Abstract
We previously demonstrated that expression of the nonproducer F12-human immunodeficiency virus type 1 (HIV-1) variant induces a block in the replication of superinfecting HIV that does not depend on the down-regulation of CD4 HIV receptors. In order to individuate the gene(s) involved in F12-HIV-induced interference, vectors expressing each of the nine F12-HIV proteins were transfected in HIV-susceptible HeLa CD4 cells. Pools of cell clones stably producing each viral protein were infected with HIV-1, and virus release was measured in terms of reverse transcriptase activity in supernatants. We hereby demonstrate that HeLa CD4 cells expressing the F12-HIV gag, vif, or nef gene were resistant, to different degrees, to infection with T-cell-line-adapted HIV-1 strains. Conversely, expression of either the tat, rev, or vpu F12-HIV gene increased the rate of HIV release, and no apparent effects on HIV replication were observed in cells expressing either the F12-HIV vpr, pol, or env gene. No variation of CD4 exposure was detected in any of the uninfected HeLa CD4 pools. These data indicate that F12-HIV homologous viral interference is the consequence of the synergistic anti-HIV effects of Gag, Vif, and Nef proteins. Retrovirus vectors expressing F12-HIV vif or nef allowed us to further establish that the expression of each mutated protein (i) inhibits the replication of clinical HIV-1 isolates as well, (ii) impairs the infectivity of the virus released by cells chronically infected with HIV-1, and (iii) limitedly to F12-HIV Vif protein, induces HIV resistance in both vif-permissive and vif-nonpermissive cells. The levels of action of F12-HIV vif and nef anti-HIV effects were also determined. We observed that HIV virions emerging from the first viral cycle on F12-HIV vif-expressing cells, although released in unaltered amounts, had a strongly reduced ability to initiate the retrotranscription process when they reinfected parental HeLa CD4 cells. Differently, we observed that expression of F12-HIV Nef protein affects the HIV life cycle at the level of viral assembling and/or release. For the first time, an inhibitory effect on the HIV life cycle in both acutely and chronically infected cells induced by mutated Vif and Nef HIV-1 proteins is described. These genes could thus be proposed as new useful reagents for anti-HIV gene therapy.
The phenomenon of homologous viral interference described for human immunodeficiency virus (HIV) as well as for animal retroviruses commonly correlates with the down-modulation of retrovirus cell receptors (51, 61, 62, 68). To date, very few models of HIV homologous viral interference independent of viral receptor exposure have been described; noninfectious pol-defective HIV virions are able to induce interference in coinfections of Jurkat cells with wild-type HIV (4). Moreover, after HIV infection at high multiplicity, CEM cells become resistant to homologous superinfection during short intervals (24 to 48 h), soon before the CD4 down-regulation takes place (69). Conversely, cells expressing the nondefective, nonproducer F12-HIV type 1 (HIV-1) variant are stably resistant to superinfection by replication-competent HIV, despite an unmodified level of CD4 exposure (16–18).
F12-HIV is an HIV-1 genome cloned from a Hut-78 cell clone infected with the supernatant of the peripheral blood lymphocytes (PBLs) of an HIV-seropositive patient. This variant is unable to code for even aberrant HIV particles as a consequence of a heavily altered structural viral protein pattern (19). Nucleotide sequence analysis failed to highlight genomic deletions, but the presence of many amino acid substitutions scattered along the whole viral genome (mainly in vif and pol genes) was observed (10).
Experimental evidence has suggested that F12-HIV-induced homologous viral interference could be the consequence of the inhibiting action of some F12-HIV viral protein(s). In fact, superinfecting HIV is able to enter into CD4+ F12-HIV-expressing cells and to retrotranscribe its genome, thus ruling out any effect of F12-HIV expression on the penetration of superinfecting HIV (17). Moreover, no encapsidation of the F12-HIV genome in virions emerging from either HIV-superinfected, F12-HIV-expressing cells (17) or cells chronically infected with HIV transduced with the F12-HIV genome (7) was observed, thereby excluding a direct role of F12-HIV genomic RNA.
To individuate the F12-HIV protein(s) involved in viral interference, vectors expressing each of the nine F12-HIV genes were separately transfected into highly HIV-susceptible HeLa CD4 cells that were subsequently infected with HIV-1. In this paper, we demonstrate that HeLa CD4 cells expressing F12-HIV Gag, Vif, or Nef protein are protected from infection with T-cell-line-adapted (TCLA) HIV-1 strains. Furthermore, expression of F12-HIV Vif or Nef protein driven by retroviral vectors is able to inhibit both the replication of T-tropic clinical HIV-1 isolates in transduced CEMss cells and the infectivity of virus released from cells chronically infected with HIV-1. Finally, the levels of action at which F12-HIV Vif or Nef protein inhibits HIV replication were determined.
MATERIALS AND METHODS
Construction of expressing vectors.
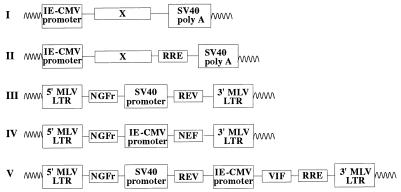
A schematic representation of the different molecular constructs used is given in Fig. 1. Intronless F12-HIV genes and rev-responsive elements (RRE) were obtained through PCR amplification from plasmid pUc/F12-HIV (10) or, to obtain wild-type vif or nef genes, from plasmid pNL4-3 (1). tat and rev genes were amplified by reverse transcriptase (RT)-PCR of total RNA extracted from parental Hut-78/F12 cells. DNA and RNA-PCR were performed as described elsewhere (55). The sequences of primers used for amplifications are reported in Table 1. The same couples of primers were used to amplify vif and nef genes from plasmids pUc/F12-HIV and pNL4-3. PCR products were purified and digested with enzymes whose recognition sequences are included on the 5′ side of each primer in Table 1. F12-HIV vpr, vpu, env, tat, and NL4-3 and F12-HIV nef genes and RRE sequences were inserted into the polylinker of the immediate-early cytomegalovirus (IE-CMV)-promoted pcDNAI (Invitrogen, San Diego, Calif.)-expressing vector (Fig. 1I). Conversely, constructs expressing F12-HIV gag and pol and either NL4-3 or F12-HIV vif (Fig. 1II) were obtained by inserting each gene in the pcDNAI vector in which RRE sequences were previously inserted in BamHI-EcoRI sites. rev-expressing retrovirus vector (Fig. 1III) was obtained by inserting F12-HIV rev cDNA in a retrovirus vector (L-NGFr) expressing the cDNA of the low-affinity human nerve growth factor receptor (NGFr) (32) truncated in its cytoplasmic domain and driven by the Moloney murine leukemia virus (MLV) long terminal repeat (LTR). The L-NGFr retrovirus vector was obtained by inserting the NGFr cDNA excised from the pUc19 vector with EcoRI and HindII sites in EcoRI and HpaI sites of the polylinker of an LXSN retroviral vector (43) previously deleted (by BamHI-NcoI cuts) from the simian virus 40 (SV40) promoter-G418 resistance cassette. F12-HIV rev cDNA was placed under the control of the SV40 promoter, and the SV40-rev cassette was finally inserted in the XhoI site of the L-NGFr retrovirus vector. Retroviral constructs expressing F12-HIV nef (Fig. 1IV) and vif (Fig. 1V) genes were obtained by inserting, in the filled XhoI site of the L-NGFr retrovirus vector, the IE-CMV promoter-nef cassette (recovered from the pcDNAI-nef-expressing vector after HhaI-NotI digestion and filling) and the IE-CMV promoter-vif-RRE cassette (obtained from the pcDNAI-vif-RRE-expressing vector by BsaI-XbaI fragment digestion and filling), respectively.
FIG. 1.
Schematic map of pcDNAI-based vectors expressing rev-independent (I) or rev-dependent (II) F12-HIV genes. The structures of rev-, nef-, and vif-expressing retrovirus vectors (III, IV, and V, respectively) are also shown.
TABLE 1.
