Abstract
Semliki Forest virus (SFV), an enveloped alphavirus, is a well-characterized paradigm for viruses that infect cells via endocytic uptake and low-pH-triggered fusion. The SFV spike protein is composed of a dimer of E1 and E2 transmembrane subunits, which dissociate upon exposure to low pH, liberating E2 and the fusogenic E1 subunit to undergo independent conformational changes. SFV fusion and infection are blocked by agents such as ammonium chloride, which act by raising the pH in the endosome and inhibiting the low-pH-induced conformational changes in the SFV spike protein. We have previously isolated an SFV mutant, fus-1, that requires more acidic pH to trigger its fusion activity and is therefore more sensitive to inhibition by ammonium chloride. The acid shift in the fusion activity of fus-1 was here shown to be due to a more acidic pH threshold for the initial dissociation of the fus-1 spike dimer, thereby resulting in a more acidic pH requirement for the subsequent conformational changes in both fus-1 E1 and fus-1 E2. Sequence analysis demonstrated that the fus-1 phenotype was due to a mutation in the E2 spike subunit, threonine 12 to isoleucine. fus-1 revertants that have regained the parental fusion phenotype and ammonium chloride sensitivity were shown to have also regained E2 threonine 12. Our results identify a region of the SFV E2 spike protein subunit that regulates the pH dependence of E1-catalyzed fusion by controlling the dissociation of the E1/E2 dimer.
Virus fusion proteins have evolved to keep their membrane fusion activities inactive on the surface of the virus membrane until specifically triggered by the conditions of infecting a host cell. In the case of viruses that fuse at the plasma membrane, fusion can be triggered by receptor binding (1, 11) and may require additional molecules that function as coreceptors, as does human immunodeficiency virus type 1 (2, 7). For those viruses that use the endocytic pathway and low pH to infect cells, fusion is specifically triggered by the mildly acidic pH within endocytic vesicles (1, 11). A well-characterized example of such a low-pH-dependent virus is the enveloped alphavirus Semliki Forest virus (SFV) (reviewed in references 16 and 30). SFV fusion is catalyzed by the virus spike protein, triggered by an endosomal pH of ≤6.2, and blocked by agents such as NH4Cl or monensin, which act by raising the pH within endosomes above the critical threshold required to trigger virus-membrane fusion.
The SFV spike protein is composed of trimers (E1/E2/E3)3, containing a dimer of two transmembrane subunits, E1 and E2, of about 50 kDa, and an associated peripheral polypeptide, E3, of about 10 kDa. E3 and E2 are synthesized as a precursor, p62, which is posttranslationally processed after a tetrabasic cleavage site, probably in the trans-Golgi network (5). E1 is the fusion-active spike protein subunit, contains the putative fusion peptide between amino acids 79 and 97 (8, 19, 23), and binds specifically to membranes upon acid-pH treatment (22). E1 forms a tight but nonconvalent dimer with p62 in the rough endoplasmic reticulum and is transported to the plasma membrane and packaged into the virus particle in association with p62/E2.
Exposure to acid pH in vitro or within the endosome triggers a defined series of conformational changes within the SFV spike protein (see references 9 and 16 for reviews). The dimeric interaction between E2 and E1 is disrupted, resulting in the loss of coimmunoprecipitation and cosedimentation on sucrose gradients. E2 becomes trypsin sensitive and exposes new antigenic epitopes, and E1 exposes new antigenic epitopes, becomes trypsin resistant, forms a stable E1 homotrimer, and associates with the target membrane. The E1 homotrimer appears to be critical for fusion, as a mutation within the fusion peptide, glycine 91 to aspartate, prevents E1 homotrimer formation and also blocks fusion and infection (19).
An SFV mutant, fus-1, with a lower pH threshold for membrane fusion was isolated by selecting for viruses resistant to in vitro fusion with RNase-filled liposomes (20). Unlike the wild-type (wt) virus pH threshold of ∼6.2, fus-1 requires treatment below ∼pH 5.5 to trigger fusion (20). As predicted from its more acidic pH dependence, fusion of fus-1 occurs within late endosomes, which have a more acidic lumenal pH than early endosomes (21, 29), and fus-1 infection is more sensitive than wt to inhibition by weak bases that raise the pH of endosomes (20).
Studies of the pH-dependent spike protein conformational changes in fus-1 revealed an interesting paradox, in that fus-1’s lower pH threshold of fusion did not correlate with a lower pH threshold for the conformational change in a single subunit. The E2 subunit of fus-1 has a lower pH threshold than wt for the binding of acid-specific monoclonal antibodies (MAbs) (18) and for conversion to trypsin sensitivity (17). Surprisingly, assays of fus-1 E1 conformational changes suggested that a more acidic pH was also required to trigger its acid-specific MAb binding (18) and conversion to trypsin resistance (29). Thus, it was unclear which subunit conferred the fus-1 phenotype, or if a double mutation could be involved.
Recently, in vitro mutagenesis of the SFV p62 cleavage site was used to block the processing of p62 to E2 and E3. These studies indicated that cleavage is important in the control of the SFV fusion reaction (13, 25, 26, 28). Viruses containing uncleaved p62 are not infectious, and the p62/E1 form of the spike protein requires much lower pH, ∼5.0, to trigger fusion (28). This lower pH threshold is due to the fact that the dissociation of the E1/p62 dimer requires a considerably lower pH than does the dissociation of the E1/E2 dimer normally found in the virus particle (28, 33). Thus, dimer dissociation seems a prerequisite for the subsequent conformational changes in E1 that lead to fusion and infection.
These findings on the p62 mutants suggested to us that the lower pH threshold of fus-1 E1 and E2 conformational changes could be due to alterations in fus-1 dimer interactions. Synthesis and processing of the fus-1 spike protein appear normal, and the assembled virus contains E2 (20). It was possible, however, that the dissociation of the fus-1 E1/E2 dimer interaction was nonetheless shifted to a more acidic pH. Such a dimer alteration could cause the lower pH thresholds of the E1 and E2 conformational changes and result in a more acidic pH threshold for fusion.
