Abstract
To study hepatitis C virus (HCV) genetic mutation during interferon (IFN) therapy, the temporal changes in HCV quasispecies heterogeneity were compared before and after treatment for nine patients infected with HCV genotype 1, including four nonresponders, four responders who relapsed after therapy, and one responder who experienced a breakthrough of viremia during therapy. Nine untreated patients with an average time between specimens of 8.4 years served as controls. Sequences from the second envelope glycoprotein gene hypervariable region 1 (HVR1) and the putative IFN sensitivity-determining region (ISDR) of the nonstructural NS5A gene were analyzed by heteroduplex mobility assays and nucleotide sequencing. A strong positive correlation was found between the percent change in a heteroduplex mobility ratio (HMR) and percent change in nucleotide sequence (r = 0.941, P < 0.001). The rate of fixation of mutations in the HVR1 was significantly higher for IFN-treated patients than for controls (6.97 versus 1.31% change in HMR/year; P = 0.02). Similarly, a higher rate of fixation of mutations was observed in the ISDR for IFN-treated patients than for untreated controls, although the result was not significant (1.45 versus 0.15 amino acid changes/year; P = 0.12). On an individual patient basis, IFN therapy was associated with measurable HVR1 and ISDR mutation in nine of nine (100%) and two of nine (22.2%) patients, respectively. Evolution to IFN-resistant ISDR sequences was observed in only one of nine IFN-treated patients. These data suggest that IFN therapy frequently exerts pressure on the HCV envelope region, while pressure on the ISDR was evident in only a subset of patients. Thus, the selection pressures evoked on HCV genotype 1 quasispecies during IFN therapy appear to differ among different patients.
Hepatitis C virus (HCV), the etiologic agent of chronic non-A, non-B hepatitis, is an enveloped, positive-stranded RNA virus classified within the flaviviridae (4). Acute HCV infection results in persistent viremia in 85 to 95% of cases, and at least 60% of infected individuals develop chronic hepatitis (1). Furthermore, approximately 20 to 30% of chronic hepatitis C cases eventually progress to cirrhosis and/or hepatocellular carcinoma, and chronic hepatitis C is now the leading indication for orthotopic liver transplantation in the United States (1).
Currently, recombinant alpha interferon (IFN), at the standard dose of 3 to 5 mU three times per week, is the most widely used treatment for chronic hepatitis C. A 6-month course of systemic IFN therapy leads to normalization of serum alanine aminotransferase levels in 40 to 50% of cases. However, biochemical relapse following discontinuation of therapy is common (6, 9, 28, 29). Virological factors including high pretreatment titers of HCV, and the viral genotype have been associated with either lack of response or relapse after therapy (6, 26, 44, 48, 53, 54).
HCV exists in infected individuals as a quasispecies which usually consists of a predominant viral variant and a variable mixture of highly related yet genetically distinct variants (35). The study of the biological role of HCV quasispecies has historically focused on hypervariable region 1 (HVR1) of the second envelope (E2) glycoprotein gene (25, 50). Numerous studies suggest that the HVR1 is a target of neutralizing antibodies and that the selection directed at this region of the E2 protein is responsible for the fixation of the apparent hypervariability (13, 24, 32, 39, 49, 56). With respect to the role of HVR1 in response to IFN therapy, previous studies have reported an association between a high level of mutation within the HVR1 and failure to respond to IFN (16, 30, 37, 38, 40, 47). In contrast, for the nonstructural NS5A gene, conservation of sequence was associated with lack of response to IFN therapy: a consensus IFN sensitivity-determining region (ISDR) sequence was associated with lack of response to IFN in Japanese patients infected with HCV genotype 1b (HCV-1b), while mutations within the ISDR were associated with response to IFN therapy (2, 11, 12). Recent studies from Europe on HCV-1b-infected patients (15, 33, 43, 55) do not support such a correlation between ISDR sequences and responses to IFN therapy.
In a recent study of North American patients infected with HCV-1, no correlation between ISDR sequences and responses to IFN therapy was found in 15 HCV-1a-infected patients, while the ISDR consensus or intermediate sequence was detected in four of five HCV-1b-infected patients who either were nonresponsive to IFN therapy or responded and then relapsed after stopping IFN therapy (27). Moreover, it has recently been shown that the NS5A gene product interacts with the IFN-induced cellular protein kinase, PKR, in an ISDR-dependent fashion, inhibiting PKR activity (14). Thus, the interaction of NS5A with PKR may represent one mechanism used by HCV to resist the antiviral activities of IFN and may explain the clinical observations of HCV-1b ISDR mutation and response to IFN therapy. Therefore, to further investigate the role of HCV genetic divergence in the development of resistance to IFN therapy, we performed a detailed analysis of the mutational frequency of the E2 HVR1 and NS5A ISDR, before and after standard IFN therapy, using a combination of heteroduplex analysis (7, 8, 21, 40, 52) and nucleotide sequencing. For comparison, we also present data on E2 HVR1 and NS5A ISDR evolution rates in nine untreated control patients.
MATERIALS AND METHODS
Patients and virologic monitoring.
Serum samples were obtained before, during, and after IFN therapy from nine patients with active HCV infections who were participating under informed consent in ongoing studies at the University of Washington Medical Center. Active HCV infection was determined by positive serologic testing for HCV antibodies (EIA2 [Abbott Laboratories] and RIBA II [Ortho Diagnostics]), by positive testing for HCV RNA by reverse transcriptase-mediated PCR (RT-PCR) using primers specific to the 5′ untranslated region, and by abnormal biochemical markers (18, 22). HCV genotype was determined by restriction fragment length polymorphism analysis of the 5′ noncoding region (5). Changes in HCV RNA levels were monitored by quantitative competitive PCR and bDNA version 2.0 (Chiron Corporation, Emeryville, Calif.) (17, 19). Nonresponse was defined as continuous detection of HCV RNA in patient serum during therapy; four patients (patients 1 to 4) were designated nonresponders. Response to IFN therapy was defined virologically as the sustained conversion of a patient to HCV RNA-negative status by RT-PCR during therapy. Patients who relapsed following discontinuation of therapy (patients 6 to 9) were designated responder-relapse, while the patient who initially responded to IFN therapy but experienced breakthrough of HCV viremia while still on IFN therapy (patient 5) was designated a responder-breakthrough. In addition, sequential samples taken on average 8.4 years apart were obtained for nine control patients who did not receive IFN therapy. Of the nine patients, five were infected with HCV-1a and four were infected with HCV-1b. Heteroduplex analysis and nucleotide sequencing results from three control patients (patients 10 to 12) are presented in detail in this report.
RNA extraction, PCR, cloning, and sequencing.