Sequences of primers for PCR amplifications
| Amplified HIV-1 region | Nucleotide sequence (5′ → 3′) | Restriction site | F12-HIV nucleotide rangea |
|---|---|---|---|
| gag | Forward CTC GGA TCC AGA GAG ATG GGT GCG AGA GCG TCA GTA TTAb | BamHI | 826–853 |
| Reverse CTC GTC GAC TTA TTG TGA CGA TGG GTC GTT | SalI | 2332–2312 | |
| pol | Forward CCC GGA TCC ATG AAA GAT TGT ACC GAG AGA CAG GCTAA | BamHI | 2036–2124 |
| Reverse CGG GTC GAC CTA ATC CTC ATC CTG TCT AC | SalI | 5137–5118 | |
| vif | Forward TCG GAT CCG GGA TTA TGG AAA ACA GATGGC AGG TG | BamHI | 5074–5101 |
| Reverse TCG TCG ACG CTC TAG TGT CCA TTC ATT GTA TG | SalI | 5662–5629 | |
| vpr | Forward CAG AAG CTT GGATAG ATG GAA CAA GCC CCA GAA GAC | HindIII | 5193–5219 |
| Reverse ATA GGA TCC CTA TGT CGA CAC CCA ATT CTG | BamHI | 5835–5815 | |
| vpu | Forward CCC GGA TCC CATGTA ATG CAA CCT ATA CAA ATA GCA ATA GTA GC | BamHI | 6096–6130 |
| Reverse CCC GAA TTC CTA CAG ATC ATC AAC ATC CCA AGG | EcoRI | 6347–6324 | |
| env | Forward CCC GAA TTC GTG GCA ATG AGA GTG AAG GAG AAA TAT GAG | EcoRI | 6257–6286 |
| Reverse CCC CTC GAG TTA TAG CAA AAT CCT TTC CAA | XhoI | 8817–8797 | |
| tat | Forward CCC GGA TCC CAA GAA ATG GAG CCA GTA GAT CCT AGA | BamHI | 5865–5891 |
| Reverse GCC GAA TTC CTA TTC CTT CGG GCC TGT CGG | EcoRI | 8446–8426 | |
| rev | Forward CCC GAA TTC TCT CCT ATG GCA GGA AGA AGC GGA GAC A | EcoRI | 6004–6031 |
| Reverse GCC CTC GAG CTA TTC TTT AGC TCC TGA CTC | XhoI | 8675–8655 | |
| nef | Forward GGA AGC TTA TAA GAT GGG TGG CAA GTG GTC AA | HindIII | 8813–8837 |
| Reverse CTC GAA TTC TCA GCA GTT CTT GAA GTA CTC C | BamHI | 9439–9418 | |
| rre | Forward AAG GAT CCG AGT CGA CAA GAG CAG TGG GAA TAG GAG CT | BamHI-SalI | 7776–7798 |
| Reverse CAG AAT TCA GCC CCA GGA GCT GTT GAT CCT TTA GGT ATC TT | EcoRI | 8028–7998 |
EMBL data library accession no. Z11530.
Restriction site sequences are underlined.
All of the open reading frames inserted in the expression vectors were sequenced by the dideoxy chain termination method with the Sequenase II kit (U.S. Biochemicals, Cleveland, Ohio) to exclude any artifactual mutations. In all HeLa CD4-transfected cells, resistance to hygromycin B was expressed in trans by the p220.2 vector (29).
Cell cultures and transfections.
HeLa CD4, Cos-1, GP+E86 (39), and AM12 (42) cells were grown in Dulbecco modified minimal essential medium supplemented with 10% decomplemented fetal calf serum (FCS) and, for HeLa CD4 cells only, 0.5 mg of G418 (70% activity; GIBCO-BRL, Gaithersburg, Md.) per ml. D10 (an Hut-78 cell clone chronically infected by an HIV-1 isolate) (19), C8166, CEMss, and H9/HTLVIIIB cells were maintained in RPMI 1640 medium supplemented with 10% decomplemented FCS. Human PBLs from healthy donors were activated for 48 h with phytohemagglutinin (PHA) and were then cultivated in RPMI 1640 medium supplemented with 20% FCS and 50 U of interleukin-2 (Roche, Nutley, N.J.) per ml.
Cotransfections of HeLa CD4 cells were performed by the calcium phosphate method (71), with a molar ratio of 10:1 between pcDNAI-based and p220.2 vectors. Hygromycin B-resistant cells, which were obtained after 10 to 14 days of selection with 0.2 mg of hygromycin B (Boehringer GmbH, Mannheim, Germany) and 0.1 mg of G418 per ml, were cloned by the end dilution method. Pools of HeLa CD4 cells separately expressing each F12-HIV gene were composed of at least five different cell clones producing the respective F12-HIV protein.
Ecotropic GP+E86 packaging cells were transfected with different L-NGFr-based retroviral vectors by the calcium phosphate method. Two days later, supernatants were used to infect AM12 amphotropic packaging cells. After an additional 4 days, NGFr-expressing AM12 cells were selected by an immunomagnetic-positive procedure as previously described (7, 41). Supernatants from NGFr-positive AM12 cells were used to infect (four cycle infections) parental HeLa CD4 cells, CEMss cells, PBLs, or D10 cells. Four days later, amphotropic infected cells were selected for NGFr expression.
HIV infections, transfections, and detection.
Supernatants from acutely infected CEMss cells were used as the source of TCLA HIV-1 strains (i.e., NL4-3 and HTLVIIIB). Titers (ranging from 106 to 107 50% tissue culture infective doses [TCID50]/ml) were measured as previously described (18) by scoring the syncytium number on C8166 cells 5 days after the infection of serially diluted virus preparations. HIV-1 clinical isolates were obtained as PBL supernatants from AIDS patients. Titers (calculated as 50% infectious doses by infecting fresh PHA-stimulated PBLs from healthy donors with serially diluted supernatants) ranged from 5 × 104 and 5 × 105 infectious doses/ml. Supernatants of Cos-1 cells transfected with the 6.9 molecular clone (21) were used as the source of a vif-deleted HIV strain whose titers were measured in nanograms of HIV p24 per milliliter by an enzyme-linked immunosorbent assay (ELISA) (Antigen Capture Assay; Abbott, North Chicago, Ill.)
HIV release was monitored by an RT assay (52) or by ELISA, whereas intracellular HIV p24 was measured by an ELISA as described elsewhere (16).
Single-cycle infections were performed by challenging HeLa CD4 cells with TCLA HIV-1 strains (previously treated with DNase as described elsewhere [66]) at a multiplicity of infection (MOI) of 2. Virus was adsorbed to cells in a small volume (0.15 ml for 24-well cultures) for 1 h at 37°C, and then, in order to eliminate possible virus carryover, free HIV was removed by several washes followed by a treatment of cell cultures with trypsin for 15 min at 37°C.
Transfections of the pNL4-3 HIV infectious molecular clone either alone (2 μg of plasmid for 5 × 104 cells) or, in transient transfection experiments, together with pcDNAI-based vif- or nef-expressing vectors in a molar ratio of 1:10 were performed by the calcium-phosphate method (71), and cell lysates and/or supernatants were assayed by an HIV p24 ELISA 48 to 72 h posttransfection.
Molecular analyses.
RNA analyses were performed on total RNA extracted with the RNA-FastI kit (Molecular Systems, San Diego, Calif.) following the manufacturer’s recommendations. Northern blots were performed as described elsewhere (55) and hybridized with random-primed, 32P-labelled F12-HIV-specific genes or T-tropic HIV coreceptor CXCR4 cDNA (20). Clones of HeLa CD4 rev-expressing cells transfected with the pcDNAI/RRE expression vector were selected on the basis of transcription of the F12-HIV RRE sequences. On the other hand, the abilities of HeLa CD4 pools to produce the respective F12-HIV or NL4-3 protein were tested by Western blotting or radioimmunoprecipitation assay (RIPA). Western blots were performed by lysing cells in phosphate-buffered saline (PBS)–1% Triton X-100. Samples of 50 μg of protein were separated through sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (Hybond-ECL; Amersham, Buckinghamshire, United Kingdom) by electroblotting. Filters were blocked overnight at 4°C by 10% nonfat dry milk–0.1% Triton X-100 in PBS. The membranes were then incubated for 1 h at room temperature (r.t.) with the specific antibodies, washed twice with PBS, and incubated for 1 h at r.t. with the appropriate dilution of the horseradish peroxidase-labelled secondary antibody (Amersham). Finally, the filters were incubated in an enhanced chemioluminescence detection reagent (Amersham) for 1 min at r.t. and were exposed for 1 to 5 min to Hyperfilm-ECL (Amersham).
RIPAs were performed as described elsewhere (19) by labelling cells for 16 h with [35S]cysteine plus [35S]methionine, except for the F12-HIV vpr-expressing HeLa CD4 pool, which was labelled with [35S]methionine plus [3H]leucine, as reported elsewhere (36).
For the detection of F12-HIV or NL4-3 proteins (by RIPA or Western blotting), specific antisera obtained from the NIH Research and Reference Program were utilized.
