Abstract
The Japanese sawyer beetle, Monochamus alternatus, is not only one of the most important wood boring pest itself, but also a major vector of the invasive pinewood nematode (PWN), which is the causal agent of the devastative pine wilt disease (PWD) and threats the global pine forest. Here, we present a near-complete genome of M. alternatus at the chromosome level. The assembled genome was 792.05 Mb with contig N50 length of 55.99 Mb, which is the largest N50 size among the sequenced Coleoptera insects currently. 99.57% of sequence was anchored onto ten pseudochromosomes (one X-chromosome and nine autosomes), and the final genome harbored only 13 gaps. BUSCO evaluation revealed the presence of 99.0% of complete core genes. Thus, our genome assembly represented the highest-contiguity genome assembly as well as high completeness in insects so far. We identified 20,471 protein-coding genes, of which 20,070 (98.04%) were functionally annotated. The genome assembly of M. alternatus provides a valuable resource for exploring the evolution of the symbiosis between PWN and the vector insects.
Subject terms: Evolutionary ecology, Molecular ecology
Background & Summary
Vector-borne plant diseases widely occur and cause severe ecological and economic losses in agricultural and forestry ecosystem. Vector insects play particularly important roles in the evolution of the pathogen dispersion and pathogenesis1. The plant parasitic nematode Bursaphelenchus xylophilus, also known as pine wood nematode (PWN), is the causal agent of the devastative pine wilt disease (PWD) and threats the global pine forest2,3. The transmission of PWN from dead pine trees to susceptible, live pine trees exclusively relied on the vector beetles belonging to species of the genus Monochamus (Coleoptera: Cerambycidae)4. During the invasion history of PWN, its vector species also shifted with the geographic locations. The primary vector in North America, native region of PWN, is Monochamus carolinensis, then changed to M. alternatus and M. salturatis in Asia and M. galloprovincialis in Europe, the invasive regions of PWN5. The vector species shift thus greatly contributes to the evolutionary ecology of PWD6. However, the underlying molecular mechanism is still far unknown due to few genomic resources of these vector insects7,8.
The Japanese swayer beetle, M. alternatus, is not only a main vector of the invasive PWN, but also one of the most important wood boring pest itself across East Asia such as China, South Korea and Japan, where is the place with the most serious PWD epidemic damage9–11. This vector beetle and PWN has formed a close symbiosis based on their high synchronization of life cycle, mediated by the chemical signals11–13. Specifically, the M. alternatus beetles prefer to select the weakened or dying trees with PWN infection to oviposit and complete the development of their offspring. The third-stage juveniles (LIII) of PWN are attracted by specific terpenes produced by mature insect larvae and aggregate around pupal chambers in diseased trees12, and fourth-stage juveniles (LIV) are attracted into the trachea of newly emerged adults by ascarosides secreted by the beetles13. The newly eclosed beetle should have a maturation feeding in healthy trees. The nematode then departs from the spiracles driven by CO2 enhanced by feeding behavior and invades new healthy trees via the feeding wounds14,15, thus starting a new cycle of infection, propagation and dispersal. While the chemical signals among the symbiosis have been well characterized, the molecular mechanism of the chemical communications remains elusive. Furthermore, as with most vector-borne diseases, vector control is the key to manage those diseases efficiently. Unfortunately, there is still lack of effective and efficient control method against this vector beetle, therefore, a high-quality reference genome is needed for both further understanding this symbiosis and its maintenance as well as new control approaches, such as genetic-engineered management strategy.
Recently, Gao et al. reported a chromosome-level genome assembly of M. alternatus based on Nanopore sequencing technology8. Here, we constructed a high-quality chromosome-scale genome of M. alternatus through combining Pacific Biosciences (PacBio) high-fidelity (HiFi), high-throughput chromosome conformation capture (Hi-C), and Illumina short-read sequencing data. Subsequently, we performed structural and functional annotation of the assembled genome through integrating transcriptome data from different tissues of M. alternatus. The high-quality reference genome of M. alternatus provides a valuable resource for exploring the evolution of coleopteran insects and the interaction mechanism between PWN and its vector insects.
A total of 99.27 Gb (127.81×) of HiFi reads (Table 1) were used to generate the primary genome assembly. Furthermore, 145.39 Gb (187.19×) of Hi-C data (Table 1) was used to anchor contigs to chromosome-level genome using the juicer and 3D-DNA pipeline. The final assembled genome is 792.5 Mb, which was very close to the estimated genome size (776.7 Mb) (Fig. 1a; Table 2) based on the distribution of k-mer frequencies, with a contig N50 size of 55.99 Mb and a scaffold N50 size of 86.21 Mb. Based on Hi-C data, 99.57% of genome sequence was successfully anchored onto ten pseudochromosomes (Fig. 1b). Realignment of Illumina genome sequencing and RNA-seq reads to the reference genome achieved average mapping rates of 98.32% and 95.53% (Table S1), respectively. MUMmer-based genome alignment indicated that our genome assembly exhibited 1:1 synteny relationship with the closely related species M. saltuarius (Fig. S1), and chromosome 7 of M. alternatus was determined as the X chromosome based on chromosomal synteny. BUSCO v5.2.216 was used to evaluate the completeness of the genome assembly of M. alternatus based on the insect_odb10 dataset. The results showed that 99.0% of complete BUSCOs were successfully captured by our genome assembly, including 98.3% of single-copy and 0.7% of duplicated BUSCOs (Table 2).