A goal of this study was to determine the molecular mechanism responsible for the lower pH thresholds of the fus-1 E1 and E2 conformational changes. Amino acid sequence differences had been reported between the prototype wt SFV strain and a plaque-purified strain derived from it (here referred to as S1J) (15). S1J was the direct parent virus to both fus-1 and a number of previously reported temperature-sensitive (ts) mutants. To map the fus-1 mutation, we expressed the fus-1 structural proteins in the context of the wt virus infectious clone (wt/ic). We then used this virus chimera, fus-1/ic, together with fus-1, wt virus, and the S1J parent virus to analyze acid-induced spike protein conformational changes and dimer dissociation and to determine the amino acid sequence of the structural proteins.
(Data in this report are from a thesis submitted by Sallie Glomb-Reinmund in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Sue Golding Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University.)
MATERIALS AND METHODS
Viruses.
Our wt virus stock was previously plaque purified from prototype wt virus obtained from the Department of Virology at the University of Helsinki and is referred to as wt in this study and others (20, 32). The parent virus to fus-1 is a plaque-purified virus stock derived at the University of Helsinki and is here referred to as S1J (15). fus-1 was isolated from S1J by mutagenesis with nitrosoguanidine and selection for resistance to fusion with RNase-filled liposomes at pH 5.5 (20). The fus-1 revertants R43 and R46 were selected for resistance to inhibition by NH4Cl, and were shown to have fusion and entry properties similar to those of the parent virus (21).
Virus stocks were prepared by propagation in BHK cells at low multiplicity of infection (20). [35S]methionine/cysteine-labeled virus was prepared in BHK cells and purified by banding on a discontinuous sucrose gradient, all as previously described (20).
Virus infectious clones.
RNA from fus-1 virus was isolated, cDNA was synthesized, and a 5-kb SalI/HindIII fragment encoding the fus-1 structural proteins was cloned into the plasmid pZ152 to give pfus-1sp, all as previously described for wt virus (23). To confirm the fus-1 phenotype and permit future mapping, the fus-1 structural protein coding sequences were subcloned into the pSP6-SFV-4 wt/ic (24) by using the following strategy. The fus-1 parent virus S1J was reported to have a sequence change of E2 His170 to Arg, which would be predicted to ablate an NdeI site, leaving a unique NdeI site at the 3′ end of E1 (31). The loss of the E2 NdeI site in pfus-1sp was confirmed by restriction digestion (data not shown). pfus-1sp was digested to completion with BglII and NdeI, and the 4,318-bp fragment encoding the fus-1 structural proteins was purified. This was ligated with the 9,970-bp BglII/NdeI fragment from wt/ic to give fus-1/ic, which contained nucleotides 6716 to 11034 derived from the fus-1 virus. All bacterial growth steps were performed at 30°C to minimize rearrangement of the infectious clone (32). Virus stocks derived from the wt/ic and fus-1/ic plasmids were prepared by RNA transcription and electroporation of BHK cells as previously described (6, 24) and used to prepare [35S]methionine/cysteine labeled virus as described above.
DNA sequence analysis.
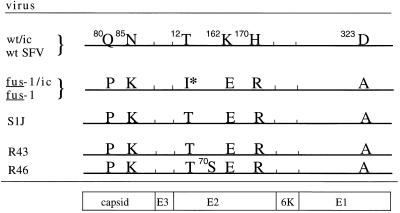
All DNA sequencing was performed by DyeDioxy-Terminator cycle sequencing in the Einstein sequencing facility, using an automated DNA sequencer (Applied Biosystems). To sequence from virus RNA, virus was grown in one 75-cm2 flask of BHK cells and pelleted for 1 h at 40,000 rpm in an SW41 rotor at 4°C. Viral RNA was isolated, reverse transcribed, and amplified by PCR, using either Vent (New England Biolabs, Inc., Beverly, Mass.) or Pfu (Stratagene, La Jolla, Calif.) polymerase as previously described (19, 32) and as recommended by the manufacturers. The amplified DNA was purified for sequencing by using a QIAquick kit (Qiagen Inc., Chatsworth, Calif.). Sequence analysis of fus-1 and fus-1/ic revealed nine single base changes compared to the sequence of the wt/ic obtained from Peter Liljeström (24). There were three silent mutations, which were at positions proline 22 in capsid (CCT→CCC), phenylalanine 258 in E2 (TTC→TTT), and cysteine 96 in E1 (TGC→TGT). The remaining mutations are summarized in Fig. 7. Note also that position 323 of E1 was an aspartic acid in both our wt virus and wt/ic, in agreement with our previous analysis (32), and that E2 position 162 was a lysine in our wt virus and wt/ic, while these positions in the original SFV sequence were reported as asparagine and glutamic acid, respectively (8).
FIG. 7.
Amino acid sequence differences among wt, mutant, and revertant virus structural proteins. The E2 isoleucine 12 change responsible for the fus-1 phenotype is marked (∗). No amino acid sequence differences were observed between wt/ic and wt SFV or between fus-1 and fus-1/ic. The positions of the SFV structural protein coding regions are marked.
Virus RNA synthesis assay.
BHK cells were grown in 24-well trays and incubated with the indicated concentrations of NH4Cl for 15 min at 37°C. Cells were then infected with virus at a multiplicity of infection of ∼1 PFU/cell in the presence of the indicated concentrations of NH4Cl for 90 min (termed the virus entry step), treated for 30 min at 37°C with 2 μg of actinomycin D (ACD) per ml and 15 mM NH4Cl to inhibit cellular RNA synthesis and secondary virus infection, and labeled with [3H]uridine in this medium for 3.5 h. To harvest, cells were washed on ice with phosphate-buffered saline (PBS), fixed with 1 ml of ice-cold 10% trichloroacetic acid (TCA) for 60 min, washed with 0.5 ml of cold 5% TCA, and lysed at room temperature with 0.5 ml of 0.1 M KOH for 30 min, and the radioactivity in the lysates was determined by liquid scintillation counting. Each experiment included control cells infected in the absence of NH4Cl during the virus entry step and then labeled as described above. Triplicate samples were run, and [3H]uridine incorporation was calculated as percent incorporation compared to control cells.
Analysis of low-pH-dependent conformational changes in virus spike proteins.