Expression of the CD4 HIV receptors was detected by direct immunofluorescence analysis with the phycoerythrin-conjugated Leu3a monoclonal antibody (Becton Dickinson, Montain View, Calif.) as described elsewhere (41), and labelled cell populations were analyzed with a cytofluorometer (Fac-scan; Becton Dickinson). DNA-PCR analyses were performed as described elsewhere (66) on lysates of cells infected with DNase-treated HIV. HIV-1-specific oligoprimers able to discriminate between products of partial (both primers recognizing a region immediately on the 5′ side of the primer binding site, the HIV genomic region where retrotranscription begins) and complete (the same forward primer but the reverse one recognizing a region in 3′ with respect to the primer binding site) retrotranscription processes were utilized. The respective sequences are as follows: oligonucleotide forward, 5′-CAGATATCCACTGACCTTTGGATGGTGC-3′ (110 to 137); oligonucleotide reverse 1, 5′-CTGAGGGATCTCTAGTTACCAGAGTC-3′ (602 to 577); oligonucleotide reverse 2, 5′-ATCTCTCTCCTTCTAGCCTCCGCTAGT-3′ (791 to 765); and oligonucleotide probe, 5′-TCTGGTTAGACCAGATCTGAGCCTGGGA-3′ (462 to 488). The numeration refers to the sequence of the HXB2 isolate (GenBank accession no. K03455), and no nucleotide variations could be found in the regions recognized by the above-described oligoprimers between the HTLVIIIB and NL4-3 strains.
RESULTS
Isolation and characterization of HeLa CD4 cell clones expressing single HIV genes.
As previously reported, structural HIV genes (i.e., gag, pol, and env) contain instability sequences that can be counteracted by the binding of the HIV Rev (and, consequently, of cellular factors) to the RRE sequences (6, 13, 22, 30, 50). Data on the stability of vif-specific RNA are more controversial (5, 25), but we observed that both wild-type and F12-HIV vif RNAs were degraded in the absence of Rev-RRE interaction (not shown). Thus, in vectors expressing rev-dependent genes, RRE sequences were added in 3′ with respect to each HIV gene (except in vector expressing the env gene that itself contains the RRE sequences). These constructs were transfected in HeLa CD4 cells homogeneously expressing the NGFr-rev retrovirus vector, whereas vectors expressing rev-independent genes (i.e., tat, vpr, vpu, and nef) were transfected in parental HeLa CD4 cells.
Hygromycin-resistant cell clones were isolated and selected on the basis of the RNA and protein expression of the F12-HIV or wild-type transfected gene (not shown). Pools of at least five different selected clones were set up and further analyzed for viral protein expression (Fig. 2 and 3).
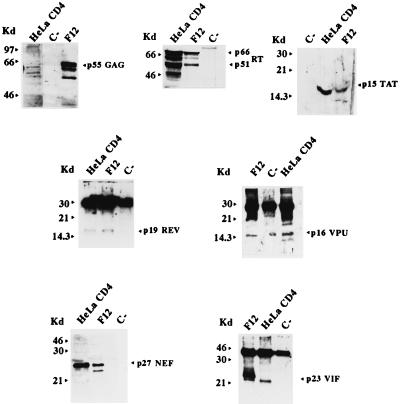
FIG. 2.
Western blot analyses of HeLa CD4 pools transfected with vectors expressing either the F12-HIV gag, pol, tat, vpu, nef, or vif gene or transduced with rev-expressing retrovirus vector. Hygromycin-resistant HeLa CD4 cells (C−) and parental Hut78/F12 cells were used as negative and positive controls, respectively. Molecular size markers (in kilodaltons) are given to the left of each panel.
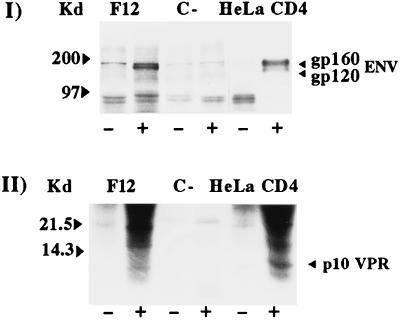
FIG. 3.
RIPA of HeLa CD4 pools transfected with env (I)- or vpr (II)-expressing vectors. Hygromycin-resistant HeLa CD4 cells (C−) and Hut-78/F12 cells were used as negative and positive controls, respectively. Cell lysates were incubated with normal (−) or hyperimmune (+) specific rabbit antisera. Molecular size markers (in kilodaltons) are given to the left of each panel.
No impairment in cell growth (not shown), expression of the CD4 receptors (Fig. 4) (except for the NL4-3 nef-expressing HeLa CD4 pool, in which CD4 down-regulation was observed [not shown]), and expression of CXCR4 (the coreceptor of the T-lymphotropic, syncytium-inducing HIV) (20) mRNA (not shown) was detected in all of the HeLa CD4 pools.
FIG. 4.
CD4 fluorescence-activated cell sorter analyses of HeLa CD4 pools transfected with vector expressing each F12-HIV protein or with the hygromycin B resistance vector only (p220.2). HeLa and parental HeLa CD4 cells were used as negative and positive controls, respectively. Dotted lines identify higher values of fluorescence intensity detected in each pool labelled with unspecific phycoerythrin-conjugated mouse immunoglobulin Gs.
rev-RRE-dependent genes.
Lower amounts of the F12-HIV Gag protein were produced in transfected HeLa CD4 cells with respect to the parental Hut-78/F12 cells (Fig. 2). In addition, no Gag cleavage products were detected (not shown), likely as the consequence of the absence of the virus-specific protease encoded in HIV by the pol gene. However, even when the F12-HIV gag gene was expressed in the context of the whole viral genome, no or a very low level of Gag cleavage was always observed (17, 19). In contrast to what was reported for other cells expressing HIV Gag or Gag-Pol polyprotein (34, 49, 58) and similarly to what was observed in parental Hut-78/F12 cells (19), no release of virus-like particles was observed in the F12-HIV gag-expressing HeLa CD4 cells (not shown).
Western blot analysis of the F12-HIV pol-expressing HeLa CD4 cells (Fig. 2) shows the presence of the RT p66-p51 heterodimer typically produced in HIV-infected cells (15). Furthermore, both the F12-HIV p31 integrase and the p15 protease (both encoded by the HIV pol gene) were also detected by RIPA (data not shown).
It is worth noting that, when expressed alone, the F12-HIV Env protein appears cleaved (Fig. 3I), while no gp160 Env cleavage products have been detected in any of the F12-HIV-expressing cells analyzed so far (16–19), despite the unmodified env cleavage site detected in the F12-HIV genome (10).
rev-RRE-independent genes.
HeLa CD4 cells homogeneously expressing the F12-HIV rev gene were obtained by transducing the cells with the L-NGFr-rev retroviral vector described above.
We were able to obtain cell clones stably expressing the Vpr protein (Fig. 3II), despite unsuccessful attempts that were previously reported (14, 33). This was probably the consequence of the premature stop codon in the F12-HIV vpr gene (10) that generates a truncated protein of 78 amino acids (instead of the 96 from the complete vpr gene), leaving out the basic domain which is directly involved in the inhibition of cell growth (14).
It is worth noting that expression of the F12-HIV nef gene also is unable to affect CD4 expression (Fig. 4).
Effects of F12-HIV proteins on HIV replication: inhibitory action induced by gag, vif, and nef gene expression.
The main goal of the present work was to determine which F12-HIV gene(s) is involved in F12-HIV-induced homologous viral interference. Thus, HeLa CD4 cells separately expressing each of the nine F12-HIV genes were infected with TCLA HIV-1 strains (i.e., NL4-3 or, in replicated experiments, HTLVIIIB) at a MOI of from 0.01 to 5. Titers of challenging virus were determined for C8166 cells and were confirmed for HeLa CD4 cells, except at the time of viral outcome, which was delayed in HeLa CD4 cells by about 4 to 6 days with respect to C8166 cells.
The appearance in infected cell cultures of both syncytia and high RT activity levels on supernatants (>5 × 104 cpm/ml) led to a progressive decrease of cell viability and, finally, to cell death. In addition, RT data concerning infected gag-, pol-, env-, and vif-expressing cells were compared to those for infected rev-expressing cells, since expression of these genes needs coexpression of Rev protein, which was per se able to accelerate the HIV replication cycle (see below).
On the basis of the observed effects on HIV replication (which were reproducible regardless of which TCLA HIV-1 strain was utilized), F12-HIV proteins could be classified as inhibiting, enhancing, or not affecting proteins.
HIV-inhibiting F12-HIV proteins.
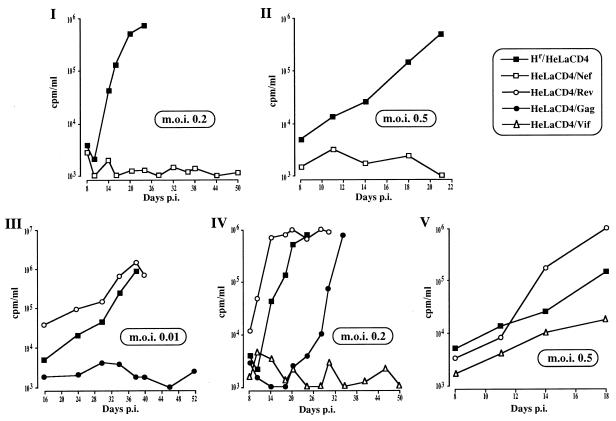
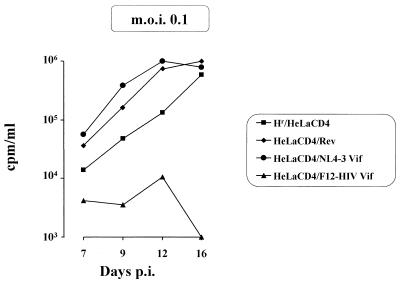
HeLa CD4 cells expressing either F12-HIV gag, vif, or nef were resistant (at different degrees) to HIV infection (Fig. 5).