Total RNA was extracted from patient sera by the single-step guanidinium method (3, 52). The HVR1 was amplified by reverse transcriptase-nested PCR and cloned as described previously (52). Nested PCR was also used to amplify the ISDR. For genotype 1a, outer primer set 5A-1a-2 (5′TGACGTCCATGCTCACTGAT and 5′GAGACTTCCGCAGGATTTCT) and inner primer set 5A-1a-1 (5′ CCTCCCATATAACAGCAGAG and 5′CGAAGGAGTCCAGAATCACC) were used. For genotype 1b, outer primer set 5A-1b-2 (5′CAGAGACGGCTAAGCGTAGG and 5′CTGGATTTCCGCAGGATCTC) and inner primer set 5A-1b-1 (5′TCCTTGGCCAGCTCTTCAGC and 5′TCCCTCTCATCCTCCTCGC) were used. ISDR sequences were amplified by hot-start nested PCR with the following cycling parameters: 30 s at 94°C, 25 s at 65°C, and 30 s at 72°C for 30 cycles with 50 pmol of each primer. PCR products were then cloned in the TA cloning vector (Invitrogen), and plasmid DNA containing HVR1 or ISDR inserts was prepared for sequencing using the QIAwell plasmid prep system (Qiagen) and sequenced by the fluorescence-based Taq dye deoxy terminator cycle sequencing system (ABI), using M13 universal primers as described previously (52). For direct sequencing of PCR products, a third primer set, 5A-1a-3 (5′TAGTCGGGCTTTTTCCACG and 5′ TAGGGTCGCAATTACCTTG) or 5A-1b-3 (5′CAGAGACGGCTAAGCGTAGG and 5′CTGGATTTCCGCAGGATCTC), was used in first-round PCR amplification, followed by either the 5A-1a-2 or 5A-1b-2 primer set in second-round PCR amplification. PCR products were gel purified and directly sequenced in both directions, using primer sets 5A-1a-1 and 5A-1b-1 for genotype 1a and 1b ISDR sequences, respectively. Sequences were analyzed with the Genetics Computer Group software. For calculation of the rate of fixation of mutation of the HVR1 and ISDR, the percent change in heteroduplex mobility ratio (HMR) (see below) per year was calculated.
Heteroduplex analysis.
The heteroduplex mobility assay is a new technique (7, 8) that we have applied for the assessment of genetic heterogeneity of HCV quasispecies (21, 40, 52). In a related technique, termed the heteroduplex tracking assay (HTA), a radiolabeled probe is hybridized to unlabeled target DNA and analyzed by nondenaturing polyacrylamide gel electrophoresis plus autoradiography (7, 8). Probe hybridized to itself (unlabeled) served as a marker for identification of homoduplexes. Hybrids with nucleotide changes relative to the probe displayed retarded mobility and were identified as heteroduplexes. To determine the total number of variants in a quasispecies population (complexity), the genetic diversity of the individual variants, and their relative abundance, clonal frequency analysis was performed as described previously (21, 40, 52). The clonal frequency analysis technique provides a detailed assessment of the level of quasispecies complexity and genetic diversity, because a large number of individual clones are simultaneously analyzed by hybridization with a patient-specific probe (21, 40, 52). In brief, PCR products from selected time points were ligated into the TA cloning vector, and individual clones were reamplified to generate clonal PCR products for heteroduplex analysis. At least 20 recombinant HVR1 or ISDR clones were subjected to clonal frequency analysis.
Quasispecies complexity was determined by counting the total number of unique gel shift patterns. Quasispecies genetic diversity was determined by deriving the average heteroduplex mobility of all clones relative to the homoduplex probe control. An HMR was calculated by dividing the distance in millimeters from the origin of the gel to the heteroduplex by the distance in millimeters from the origin to the homoduplex control. In cases where both strands of the heteroduplex were clearly distinguishable, the average of the distance of each strand of the heteroduplex was used to calculate heteroduplex mobility (40). The HMRs for all variants in the population were averaged to provide the final HMR value. To estimate the percent genetic change within the HVR1 and ISDR between two time points the percent change in HMR was calculated as (HMRtime 2 − HMRtime 1/HMRtime 1) × 100, where HMRtime 1 and HMRtime 2 represent the HMRs from pre-IFN and post-IFN therapy time points, respectively. For the untreated control patients, HMRtime 1 and HMRtime 2 represent the two time points at which serum was collected. To follow the temporal changes in the total quasispecies population during IFN therapy, HTA (7, 8) was performed with a radiolabeled probe hybridized separately to heterogeneous PCR products amplified from patient serum before, during, and after IFN therapy as described previously (21, 40, 52). For some patients, the entire heterogeneous PCR product was labeled by subjecting the second-round PCR products to an additional three cycles of PCR in the presence of [α-33P]dATP and 33 μmol of each deoxynucleoside triphosphate. Probes were then mixed with target at the ratio of 1:100 in annealing buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.8], 2 mM EDTA), denatured in boiling water for 2 min, placed immediately on ice for 10 min, and then incubated at 55°C for 10 min to form heteroduplexes (34). The resulting reaction products were electrophoresed in 5% neutral polyacrylamide gels, dried, and scanned with a Molecular Dynamics (Sunnyvale, Calif.) PhosphorImager.
Statistical analyses.
Student’s t tests were used to compare the differences between HMRs at different time points and between HVR1 and ISDR rates of fixation of mutations, while linear regression was used to determine the correlation between percent change in HMR and percent change in nucleotides.
RESULTS
Assessment of HCV infection during IFN therapy.
The virological and clinical features of the nine patients who were treated with IFN and three of the nine untreated control patients are shown in Table 1. There was no significant difference in pretreatment HCV titers between any of the response groups.
TABLE 1.
Clinical and virologic features of the 12 patients in this studya
| Patient | HCV genotype used for infection | Dose of IFNb | Response | Time point | HCV RNA titerc (log eq/ml) |
|---|---|---|---|---|---|
| 1 | 1b | 3 mU tiw | NR | Pre-Rx | 7.7 |
| During-Rx | 7.3 | ||||
| Post-Rx | 7.9 | ||||
| 2 | 1a | 3 mU tiw | NR | Pre-Rx | 7.2 |
| During-Rx | 6.0 | ||||
| Post-Rx | 7.3 | ||||
| 3 | 1a | 3 mU tiw | NR | Pre-Rx | 7.0 |
| During-Rx | 6.2 | ||||
| Post-Rx | 7.3 | ||||
| 4 | 1a | 3 mU tiw | NR | Pre-Rx | 7.3 |
| Post-Rx | 7.5 | ||||
| 5 | 1a | 5 mU tiw | RB | Pre-Rx | 7.7 |
| Breakthrough | 7.1 | ||||
| Post-Rx | 7.7 | ||||
| 6 | 1a | 5 mU tiw | RR | Pre-Rx | 7.4 |
| Post-Rx | Neg | ||||
| Relapse | 6.3 | ||||
| 7 | 1a | 3 mU tiw, 3 mo | RR | Pre-Rx | 6.4 |
| 3 mU daily, 3 mo | Post-Rx | Neg | |||
| Relapse | 6.1 | ||||
| 8 | 1a | 5 mU tiw, 3 mo | RR | Pre-Rx | 7.3 |
| 5 mU daily, 3 mo | Post-Rx | Neg | |||
| Relapse | 7.5 | ||||
| 9 | 1b | 5 mU tiw | RR | Pre-Rx | 7.7 |
| Post-Rx | Neg | ||||
| Relapse | 7.4 | ||||
| 10 | 1b | NA | Control | Time 0 | 6.6 |
| Time 12 yr | 6.7 | ||||
| 11 | 1a | NA | Control | Time 0 | 5.4 |
| Time 9 yr | 7.6 | ||||
| 12 | 1b | NA | Control | Time 0 | 6.8 |
| Time 13 yr | 5.8 |
Patients 1 to 4 were nonresponders (NR), while patient 5 was a responder who experienced virologic breakthrough (RB) during therapy (Rx). Patients 6 to 9 were responder-relapsers (RR) who were negative at the end of therapy but relapsed following cessation of therapy. Patients 10 to 12 represent untreated control patients.