The overall method to assess conformational changes in the virus spike protein involved treating [35S]methionine/cysteine-labeled virus at the indicated pH in the presence of 1 mM liposome target membranes (cholesterol/phospholipid ratio of 1:2) as indicated. Liposomes were prepared from purified phospholipids and cholesterol as previously published (22). The virus mixture was then neutralized, and conformational changes were evaluated. The pH dependence of E1/E2 dimer dissociation was monitored by solubilizing virus samples with 1.0 to 0.2% Nonidet P-40 (NP-40) prior to neutralization and sedimenting them on 5 to 20% (wt/wt) sucrose gradients containing 0.1% NP-40 in a Beckman SW41 rotor at 39,000 rpm for 22 h at 4°C (6, 33). The exposure of acid conformation-specific epitopes was determined by immunoprecipitation with previously characterized acid conformation-specific MAbs (18). Formation of the E1 homotrimer was evaluated by incubating samples in sodium dodecyl sulfate (SDS) gel sample buffer for 3 min at 30°C and then analyzing them by SDS-polyacrylamide gel electrophoresis (PAGE) (19, 35). E1 conversion to trypsin resistance was assayed by digestion of samples with 200 μg of tosylsulfonyl phenylalanyl chloromethyl ketone-trypsin per ml in 1% Triton X-100 in PBS containing Ca2+ and Mg2+ (PBS2+) for 10 min at 37°C, followed by TCA precipitation and SDS-PAGE analysis (19, 29). Quantitation of SDS-gel assays was performed by phosphorimaging and ImageQuant 3.3 software (Molecular Dynamics, Sunnyvale, Calif.).
Analysis of low-pH-dependent conformational changes in virus spike protein ectodomains.
Soluble monomeric ectodomain fragments of the E1 and E2 subunits, termed E1* and E2*, were prepared by digestion with proteinase K in Triton X-114 and purified by phase separation and concanavalin A chromatography as previously described (17, 22). Samples were pH treated in the presence of 1 mM liposomes (cholesterol/phospholipid ratio of 1:1), and conformational changes were evaluated by immunoprecipitation with acid conformation-specific MAbs and by solubilization in SDS-gel sample buffer for 3 min at 30°C and SDS-PAGE to visualize formation of the E1* homotrimer (22).
RESULTS
fus-1 cloning and infectious clone construction.
Little was known about potential amino acid sequence differences between fus-1 and the wt and S1J strains except that fus-1 had no alterations in the sequence of the putative fusion peptide on the E1 subunit (23). We chose to substitute the cloned fus-1 structural proteins into wt/ic, reasoning that by this strategy we would obtain a clone that should express the fus-1 phenotype and enable further mapping if necessary. cDNA was prepared from fus-1 viral RNA, and an ∼4-kb fragment encoding the fus-1 structural proteins was subcloned into wt/ic to generate fus-1/ic. This virus chimera contains the C-terminal third of the fus-1 nonstructural protein 4, the complete fus-1 capsid, E3, E2, and 6K sequences, and the sequence of fus-1 E1 from the amino terminus to a point about 24 residues above the C-terminal transmembrane domain. This clone was used to produce fus-1/ic virus stock and radiolabeled virus, which were used to establish fusion phenotype and characterize spike protein conformational changes.
Spike protein conformational changes in fus-1 and fus-1/ic.
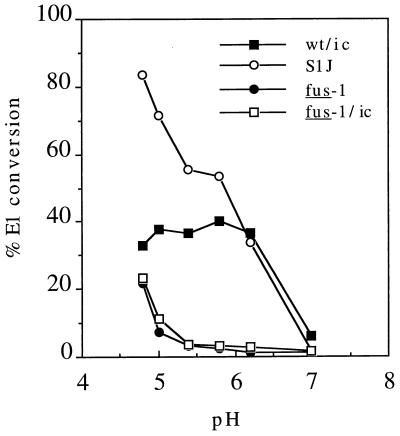
Our laboratory and others have isolated MAbs that are specific for the acid conformation of the E1 or E2 spike subunits (18, 35). Exposure of both the E1 and E2 acid-induced epitopes is shifted to ∼pH 5.3 for fus-1, compared to the wt threshold of pH 6.2 (18). Kinetic studies of wt virus show the E2 conversion happens postfusion whereas E1 reactivity occurs prior to membrane fusion (4, 14). To characterize fus-1/ic and to define the phenotype of the S1J parent virus, we used MAb E1a-1 to compare the pH dependence of E1 conversion in wt SFV, fus-1, fus-1/ic, and S1J. End point assays based on 10 min of virus pH treatment at 37°C were performed. The exposure of the E1a-1 epitope for wt SFV had a threshold of pH 6.2, while fus-1 and fus-1/ic both had a shifted threshold of ∼pH 5.0 (Fig. 1). The S1J pH threshold of 6.2 was similar to that of wt SFV. The maximum extent of epitope exposure for wt virus was about 40%, similar to published reports (4), while that of S1J was somewhat higher. E1 may not completely convert due to the simultaneous acid inactivation of the virus fusion activity (4). Both fus-1 and fus-1/ic were inefficiently precipitated even at pH 4.8. Similar shifts in the fus-1 pH threshold were observed with MAb anti-E1" (data not shown).
FIG. 1.
pH dependence of E1 epitope exposure for wt and mutant viruses. 35S-labeled wt/ic, S1J, fus-1, and fus-1/ic virus preparations were treated at the indicated pH for 10 min at 37°C, adjusted to neutral pH, solubilized in lysis buffer containing 1% Triton X-100, and immunoprecipitated with MAb E1a-1, an acid conformation-specific antibody. E1 precipitated by the MAb was quantitated by SDS-PAGE and phosphorimaging and then compared to the total E1 precipitated by a rabbit polyclonal antibody. Data represent the mean of three separate experiments for each virus. The standard deviations ranged from 1 to 30%.
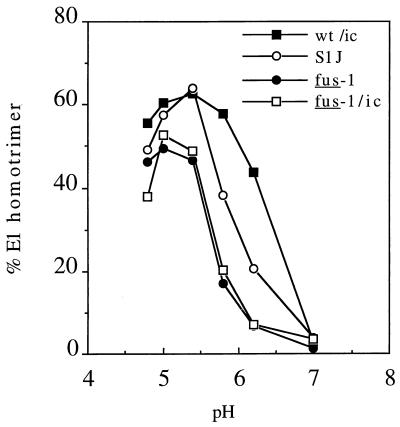
A key intermediate in SFV fusion is the formation of an E1 homotrimer, which occurs following treatment of wt virus at pH 6.2 or below (19, 35). The pH dependence of E1 homotrimer formation for wt SFV, S1J, fus-1, and fus-1/ic was determined by pH treatment of 35S-labeled virus for 10 min at 37°C, followed by mild SDS solubilization to maintain the homotrimer and SDS-PAGE analysis (Fig. 2). wt SFV homotrimer formation had a pH threshold of 6.2, whereas fus-1 and fus-1/ic homotrimer formation were shifted to a pH threshold of ∼5.8. S1J homotrimer formation was initiated at pH 6.2 but displayed an intermediate pH dependence compared to wt SFV and fus-1. Similar results were obtained in an alternative assay based on the resistance of the E1 homotrimer to trypsin digestion (data not shown). Interestingly, the electrophoretic migration of S1J, fus-1, and fus-1/ic homotrimers on SDS-gels was slightly faster than that of wt virus (data not shown). As will be evident from the sequence analysis below, this difference in migration is unlikely to be due to a change in E1’s molecular weight or posttranslational processing and may suggest altered folding of the E1 homotrimer in S1J and fus-1.
