Abstract
Qingyuan partridge chicken (QYM) is a highly regarded native breed in China, highly esteemed for its exceptional breeding characteristics. However, the investigation into the selection signatures and its strains remains largely unexplored. In this study, blood sampling, DNA extracting, and high-depth resequencing were performed in 27 QYMs. Integrating the genomic data of 14 chicken (70 individuals) breeds from other researches, to analyze the genetic structure, selection signatures, and effects of selective breeding within QYM and its 3 strains (QYMA, QYMB, and QYMC). Population structure analysis revealed an independent QYM cluster, which exhibited distinct from other breeds, with each of its 3 strains displaying distinct clustering patterns. Linkage disequilibrium analysis highlighted QYMB's notably slower decay rate, potentially influenced by selection pressure from various production indicators. Examination of selection signatures uncovered genes and genetic mechanisms associated with genomic changes resulting from extensive selective breeding within the QYM and its strains. Intriguingly, diacylglycerol kinase beta (DGKB) and catenin alpha 2 (CTNNA2) were identified as commonly selected genes across the 3 QYM strains, linked to energy metabolism, muscle development, and fat metabolism. Our research validates the substantial impact of selective breeding on QYM and its strains, concurrently identifying genomic regions and signaling pathways associated with their distinctive characters. This research also establishes a fundamental framework for advancing yellow-feathered broiler breeding strategies.
Key words: Qingyuan partridge chicken, whole-genome resequencing, selective breeding, selection signatures, SNP
INTRODUCTION
Livestock production in general and domestic chicken production in particular plays a vital socio economic role for people living in low income countries of Africa and Asia (Mohammadifar et al., 2014; Moazeni et al., 2016a). Domestic chickens are widely distributed avian species around the world, due to their short generation interval and adaptability in a wide range of agro ecologies (Moazeni et al., 2016b; Mohammadifar and Mohammadabadi, 2018; Khabiri et al., 2022). The domestic chickens provide high quality protein and income for the poor rural households and are the most widely kept livestock species in the world (Mohammadabadi et al. 2010; Mohammadifar and Mohammadabadi, 2018). This is due to the presence of the valuable traits of chicken like disease resistance, adaptation to harsh environments and ability to utilize poor quality feeds (Shahdadnejad et al., 2016; Khabiri et al., 2023).
Qingyuan partridge chicken (QYM) is a famous Chinese yellow-feathered broilers breed (Kang et al., 2023). It is known as one of the top 10 breeds in Guangdong Province, China. It is renowned for its exceptional meat quality, appealing taste, and distinctive flavor. In China, consumers demonstrate a preference for poultry meat that possesses both high quality and delectable flavor (Sarsenbek et al., 2013; Du et al., 2017). Additionally, there is a preference for chickens with uniform appearance. Therefore, QYM, which fulfills market demands, holds significant importance in the consumer market for yellow-feathered broilers, is considered an excellent breeding material (Guan et al., 2013). White-feathered broilers exemplify the breed of broilers that exhibit rapid growth, characterized by large body size, efficient feed conversion ratio, and high meat production efficiency. Native chickens are considered to be a prime example of high-quality broilers, with rich meat flavor, dense meat quality, and high nutrient content (Kang et al., 2023; Li et al., 2023). However, it is worth noting that their growth rate and feed conversion rate are comparatively lower when compared to white-feather broilers. In breeding production, the grandparents of broiler supporting lines usually include white-feather broiler breeds with fast growth rate and commercial layer breeds. As a result, the commercial generation mixes most of the genetic components from commercial breeds, and its meat quality and flavor cannot entirely meet the consumer demands. In order to solve this problem, 3 QYM strains were selectively bred over the course of 17 generations. The strains encompass the A strain (QYMA), distinguished by notable traits including substantial weight and rapid growth. B strain (QYMB) exhibits grain-saving properties and unitary balance. C strain (QYMC) exhibits a high level of reproduction and a uniform appearance. These 3 strains can serve as exceptional supporting lines for addressing issues commonly found in traditional supporting lines, such as poor meat quality, unsatisfactory meat flavor, and inconsistent appearance. The focus of this research revolves around the ambiguous impact of artificial domestication and selective breeding on QYM and its strains, compounded by the unexplored genetic information indirectly influenced by artificial selection.
The extensive utilization of whole-genome resequencing (WGRS) technology in livestock and poultry breeding has significantly contributed to delineating genetic relationships and admixture levels among populations. This technology serves as a crucial tool for investigating selectively advantageous genes linked to specific phenotypic traits, thereby establishing a foundational molecular basis essential for understanding distinctive features across diverse breeds (Chen et al., 2021; Huang et al., 2021). Exploration into the genetic composition of various chicken breeds has unveiled instances of genetic introgression among different variants of Xinjiang local chickens (Azimu et al., 2018), with potential analogous events occurring between feral chickens and the wild chickens populations (Cerezo et al., 2023). Additionally, previous research documents genetic exchange between WenChang chickens and breeds from various provinces (Tian et al., 2023). Current research endeavors focus on meticulously exploring and analyzing primary dominant ancestry components inherent in Yellow-feathered chickens across diverse regions (Huang et al., 2020). Considerable research attention has been devoted to the genomic signatures of selective breeding in poultry. The use of whole-genome resequencing has been pivotal in pinpointing specific genetic traits associated with Yeonsan Ogye, particularly those related to immunity and egg development (Cho et al., 2022). Investigations into selective traits of yellow-feathered broilers have sparked significant interest. Reports have detailed the determination of genomic signatures in Wuhua yellow chickens through whole-genome resequencing, revealing genes related to disease resistance, plumage pigmentation, and black feather attributes (Weng et al., 2020). Simultaneously, the exploration of environmental pressures has highlighted Tibetan chickens as a significant subject of investigation. Studies indicate discernible regions under selection within Tibetan chicken genomes, encompassing economically vital traits when facing environmental pressures (Li et al., 2019; Shi et al., 2023). Furthermore, foundational groundwork in the realm of molecular research within waterfowl has been established through an analysis of population structure and selective traits across diverse experimental duck breeds (Li et al., 2023). Moreover, the epigenome comprising different mechanisms for example, DNA methylation, remodeling, histone tail modifications, chromatin microRNAs and long non-coding RNAs, interact with environ-mental factors like nutrition, pathogens, climate to influence the expression profile of genes and the emergence of specific pheno-types (Barazandeh et al. 2016a; Masoudzadeh et al., 2020). Multi-level interactions between the genome, epigenome and environmental factors might occur (Mohamadipoor et al., 2021; Ahmadabadi et al., 2023). Furthermore, numerous lines of evidence suggest the influence of epigenome variation on health and production (Barazandeh et al. 2016b; Bordbar et al., 2022; Safaei et al. 2022). The expression of eukaryotic genes is temporarily and multidimensionally controlled (Shahsavari et al., 2022). Only a relatively small set of the entire genome is expressed in each type of tissue, and the expression of genes depends on the stage of development (Mohammadabadi et al., 2021; Shokri et al., 2023). Therefore, gene expression in eukaryotes is specific to each tissue (Mohammadinejad et al., 2022). Also, the amount of gene products that are made in the same tissue as well as in other tissues that make up that product, regulates the expression of that gene (Mohammadabadi et al., 2023; Shokri et al., 2023). One of the basic activities in domestic animals is the study of genes and proteins related to economic traits and their study at the cellular or chromosomal level (Safaei et al., 2023).
In this study, we conducted an analysis of the population structure and selection signatures within the QYM population, employing high-depth WGRS. Our aim was to systematically evaluate the quality of domestication. Specifically, we sought to scientifically estimate the selective breeding effects evident in 3 distinct strains of QYM and to comprehensively elucidate the genetic mechanisms underlying domestication or selective breeding. The selection signatures of QYM have not been reported, and this research is the first to mine the selection signatures of QYM and evaluate its selective breeding effect. This research contributes to providing a theoretical framework for the breeding of yellow-feathered broilers and facilitates an in-depth genetic perspective on the selective breeding effects observed across the 3 QYM strains.