Dose of IFN was for 6 months unless otherwise stated. tiw, three times per week; NA, not applicable.
Viral RNA was quantitated by bDNA assay, and whenever bDNA was negative, quantitative PCR was performed to determine the residual quantity of viral RNA. Neg, negative for HCV RNA by RT-PCR.
Utility of heteroduplex analysis to quantitate changes in quasispecies genetic diversity over time.
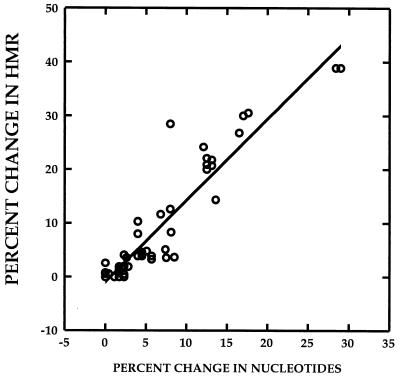
We have previously demonstrated that the extent of HCV quasispecies genetic diversity, expressed as an HMR (see Materials and Methods), is proportional to the nucleotide sequence differences between any probe and target molecule (40). Figure 1 depicts a strong correlation (r = 0.941, P < 0.01) between the percent change in nucleotide sequence between two quasispecies variants from two different time points and the percent change in HMR for the two variants, defined as (HMRtime 2 − HMRtime 1/ HMRtime 1) × 100. The data were derived by analysis of HCV heteroduplex mobilities for 60 DNA specimens amplified from the HVR1 (n = 48) and the ISDR (n = 12). These data indicate that it is possible to estimate the extent of change or evolution within the HVR1 and ISDR between any two time points in a given patient by measuring the percent change in HMR.
FIG. 1.
Estimation of percent change in genetic diversity between two time points, using heteroduplex analysis. The correlation between percent change in HMR and percent change in nucleotide sequence of quasispecies major variants between two time points is depicted. The r value for the correlation was 0.941 (P < 0.01). The data were derived from 60 paired specimens for which both HMR and nucleotide sequence data were available. Of the 60 measurements, 12 were derived from pairs of ISDR clones and 48 were derived from pairs of HVR1 clones.
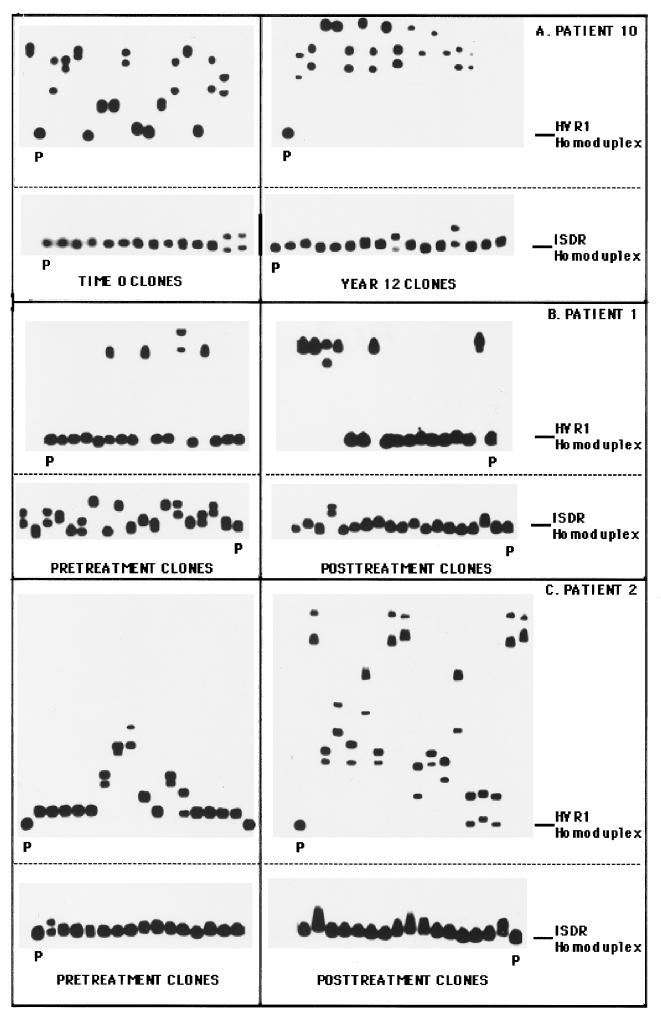
Figure 2 illustrates the changes in the clonal frequency of HCV quasispecies among three selected patients, determined by the clonal frequency analysis technique. In each case, mutation patterns in the HVR1 and ISDR were compared between two time points. Figure 2A depicts the changes in the HVR1 and ISDR for control patient 10 over a time interval of 12 years. For this patient, the HVR1 evolved extensively over 12 years, with the HMR decreasing from 0.881 to 0.779 (P < 0.01) between the two time points, indicating an increase in overall genetic diversity (Fig. 2A, top). The ISDR, however, remained genetically stable during this interval, as evidenced by similar clonal frequency analysis patterns and similar HMRs (0.992 versus 0.990, P = 0.08) at the two time points (Fig. 2A, bottom).
FIG. 2.
