding metagenomic approaches for microplastic degrading organisms. Polym. Test. 2023;128 doi: 10.1016/j.polymertesting.2023.108223. [DOI] [Google Scholar]
- 37.Shaji A., Kamalesh R., Dinakarkumar Y., Saravanan A., Arokiyaraj S., Palaniappan Mani H., Madhuri Veera H., Babu Muthu D., Ramakrishnan G., Romauld S. Microbial degradation of marine plastic debris: a comprehensive review on the environmental effects, disposal, and biodegradation. Biochem. Eng. J. 2024;201 doi: 10.1016/j.bej.2023.109133. [DOI] [Google Scholar]
- 38.Zhang K., Su J., Xiong X., Wu X., Wu C., Liu J. Microplastic pollution of lakeshore sediments from remote lakes in Tibet plateau, China. Environ. Pollut. 2016;219:450–455. doi: 10.1016/j.envpol.2016.05.048. [DOI] [PubMed] [Google Scholar]
- 39.Winkler A., Santo N., Ortenzi M.A., Bolzoni E., Bacchetta R., Tremolada P. Does mechanical stress cause microplastic release from plastic water bottles? Water Res. 2019;166 doi: 10.1016/j.watres.2019.115082. [DOI] [PubMed] [Google Scholar]
- 40.Hebner T.S., Maurer-Jones M.A. Characterizing microplastic size and morphology of photodegraded polymers placed in simulated moving water conditions. Environ. Sci.: Process. Impacts. 2020;22:398–407. doi: 10.1039/C9EM00475K. [DOI] [PubMed] [Google Scholar]
- 41.He J., Han L., Wang F., Ma C., Cai Y., Ma W., Genbo Xu E., Xing B., Yang Z. Photocatalytic strategy to mitigate microplastic pollution in aquatic environments: promising catalysts, efficiencies, mechanisms, and ecological risks. Crit. Rev. Environ. Sci. Technol. 2023;4:504–526. doi: 10.1080/10643389.2022.2072658. [DOI] [Google Scholar]
- 42.Ariza-Tarazona M.C., Villarreal-Chiu J.F., Barbieri V., Siligardi C., Cedillo-Gonzalez E.I. New strategy for microplastic degradation: green photocatalysis using a protein-based porous N-TiO2 semiconductor. Ceram. 2019;45(7):9618–9624. doi: 10.1016/j.ceramint.2018.10.208. [DOI] [Google Scholar]
- 43.Liu P., Qian L., Wang H., Zhan X., Lu K., Gu C., Gao S. New Insights into the aging behavior of microplastics accelerated by advanced oxidation processes. Environ. Sci. Technol. 2019;53(7):3579–3588. doi: 10.1021/acs.est.9b00493. [DOI] [PubMed] [Google Scholar]
- 44.Mao R., Lang M., Yu X., Wu R., Yang X., Guo X. Aging mechanism of microplastics with UV irradiation and its effects on the adsorption of heavy metals. J. Hazard Mater. 2020;393 doi: 10.1016/j.jhazmat.2020.122515. [DOI] [PubMed] [Google Scholar]
- 45.Padervand M., Lichtfouse E., Robert D., Wang C. Removal of microplastics from the environment. A review. Environ. Chem. Lett. 2020;18(3):807–828. doi: 10.1007/s10311-020-00983-1. [DOI] [Google Scholar]
- 46.Miri S., Saini R., Davoodi S.M., Pulicharla R., Brar S.K., Magdouli S. Biodegradation of microplastics: better late than never. Chemosphere. 2022;286 doi: 10.1016/j.chemosphere.2021.131670. [DOI] [PubMed] [Google Scholar]
- 47.Sun Q., Li J., Wang C., Chen A., You Y., Yang S., Liu H., Jiang G., Wu Y., Li Y. Research progress on distribution, sources, identification, toxicity, and biodegradation of microplastics in the ocean, freshwater, and soil environment. Front. Environ. Sci. Eng. 2022;16(1):1. doi: 10.1007/s11783-021-1429-z. [DOI] [Google Scholar]
- 48.Chen J., Wu J., Sherrell P.C., Chen J., Wang H., Zhang W., Yang J. How to build a microplastics-free environment: strategies for microplastics degradation and plastics recycling. Adv. Sci. 2022;9 doi: 10.1002/advs.202103764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corcoran P.L. In: Handbook of Microplastics in the Environment. Rocha-Santos T., Costa M., Mouneyrac C., editors. Springer International Publishing; 2020. Degradation of microplastics in the environment; pp. 1–12. [DOI] [Google Scholar]
- 50.Rodríguez-Narvaez O.M., Goonetilleke A., Perez L., Bandala E.R. Engineered technologies for the separation and degradation of microplastics in water: a review. Chem. Eng. J. 2021;414 doi: 10.1016/j.cej.2021.128692. [DOI] [Google Scholar]
- 51.Zhou D., Chen J., Wu J., Yang J., Wang H. Biodegradation and catalytic-chemical degradation strategies to mitigate microplastic pollution. Sustain. Mater. Technol. 2021;28 doi: 10.1016/j.susmat.2021.e00251. [DOI] [Google Scholar]
- 52.Boschi A., Scieuzo C., Salvia R., Arias C.F., Peces Perez R., Bertocchini F., Falabella P. Beyond microbial biodegradation: plastic degradation by Galleria mellonella. J. Polym. Environ. 2023 doi: 10.1007/s10924-023-03084-6. In press. [DOI] [Google Scholar]
- 53.Horton A.A., Walton A., Spurgeon D.J., Lahive E., Svendsen C. Microplastics in freshwater and terrestrial environments: evaluating the current understanding to identify the knowledge gaps and future research priorities. Sci. Total Environ. 2017;586:127–141. doi: 10.1016/j.scitotenv.2017.01.190. [DOI] [PubMed] [Google Scholar]
- 54.Hartmann N.B., Hüffer T., Thompson R.C., Hassellöv M., Verschoor A.J., Daugaard A.E., Rist S., Karlsson T., Brennholt N., Cole M., Herrling M.P., Hess M.C., Ivleva N.P., Lusher A.L., Wagner M. Are we speaking the same language? Recommendations for a definition and categorization framework for plastic debris. Environ. Sci. Technol. 2019;53(3):1039–1047. doi: 10.1021/acs.est.8b05297. [DOI] [PubMed] [Google Scholar]
- 55.Feldman D. Polymer weathering: photo-oxidation. J. Polym. Environ. 2002;10:163–173. doi: 10.1023/A:1021148205366. [DOI] [Google Scholar]
- 56.Bacha A.U.R., Nabi I., Zhang L. Mechanisms and the engineering approaches for the degradation of microplastics. ACS ES&T Eng. 2021;1(11):1481–1501. doi: 10.1021/acsestengg.1c00216. [DOI] [Google Scholar]
- 57.Viel T., Manfra L., Zupo V., Libralato G., Cocca M., Costantini M. Biodegradation of plastics induced by marine organisms: future perspectives for bioremediation approaches. Polymers. 2023;15:2673. doi: 10.3390/polym15122673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paço A., Duarte K., da Costa J.P., Santos P.S., Pereira R., Pereira M.E., Freitas A.C., Duarte A.C., Rocha-Santos T.A. Biodegradation of polyethylene microplastics by the marine fungus Zalerion maritimum. Sci. Total Environ. 2017;586:10–15. doi: 10.1016/j.scitotenv.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 59.Kumar M., Xiong X., He M., Tsang D.C., Gupta J.K., Khan E., Harrad S., Hou D., Ok Y.S., Bolan N.S. Microplastics as pollutants in agricultural soils. Environ. Pollut. 2020;265 doi: 10.1016/j.envpol.2020.114980. [DOI] [PubMed] [Google Scholar]