MATERIALS AND METHODS
Ethics Statement
The animal experiment performed in this study satisfied the requirements of the Institutional Animal Care and Use Committee at the South China Agricultural University (approval ID: 2023F273).
Sample Information
QYM population comprised 27 individuals and was categorized into 3 strains: QYMA (N = 9, female), QYMB (N = 9, female), and QYMC (N = 9, female). All QYM samples were collected from QINGYUAN FENGXIANG MAJI development Co. Ltd, Qingyuan City, Guangdong Province, China. Firstly, blood samples were collected from all QYM individuals using the wing vein puncture method for blood collection. Subsequently, DNA extraction was carried out on all blood samples utilizing a DNA extraction kit. Lastly, DNA concentration and purity were evaluated using a spectrophotometer. 30× whole-genome resequencing was performed on all DNA samples from QYM population, and combined with the previous sequencing data of our research group (Luo et al., 2020), a flock consisting of 15 chicken breeds (N = 97) was generated. The flock consisted of 2 commercial breeds (5 White Leghorn chickens [LH] and 5 White Recessive Rock chickens [WRR]), 1 ancestral breed (5 red jungle fowl [RJF]), and 12 Chinese indigenous breeds (27 Qingyuan Partridge chickens [QYM], 5 Huiyang Bearded chickens [BC], 5 Beijing You chickens [BJY], 5 Jining Bairi chickens [BR], 5 Luxi Gamecock chickens [LX], 5 Liyang chickens [LY], 5 Silkies [S], 5 Tulufan Gamecock chickens [TLF], 5 Wannan Yellow chickens [WN], 5 Xinghua chickens [X], 5 Huanglang chickens [XH], and 5 Yunyang Da chickens [YY]). Detailed information is available in Table S1. Libraries for each QYM individual were constructed (Paired end, 2 × 150 bp) using the DNBSEQ-T7 platform (Hunan, China).
Quality Control and Mapping
FASTP (v0.20.1) was used to filter the data in the following aspects: 1) ead pairs with a quality value (Q) of ≤20 bases, accounting for more than 50% of the total bases, were removed, 2) the adapter fragment in reads were removed, 3) reads containing an excessive number of N-bases were filtered, with those containing more than 5 Ns were removed, 4) reads with a length less than 100 were filtered. Clean Reads after preliminary quality control were mapped to the reference genome GRCg7b (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_016699485.2/) using the Burrows-Wheeler Aligner (v0.7.17) genome alignment software.
Variant Identification
Based on the BAM file after mapping, Genome Analysis Toolkit (GATK) version 4.1.7 is used to identify and call all single nucleotide polymorphisms (SNP) and insertions/deletions (INDEL) across the entire genome of each individual sample. This process resulted in the GVCF file for each sample. The GATK tool CombineGVCFs was utilized to merge the GVCF of all individual samples. All the merged variants were filtered in GATK using the following criteria: “QD < 2.0 || FS > 60.0 || MQ < 40.0 || SOR > 3.0 || MQRankSum < -12.5 || The standard ReadPosRankSum < -8.0”. To obtain high-quality SNPs, the SNPs should be provided to VCFtools, according to 1) max-missing 0.1, 2) -maf 0.05 as the standard. Finally, autosomal SNPs were extracted for subsequent analysis.
Population Structure and Linkage Disequilibrium Analysis
The genetic distance was calculated using PLINK1.9, and the phylogenetic tree was constructed by neighbor-joining method, which was visualized using iTOL (https://itol.embl.de/). Linkage disequilibrium-based pruning of autosomal SNPs in chickens was conducted using PLINK 1.9. The pruned SNPs were used for principal component analysis (PCA) and genetic composition analysis. Admixture v1.3.0 (Alexander et al., 2009) is used for genetic composition analysis (ranging from K = 2 to K = 10). Utilize the R to visualize the results. Linkage disequilibrium (LD) decay was computed for each breed using PopLDdecay software (Zhang et al., 2019).
Selective Sweep and Enrichment Analysis
Fixation index (FST) and nucleotide diversity (π) were used to identify the selected genomic regions of QYM population and different QYM strains. VCFtools (v0.1.15) was used to calculate the FST and π of the window (window size is 20 Kb and the step size is 10 Kb). In addition, to ensure the precision of detection, Windows with a SNP count of less than 5 were removed. The π value of the selected window is represented by the logarithm base 2 of the π ratio. The selected regions are determined by the intersection window of the top 5% of the FST value and the log2 (π ratio) value. Finally, the functional enrichment analysis of genes in the candidate regions was conducted using Gene Ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
RESULTS
WGRS and Descriptive Statistics
The DNA extraction from all blood samples was successful, yielding DNA samples with an average volume of 60 μL, a mean concentration of 121.90 ng/μL, and an average DNA quantity of 7.31 μL (Table S2).
After the WGRS data quality control on all QYM individuals, the average data size for each individual was 36.49Gb. The average Q30 was 95.26%. The average mapped reads rate was 99.81%. The average Map Depth was 34.01. Lastly, the average coverage rate was 96.83% (Table S3).
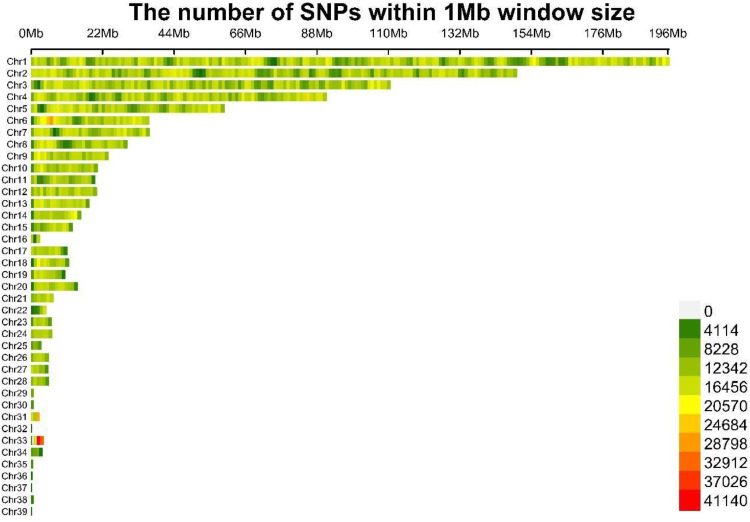
After filtering the population quality control data for QYM, a total of 13,094,506 high-quality SNPs were identified. The distribution of SNPs on 39 autosomes was depicted in Figure 1. It was evident from the figure that SNPs exhibited a uniform distribution across the chromosomes.
Figure 1.
Plots of autosomal SNP density in QYM flock.
Population Structure and Linkage Disequilibrium Analysis
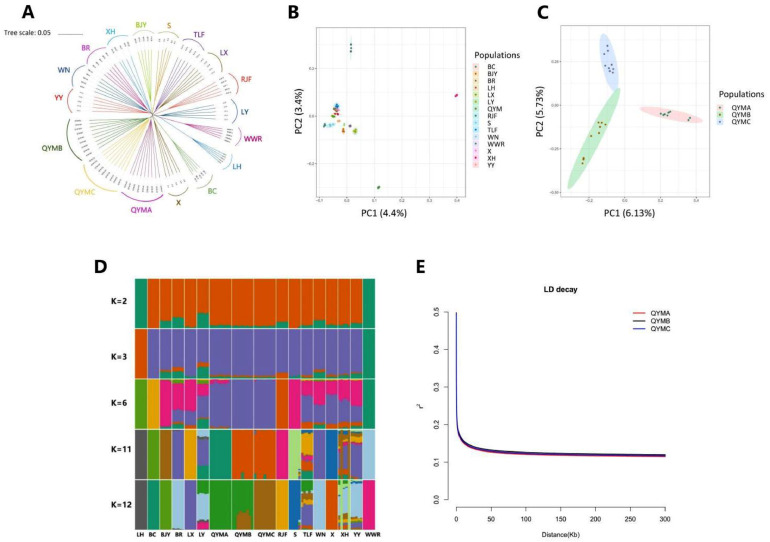
In order to evaluate the genetic relationship between QYM and other breeds, a neighbor-joining tree was constructed by autosomal variation including 13,094,506 SNPs. Principal component analysis and admixture analysis were performed using the 1,341,880 SNPs obtained after LD-based pruned.
