Abstract
This study introduces a novel approach for integrating sensitive patient information within medical images with minimal impact on their diagnostic quality. Utilizing the mask region-based convolutional neural network for identifying regions of minimal medical significance, the method embeds information using discrete cosine transform-based steganography. The focus is on embedding within “insignificant areas”, determined by deep learning models, to ensure image quality and confidentiality are maintained. The methodology comprises three main steps: neural network training for area identification, an embedding process for data concealment, and an extraction process for retrieving embedded information. Experimental evaluations on the CHAOS dataset demonstrate the method’s effectiveness, with the model achieving an average intersection over union score of 0.9146, indicating accurate segmentation. Imperceptibility metrics, including peak signal-to-noise ratio, were employed to assess the quality of stego images, with results showing high capacity embedding with minimal distortion. Furthermore, the embedding capacity and payload analysis reveal the method’s high capacity for data concealment. The proposed method outperforms existing techniques by offering superior image quality, as evidenced by higher peak signal-to-noise ratio values, and efficient concealment capacity, making it a promising solution for secure medical image handling.
Keywords: Medical image security, Data hiding techniques, Mask-RCNN, Deep learning in medical imaging, High-capacity embedding, DCT-based steganography
Subject terms: Image processing, Machine learning, Software
Introduction
The medical information of patients must be protected from unauthorized access. Generally, medical information security refers to the rules of security policies that strictly ethical rights and privacy of the patient and must be concerned by entities. There are many widely used standards and tools for protecting personal medical information such as the ISO27799 (Security Management in Health Using ISO/IEC/27799)1, Cryptography2, and Steganography3 techniques. ISO27799 is a standard that provides security management guidelines for health organizations to protect medical information. Cryptography is data protection using an encryption provider which converts medical information into unintelligible text using a secure key. Steganography can be defined as the use of a host (container) data to hide or embed a piece of information that is hidden directly in media content, in such a way that it is imperceptible to a human observer but can be detected/extracted easily with a computer4. Steganography is employed in various useful applications, such as in Medical Imaging Systems where a separation is considered necessary between patients’ medical files and their personal information for the sake of confidentiality5.
Medical image steganography aims at delivering a modified medical image to secure the transfer of hidden information inside with little perception of third-party. Medical image steganography is an emerging field of research that aims to embed secret information into medical images in a way that is imperceptible to the human eye, while ensuring the integrity and confidentiality of the medical data. The primary motivation behind medical image steganography is to protect patient privacy by preventing unauthorized access to sensitive medical information. High-capacity data hiding techniques, such as deep learning based models, have been developed to address the limitations of traditional steganography methods, which have limited embedding capacity and are vulnerable to attacks. The RCNN-Mask model is a deep learning-based approach that can effectively embed a large amount of secret data into medical images, while maintaining the visual quality of the images and ensuring robustness against attacks.
Deep learning has emerged as a powerful tool for data hiding approaches, enabling the development of high-capacity, robust and imperceptible steganography techniques. Deep learning-based approaches leverage the power of neural networks to learn the optimal mapping between cover images and secret data, allowing for the generation of stego-images that are visually indistinguishable from their cover counterparts. These techniques are designed to embed secret data into the cover images in a way that is robust against attacks, such as image processing or compression, while maintaining the quality of the stego-images. Furthermore, deep learning-based approaches can be adapted to different types of data, including images, audio, and video, allowing for the development of versatile and flexible steganography techniques. Overall, deep learning-based data hiding approaches have revolutionized the field of steganography, enabling the development of advanced and effective techniques for data protection and privacy.
The proposed high-capacity data hiding for medical images based on the RCNN-mask model is a novel steganography technique that aims to embed a large amount of secret data into medical images while maintaining their visual quality and ensuring robustness against attacks. The RCNN-mask model, which is a deep learning-based object detection and segmentation model, is used to identify and segment regions of interest in the cover image, which are then used to embed the secret data. The proposed technique leverages the high-capacity of the RCNN-mask model to embed a large amount of secret data into the cover image, while ensuring that the stego-image remains imperceptible to the human eye. Experimental results show that the proposed technique achieves high embedding capacity and low distortion, as well as robustness against various attacks, such as JPEG compression and cropping. Overall, the proposed high-capacity data hiding for medical images based on the RCNN-mask model is a promising steganography technique that can be used to protect sensitive medical data while ensuring its confidentiality and integrity.
The advent of digital imaging in medical diagnostics has revolutionized healthcare, enabling the storage, sharing, and detailed analysis of medical images. However, this digital transition also introduces significant challenges in ensuring the security and privacy of sensitive patient information embedded within these images. Existing methods for data hiding within medical images often struggle to achieve a harmonious balance between embedding capacity, image integrity, and robustness against attacks. Specifically, traditional techniques tend to compromise on visual quality to increase payload capacity or fail to adequately protect embedded data against common image processing attacks, such as noise addition or compression. Furthermore, many current approaches lack the sophistication to selectively embed information in regions that do not compromise the diagnostic value of medical images, leading to potential risks in clinical interpretation. These technical gaps underscore the need for an advanced data hiding method that not only supports high-capacity embedding without degrading image quality but also ensures the robust protection of embedded data, all while preserving the diagnostic usability of medical images. Addressing these challenges requires a nuanced understanding of both medical image processing and security principles, guiding the development of a method that adeptly navigates the trade-offs inherent in secure medical data handling.
In the era of digital healthcare, the significance of medical images has grown exponentially as essential components of patient records and medical research data. Recent reports highlight a substantial increase in the utilization of digital medical images across various healthcare settings, underscoring their critical role in enhancing diagnostic accuracy, facilitating telemedicine, and supporting advanced medical research. However, this reliance on digital imaging also brings to the fore concerns regarding the security and privacy of sensitive patient information. Studies such as “HIDEmarks: Hiding multiple marks for robust medical data sharing using IWT-LSB”6 and “Robust copyright protection technique with high-embedding capacity for color images”7 have shed light on innovative approaches to protect this data, yet gaps remain in ensuring these methods meet the nuanced demands of medical imaging. These works emphasize the urgent need for robust, high-capacity data hiding techniques that can safeguard patient information without compromising the diagnostic integrity of medical images. Motivated by these challenges, our work aims to bridge these gaps by introducing a sophisticated data hiding method tailored for the unique requirements of medical imaging. By leveraging the latest advancements in deep learning and steganography, we strive to offer a solution that upholds the confidentiality of embedded data while ensuring the visual and diagnostic quality of medical images remains intact.