Clonal frequency analysis of the temporal changes in HCV quasispecies in the HVR1 and ISDR. The autoradiograms on the left represent clonal frequency analysis of the HVR1 or the ISDR derived from time zero (for patient 10) or pretreatment (patients 1 and 2) time point, while the autoradiograms on the right represent clonal frequency analysis of the HVR1 or ISDR derived from the year 12 (patient 10) or posttreatment (patients 1 and 2) time point. Radiolabeled HVR1 or ISDR probes corresponding to pretreatment quasispecies major variants were hybridized to HVR1 or ISDR PCR products derived from individual recombinant HVR1 or ISDR molecules, and heteroduplex analysis was performed as described in Materials and Methods. The position of the homoduplex is indicated with a line, while the reference homoduplex probe is labeled P. (A) Changes in the HVR1 and ISDR for control patient 10 with a time interval of 12 years between specimens; (B) profile of changes in the HVR1 and ISDR before and after IFN therapy (50 weeks) for nonresponsive patient 1; (C) profile of changes in the HVR1 and ISDR before and after IFN therapy (49 weeks) for nonresponsive patient 2.
Figure 2B illustrates the changes in the HVR1 and ISDR before and after IFN therapy for an IFN nonresponder (patient 1). In the HVR1, two predominant variant populations were detected at both time points for patient 1 (Fig. 2B, top). One variant formed slowly migrating heteroduplexes, indicating substantial divergence from the other variant. Upon sequencing, we found that variants 1 and 2 differed by 31 nucleotides and 16 amino acids (data not shown). In direct contrast to the HVR1, patient 1 displayed genetic heterogeneity within the ISDR in pretreatment serum, with a complexity of eight unique gel shifts variants (Fig. 2B, bottom). During IFN therapy, the heterogeneity of the ISDR quasispecies population lessened, so that at 32 weeks only three unique ISDR variants were detected. The HMR increased from pretreatment to the posttreatment time point (0.954 versus 0.988, P < 0.01), indicating a reduction in overall ISDR genetic diversity during therapy. These experiments indicated that for patient 1, the HCV quasispecies changed less extensively in the HVR1 than the ISDR during IFN. Furthermore, nonresponse to IFN therapy was associated with homogenization of the ISDR quasispecies population toward the intermediate type ISDR sequence associated with IFN resistance (12) (see Fig. 5).
FIG. 5.
Direct sequencing of the ISDR before and after IFN therapy. ISDR sequences are shown for IFN-treated patients 1 to 9 and untreated control patients 10 to 12. (A) Alignment of the ISDR from genotype 1a-infected patients relative to the ISDR of the prototype genotype 1a strain of HCV, HCV-1 (accession no. M62321). The underlines represent the three amino acid changes in the putative ISDR of genotype 1a that differ from the prototype genotype 1b ISDR. (B) Alignment of the ISDR from genotype 1b-infected patients relative to the consensus ISDR associated with IFN resistance from the prototype genotype 1b strain of HCV, HCV-J (accession no. D90208). For patient 5, BT represents the breakthrough time point. preMV1 and preMV2 represent the two ISDR variants detected in pretreatment (pre) serum of patient 1. post, posttreatment.
Figure 2C (top) indicates that for nonresponsive patient 2, HVR1 quasispecies heterogeneity increased following IFN therapy relative to the pretreatment time point, as evidenced by slowly migrating heteroduplexes in the posttherapy autoradiogram. The HMR decreased from 0.9303 to 0.725 (P < 0.01), indicating an overall increase in quasispecies genetic diversity. For this patient, one of the major HVR1 variants present at 49 weeks was derived from a minor HVR1 variant present in pretreatment serum, which suggested that selective expansion of a minor variant occurred during IFN therapy (data not shown). The ISDR quasispecies population in nonresponsive patient 2 appeared relatively unchanged by IFN therapy, as shown by similar clonal frequency patterns before and after therapy (Fig. 2C, bottom). The HMR of the ISDR remained unchanged between pretreatment and posttreatment time points (0.995 versus 0.995, P = 0.964). Therefore, these results indicate that for patient 2, evolution of the HVR1 occurred during IFN therapy, while the ISDR remained relatively unchanged.
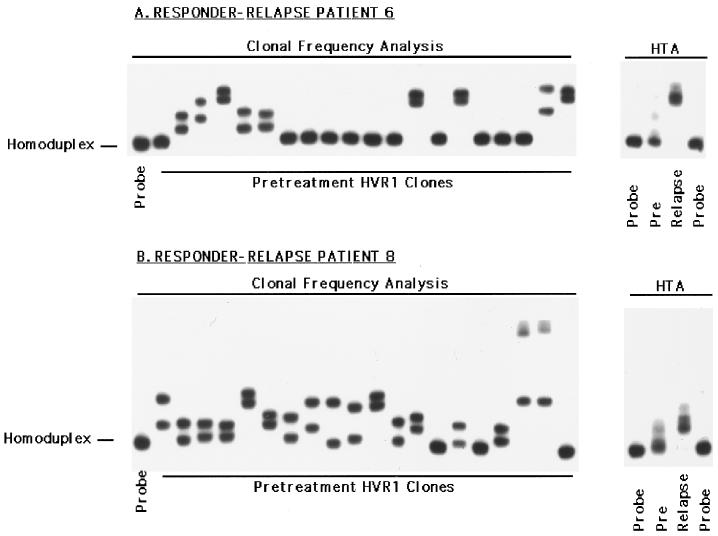
Changes in the HVR1 in responder-relapse patients.
Figure 3 depicts the changes in the HVR1 observed during IFN therapy for responder-relapse patients 6 and 8, using a combination of the HTA and clonal frequency analysis. The right side of Fig. 3A illustrates the HVR1 quasispecies profile detected by HTA for patient 6; the clonal frequency analysis from the pretreatment time point is shown the left. HTA clearly demonstrates a major change in the HVR1 quasispecies profile of this patient which was associated with virologic relapse, while the clonal frequency analysis indicates considerable genetic heterogeneity within this region at the onset of therapy (HMR = 0.968, complexity = 5 unique variants). Similarly for patient 8, virologic relapse was associated with a clear change in the HVR1 quasispecies distribution. Furthermore, there was extensive pretreatment HVR1 quasispecies genetic heterogeneity (Fig. 3B), as determined by the extent (HMR = 0.972) and number of unique gel shifts (complexity = 11 unique variants). Responder-relapse patient 7 also displayed major changes in the HVR1 coincident with virologic relapse, while patient 9 displayed changes in the proportion of minor HVR1 quasispecies variants (Fig. 4 and data not shown).
FIG. 3.
Assessment of pretreatment HVR1 quasispecies heterogeneity by clonal frequency analysis (left) and analysis of the temporal changes in HVR1 quasispecies by HTA (right) in responder-relapse patients 6 (A) and 8 (B). For clonal frequency analysis, representative pretreatment HVR1 clones were hybridized to a patient-specific, radiolabeled HVR1 probe and subjected to heteroduplex analysis. For HTA, heterogeneous HVR1 PCR products from the indicated time points were subjected to heteroduplex analysis.