The neighbor-joining tree analysis showed that the group clustering aligned with the breed information, and there were no instances of mixed individuals (Figure 2A). The effects of grouping based on variety are clearly significant, and the phylogenetic relationship is evident. Qingyuan Partridge chickens population can be classified as a distinct category, with the closest genetic distance to the X population, followed by BC population. Leghorn chickens and WWR are representative of commercial breeds that initially formed 1 clade and later developed into 2 separate clades. It is noteworthy that QYM can form a clade. The neighbor-joining tree shows that it forms 3 clades at similar genetic distances, corresponding to the 3 strains in the QYM flock: QYMA, QYMB, and QYMC.
Figure 2.
Genetic background analysis. (A) Neighbor-joining tree constructed using genetic distances between individuals. (B) PCA results of all chickens. (C) PCA results of 3 strains from QYM population. (D) Genetic structure of 15 flocks by ADMIXTURE program. (E) LD decay curves for 3 strains from QYM population.
The PCA result was consistent with the neighbor-joining tree. Principal component 1 (PC1, accounting for 4.4% of the total variation) and principal component 2 (PC2, accounting for 3.4% of the total variation) clearly demonstrate the clustering of all individuals (Figure 2B). All samples can be divided into 4 clusters: 2 commercial breeds (LH and WWR), one representative of an evolutionary ancestor (RJF), and a Chinese native population (12 flocks including QYM). Among the populations, QYM was distinct from the others, except for its close relationship with X. Principal component analysis results within QYM population revealed that PC1 (6.13% of the total variation) and PC2 (5.73% of the total variation) showed the genetic relationships among the 3 QYM strains. QYMA, QYMB, and QYMC can be visibly divided into 3 clusters based on PC1 and PC2. The results of all PCA are consistent with the results of the neighbor-joining tree (Figure 2C).
To assess the degree of genetic confounding in QYM population, an admixture analysis was conducted (Figure 2D and Figure S2). When K = 2, the cross-validation error is minimum (Figure S1), 2 commercial populations have different genetic components compared to other populations, and there is a significant separation between commercial breeds and native breeds. It is evident that the genetic composition of the QYM population had a minimal contribution from the genetic components of commercial breeds, and there was no significant introgression. When K = 3, LH and WWR, and subsequently analyze the mixing proportion of 2 different commercial varieties. At K = 6, the QYMB strain was delineated. At K = 11, the QYMA strain formed a distinct cluster. Similarly, at K = 12, the QYMC strain emerged as a separate group. The admixture analysis is consistent with the results of the neighbor-joining tree and principal component analysis.
Linkage disequilibrium decay plots were generated for all breeds based on 13,094,506 SNPs (Figure S3). The results showed that there were differences in decay rate between different breeds. The decay rate of BC was observed to be low, indicating a significant level of LD among SNPs of BC, followed by 2 commercial breeds, WWR and LH. In addition to QYM, LD decay rate was found to be similar among the other 10 Chinese native breeds. LD decay rate was found to be the fastest in QYM population that underwent high-depth sequencing. The LD decay rate of the 3 QYM strains was almost the same, with slight discrepancies observed in the decay rate (Figure 2E). QYMB shows a greater level of LD and a slower rate of decay compared to QYMC and QYMA. QYMA has the lowest level of LD and the fastest decay rate.
Genome-Wide Selective Sweeps and Enrichment Analyses
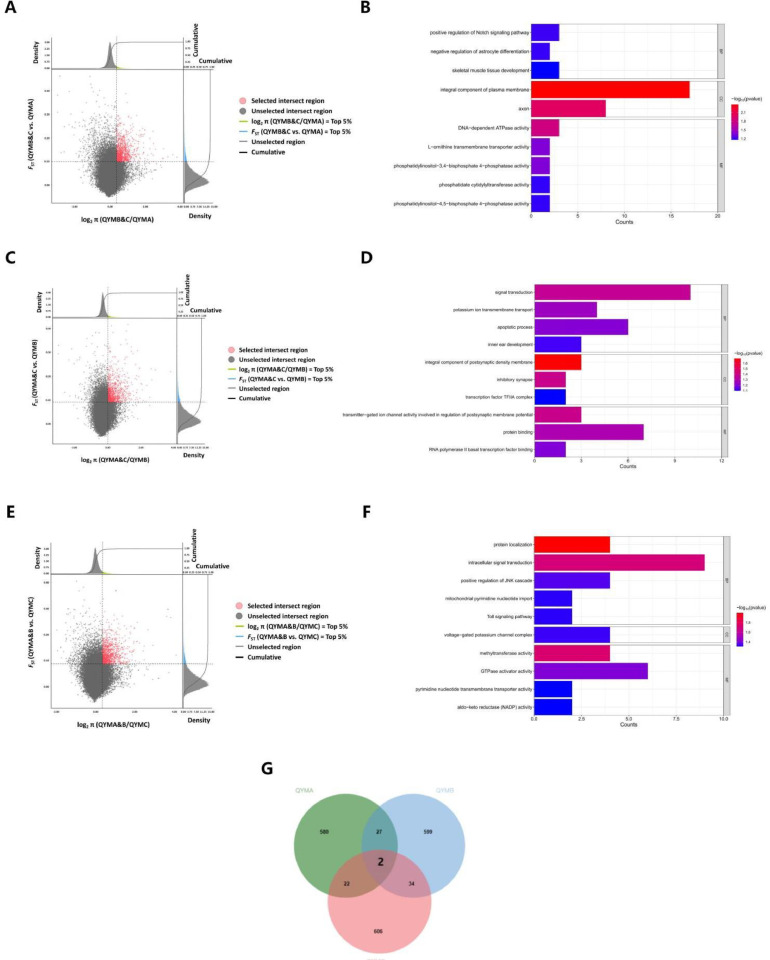
Selective Sweep and Enrichment Analysis of QYM Population. In order to explore the selection signatures of QYM, 14 breeds other than QYM were used as the background pool, and the selected regions of the QYM genome were identified by integrating FST and π ratio. The threshold for screening significant regions as potential selected regions is determined by taking the top 5% of the results of FST, log2 (π ratio). The selection criteria for the 2 methods were defined as FST ≥ 0.0626 and log2 (π ratio) ≥ 0.2780. By combining the 2 methods for selective sweep analysis (Figure 3A, Table S4), 1,215 selected regions were identified in QYM population. Chromosome 1 had the most selected regions, with a total of 381 regions. Following closely, chromosome 2 displayed the second most selected regions, with a count of 190 regions. The chromosome with the third highest number of regions is chromosome 3 (173 regions) (Figure S4A). A total of 685 genes were identified and annotated from the selected regions obtained by selective sweep analysis. Functional enrichment analysis was performed on the genes within the selected regions of QYM, Gene ontology analysis revealed enrichment in terms associated with lipid and fat metabolism. For example, steroid binding (GO:0005496) and the positive regulation of fatty acid metabolic process (GO:0045923) (Figure 3B, Table S5). Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis enriched oocyte meiosis, regulation of actin cytoskeleton, progesterone mediated oocyte maturation, and other signaling pathways are associated with cytoskeleton and reproduction (Figure 3C).
Figure 3.
Identification of selection signatures in the QYM population. (A) Genome-wide selective sweep analysis of QYM flock. The horizontal coordinate represents the log2 (π ratio) values of the other 14 populations compared to the QYM population, corresponding to the cumulative density plot above. The vertical coordinate represents the FST of the other 14 populations versus the QYM population, corresponding to the cumulative density plot on the right. (B) GO enrichment analysis for genes annotated within selected regions in the whole genome of QYM population (Top 10). (C) KEGG pathway enrichment analysis for genes annotated within selected regions in the whole genome of QYM population.