FIG. 2.
pH dependence of wt and mutant E1 homotrimer formation. 35S-labeled wt/ic, S1J, fus-1, and fus-1/ic virus preparations were treated at the indicated pH for 10 min at 37°C in the presence of liposomes, adjusted to neutral pH, and solubilized in SDS sample buffer for 3 min at 30°C. Samples were analyzed by SDS-PAGE and quantitated by phosphorimaging. Results are shown as percent E1 in the homotrimer band compared to total E1 and represent the mean for three experiments, with standard deviations between 1 and 33%.
In vivo virus entry phenotypes.
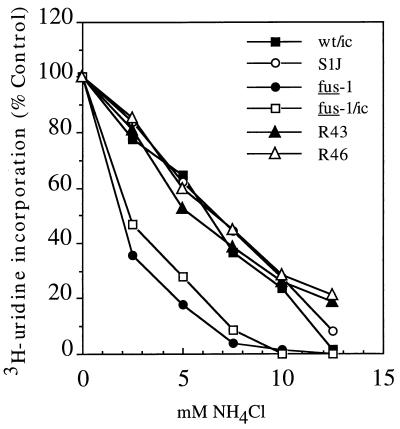
The sensitivity of virus infection to NH4Cl, a weak base that neutralizes endosomal acidity, correlates with the pH required to trigger viral membrane fusion. The lower the pH threshold for fusion, the more sensitive virus infection is to NH4Cl, as was clearly demonstrated for fus-1 compared to the parental S1J virus (20). In addition, resistance of virus infection to NH4Cl was previously used to select for fus-1 revertants, which were demonstrated to have fusion properties similar to those of S1J (21). We assayed the NH4Cl sensitivity of fus-1/ic infection and compared it in parallel with that of fus-1, wt SFV, S1J, and two fus-1 revertants, R43 and R46 (Fig. 3). Infection was quantitated by measuring the ACD-insensitive incorporation of [3H]uridine into viral RNA. Both fus-1 and fus-1/ic were much more sensitive than wt SFV to NH4Cl, displaying half-maximal inhibition at ∼2.5 mM NH4Cl compared to ∼6.5 mM NH4Cl for wt SFV. S1J, R43, and R46 were found to have similar NH4Cl sensitivities as wt SFV, suggesting similar membrane fusion thresholds that are significantly higher than that of fus-1. Taken together, these results and the previous studies of S1J cell-cell fusion (20) indicate that S1J has a membrane fusion threshold resembling that of wt SFV, suggesting that the amount of homotrimer generated by pH 6.2 treatment is sufficient to trigger S1J fusion. In contrast, the substantially more acidic pH required for fus-1 and fus-1/ic E1 conformational changes leads to a significantly more acidic fusion threshold.
FIG. 3.
Sensitivity of infection by wt, mutant, and revertant viruses to inhibition by NH4Cl. BHK cells were pretreated with the indicated concentrations of NH4Cl for 15 min at 37°C and then infected with wt/ic, S1J, fus-1, fus-1/ic, R43 revertant, or R46 revertant at 1 PFU/cell for 1.5 h in the continued presence of NH4Cl. Following infection, the cells were treated with 2 μg of ACD per ml and 15 mM NH4Cl for 30 min and then labeled with [3H]uridine for 3.5 h in the continued presence of ACD and 15 mM NH4Cl. Infection was quantitated as the percent [3H]uridine incorporation compared to controls infected in the absence of NH4Cl. Background incorporation by uninfected cells was subtracted from all points. Data represent the mean of three separate experiments for each virus, with standard deviations between 1 and 20%.
Role of E1/E2 dimer dissociation in the fus-1 phenotype.
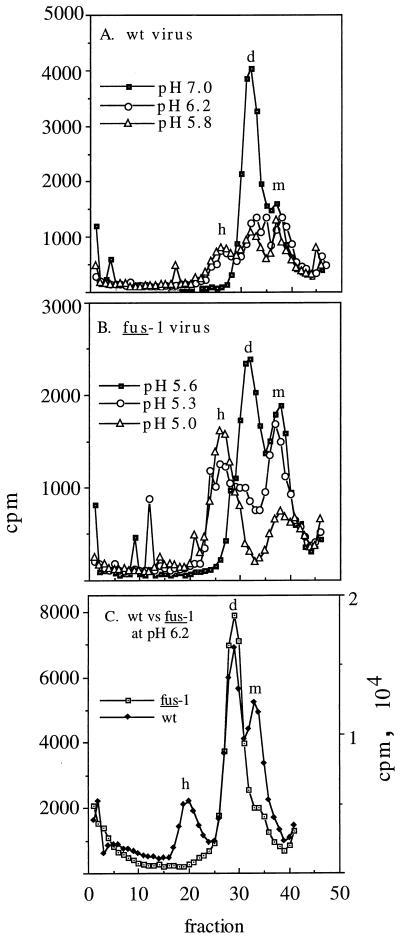
Following acid treatment, the first spike protein conformational change detected is a weakening of the tight E1/E2 heterodimer interaction, resulting in the loss of both coimmunoprecipitation and cosedimentation on sucrose gradients. For wt virus, this conformational change has a threshold of ∼pH 7.0 (33) and occurs with kinetics considerably faster than those of E1 homotrimerization and conversion to MAb reactivity (14). We tested the pH required to dissociate the fus-1 dimer in order to determine if an altered dimer pH threshold might be responsible for the acid-shifted phenotype of the E1 and E2 subunits. Coimmunoprecipitation assays were not successful, as fus-1 was poorly recognized by our panel of anti-E2 and anti-E1 MAbs (data not shown). We therefore used sucrose gradient sedimentation analysis of pH-treated, NP-40-solubilized virus to characterize the oligomeric structure of the E1 and E2 subunits of the spike protein (6, 33). 35S-labeled wt or fus-1 was pH treated for 10 min at 37°C, solubilized, neutralized, and analyzed by sedimentation on 5 to 20% sucrose gradients at pH 7.0. For wt SFV, the expected dimer and monomer peaks were observed in pH 7.0-treated virus, while in virus exposed to pH 6.2 or 5.8, the dimer peak decreased, and a homotrimer peak was formed (Fig. 4A). In contrast, fus-1 virus maintained clear dimer and monomer peaks even following treatment as low as pH 5.6. Treatment at pH 5.3 or below was required to completely dissociate the dimer peak and trigger the formation of a homotrimer peak (Fig. 4B). While the wt virus showed considerable dimer dissociation and homotrimer formation at pH 6.2, the fus-1 mutant showed almost no dimer dissociation and no homotrimer following treatment at pH 6.2 (Fig. 4C). Aliquots of the peaks were analyzed by SDS-PAGE, and in keeping with the published reports (34, 35), the results confirmed that the dimer peak contained E1 and E2, while the monomer peak was primarily E2 and the homotrimer peak was primarily E1 (data not shown).