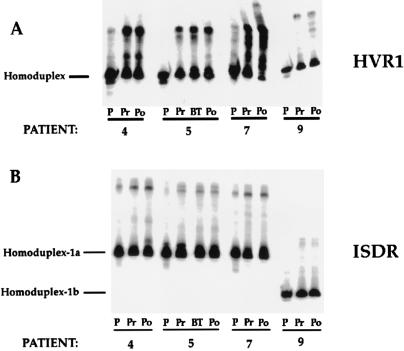
FIG. 4.
HTA for patients 4, 5, 7, and 9 for the HVR1 and ISDR. Pretreatment and posttreatment HVR1 and ISDR sequences were analyzed by HTA using the corresponding patient’s heterogeneous pretreatment PCR product as a radiolabeled probe. Lanes: P, 33P-radiolabeled probe; Pr, Po, and BT, pretreatment, posttreatment, and breakthrough time points, respectively. Note that for the ISDR, two different-size PCR products were analyzed, one corresponding to the ISDR from genotype 1a-infected patients and the other corresponding to the ISDR from genotype 1b-infected patients.
Changes in the HVR1 and ISDR in patients 4, 5, 7, and 9.
Figure 4 illustrates HTA of the HVR1 and ISDR derived from the pretreatment and posttreatment time points for patients 4, 5, 7, and 9. As shown in Fig. 4A, HVR1 sequences did not change significantly for nonresponder patient 4 or for responder-breakthrough patient 5. However, responder-relapse patient 7 displayed a gel shift pattern that was consistent with a change in the predominant HVR1 quasispecies variant during virologic relapse. Responder-relapse patient 9 also displayed changes in the proportions of minor HVR1 quasispecies variants during virologic relapse, but the predominant HVR1 quasispecies variant remained stable. In contrast to the mutations observed in the HVR1, no significant changes could be seen in the ISDR for patients 4, 7, and 9 following IFN therapy compared to the pretreatment quasispecies (Fig. 4B).
Direct sequencing analysis of the ISDR.
Direct sequencing of the PCR products from pretreatment and posttreatment time points confirmed the relative stasis of the ISDR during IFN therapy in most patients (Fig. 5). As described by Enomoto and colleagues (11), patients with ISDR sequences identical to the consensus HCV-1b sequence, HCV-J, were generally nonresponsive to IFN therapy. Eighty-seven percent of patients with one to three mutations in the ISDR (classified as intermediate-type sequences) were nonresponsive to IFN therapy, while most patients with four or more mutations in the ISDR relative to the consensus sequence were responsive to therapy. As shown in Fig. 5, nonresponsive patient 1 had two major ISDR variants in pretreatment serum; one (preMV2) had amino acid mutations consistent with it being classified as IFN sensitive, while the other major variant (preMV1) would be classified as intermediate type in the Enomoto classification scheme (12). Following IFN therapy, only intermediate-type ISDR sequences (preMV1) remained. Nonresponsive patients 2 to 4 all had ISDR sequences which did not change following IFN therapy. Three amino acid changes in the ISDR were detected at virologic relapse in responder-breakthrough patient 5. These changes were maintained until the end of therapy. Responder-relapse patients 6 to 9 all had ISDR sequences which did not change during IFN therapy. Control patients 10 and 11 each had ISDR sequences that differed by one amino acid between the 12- and 9-year follow-up specimens, respectively. Remarkably, the sequence of the ISDR in patient 12 changed significantly during the 13-year time period, accumulating nine amino acid changes and one deletion at codon 2218.
Rate of fixation of mutations in the HVR1 and ISDR.
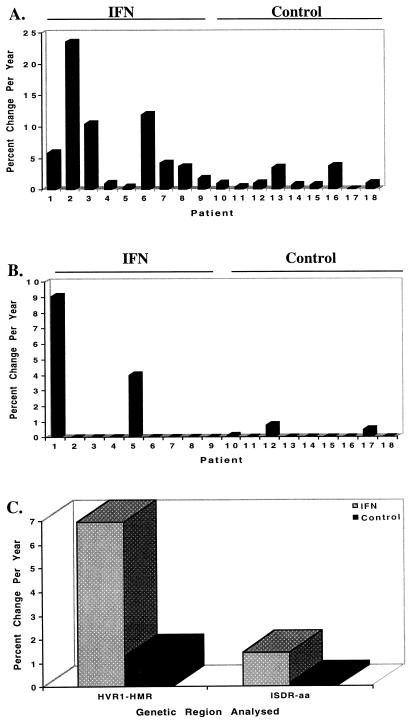
Due to the strong correlation between the percent change in HMR and percent nucleotide change between two time points, we were able to calculate the rate of fixation of mutations for the HVR1 and ISDR for our patient population. For the IFN-treated patients, the rate of fixation of mutation of the HVR1 was higher than that of the ISDR (6.97% versus 1.09% change in HMR/year, P = 0.019). The rate of fixation of mutation of the HVR1 was higher for IFN-treated patients than for all nine untreated control patients (6.97% versus 1.31% change in HMR/year, P = 0.02). Similarly, the rate of fixation of mutations in the ISDR was approximately 10-fold higher for IFN-treated patients than for untreated control patients (1.45% versus 0.15% percent amino acid changes/year), although the results for ISDR did not reach statistical significance (P = 0.12). IFN therapy was associated with detectable HVR1 and ISDR mutation in nine of nine (100%) and two of nine (22.2%) patients, respectively. These data are depicted graphically for individual IFN-treated and control patients in Fig. 6A and B and for both patient populations in Fig. 6C.
FIG. 6.
Summary of the changes in rate of fixation of mutations in the HVR1 and ISDR for IFN-treated patients 1 to 9 and untreated control patients 10 to 18. (A) Changes in the HVR1, expressed as the percent change in HMR per year; (B) changes in the ISDR, expressed as the percent change in amino acids per year; (C) summary graph comparing changes in both the HVR1 and ISDR for the IFN-treated and control patient populations. For the HVR1, the average values for IFN-treated and control patients were 6.97 and 1.31% percent change in HMR per year, respectively (P = 0.019); for the ISDR, the average values for IFN-treated and control patients were 1.45 and 0.15% change in amino acids per year, respectively (P = 0.12).
DISCUSSION
The effect of IFN therapy on HCV quasispecies is currently an important and controversial topic. This study presents detailed analysis of two regions of HCV-1 (HVR1 and ISDR) in nine patients before and after IFN therapy and in nine untreated control patients. The most controversial issue related to HCV quasispecies and IFN therapy pertains to the putative ISDR first described for genotype 1b isolates from Japan by Enomoto and colleagues (11). Although several studies have confirmed this report (2, 12), others do not support such an association (15, 33, 43, 55). Mutation of the HVR1 during natural HCV infection is well documented (13, 24, 25, 32, 39, 49, 50, 56), and several groups including our own have reported a significant inverse correlation between the extent of HVR1 divergence in pretreatment sera and subsequent response to IFN therapy (16, 30, 37, 38, 40, 47). Previous reports have demonstrated changes in HVR1 coincident with biochemical (10, 36) and virologic (38) relapse following cessation of IFN therapy. However, the present study is the first to quantify the rate of HVR1 divergence during therapy in direct comparison with a genotype-matched control population.