Table 1.
Summary of genome sequencing data for Monochamus alternatus.
| Sequencing strategy | Platform | Usage | Insertion size | Total data (Gb) | Sequence coverage (×) |
|---|---|---|---|---|---|
| Short-reads | Illumina | Genome survey | 250 bp | 43.05 | 55.43 |
| HiFi | PacBio Sequel II | Assembly | 15 kb | 99.27 | 127.81 |
| Hi-C | Illumina | Hi-C assembly | 350 bp | 145.39 | 187.19 |
Fig. 1.
Genome size estimation and heatmap of genome-wide Hi-C interaction. (a) K-mer frequency analysis was performed for genome size estimation of M. alternatus using GCE (v1.0.2) based on Illumina genome sequencing data. The second peak with depth at 48 represents the main peak, and the first peak with depth at 24 indicates the heterozygous peak. (b) The heatmap shows the scaffolding result of the M. alternatus genome based on the juicer and 3ddna pipeline. The first ten blue rectangles represents ten pseudochromosomes.
Table 2.
Summary statistics of Monochamus alternatus genome assembly.
| Monochamus alternatus | |
|---|---|
| Genome assembly | |
| Estimated genome size (Mb) | 776.7 |
| Assembled genome size (Mb) | 792.05 |
| Contig N50 (Mb) | 55.99 |
| Number of contigs | 33 |
| Scaffold N50 (Mb) | 86.21 |
| Number of scaffolds | 14 |
| GC content (%) | 32.37 |
| Anchoring rate (%) | 98% |
| Complete BUSCO of genome assembly | 99.00% |
| Gene annotation | |
| Number of protein-coding genes | 20,471 |
| Average gene length (bp) | 8161.45 |
| Average CDS length (bp) | 1175.53 |
| Average exon length (bp) | 282.41 |
| Average intron length (bp) | 2207.48 |
| Average exon number | 4.16 |
| Complete BUSCO of gene annotations | 97.60% |
Additionally, we made a comparison of contig size between M. alternatus and other coleopteran insects with publicly available genome assemblies in NCBI database. Compared with 116 other chromosome-level genome assemblies of Coleoptera insects, the genome assembly of M. alternatus showed the highest quality among the coleopteran insects at least in terms of contig N50 size (Fig. 2a; Table S2). Tree map representation was utilized to display contig size and gap number for each chromosome (Fig. 2b), indicating that four chromosomes (i.e. chr1, chr5, chr6, and chr8) were gap-free and the remaining six chromosomes harbored no more than three gaps for each. Moreover, we examined whether telomeres and centromeres were present in our assembled genome. The results showed that telomeric regions could be detected on both ends of nine chromosomes, and the telomeric region was identified on the single end of chromosome 7 (Fig. 2c; Table S3). As with centromeres, one candidate centromeric region was identified for each of ten chromosomes (Fig. 2c; Table S4). Together, we obtained a high-quality genome assembly of M. alternatus with high contiguity and high completeness.
Fig. 2.
Comparison of the M. alternatus genome assembly with other sequenced Coleoptera insects and contig size tree map of Monochamus alternatus genome. (a) Scatter plot showing the contig N50 distribution of genome assemblies for M. alternatus and other Coleoptera insects publicly available. The assembly reports for Coleoptera insects were collected from NCBI datasets, and only chromosome-level genome assemblies were used for comparison. X-axis indicates the contig N50 size in megabases (Mb), and Y-axis denotes the genome size in Mb. The vertical dotted lines in orange and red represent the cutoff of 10 Mb and 20 Mb, respectively. Currently, a total of seven public Coleoptera insect genome assemblies showed contig N50 size ≥ 20 Mb. Compared with these available genome assemblies of Coleoptera insects, the M. alternatus genome assembly exhibited the highest contiguity level. (b) Tree map representation of the distribution of chromosome sizes and corresponding contig sizes. Four chromosomes chr1, chr5, chr6, and chr8 were composed of a single contig, the remaining six chromosomes harbored no more than three gaps for each. (c) Identification of telomeres and centromeres in the M. alternatus genome assembly. The telomeres were identified using the TeloExplorer module within quarTeT with ‘AACCT’ as the repeat monomer. The CentroMiner module within quarTeT was used for identification of centromeric regions.