The motivation behind this work is to address the growing need for robust data security in medical imaging without compromising the diagnostic value of medical images. As medical imaging plays a crucial role in diagnosis and treatment planning, ensuring the confidentiality and integrity of patient data embedded within these images is paramount. As medical images are shared and stored electronically, the risk of unauthorized access and potential misuse of sensitive patient data escalates. Our work aims to address this challenge by providing a secure method for embedding patient information directly into medical images without impairing their diagnostic value. The key contributions of our study are summarized as follows:
We introduce a novel data hiding technique based on the mask region-based convolutional neural network (Mask-RCNN) for identifying medically insignificant regions within DICOM images for secure data embedding.
The proposed method utilizes discrete cosine transform (DCT)-based steganography to embed patient information discreetly, ensuring the integrity and diagnostic quality of medical images are maintained.
Our approach is demonstrated to achieve high embedding capacity and maintain image quality through rigorous evaluation on DICOM images, outperforming existing methods in terms of robustness, payload capacity, and image fidelity.
The manuscript is organized as follows: “Related work” provides a detailed review of related work, highlighting the gap our study aims to fill. “The proposed method” describes the methodology, including the use of Mask-RCNN for region identification and the DCT-based steganography technique for data embedding. “Experiments” presents the experimental setup, including the dataset used, evaluation metrics, and comparative analysis with existing methods. “Robustness analysis” discusses the results, emphasizing the efficacy and benefits of our approach. Finally, “Conclusion” concludes the paper with a summary of our findings and potential directions for future research.
Related work
Steganography’s significance is rooted in its ability to conceal the presence or absence of concealed information from unintended recipients, in stark contrast to cryptography, where decryption alone reveals the message. In this section, we delve into pivotal endeavors concerning safeguarding medical data through steganography, while also exploring contemporary methodologies integrating deep learning for steganographic applications over recent years.
Medical image steganography
In8, Bozhidar et al. presented an innovative steganography method termed BOOST, designed to conceal user data within medical images. Their approach unfolded in two distinct stages: Initially, the confidential patient data underwent encryption using a novel “pseudorandom generator based on the nuclear spin generator” technique, resulting in encrypted data. This encrypted output was subsequently transcribed into a binary sequence using an ASCII table. In the subsequent step, this binary sequence found its place within the least significant bit of the non-black pixels in the image. Notably, their method achieved remarkable results, boasting PSNR values surpassing 113 dB, all while accommodating a payload capacity of 0.74 bits per pixel. The substantial payload capacity emphasizes the potential for real-world applications. However, it is important to consider the computational overhead of these encryption and embedding processes, especially when dealing with large medical image datasets.
In9, Romany et al. introduced an encompassing steganography method that amalgamates several techniques for robust data hiding within medical images. They proposed the application of RSA encryption for safeguarding sensitive information, the Ripplet Transform for image manipulation, and LSB substitution for embedding secret data. An adaptive genetic-algorithm-based optimum pixel adjustment process (OPAP) was implemented to enhance imperceptibility by fine-tuning the stego image. This comprehensive approach demonstrated resilience against RS attacks and established that Discrete Ripplet Transform (DRT) yielded superior results in comparison to Integer Wavelet Transform (IWT). Notably, the achieved PSNR values ranged from 49 to 56 dB, indicating a trade-off between visual quality and payload capacity.
In10, Songul and Engin presented an innovative steganography technique termed “Genetic Algorithm-Optimum Pixel Similarity”. This approach leverages pixel similarity and LSB embedding to seamlessly integrate a substantial amount of data, specifically 10,000 characters, into 256 256 medical images. What sets this method apart is its ability to achieve embedding without resorting to data compression techniques. The fitness function for the genetic algorithm is adopted from PSNR, with random selection as the key method. Impressively, the average PSNR achieved was recorded at 47.41 dB, highlighting the delicate balance between imperceptibility and embedding capacity.
Partha et al. explored patient data protection in11 through a novel steganographic method, employing support vector machine (SVM) and discrete wavelet transform (DWT). The SVM was utilized for the recognition of regions of interest (ROI) and non-ROI (NROI) within medical images. RGB components were subjected to IWT, and a circular array technique facilitated the integration of confidential information within NROI pixels. Impressively, this approach yielded an average PSNR value of 64 dB, showcasing its potential for robust and secure patient data embedding. In another study12, a robust and reversible data hiding scheme was proposed, involving a support vector neural network (SVNN) classifier and the contourlet transform method. The SVNN was trained to identify suitable pixels for concealment, with the HL band of the CT coefficient serving as the container for hidden data. The method was rigorously analyzed with and without noise, demonstrating exceptional results with a PSNR value of up to 89.3253 dB, outperforming the SVNN-wavelet approach from13.
In14, an innovative approach was introduced that encoded patient data using enhanced Huffman compression coding for enhanced payload capacity and security. This encoded data was then concealed within medical images using pixels contrast (PC) and the Henon map algorithm. The study evaluated outcomes based on histogram analysis, PSNR, and SIMM metrics, with achieved PSNR values ranging between 70 and 71 dB. A novel steganography technique was proposed in15, utilizing a combination of a 3-D chaotic system, one-particle quantum walk (QW), and particle swarm optimization (PSO). This intricate methodology ensured the privacy of medical data by generating sequences for PSO through chaotic systems and QW, which were then utilized to replace confidential medical images with concealed data. Despite its high visual quality, this technique achieved an average PSNR of 44.1 dB, reflecting inherent limitations in data capacity. In16, the authors introduced a technique for securely compressing 2D medical images, such as MRI, CT, and ultrasound scans, to efficiently manage storage space while maintaining image integrity and privacy. The method employs a multi-level compression strategy using a dictionary mechanism, combined with a 256-bit symmetric key encryption based on a hashing technique to ensure data security. Additionally, the fuzzy trapezoidal correlation method is utilized for accurately reconstructing the original image from its compressed state, ensuring minimal quality loss. The approach has demonstrated significant reductions in image size (up to 58).