Selective Sweep and Enrichment Analysis of 3 Strains From QYM Population
In order to explore the selection signatures of the 3 QYM strains, selective sweep analyses were performed on each strain using a consistent identification method to detect the selected regions of QYMA, QYMB, and QYMC. FST and log2 (π ratio) values were computed for 3 combined forms: 1) QYMB&C vs. QYMA, 2) QYMA&C vs. QYMB, 3) QYMA&B vs. QYMC. The candidate region for selected genes of the 3 strains was determined by taking the intersection of the top 5% regions of FST and the top 5% regions of log2 (π ratio).
1) QYMB&C vs. QYMA. 1042 selected regions were identified in QYMA strain (Figure 4A, Table S6). Among these regions, chromosome 1 had the highest number of selected regions (281), followed by chromosome 4 (172) and chromosome 2 (166). These selected regions contain a total of 632 genes (Figure S4B). Functional enrichment analysis was performed on the genes within the QYMA selected regions, and subsequent GO and KEGG enrichment analysis revealed pathways associated with skeletal muscle metabolism. For example, skeletal muscle tissue development (GO:0007519), phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity (GO:0016316), phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity (GO:0034597), phosphatidylinositol signaling system signal pathways. Related to lipid metabolism is phosphatidate cytidylyltransferase activity (GO:0004605). DNA-dependent ATPase activity (GO:0008094) is related to energy production. negative regulation of intracellular estrogen receptor signaling pathway (GO:0033147) is found to be associated with estrogen (Figure 4B, Table 1, Table S7).
Figure 4.
Identification of selection signatures in the 3 strains from QYM population. (A) Genome-wide selective analysis sweep of QYMA strain. The horizontal coordinate represents the log2 (π ratio) values of the QYMB and QYMC compared to the QYMA, corresponding to the cumulative density plot above. The vertical coordinate represents the FST of the QYMB and QYMC compared to the QYMA, corresponding to the cumulative density plot on the right. (B) GO enrichment analysis for genes annotated within selected regions in the whole genome of QYMA strain (Top 10). (C) Genome-wide selective sweep analysis of QYMB strain. The horizontal coordinate represents the log2 (π ratio) values of the QYMA and QYMC compared to the QYMB, corresponding to the cumulative density plot above. The vertical coordinate represents the FST of the QYMA and QYMC compared to the QYMB, corresponding to the cumulative density plot on the right. (D) GO enrichment analysis for genes annotated within selected regions in the whole genome of QYMB strain (Top 10). (E) Genome-wide selective sweep analysis of QYMC strain. The horizontal coordinate represents the log2 (π ratio) values of the QYMA and QYMB compared to the QYMC, corresponding to the cumulative density plot above. The vertical coordinate represents the FST of the QYMA and QYMB compared to the QYMC, corresponding to the cumulative density plot on the right. (F) GO enrichment analysis for genes annotated within selected regions in the whole genome of QYMC strain (Top 10). (G) GO enrichment analysis for genes annotated within selected regions in the whole genome of QYMC strain (Top 10).
Table 1.
KEGG pathway enrichment analysis for genes annotated within selected regions in the whole genome of QYMA strain.
| Pathway | Gene count | Gene ID of ensemble |
|---|---|---|
| Phosphatidylinositol signaling system | 6 | ENSGALG00010020362, ENSGALG00010009381, ENSGALG00010029209, ENSGALG00010013617, ENSGALG00010015669, ENSGALG00010004094 |
| Notch signaling pathway | 4 | ENSGALG00010020053, ENSGALG00010019700, ENSGALG00010019724 |
2) QYMA&C vs. QYMB. Results of selective sweep analysis showed that QYMB had 1036 selected regions (Figure 4C, Table S8), and the top 3 chromosomes with the most selected regions were chromosome 1 (289 regions). Chromosome 2 (178 regions) and chromosome 3 (122 regions) (Figure S4C). A total of 663 genes were annotated in all selected regions. Functional enrichment analysis of genes within the QYMB selection region was performed. These candidate genes have been identified as being associated with various organism development processes such as protein binding (GO:0005515), apoptotic process (GO:0006915), and metabolic pathways (Figure 4D, Table 2, Table S9).
Table 2.
KEGG pathway enrichment analysis for genes annotated within selected regions in the whole genome of QYMB strain.
| Pathway | Gene count | Gene ID of ensemble |
|---|---|---|
| Adherens junction | 8 | ENSGALG00010002706, ENSGALG00010024013, ENSGALG00010005596, ENSGALG00010005035, ENSGALG00010001686, ENSGALG00010016779, ENSGALG00010014030, ENSGALG00010013604 |
| Mucin type O-glycan biosynthesis | 4 | ENSGALG00010022247, ENSGALG00010009777, ENSGALG00010029378, ENSGALG00010024725 |
| Metabolic pathways | 38 | ENSGALG00010022796, ENSGALG00010019982, ENSGALG00010020311, ENSGALG00010024338, ENSGALG00010020275, ENSGALG00010015663, ENSGALG00010007439, ENSGALG00010006922, ENSGALG00010005437, ENSGALG00010005610, ENSGALG00010021805, ENSGALG00010007436, ENSGALG00010026839, ENSGALG00010003933, ENSGALG00010005395, ENSGALG00010009777, ENSGALG00010012650, ENSGALG00010007432, ENSGALG00010024251, ENSGALG00010024011, ENSGALG00010024132, ENSGALG00010010953, ENSGALG00010023237, ENSGALG00010022247, ENSGALG00010000797, ENSGALG00010020346, ENSGALG00010001728, ENSGALG00010009091, ENSGALG00010000792, ENSGALG00010024785, ENSGALG00010024725, ENSGALG00010001820, ENSGALG00010013019, ENSGALG00010004372, ENSGALG00010029378, ENSGALG00010025230, ENSGALG00010004094, ENSGALG00010021390 |
3) QYMA&B vs. QYMC. QYMC has 1064 selected regions (Figure 4E, Table S10), among which chromosome 2 has the most selected regions (259 regions). The second is chromosome 1, with 249 regions. The third is chromosome 4, with 122 regions (Figure S4D). All of the selected regions were annotated to a total of 665 genes. Functional enrichment analysis was performed on the genes within the selected regions of QYMC, which were enriched to such as negative regulation of Wnt signaling pathway (GO:0030178), positive regulation of JNK cascade (GO:0046330), methyltransferase activity (GO:0008168), mitochondrial pyrimidine nucleotide import (GO:1990519) and other terms related to growth and development, pigmentation, epigenetic regulation, and energy metabolism (Figure 4F, Table S11).
Combining the results of 3 QYM strains, it was found that QYMA exhibited the lowest number of annotated genes in the specified region. Additionally, there was minimal variation in the number of selected genes between QYMB and QYMC. Two specific genes, namely diacylglycerol kinase beta (DGKB) and catenin alpha 2 (CTNNA2), were selected in 3 strains of QYM population (Figure 4G).
DISCUSSION
Qingyuan Partridge chickens is a renowned native chicken breed in China and is considered a valuable genetic resource for poultry breeding. The QYM has been selectively bred to cultivate strains that fulfill particular production requirements, with an emphasis on promoting growth and development, unitary balance, fertility while preserving their inherent advantages. Whole-genome resequencing technology relies on the utilization of a known reference genome specific to a particular species. This technological advancement involves the comprehensive sequencing of the complete genome of various individuals within a particular species, thereby facilitating the identification and analysis of genetic variations among different individuals or populations (Ley et al., 2008; Daetwyler et al., 2014). The study conducted whole-genome resequencing on all individuals of QYM and performed population structure analysis and LD level analysis to evaluate the breeding effects of the QYM breed and its strains from a genomic perspective. Furthermore, genome-wide selective sweeps and enrichment analyses were conducted on the QYM breed and its three strains to gain a deeper understanding of the genetic mechanisms involved in targeted breeding. These analyses provide insights from the perspective of life sciences.