Besides, we made a comparison of quality metrics between two genome assemblies generated by PacBio HiFi (hereafter referred to as HiFi assembly) and Nanopore sequencing technologies (hereafter referred to as Nanopore assembly). Firstly, we recalculated the N50 metrics of scaffold-level and contig-level genome using the same software assembly-stats to exclude the bias caused by different approaches. The HiFi assembly possessed higher quality in contig N50 size (55.99 Mb) compared with the Nanopore assembly (15.77 Mb) (Table S5). Secondly, significant difference existed between two assemblies in terms of the HiC interaction heatmap. Dozens of gaps (72) were detected in the Nanopore assembly8, while less gaps (13) were found in the HiFi assembly (Fig. 1b). Thirdly, the HiFi assembly (99.57%) possessed higher chromosome anchoring rate compared with the Nanopore assembly (Table S5). Fourthly, the Nanopore assembly showed higher genome BUSCO score, while the HiFi assembly showed higher gene set BUSCO score (Table S5). Fifthly, the HiFi assembly showed higher mapping rates against all RNA-seq samples generated by two studies (Table S1). Finally, telomeres were undetectable in all chromosomes of the Nanopore assembly based on the monomer ‘TTAGG/CCTAA’, while telomeres were present in both ends of nine chromosomes and single end of the remaining one of the HiFi assembly (Table S5). Thus, the HiFi assembly showed advancement in contiguity compared with the Nanopore assembly, representing a near telomere-to-telomere (T2T) assembly of M. alternatus.
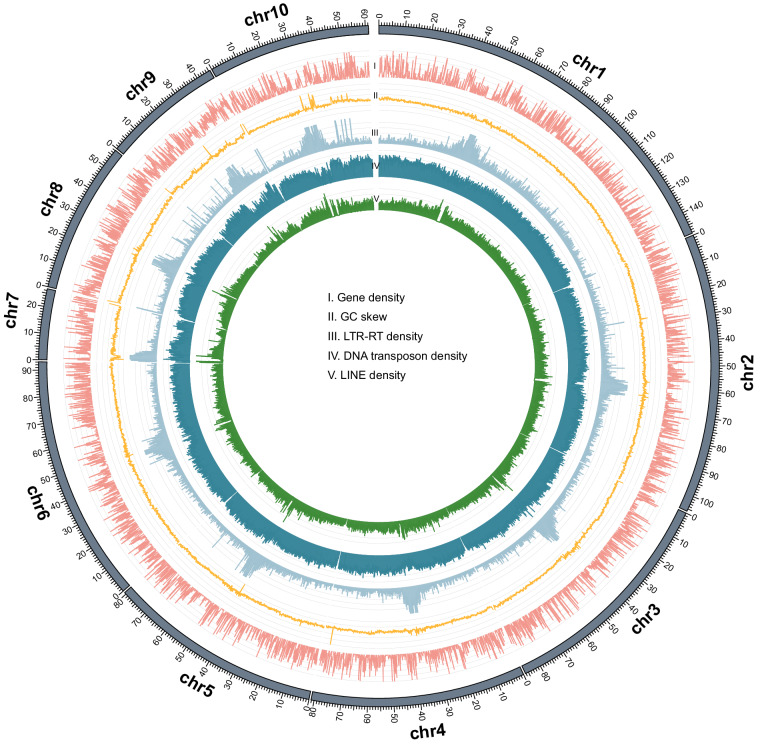
The repeat sequences in M. alternatus were annotated using the RepeatMasker pipeline. In total, 58.23% of the M. alternatus genome was composed of repeat sequences. Among them, DNA transposons (35.81%) and long interspersed nuclear elements (LINEs, 9.45%) represent top two richest repeat types, and long terminal repeat retrotransposons (LTR-RTs), Penelope and short interspersed nuclear elements (SINEs) occupied 7.88%, 0.63% and 0.15% of genome sequence (Table 3). Based on the masked genome, we predicted protein-coding genes through combining three approaches, finally yielding 20,471 consensus protein coding genes (Table 2). BUSCO assessment showed that 97.6% of complete BUSCOs were present in the predicted gene set. Additionally, the canonical non-coding RNAs in M. alternatus were identified using different methods, including 1384 ribosomal RNAs (rRNAs), 540 transfer RNAs (tRNAs), 67 microRNAs (miRNAs), and 77 small nuclear RNAs (snRNAs) (Table 4). The landscape of M. alternatus genome assembly and gene annotations was presented as a Circos plot (Fig. 3). The functions of protein-coding genes were annotated against multiple database, such as SwissProt, InterPro, Pfam, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG). The results indicated that 98.04% of coding genes could be functionally annotated by at least one public database and transcriptome (Table 5), suggesting the high confidence of our gene annotation.
Table 3.
Summary of repeat sequences in the genome assembly of M. alternatus.
| Class | Number | Length | Percentage (%) |
|---|---|---|---|
| LTR | |||
| Gypsy | 29,600 | 24,111,649 | 3.05 |
| Copia | 1339 | 975,226 | 0.12 |
| Other | 98,671 | 37,100,109 | 4.71 |
| Non-LTR retroelements | |||
| LINE | 213,558 | 74,577,309 | 9.45 |
| SINE | 12,366 | 1,166,742 | 0.15 |
| Penelope | 14,938 | 5,010,542 | 0.63 |
| DNA transposons | 896,596 | 282,736,732 | 35.81 |
| Simple repeats | 106,733 | 7,985,424 | 1.01 |
| Low complexity | 18,173 | 885,814 | 0.11 |
| Unclassified | 146,883 | 26,928,207 | 3.41 |
| Total | 459,696,228 | 58.23 | |
Table 4.