The steganography technique presented by Hashim et al. in17 targeted data security during transmission within an IoT framework. Encrypted patient data was divided into blocks and concealed within medical images using the Henon map parameters for random pixel selection. This technique demonstrates effective use of steganography mechanisms for IoT data security. Prasanth et al. introduced an invisible watermarking scheme in18 for embedding patient information into EGG signals for telemedicine applications. A QR code of patient data was decomposed and utilized for watermarking EGG signals. This intricate approach provided a unique approach to securing medical information within telemedicine applications.
Arunkumar et al. proposed a novel technique in19 for secure medical image transmission. The medical image was encrypted using the logistic chaotic map, followed by embedding using an embedding distortion measure based co-accurate matrix. The method prioritized security and yielded high visual quality while ensuring secure image transmission. The secure steganography method outlined in20 incorporated a shell matrix and LSB for enhanced data security. While the method exhibited high payload capacity, it required substantial computational resources for high-resolution images, thereby impacting complexity. The authors, in21, introduced a novel approach involving a genetic algorithm to enhance PSNR levels in Stego images. The technique aimed to cover a medical image with a natural image, utilizing a combination of mechanisms such as one-point crossover, random resetting mutation, and tournament selection. While the method achieved infinite PSNR and SSIM values without causing distortion, its complexity remained a significant consideration.
Research in the field of medical image data hiding has laid a solid foundation but has also encountered several challenges that our study aims to address. Notably, existing techniques often struggle with balancing the trade-off between embedding capacity, robustness against attacks, and preserving image quality. To mitigate this challenge, our proposed approach leverages advanced deep learning architectures, such as Mask-RCNN, to precisely identify regions within medical images suitable for data embedding while minimizing the risk of diagnostic information loss. Moreover, we employ sophisticated embedding techniques, such as the discrete cosine transform (DCT), to embed data in frequency components least perceptible to the human eye, thereby ensuring both robustness and imperceptibility. Additionally, our model incorporates adaptive selection mechanisms to intelligently prioritize embedding in regions with optimal texture and complexity characteristics, further enhancing the security and reliability of the embedding process. By addressing these key challenges, our approach represents a significant advancement in the field, offering a more balanced and effective solution for high-capacity data hiding in medical images.
The proposed method
Overview
The proposed model innovatively integrates the mask region-based convolutional neural network (Mask-RCNN) with discrete cosine transform (DCT) for high-capacity data hiding within medical images. At its core, the model employs Mask-RCNN, a state-of-the-art deep learning framework known for its precision in instance segmentation tasks. This model is adept at identifying and segmenting regions within medical images that are medically less significant, thereby earmarking them as potential areas for secure data embedding. The choice of Mask-RCNN is motivated by its dual capability to classify individual pixels in an image while precisely delineating the boundaries of objects, making it an ideal candidate for isolating regions where embedding can occur without affecting the diagnostic value of the image.
Once these regions are identified, the model utilizes the discrete cosine transform (DCT), a cornerstone technique in signal processing that transforms spatial domain data into frequency domain. This transformation facilitates the embedding of sensitive information into the frequency components of the image, specifically targeting the mid-frequency components. This choice is strategic; the human eye is less sensitive to changes in these components, ensuring that the embedding remains imperceptible. Moreover, DCT provides a mechanism to adjust the embedding intensity, allowing for a flexible trade-off between embedding capacity and image quality.
The embedding process is further refined through an adaptive selection mechanism, which evaluates the suitability of identified regions based on their texture and complexity, ensuring that data is embedded in areas where it is least likely to be detected or affect image quality. This nuanced approach, combining Mask-RCNN’s segmentation prowess with DCT’s embedding efficiency, represents a significant technical advancement in the field of medical image security.
This study’s contribution lies in the creation of DICOM files that seamlessly integrate patient information into medical images with an exceedingly minimal impact-almost inconsequential-in order to safeguard against misdiagnosis, all achieved through the application of steganography principles. The devised approach involves concealing patient data within areas of the medical image that hold marginal relevance. Here, “insignificant areas” refer to regions devoid of crucial medical data, such as the black segments found in grayscale DICOM images. The identification of these areas is facilitated by deep learning (DL) models, which effectively discern non-essential regions within the original images. Subsequently, sensitive medical information is discreetly embedded within these inconspicuous regions using DCT-based steganography. A comprehensive visual representation of the proposed methodology is depicted in Fig. 1, outlining three fundamental stages: neural network training, embedding, and extraction.
Figure 1.
Overview of the proposed steganography method.
Neural network training
The key-concept in the proposed method is the detection of insignificant areas in medical DICOM images which will be exploited to conceal sensitive information. We assume that the best way to detect these regions is by correctly detecting the main objects in the image. In the literature, CNN-based methods outperform traditional techniques in the detection and segmentation of objects inside images. Thus, we adopt Mask-RCNN architecture22, as one of the efficient techniques especially in the field of medical images. This architecture is proposed to detect the main objects which represent the significant area that should be kept safe during information embedding. To train the Mask-RCNN model to obtain binary segmentation masks, we use the architecture depicted in Fig. 2 on various DICOM files datasets. This architecture is divided into two stages:
Figure 2.
Mask R-CNN architecture.
First stage: it takes an image and produces a feature map and regions proposals. The feature map is obtained through a series of operations conducted on the original image by a backbone constructed from CNN layers (ResNet and feature pyramid). Region proposal network takes the feature map and produces regions that may contain objects.
Second stage: it consists of aligning regions of interests (RoIalign). It takes as input feature map and region proposals and generates as output the fixed size regions of interest from region proposals, and three parallel branches for predicting: object category, instance bounding-box, and binary segmentation masks.
This architecture is trained on various datasets to determine the binary segmentation mask where 1’s represents object pixels (significant region) and 0’s represents the background of the image (i.e. the insignificant area). The loss function used to train this model is defined by Eq. (1).
| 1 |
where is the classification loss, is the bounding box regression loss, and is the mask Loss.