- 60.Roy P., Mohanty A.K., Misra M. Microplastics in ecosystems: their implications and mitigation pathways. Environ. Sci.: Adv. 2022;1:9–29. doi: 10.1039/D1VA00012H. [DOI] [Google Scholar]
- 61.Hadian-Ghazvini S., Hooriabad Saboor F., Safaee Ardekani L. In: Microplastics Pollution in Aquatic Media. Environmental Footprints and Eco-Design of Products and Processes. Sillanpää M., Khadir A., Muthu S.S., editors. Springer; Singapore: 2022. Bioremediation techniques for microplastics removal; pp. 327–377. [DOI] [Google Scholar]
- 62.Correia Prata J., da Costa J.P., Lopes I., Duarte A.C., Rocha-Santos T. Effects of microplastics on microalgae populations: a critical review. Sci. Total Environ. 2019;665:400–405. doi: 10.1016/j.scitotenv.2019.02.132. [DOI] [PubMed] [Google Scholar]
- 63.Jakubowicz I. Evaluation of degradability of biodegradable polyethylene (PE) Polym. Degrad. Stabil. 2003;80(1):39–43. doi: 10.1016/S0141-3910(02)00380-4. [DOI] [Google Scholar]
- 64.Chamas A., Moon H.H., Zheng J., Qiu Y., Tabassum T., Jang J.H., Abu-Omar M.M., Scott S.L., Suh S. Degradation rates of plastics in the environment. ACS Sustain. Chem. Eng. 2020;8:3494–3511. doi: 10.1021/acssuschemeng.9b06635. [DOI] [Google Scholar]
- 65.Lv M., Jiang B., Xing Y., Ya H., Zhang T., Wang X. Recent advances in the breakdown of microplastics: strategies and future prospectives. Environ. Sci. Pollut. Res. 2022;29:65887–65903. doi: 10.1007/s11356-022-22004-0. [DOI] [PubMed] [Google Scholar]
- 66.Balani K., Verma V., Agarwal A., Narayan R. first ed. The American Ceramic Society; 2015. Biosurfaces: A Materials Science and Engineering Perspective. Published 2015 by John Wiley & Sons, Inc. [Google Scholar]
- 67.Ivleva N.P. Chemical analysis of microplastics and nanoplastics: challenges, advanced methods, and perspectives. Chem. Rev. 2021;121(19):11886–11936. doi: 10.1021/acs.chemrev.1c00178. [DOI] [PubMed] [Google Scholar]
- 68.Jaiswal S., Sharma B., Shukla P. Integrated approaches in microbial degradation of plastics. Environ. Technol. Innov. 2020;17 doi: 10.1016/j.eti.2019.100567. [DOI] [Google Scholar]
- 69.Serrano-Ruiz H., Martin-Closas L., Pelacho A.M. Biodegradable plastic mulches: impact on the agricultural biotic environment. Sci. Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141228. [DOI] [PubMed] [Google Scholar]
- 70.Amaral-Zettler L.A., Zettler E.R., Mincer T.J. Ecology of the plastisphere. Nat. Rev. Microbiol. 2020;18:139–151. doi: 10.1038/s41579-019-0308-0. [DOI] [PubMed] [Google Scholar]
- 71.Lear G., Kingsbury J.M., Franchini S., Gambarini V., Maday S.D., Wallbank J.A., Weaver L., Pantos O. Plastics and the microbiome: impacts and solutions. Environ. Microbiol. 2021;16:2. doi: 10.1186/s40793-020-00371-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guasch H., Bernal S., Bruno D., Almroth B.C., Cochero J., Corcoll N., Cornejo D., Gacia E., Kroll A., Lavoie I., Ledesma J.L., Lupon A., Margenat H., Morin S., Navarro E., Ribot M., Riis T., Schmitt-Jansen M., Tlili A., Martí E. Interactions between microplastics and benthic biofilms in fluvial ecosystems: knowledge gaps and future trends. Freshw. Sci. 2022;41:3. doi: 10.1086/721472. [DOI] [Google Scholar]
- 73.Denis B., Pérez O.A., Lizardi-Jiménez M.A., Dutta A. Numerical evaluation of direct interfacial uptake by a microbial consortium in an airlift bioreactor. Int. Biodeterior. Biodegrad. 2016;119:542–551. doi: 10.1016/j.ibiod.2016.08.012. [DOI] [Google Scholar]
- 74.Varjani S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017;223:277–286. doi: 10.1016/j.biortech.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 75.Koshti R., Mehta L., Samarth N. Biological recycling of polyethylene terephthalate: a mini-review. J. Polym. Environ. 2018;26(8):3520–3529. doi: 10.1007/s10924-018-1214-7. [DOI] [Google Scholar]
- 76.Narciso-Ortiz L., Coreño-Alonso A., Mendoza-Olivares D., Lucho-Constantino C.A., Lizardi-Jiménez M.A. Baseline for plastic and hydrocarbon pollution of rivers, reefs, and sediment on beaches in Veracruz State, México, and a proposal for bioremediation. Environ. Sci. Pollut. Res. 2020;18:23035–23047. doi: 10.1007/s11356-020-08831-z. [DOI] [PubMed] [Google Scholar]
- 77.Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y., Toyohara K., Miyamoto K., Kimura Y. A bacterium that degrades and assimilates poly(ethylene terephthalate) Science. 2016;351:1196–1199. doi: 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- 78.Zhang Z., Peng H., Yang D., Zhang G., Zhang J., Ju F. Polyvinyl chloride degradation by a bacterium isolated from the gut of insect larvae. Nat. Commun. 2022;13:5360. doi: 10.1038/s41467-022-32903-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cazaudehore G., Monlau F., Gassie C., Lallement A., Guyoneaud R. Active microbial communities during biodegradation of biodegradable plastics by mesophilic and thermophilic anaerobic digestion. J. Hazard Mater. 2023;443 doi: 10.1016/j.jhazmat.2022.130208. [DOI] [PubMed] [Google Scholar]
- 80.Moog D., Schmitt J., Senger J., Zarzycki J., Rexer K.-H., Linne U., Erb T., Maier U.G. Using a marine microalga as a chassis for polyethylene terephthalate (PET) degradation. Microb. Cell Factories. 2019;18:171. doi: 10.1186/s12934-019-1220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim J.W., Park S.B., Tran Q.G., Cho D.H., Chou D.Y., Lee Y.J., Kim H.S. Functional expression of polyethylene terephthalate-degrading enzyme (PETase) in green microalgae. Microb. Cell Factories. 2020;19:97. doi: 10.1186/s12934-020-01355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Z., Xiao Y., Weber G., Wei R., Wang Z. Yeast cell surface display of bacterial PET hydrolase as a sustainable biocatalyst for the degradation of polyethylene terephthalate. Methods Enzymol. 2021;648:457–477. doi: 10.1016/bs.mie.2020.12.030. [DOI] [PubMed] [Google Scholar]