The present results can be summarized as follows. IFN therapy was associated with an increased rate of fixation of mutations in both the HVR1 and the ISDR of HCV-1 compared to the same regions analyzed in genotype-matched untreated control patients. The results were statistically significant for the HVR1 but not for the ISDR, even though the mean rate of ISDR divergence was 10-fold greater in treated patients than in controls. Similar to previous studies, we observed significant HVR1 divergence in two of four IFN nonresponders, the one responder breakthrough patient, and all four responder-relapse patients, further supporting the hypothesis that IFN partially acts through immunomodulatory mechanisms in chronic hepatitis C. The lack of statistical significance related to ISDR divergence may be a consequence of the small sample size (nine patients). However, accelerated genetic divergence of the ISDR was observed in only two of nine treated patients; in the remaining seven patients, the ISDR quasispecies master sequence remained stable during the 6-month course of IFN therapy (Fig. 5 and 6B). The most likely explanation for these results is that IFN exerts selective pressure on the ISDR of only a subset of patients with HCV-1 infection, which would be consistent with the controversial findings in clinical studies from Japan and Europe. It has been postulated that the differences in study outcome possibly reflects geographic differences in viral or host factors (23). It is noteworthy that one of the two patients with divergent ISDR sequences during therapy was infected with genotype 1a and the other was infected with genotype 1b; thus, our findings may not be related to HCV subtype. Although mutation in the HVR1 has been clearly linked to viral persistence in humans via antibody escape mechanisms (13, 32, 51), the selective forces acting on the ISDR could be immunological, as is postulated for HVR1, or could reflect molecular interactions with host cell proteins, as suggested by studies which demonstrate interaction of NS5A with the cellular protein kinases, PKR (14), and a member of the CMGC kinase family (41) (required for phosphorylation of NS5A [46]). Further studies will help better define the selective forces acting on the ISDR.
This study demonstrates that HVR1 and ISDR show different patterns of evolution under IFN pressure. In one subject (patient 1) who was infected with HCV-1b, we saw no significant effect of IFN on HVR1 sequences, as two major variants were consistently observed in roughly equal proportions in the patient’s serum before, during, and after therapy. Surprisingly, however, we observed striking alterations in the ISDR during therapy. Before treatment, many genetic variants existed in the ISDR, and the work of Enomoto et al. (11, 12) indicated that a majority had sequences associated with IFN sensitivity. During IFN therapy, this region became genetically more homogeneous, with a reduction in genetic complexity and diversity. The genetic variants which emerged during IFN therapy had the intermediate-type ISDR sequences associated with IFN resistance. This finding suggests that IFN induced a selective pressure on the ISDR toward the IFN-resistant phenotype and that IFN was not exerting selective pressure on the HVR1 in this case. However, it must be stressed that patient 1 was the only patient who displayed changes in the quasispecies distribution of the ISDR toward the IFN-resistant motif. The finding of nine amino acid changes and a single amino acid deletion at codon 2218 in the ISDR of untreated control patient 12 (Fig. 5) suggests that the ISDR diversification may occur as a result of other unidentified selective pressures. In this control patient, genetic change in the ISDR were associated with a 10-fold decrease in HCV RNA titers, suggesting the mutation may have reduced the replicative capacity of the virus.
The findings reported herein suggest that the ISDR locus per se does not function in a manner consistent with a major role in mediating IFN resistance in the majority of patients from our geographic area. This is in accord with recent studies from Europe (15, 33, 42, 55) and Japan, which described patients who had consensus IFN-resistant ISDR sequences yet still responded to IFN therapy (2). Furthermore, we have recently demonstrated that there is no particular ISDR sequence associated with response or nonresponse in HCV-1a-infected patients receiving IFN therapy, while there may be such an association for HCV-1b infections in patients from our geographic area (27). Thus, although the ISDR may modulate in part the response to IFN, as suggested by the inhibition of the IFN-induced protein kinase, PKR, by NS5A (14), it is possible that other domains of the NS5A protein possess functions which directly or indirectly influence response to IFN therapy. In this regard, recent studies indicate that the amino-terminal region of NS5A has a transcriptional activation function in yeast (31, 45). Thus, future clinical and molecular studies should be aimed at the entire coding region of NS5A in addition to other HCV genes.
The use of clonal frequency analysis in this study allowed multiple quasispecies clonal variants to be assayed simultaneously, which provided an accurate assessment of the overall level of quasispecies heterogeneity (reviewed in reference 20). This technique also identified certain HVR1 minor quasispecies variants which persisted during IFN therapy and reappeared as major quasispecies variants at the end of therapy (e.g., patient 2). This observation attests to the sensitivity of heteroduplex analysis in the detection of minor quasispecies variants and suggests that IFN therapy induced selective expansion of a minor quasispecies population. Using heteroduplex analysis, Gretch et al. also found emergence of minor quasispecies variants in liver transplant recipients with asymptomatic HCV infections (21).
The clinical importance of the current study is the demonstration of significant alterations in the HCV genome in nonresponsive or relapse patients infected with HCV-1 who were treated with IFN via the standard thrice-weekly regimen. Our results suggest a comparison of the antiviral efficacy of IFN when given via daily versus intermittent dosing regimens, or when given as monotherapy compared to combination therapy, to a cohort of matched subjects, i.e., those with similar HCV RNA levels and HCV genotypes at study initiation. Such combined clinical-molecular studies should prove useful for determining the mechanisms of action of therapeutic agents like IFN and for further optimizing antiviral therapy for chronic hepatitis C.
ACKNOWLEDGMENTS
We thank Jeff Wilson, Anthony Marquardt, Corazon dela Rosa, and Maureen Guajardo for technical assistance, Colleen Lasley and Jean Moore for assistance with preparation of the manuscript, and Jean-Michel Pawlotsky and Michael Katze for helpful discussions.
D.R.G. was partially supported by NIH grants AI41320-02 and AI39049-02; J.I.M. was supported by NIH grants AI27757 and AI32885. This research was partially funded by grants to D.R.G. from the University of Washington Royalty Research Fund and by a nonrestricted educational grant from Schering Plough.