FIG. 5.
RT activities (measured in counts per minute per milliliter of supernatant and normalized for 106 cells after background subtraction) at different days post-HIV infection (p.i.) of nef-expressing HeLa CD4 cells (MOIs of 0.2 and 0.5 [I and II, respectively]); gag- or rev-expressing HeLa CD4 cells (MOI of 0.01 [III]); gag-, vif-, or rev-expressing HeLa CD4 cells (MOI of 0.2 [IV]); and vif- or rev-expressing HeLa CD4 cells (MOI of 0.5 [V]). As a control, hygromycin-resistant (Hr) HeLa CD4 cells were utilized. Challenging virus was adsorbed in a small volume (e.g., 0.15 ml in 24-well plates) in semiconfluent cell cultures for 1 h at 37°C, and then cells were refed with fresh complete medium. Data from a representative of three different experiments are reported.
The most effective anti-HIV F12-HIV protein was Nef. In fact, no RT activity was observed in the supernatants of nef-expressing HeLa CD4 cells infected at a MOI of either 0.01 (not shown), 0.2 (Fig. 5I), or 0.5 (Fig. 5II). The inhibitory action was overcome by increasing the MOI to 5 (not shown). The effect of F12-HIV nef expression on the replication of challenging HIV could not be adequately compared with that of wild-type Nef protein, whose expression, in contrast to that observed for F12-HIV nef-expressing cells, induces CD4 down-regulation in both CEMss and HeLa CD4 cell lines (not shown).
F12-HIV Vif protein seems slightly less effective than Nef protein in protecting cells from TCLA HIV-1 infection. At a MOI of 0.2, no RT activity was detected in supernatants of HIV-infected, vif-expressing cells for up to 50 days postinfection (Fig. 5IV). A limited viral spread was detected when the MOI was increased to 0.5 (Fig. 5V), whereas the HIV-inhibiting effect was overcome at a MOI of 1.5, even if the viral outcome was delayed with respect to control rev-expressing cells (not shown). In contrast, in cells expressing the wild-type NL4-3 Vif protein, HIV replicated as efficiently as in control cells (Fig. 6).
FIG. 6.
RT activities in supernatants at different days post-HIV infection (p.i.) (MOI of 0.1) of rev-, NL4-3 vif-, or F12-HIV vif-expressing HeLa CD4 cells. As control, hygromycin-resistant (Hr) HeLa CD4 cells were utilized. Challenge with HIV was performed as described in the legend to Fig. 5. Data from a representative of two different experiments are reported.
When F12-HIV gag-expressing HeLa CD4 cells were infected at a MOI of 0.01, the RT activities of supernatants remained at background levels until 52 days postinfection (Fig. 5III). By increasing the MOI to 0.2, both syncytia and RT activity in the supernatants were detectable starting at 29 days postinfection, i.e., with a clear delay compared to rev-expressing and control hygromycin-resistant HeLa CD4 cells (Fig. 5IV). Conversely, no inhibitory effect was observed when gag-expressing cells were infected at a MOI of 0.5 (not shown).
In an attempt to evaluate a possible negative effect on the HIV replication of F12-HIV RRE sequences coupled with rev expression, we transfected HeLa CD4 rev-expressing cells with a pcDNAI vector transcribing the F12-HIV RRE sequences. We observed that in rev-RRE-coexpressing clones, HIV can replicate even better than in HeLa CD4/rev cells (data not shown), whatever MOI is used. Therefore, we may exclude that the inhibition of the HIV life cycle observed in both gag- and vif-expressing cells was the consequence of an unspecific effect of rev-RRE coexpression.
F12-HIV proteins enhancing or not affecting HIV.
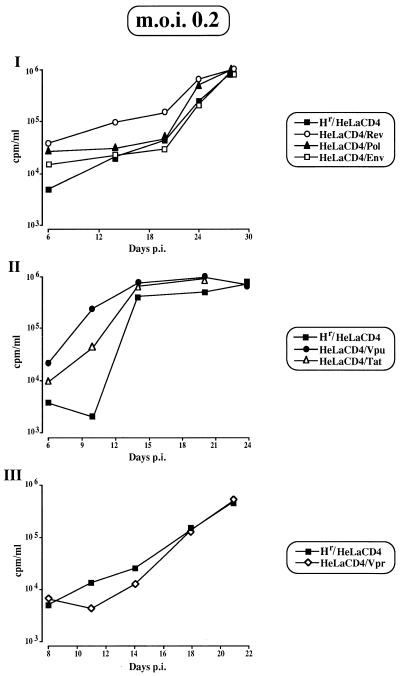
Expression in HeLa CD4 cells of either F12-HIV rev, tat, or vpu led to an increased rate in HIV release at any MOI used (Fig. 7I and II [in which data from the infection with a MOI of 0.2 are reported]). This positive effect was not surprising, considering that (i) no amino acid substitutions with respect to replication-competent HIV strains have been detected in any of these three genes (10); (ii) Rev and Tat proteins play essential roles in the progression of the HIV life cycle; and (iii) data about the increase of HIV release in vpu-expressing cells have already been reported elsewhere (56).
FIG. 7.
RT activities in supernatants at different days post-HIV infection (p.i.) (MOI of 0.2) of rev-, env-, or pol-expressing HeLa CD4 cells (I); tat- or vpu-expressing HeLa CD4 cells (II); and vpr-expressing HeLa CD4 cells (III). As a control, hygromycin-resistant (Hr) HeLa CD4 cells were utilized. Challenge with HIV was performed as described in the legend to Fig. 5. Data from a representative of two different experiments are reported.
Conversely, expression of F12-HIV pol or env (Fig. 7I) or vpr (Fig. 7III) seems to have no influence on HIV replication at any MOI utilized.
F12-HIV vif or nef expression also inhibits replication of T-tropic HIV-1 clinical isolates.
To establish whether the above-described anti-HIV effects also operate against HIV-1 clinical isolates, HeLa CD4 cells were separately challenged with supernatants obtained from PBLs of 21 patients containing viral isolates characterized as T-tropic HIV strains. Even if a minority of these HIV isolates were able to penetrate parental HeLa CD4 cells (as assessed by infecting HeLa CD4-LTR-β-gal cells (67a), in no case were levels of HIV replication sufficient to be detected by RT or ELISA achieved.
To overcome this difficulty, we challenged CEMss cells (a cell line that may well support the replication of T-tropic HIV clinical isolates) transduced with retroviral vectors coexpressing F12-HIV Vif or Nef protein (as assessed by Western blots; not shown) and, as a selection marker, a truncated form of the human NGFr (Fig. 1). Unfortunately, the inability to produce detectable amounts of intracellular F12-HIV Gag protein through the retroviral vector strategy hampered the possibility of evaluating the effect of this protein on the replication of HIV-1 clinical isolates.
According to the results obtained in transfected HeLa CD4 cells, the expression of F12-HIV Vif or Nef protein in CEMss cells inhibits the replication of TCLA HIV-1 strains (Table 2). The antiviral effect appeared more pronounced in nef-expressing cells with respect to vif-expressing cells and was independent of CD4 exposure (data not shown).
TABLE 2.
Percentages of inhibition of HIV release in acutely infected (MOI of 0.1) CEMss cells transduced with F12-HIV vif- or nef-expressing retroviral vectora
| Cells | % of inhibition on the following days postinfection:
|
||
|---|---|---|---|
| 11 | 14 | 19 | |
| CEMss/vif | 94.1 | 93.7 | 97.8 |
| CEMss/nef | 98.9 | 99.7 | 99.8 |
Percentages of inhibition with respect to HIV-infected CEMss cells transduced with the empty L-NGFr retroviral vector (CEMss L-NGFr). RT values in supernatants of infected CEMss L-NGFr cells (expressed in counts per minute per milliliter, normalized for 106 cells and after background subtraction) were 7 × 105, 1.3 × 106, and 7.5 × 105 at days 11, 14, and 19 postinfection, respectively. Data from a representative of two independent experiments are reported.
Fully NGFr-positive CEMss cell populations were separately infected with three selected T-tropic HIV-1 clinical isolates at a MOI of 1, as calculated by infecting fresh PHA-stimulated PBLs from healthy donors. The results reported in Fig. 8 demonstrate that the anti-HIV effects of F12-HIV Vif or Nef proteins were fully operative even when cells were challenged with HIV-1 clinical isolates. Of note, and in contrast to what was observed when cells were challenged with TCLA HIV-1 strains, in this case the expression of F12-HIV Vif protein may induce an anti-HIV state as effective as that induced by F12-HIV Nef protein.