Summary of non-coding RNAs annotated in the M. alternatus genome.
| Type | Copy number | Average length (bp) | Total length (bp) |
|---|---|---|---|
| miRNA | 67 | 79.78 | 5345 |
| tRNA | 540 | 75.36 | 40,696 |
| rRNA | |||
| 28S | 39 | 2071.79 | 80,800 |
| 18S | 53 | 1030.62 | 54,623 |
| 5.8S | 101 | 119.16 | 3100 |
| 5S | 1191 | 118 | 12,035 |
| snRNA | |||
| CD-box | 20 | 143.35 | 2867 |
| HACA-box | 3 | 139 | 417 |
| Splicing | 54 | 148.52 | 8020 |
Fig. 3.
Overview of the genome landscape of the Monochamus alternatus genome assembly. In the Circos plot, the outmost track denotes the chromosomal ideograms (scale = 1 Mb). From the outer to the inner tracks, the density of protein-coding genes (I), GC skew (II), LTR-RTs (III), DNA transposons (IV), and LINEs (V) on each chromosome was calculated in nonoverlapping 100-kb windows and displayed.
Table 5.
Functional annotation of the predicted genes in M. alternatus.
| Type | Number | Percentage (%) |
|---|---|---|
| SwissProt | 10,527 | 51.42 |
| RNA-seq | 19,181 | 93.7 |
| InterPro | 16,317 | 79.71 |
| Pfam | 10,727 | 52.4 |
| GO | 8975 | 43.84 |
| KEGG | 12,426 | 60.7 |
| Annotated | 20,070 | 98.04 |
| Unannotated | 401 | 1.96 |
| Total | 20,471 | 100 |
We selected thirteen coleopteran insect species to perform phylogenomic analysis. The phylogenetic tree was reconstructed from 680 strict single-copy orthologous genes using OrthoFinder. The results indicated that M. alternatus was most closely related to the Asian longhorned beetle Anoplophora glabripennis, and they diverged from each other approximately 25 million years ago (Mya) (Fig. 4a). CAFÉ analysis indicated that 759 gene families were significantly expanded in M. alternatus compared to the most recent common ancestor (Fig. 4a). We made a comparison of gene families across seven Coleoptera insects, and found that 5441 orthogroups (OGs) were highly conserved in these beetles, and 392 OGs were species-specific in M. alternatus (Fig. 4b). Functional enrichment analysis showed that the expanded gene families in M. alternatus were significantly enriched in many physiological processes, such as transcription factors (K09427; BH-adjusted p-value = 1.43 × 10−12), membrane trafficking (K21440; BH-adjusted p-value = 1.89 × 10−9), Toll and Imd signaling pathway (K20674; BH-adjusted p-value = 4.23 × 10−5), apoptosis (K20015; BH-adjusted p-value = 1.22 × 10−7), and insect hormone biosynthesis (K10719; BH-adjusted p-value = 3.99 × 10−9) (Fig. 4c).
Fig. 4.
Phylogenetic tree of Monochamus alternatus and other Coleoptera insects and evolutionary analysis of gene families. (a) The phylogenetic tree of M. alternatus and other Coleoptera insects. The maximum likelihood tree was constructed using RAxML with Drosophila melanogaster as the outgroup based on 680 single-copy orthologs. The species divergence time was estimated using r8s. The numbers of expanded and contracted gene families revealed by CAFÉ analysis are shown above each species name. The stacked bar plots on the right represent the distribution of different types of genes in corresponding species, including single-copy, multi-copy, unique, and unclustered genes. (b) Upset plot showing the common and unique orthogroups identified in various Coleoptera insects. (c) KEGG enrichment analysis result of gene families that were significantly expanded in M. alternatus compared to the recent ancestor.
Methods
Insect rearing, sample collection, and genome sequencing
Last instar larval M. alternatus were collected from host trees of Pinus massoniana, in Fuyang, Zhejiang province in late autumn in 2016. They were reared for approximately twenty generations in laboratory. The larva and pupa were fed on artificial diet in a 10-ml tube at a 12:12 h light:dark (L:D) cycle at 25 °C placed in a climate chamber. Fresh diet was provided every week3. The adults were fed on the fresh pine branches for sexual maturation and laid eggs under the back of pine logs (6–10 cm in diameter and 30 cm in length). One week later, the logs were debarked and the hatched larva were collected for next generation rearing. One newly emerged adult male without feeding was prepared for PacBio HiFi sequencing and Hi-C sequencing. The beetle’s gut was removed and body surface was cleaned by 75% ethanol to avoid microbial contamination.
Whole-Genome Sequencing (WGS) was used to estimate the genomic characteristics of M. alternatus. Illumina paired-end library was constructed and sequenced on Illumina HiSeq 2500 platform. A total of 43.05 Gb of clean data was generated (Table 1).
Genomic DNA for PacBio HiFi sequencing was prepared by the CTAB method and followed by purification with QIAGEN Blood & Cell Culture DNA Midi Kit (QIAGEN, USA) according to the manufactural procedure. The library was constructed Sequel II HiFi (CCS) method. SMRTbell target size libraries were constructed for sequencing according to the standard protocol of PacBio (Pacific Biosciences, CA USA) using 15 kb preparation solutions, and sequenced on PacBio Sequel II platform with Sequencing Primer V2 and Sequel II Binding Kit 2.0 in Grandomics Biosciences Co., Ltd (Wuhan, China).