- 83.Hachisuka S.I., Nishii T., Yoshida S. Development of a targeted gene disruption system in the poly(ethylene terephthalate)-degrading bacterium Ideonella sakaiensis and its applications to PETase and MHETase genes. Appl. Environ. Microbiol. 2021;87(18) doi: 10.1128/AEM.00020-21. 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brandenberg O.F., Schubert O.T., Kruglyak L. Towards synthetic PETtrophy: engineering Pseudomonas putida for concurrent polyethylene terephthalate (PET) monomer metabolism and PET hydrolase expression. Microb. Cell Factories. 2022;21:119. doi: 10.1186/s12934-022-01849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Z., Wang Y., Cheng Y., Wang X., Tong S., Yang H., Wang Z. Efficient biodegradation of highly crystallized polyethylene terephthalate through cell surface display of bacterial PETase. Sci. Total Environ. 2020;709 doi: 10.1016/j.scitotenv.2019.136138. [DOI] [PubMed] [Google Scholar]
- 86.Tournier V., Topham C.M., Gilles A., David B., Folgoas C., Moya-Leclair E., Kamionka E., Desrousseaux M.-L., Texier H., Gavalda S., Cot M., Guémard E., Dalibey M., Nomme J., Cioci G., Barbe S., Chateau M., André I., Duquesne S., Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020;580:216–219. doi: 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- 87.Maity W., Maity S., Bera S., Roy A. Emerging roles of PETase and MHETase in the biodegradation of plastic wastes. Appl. Biochem. Biotechnol. 2021;193(8):2699–2716. doi: 10.1007/s12010-021-03562-4. [DOI] [PubMed] [Google Scholar]
- 88.Yoshida S., Hiraga K., Taniguchi I., Oda K. Ideonella sakaiensis, PETase, and MHETase: from identification of microbial PET degradation to enzyme characterization. Methods Enzymol. 2021;648:187–205. doi: 10.1016/bs.mie.2020.12.007. [DOI] [PubMed] [Google Scholar]
- 89.Puspitasari N., Tsai S.L., Lee C.K. Class I hydrophobins pretreatment stimulates PETase for monomers recycling of waste PETs. Int. J. Biol. Macromol. 2021;15(176):157–164. doi: 10.1016/j.ijbiomac.2021.02.026. [DOI] [PubMed] [Google Scholar]
- 90.von Haugwitz G., Han X., Pfaff L., Li Q., Wei H., Gao J., Methling K., Ao Y., Brack Y., Mican J., Feiler C.G., Weiss M.S., Bednar D., Palm G.J., Lalk M., Lammers M., Damborsky J., Weber G., Liu W., Bornscheuer U.T., Wei R. Structural insights into (Tere)phthalate-Ester hydrolysis by a carboxylesterase and its role in promoting PET depolymerization. ACS Catal. 2022;12(24):15259–15270. doi: 10.1021/acscatal.2c03772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barone G.D., Ferizović D., Biundo A., Lindblad P. Hints at the applicability of microalgae and cyanobacteria for the biodegradation of plastics. Sustainability. 2020;12 doi: 10.3390/su122410449. [DOI] [Google Scholar]
- 92.Atanasova N., Stoitsova S., Paunova-Krasteva T., Kambourova M. Plastic degradation by extremophilic bacteria. Int. J. Mol. Sci. 2021;22(11):5610. doi: 10.3390/ijms22115610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barone G.D., Cernava T., Ullmann L., Liu J., Lio E., Germann A.T., Nakielski A., Russo D.A., Chavkin T., Knufmann K., Tripodi F., Coccetti P., Secundo F., Fu P., Pflegeri B., Axmann I.M., Lindblad P. Recent developments in the production and utilization of photosynthetic microorganisms for food applications. Heliyon. 2023;9(4) doi: 10.1016/j.heliyon.2023.e14708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rasala B.A., Mayfield S.P. Photosynthetic biomanufacturing in green algae; production of recombinant proteins for industrial, nutritional, and medical uses. Photosynth. Res. 2015;123(3):227–239. doi: 10.1007/s11120-014-9994-7. [DOI] [PubMed] [Google Scholar]
- 95.Acién F.G., Gomez-Serrano C., Morales-Amaral M.M., Fernández Sevilla J.M., Molina-Grima E. Wastewater treatment using microalgae: how realistic a contribution might it be to significant urban wastewater treatment? Appl. Microbiol. 2016;100:9013–9022. doi: 10.1007/s00253-016-7835-7. [DOI] [PubMed] [Google Scholar]
- 96.Plöhn M., Spain O., Sirin S., Silva M., Escudero-Oñate C., Ferrando-Climent L., Allahverdiyeva Y., Funk C. Wastewater treatment by microalgae. Physiol. Plantarum. 2021;173(2):568–578. doi: 10.1111/ppl.13427. [DOI] [PubMed] [Google Scholar]
- 97.Michalak I., Chojnacka K., Witek-Krowiak A. State of the art for the biosorption process—a review. Appl. Biochem. 2013;170:1389–1416. doi: 10.1007/s12010-013-0269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Spain O., Plöhn M., Funk C. The cell wall of green microalgae and its role in heavy metal removal. Physiol. Plantarum. 2021;173(2):526–535. doi: 10.1111/ppl.13405. [DOI] [PubMed] [Google Scholar]
- 99.Blázquez-Sánchez P., Engelberger F., Cifuentes-Anticevic J., Sonnendecker C., Griñén A., Reyes J., Díez B., Guixé V., Richter P.K., Zimmermann W., Ramírez-Sarmiento C.A. Antarctic polyester hydrolases degradealiphatic and aromatic polyesters at moderate temperatures. Appl. Environ. Microbiol. 2022;88 doi: 10.1128/AEM.01842-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roberts C., Edwards S., Vague M., León-Zayas R., Scheffer H., Chan G., Swartz N.A., Mellies J.L. Environmental consortium containing Pseudomonas and Bacillus species synergistically degrades polyethylene terephthalate plastic. mSphere. 2020;6 doi: 10.1128/msphere.01151-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wallace P.W., Haernvall K., Ribitsch D., Zitzenbacher S., Schittmayer M., Steinkellner G., Gruber K., Guebitz G.M., Birner-Gruenberger R. PpEst is a novel PBAT degrading polyesterase identified by proteomic screening of Pseudomonas pseudoalcaligenes. Appl. Microbiol. Biotechnol. 2017;101(6):2291–2303. doi: 10.1007/s00253-016-7992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gewert B., Plassmann M.M., MacLeod M. Pathways for degradation of plastic polymers floating in the marine environment. Environ. Sci.: Process. Impacts. 2015;17(9):1513–1521. doi: 10.1039/C5EM00207A. [DOI] [PubMed] [Google Scholar]