FIG. 8.
RT activities in supernatants of CEMss cells transduced with retrovirus vectors expressing F12-HIV Vif or Nef protein at different days after infection (p.i.) (MOI of 1) with three different T-tropic HIV-1 clinical isolates. As a control, CEMss cells transduced with the empty L-NGFr retrovirus vector were also infected. A total of 106 cells were incubated for 24 h with the appropriate volumes of supernatants containing the challenging virus and were then washed and resuspended in fresh medium. Data from a representative of two different experiments are reported.
F12-HIV vif or nef gene expression decreases the infectivity of HIV-1 released from chronically infected cells.
We already observed that the expression of the whole F12-HIV genome was able to block the infectivity of virions released by cells chronically infected with HIV-1 (7). Attempting to test whether the above-described anti-HIV effects of F12-HIV Vif or Nef protein could be reproduced in chronically infected cells, retroviral vectors expressing the relative F12-HIV genes were utilized to transduce a Hut-78 cell clone (D10) chronically infected with HIV-1 (19). As a control, D10 cells were also transduced with the empty L-NGFr retroviral vector. Supernatants from equal numbers of homogeneously expressing NGFr cells were tested in terms of amounts of retroviral particles by RT assaying and in terms of HIV infectivity by viral titration. As shown in Table 3, whereas no significant variations in RT activity were detected, a strong impairment in the infectivity of the released virus could be readily observed (more than 95 and 99% of reduction in vif- and nef-expressing D10 cells, respectively). Thus, the ability of both F12-HIV Vif and Nef proteins to inhibit HIV infectivity in a model of HIV chronic infection was demonstrated.
TABLE 3.
Inhibition of viral infectivity of HIV released from cells chronically infected with HIV transduced with F12-HIV vif- or nef-expressing retrovirus vectora
| Cells | RTb | Viral infectivityc |
|---|---|---|
| D10 | 3.7 × 105 | 3 × 104 |
| D10/L-NGFr | 5.3 × 105 | 2 × 104 |
| D10/vif | 6.6 × 105 | 9 × 102 |
| 95.5d | ||
| D10/nef | 3.8 × 105 | 1.1 × 102 |
| 99.5d |
Data from a representative of three independent experiments are reported.
Values are expressed in counts per minute per milliliter and normalized for 106 cells after background subtraction.
Values are expressed as TCID50 per milliliter × 10−6 cells.
Percentages of inhibition with respect to L-NGFr-transduced D10 cells are indicated.
At which level does F12-HIV Vif or Nef protein interfere with the HIV replicative cycle? (i) F12-HIV Vif.
To investigate at which level the anti-HIV effects of F12-HIV Vif protein act, we followed the fate of HIV infecting F12-HIV vif-expressing cells through single-cycle infection experiments.
HeLa CD4 cells expressing F12-HIV Vif protein were infected at high multiplicity (MOI of 2) with DNase-pretreated preparations of strain NL4-3 (superimposable results were obtained by using the HTLVIIIB strain) and, after 1 h of virus absorption, cells were treated with trypsin for 15 min at 37°C. No differences in the kinetics of the first replication cycle of challenging HIV were observed between control and F12-HIV vif-expressing cells, as assessed either by analyzing the retrotranscription processes through PCR analysis, by measuring the amounts of neosynthesized intracellular HIV p24 viral protein, or by estimating the level of viral release in supernatants (data not shown). Thus, we investigated whether an impaired infectivity of HIV released after the first replicative cycle in F12-HIV vif-expressing cells could be on the basis of the anti-HIV effect. To verify this, equal amounts of HIV obtained after the first replication cycle in control or F12-HIV vif-expressing cells were utilized to challenge parental HeLa CD4 cells. As assessed by PCR analyses (Fig. 9I), products of the HIV early retrotranscription process were found in lower amounts in cells infected with supernatant from F12-HIV vif-expressing cells compared to those detectable in cells infected with supernatants from control cells. However, at later times (48 to 72 h after the infection), this difference was no longer detectable (not shown).
FIG. 9.
(I) PCR analysis of lysates of HeLa CD4 cells infected with supernatants obtained from a single HIV replication cycle in F12-HIV rev- or vif-expressing HeLa CD4 cells and, as controls, in parental HeLa CD4 or in CD4-negative HIV-resistant HeLa cells. Semiconfluent cell cultures were infected with HIV (MOI of 2), and, after 1 h of adsorption, free virus was removed by several washings followed by trypsin treatment (see Materials and Methods). After an additional 16 h of incubation, supernatants were collected and titrated for HIV p24 content. Levels of HIV p24 in supernatants of challenged HeLa cells were consistently below the sensitivity threshold of the ELISA. After DNase treatment, volumes of each supernatant normalized for HIV p24 content (except for the supernatant from infected HeLa cells) were used to infect parental HeLa CD4 cells (200 pg for 5 × 104 cells). These cells were finally harvested at both 6 and 16 h after infection and lysed. Samples corresponding to 2 × 104 cells were amplified at the same time with HIV-LTR (forward and reverse 1) (A)- and β-globin (B)-specific primers. The HIV-challenged cell types from which the supernatants were obtained to reinfect HeLa CD4 cells are indicated above each lane. Times (in hours) of cell collection after infection (p.i.) of HeLa CD4 cells are indicated at the bottom. Molecular sizes (in base pairs) of amplification products are shown. (II) PCR analysis of lysates of HeLa CD4 cells infected with DNase-treated supernatants obtained 48 h after cotransfections on rev-expressing HeLa CD4 cells of infectious molecular clone pNL4-3 HIV-1, together with vectors expressing wild-type or F12-HIV Vif protein. Infections and PCR analyses were performed as described above. Samples corresponding to 2 × 104 cells were amplified at the same time with HIV-LTR (A)- and β-globin (B)-specific primers. The plasmids transfected in rev-expressing HeLa CD4 cells giving rise to the HIV used to reinfect HeLa CD4 cells are indicated above each lane. C+, amplification products obtained from a lysate of 104 CEMss cells acutely infected with strain NL4-3. Times (in hours) of cell collection after infection (p.i.) of HeLa CD4 cells are indicated at the bottom. Molecular sizes (in base pairs) of amplification products are shown.
These results indicate that the anti-HIV effect observed in F12-HIV vif-expressing cells could be the consequence of a progressive impairment in the infectivity of challenging HIV, as soon as it replicates in cells expressing the F12-HIV Vif protein.
Furthermore, to definitely rule out the possibility that our results could be (even in part) the product of artifacts due to the use of stable cell clones, we reproduced through transient transfection experiments the mechanistic analyses described above. Therefore, an uncloned HeLa CD4 cell population expressing the rev gene was cotransfected with pcDNAI-based vectors expressing the wild-type or F12-HIV vif gene together with the pNL4-3 HIV infectious molecular clone with a molar ratio of 10:1, so that the viral genome could be preferentially expressed by the cells receiving the cotransfected plasmid too. Along with that already observed in single-cycle infections, no significant variations in HIV release were detected in cell cultures transfected with vector expressing the vif mutant with respect to those transfected with either the control vector or the vector expressing the wild-type Vif protein (data not shown). However, HIV emerging from cells transfected with F12-HIV Vif protein shows the characteristic delayed kinetics of retrotranscription when it reinfects parental HeLa CD4 cells (Fig. 9II). Thus, we may virtually exclude that the use of stably transfected cell clones led to misinterpretations about the effects of F12-HIV Vif protein on HIV replication.
Most of the above-described results for F12-HIV vif-induced anti-HIV effects were produced in vif-permissive cells (HeLa CD4 and CEMss), i.e., cells able to complement the HIV Vif function(s) (23). In order to add more insights about the mechanism of action of F12-HIV vif-induced HIV inhibition, we extend our observations to vif-nonpermissive cells, i.e., cells in which the Vif protein is absolutely required for HIV replication (e.g., PBLs and H9) (23). Therefore, PBLs were transduced with F12-HIV vif-expressing retrovirus vector and, after the NGFr-based immunoselection, were infected with the NL4-3 HIV-1 strain (MOI of 0.1). As shown in Fig. 10, expression of the Vif protein mutant was able to induce a strong inhibition of HIV replication even in vif-nonpermissive cells, indicating that the lack of vif complementing activity apparently does not influence the HIV resistance phenotype induced by F12-HIV vif expression.
FIG. 10.