Embedding process
The process of embedding, also known as the concealment process, encompasses a series of sequential steps aimed at seamlessly integrating confidential patient-related data into DICOM images, ultimately giving rise to a Stego image. To elaborate, the focal point of this embedding is the inconspicuous area previously identified using the Mask-RCNN model. This entire procedure can be delineated into three distinct phases, outlined in the subsequent subsections.
Sensitive information preprocessing
In this first phase, the sensitive patient information is retrieved from the DICOM file and transformed into a QR code image via QR Code generator. Figure 3 depicts some examples of patient information with their corresponding QR codes generated using the Zxing library23.
Figure 3.
Examples of patient data with their corresponding QR codes.
The Generated QR code image is then normalized using Eq. (2) to convert the values within the image to a range between 0 and 1. After that, the normalized image is adjusted using Eq. (3), where the value of is fixed experimentally ( = 0.02). The adjusted image is then divided into 4 4 mini matrices called blocks () that are hidden one by one during the concealment phase.
| 2 |
| 3 |
Cover preprocessing
The cover, derived from the DICOM file, is subjected to a dual stage preprocessing procedure. Initially, the mask-RCNN model is employed in inference mode to pinpoint the inconsequential regions within the cover, which yields a binary segmentation mask. Subsequently, given that DICOM images are frequently encoded in 16-bit grayscale, the cover is normalized using Eq. (2) to effectively remap its values within the [0, 1] interval.
The resulting mask obtained from the first preprocessing step is utilized to determine the indices corresponding to the insignificant areas within the normalized cover image, referred to us as () image. These insignificant areas are subsequently divided into blocks with dimensions of 4 4, denoted as (). The transformation coefficients of () are then computed using the two-dimensional discrete cosine transform function (2D-DCT)24, resulting in the generation of () blocks. The 2D-DCT for a matrix I (with dimensions M N) is calculated using the formula specified in Eq. (4)
| 4 |
where
(m, n) and I(m, n) correspond to the position values and the pixel value at position (m, n) in the spatial domain respectively. C(u, v) isthe corresponding position value and the frequency coefficient at position (u, v) in the transform domain.
Information concealment
During this stage, the 4 4 blocks originating from the cover, denoted as (), and the 4 4 blocks representing the secret message, referred to as () and generated in the prior phase, are merged together-specifically, () is concealed within ()-resulting in the formation of the corresponding block () within the Stego image. This concealment process is executed through the application of Eq. (5).
| 5 |
where
: Represents the block of the Stego image resulting from the concealment process.
- IDCT2 is the inverse two-dimensional discrete cosine transform function (2D-IDCT). This function is explained by Eq. (6).
6 - Qun: Denotes the function that integrates the block into and generates the pre-Stego block. This block is computed using the formula depicted in Eq. (7).
where:7 - i,j: are respectively the i-th and j-th ligne and column in the block.
- : is the number of intervals that satisfy the cover coefficients on the interval of [0, 4].
- .
Subsequently, the inverse two-dimensional discrete cosine transform function (2D-IDCT) is employed on , yielding the ultimate that characterizes the Stego blocks. Following this, the Stego blocks are amalgamated to forge the Stego image, which subsequently undergoes de-normalization to confine values within the [0, 65535] range. This critical step guarantees that the Stego image adheres to the 16-bit DICOM file format, conserving the initial encoding scheme. An illustrative demonstration of the concealment process can be found in Fig. 4. Algorithm 1 prescribes the sequential steps that constitute the embedding process.
Figure 4.

A visual illustration of the hiding technique when = 8.
Algorithm 1.
The embedding algorithm.
Extraction process
The extraction process serves as the reverse of the concealment process, with the aim of recovering the patient data hidden within Stego images. This retrieval is exclusively authorized for users possessing the requisite key. The extraction unfolds across two distinct phases: mask generation and the extraction process.
In the mask generation phase, the Stego image is subjected to the Mask R-CNN model operating in inference mode. This operation generates a binary segmentation mask tailored to pinpoint insignificant regions. This mask, a critical tool, facilitates the identification and indexing of these areas-precisely where the secret data has been concealed.
Moving to the extraction process, an essential preliminary step involves normalizing the Stego image to ensure pixel values are confined within the [0, 1] range. Leveraging the binary mask associated with the Stego image, the positioning of pixels utilized for concealment is discerned. This determination is pivotal in the creation of the insignificant Stego matrix. Subsequently, the matrix is partitioned into blocks and subjected to the Discrete Cosine Transform (DCT) function, transitioning it into the frequency domain. The outcome is a set of transformation coefficients. By employing the inverse merge operation (IQun), these coefficients facilitate the extraction of the mini block corresponding to the QR code, denoted as . The extraction equation is formally expressed as detailed in Eq. (8)
| 8 |
where the inverse merge equation (IQun) is calculated by Eq. (9).
| 9 |
Finally, the resulting blocks are then concatenated to produce the QR code of the patient information. The reverse process is summarized by Algorithm 2
Algorithm 2.
The extraction algorithm.
Experiments
CHAOS dataset
CHAOS dataset aims to segment abdominal organs (liver, kidneys, and Spleen) using CT and MRI data25. It consists of two datasets, Each one corresponding to a series of DICOM images. The first comprises CT images of 40 different patients with a healthy liver. The patient orientation and alignment are the same for all the data sets. The data consists of 16-bit DICOM images with a resolution of , an x–y spacing of 0.7–0.8 mm, and an inter-slice distance (ISD) of 3–3.2 mm. The second database includes 120 DICOM data sets from two different MRI sequences, each of which is being routinely performed to scan the abdomen using different radiofrequency pulse and gradient combinations. The data sets are acquired by a 1.5 T Philips MRI, which produces 12 bit DICOM images having a resolution of . The ISDs vary between 5.5 and 9 mm (average 7.84 mm), x–y spacing is between 1.36 and 1.89 mm (average 1.61 mm) and the number of slices is between 26 and 50. we have randomly selected 1200 DICOM images from the CHAOS dataset. These images are divided into 1023 images (11 patient images) for training, 199 images for validation (3 patient images) 10 images for testing. We manually created the annotation of these images using VIA Annotation Software26, with the intention to make a semantic segmentation to separate the foreground that we consider as Significant area from the background that we consider as insignificant area.