REFERENCES
- 1.Anonymous. National Institutes of Health Consensus Development Conference Panel statement: management of hepatitis C. Hepatology. 1997;26:2S–10S. doi: 10.1002/hep.510260701. [DOI] [PubMed] [Google Scholar]
- 2.Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase H, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. doi: 10.1002/hep.510250342. [DOI] [PubMed] [Google Scholar]
- 3.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 4.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 5.Davidson F, Simmonds P, Ferguson J C, Jarvis L M, Dow B C, Follett E A, Seed C R, Krusius T, Lin C, Medgyesi G A, et al. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J Gen Virol. 1995;76:1197–1204. doi: 10.1099/0022-1317-76-5-1197. [DOI] [PubMed] [Google Scholar]
- 6.Davis G L, Balart L A, Schiff E R, Lindsay K, Bodenheimer H C, Jr, Perrillo R P, Carey W, Jacobson I M, Payne J, Dienstag J L, et al. Treatment of chronic hepatitis C with recombinant interferon alfa. A multicenter randomized, controlled trial. Hepatitis Interventional Therapy Group. N Engl J Med. 1989;321:1501–1506. doi: 10.1056/NEJM198911303212203. [DOI] [PubMed] [Google Scholar]
- 7.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen W H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 9.Di Bisceglie A M, Martin P, Kassianides C, Lisker Melman M, Murray L, Waggoner J, Goodman Z, Banks S M, Hoofnagle J H. Recombinant interferon alfa therapy for chronic hepatitis C. A randomized, double-blind, placebo-controlled trial. N Engl J Med. 1989;321:1506–1510. doi: 10.1056/NEJM198911303212204. [DOI] [PubMed] [Google Scholar]
- 10.Enomoto N, Kurosaki M, Tanaka Y, Marumo F, Sato C. Fluctuation of hepatitis C virus quasispecies in persistent infection and interferon treatment revealed by single-strand conformation polymorphism analysis. J Gen Virol. 1994;75:1361–1369. doi: 10.1099/0022-1317-75-6-1361. [DOI] [PubMed] [Google Scholar]
- 11.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A gene. J Clin Invest. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 13.Farci P, Alter H J, Wong D C, Miller R H, Govindarajan S, Engle R, Shapiro M, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gale M J, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 15.Germanidis G, Pellerin M, Bastie A, Stuyver L, Duverlie G, Darthuy F, Remire J, Duval J, Dhumeaux D, Pawlotsky J M. Study of the genetic heterogeneity of the NS5A region of the HCV-1b genome and evolution under interferon alfa therapy. Hepatology. 1996;24:264A. [Google Scholar]
- 16.Gonzalez-Peralta R P, Qian K, She J Y, Davis G L, Ohno T, Mizokami M, Lau J Y N. Clinical implications of viral quasispecies heterogeneity in chronic hepatitis C. J Med Virol. 1996;49:242–247. doi: 10.1002/(SICI)1096-9071(199607)49:3<242::AID-JMV14>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 17.Gretch D, Corey L, Wilson J, dela Rosa C, Willson R, Carithers R, Jr, Busch M, Hart J, Sayers M, Han J. Assessment of hepatitis C virus RNA levels by quantitative competitive RNA polymerase chain reaction: high-titer viremia correlates with advanced stage of disease. J Infect Dis. 1994;169:1219–1225. doi: 10.1093/infdis/169.6.1219. [DOI] [PubMed] [Google Scholar]
- 18.Gretch D, Lee W, Corey L. Use of aminotransferase, hepatitis C antibody, and hepatitis C polymerase chain reaction RNA assays to establish the diagnosis of hepatitis C virus infection in a diagnostic virology laboratory. J Clin Microbiol. 1992;30:2145–2149. doi: 10.1128/jcm.30.8.2145-2149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gretch D R, dela Rosa C, Carithers R L, Willson R A, Williams B, Corey L. Assessment of hepatitis C viremia using molecular amplification technologies: correlations and clinical implications. Ann Intern Med. 1995;123:321–329. doi: 10.7326/0003-4819-123-5-199509010-00001. [DOI] [PubMed] [Google Scholar]
- 20.Gretch D R, Polyak S J Groupe Français d-Etudes Moleculaires des Hépatites (GEMHEP), editors . Proceedings of the Hepatitis C virus GEMHEP Conference. Paris, France: John Libbey Eurotext; 1997. The quasispecies nature of hepatitis C virus: research methods and biological implications; pp. 57–69. [Google Scholar]
- 21.Gretch D R, Polyak S J, Wilson J J, Carithers R L, Perkins J D, Corey L. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J Virol. 1996;70:7622–7631. doi: 10.1128/jvi.70.11.7622-7631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gretch D R, Wilson J J, Carithers R L, dela Rosa C, Han J H, Corey L. Detection of hepatitis C virus RNA: comparison of one-stage polymerase chain reaction (PCR) with nested-set PCR. J Clin Microbiol. 1993;31:289–291. doi: 10.1128/jcm.31.2.289-291.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herion D, Hoofnagle J H. The interferon sensitivity determining region: all hepatitis C virus isolates are not the same. Hepatology. 1997;25:769–771. doi: 10.1002/hep.510250346. [DOI] [PubMed] [Google Scholar]
- 24.Higashi Y, Kakumu S, Yoshioka K, Wakita T, Mizokami M, Ohba K, Ito Y, Ishikawa T, Takayanagi M, Nagai Y. Dynamics of genome change in the E2/NS1 region of hepatitis C virus in vivo. Virology. 1993;197:659–668. doi: 10.1006/viro.1993.1641. [DOI] [PubMed] [Google Scholar]
- 25.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 26.Hino K, Sainokami S, Shimoda K, Iino S, Wang Y, Okamoto H, Miyakawa Y, Mayumi M. Genotypes and titers of hepatitis C virus for predicting response to interferon in patients with chronic hepatitis C. J Med Virol. 1994;42:299–305. doi: 10.1002/jmv.1890420318. [DOI] [PubMed] [Google Scholar]
- 27.Hofgärtner W T, Polyak S J, Sullivan D G, Carithers R L, Gretch D R. Mutations in the NS5A gene of hepatitis C virus in North American patients infected with HCV genotype 1a or 1b. J Med Virol. 1997;53:118–126. doi: 10.1002/(sici)1096-9071(199710)53:2<118::aid-jmv3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 28.Hoofnagle J H, Mullen K D, Jones D B, Rustgi V, Di Bisceglie A, Peters M, Waggoner J G, Park Y, Jones E A. Treatment of chronic non-A,non-B hepatitis with recombinant human alpha interferon. A preliminary report. N Engl J Med. 1986;315:1575–1578. doi: 10.1056/NEJM198612183152503. [DOI] [PubMed] [Google Scholar]
- 29.Iino S, Hino K, Yasuda K. Current state of interferon therapy for chronic hepatitis C. Intervirology. 1994;37:87–100. doi: 10.1159/000150362. [DOI] [PubMed] [Google Scholar]