- 103.Ali S.S., Elsamahy T., Al-Tohamy R., Zhu D., Mahmoud Y.A., Koutra E., Metwally M.A., Kornaros M., Sun J. Plastic wastes biodegradation: mechanisms, challenges and future prospects. Sci. Total Environ. 2021;780 doi: 10.1016/j.scitotenv.2021.146590. [DOI] [PubMed] [Google Scholar]
- 104.Lear G., Maday S.D.M., Gambarini V., Northcott G., Abbel R., Kingsbury J.M., Weaver L., Wallbank J.A., Pantos O. Microbial abilities to degrade global environmental plastic polymer waste are overstated. Environ. Res. Lett. 2022;17 doi: 10.1088/1748-9326/ac59a7. [DOI] [Google Scholar]
- 105.Magnin A., Pollet E., Phalip V., Avérous L. Evaluation of biological degradation of polyurethanes. Biotechnol. Adv. 2020;39 doi: 10.1016/j.biotechadv.2019.107457. [DOI] [PubMed] [Google Scholar]
- 106.Solano G., Rojas-Gätjens D., Rojas-Jimenez K., Chavarría M., Romero R.M. Biodegradation of plastics at home composting conditions. Environ. Chall. 2022;7 doi: 10.1016/j.envc.2022.100500. [DOI] [Google Scholar]
- 107.Sangale M.K., Shahnawaz M., Ade A.B. Potential of fungi isolated from the dumping sites mangrove rhizosphere soil to degrade polythene. Sci. Rep. 2019;9:5390. doi: 10.1038/s41598-019-41448-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kumari A., Rajput V.D., Mandzhieva S.S., Rajput S., Minkina T., Kaur R., Sushkova S., Kumari P., Ranjan A., Kalinitchenko V.P., Glinushkin A.P. Microplastic pollution: an emerging threat to terrestrial plants and insights into its remediation strategies. Plants. 2022;11:340. doi: 10.3390/plants11030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mishra A., Gupta J., Kumari T., Pal R., Thakur I.S. Unravelling the attributes of novel cyanobacteria Jacksonvillea sp. ISTCYN1 by draft genome sequencing. Bioresour. Technol. 2021;337 doi: 10.1016/j.biortech.2021.125473. [DOI] [PubMed] [Google Scholar]
- 110.Zettler E.R., Mincer T.J., Amaral-Zettler L.A. Life in the "plastisphere": microbial communities on plastic marine debris. Environ. Sci. Technol. 2013;47(13):7137–7146. doi: 10.1021/es401288x. [DOI] [PubMed] [Google Scholar]
- 111.Nava V., Matias M.G., Castillo-Escrivà A., Messyasz B., Leoni B. Microalgae colonization of different microplastic polymers in experimental mesocosms across an environmental gradient. Global Change Biol. 2022;28:1402–1413. doi: 10.1111/gcb.15989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fazey F.M.C., Ryan P.G. Debris size and buoyancy influence the dispersal distance of stranded litter. Mar. Pollut. Bull. 2016;110(1):371–377. doi: 10.1016/j.marpolbul.2016.06.039. [DOI] [PubMed] [Google Scholar]
- 113.Kalogerakis N., Karkanorachaki K., Kalogerakis G.C., Triantafyllidi E.I., Gotsis A.D., Partsinevelos P., Fava F. Microplastics generation: onset of fragmentation of polyethylene films in marine environment mesocosms. Front. Mar. Sci. 2017;4:84. doi: 10.3389/fmars.2017.00084. [DOI] [Google Scholar]
- 114.De Tender C.A., Devriese L.I., Haegeman A., Maes S., Ruttink T., Dawyndt P. Bacterial community profiling of plastic litter in the Belgian part of the North Sea. Environ. Sci. Technol. 2015;49(16):9629–9638. doi: 10.1021/acs.est.5b01093. [DOI] [PubMed] [Google Scholar]
- 115.Bryant J.A., Clemente T.M., Viviani D.A., Fong A.A., Thomas K.A., Kemp P., Karl D.M., White A.E., DeLong E.F. Diversity and activity of communities inhabiting plastic debris in the north pacific gyre. mSystems. 2016;1(3) doi: 10.1128/mSystems.00024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dussud C., Meistertzheim A.L., Conan P., Pujo-Pay M., George M., Fabre P., Coudane J., Higgs P., Elineau A., Pedrotti M.L., Gorsky G., Ghiglione J.F. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ. Pollut. 2018;236:807–816. doi: 10.1016/j.envpol.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 117.Ras M., Steyer J.P., Bernard O. Temperature effect on microalgae: a crucial factor for outdoor production. Rev. Environ. Sci. Biotechnol. 2013;12:153–164. doi: 10.1007/s11157-013-9310-6. [DOI] [Google Scholar]
- 118.Gonçalves A.L., Pires J.C., Simoes M. The effects of light and temperature on microalgal growth and nutrient removal: an experimental and mathematical approach. RSC Adv. 2016;6:22896–22907. doi: 10.1039/C5RA26117A. [DOI] [Google Scholar]
- 119.Sobolewska E., Borowski S., Nowicka-Krawczyk P., Jurczak T. Growth of microalgae and cyanobacteria consortium in a photobioreactor treating liquid anaerobic digestate from vegetable waste. Sci. Rep. 2023;13(1) doi: 10.1038/s41598-023-50173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sarmah P., Rout J. Algal colonization on polythene carry bags in a domestic solid waste dumping site of Silchar town in Assam. Phykos. 2018;48:67–77. doi: 10.1016/B978-0-12-819311-2.00022-X. [DOI] [Google Scholar]
- 121.Bhuyar P., Sundararaju S., Feng H., Rahim M.H., Muniyasamy S., Maniam G.P., Govindan N. Evaluation of microalgae's plastic biodeterioration property by a consortium of Chlorella sp. and cyanobacteria sp. Environ. Res. Eng. Manag. 2021;77(3):86–98. doi: 10.5755/j01.erem.77.3.25317. [DOI] [Google Scholar]
- 122.MacLean J., Mayanna S., Benning L.G., Horn F., Bartholomäus A., Wiesner Y., Wagner D., Liebner S. The terrestrial plastisphere: diversity and polymer-colonizing potential of plastic-associated microbial communities in soil. Microorganisms. 2021;9:1876. doi: 10.3390/microorganisms9091876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sarmah P., Rout J. In: Advances in Cyanobacterial Biology. Singh P.K., Kumar A., Singh V.K., Shrivastava A.K., editors. Elsevier; 2020. Role of algae and cyanobacteria in bioremediation: prospects in polyethylene biodegradation; pp. 333–349. [DOI] [Google Scholar]