Mask RCNN model training and evaluation
We leveraged transfer learning to prepare a Mask-RCNN model that detects insignificant areas in DICOM images. We started by fine-tuning the pre-trained weights of the MS COCO model27, which is a large object detection and instance segmentation dataset that comprises 328k images with 91 labeled categories. To fine-tune this model, we used the implementation of MASK-RCNN proposed by Matterport in28 and we started the training on the CHAOS dataset with the MS COCO weights to produce a variation of the network that targets our detection goals.
Table 1 presents the configuration details for training our variant of the Mask-RCNN model. The parameters listed in the table include the backbone architecture, which is the ResNet101 architecture in this case. The batch size, which is the number of images used in each training iteration, is set to 4. The Feature Pyramid Network (FPN) used for classification is a fully connected layer with a size of 1024. The learning rate and momentum parameters are set to 0.001 and 0.9 respectively. And to prevent overfitting, we set the weight decay parameter to 0.0001. The RPN Train Anchors per Image parameter, which refers to the number of anchors used in the region proposal network (RPN) during training, is set to 256. and the Images Per GPU parameter is set to 4, indicating that each GPU processes 4 images at a time. The Steps Per Epoch parameter is set to 10, and the Validation Steps parameter is set to 50. These parameters control the training process and the number of training and validation iterations.
Table 1.
Mask R-CNN configuration.
| Parameter | Value |
|---|---|
| Backbone | resnet101 |
| BATCH_SIZE | 4 |
| FPN_CLASSIF_FC_LAYERS_SIZE | 1024 |
| PU_COUNT | 1 |
| IMAGES_PER_GPU | 4 |
| LEARNING_MOMENTUM | 0.9 |
| LEARNING_RATE | 0.001 |
| RPN_TRAIN_ANCHORS_PER_IMAG | 256 |
| STEPS_PER_EPOCH | 10 |
| VALIDATION_STEPS | 50 |
| WEIGHT_DECAY | 0.0001 |
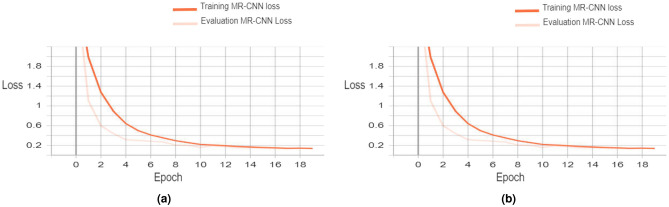
The training was conducted on a machine empowered by Core I7 and 10th generation processor, Intel UHD graphics, and 16 GB of RAM. The training is done in 28 epochs (8 epochs for the head and 20 epochs to fine-tune all layers). The histograms of the training and validation losses are presented in Fig. 5. Sub-figure (a) displays the general training and validation losses, while sub-figure (b) displays the losses of the MR-CNN mask training and validation. The MR-CNN general loss is recorded as 0.1291 at the end of the training, and the MR-CNN mask loss is noted as 0.0450.
Figure 5.
Model training and validation losses.
We evaluated the overlap between the annotated and the generated masks of the validation dataset using the Intersection Over Union (IoU) metric29. IoU is calculated using Eq. (10):
| 10 |
A lower value of IoU indicates inadequate prediction (i.e. poor prediction), whereas a value of 1 represents an entirely accurate prediction. The validation process yielded an Average IoU of 0.9146, signifying that the model can be safely used.
Imperceptibility measurement
The second part of the experiment was dedicated to the embedded process that uses the trained MR-CNN model. We tested the process on 10 images. After extracting patient information from the DICOM files and transforming them into QR codes, we applied the embedding method described above to conceal the QR codes (Message) in the Cover images to obtain Stego images as a first step. Then, we applied the extraction process to retrieve secret messages from the Stego images as a second step.
Table 2 shows the obtained results (the cover, message, Stego, and the Retrieved message) for a sample of 4 DICOM files. Based on visual inspection, it appears that there is no discernible disparity between the original (cover and message) and generated images (Stego and retrieved Qr code). However, we used the PSNR (peak signal to noise ratio) metric to evaluate the visual quality of the generated images and The CNN (Normalized Correlation Coefficient) metric to check the similarity between the cover mask and the stego mask obtained by the MR-CNN model, which affects the correctness of the extraction process.
Table 2.
Comparative results showcasing cover, original QR code (message), stego image, and retrieved QR code.
| The cover | The message | The stego | The retrieved message | |
|---|---|---|---|---|
| CT 1 |  |
 |
 |
 |
| CT 2 |  |
 |
 |
 |
| MRI 1 |  |
 |
 |
 |
| MRI 2 |  |
 |
 |
 |
Furthermore, PSNR is calculated in decibels between two images using Eq. (12):
| 11 |
| 12 |
where M and N are the numbers of rows and columns in the input images. R is the maximum fluctuation in the input image data type. Typical values for the PNSR for 16-bit data are between 30 and 80 dB.
Moreover, NCC is used in our case to measure the robustness of the model and estimate the difference between the cover mask and the Stego mask, NCC value adjacent to 1 implies that the two masks are similar. NCC formula is given in Eq. (13):
| 13 |
where and are the mean pixel values of the cover mask and the Stego mask, respectively.
Table 3 presents the NCC and PSNR values for the tested DICOM images, along with their image sizes and scan types. The NCC values listed in the table fall within the range of 0.83–1 for all images, signifying that our MR-CNN models can predict identical masks from both the Cover and Stego images. This successful prediction enables the accurate detection of the insignificant area where concealment operations take place. The PNSR values depicted in Table 3, in case of the embedding parameters and are set to 1000 and 0.02 respectively, are ranging between 70.45 and 82.38, indicating that our method effectively conceals sensitive patient information with a high level of imperceptibility.
Table 3.