FIG. 4.
Sucrose gradient sedimentation profiles of wt and fus-1 viral spike proteins. 35S-labeled wt (A and C) or fus-1 (B and C) virus was mixed with 1 mM liposomes, treated at the indicated pH for 10 min at 37°C, solubilized in 1.0 to 0.2% NP-40, and adjusted to pH 7.0. The samples were analyzed by centrifugation on 5 to 20% (wt/wt) sucrose gradients in buffer containing 0.1% NP-40. Gradients were centrifuged 22 h at 4°C in an SW41 rotor at 39,000 rpm and fractionated, and radioactivity was quantitated by scintillation counting. Fraction 1 represents the bottom of the gradient. The positions of the monomer (m), dimer (d), and E1 homotrimer (h) peaks are indicated. Recoveries ranged from 53 to 29%.
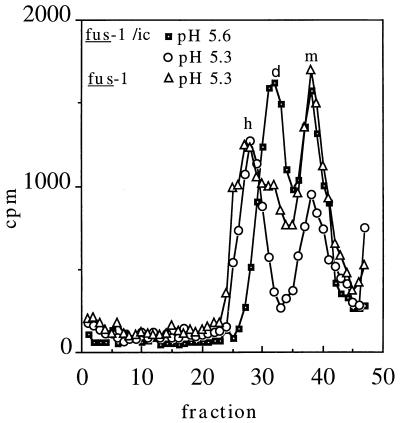
Radiolabeled virus prepared from the fus-1/ic construct was used to test if the phenotype of the cloned mutant was identical to that of fus-1 virus (Fig. 5). Similar to fus-1, fus-1/ic showed a clear spike protein dimer peak following treatment as low as pH 5.6, and dimer dissociation and homotrimer formation following treatment at pH 5.3. Thus, these results indicate that dissociation of the fus-1 E1/E2 dimer is indeed significantly acid shifted. Similar to the mild acid shift of S1J homotrimer formation, the pH dependence of S1J dimer dissociation was also somewhat acid shifted from that of wt SFV (data not shown).
FIG. 5.
Sucrose gradient sedimentation profiles of fus-1 and fus-1/ic viral spike proteins. 35S-labeled fus-1 or fus-1/ic virus was mixed with 1 mM liposomes and treated at the indicated pH for 10 min at 37°C. Samples were solubilized, neutralized, and analyzed by sucrose gradient sedimentation as for Fig. 4. Fraction 1 represents the bottom of the gradient. The positions of the monomer (m), dimer (d), and E1 homotrimer (h) peaks are indicated. Recoveries ranged from 32 to 63%.
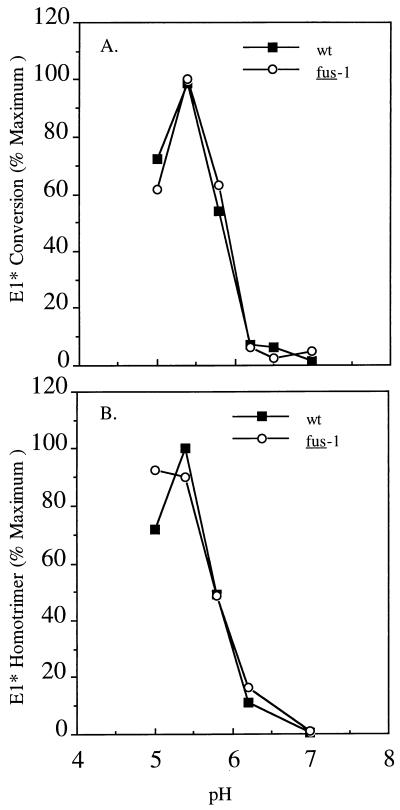
If the pH shift in fus-1 dimer dissociation is responsible for the acid shift in its E1 conformational changes, then predissociation of the dimer should permit the fus-1 E1 subunit to undergo conformational changes with the same pH dependence as that of wt E1. To assay such a monomeric form of E1, we prepared the E1 ectodomain fragment of the protein, E1*. E1* is a soluble form of E1 in which the protein transmembrane domain has been removed by proteinase K cleavage (17). This form of E1 undergoes similar acid-induced conformational changes as the intact form of the protein, including epitope exposure and formation of a trypsin-resistant homotrimer (17, 22). However, E1* is monomeric and no longer associated with E2 (17, 22). The pH dependence of wt E1* and that of fus-1 E1* were compared by measuring reactivity with an acid conformation-specific MAb, and the proteins were found to have identical pH thresholds of ∼pH 5.8 (Fig. 6A). The pH required to trigger E1* homotrimer formation was also determined for wt SFV and fus-1 and found to be ∼5.8 for both proteins (Fig. 6B). These data indicate that as a monomer not associated with the E2 subunit, fus-1 E1* can undergo its pH-dependent conformational changes with the same pH threshold as wt SFV E1* and support the conclusion that the fus-1 defect involves a pH shift in the dissociation of the E1/E2 dimer.