- 124.Chia W.Y., Tang D.Y.Y., Khoo K.S., Lup A.N.K., Chew K.W. Nature's fight against plastic pollution: algae for plastic biodegradation and bioplastics production. Environ. Sci. Ecotechnol. 2020;4 doi: 10.1016/j.ese.2020.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Khan M.J., Singh R., Shewani K., Shukla P., Bhaskar P.V., Joshi K.B., Vinayak V. Exopolysaccharides directed embellishment of diatoms triggered on plastics and other marine litter. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-74801-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sun Y., Shi J., Wang X., Ding C., Wang J. Deciphering the mechanisms shaping the plastisphere microbiota in soil. mSystems. 2022;7(4) doi: 10.1128/msystems.00352-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sarmah P., Rout J. Colonisation of Oscillatoria on submerged polythenes in domestic sewage water of Silchar town, Assam (India) J. Algal Biomass Utln. 2017;8(4):135–144. [Google Scholar]
- 128.Das G., Bordoloi N.K., Rai S.K., Mukherjee A.K., Karak N. Biodegradable and biocompatible epoxidized vegetable oil modified thermostable poly(vinyl chloride): thermal and performance characteristics post biodegradation with Pseudomonas aeruginosa and Achromobacter sp. J. Hazard Mater. 2012;209–210:434–442. doi: 10.1016/j.jhazmat.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 129.Groh K.J., Backhaus T., Carney-Almroth B., Geueke B., Inostroza P.A., Lennquist A., Leslie H.A., Maffini M., Slunge D., Trasande L., Warhurst A.M., Muncke J. Overview of known plastic packaging-associated chemicals and their hazards. Sci. Total Environ. 2019;651:3253–3268. doi: 10.1016/j.scitotenv.2018.10.015. [DOI] [PubMed] [Google Scholar]
- 130.Kotova I.B., Taktarova Y.V., Tsavkelova E.A., Egorova M.A., Malakhova D.V., Shirinkina L.I., Sokolova T.G., Bonch-Osmolovskaya E.A. Microbial degradation of plastics and approaches to make it more efficient. Microbiology. 2021;90:671–701. doi: 10.1134/S0026261721060084. [DOI] [Google Scholar]
- 131.Brdlík P., Novák J., Borůvka M., Běhálek L., Lenfeld P. The influence of plasticizers and accelerated ageing on biodegradation of PLA under controlled composting conditions. Polymers. 2023;15(1):140. doi: 10.3390/polym15010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wright R.J., Bosch R., Gibson M.I., Christie-Oleza J.A. Plasticizer degradation by marine bacterial isolates: a proteogenomic and metabolomic characterization. Environ. Sci. Technol. 2020;54(4):2244–2256. doi: 10.1021/acs.est.9b05228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Paladhi A.G., Vallinayagam S., Rajendran S., Rathinam V., Sharma V.K. In: Microbes and Microbial Biotechnology for Green Remediation. Malik J.A., editor. Elsevier; 2022. Chapter 30 - microalgae: A promising tool for plastic degradation; pp. 575–587. [DOI] [Google Scholar]
- 134.Benavides Fernández C.D., Guzmán Castillo M.P., Quijano Pérez S.A., Carvajal Rodríguez L.V. Microbial degradation of polyethylene terephthalate: a systematic review. SN Appl. Sci. 2022;4:263. doi: 10.1007/s42452-022-05143-4. [DOI] [Google Scholar]
- 135.Athiappan M., Dinesh Kumar S., Umamaheswari S., Rajaprabu M. In: Chapter 13 - Rhizosphere Engineering through Pesticides-Degrading Beneficial Bacteria. Chandra Dubey R., Kumar P., editors. Academic Press; 2022. (Rhizosphere Engineering). 239–257. [DOI] [Google Scholar]
- 136.Magalhães R.P., Cunha J.M., Sousa S.F. Perspectives on the role of enzymatic biocatalysis for the degradation of plastic PET. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222011257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Samak N.A., Jia Y., Sharshar M.M., Mu T., Yang M., Peh S., Xing J. Recent advances in biocatalysts engineering for polyethylene terephthalate plastic waste green recycling. Environ. Int. 2020;145 doi: 10.1016/j.envint.2020.106144. [DOI] [PubMed] [Google Scholar]
- 138.Lin P.P., Mi L., Moriok A.H., Yoshino M.M., Konishi S., Xu S.C., Papanek B.A., Riley L.A., Guss A.M., Liao J.C. Consolidated bioprocessing of cellulose to isobutanol using Clostridium thermocellum. Metab. Eng. 2015;31:44–52. doi: 10.1016/j.ymben.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 139.Qi X., Yan W., Cao Z., Ding M., Yuan Y. Current advances in the biodegradation and bioconversion of polyethylene terephthalate. Microorganisms. 2022;10:39. doi: 10.3390/microorganisms10010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu C., Shi C., Zhu S., Wei R., Yin C.-C. Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis. Biochem. Biophys. Res. Commun. 2019;508:289–294. doi: 10.1016/j.bbrc.2018.11.148. [DOI] [PubMed] [Google Scholar]
- 141.Han X., Liu W., Huang J.-W., Ma J., Zheng Y., Ko T.-P., Xu L., Cheng Y.-S., Chen C.-C., Guo R.-T. Structural insight into catalytic mechanism of PET hydrolase. Nat. Commun. 2017;8(1):2106. doi: 10.1038/s41467-017-02255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Joo S., Cho I.J., Seo H., Son H.F., Sagong H.-Y., Shin T.J., Choi S.Y., Lee S.Y., Kim K.-J. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat. Commun. 2018;9(1):382. doi: 10.1038/s41467-018-02881-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Urbanek A.K., Kosiorowska K.E., Mirończuk A.M. Current knowledge on polyethylene terephthalate degradation by genetically modified microorganisms. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.771133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Patel A., Matsakas L., Rova U., Christakopoulos P. A perspective on biotechnological applications of thermophilic microalgae and cyanobacteria. Bioresour. Technol. 2019;278:424–434. doi: 10.1016/j.biortech.2019.01.063. [DOI] [PubMed] [Google Scholar]
- 145.Cui Y., Chen Y., Liu X., Dong S., Tian Y., Qiao Y., Mitra R., Han J., Li C., Han X., Liu W.m, Chen Q., Wei W., Wang X., Du W., Tang S., Xiang H., Liu H., Liang Y., Houk K.N., Wu B. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy. ACS Catal. 2021;11(3):1340–1350. doi: 10.1021/acscatal.0c05126. [DOI] [Google Scholar]
- 146.Lu H., Diaz D.J., Czarnecki N.J., Zhu C., Kim W., Shroff R., Acosta D.J., Alexander B.R., Cole H.O., Zhang Y., Lynd N., Ellington A.D., Alper H.S. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature. 2022;604:662–667. doi: 10.1038/s41586-022-04599-z. [DOI] [PubMed] [Google Scholar]