The NCC and PSNR values for the tested DICOM images.
| Patient ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Scan type | CT | CT | CT | CT | CT | MR | MR | MR | MR | MR |
| Image size (Pixel) | 512 × 512 | 512 × 512 | 512 × 512 | 512 × 512 | 512 × 512 | 256 × 256 | 256 × 256 | 512 × 512 | 256 × 256 | 256 × 256 |
| NCC | 1.00 | 1.00 | 0.83 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 0.99 | 0.99 |
| PSNR (dB) | 116.07 | 113.69 | 115.53 | 116.57 | 116.02 | 107.47 | 110.3 | 117.4 | 110.3 | 110.33 |
To examine the effect of the coefficient on the final quality of the Stego, we measured the PSNR by variating the embedding parameter . Table 4 shows the PSNR value obtained.
Table 4.
The average results of the MSE values between the Cover and Stego DICOM.
| Scan type | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 100 | 200 | 300 | 400 | 500 | 600 | 700 | 800 | 900 | 1000 | |
| CT | |||||||||||
| 1 | 75.051 | 95.955 | 102.01 | 105.557 | 108.094 | 110.039 | 111.578 | 112.993 | 114.129 | 115.225 | 116.074 |
| 2 | 72.661 | 93.551 | 99.64 | 103.166 | 105.729 | 107.661 | 109.222 | 110.618 | 111.748 | 112.888 | 113.695 |
| 3 | 74.555 | 95.424 | 72.483 | 101.500 | 105.029 | 107.563 | 109.494 | 112.45 | 113.585 | 114.696 | 115.532 |
| 4 | 75.136 | 96.313 | 102.442 | 105.998 | 108.55 | 110.517 | 112.07 | 113.444 | 114.644 | 115.723 | 116.571 |
| 5 | 75.076 | 95.914 | 101.994 | 105.526 | 108.067 | 109.995 | 111.557 | 112.945 | 114.076 | 115.191 | 116.022 |
| Averag | 74.4958 | 95.4314 | 95.7138 | 104.3494 | 107.0938 | 109.155 | 110.7842 | 112.49 | 113.6364 | 114.7446 | 115.5788 |
| MRI | |||||||||||
| 1 | 66.494 | 87.347 | 93.425 | 96.949 | 99.500 | 101.447 | 102.984 | 104.394 | 105.533 | 106.643 | 107.473 |
| 2 | 69.389 | 90.217 | 96.281 | 99.803 | 102.345 | 104.283 | 105.823 | 107.227 | 108.365 | 109.476 | 110.304 |
| 3 | 76.472 | 97.313 | 103.385 | 106.904 | 109.447 | 111.386 | 112.925 | 114.321 | 115.472 | 116.581 | 117.400 |
| 4 | 69.389 | 90.217 | 96.281 | 99.803 | 102.345 | 104.283 | 105.823 | 107.227 | 108.365 | 109.476 | 110.304 |
| 5 | 69.39 | 90.222 | 96.29 | 99.816 | 102.362 | 104.303 | 105.847 | 107.253 | 108.393 | 109.505 | 110.333 |
| Averag | 70.2268 | 91.0632 | 97.1324 | 100.655 | 103.1998 | 105.1404 | 106.6804 | 108.0844 | 109.2256 | 110.3362 | 111.1628 |
Table 4 shows the obtained average results of the MSE values between the Cover and Stego DICOM images of 20 patients for different b values. The average of the MSE values was between 2049.46 and 6152.98 for b = 500.While the minimum and maximum value of MSE was recorded for b = 1000 and b = 100, respectively, which prove that the largest value of b produces a high-quality Stego image. Table 4 shows the resultant average of PSNR values to compare between Cover and Stego images in decibel (dB). The average PSNR values ranged between 80.22 and 84.96 dB for b = 500. The minimum and maximum value of PSNR are 76.35 dB (patient 2 and b = 100) and 85.39 dB (patient 14 and b = 1000) respectively. Generally, PSNR higher values refer to the invisibility of higher quality.
Capacity and payload
Table 5 presents a detailed analysis of capacity and payload values for both CT (computed tomography) and MRI (magnetic resonance imaging) images across ten different patient IDs. These values provide crucial insights into the performance and efficiency of a data hiding technique when applied to medical images. It’s essential to consider these capacity and payload values when designing and implementing data hiding techniques for medical images, as they provide insights into the trade-off between data capacity and image quality in the context of medical data security. Capacity refers to the amount of secret data that can be embedded within the medical image while maintaining the image’s visual quality and integrity. For CT images, the capacity ranges from 0.48 to 0.72, with an average capacity of 0.54. This indicates that, on average, approximately 54% of the image can be utilized to hide secret data without significant degradation in image quality. For MRI images, the capacity varies from 0.49 to 0.63, with an average capacity of 0.58. MRI images show a slightly lower but still substantial capacity, with approximately 58% of the image available for data embedding. Payload refers to the amount of secret data that is successfully embedded within the image. It is a critical metric as it indicates how much data can be reliably hidden within the image. For CT images, the payload values range from 0.02 to 0.10, with an average payload of 0.05. This suggests that, on average, 5% of the image can be effectively used to conceal secret data. For MRI images, the payload varies between 0.02 and 0.10, with an average payload of 0.05, mirroring the payload results of CT images.
Table 5.
The resultant capacity and payload.
| CT images | MRI images | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient id | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Average |
| Capacity | 0.5 | 0.54 | 0.50 | 0.48 | 0.49 | 0.63 | 0.61 | 0.54 | 0.72 | 0.58 | 0.54 |
| Payload | 0.031 | 0.037 | 0.031 | 0.032 | 0.031 | 0.09 | 0.10 | 0.02 | 0.08 | 0.10 | 0.05 |
Robustness analysis
A critical aspect of our proposed data hiding technique for medical images is its robustness against various types of noise attacks. To evaluate the resilience of our method, we subjected the stego images to three common noise attacks: Gaussian noise, uniform noise, and salt and pepper noise. The robustness was assessed by retrieving the embedded QR code from the noise-affected stego images and measuring the similarity and visual quality through normalized cross-correlation (NCC) and peak signal-to-noise ratio (PSNR), respectively.