(I) Amounts of HIV p24 in supernatants of PHA-activated human PBLs transduced with F12-HIV Vif-expressing retrovirus vector at different days after infection (p.i.) (MOI of 0.1) with the NL4-3 HIV-1 strain. As a control, PBLs transduced with the empty L-NGFr retrovirus vector were also infected. Challenging virus was adsorbed to 2 × 105 cells in 50 μl for 2 h at 37°C, and then cells were washed and refed with 1 ml of complete medium. (II) RT activities in supernatants of CEMss cells transduced with F12-HIV vif-expressing retrovirus vector at different days after infection (p.i.) with the vif-deleted HIV clone 6.9 (viral input, 2 and 20 ng of HIV p24/106 cells). As a control, CEMss cells transduced with the empty L-NGFr retrovirus vector were infected under the same conditions. Challenge with HIV was performed as already described for CEMss cells. Data from a representative of three different experiments are reported.
Finally, in order to test a possible effect in vif-permissive cells on HIV replication of F12-HIV Vif protein in the absence of the wild-type counterpart, CEMss cells transduced with F12-HIV vif-expressing retroviral vector were infected with 2 or 20 ng/106 cells of a vif-deleted HIV strain (clone 6.9), which is able to replicate in vif-permissive cells only (21). The results reported in Fig. 10II indicate that the expression of the F12-HIV Vif protein inhibits the replication of the vif-deleted HIV also, thus suggesting that the effect of the vif complementing activity could be negatively influenced by expression of the Vif protein mutant.
(ii) F12-HIV nef.
Single-cycle infection experiments similar to those described above were set up on F12-HIV nef-expressing HeLa CD4 cells. We assessed that the expression of the F12-HIV Nef protein does not interfere with the early steps of viral infection. In fact, no inhibition of either retrotranscription process (as observed by PCR analysis [not shown]) or intracellular HIV p24 protein production (which, in contrast, at the latest times after infection considered, was enhanced by almost twofold in F12-HIV nef-expressing cells [Fig. 11I]) was observed. Conversely, a strong decrease in HIV p24 levels was detected in supernatants from F12-HIV nef-expressing cells with respect to those from control cells (Fig. 11II). No significant variations in infectivity between the virus released from F12-HIV nef-expressing cells and that from control ones were observed by titrating equal amounts (as measured in picograms of HIV p24) of HIV (not shown).
FIG. 11.
Amounts of intracellular (I) and supernatant (II) HIV p24 in F12-HIV nef-expressing cells in a single-cycle infection experiment. F12-HIV nef and, as a control, hygromycin-resistant (Hr) HeLa CD4 cells were infected with HIV (MOI of 2) and then treated as described in the legend to Fig. 9. Values reported were obtained from cultures of 5 × 104 cells in 0.5 ml that were tested at different hours postinfection (p.i.). Results from a representative of three different experiments are reported.
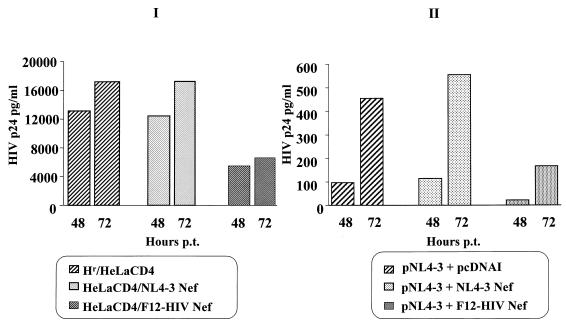
In order to verify whether the above-described inhibitory effect on HIV release could be detectable in cells expressing wild-type Nef protein, a pool of HeLa CD4 cell clones expressing the NL4-3 Nef protein was obtained. Cells expressing F12-HIV or NL4-3 Nef protein were transfected with the pNL4-3 HIV-1 infectious molecular clone, and supernatant HIV p24 levels were measured 48 and 72 h after transfection. As shown in Fig. 12I, no apparent effects on HIV release could be detected in supernatants from cells expressing wild-type nef, whereas F12-HIV nef-expressing cells, in agreement with single-cycle infection experiments (see above), released strongly reduced amounts of HIV p24 protein. Conversely, no variations in intracellular HIV p24 levels among different transfected cells were observed (not shown).
FIG. 12.
(I) Amounts of HIV p24 in supernatants of wild-type (NL4-3) or F12-HIV nef-expressing cells after transfection with the infectious molecular clone HIV-1 NL4-3. A total of 5 × 104 cells were seeded in 24-well plates and, after 24 h, transfected in triplicate with 2 μg of the pNL4-3 plasmid. After an overnight incubation, cell cultures were washed and complete medium was added. Amounts of HIV p24 protein (whose levels 24 h posttransfection were, under all conditions, below the sensitivity threshold of the ELISA) were measured at the indicated times posttransfection (p.t.). (II) Amounts of HIV p24 in supernatants of HeLa CD4 cells cotransfected with the infectious molecular clone HIV-1 pNL4-3 together with vectors expressing wild-type or F12-HIV Nef proteins. A total of 2 × 105 cells were seeded in 6-well plates and after 24 h were cotransfected in duplicate with 1 μg of infectious molecular clone pNL4-3 and a 10-fold molar excess of pcDNAI-based nef-expressing vectors. Measurements of HIV p24 protein were performed at the indicated times posttransfection (p.t.). Values from a representative of three different experiments are reported.
Taken together, these data indicate that the HIV inhibitory effect observed by infecting F12-HIV nef-expressing cells is seemingly the result of a detrimental action that the Nef-mutated protein exerts on a late step in the HIV life cycle (i.e., assembling and/or release).
Similarly to the above-described results obtained with F12-HIV vif-expressing constructs, in order to reproduce in a transient transfection system the mechanistic analyses performed in F12-HIV cells stably expressing Nef, pcDNAI-based vectors expressing wild-type or F12-HIV nef genes were cotransfected with the pNL4-3 infectious molecular clone (molar ratio, 10:1) on HeLa CD4 cells. As shown in Fig. 12II, we fairly reproduced the inhibition of HIV release induced by F12-HIV nef expression already observed in stable cell clones. Thus, also in the case of Nef mutant protein, we may exclude that results on HIV inhibition could be generated by artifacts due to the cell clone selection.
DISCUSSION
A number of HIV mutagenized viral proteins able to inhibit the replication of wild-type HIV have been described elsewhere (28, 38, 46, 67). An anti-HIV effect induced by the expression of the wild-type HIV Env glycoprotein has also been reported (63, 64); the latter is, however, correlated with the masking of the CD4 receptors, rather than with a direct negative action on the viral life cycle. Furthermore, wild-type nef gene expression can induce the protection of host cells from HIV infection as the consequence of the down-regulation of CD4 HIV receptors (3, 24). Negative trans-dominant effects on HIV viral replication induced by mutagenized forms of Gag HIV proteins have also been reported (67). These results were obtained by infecting with HIV HeLa CD4 cells stably expressing mutagenized gag genes inserted in an env−/nef− HIV genome. Conversely, the data here presented describe the inhibitory effects on HIV replication induced by a full-length, naturally occurring F12-HIV Gag protein. The F12-HIV gag gene presents 10 amino acidic substitutions that have no counterparts in any other gag gene from HIV-1 genomes sequenced so far. In particular, two substitutions reside in p17 (five on p24 and three on p15 regions) (Fig. 13), and they do not overlap those reported in the already described HIV-inhibiting gag mutants (67).
FIG. 13.
Amino acid sequences of F12-HIV Gag, Vif, and Nef proteins. The amino acid substitutions typical (i.e., not detectable in the sequences of any T-tropic HIV-1 isolates [HXB2, BRU, SF2, NL4-3, PV22, and MN] used to construct the reference consensus sequence) of the F12-HIV genes are indicated.
Considering that higher levels of the p55 Gag protein were detected in cells expressing the entire F12-HIV genome with respect to those coexpressing rev and gag genes only, it is likely that the data here presented underestimate the inhibitory effect of F12-HIV Gag protein expressed in the context of the whole viral genome.
The function of wild-type Vif protein was originally correlated with the infectivity of HIV particles. Recently, it has been shown that Vif associates with viral core structures (37) and that defects in Gag processing have been observed in PBLs infected with vif-defective HIV (57). In general, cells that do not sustain the replication of vif-defective HIV strains are defined as vif-nonpermissive cells (e.g., PBLs and H9), whereas cell lines in which Vif protein is dispensable for HIV replication are defined as vif-permissive cells (e.g., C8166, HeLa CD4, and CEMss). This was explained by admitting the presence of a vif cellular homolog in vif-permissive cells or, alternatively, of a vif-responsive HIV inhibitor in vif-nonpermissive ones (23, 54). vif is the most mutated F12-HIV gene, bearing 31 point mutations leading to 14 amino acid substitutions characteristic of the F12-HIV genome (Fig. 13). In this paper, we originally show an inhibitory effect induced by a Vif mutant protein in vif-permissive cells as well as in vif-nonpermissive ones. Thus, the effectiveness of F12-HIV Vif protein in inhibiting HIV replication seems independent of the ability of challenged cells to complement the wild-type vif function(s). Furthermore, inhibition of the replication of vif-deleted HIV in vif-permissive cells constitutively expressing the F12-HIV Vif protein may suggest that this mutant protein is able to negatively influence the vif-complementing activity. Taken together, the data presented here disclose the possibility that F12-HIV Vif protein may counteract both wild-type protein and, possibly, the vif cellular homolog function(s).