- 147.Austin H.P., Allen M.D., Donohoe B.S., Rorrer N.A., Kearns F.L., Silveira R.L., Pollard B.C., Dominick G., Duman R., El Omari K., Mykhaylyk V., Wagner A., Michener W.E., Amore A.D., Skaf M.S., Crowley M.F., Thorne A.W., Johnson C.W., Woodcock H.L., McGeehan J.E., Beckham G.T. Characterization and engineering of a plastic degrading aromatic polyesterase. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E4350–E4357. doi: 10.1073/pnas.1718804115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Priya A., Dutta K., Daverey A. A comprehensive biotechnological and molecular insight into plastic degradation by microbial community. J. Chem. Technol. Biotechnol. 2022;97:381–390. doi: 10.1002/jctb.6675. [DOI] [Google Scholar]
- 149.Deng X., Fei X., Li Y. The effects of nutritional restriction on neutral lipid accumulation in Chlamydomonas and Chlorella. Afr. J. Microbiol. Res. 2011;5:260–270. doi: 10.5897/AJMR10.557. [DOI] [Google Scholar]
- 150.Zhao P., Gu W., Wu S., Huang A., He L., Xie X., Gao S., Zhang B., Niu J., Lin A.P. Silicon enhances the growth of Phaeodactylum tricornutum Bohlin under green light and low temperature. Sci. Rep. 2014;4:3958. doi: 10.1038/srep03958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu X., Xie H., Roussou S., Lindblad P. Current advances in engineering cyanobacteria and their applications for photosynthetic butanol production. Curr. Opin. Biotechnol. 2022;73:1–8. doi: 10.1016/j.copbio.2021.07.014. [DOI] [PubMed] [Google Scholar]
- 152.Sonune N. In: Rhizobiont in Bioremediation of Hazardous Waste. Vivek K., Ram P., Manoj K., editors. Springer; Singapore: 2021. Microbes: a potential tool for bioremediation; pp. 391–407. [DOI] [Google Scholar]
- 153.Ayilara M.S., Babalola O.O. Bioremediation of environmental wastes: the role of microorganisms. Front. Agron. 2023;5 doi: 10.3389/fagro.2023.1183691. [DOI] [Google Scholar]
- 154.Tournier V., Topham C.M., Gilles A., David B., Folgoas C., Moya-Leclair E., Kamionka E., Desrousseaux M.-L., Texier H., Gavalda S., Cot M., Guémard E., Dalibey M., Nomme J., Cioci G., Barbe S., Chateau M., André I., Duquesne S., Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020;580:216–219. doi: 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- 155.Mesbah N.M. Industrial biotechnology based on enzymes from extreme environments. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.870083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Arnal G., Anglade J., Gavalda S., Tournier V., Chabot N., Bornscheuer U.T., Weber G., Marty A. Assessment of four engineered PET degrading enzymes considering large-scale industrial applications. ACS Catal. 2023;13(20):13156–13166. doi: 10.1021/acscatal.3c02922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Businesswire . Business Wire; 2023. Carbios Demonstrates Superior Performance of its Enzyme in World-Renowned Scientific Publication.https://www.businesswire.com/news/home/20231011889968/en/Carbios-Demonstrates-Superior-Performance-of-Its-Enzyme-in-World-Renowned-Scientific-Publication [Google Scholar]
- 158.Zheng J., Suh S. Strategies to reduce the global carbon footprint of plastics. Nat. Clim. Change. 2019;9(5):374–378. doi: 10.1038/s41558-019-0459-z. [DOI] [Google Scholar]
- 159.Guo X., Wang J. The chemical behaviors of microplastics in marine environment: a review. Mar. Pollut. Bull. 2019;142:1–14. doi: 10.1016/j.marpolbul.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 160.Rida Abelouah M., Ben-Haddad M., Hajji S., De-la-Torre G.E., Aziz T., Abou Oualid J., Banni M., Ait Alla A. Floating microplastics pollution in the Central Atlantic Ocean of Morocco: insights into the occurrence, characterization, and fate. Mar. Pollut. Bull. 2022;182 doi: 10.1016/j.marpolbul.2022.113969. [DOI] [PubMed] [Google Scholar]
- 161.Nabi I., Bacha A.-U.-R., Li K., Cheng H., Wang T., Liu Y., Ajmal S., Yang Y., Feng Y., Zhang L. Complete photocatalytic mineralization of microplastic on TiO2 nanoparticle film. iScience. 2020;23(7) doi: 10.1016/j.isci.2020.101326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang C., Xian Z., Jin X., Liang S., Chen Z., Pan B., Wu B., Ok Y.S., Gu C. Photo-aging of polyvinyl chloride microplastic in the presence of natural organic acids. Water Res. 2020;183 doi: 10.1016/j.watres.2020.116082. [DOI] [PubMed] [Google Scholar]
- 163.Wu X., Liu P., Shi H., Wang H., Huang H., Shi Y., Gao S. Photo aging and fragmentation of polypropylene food packaging materials in artificial seawater. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116456. [DOI] [PubMed] [Google Scholar]
- 164.Cai L., Wang J., Peng J., Wu Z., Tan X. Observation of the degradation of three types of plastic pellets exposed to UV irradiation in three different environments. Sci. Total Environ. 2018;628–629:740–747. doi: 10.1016/j.scitotenv.2018.02.079. [DOI] [PubMed] [Google Scholar]
- 165.Shi Y., Liu P., Wu X., Shi H., Huang H., Wang H., Gao S. Insight into chain scission and release profiles from photodegradation of polycarbonate microplastics. Water Res. 2021;195 doi: 10.1016/j.watres.2021.116980. [DOI] [PubMed] [Google Scholar]
- 166.Jiang J.Q. Occurrence of microplastics and its pollution in the environment: a review. Sustain. Prod. Consum. 2018;13:16–23. doi: 10.1016/j.spc.2017.11.003. [DOI] [Google Scholar]
- 167.Luo H., Zhao Y., Li Y., Xiang Y., He D., Pan X. Aging of microplastics affects their surface properties, thermal decomposition, additives leaching and interactions in simulated fluids. Sci. Total Environ. 2020;714 doi: 10.1016/j.scitotenv.2020.136862. [DOI] [PubMed] [Google Scholar]
- 168.Lee Q.Y., Li H. Photocatalytic degradation of plastic waste: a mini review. Micromachines. 2021;12(8):907. doi: 10.3390/mi12080907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.He J., Lyu P., Jiang B., Chang S., Du H., Zhu J., Li H. A novel amorphous alloy photocatalyst (NiB/In2O3) composite for sunlight-induced CO2 hydrogenation to HCOOH. Appl. Catal., B. 2021;298 doi: 10.1016/j.apcatb.2021.120603. [DOI] [Google Scholar]