Principal component analysis (Wen et al., 2011) and phylogenetic tree are effective tools for depicting the genetic evolutionary relationships among populations, while admixture analysis provides insights into the genetic composition of individual organisms (Chimusa et al., 2020; Hollox et al., 2022; Shriner, 2023). In this study, we discovered that the QYM population exhibits significant genetic differences from other breeds due to long-term domestication. The three strains within the QYM population exhibit notable distinctions. Additionally, we found that the QYM population is genetically closer to the X population, which can be attributed to both populations are believed to have originated from the same province. Native chicken breeds utilized by chicken farming enterprises for production contribute to the close genetic relationship between the QYM and X populations. The findings from the admixture analysis suggest that the QYM population has successfully preserved the purity of its strains through artificial selection, effectively preserving the breed's advantages. The QYM population has a limited degree of genetic admixture from commercial breeds, which can be attributed to the historical lack of conserving local breeds during the initial phases of the poultry industry. As a result, commercial breed genetics have infiltrated local breeds extensively. Therefore, it is crucial to strengthen conservation efforts for the QYM population in the future.
Linkage disequilibrium is a phenomenon characterized by the decay of LD due to separation and recombination between loci. This decay is most readily observed in loci that are closely linked, a phenomenon commonly referred to as “linkage disequilibrium” (Baird, 2015; Kemppainen et al., 2015). In the context of long-term adaptive evolution, the genome experiences changes in the LD of structural features (Larson et al., 2014; Qanbari et al., 2014). The level of LD in QYM is significantly lower compared to other breeds. The difference can be attributed to the variation in sequencing depth between QYM samples and samples from other breeds. QYM samples were sequenced at a depth of 30×, while the sequencing depth for other breeds was only 10×. This discrepancy in sequencing depth may have contributed to the observed disparity in LD levels. Sequencing depth refers to the proportion of the total number of bases acquired through sequencing in relation to the size of the genome, or the average frequency at which each base is detected in the genome (Sims et al., 2014). The factor in question holds significant importance as it greatly influences the process of whole-genome resequencing. A greater sequencing depth has been shown to enhance the identification of novel variants and enhance the coverage of the genome (Jiang et al., 2019). Therefore, the QYM method offers a more extensive coverage and detects a higher number of SNPs, resulting in a reduced level of linkage. Additionally, the QYM population has a significantly larger sample size compared to other breeds. These unforeseen errors have led to an imprecise evaluation of the variations in LD levels between QYM and other breeds. Among the three QYM strains with equal sequencing depth and sample size, QYMB exhibits the highest level of LD. This observation implies that the B strain has experienced the most pronounced selection pressure during directional breeding, potentially attributable to the concurrent selection for traits such as feed efficiency, reproduction, and growth. The available evidence strongly suggests a positive breeding effect of QYM and its 3 strains.
FST and π are well-established and dependable techniques utilized for the analysis of genome-wide selective sweeps (Sun et al., 2020; Li et al., 2023; Xue et al., 2023). Qingyuan Partridge chicken contains the selected gene SMURF2, which plays a significant role in the regulation of cell growth, differentiation, and development. Importantly, it plays a crucial role in the hypertrophy, differentiation, and development of skeletal muscle in chickens (Cai et al., 2021). The selected genes MYL12A and MYL12B are members of the myosin regulatory light chain family and are important components of the cellular cytoskeleton (Takashima, 2009). RPS6KA2 has been identified as another significant factor in the ovulatory cascade (Akin et al., 2021). The selected gene PPARD plays a significant role in regulating expression and influencing adipocyte differentiation in the fat metabolism pathway (Shi et al., 2013). SNPs in the 5′ regulatory region of PPARD are strongly correlated with fat deposition traits in a local Chinese pig population (Zhang et al., 2015). Some researchers have additionally proposed that PPARD interacts with lncRNA, playing a role in regulating lipid metabolism, adipocyte proliferation, and differentiation in the abdominal fat of Gushi Chickens (Zhai et al., 2021). The selection signatures of QYM breed, including muscle development, fat metabolism, and reproductive performance, can be due to consumer demand for tender and flavorful yellow-feathered chicken meat. Additionally, poultry producers selectively breed hens with superior egg-laying abilities, resulting in the unique and high-performing QYM breed through long-term artificial selection.
The selection signatures of the three QYM strains indicates that the breeding goal of QYMA, characterized by its large weight and rapid growth, is achieved through the regulation of skeletal muscle development, energy production and lipid metabolism. The breeding goal of QYMB is to achieve decreased feed intake and balancing various indicators by regulating apoptosis and metabolism. The breeding goal of QYMC is characterized by excellent reproductive ability and stable appearance, is achieved through the regulation of methylation processes and pigment deposition pathways.In the case of the QYMC strain, DNA methylation has been reported to play a crucial role and mechanism in oocyte cells (Sendzikaite et al., 2019). Betaine, an essential methyl donor, can enhance egg production in laying hens (Omer et al., 2018). Betaine can impact liver lipid synthesis and modify the methylation status of transport-related genes and their promoters, thereby affecting their expression and improving the egg-laying rate in hens (Omer et al., 2020). Methylation has been found to be associated with chicken embryonic gonadal sex differentiation (Li et al., 2022). m6A may have a significant role in regulating chicken ovarian development. It has been reported that a decrease in H3K4 methylation levels can reduce transcriptional activity and increase DNA methylation in oocyte cells (Jia et al., 2023), thereby interfering with their developmental potential and female fertility capacity (Mei et al., 2023). We observed that candidate genes in the QYMC strain are enriched in the Wnt pathway and JNK pathway, which are important pathways regulating melanin production (Liu et al., 2002; Yun et al., 2019). Effective regulation of melanin production can be achieved by using Wnt or JNK inhibitors or activators (Takeda et al., 2000; Kim et al., 2020).
It is noteworthy that DGKB and CTNNA2 are 2 genes that have been collectively selected in the directed breeding of the QYM 3 strains. There is evidence indicating that DGKB significantly influences glucose metabolism (Renstrom et al., 2011; van de Bunt et al., 2015) and has also been found to be associated with fat deposition (Xing et al., 2019). Recent research has indicated that DGKB has been identified through GWAS as potentially related to bone and muscle development in Chinese gamecocks populations (Ren et al., 2023). Regarding CTNNA2, it has been reported to be associated with milk fat production in cows (Laodiim et al., 2023) and is also implicated in lipid metabolism (Schooling, 2016). CTNNA2 emerges as the highest hub gene in the interaction network and makes a significant contribution to muscle development (Malik et al., 2015).
CONCLUSIONS
In summary, the genomic characteristics of chicken breeds and strains have been shaped by artificial selection during their formation. We conducted breeding effect and genetic characteristics of QYM and its 3 strains based on SNPs. The QYM population is clearly distinct from other breeds, with its 3 strains categorized into 3 distinct branches. Selective genes associated with energy metabolism, muscle development, fat metabolism, and pigment synthesis pathways were identified in QYM and its 3 strains. DGKB and CTNNA2 were identified as commonly selected genes in all 3 strains, and were annotated to be involved in muscle development and fat deposition.
ACKNOWLEDGMENTS
This work was supported by STI2030—Major Projects (2023ZD04064).
DISCLOSURES
The authors declare no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2024.103626.