FIG. 6.
pH dependence of E1* conformational changes. (A) Reactivity with MAb E1a-1. 35S-labeled wt and fus-1 ectodomains were treated at the indicated pH for 10 min at 37°C in the presence of 1 mM liposomes, adjusted to neutral pH, solubilized in lysis buffer containing 1% Triton X-100, and immunoprecipitated with MAb E1a-1. E1* precipitation was quantitated by SDS-PAGE and phosphorimaging and compared to total E1* precipitated by a rabbit polyclonal antibody. Data represent the mean of three separate experiments. (B) E1* homotrimer formation. 35S-labeled wt SFV and fus-1 ectodomains were treated at the indicated pH for 10 min at 37°C in the presence of liposomes, adjusted to neutral pH, and solubilized in SDS sample buffer for 3 min at 30°C. E1 homotrimer formation was quantitated by SDS-PAGE and phosphorimaging. Data represent means of three separate experiments.
Determination of the mutation responsible for the fus-1 phenotype.
As described above, the infectious clone encoding the fus-1 structural proteins reproduced the fus-1 phenotype in all assays, indicating that the mutation(s) responsible for the fus-1 defect was contained within the subcloned NdeI-BglII fragment. The sequence of this fragment in fus-1/ic was compared to that of wt/ic (obtained from Peter Liljeström). Six amino acid changes were observed. All changes in fus-1/ic were confirmed to be present in the fus-1 virus stock by reverse transcription of RNA from fus-1 virus–PCR amplification (RT-PCR) and sequencing of selected regions. The sequences at these positions in wt/ic and in our wt virus stock were confirmed by sequence analysis of the clone or RT-PCR and sequencing of virus RNA.
Figure 7 lists the amino acid sequence differences found between the fus-1/ic and fus-1 virus sequences and the wt/ic and wt virus sequences. The fus-1 E3 and 6K sequences were found to be identical to the wt sequence. The fus-1 capsid sequence contained two amino acid changes, glutamine 80 to proline (CAG→CCG) and asparagine 85 to lysine (AAC→AAA). Three changes, threonine 12 to isoleucine (ACA→ATA), lysine 162 to glutamate (AAG→GAG), and histidine 170 to arginine (CAT→CGT), were observed in E2. One change, aspartic acid 323 to alanine (GAC→GCC), occurred in E1.
Since fus-1 was derived from S1J, some of the six amino acid changes could be due to its genetic background. Although S1J and wt SFV came from the same original SFV isolate and have similar passage histories, the two wt stocks are different plaque-purified isolates (20, 31). Partial sequence analysis of S1J had previously revealed two amino acid differences from the published wt SFV sequence, E1 D323→A, and E2 H170→R (31). Therefore, these two amino acid changes detected in fus-1 were due to its S1J background. RT-PCR and sequence analysis of S1J RNA were performed, and the results demonstrated that all of the fus-1/ic amino acid alterations were due to background changes in the parental S1J virus, with the single exception of E2 T12→I (Fig. 7). This result suggested that the fus-1 pH shift phenotype was due to the substitution of isoleucine at E2 position 12, a position that is threonine in both wt SFV and S1J.
To confirm that the E2 T12→I amino acid change was in fact the specific mutation responsible for the fus-1 phenotype, we analyzed the sequences of two fus-1 revertants, R43 and R46, in the regions containing the six amino acid changes from wt SFV (Fig. 7). The sequence of R43 was identical to that of the S1J parent and had reverted to threonine at E2 position 12. The sequence of R46 was identical to S1J at all six positions, including E2 T12, but in addition contained an extra mutation of alanine to serine at E2 position 70. This mutation presumably does not affect the R46 fusion phenotype, as its membrane fusion and NH4Cl sensitivity were identical to those of S1J, R43, and wt SFV (Fig. 3 and reference 21). Thus, fus-1 contains a critical amino acid change, E2 I12, which reverts to the parental T12 in two fus-1 revertants, identifying E2 I12 as the genetic alteration producing the acid-shifted fusion phenotype of fus-1.
DISCUSSION
The SFV mutant fus-1 has been used in cell biological studies as a probe for the pH of compartments of the endocytic pathway (20, 21, 29). The mutant was also important in establishing the correlation between the pH threshold of SFV fusion and the sensitivity of virus infection to various concentrations of weak bases, ionophores, or specific inhibitors of the vacuolar proton pump such as bafilomycin (10, 20). The finding that an SFV mutant with a lower pH threshold for in vitro fusion also has a lower pH threshold for spike protein conformational changes and a higher sensitivity to agents that neutralize endosomal acidity provided strong evidence for the key role of low pH in triggering fusion during SFV infection. In spite of its use in such cell biology and virology systems, however, the genetic alteration in fus-1 was unknown, and its phenotype of lower pH thresholds for the conformational changes in both the E1 and E2 subunits was puzzling. Recent studies demonstrated that mL, an engineered virus with a p62 cleavage mutation, had a more acidic pH threshold for virus fusion and E1 conformational changes, due to a more acidic pH threshold for dimer dissociation (28). The similarities in the phenotypes of fus-1 and mL suggested that the fus-1 mutation could be acting to shift the pH of dimer dissociation. Kinetic studies of wt dimer dissociation indicate that it occurs prior to E1 epitope exposure and homotrimer formation, in keeping with its possible role in regulating these conformational changes (14). Our results revealed that the fus-1 E1 conformational changes all had a more acidic pH threshold than wt and that the dissociation of the fus-1 E1/E2 dimer was also shifted to a more acidic pH. In contrast, monomeric E1* from fus-1 had the same pH threshold as wt E1* for MAb reactivity and homotrimer formation, indicating that the pH shift in E1 conformational changes was lost once the fus-1 E1/E2 dimer interaction was disrupted. Sequence analysis of fus-1 and two fus-1 revertants demonstrated that the relevant amino acid alteration in fus-1 was a change of E2 T12→I. Thus, the substitution of isoleucine on E2 acted to stabilize the dimer interaction between the fus-1 E1 and E2 subunits and regulated the pH dependence of E1-catalyzed fusion.
In addition to the T12→I change, fus-1 has five other changes from our wt SFV strain, all of which are background changes found in its parent strain, S1J. S1J is similar to our plaque-purified wt strain in the pH dependence of cell-cell fusion, NH4Cl sensitivity, E1 MAb reactivity, and E1 homotrimer formation (this study and reference 20). In previous in vivo studies, we characterized wt, fus-1, and revertant RNA release into the cytoplasm following fusion in the endosome (21). Since endosomes become more acidic with time (27), viruses with a lower pH threshold of fusion do not fuse until later in the endocytic pathway and show a slower release of virus RNA. The half-time of wt RNA penetration was ∼15 min, and that of fus-1 was ∼45 min. The half-time of revertant RNA penetration was ∼25 min. This difference in the kinetics of RNA release between wt and revertants may reflect S1J background differences, which merit further investigation.