- 170.He J., Lyu P., Li D., Cheng C., Chang S., Qin L., Zheng C., Zhu J. Bio-alcohol induced self-assembly of heterojunctioned TiO2/WO3 composites into a hierarchical yolk-shell structure for photocatalysis. Chem. Commun. 2021;57(56):6883–6886. doi: 10.1039/D1CC01975A. [DOI] [PubMed] [Google Scholar]
- 171.Liu H., Feng Y., Shao J., Chen Y., Wang Z.L., Li H., Chen X., Bian Z. Self-cleaning triboelectric nanogenerator based on TiO2 photocatalysis. Nano Energy. 2020;70 doi: 10.1016/j.nanoen.2020.104499. [DOI] [Google Scholar]
- 172.Mu C., Zhang Y., Cui W., Liang Y., Zhu Y. Removal of bisphenol A over a separation free 3D Ag3PO4-graphene hydrogel via an adsorption-photocatalysis synergy. Appl. Catal., B. 2017;212:41–49. doi: 10.1016/j.apcatb.2017.04.018. [DOI] [Google Scholar]
- 173.Tofa T.S., Ye F., Kunjali K.L., Dutta J. Enhanced visible light photodegradation of microplastic fragments with plasmonic platinum/Zinc oxide nanorod photocatalysts. Catalysts. 2019;9(10):819. doi: 10.3390/catal9100819. [DOI] [Google Scholar]
- 174.Ouyang Z., Yang Y., Zhang C., Zhu S., Qin L., Wang W., He D., Zhou Y., Luo H., Qin F. Recent advances in photocatalytic degradation of plastics and plastic-derived chemicals. J. Mater. 2021;9(23):13402–13441. doi: 10.1039/d0ta12465f. [DOI] [Google Scholar]
- 175.Wang L., Jiang S., Gui W., Li H., Wu J., Wang H., Yang J. Photocatalytic upcycling of plastic waste: mechanism, integrating modus, and selectivity. Small Struct. 2023;4 doi: 10.1002/sstr.202300142. [DOI] [Google Scholar]
- 176.Uekert T., Kuehnel M.F., Wakerley D., Reisner E. Plastic waste as a feedstock for solar-driven H2 generation. Energy Environ. Sci. 2018;11:2853–2857. doi: 10.1039/C8EE01408F. [DOI] [Google Scholar]
- 177.Chen J., Zhang L., Wang L., Kuang M., Wang S., Yang J. Toward carbon neutrality: selective conversion of waste plastics into value-added chemicals. Matter. 2023;6(10):3322–3347. doi: 10.1016/j.matt.2023.07.025. [DOI] [Google Scholar]
- 178.Cao B., Wan S., Wang Y., Guo H., Ou M., Zhong Q. Highly-efficient visible-light-driven photocatalytic H2 evolution integrated with microplastic degradation over MXene/ZnxCd1xS photocatalyst. J. Colloid Interface Sci. 2022;605:311–319. doi: 10.1016/j.jcis.2021.07.113. [DOI] [PubMed] [Google Scholar]
- 179.Jiao X., Zheng K., Chen Q., Li X., Li Y., Shao W., Xu J., Zhu J., Pan Y., Sun Y., Xie Y. Photocatalytic conversion of waste plastics into C2 fuels under simulated natural environment conditions. Angew. Chem. Int. 2020;59(36):15497–15501. doi: 10.1002/anie.201915766. [DOI] [PubMed] [Google Scholar]
- 180.Singh Jadaun J., Bansal S., Sonthalia A., Rai A.K., Singh S.P. Biodegradation of plastics for sustainable environment. Bioresour. Technol. 2022;347 doi: 10.1016/j.biortech.2022.126697. [DOI] [PubMed] [Google Scholar]
- 181.Bourgade B., Stenjö K. Synthetic biology in marine cyanobacteria: advances and challenges. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.994365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Meng X., Liu L., Chen X. Bacterial photosynthesis: state-of-the-art in light-driven carbon fixation in engineered bacteria. Curr. Opin. Microbiol. 2022;69 doi: 10.1016/j.mib.2022.102174. [DOI] [PubMed] [Google Scholar]
- 183.Hu J., Meng W., Su Y., Qian C., Fu W. Emerging technologies for advancing microalgal photosynthesis and metabolism toward sustainable production. Front. Mar. Sci. 2023;13 doi: 10.3389/fmars.2023.1260709. [DOI] [Google Scholar]
- 184.Markley A.L., Begemann M.B., Clarke R.E., Gordon G.C., Pfleger B.F. Synthetic biology toolbox for controlling gene expression in the cyanobacterium synechococcus sp. Strain PCC 7002. ACS Synth. Biol. 2015;4(5):595–603. doi: 10.1021/sb500260k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Englund E., Liang F., Lindberg P. Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium synechocystis sp. PCC 6803. Sci. Rep. 2016;6 doi: 10.1038/srep36640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li S., Sun T., Xu C., Chen L., Zhang W. Development and optimization of genetic toolboxes for a fast-growing cyanobacterium synechococcus elongatus UTEX 2973. Metab. Eng. 2018;48:163–174. doi: 10.1016/j.ymben.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 187.Liu D., Pakrasi H.B. Exploring native genetic elements as plug-in tools for synthetic biology in the cyanobacterium synechocystis sp. PCC 6803. Microb. Cell Factories. 2018;17:48. doi: 10.1186/s12934-018-0897-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Miao R., Jahn M., Shabestary K., Hudson E.P. 2023. CRISPR Interference Screens Reveal Tradeoffs between Growth Rate and Robustness in Synechocystis Sp. PCC 6803 across Trophic Conditions. BioRxiv 528328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jüttner F., Hans R. The reducing capacities of cyanobacteria for aldehydes and ketones. Appl. Microbiol. Biotechnol. 1986;25(1):52–54. doi: 10.1007/BF00252512. [DOI] [Google Scholar]
- 190.Nakamura K., Yamanaka R. Light-mediated regulation of asymmetric reduction of ketones by a cyanobacterium. Tetrahedron Asymmetry. 2002;13:2529–2533. doi: 10.1016/S0957-4166(02)00683-3. [DOI] [Google Scholar]
- 191.Shimoda K., Kubota N., Hamada H., Kaji M., Hirata T. Asymmetric reduction of enones with synechococcus sp. PCC 7942. Tetrahedron Asymmetry. 2004;15(11):1677–1679. doi: 10.1016/j.tetasy.2004.04.024. [DOI] [Google Scholar]
- 192.Assil-Companioni L., Büchsenschütz H.C., Solymosi D., Dyczmons-Nowaczyk N.G., Bauer K.K.F., Wallner S., Macheroux P., Allahverdiyeva Y., Nowaczyk M.M., Kourist R. Engineering of NADPH supply boosts photosynthesis-driven biotransformations. ACS Catal. 2020;10(20):11864–11877. doi: 10.1021/acscatal.0c02601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tüllinghoff A., Uhl M., Nintzel F.E.H., Schmid A., Bühler B., Toepel J. Maximizing photosynthesis-driven Baeyer–Villiger oxidation efficiency in recombinant Synechocystis sp. PCC6803. Front. Catal. 2022;1:1–14. doi: 10.3389/fctls.2021.780474. [DOI] [Google Scholar]