To anchor hybrid scaffolds onto the chromosome, genomic DNA was extracted for the Hi-C library from the same beetle as HiFi. Then, we constructed the Hi-C library and obtained sequencing data via the Illumina DNBSEQ-T7 platform in Grandomics Biosciences Co., Ltd (Wuhan, China).
For transcriptome sequencing, the total RNA of each sample from development stage (egg, late larvae and early pupae) or adult tissue (gut, muscle, brain and trachea) was extracted using TRIzol (Thermo Fisher Scientific, USA). Paired-end libraries were constructed NEBNext®Ultra™ RNA Library Prep Kit for Illumina®(NEB, USA) following manufacturer’s recommendations. The library preparations were then paired-end sequenced on an Illumina HiSeq. 2500 platform. A total of 45.04 Gb of data was generated.
Genome assembly and quality evaluation
To examine the genome feature of M. alternatus, GCE v1.0.217 was used for k-mer frequency analysis with default parameters. To achieve high-quality genome assembly, Hifiasm (v0.16.1-r375)18 was used to de novo assemble the M. alternatus genome based on PacBio HiFi reads with default parameters. Then, the Hi-C reads were employed to scaffold contigs onto chromosomes through sorting, orientation, and ordering using Juicer v1.619 and 3D-DNA v17012320 to generate the final version of genome assembly. Benchmarking Universal Single-Copy Orthologs (BUSCO v5.2.2)16 was performed to assess the quality of the genome assembly using the insecta_odb10 dataset.
Identification of telomeres and centromeres
Both telomeres and centromeres were identified using quarTeT21. Telomeres were predicted using quarTeT TeloExplorer module with the parameters: ‘-c other -m 100’. Centromeres were predicted using quarTeT CentroMiner module with the parameters: ‘-n 100 -m 200 -s 0.8 -d 10 -e 0.00001 -g 50000 -i 100000 –trf 2 7 7 80 10 50 -r 3’. The chromosomal distribution of telomeres and centromeres was visualized using the R package chromoMap v4.1.1222.
Chromosomal synteny analysis and identification of sex chromosomes
The whole-genome synteny analysis between M. alternatus and the closely related species M. saltuarius was performed using MUMmer v4.0.0beta223 with default parameters. The chromosome that was syntenic with chrX in M. saltuarius was defined as the X chromosome in M. alternatus.
Repeat sequence and non-coding RNA annotation
Firstly, a de novo repeat library was constructed by combining RepeatModeler v1.0.11 (http://www.repeatmasker.org/RepeatModeler) with LTR_retriever v2.9.024, which integrates the LTR discovery result from LTR_finder v1.0.725 and LTRharvest v1.5.926. TRF v4.10.027 was used for identification of tandem repeats. Then, the repeat sequences in the M. alternatus genome was identified and masked using RepeatMasker v4.0.7 (http://www.repeatmasker.org/RepeatMasker) against the species-specific de novo repeat library and RepBase library v26.03. Additionally, tRNAscan-SE28 was used to predict tRNA genes. Other non-coding RNAs such as rRNA, miRNA, and snRNA were annotated using INFERNAL v1.129 through search against the Rfam database v9.130.
Gene prediction and functional annotation
To predict the protein-coding genes in the M. alternatus genome, we employed a strategy integrating ab initio prediction, homology searching and transcriptome-based approaches. For transcriptome-based prediction, HISAT v2.2.131 was used to align the RNA-seq data to the reference genome, and StringTie v2.1.631 was used for transcript assembly. Subsequently, TransDecoder v5.5.0 (https://github.com/TransDecoder/TransDecoder) was employed to estimate the potential open reading frames (ORFs). For the homology-based approaches, the protein sequences from seven Coleoptera insects and Drosophila melanogaster were downloaded from public database and GenomeThreader v1.7.132 was used for homology search. For ab initio prediction, AUGUSTUS v3.4.033 was employed for gene prediction based on the species-specific gene model. Finally, the EVidenceModeler pipeline34 was employed to generate a set of protein-coding genes through combining different sources of evidence. To maintain the confidence of predicted genes, we retained only gene models that had at least one supporting evidence from homologous proteins of closely related species, InterProScan domain and RNA-seq data. For functional annotation, we searched against the SwissProt protein database and InterPro database using DIAMOND v2.1.718 (E-value = 1e-5) and InterProScan v5.21.6035, respectively. The assignment of KEGG orthology (KO) terms was conducted through search against the hidden Markov model (KOfam) database using KofamScan v1.3.036 with default parameters.