Studies of the level of action of the F12-HIV Vif-induced anti-HIV effect indicate that (i) in the first infection cycle, the presence of the F12-HIV Vif protein does not influence HIV replication and (ii) the replication efficiency of HIV emerging from F12-HIV vif-expressing cells after the first replication cycle is markedly reduced, as demonstrated by the consistent inhibition in the early retrotranscription step observed by PCR analysis of infected vif-permissive cells.
The following hypotheses may be considered. (i) The observed reduced amounts of retrotranscription products may have derived from an impaired ability in the cell entry of HIV obtained from F12-HIV vif-expressing cells. This could be the consequence of an altered viral assembly in producer cells, as was already observed through infection with HIV coding for mutagenized Vif proteins (8, 31, 54, 57). (ii) Alternatively (or in addition), F12-HIV Vif protein is encapsidated in (at least) part of the emerging HIV and interferes with the early replication step in subsequent viral cycles. This could be conceivable by admitting a role for Vif protein in the early retrotranscription process, as has been suggested by the evidence that vif− HIV strains are inhibited in the early replication steps when they infect cells unable to support the vif deficiency (vif-nonpermissive cells) (59, 70).
The inhibition of retrotranscription was observed in parental HeLa CD4 cells at early times after infection but was not still effective at later times. This was probably due to the replication of HIV escaping (in view of the high MOI used) the F12-HIV Vif-induced inhibition effect during the first replication cycle or, alternatively, to the effect of intracellular vif complementing factor(s) overcoming the possible negative trans-dominant effect of F12-HIV Vif protein. Similar mechanisms could be based on the reduced but not abolished infectivity of retrovirions released by cells chronically infected with HIV-1 that were transduced with F12-HIV vif-expressing retrovirus vector.
We previously published that when the vif gene of the syncytium-inducing, rapid-high NL4-3 strain was replaced by the F12-HIV vif one, the phenotype of the resulting chimeric provirus transfected in vif-permissive cells (i.e., HeLa CD4 and SupT1) was dramatically mutated in a slow-low, non-syncytium-inducing HIV (11). This observation, together with the results reported here, demonstrates that expression of the F12-HIV vif gene is able to negatively control HIV replication either in cis or in trans configurations.
It has been reported that Nef protein expression (i) down-regulates the CD4 HIV receptors (3), (ii) increases both proviral DNA synthesis (2) and HIV infectivity (45, 60), and (iii) is involved in SIV-induced AIDS pathogenesis (35). The F12-HIV nef gene shows 19 point mutations resulting in 13 amino acid substitutions (10), 3 of which have no counterparts in any nef gene sequenced so far (Fig. 13). The phenotype induced by the F12-HIV Nef protein seems quite different from that described for wild-type Nef. In fact, not only is F12-HIV Nef unable to down-regulate CD4 receptors, but in HeLa CD4 cells it induces a strong inhibition rather than an increase in HIV infectivity. To our knowledge, this is the first report concerning a negative effect of the Nef protein on the HIV life cycle acting through a mechanism independent of CD4 down-regulation. The results from single-cycle infection experiments demonstrated that the F12-HIV Nef protein exerts its anti-HIV activity in a step subsequent to viral protein synthesis. It has been proposed that wild-type Nef increases HIV viral infectivity acting at the level of viral core formation (2, 44). Our data about the F12-HIV Nef level of action seem compatible with this model, by admitting that the mutated Nef protein is able to act in the appropriate HIV replication step, but likely in a wrong manner. The already reported evidence that Nef could be dispensable for HIV replication in immortalized cell lines (45, 60) may enforce the hypothesis that F12-HIV Nef protein exerts its HIV inhibitory effect through a direct or indirect interaction with viral proteins other than wild-type Nef. This was also suggested by the evidence that expression of the nef mutant inhibits replication of HTLVIIIB, an HIV-1 strain defective in nef expression. The recently described ability of wild-type Nef protein to induce phosphorylation of HIV matrix Gag protein through binding of a cellular kinase (65) could be considered a demonstration of the possibility that the Nef protein influences the structure of other HIV proteins.
HIV-1 emerging from chronically infected cells expressing F12-HIV Nef protein was impaired in its infectivity in spite of essentially unvaried amounts of released retrovirions. These data may apparently contrast with the evidence that in acute HIV infection, an inhibition at the level of virus assembling and/or release could be observed in F12-HIV nef-expressing cells, without any evident impairment in the infectivity of released virus. It is possible that different models of virus-cell interactions (i.e., acute versus chronic infections) allowed detection of different steps during which F12-HIV nef expression could inhibit HIV replication. In particular, in cells chronically infected with HIV, the F12-HIV Nef protein may induce an anti-HIV effect through its encapsidation in released virions (as demonstrated for wild-type Nef proteins) (44, 48), thus exerting its anti-HIV effect on target cells.
The block of HIV replication induced by F12-HIV Vif or Nef protein persists at MOIs of 0.2 and 0.5 TCID50/cell, respectively. This is a substantial improvement with respect to the higher MOI at which viral interference was observed in HeLa CD4 clones expressing the full-length F12-HIV genome (0.017 TCID50/cell) (17), which indeed also expresses HIV-enhancing proteins such as Tat, Rev, and Vpu.
In conclusion, the results regarding the negative effects of either Gag, Vif, or Nef protein in HIV replication seem interesting since they (i) definitively demonstrate the involvement of these proteins in F12-HIV-induced homologous viral interference, (ii) could represent a model by which to study the already largely unknown functions that Vif and Nef proteins play in the HIV life cycle, and (iii) allow us to consider F12-HIV gag, vif, and nef genes as new candidates for experiments in anti-HIV gene therapy. This perspective is strongly encouraged by the evidence that the anti-HIV effect of F12-HIV Vif or Nef protein is not restricted to laboratory-adapted HIV-1 strains but is fully operative also when cells are challenged with T-tropic HIV-1 clinical isolates. Moreover, the ability of retroviral constructs expressing F12-HIV Vif or Nef protein to inhibit the viral infectivity of HIV released from already infected cells is a very important feature regarding the attempt to perform gene therapy experiments in cells also from HIV-infected AIDS patients.
ACKNOWLEDGMENTS
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: antiserum to HIV-1 Gag from Michael Phelan; antiserum to HIV-1 RT from Division of AIDS, NIAID; antiserum to HIV-1 protease C-terminal peptide from Bruce Korant; antiserum to HIV-1 integrase from Duane P. Grandgenett (27); antiserum HT3 to HIV-1 Env (12, 40, 53); antiserum to HIV-1 Rev from David Rekosh and Marie Louise Hammarskjold; antiserum to HIV-1 Tat and antiserum to HIV-1 Vpr (25, 36) from Bryan Cullen; antiserum to Vpu from Frank Maldarelli and Klaus Strebel; antiserum to HIV-1 Vif from Dana Gabuzda (26); and antiserum to HIV-1 Nef from L. Ratner (9, 47). We are grateful to Elisa Vicenzi and Guido Poli, Dibit Institute, Milan, Italy, who kindly provided the T-tropic HIV-1 clinical isolates, and to Genoveffa Franchini, National Institutes of Health, Bethesda, Md., and B. Ensoli, Laboratory of Virology, Istituto Superiore di Sanità, Rome, Italy, for their generous gift of the vif-deleted 6.9 HIV molecular clone. We are indebted to Cristiana Chelucci, Laboratory of Hematology and Oncology, Istituto Superiore di Sanità, Rome, Italy, for critical reading of the manuscript and to A. Lippa and F. M. Regini for excellent editorial assistance.