In Table 6, the NCC values obtained under Gaussian noise, uniform noise, and salt and pepper noise attacks were 0.3188, 0.3150, and 0.3193, respectively. These results indicate a moderate level of correlation between the original and retrieved QR codes post-attack, demonstrating the method’s capability to withstand noise perturbations to a certain extent. Moreover, the PSNR values remained above 53 dB across all noise types, suggesting that the visual quality of the stego images is preserved well above acceptable thresholds, even in the presence of noise. This is significant as it ensures that the diagnostic value of medical images is not compromised due to embedding and subsequent noise attacks.
Table 6.
The robustness evaluation results under the noise attacks.
| Stego attack | Gaussian noise | Uniform noise | Salt and pepper noise |
|---|---|---|---|
| The retrieved QR code |  |
 |
 |
| NCC | 0.3188 | 0.3150 | 0.3193 |
| PSNR (dB) | 53.36 | 53.33 | 53.39 |
In developing our data hiding approach for medical images based on the Mask-RCNN model, we meticulously balanced the trade-off between robustness, visual quality, and payload capacity. This delicate equilibrium ensures that while concealing data within the medical images, the method maintains resilience against various attacks, preserves high visual fidelity, and accommodates a significant payload for information embedding. By optimizing the embedding process and incorporating error correction coding techniques, we mitigate the risk of information loss and maintain the integrity of the stego images, even in the presence of noise or other forms of interference. Furthermore, careful selection of embedding parameters and compression algorithms allows us to strike an optimal balance between concealing capacity and visual imperceptibility, ensuring that the embedded data remains imperceptible to the human eye while maximizing the amount of information that can be securely hidden within the images. This careful consideration of trade-offs empowers our method to deliver robust and high-quality stego images suitable for secure transmission and storage of sensitive medical data.
Comparison
Table 7 provides a comprehensive comparison between the proposed data hiding method and several other existing techniques, highlighting key parameters such as image size, embedding capacity, payload, and best PSNR (peak signal-to-noise ratio). Bozhidar and Borislav’s8 method employs a Nuclear Spin Generator on images, achieving a relatively high embedding capacity of 0.75 Bpp. It successfully hides 83,883 bits of data while maintaining a remarkable PSNR of 113.50, indicating good image quality preservation. Subhadip20 and his team utilize a combination of LSB (Least Significant Bit) and a shell matrix technique on images, resulting in a much higher embedding capacity of 3 Bpp. This approach allows the concealment of a substantial 786,432 bits of data. However, the PSNR of 48.42 indicates some loss in image quality compared to the previous method. Akshay30 and his collaborators employ deep learning on images, achieving an impressive embedding capacity of 24 Bpp. While the exact payload is not specified, this approach prioritizes data capacity over PSNR, which is lower at 37.55, indicating some visual quality degradation. Atique31 and his team also apply deep learning, but on images, resulting in an embedding capacity of 8 Bpp. They manage to conceal 89,910 bits of data with a PSNR of 36.58, indicating some trade-off between capacity and image quality. In contrast, the proposed method operates on images using deep learning and achieves a reasonable embedding capacity of 0.50 Bpp, which allows the concealment of 131,524 bits of data. Notably, it outperforms the other methods in terms of PSNR, attaining an impressive 115.53, signifying exceptional image quality preservation. In summary, the proposed method strikes a balance between embedding capacity and image quality, achieving a competitive capacity while maintaining outstanding PSNR, making it a promising choice for data hiding in medical images.
Table 7.
Compare the proposed method with other methods.
| Method | Image size | Technique | Embedding capacity (Bpp) | Payload (bits) | Best PSNR |
|---|---|---|---|---|---|
| Bozhidar and Borislav8 | 336 336 | Nuclear spin generator | 0.75 | 83,883 | 113.50 |
| Subhadip et al.20 | 256 256 | LSB and shell matrix | 3 | 786,432 | 48.42 |
| Akshay et al.30 | 128 128 | Deep learning | 24 | – | 37.55 |
| Atique et al.31 | 300 300 | Deep learning | 8 | 89,910 | 36.58 |
| Proposed method | 512 512 | Deep learning | 0.50 | 131,524 | 115.53 |
Significant values are in [bold].
Conclusion
In conclusion, this study has presented a novel approach to high-capacity data hiding for medical images based on the Mask-RCNN model. The Mask RCNN, as a deep learning segmentation model, is leveraged to identify medically less significant regions in DICOM images, where secret information, represented as a QR code, were embedded using DCT data hiding capabilities. The Mask RCNN model was fine-tuned and pretrained to consistently identify the same embedding regions in both original and Stego images, enabling reliable steganalysis. The necessity of this study is underscored by the ever-increasing reliance on digital medical imaging and the paramount importance of maintaining patient confidentiality in such a context.
The experimental outcomes, validated by well-recognized objective evaluation metrics such as peak signal-to-noise ratio (PSNR) and normalized cross-correlation (NCC), demonstrate our method’s superiority in achieving a delicate balance between embedding capacity, robustness against noise, and imperceptibility. The results demonstrated the effectiveness of the proposed technique in ensuring high capacity data embedding in medical images with low distortion. The introduced method has a minimum PSNR of 70 and a maximum PNSR of 115 which is large enough for the naked eye to not distinguish between the cover image and its stego image. Furthermore, this method demonstrates superior performance in comparison to its competitors.
The future work will delve into enhancing the robustness and scalability of our approach to accommodate diverse medical imaging modalities and data formats. Additionally, we aim to explore the integration of advanced encryption techniques to fortify the security of embedded data further. Furthermore, considering the dynamic nature of cybersecurity threats, continual evaluation and adaptation of our model will be essential to ensure its effectiveness in real-world clinical settings.
Author contributions
A.E. designed the scheme. H.S. and O.T. conducted the experiments and analyzed the results. All authors contributed to the writing and discussion of the paper. All authors reviewed the manuscript.
Data availability
nThe datasets generated and/or analysed during the current study are available from the corresponding authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fraser, R. Iso 27799: Security management in health using ISO/IEC 17799. In Canadian Institute for Health Information (CIHI) Partnership Conference, June 2006 (2006).