Reconstruction of phylogenetic tree of Coleoptera insect species
To perform phylogenomic analysis, the genome assemblies and gene annotations of thirteen Coleoptera insects and an outgroup D. melanogaster were retrieved from several public database, such as A. glabripennis (EnsemblMetazoa: Agla_1.0.48)37, Aethina tumida (NCBI RefSeq: GCF_024364675.1)38, Altica viridicyanea (NGDC GWH: GWHAMMQ00000000)39, Callosobruchus maculatus (NCBI GenBank: GCA_900659725.1)40, Dendroctonus ponderosae (NCBI GenBank: GCA_020466635.2)41, Dendroctonus valens42, Diabrotica virgifera (NCBI RefSeq: GCF_003013835.1)43, Gonioctena quinquepunctata (NCBI GenBank: GCA_018342105.1)44, Leptinotarsa decemlineata (i5k: OGSv1.2_GCF_000500325)45, Nebria riversi46, Photinus pyralis (NCBI RefSeq: GCF_008802855.1)47, Sitophilus oryzae (NCBI RefSeq: GCF_002938485.1)48, and Tribolium castaneum (EnsemblMetazoa: Tcas5.2.48)49. For each gene, only the longest transcript was kept for downstream analysis. OrthoFinder v2.5.450 was performed to identify orthologs and paralogs using DIAMOND v2.1.7 with default parameters. To infer the phylogeny of these insects, multiple sequence alignments of single-copy orthologous genes were performed using MAFFT v7.49051 with default parameters. The alignment results were concatenated to form a super-sequence for each species and trimmed using trimAl v1.252 with the “-gappyout” parameter. The optimal amino acid substitution model was estimated using ProtTest v3.4.253. Then, RAxML v8.2.1054 was employed to construct the phylogenetic tree using the maximum likelihood method with LG + I + G + F model and 1000 bootstrap replicates. Species divergence age was adopted from the TimeTree55 database: D. valens vs D. ponderosae 21.6 Mya; C. maculatus vs D. virgifera 79–221 Mya; T. castaneum vs D. melanogaster 195–361.6 Mya; and A. glabripennis vs L. decemlineata 89.7–220.9 Mya. R8s v1.8156 was used for calibrating the species divergence time. The species tree was visualized using FigTree v1.4.2.
Gene family expansion and contraction
The count matrix of gene family for each species was obtained from OrthoFinder analysis. The matrix table and the ultrametric tree were taken as inputs to analyze the expansion and contraction of gene families using CAFE v4.2.157.
Data Records
Raw data from PacBio HiFi (CRR1002983)58, Hi-C (CRR1002984)59 and Illumina (CRR1002982)60 genome sequencing and RNA-seq data (CRR1003137-CRR1003143)61–67 have been deposited in the Genome Sequence Archive (GSA, https://ngdc.cncb.ac.cn/gsa) at the National Genomic Data Center (NGDC)68. The genome assembly has been deposited in the Genome Warehouse (GWH, https://ngdc.cndb.ac.cn/gwh)69 at NGDC under the accession number of GWHEQWP00000000. All data were associated with the BioProject PRJCA022378. The genome sequence and raw reads have also been deposited in GenBank (JBBBDW000000000.170) and Sequence Read Archive (SRA, SRR2824870271 for PacBio HiFi, SRR2824870172 for Hi-C, SRR2824870373 for Illumina, and SRR28248694-SRR2824870074–80 for RNA-seq data) at National Center for Biotechnology Information (NCBI) under BioProject PRJNA1084890.
Technical Validation
Two distinct methods were employed to assess the completeness and accuracy of the M. alternatus genome. First, employing the insecta_odb10 datasets, the BUSCO analysis demonstrated successful identification of 99.0% of core genes as complete. Second, the realignment of Illumina genome sequencing and RNA-seq reads to the M. alternatus genome resulted in mapping rates of 98.32% and 95.53%, respectively. To further appraise the comprehensiveness and accuracy of gene prediction, BUSCO analysis was conducted based on the Insecta datasets, yielding a complete BUSCO score of 97.6%.
Supplementary information
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2021YFC2600100) and the National Natural Science Foundation of China (32070486 and 32088102).
Author contributions
B.Z. and J.S. conceived the idea. L.X., B.L. and D.Y. analyzed the data. B.Z., L.X. and D.Y. drafted the first version of the manuscript. B.L. and J.S. revised the manuscript. X.T. prepared the materials. B.Z. and J.S. supervised the project. All authors have read and approved the final version of the manuscript.
Code availability
No specific script was utilized in this study. The codes and pipelines used for genome sequencing data analysis were performed following the instructions of corresponding bioinformatics tools. The version and parameters of the software have been included in the Methods section.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Longsheng Xing, Bo Liu, Dunyang Yu.
Contributor Information
Jianghua Sun, Email: sunjh@hbu.edu.cn.
Bin Zhang, Email: binzhang@hbu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-024-03150-1.