In previous unpublished experiments, the cDNA encoding the fus-1 structural proteins was expressed transiently in COS cells via a simian virus 40 vector. Surprisingly, although this system faithfully reproduces the phenotype of other SFV fusion mutants (13, 19, 23), the fus-1 spike protein did not exhibit an acid-shifted pH threshold for COS cell fusion. This same cDNA showed a markedly acid-shifted fusion threshold when here expressed in the context of the SFV infectious clone. These results could be due to technical limitations of the syncytium assay, which can be affected by such variables as spike protein density (36). Alternatively, a requirement for the nucleocapsid or assembled virus particle may be necessary for expression of the fus-1 phenotype.
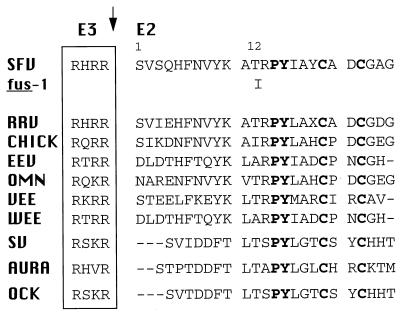
The alphavirus structural proteins are ∼51% identical in the E1 subunit and ∼42% identical in the E2 subunit, and they show striking conservation of residues such as cysteine and proline that are of particular importance in conferring secondary structure, suggesting overall conservation of spike structure (30). The fus-1 T12→I substitution occurs close to the p62 cleavage site within the amino-terminal domain of E2 and is predicted to cause an increase in the hydrophobicity of this region and the loss of the hydrogen bonding capability of threonine. Figure 8 shows the amino acid sequences of several different alphaviruses in the E2 T12 region. The region contains invariant proline at position 14, tyrosine at position 15, and cysteine residues at 19 and 22. Seven of ten alphaviruses contain threonine at position 12, while eastern and western equine encephalitis viruses contain alanine at position 12. Interestingly, like fus-1, Chickungunya virus has isoleucine at position 12. The fusion pH threshold for this alphavirus is not known.
FIG. 8.
Sequence comparison of the E2 T12 region in alphaviruses. Amino acid numbering is given for SFV E2, starting with residue 1 at the amino terminus. The tetrabasic cleavage site which generates E3 and E2 is boxed, invariant residues in E2 are shown in bold, and sequence gaps are shown as dashes. The sequence of this region of fus-1 is identical to that of wt SFV except for the T12→I change. RRV, Ross River virus; CHICK (Chickungunya virus) (NCBI accession no. 576465); EEV, eastern equine encephalitis virus; OMN, O’Nyong-nyong virus; VEE, Venezuelan equine encephalitis virus; WEE, western equine encephalitis virus; SV, Sindbis virus; AURA, Aura virus; OCK, Ockelbo virus, as listed in references 16 and 30.
Our results indicate that isoleucine at the E2 12 position leads to increased stability of the E1/E2 dimer. Additional examples of a potential role for isoleucine in the stabilization of protein-protein interactions are suggested by analysis of influenza virus hemagglutinin (HA) mutants selected for a higher pH threshold of fusion. HA is a trimer containing three copies of HA1, the receptor binding domain, and HA2, which makes up the stem region and contains the transmembrane domain and N-terminal fusion peptide (see references 12 and 37 for reviews). HA mutants that fuse at higher pH fall into two classes. One group contains mutants that map within or close to the fusion peptide and act to destabilize its normally buried position in the HA trimer. Among such destabilizing mutants is a change of the wt isoleucine at HA2 6, within the fusion peptide, to methionine. The second class of fusion mutants localizes to residues in the trimer interface that stabilize inter-HA interactions. A mutant at position HA2 81 changes a wt isoleucine to serine, resulting in a less stable HA trimer and a higher fusion threshold. Thus, these two examples in a pH-dependent fusion protein with well-characterized structure suggest that the presence of an isoleucine may act to stabilize subunit interactions, similar to our model for the effect of the fus-1 mutation.
The currently available information suggests that several different types of mutations can affect the pH threshold of alphavirus fusion via different mechanisms. As discussed here, the fus-1 mutation and mutations that prevent p62 cleavage decrease the fusion pH threshold by increasing the stability of the E1/E2 dimer interaction. Mutations within the putative E1 fusion peptide can also cause a more acidic pH threshold for fusion (19, 23). In contrast to fus-1, such fusion peptide mutants have an E1/E2 dimer interaction less stable than that of wt virus but have a marked decrease in the pH threshold and efficiency with which E1 subsequently trimerizes and associates with the target membrane (19). A Sindbis virus neurovirulence mutant has two mutations in different regions within the E1 subunit, valine 72→alanine and glycine 313→aspartate, which cause a more acidic fusion threshold by an unknown mechanism (3). Taken together, results from these alphavirus fusion mutants suggest that various domains of the spike protein are involved in the overall control of the protein’s response to acid pH and that these domains play different roles in the fusion reaction. Our results with fus-1 identify a domain of the SFV E2 spike subunit that controls fusion pH dependence and indicate that a mutation in this region acts indirectly on E1 by its effect on the dimer stability.
ACKNOWLEDGMENTS
We thank Matthew Klimjack and Anna Ahn for technical assistance, the members of our laboratory for helpful discussions and suggestions, and Duncan Wilson and the members of our laboratory for critical reading of the manuscript. We thank Jack Lenz for very helpful advice on the use of the revertants, and we thank Peter Liljeström and Henrik Garoff for providing pSP6-SFV4.
This work was supported by grants to M.K. from the Public Health Service (GM52929) and the Hirschl Charitable Trust, by a Jack K. and Helen B. Lazar fellowship in cell biology, and by Cancer Center core support grant NIH/NCI P30-CA13330. S.G.-R. was supported by NIH training grant 2T32 CA09173-15.