- 194.Erdem E., Malihan-Yap L., Assil-Companioni L., Grimm H., Barone G.D., Serveau-Avesque C., Amouric A., Duquesne K., de Berardinis V., Alladverdiyeva Y., Alphand V., Kourist R. Photobiocatalytic oxyfunctionalization with high reaction rate using a Baeyer–Villiger Monooxygenase from Burkholderia xenovorans in metabolically engineered cyanobacteria. ACS Catal. 2022;12:66–72. doi: 10.1021/acscatal.1c04555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mascia F., Pereira S.B., Pacheco C.C., Oliveira P., Solarczek J., Schallmey A., Kourist R., Alphand V., Tamagnini P. Light-driven hydroxylation of testosterone by Synechocystis sp. PCC 6803 expressing the heterologous CYP450 monooxygenase CYP110D1. Green Chem. 2022;24:6156–6167. doi: 10.1039/D1GC04714K. [DOI] [Google Scholar]
- 196.Jurkaš V., Weissensteiner F., De Santis P., Vrabl S., Sorgenfrei F.A., Bierbaumer S., Kara S., Kourist R., Wangikar P.P., Christoph C.K., Kroutil W. Transmembrane shuttling of photosynthetically produced electrons to propel extracellular biocatalytic redox reactions in a modular fashion. Angew. Chem. Int. Ed. 2022 doi: 10.1002/anie.202207971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Barone G.D., Hubáček M., Malihan-Yap L., Grimm H.C., Nikkanen L., Pacheco C.C., Tamagnini P., Allahverdiyeva Y., Kourist R. Towards the rate limit of heterologous biotechnological reactions in recombinant cyanobacteria. Biotechnol. Biofuels. 2023;16:4. doi: 10.1186/s13068-022-02237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Schmermund L., Jurkaš V., Özgen F.F., Barone G.D., Büchsenschütz H.C., Winkler C.K., Schmidt S., Kourist R., Kroutil W. Photo-Biocatalysis: biotransformations in the presence of light. ACS Catal. 2019;9:4115–4144. doi: 10.1021/acscatal.9b00656. [DOI] [Google Scholar]
- 199.Grimm H.C., Erdem E., Kourist R. In: The Autotrophic Biorefinery: Raw Materials from Biotechnology. Kourist R., Schmidt S., editors. De Gruyter; 2021. Chapter 8 - biocatalytic applications of autotrophic organisms; pp. 207–246. [DOI] [Google Scholar]
- 200.Böhmer S., Marx C., Gómez-Baraibar Á., Nowaczyk M.M., Tischler D., Hemschemeier A., Happe T. Evolutionary diverse Chlamydomonas reinhardtii old yellow enzymes reveal distinctive catalytic properties and potential for whole-cell biotransformations. Algal Res. 2020;50 doi: 10.1016/j.algal.2020.101970. [DOI] [Google Scholar]
- 201.Wicaksono W.A., Erschen S., Krause R., Müller H., Cernava T., Berg G. Enhanced survival of multi-species biofilms under stress is promoted by low-abundant but antimicrobial-resistant keystone species. J. Hazard Mater. 2022;422 doi: 10.1016/j.jhazmat.2021.126836. [DOI] [PubMed] [Google Scholar]
- 202.Sun X.-L., Xiang H., Xiong H.Q., Fang Y.-C., Wang Y. Bioremediation of microplastics in freshwater environments: a systematic review of biofilm culture, degradation mechanisms, and analytical methods. Sci. Total Environ. 2023;863 doi: 10.1016/j.scitotenv.2022.160953. [DOI] [PubMed] [Google Scholar]
- 203.Amaral-Zettler L.A., Ballerini T., Zettler E.R., Abdala Asbun A., Adame A., Casotti R., Dumontet B., Donnarumma V., Engelmann J.C., Frère L., Mansui J., Philippon M., Pietrelli L., Sighicelli M. Diversity and predicted inter- and intra-domain interactions in the Mediterranean Plastisphere. Environ. Pollut. 2021;286 doi: 10.1016/j.envpol.2021.117439. [DOI] [PubMed] [Google Scholar]
- 204.Joint I., Tait K., Callow M.E., Callow J.A., Milton D., Williams P., Cámara M. Cell-to-cell communication across the prokaryote-eukaryote boundary. Science. 2002;298(5596):1207. doi: 10.1126/science.1077075. [DOI] [PubMed] [Google Scholar]
- 205.Croft M.T., Lawrence A.D., Raux-Deery E., Warren M.J., Smith A.G. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;43(7064):90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 206.Goecke F., Labes A., Wiese J., Imhoff J.F. Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 2010;409:267–299. doi: 10.3354/meps08607. [DOI] [Google Scholar]
- 207.Krug L., Morauf C., Donat C., Müller H., Cernava T., Berg G. Plant growth-promoting methylobacteria selectively increase the biomass of biotechnologically relevant microalgae. Front. Microbiol. 2020;11:427. doi: 10.3389/fmicb.2020.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Le Bail A., Billoud B., Kowalczyk N., Kowalczyk M., Gicquel M. Auxin metabolism and function in the multicellular brown alga Ectocarpus siliculosus. Plant Physiol. 2010;153:128–144. doi: 10.1104/pp.109.149708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chu R., Li S., Zhu L., Yin Z., Hu D., Liu C., Mo F. A review on co-cultivation of microalgae with filamentous fungi: efficient harvesting, wastewater treatment and biofuel production. Renew. Sustain. Energy Rev. 2021;139 doi: 10.1016/j.rser.2020.110689. [DOI] [Google Scholar]
- 210.Leng L., Li W., Chen J., Leng S., Chen J., Wei L., Peng H., Li J., Zhou W., Huang H. Co-culture of fungi-microalgae consortium for wastewater treatment: a review. Bioresour. Technol. 2021;330 doi: 10.1016/j.biortech.2021.125008. [DOI] [PubMed] [Google Scholar]
- 211.Krug L., Erlacher A., Markut K., Berg G., Cernava T. The microbiome of alpine snow algae shows a specific inter-kingdom connectivity and algae-bacteria interactions with supportive capacities. ISME J. 2020;14:2197–2210. doi: 10.1038/s41396-020-0677-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Niu L., Chen Y., Li Y., Wang Y., Shen J., Wang L., Zhang W., Zhang H., Zhao Bo. Diversity, abundance and distribution characteristics of potential polyethylene and polypropylene microplastic degradation bacterial communities in the urban river. Water Res. 2023;232 doi: 10.1016/j.watres.2023.119704. [DOI] [PubMed] [Google Scholar]
- 213.Tzintzun-Camacho O., Loera O., Ramírez-Saad H., Gutiérrez-Rojas M. Comparison of mechanisms of hexadecane uptake among pure and mixed cultures derived from a bacterial consortium. Int. Biodeterior. Biodegrad. 2012;70:1–7. doi: 10.1016/j.ibiod.2012.01.009. [DOI] [Google Scholar]
- 214.Zeng C., Ding F., Zhou J., Dong W., Cui Z., Yan X. Biodegradation of Poly(ethylene terephthalate) by Bacillus safensis YX8. Int. J. Mol. Sci. 2023;24(22) doi: 10.3390/ijms242216434. [DOI] [PMC free article] [PubMed] [Google Scholar]