References
- 1.Eigenbrode SD, Bosque-Perez NA, Davis TS. Insect-Borne Plant Pathogens and Their Vectors: Ecology, Evolution, and Complex Interactions. Annu Rev Entomol. 2018;63:169–191. doi: 10.1146/annurev-ento-020117-043119. [DOI] [PubMed] [Google Scholar]
- 2.Zhou J, et al. Immune tolerance of vector beetle to its partner plant parasitic nematode modulated by its insect parasitic nematode. FASEB J. 2018;32:4862–4877. doi: 10.1096/fj.201800247R. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, et al. miR-31-5p regulates cold acclimation of the wood-boring beetle Monochamus alternatus via ascaroside signaling. BMC Biol. 2020;18:184. doi: 10.1186/s12915-020-00926-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linit MJ. Nematode-vector relationships in the pine wilt disease system. Journal of Nematology. 1988;20:227–235. [PMC free article] [PubMed] [Google Scholar]
- 5.Akbulut S, Stamps WT. Insect vectors of the pinewood nematode: a review of the biology and ecology of Monochamus species. Forest Pathology. 2011;42:89–99. doi: 10.1111/j.1439-0329.2011.00733.x. [DOI] [Google Scholar]
- 6.Kirino H, Maehara N, Shinya R. How did Bursaphelenchus nematodes acquire a specific relationship with their beetle vectors. Monochamus? Front Physiol. 2023;14:1209695. doi: 10.3389/fphys.2023.1209695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu N, et al. Chromosome-level genome assembly of Monochamus saltuarius reveals its adaptation and interaction mechanism with pine wood nematode. Int J Biol Macromol. 2022;222:325–336. doi: 10.1016/j.ijbiomac.2022.09.108. [DOI] [PubMed] [Google Scholar]
- 8.Gao YF, et al. Chromosome-level genome assembly of the Japanese sawyer beetle Monochamus alternatus. Sci Data. 2024;11:199. doi: 10.1038/s41597-024-03048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi F, Yamane A, Ikeda T. The Japanese pine sawyer beetle as the vector of pine wilt disease. Annu Rev Entomol. 1984;29:115–135. doi: 10.1146/annurev.en.29.010184.000555. [DOI] [Google Scholar]
- 10.Futai K. Pine wood nematode, Bursaphelenchus xylophilus. Annu Rev Phytopathol. 2013;51:61–83. doi: 10.1146/annurev-phyto-081211-172910. [DOI] [PubMed] [Google Scholar]
- 11.Zhao L, Mota M, Vieira P, Butcher RA, Sun J. Interspecific communication between pinewood nematode, its insect vector, and associated microbes. Trends Parasitol. 2014;30:299–308. doi: 10.1016/j.pt.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Zhao L, et al. Chemical signals synchronize the life cycles of a plant-parasitic nematode and its vector beetle. Curr Biol. 2013;23:2038–2043. doi: 10.1016/j.cub.2013.08.041. [DOI] [PubMed] [Google Scholar]
- 13.Zhao L, et al. Ascarosides coordinate the dispersal of a plant-parasitic nematode with the metamorphosis of its vector beetle. Nat Commun. 2016;7:12341. doi: 10.1038/ncomms12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, et al. Chemical signals of vector beetle facilitate the prevalence of a native fungus and the invasive pinewood nematode. Journal of Nematology. 2017;49:341–347. doi: 10.21307/jofnem-2017-081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Wickham JD, Zhao L, Sun J. CO2 drives the pine wood nematode off its insect vector. Curr Biol. 2019;29:R619–R620. doi: 10.1016/j.cub.2019.05.033. [DOI] [PubMed] [Google Scholar]
- 16.Manni M, Berkeley MR, Seppey M, Simao FA, Zdobnov EM. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021;38:4647–4654. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, B. et al. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. arXiv (2013).
- 18.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 19.Durand NC, et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudchenko O, et al. De novo asssembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y, et al. quarTeT: a telomere-to-telomere toolkit for gap-free genome assembly and centromeric repeat identification. Hortic. Res. 2023;10:uhad127. doi: 10.1093/hr/uhad127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anand L, Rodriguez Lopez CM. ChromoMap: an R package for interactive visualization of multi-omics data and annotation of chromosomes. BMC bioinformatics. 2022;23:33. doi: 10.1186/s12859-021-04556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcais G, et al. MUMmer4: A fast and versatile genome alignment system. PLoS Comput Biol. 2018;14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ou S, Jiang N. LTR_retriever: A highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellinghaus D, Kurtz S, Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC bioinformatics. 2008;9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalvari I, et al. Non-coding RNA analysis using the Rfam database. Curr. Protoc. Bioinformatics. 2018;62:e51. doi: 10.1002/cpbi.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pertea M, Kim D, Pertea GM, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gremme G, Brendel V, Sparks ME, Kurtz S. Engineering a software tool for gene structure prediction in higher organisms. Inform. Software Tech. 2005;47:965–978. doi: 10.1016/j.infsof.2005.09.005. [DOI] [Google Scholar]
- 33.Stanke M, et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philip, J. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics, 1236-1240 (2014). [DOI] [PMC free article] [PubMed]
- 36.Aramaki T, et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36:2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna DD, et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle-plant interface. Genome Biol. 2016;17:227. doi: 10.1186/s13059-016-1088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang, Q. et al. Identification of sex chromosomes and primary sex ratio in the small hive beetle, a worldwide parasite of honey bees. Gigascience12 (2022). [DOI] [PMC free article] [PubMed]