REFERENCES
- 1.Bentz J. Viral fusion mechanisms. Boca Raton, Fla: CRC Press; 1993. pp. 1–529. [Google Scholar]
- 2.Binley J, Moore J P. HIV-cell fusion: the viral mousetrap. Nature. 1997;387:346–348. doi: 10.1038/387346a0. [DOI] [PubMed] [Google Scholar]
- 3.Boggs W M, Hahn C S, Strauss E G, Strauss J H, Griffin D E. Low pH-dependent Sindbis virus-induced fusion of BHK cells: differences between strains correlate with amino acid changes in the E1 glycoprotein. Virology. 1989;169:485–488. doi: 10.1016/0042-6822(89)90178-5. [DOI] [PubMed] [Google Scholar]
- 4.Bron R, Wahlberg J M, Garoff H, Wilschut J. Membrane fusion of Semliki Forest virus in a model system: correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 1993;12:693–701. doi: 10.1002/j.1460-2075.1993.tb05703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.deCurtis I, Simons K. Dissection of Semliki Forest virus glycoprotein delivery from the trans-Golgi network to the cell surface in permeabilized BHK cells. Proc Natl Acad Sci USA. 1988;85:8052–8056. doi: 10.1073/pnas.85.21.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duffus W A, Levy-Mintz P, Klimjack M R, Kielian M. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J Virol. 1995;69:2471–2479. doi: 10.1128/jvi.69.4.2471-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 8.Garoff H, Frischauf A-M, Simons K, Lehrach H, Delius H. Nucleotide sequence of cDNA coding for Semliki Forest virus membrane glycoproteins. Nature. 1980;288:236–241. doi: 10.1038/288236a0. [DOI] [PubMed] [Google Scholar]
- 9.Garoff H, Wilschut J, Liljestrom P, Wahlberg J M, Bron R, Suomalainen M, Smyth J, Salminen A, Barth B U, Zhao H. Assembly and entry mechanisms of Semliki Forest virus. Arch Virol. 1994;9:329–338. doi: 10.1007/978-3-7091-9326-6_33. [DOI] [PubMed] [Google Scholar]
- 10.Glomb-Reinmund, S., and M. Kielian. 1998. Unpublished data.
- 11.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 12.Hughson F M. Structural characterization of viral fusion proteins. Curr Biol. 1995;5:265–274. doi: 10.1016/s0960-9822(95)00057-1. [DOI] [PubMed] [Google Scholar]
- 13.Jain S K, DeCandido S, Kielian M. Processing of the p62 envelope precursor protein of Semliki Forest virus. J Biol Chem. 1991;266:5756–5761. [PubMed] [Google Scholar]
- 14.Justman J, Klimjack M R, Kielian M. Role of spike protein conformational changes in fusion of Semliki Forest virus. J Virol. 1993;67:7597–7607. doi: 10.1128/jvi.67.12.7597-7607.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keränen S, Kääriäinen L. Isolation and basic characterization of temperature-sensitivity mutants from Semliki Forest virus. Acta Pathol Microbiol Scand Sect B. 1974;82:810–820. doi: 10.1111/j.1699-0463.1974.tb02378.x. [DOI] [PubMed] [Google Scholar]
- 16.Kielian M. Membrane fusion and the alphavirus life cycle. Adv Virus Res. 1995;45:113–151. doi: 10.1016/s0065-3527(08)60059-7. [DOI] [PubMed] [Google Scholar]
- 17.Kielian M, Helenius A. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J Cell Biol. 1985;101:2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kielian M, Jungerwirth S, Sayad K U, DeCandido S. Biosynthesis, maturation, and acid activation of the Semliki Forest virus fusion protein. J Virol. 1990;64:4614–4624. doi: 10.1128/jvi.64.10.4614-4624.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kielian M, Klimjack M R, Ghosh S, Duffus W A. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J Cell Biol. 1996;134:863–872. doi: 10.1083/jcb.134.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kielian M C, Keränen S, Kääriäinen L, Helenius A. Membrane fusion mutants of Semliki Forest virus. J Cell Biol. 1984;98:139–145. doi: 10.1083/jcb.98.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kielian M C, Marsh M, Helenius A. Kinetics of endosome acidification detected by mutant and wild-type Semliki Forest virus. EMBO J. 1986;5:3103–3109. doi: 10.1002/j.1460-2075.1986.tb04616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klimjack M R, Jeffrey S, Kielian M. Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J Virol. 1994;68:6940–6946. doi: 10.1128/jvi.68.11.6940-6946.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy-Mintz P, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobigs M, Garoff H. Fusion function of the Semliki Forest virus spike is activated by proteolytic cleavage of the envelope glycoprotein precursor p62. J Virol. 1990;64:1233–1240. doi: 10.1128/jvi.64.3.1233-1240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobigs M, Wahlberg J M, Garoff H. Spike protein oligomerization control of Semliki Forest virus fusion. J Virol. 1990;64:5214–5218. doi: 10.1128/jvi.64.10.5214-5218.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellman I, Fuchs R, Helenius A. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- 28.Salminen A, Wahlberg J M, Lobigs M, Liljeström P, Garoff H. Membrane fusion process of Semliki Forest virus II: cleavage-dependent reorganization of the spike protein complex controls virus entry. J Cell Biol. 1992;116:349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid S L, Fuchs R, Kielian M, Helenius A, Mellman I. Acidification of endosome subpopulations in wild-type Chinese hamster ovary cells and temperature-sensitive acidification-defective mutants. J Cell Biol. 1989;108:1291–1300. doi: 10.1083/jcb.108.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Syvaoja P, Peranen J, Suomalainen M, Keränen S, Kääriäinen L. A single amino acid change in E3 of ts1 mutant inhibits the intracellular transport of SFV envelope protein complex. Virology. 1990;179:658–666. doi: 10.1016/0042-6822(90)90133-c. [DOI] [PubMed] [Google Scholar]
- 32.Vashishtha M, Phalen T, Marquardt M T, Ryu J S, Ng A, Kielian M. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J Cell Biol. 1998;140:91–99. doi: 10.1083/jcb.140.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahlberg J M, Boere W A M, Garoff H. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J Virol. 1989;63:4991–4997. doi: 10.1128/jvi.63.12.4991-4997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahlberg J M, Bron R, Wilschut J, Garoff H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J Virol. 1992;66:7309–7318. doi: 10.1128/jvi.66.12.7309-7318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahlberg J M, Garoff H. Membrane fusion process of Semliki Forest virus I: low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol. 1992;116:339–348. doi: 10.1083/jcb.116.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White J M. Viral and cellular membrane fusion proteins. Annu Rev Phys. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- 37.Wiley D C, Skehel J J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]