Appendix. Supplementary materials
REFERENCES
- Akin N., AH L.e., Ha U., Romero S., Sanchez F., Pham T.D., Nguyen M., Anckaert E., Ho T.M., Smitz J., Vuong L.N. Positive effects of amphiregulin on human oocyte maturation and its molecular drivers in patients with polycystic ovary syndrome. Hum. Reprod. 2021;37:30–43. doi: 10.1093/humrep/deab237. [DOI] [PubMed] [Google Scholar]
- Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimu W., Manatbay B., Li Y., Kaimaerdan D., Wang H.E., Reheman A., Muhatai G. Genetic diversity and population structure analysis of eight local chicken breeds of Southern Xinjiang. Br. Poult. Sci. 2018;59:629–635. doi: 10.1080/00071668.2018.1523537. [DOI] [PubMed] [Google Scholar]
- Baird S.J. Exploring linkage disequilibrium. Mol. Ecol. Resour. 2015;15:1017–1019. doi: 10.1111/1755-0998.12424. [DOI] [PubMed] [Google Scholar]
- Barazandeh A., Mohammadabadi M.R., Ghaderi-Zefrehei M., Nezamabadipour H. Predicting CpG islands and their relationship with genomic feature in cattle by hidden markov model algorithm. Iranian J. Appl. Anim. Sci. 2016;6:571–579. [Google Scholar]
- Barazandeh A., Mohammadabadi M.R., Ghaderi-Zefrehei M., Nezamabadipour H. Genome-wide analysis of CpG islands in some livestock genomes and their relationship with genomic features. Czech J. Anim. Sci. 2016;61:487. [Google Scholar]
- Bordbar F., Mohammadabadi M., Jensen J., Xu L., Li J., Zhang L. Identification of candidate genes regulating carcass depth and hind leg circumference in simmental beef cattle using Illumina Bovine Beadchip and next-generation sequencing. Animals. 2022;12:1103. doi: 10.3390/ani12091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B., Li Z., Ma M., Zhang J., Kong S., Abdalla B.A., Xu H., Jebessa E., Zhang X., Lawal R.A., Nie Q. Long noncoding RNA SMUL suppresses SMURF2 production-mediated muscle atrophy via nonsense-mediated mRNA decay. Mol. Ther. Nucleic. Acids. 2021;23:512–526. doi: 10.1016/j.omtn.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo M.L.M., Lopez S., van Dorp L., Hellenthal G., Johnsson M., Gering E., Henriksen R., Wright D. Population structure and hybridisation in a population of Hawaiian feral chickens. Heredity (Edinb) 2023;130:154–162. doi: 10.1038/s41437-022-00589-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Huang Y., Wang Z., Teng S., Hanif Q., Lei C., Sun J. Whole-genome resequencing reveals diversity and selective signals in Longlin goat. Gene. 2021;771 doi: 10.1016/j.gene.2020.145371. [DOI] [PubMed] [Google Scholar]
- Chimusa E.R., Defo J., Thami P.K., Awany D., Mulisa D.D., Allali I., Ghazal H., Moussa A., Mazandu G.K. Dating admixture events is unsolved problem in multi-way admixed populations. Brief. Bioinform. 2020;21:144–155. doi: 10.1093/bib/bby112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y., Kim J.Y., Kim N. Comparative genomics and selection analysis of Yeonsan Ogye black chicken with whole-genome sequencing. Genomics. 2022;114 doi: 10.1016/j.ygeno.2022.110298. [DOI] [PubMed] [Google Scholar]
- Daetwyler H.D., Capitan A., Pausch H., Stothard P., van Binsbergen R., Brondum R.F., Liao X., Djari A., Rodriguez S.C., Grohs C., Esquerre D., Bouchez O., Rossignol M.N., Klopp C., Rocha D., Fritz S., Eggen A., Bowman P.J., Coote D., Chamberlain A.J., Anderson C., VanTassell C.P., Hulsegge I., Goddard M.E., Guldbrandtsen B., Lund M.S., Veerkamp R.F., Boichard D.A., Fries R., Hayes B.J. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 2014;46:858–865. doi: 10.1038/ng.3034. [DOI] [PubMed] [Google Scholar]
- Du Y.F., Ding Q.L., Li Y.M., Fang W.R. Identification of differentially expressed genes and pathways for myofiber characteristics in soleus muscles between chicken breeds differing in meat quality. Anim. Biotechnol. 2017;28:83–93. doi: 10.1080/10495398.2016.1206555. [DOI] [PubMed] [Google Scholar]
- Guan R., Lyu F., Chen X., Ma J., Jiang H., Xiao C. Meat quality traits of four Chinese indigenous chicken breeds and one commercial broiler stock. J. Zhejiang. Univ. Sci. B. 2013;14:896–902. doi: 10.1631/jzus.B1300163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollox E.J., Zuccherato L.W., Tucci S. Genome structural variation in human evolution. Trends. Genet. 2022;38:45–58. doi: 10.1016/j.tig.2021.06.015. [DOI] [PubMed] [Google Scholar]
- Huang M., Zhang H., Wu Z.P., Wang X.P., Li D.S., Liu S.J., Zheng S.M., Yang L.J., Liu B.B., Li G.X., Jiang Y.C., Chen H., Ren J. Whole-genome resequencing reveals genetic structure and introgression in Pudong White pigs. Animal. 2021;15 doi: 10.1016/j.animal.2021.100354. [DOI] [PubMed] [Google Scholar]
- Huang X., Otecko N.O., Peng M., Weng Z., Li W., Chen J., Zhong M., Zhong F., Jin S., Geng Z., Luo W., He D., Ma C., Han J., Ommeh S.C., Zhang Y., Zhang X., Du B. Genome-wide genetic structure and selection signatures for color in 10 traditional Chinese yellow-feathered chicken breeds. BMC. Genomics. 2020;21:316. doi: 10.1186/s12864-020-6736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari Ahmadabadi S.A.A., Askari-Hemmat H., Mohammadabadi M., Asadi M., Mansouri M. The effect of Cannabis seed on DLK1 gene expression in heart tissue of Kermani lambs. Agric. Biotechnol. J. 2023;15:217–234. [Google Scholar]
- Jia C., Zhang M., Liu X., Xu W., Xiong Y., Huang R., Li M., Li M. Transcriptome-wide m6A methylation profiling of Wuhua yellow-feathered chicken ovary revealed regulatory pathways underlying sexual maturation and low egg-laying performance. Front. Genet. 2023;14 doi: 10.3389/fgene.2023.1284554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Jiang Y., Wang S., Zhang Q., Ding X. Optimal sequencing depth design for whole genome re-sequencing in pigs. BMC. Bioinformatics. 2019;20:556. doi: 10.1186/s12859-019-3164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Ren M., Li S., Lu Y., Deng X., Zhang Z., Gan J., Wei J., Hua G., Yu H., Li H. Estimation of genetic parameters for important traits using a multi-trait model in late-feathering Qingyuan partridge hens in China. J. Anim. Breed. Genet. 2023;140:158–166. doi: 10.1111/jbg.12739. [DOI] [PubMed] [Google Scholar]
- Kemppainen P., Knight C.G., Sarma D.K., Hlaing T., Prakash A., Maung M.Y., Somboon P., Mahanta J., Walton C. Linkage disequilibrium network analysis (LDna) gives a global view of chromosomal inversions, local adaptation and geographic structure. Mol. Ecol. Resour. 2015;15:1031–1045. doi: 10.1111/1755-0998.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabiri A., Toroghi R., Mohammadabadi M., Tabatabaeizadeh S. Cloning and nucleotide sequencing of the complete matrix protein of Newcastle disease virus subgenotype VII. 1.1 prevalence in broiler flocks of northeastern Iran. Modern Genetic J. 2022;17:113–125. [Google Scholar]