- 2.Katz J, Lindell Y. Introduction to Modern Cryptography: Principles and Protocols. Chapman and hall/CRC; 2007. [Google Scholar]
- 3.Cox I, Miller M, Bloom J, Fridrich J, Kalker T. Digital Watermarking and Steganography. Morgan kaufmann; 2007. [Google Scholar]
- 4.Yahya A. Steganography Techniques for Digital Images. Springer; 2019. [Google Scholar]
- 5.Hua C, et al. Steganography for medical record image. Comput. Biol. Med. 2023;165:107344. doi: 10.1016/j.compbiomed.2023.107344. [DOI] [PubMed] [Google Scholar]
- 6.Singh OP, et al. Hidemarks: Hiding multiple marks for robust medical data sharing using IWT-LSB. Multimed. Tools Appl. 2023;20:1–19. [Google Scholar]
- 7.Mahto D, Singh A, Singh K, Singh O, Agrawal A. Robust copyright protection technique with high-embedding capacity for color images. ACM Trans. Multimed. Comput. Commun. Appl. 2023;20:20. [Google Scholar]
- 8.Stoyanov B, Stoyanov B. Boost: Medical image steganography using nuclear spin generator. Entropy. 2020;22:501. doi: 10.3390/e22050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansour RF, Abdelrahim EM. An evolutionary computing enriched rs attack resilient medical image steganography model for telemedicine applications. Multidimension. Syst. Signal Process. 2019;30:791–814. doi: 10.1007/s11045-018-0575-3. [DOI] [Google Scholar]
- 10.Karakus S, Avci E. A new image steganography method with optimum pixel similarity for data hiding in medical images. Med. Hypotheses. 2020;139:109691. doi: 10.1016/j.mehy.2020.109691. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhuri P, Pal P, Si T. A novel steganographic technique for medical image using SVM and IWT. Multimed. Tools Appl. 2023;20:1–20. doi: 10.1007/s11042-022-14301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reshma V, Vinod Kumar R, Shahi D, Shyjith M. Optimized support vector neural network and contourlet transform for image steganography. Evol. Intell. 2020;20:1–17. [Google Scholar]
- 13.Vk R, Rs VK. Pixel prediction-based image steganography by support vector neural network. Comput. J. 2021;64:731–748. doi: 10.1093/comjnl/bxaa017. [DOI] [Google Scholar]
- 14.Hashim MM, Mahmood AA, Mohammed MQ. A pixel contrast based medical image steganography to ensure and secure patient data. Int. J. Nonlinear Anal. Appl. 2021;12:1885–1904. [Google Scholar]
- 15.Abd-El-Atty B. A robust medical image steganography approach based on particle swarm optimization algorithm and quantum walks. Neural Comput. Appl. 2022;20:1–13. [Google Scholar]
- 16.Srinivasu PN, Norwawi N, Amiripalli SS, Deepalakshmi P. Secured compression for 2d medical images through the manifold and fuzzy trapezoidal correlation function. Gazi Univ. J. Sci. 2021;35:1372–1391. doi: 10.35378/gujs.884880. [DOI] [Google Scholar]
- 17.Hashim, M. M., Rhaif, S. H., Abdulrazzaq, A. A., Ali, A. H. & Taha, M. S. Based on iot healthcare application for medical data authentication: Towards a new secure framework using steganography. In IOP Conference Series: Materials Science and Engineering, vol. 881, 012120 (IOP Publishing, 2020).
- 18.Sanivarapu PV, Rajesh KN, Reddy NR, Reddy NCS. Patient data hiding into ecg signal using watermarking in transform domain. Phys. Eng. Sci. Med. 2020;43:213–226. doi: 10.1007/s13246-019-00838-2. [DOI] [Google Scholar]
- 19.Arunkumar S, Subramaniyaswamy V, Logesh R. Hybrid robust image steganography approach for the secure transmission of biomedical images in cloud. EAI Endorsed Trans. Pervas. Health Technol. 2019;5:e1–e1. [Google Scholar]
- 20.Mukherjee S, Mukhopadhyay S, Sarkar S. A shell-matrix-based image steganography technique for multimedia security and covert communication. Innov. Syst. Softw. Eng. 2022;20:1–16. [Google Scholar]
- 21.Vazquez E, et al. Confidentiality in medical images through a genetic-based steganography algorithm in artificial intelligence. Front. Robot. AI. 2022;9:1031299. doi: 10.3389/frobt.2022.1031299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, K., Gkioxari, G., Dollar, P. & Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision (ICCV) (2017).
- 23.Richter, J. Qr code generator 1.1 based on zxing (2023).
- 24.Balaska N, Belmeguenai A, Goutas A, Ahmida Z, Boumerdassi S. Securing medical data by combining encryption and robust blind medical image watermarking based on zaslavsky chaotic map and dct coefficients. SN Comput. Sci. 2022;3:1–17. doi: 10.1007/s42979-021-01012-w. [DOI] [Google Scholar]
- 25.Kavur AE, et al. CHAOS challenge—combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 2021;69:101950. doi: 10.1016/j.media.2020.101950. [DOI] [PubMed] [Google Scholar]
- 26.Dutta, A. & Zisserman, A. The via annotation software for images, audio and video. In Proceedings of the 27th ACM International Conference on Multimedia, 2276–2279 (2019).
- 27.Lin, T.-Y. et al. Microsoft coco: Common objects in context. In Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6–12, 2014, Proceedings, Part V 13, 740–755 (Springer, 2014).
- 28.Abdulla, W. Mask r-cnn for object detection and instance segmentation on keras and tensorflow. https://github.com/matterport/Mask_RCNN (2017).
- 29.Rezatofighi, H. et al. Generalized intersection over union: A metric and a loss for bounding box regression. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 658–666 (2019).
- 30.Kumar A, Rani R, Singh S. Encoder–decoder architecture for image steganography using skip connections. Proced. Comput. Sci. 2023;218:1122–1131. doi: 10.1016/j.procs.2023.01.091. [DOI] [Google Scholar]
- 31.ur Rehman, A., Rahim, R., Nadeem, S. & ul Hussain, S. End-to-end trained cnn encoder-decoder networks for image steganography. In Computer Vision—ECCV 2018 Workshops: Munich, Germany, September 8–14, 2018, Proceedings, Part IV 15, 723–729 (Springer, 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
nThe datasets generated and/or analysed during the current study are available from the corresponding authors upon request.