- 39.Xue HJ, et al. The draft genome of the specialist flea beetle Altica viridicyanea (Coleoptera: Chrysomelidae) BMC Genomics. 2021;22:243. doi: 10.1186/s12864-021-07558-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnqvist, G. et al. A chromosome-level assembly of the seed beetle Callosobruchus maculatus genome with annotation of its repetitive elements. G3 (Bethesda) (2023). [DOI] [PMC free article] [PubMed]
- 41.Keeling CI, et al. Chromosome-level genome assembly reveals genomic architecture of northern range expansion in the mountain pine beetle, Dendroctonus ponderosae Hopkins (Coleoptera: Curculionidae) Mol Ecol Resour. 2022;22:1149–1167. doi: 10.1111/1755-0998.13528. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z, et al. Chromosome-level genome assembly and population genomic analyses provide insights into adaptive evolution of the red turpentine beetle, Dendroctonus valens. BMC Biol. 2022;20:190. doi: 10.1186/s12915-022-01388-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coates BS, et al. A draft Diabrotica virgifera virgifera genome: insights into control and host plant adaption by a major maize pest insect. BMC Genomics. 2023;24:19. doi: 10.1186/s12864-022-08990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukicheva, S., Flot, J. F. & Mardulyn, P. Genome assembly of the cold-tolerant leaf beetle Gonioctena quinquepunctata, an important resource for studying its evolution and reproductive barriers between species. Genome Biol. Evol. 13 (2021). [DOI] [PMC free article] [PubMed]
- 45.Schoville SD, et al. A model species for agricultural pest genomics: the genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae) Sci. Rep. 2018;8:1931. doi: 10.1038/s41598-018-20154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weng YM, Francoeur CB, Currie CR, Kavanaugh DH, Schoville SD. A high-quality carabid genome assembly provides insights into beetle genome evolution and cold adaptation. Mol. Ecol. Resour. 2021;21:2145–2165. doi: 10.1111/1755-0998.13409. [DOI] [PubMed] [Google Scholar]
- 47.Fallon, T. R. et al. Firefly genomes illuminate parallel origins of bioluminescence in beetles. Elife7 (2018). [DOI] [PMC free article] [PubMed]
- 48.Parisot N, et al. The transposable element-rich genome of the cereal pest Sitophilus oryzae. BMC Biol. 2021;19:241. doi: 10.1186/s12915-021-01158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herndon N, et al. Enhanced genome assembly and a new official gene set for Tribolium castaneum. BMC Genomics. 2020;21:47. doi: 10.1186/s12864-019-6394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 56.Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 57.Mira VH, Thomas GWC, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 58.2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014182/CRR1002983
- 59.2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014182/CRR1002984
- 60.2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014182/CRR1002982
- 61.2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003137
- 62.2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003138
- 63.2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003139
- 64.2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003140
- 65.2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003141
- 66.2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003142
- 67.2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003143
- 68.Chen T, et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genomics Proteomics Bioinformatics. 2021;19:578–583. doi: 10.1016/j.gpb.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen M, et al. Genome Warehouse: A Public Repository Housing Genome-scale Data. Genomics Proteomics Bioinformatics. 2021;19:584–589. doi: 10.1016/j.gpb.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang B. 2024. Monochamus alternatus isolate DY-2024, whole genome shotgun sequencing project. GenBank. JBBBDW000000000.1
- 71.2024. NCBI Sequence Read Archive. SRR28248702
- 72.2024. NCBI Sequence Read Archive. SRR28248701
- 73.2024. NCBI Sequence Read Archive. SRR28248703
- 74.2024. NCBI Sequence Read Archive. SRR28248694
- 75.2024. NCBI Sequence Read Archive. SRR28248695
- 76.2024. NCBI Sequence Read Archive. SRR28248696
- 77.2024. NCBI Sequence Read Archive. SRR28248697
- 78.2024. NCBI Sequence Read Archive. SRR28248698
- 79.2024. NCBI Sequence Read Archive. SRR28248699
- 80.2024. NCBI Sequence Read Archive. SRR28248700
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014182/CRR1002983
- 2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014182/CRR1002984
- 2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014182/CRR1002982
- 2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003137
- 2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003138
- 2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003139
- 2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003140
- 2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003141
- 2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003142
- 2023. NGDC Genome Sequence Archive (GSA) https://ngdc.cncb.ac.cn/gsa/browse/CRA014193/CRR1003143
- Zhang B. 2024. Monochamus alternatus isolate DY-2024, whole genome shotgun sequencing project. GenBank. JBBBDW000000000.1
- 2024. NCBI Sequence Read Archive. SRR28248702
- 2024. NCBI Sequence Read Archive. SRR28248701
- 2024. NCBI Sequence Read Archive. SRR28248703
- 2024. NCBI Sequence Read Archive. SRR28248694
- 2024. NCBI Sequence Read Archive. SRR28248695
- 2024. NCBI Sequence Read Archive. SRR28248696
- 2024. NCBI Sequence Read Archive. SRR28248697
- 2024. NCBI Sequence Read Archive. SRR28248698
- 2024. NCBI Sequence Read Archive. SRR28248699
- 2024. NCBI Sequence Read Archive. SRR28248700
Supplementary Materials
Data Availability Statement
No specific script was utilized in this study. The codes and pipelines used for genome sequencing data analysis were performed following the instructions of corresponding bioinformatics tools. The version and parameters of the software have been included in the Methods section.