- 30.Kanazawa Y, Hayashi N, Mita E, Li T, Hagiwara H, Kasahara A, Fusamoto H, Kamada T. Influence of viral quasispecies on effectiveness of interferon therapy in chronic hepatitis C patients. Hepatology. 1994;20:1121–1130. [PubMed] [Google Scholar]
- 31.Kato N, Lan K H, Ono-Nita S K, Shiratori Y, Omata M. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol. 1997;71:8856–8859. doi: 10.1128/jvi.71.11.8856-8859.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khorsi H, Castelain S, Wyseur A, Izopet J, Canva V, Rombout A, Capron D, Capron J P, Lunel F, Stuyver L, Duverlie G. Mutations of hepatitis C virus 1b NS5A 2209-2248 amino acid sequence do not predict the response to recombinant interferon-alfa therapy in French patients. J Hepatol. 1997;27:72–77. doi: 10.1016/s0168-8278(97)80282-6. [DOI] [PubMed] [Google Scholar]
- 34.Liu S-L, Schacker T, Musey L, Shriner D, McElrath M J, Corey L, Mullins J I. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J Virol. 1997;71:4284–4295. doi: 10.1128/jvi.71.6.4284-4295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gómez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizokami M, Lau J Y, Suzuki K, Nakano T, Gojobori T. Differential sensitivity of hepatitis C virus quasispecies to interferon-alpha therapy. J Hepatol. 1994;21:884–886. doi: 10.1016/s0168-8278(94)80254-8. [DOI] [PubMed] [Google Scholar]
- 37.Moribe T, Hayashi N, Kanazawa Y, Mita E, Fusamoto H, Negi M, Kaneshige T, Igimi H, Kamada T, Uchida K. Hepatitis C viral complexity detected by single-strand conformation polymorphism and response to interferon therapy. Gastroenterology. 1995;108:789–795. doi: 10.1016/0016-5085(95)90452-2. [DOI] [PubMed] [Google Scholar]
- 38.Okada S, Akahane Y, Suzuki H, Okamoto H, Mishiro S. The degree of variability in the amino terminal region of the E2/NS1 protein of hepatitis C virus correlates with responsiveness to interferon therapy in viremic patients. Hepatology. 1992;16:619–624. doi: 10.1002/hep.1840160302. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto H, Kojima M, Okada S, Yoshizawa H, Iizuka H, Tanaka T, Muchmore E E, Peterson D A, Ito Y, Mishiro S. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology. 1992;190:894–899. doi: 10.1016/0042-6822(92)90933-g. [DOI] [PubMed] [Google Scholar]
- 40.Polyak S J, Faulkner G, Carithers R L, Corey L, Gretch D R. Assessment of hepatitis C virus quasispecies heterogeneity by gel shift analysis: correlation with response to interferon therapy. J Infect Dis. 1997;175:1101–1107. doi: 10.1086/516448. [DOI] [PubMed] [Google Scholar]
- 41.Reed K E, Xu J, Rice C M. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Squadrito G, Leone F, Sartori M, Nalpas B, Berthelot P, Raimondo G, Pol S, Brechot C. HCV NS5A mutations and response of chronic hepatitis to alpha interferon. Hepatology. 1996;24:155A. doi: 10.1053/gast.1997.v113.pm9247477. [DOI] [PubMed] [Google Scholar]
- 43.Squadrito G, Leone F, Sartori M, Nalpas B, Berthelot P, Raimondo G, Pol S, Brechot C. Mutations in the nonstructural 5A region of hepatitis C virus and response of chronic hepatitis C to interferon alfa. Gastroenterology. 1997;113:567–572. doi: 10.1053/gast.1997.v113.pm9247477. [DOI] [PubMed] [Google Scholar]
- 44.Takada N, Takase S, Takada A. Effects of genotypes of hepatitis C virus on interferon treatment for chronic type C hepatitis. Gastroenterol Jpn. 1993;28:268–275. doi: 10.1007/BF02779230. [DOI] [PubMed] [Google Scholar]
- 45.Tanimoto A, Ide Y, Arima N, Sasaguri Y, Padmanabhan R. The amino terminal deletion mutants of hepatitis C virus nonstructural protein NS5A function as transcriptional activators in yeast. Biochem Biophys Res Commun. 1997;236:360–364. doi: 10.1006/bbrc.1997.6967. [DOI] [PubMed] [Google Scholar]
- 46.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toyoda H, Kumada T, Nakano S, Takeda I, Sugiyama K, Osada T, Kiriyama S, Sone Y, Kinoshita M, Hadama T. Quasispecies nature of hepatitis C virus and response to alpha interferon: significance as a predictor of direct response to interferon. J Hepatol. 1997;26:6–13. doi: 10.1016/s0168-8278(97)80002-5. [DOI] [PubMed] [Google Scholar]
- 48.Tsubota A, Chayama K, Arase Y, Koida I, Saitoh S, Ikeda K, Iwasaki S, Matsumoto T, Kobayashi M, Kumada H. Factors useful in predicting the response to interferon therapy in chronic hepatitis C. J Gastroenterol Hepatol. 1993;8:535–539. doi: 10.1111/j.1440-1746.1993.tb01648.x. [DOI] [PubMed] [Google Scholar]
- 49.van Doorn L J, Capriles I, Maertens G, DeLeys R, Murray K, Kos T, Schellekens H, Quint W. Sequence evolution of the hypervariable region in the putative envelope region E2/NS1 of hepatitis C virus is correlated with specific humoral immune responses. J Virol. 1995;69:773–778. doi: 10.1128/jvi.69.2.773-778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q L, Houghton M. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 51.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, Brunetto M, Barr P J, Miyamura T, McHutchinson J, Houghton M. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson J J, Polyak S J, Day T D, Gretch D R. Characterization of simple and complex hepatitis C virus quasispecies by heteroduplex gel shift analysis: correlation with nucleotide sequencing. J Gen Virol. 1995;76:1763–1771. doi: 10.1099/0022-1317-76-7-1763. [DOI] [PubMed] [Google Scholar]
- 53.Yoshioka K, Kakumu S, Wakita T, Ishikawa T, Itoh Y, Takayanagi M, Higashi Y, Shibata M, Morishima T. Detection of hepatitis C virus by polymerase chain reaction and response to interferon-alpha therapy: relationship to genotypes of hepatitis C virus. Hepatology. 1992;16:293–299. doi: 10.1002/hep.1840160203. [DOI] [PubMed] [Google Scholar]
- 54.Yun Z B, Reichard O, Chen M, Lundeberg J, Norkrans G, Fryden A, Sonnerborg A, Weiland O. Serum hepatitis C virus RNA levels in chronic hepatitis C—importance for outcome of interferon alfa-2b treatment. Scand J Infect Dis. 1994;26:263–270. doi: 10.3109/00365549409011794. [DOI] [PubMed] [Google Scholar]
- 55.Zeuzem S, Lee J H, Roth W K. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25:740–744. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]
- 56.Ziebert A, Schreier E, Roggendorf M. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology. 1995;208:653–661. doi: 10.1006/viro.1995.1196. [DOI] [PubMed] [Google Scholar]