- Khabiri A., Toroghi R., Mohammadabadi M., Tabatabaeizadeh S.E. Introduction of a Newcastle disease virus challenge strain (sub-genotype VII. 1.1) isolated in Iran. Vet. Res. Forum. 2023;14:221. doi: 10.30466/vrf.2022.548152.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Hong A.R., Kim Y.H., Yoo H., Kang S.W., Chang S.E., Song Y. JNK suppresses melanogenesis by interfering with CREB-regulated transcription coactivator 3-dependent MITF expression. Theranostics. 2020;10:4017–4029. doi: 10.7150/thno.41502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laodiim T., Koonawootrittriron S., Elzo M.A., Suwanasopee T., Jattawa D., Sarakul M. Genetic factors influencing milk and fat yields in tropically adapted dairy cattle: insights from QTL analysis and gene associations. Anim. Biosci. 2023;37:576–590. doi: 10.5713/ab.23.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G., Piperno D.R., Allaby R.G., Purugganan M.D., Andersson L., Arroyo-Kalin M., Barton L., Climer V.C., Denham T., Dobney K., Doust A.N., Gepts P., Gilbert M.T., Gremillion K.J., Lucas L., Lukens L., Marshall F.B., Olsen K.M., Pires J.C., Richerson P.J., Rubio D.C.R., Sanjur O.I., Thomas M.G., Fuller D.Q. Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6139–6146. doi: 10.1073/pnas.1323964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley T.J., Mardis E.R., Ding L., Fulton B., McLellan M.D., Chen K., Dooling D., Dunford-Shore B.H., McGrath S., Hickenbotham M., Cook L., Abbott R., Larson D.E., Koboldt D.C., Pohl C., Smith S., Hawkins A., Abbott S., Locke D., Hillier L.W., Miner T., Fulton L., Magrini V., Wylie T., Glasscock J., Conyers J., Sander N., Shi X., Osborne J.R., Minx P., Gordon D., Chinwalla A., Zhao Y., Ries R.E., Payton J.E., Westervelt P., Tomasson M.H., Watson M., Baty J., Ivanovich J., Heath S., Shannon W.D., Nagarajan R., Walter M.J., Link D.C., Graubert T.A., DiPersio J.F., Wilson R.K. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Li Y., Li M., Che T., Tian S., Chen B., Zhou X., Zhang G., Gaur U., Luo M., Tian K., He M., He S., Xu Z., Jin L., Tang Q., Dai Y., Xu H., Hu Y., Zhao X., Yin H., Wang Y., Zhou R., Yang C., Du H., Jiang X., Zhu Q., Li M. Population genomics identifies patterns of genetic diversity and selection in chicken. BMC. Genomics. 2019;20:263. doi: 10.1186/s12864-019-5622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang X., Wang X., Sun C., Zheng J., Li J., Yi G., Yang N. The m6A methylation regulates gonadal sex differentiation in chicken embryo. J. Anim. Sci. Biotechnol. 2022;13:52. doi: 10.1186/s40104-022-00710-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Mei H., Liu Y., Li Z., Qamar H., Yu M., Ma X. Dietary supplementation with rutin alters meat quality, fatty acid profile, antioxidant capacity, and expression levels of genes associated with lipid metabolism in breast muscle of Qingyuan partridge chickens. Foods. 2023;12:2302. doi: 10.3390/foods12122302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li Y., Semenov M., Han C., Baeg G.H., Tan Y., Zhang Z., Lin X., He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- Luo W., Luo C., Wang M., Guo L., Chen X., Li Z., Zheng M., Folaniyi B.S., Luo W., Shu D., Song L., Fang M., Zhang X., Qu H., Nie Q. Genome diversity of Chinese indigenous chicken and the selective signatures in Chinese gamecock chicken. Sci. Rep. 2020;10:14532. doi: 10.1038/s41598-020-71421-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A., Lee E.J., Jan A.T., Ahmad S., Cho K.H., Kim J., Choi I. Network analysis for the identification of differentially expressed hub genes using Myogenin Knock-down muscle satellite cells. PLoS. One. 2015;10 doi: 10.1371/journal.pone.0133597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoudzadeh S.H., Mohammadabadi M.R., Khezri A., Kochuk-Yashchenko O.A., Kucher D.M., Babenko O.I., Bushtruk M.V., Tkachenko S.V., Stavetska R.V., Klopenko N.I., Oleshko V.P., Tkachenko M.V., Titarenko I.V. Dlk1 gene expression in different tissues of lamb. Iranian J Appl. Anim. Sci. 2020;10:669–677. [Google Scholar]
- Mei N.H., Guo S.M., Zhou Q., Zhang Y.R., Liu X.Z., Yin Y., He X., Yang J., Yin T.L., Zhou L.Q. H3K4 methylation promotes expression of mitochondrial dynamics regulators to ensure oocyte quality in mice. Adv. Sci. (Weinh) 2023;10 doi: 10.1002/advs.202204794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazeni S., Mohammadabadi M.R., Sadeghi M., Shahrbabak H.M., Koshkoieh A.E., Bordbar F. Association between UCP gene polymorphisms and growth, brreeding value of growth and reproductive traits in Mazandaran indigenous chicken. Open J. Anim. Sci. 2016;6:1–8. [Google Scholar]
- Moazeni S.M., Mohammadabadi M.R., Sadeghi M., Moradishahrbabak H. Association of the melanocortin-3(MC3R) receptor gene with growth and reproductive traits in Mazandaran indigenous chicken. J. Livest. Sci. Technol. 2016;4:51–56. [Google Scholar]
- Mohamadipoor L., Mohammadabadi M., Amiri Z., Babenko O., Stavetska R., Kalashnik O., Kucher D., Kochuk-Yashchenko O., Asadollahpour H. Signature selection analysis reveals candidate genes associated with production traits in Iranian sheep breeds. BMC. Vet. Res. 2021;17:1–9. doi: 10.1186/s12917-021-03077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadabadi M., Masoudzadeh S.H., Khezri A., Kalashnyk O., Stavetska R.V., Klopenko N.I., Oleshko V.P., Tkachenko S.V. Fennel (Foeniculum vulgare) seed powder increases delta-like non-canonical notch ligand 1 gene expression in testis, liver, and humeral muscle tissues of growing lambs. Heliyon. 2021;7:e08542. doi: 10.1016/j.heliyon.2021.e08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadabadi M., Golkar A., Hesni M.A. The effect of fennel (Foeniculum vulgare) on insulin-like growth factor 1 gene expression in the rumen tissue of Kermani sheep. Agric. Biotechnol. J. 2023;15:239–256. [Google Scholar]
- Mohammadabadi M.R., Nikbakhti M., Mirzaee H.R., Shandi A., Saghi D.A., Romanov M.N., Moiseyeva I.G. Genetic variability in three native Iranian chicken populations of the Khorasan province based on microsatellite markers. Russ. J. Genet. 2010;46:505–509. [PubMed] [Google Scholar]
- Mohammadifar A., Faghih Imani S.A., Mohammadabadi M.R., Soflaei M. The effect of TGFb3 gene on phenotypic and breeding values of body weight traits in Fars native fowls. J. Agric. Biotech. 2014;5:125–136. [Google Scholar]
- Mohammadifar A., Mohammadabadi M.R. Melanocortin-3 receptor (mc3r) gene association with growth and egg production traits in Fars indigenous chicken. Malays. Appl. Biol. 2018;47:85–90. [Google Scholar]
- Mohammadinejad F., Mohammadabadi M., Roudbari Z., Sadkowski T. Identification of key genes and biological pathways associated with skeletal muscle maturation and hypertrophy in Bos taurus, Ovis aries, and Sus scrofa. Animals. 2022;12:3471. doi: 10.3390/ani12243471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer N.A., Hu Y., Idriss A.A., Abobaker H., Hou Z., Yang S., Ma W., Zhao R. Dietary betaine improves egg-laying rate in hens through hypomethylation and glucocorticoid receptor-mediated activation of hepatic lipogenesis-related genes. Poult. Sci. 2020;99:3121–3132. doi: 10.1016/j.psj.2020.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer N.A., Hu Y., Hu Y., Idriss A.A., Abobaker H., Hou Z., Dong H., Zhao R. Dietary betaine activates hepatic VTGII expression in laying hens associated with hypomethylation of GR gene promoter and enhanced GR expression. J. Anim. Sci. Biotechnol. 2018;9:2. doi: 10.1186/s40104-017-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qanbari S., Simianer H. Mapping signatures of positive selection in the genome of livestock. Livest. Sci. 2014;166:133–143. [Google Scholar]
- Ren X., Guan Z., Zhao X., Zhang X., Wen J., Cheng H., Zhang Y., Cheng X., Liu Y., Ning Z., Qu L. Systematic selection signature analysis of chinese gamecocks based on genomic and transcriptomic data. Int. J. Mol. Sci. 2023;24:5868. doi: 10.3390/ijms24065868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renstrom F., Shungin D., Johansson I., Florez J.C., Hallmans G., Hu F.B., Franks P.W. Genetic predisposition to long-term nondiabetic deteriorations in glucose homeostasis: ten-year follow-up of the GLACIER study. Diabetes. 2011;60:345–354. doi: 10.2337/db10-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaei S.M.H., Dadpasand M., Mohammadabadi M., Atashi H., Stavetska R., Klopenko N., Kalashnyk O. An origanum majorana leaf diet influences myogenin gene expression, performance, and carcass characteristics in lambs. Animals. 2022;13:14. doi: 10.3390/ani13010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaei S.M.H., Mohammadabadi M., Moradi B., Kalashnyk O., Klopenko N., Babenko O., Borshch O.O., Afanasenko V. Role of fennel (Foeniculum vulgare) seed powder in increasing testosterone and igf1 gene expression in the testis of lamb. Gene Expr. 2023 [Google Scholar]
- Sarsenbek A., Wang T., Zhao J.K., Jiang W. Comparison of carcass yields and meat quality between Baicheng-you chickens and arbor acres broilers. Poult. Sci. 2013;92:2776–2782. doi: 10.3382/ps.2012-02841. [DOI] [PubMed] [Google Scholar]
- Schooling C.M. Plasma levels of vitamin K and the risk of ischemic heart disease: a Mendelian randomization study. J. Thromb. Haemost. 2016;14:1211–1215. doi: 10.1111/jth.13332. [DOI] [PubMed] [Google Scholar]
- Sendzikaite G., Kelsey G. The role and mechanisms of DNA methylation in the oocyte. Essays Biochem. 2019;63:691–705. doi: 10.1042/EBC20190043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahdadnejad N., Mohammadabadi M.R., Shamsadini M. Typing of clostridium perfringens isolated from broiler chickens using multiplex PCR. Genetics Third Millennium. 2016;14:4368–4374. [Google Scholar]
- Shahsavari M., Mohammadabadi M., Khezri A., Borshch O., Babenko O., Kalashnyk O., Afanasenko V., Kondratiuk V. Effect of fennel (foeniculum vulgare) seed powder consumption on insulin-like growth factor 1 gene expression in the liver tissue of growing lambs. Gene Expr. 2022;21:21–26. [Google Scholar]
- Shi H., Luo J., Zhu J., Li J., Sun Y., Lin X., Zhang L., Yao D., Shi H. PPAR gamma regulates genes involved in triacylglycerol synthesis and secretion in mammary gland epithelial cells of dairy goats. PPAR Res. 2013;2013 doi: 10.1155/2013/310948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Shao D., Yang L., Liang Q., Han W., Xue Q., Qu L., Leng L., Li Y., Zhao X., Dong P., Walugembe M., Kayang B.B., Muhairwa A.P., Zhou H., Tong H. Whole genome analyses reveal novel genes associated with chicken adaptation to tropical and frigid environments. J. Adv. Res. 2023;47:13–25. doi: 10.1016/j.jare.2022.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokri S., Khezri A., Mohammadabadi M., Kheyrodin H. The expression of MYH7 gene in femur, humeral muscle and back muscle tissues of fattening lambs of the Kermani breed. Agric. Biotechnol. J. 2023;15:21. [Google Scholar]
- Shriner D. Overview of admixture mapping. Curr. Protoc. 2023;3:e677. doi: 10.1002/cpz1.677. [DOI] [PubMed] [Google Scholar]
- Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- Sun T., Huang G.Y., Wang Z.H., Teng S.H., Cao Y.H., Sun J.L., Hanif Q., Chen N.B., Lei C.Z., Liao Y.Y. Selection signatures of Fuzhong Buffalo based on whole-genome sequences. BMC. Genomics. 2020;21:674. doi: 10.1186/s12864-020-07095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S. Phosphorylation of myosin regulatory light chain by myosin light chain kinase, and muscle contraction. Circ. J. 2009;73:208–213. doi: 10.1253/circj.cj-08-1041. [DOI] [PubMed] [Google Scholar]
- Takeda K., Takemoto C., Kobayashi I., Watanabe A., Nobukuni Y., Fisher D.E., Tachibana M. Ser298 of MITF, a mutation site in Waardenburg syndrome type 2, is a phosphorylation site with functional significance. Hum. Mol. Genet. 2000;9:125–132. doi: 10.1093/hmg/9.1.125. [DOI] [PubMed] [Google Scholar]
- Tian S., Li W., Zhong Z., Wang F., Xiao Q. Genome-wide re-sequencing data reveals the genetic diversity and population structure of Wenchang chicken in China. Anim. Genet. 2023;54:328–337. doi: 10.1111/age.13293. [DOI] [PubMed] [Google Scholar]
- van de Bunt M., Manning F.J., Dai X., Barrett A., Grey C., Li L., Bennett A.J., Johnson P.R., Rajotte R.V., Gaulton K.J., Dermitzakis E.T., MacDonald P.E., McCarthy M.I., Gloyn A.L. Transcript expression data from human islets links regulatory signals from genome-wide association studies for type 2 diabetes and glycemic traits to their downstream effectors. PLoS. Genet. 2015;11 doi: 10.1371/journal.pgen.1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S.H., Lu Z.S. Factors affecting the effective number of tests in genetic association studies: a comparative study of three PCA-based methods. J. Hum. Genet. 2011;56:428–435. doi: 10.1038/jhg.2011.34. [DOI] [PubMed] [Google Scholar]
- Weng Z., Xu Y., Li W., Chen J., Zhong M., Zhong F., Du B., Zhang B., Huang X. Genomic variations and signatures of selection in Wuhua yellow chicken. PLoS. One. 2020;15 doi: 10.1371/journal.pone.0241137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing K., Wang K., Ao H., Chen S., Tan Z., Wang Y., Xitong Z., Yang T., Zhang F., Liu Y., Ni H., Sheng X., Qi X., Wang X., Guo Y., Wang C. Comparative adipose transcriptome analysis digs out genes related to fat deposition in two pig breeds. Sci. Rep. 2019;9:12925. doi: 10.1038/s41598-019-49548-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z., Wang L., Tian Y., Yang Y., Li P., Yang G., Lu H., Wang S., Zeng W., Zhang T. A genome-wide scan to identify signatures of selection in Lueyang black -bone chicken. Poult. Sci. 2023;102 doi: 10.1016/j.psj.2023.102721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C.Y., Hong S.D., Lee Y.H., Lee J., Jung D.E., Kim G.H., Kim S.H., Jung J.K., Kim K.H., Lee H., Hong J.T., Han S.B., Kim Y. Nuclear entry of CRTC1 as druggable target of acquired pigmentary disorder. Theranostics. 2019;9:646–660. doi: 10.7150/thno.30276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B., Zhao Y., Fan S., Yuan P., Li H., Li S., Li Y., Zhang Y., Huang H., Li H., Kang X., Li G. Differentially expressed lncRNAs related to the development of abdominal fat in Gushi Chickens and their interaction regulatory network. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.802857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Dong S.S., Xu J.Y., He W.M., Yang T.L. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35:1786–1788. doi: 10.1093/bioinformatics/bty875. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Gao T., Hu S., Lin B., Yan D., Xu Z., Zhang Z., Mao Y., Mao H., Wang L., Wang G., Xiong Y., Zuo B. The functional SNPs in the 5′ regulatory region of the porcine PPARD gene have significant association with fat deposition traits. PLoS. One. 2015;10 doi: 10.1371/journal.pone.0143734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.