This work was supported by grants from AIDS Project of the Ministry of Health, Rome, Italy.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiken C, Konner J, Landau N R, Lenburg M E, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 4.Bernier R, Tremblay M. Homologous interference resulting from the presence of defective particles of human immunodeficiency virus type 1. J Virol. 1995;69:291–300. doi: 10.1128/jvi.69.1.291-300.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanc D, Patience C, Schulz T F, Weiss R, Spire B. Transcomplementation of Vif-HIV-1 mutants in CEM cell suggests that Vif affects late steps of the viral life cycle. Virology. 1993;193:186–192. doi: 10.1006/viro.1993.1114. [DOI] [PubMed] [Google Scholar]
- 6.Bogerd H P, Fridell R A, Madore S, Cullen B R. Identification of a novel cellular co-factor for the Rev/Rex class of regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 7.Bona R, d’Aloja P, Olivetta E, Modesti A, Modica A, Ferrari G, Verani P, Federico M. Aberrant, noninfectious HIV-1 particles are released by chronically infected human T-cells transduced with a retroviral vector expressing an interfering HIV-1 variant. Gene Ther. 1997;4:1085–1092. doi: 10.1038/sj.gt.3300501. [DOI] [PubMed] [Google Scholar]
- 8.Borman A M, Quillent C, Charneau P, Dauguet C, Clavel F. Human immunodeficiency virus type 1 vif mutant particles from restrictive cells: role of Vif in correct particle assembly and infectivity. J Virol. 1995;69:2058–2067. doi: 10.1128/jvi.69.4.2058-2067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant M L, Ratner L, Duronio R J, Kishore N S, Devadas B, Adams S P, Grodon J I. Incorporation of 12-methoxydodecanoate into the human immunodeficiency virus Gag polyprotein precursor inhibits its proteolytic processing and virus production in a chronically infected human lymphoid cell line. Proc Natl Acad Sci USA. 1991;88:2055–2059. doi: 10.1073/pnas.88.6.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlini F, Federico M, Equestre M, Ricci S, Ratti G, Zibai Qi, Verani P, Rossi G B. Sequence analysis of an HIV-1 proviral DNA from a nonproducer chronically infected Hut-78 cellular clone. J Viral Dis. 1992;1:40–55. [Google Scholar]
- 11.Carlini F, Nicolini A, d’Aloja P, Federico M, Verani P. The non-producer phenotype of the human immunodeficiency virus type 1 provirus F12/HIV-1 is the result of multiple genetic variations. J Gen Virol. 1996;77:2009–2013. doi: 10.1099/0022-1317-77-9-2009. [DOI] [PubMed] [Google Scholar]
- 12.Crise B, Rose J K. Human immunodeficiency virus type 1 glycoprotein precursor retains CD4-p56 lck complex in the endoplasmic reticulum. J Virol. 1992;66:2296–2301. doi: 10.1128/jvi.66.4.2296-2301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daly T J, Cook K S, Gray G S, Maione T E, Rusche J R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature. 1989;342:816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- 14.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N R. Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.di Marzo Veronese F, Copeland T D, De Vico A L, Rahman R, Oroszlan S, Gallo R C, Sarnagadharan M G. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986;231:1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]
- 16.Federico M, Nappi F, Ferrari G, Chelucci C, Mavilio F, Verani P. A nonproducer, interfering human immunodeficiency virus (HIV) type 1 provirus can be transduced through a murine leukemia virus-based retroviral vector: recovery of an anti-HIV mouse/human pseudotype retrovirus. J Virol. 1995;69:6618–6626. doi: 10.1128/jvi.69.11.6618-6626.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Federico M, Nappi F, Bona R, d’Aloja P, Verani P, Rossi G B. Full expression of transfected nonproducer interfering HIV-1 proviral DNA abrogates susceptibility of human HeLa CD4+ cells to HIV. Virology. 1995;206:76–84. doi: 10.1016/s0042-6822(95)80021-2. [DOI] [PubMed] [Google Scholar]
- 18.Federico M, Taddeo B, Carlini F, Nappi F, Verani P, Rossi G B. A recombinant retrovirus carrying a nonproducer human immunodeficiency virus (HIV) type 1 variant induces resistance to superinfecting HIV. J Gen Virol. 1993;74:2099–2110. doi: 10.1099/0022-1317-74-10-2099. [DOI] [PubMed] [Google Scholar]
- 19.Federico M, Titti F, Buttò S, Orecchia A, Carlini F, Taddeo B, Macchi B, Maggiano N, Verani P, Rossi G B. Biologic and molecular characterization of producer and nonproducer clones from Hut-78 cells infected with a patient HIV isolate. AIDS Res Hum Retroviruses. 1989;5:385–396. doi: 10.1089/aid.1989.5.385. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y, Broder C C, Kennedy P, Berger E. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 21.Fisher A G, Ensoli B, Ivanoff L, Chamberlain M, Petteway S, Ratner L, Gallo R C, Wong-Staal F. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237:888–893. doi: 10.1126/science.3497453. [DOI] [PubMed] [Google Scholar]
- 22.Fritz C C, Zapp M L, Green M R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 23.Gabuzda D H, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine W A, Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia J V, Alfano J, Miller A D. The negative effect of human immunodeficiency virus type 1 Nef on cell surface CD4 expression is not species specific and requires the cytoplasmic domain of CD4. J Virol. 1993;67:1511–1516. doi: 10.1128/jvi.67.3.1511-1516.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrett E D, Tiley L S, Cullen B R. Rev activates expression of the human immunodeficiency virus type 1 vif and vpr gene products. J Virol. 1991;65:1653–1657. doi: 10.1128/jvi.65.3.1653-1657.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goncalves J, Jallepalli P, Gabuzda D H. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J Virol. 1994;68:704–712. doi: 10.1128/jvi.68.2.704-712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grandgenett D P, Goodarzi G. Folding of the multidomain human immunodeficiency virus type-I integrase. Protein Sci. 1994;3:888–897. doi: 10.1002/pro.5560030604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green M, Ishino M, Loewenstein P M. Mutational analysis of HIV-1 Tat minimal domain peptides: identification of trans-dominant mutants that suppress HIV-LTR-driven gene expression. Cell. 1989;58:215–223. doi: 10.1016/0092-8674(89)90417-0. [DOI] [PubMed] [Google Scholar]
- 29.Haurer C A, Getty R, Tyrocinski M L. Epstein-Barr virus episome-based promoter function in human myeloid cells. Nucleic Acids Res. 1989;17:1989–2003. doi: 10.1093/nar/17.5.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heaphy S, Dingwall C, Ernberg I, Gait M J, Green S M, Karn J, Lowe A D, Singh M, Skinner M A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990;60:685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- 31.Hoglund S, Ohagen A, Lawrence K, Gabudza D. Role of Vif during packing of the core of HIV-1. Virology. 1994;201:349–355. doi: 10.1006/viro.1994.1300. [DOI] [PubMed] [Google Scholar]
- 32.Johnson D, Lanahan A, Buck C R, Seghal A, Morgan C, Mercer E, Bothwell M, Chao M. Expression and structure of the human NGF receptor. Cell. 1986;47:545–554. doi: 10.1016/0092-8674(86)90619-7. [DOI] [PubMed] [Google Scholar]
- 33.Jowett J B M, Planelles V, Poon B, Shah N P, Chen M-L, Chen I S Y. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karacostas V, Nagashima K, Gonda M A, Moss B. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc Natl Acad Sci USA. 1989;86:8964–8967. doi: 10.1073/pnas.86.22.8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kestler H W, Ringler D J, Mori K, Panicali D L, Sehegal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 36.Lavallée C, Cohen E A. HIV-1 HxBc2 strain encodes a truncated vpr gene product of 78 amino acids. J Acquired Immune Defic Syndr. 1993;6:529–539. doi: 10.1097/00126334-199305000-00014. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, Wu X, Newman M, Shaw G M, Hahn B H, Kappes J C. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J Virol. 1995;69:7630–7638. doi: 10.1128/jvi.69.12.7630-7638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malim M H, Freimuth W W, Liu J, Boyle T J, Lyerly H K, Cullen B R, Nabel G J. Stable expression of transdominant Rev protein in human T cells inhibits human immunodeficiency virus replication. J Exp Med. 1992;176:1197–1201. doi: 10.1084/jem.176.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markowitz D, Goff S, Bank A. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsushita S M, Robert-Guroff M, Koito A, Hattori T, Hoshino H, Javaherian K, Takatsuki K, Putney S. Characterization of a human immunodeficiency virus neutralizing monoclonal antibody and mapping of the neutralizing epitope. J Virol. 1988;62:2107–2114. doi: 10.1128/jvi.62.6.2107-2114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mavilio F, Ferrari G, Rossini S, Nobili N, Bonini C, Casorati G, Traversari C, Bordignon C. Peripheral blood lymphocytes as target cells of retroviral vector-mediated gene transfer. Blood. 1994;83:1988–1997. [PubMed] [Google Scholar]
- 42.Miller A D, Buttimore C. Redesign of retrovirus packaging cell line to avoid recombinant leading to helper virus production. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–982. [PMC free article] [PubMed] [Google Scholar]
- 44.Miller A D, Warmerdam M T, Page K A, Feinberg M B, Greene W C. Expression of the human immunodeficiency virus type 1 (HIV-1) nef gene during HIV-1 production increases progeny particle infectivity independently of gp160 or viral entry. J Virol. 1995;69:579–584. doi: 10.1128/jvi.69.1.579-584.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller M D, Warmerdam M T, Gaston I, Greene W C, Feinberg M B. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modesti N, Garcia J, Debouck C, Peterlin M, Gaynor R. Trans-dominant Tat mutants with alterations in the basic domain inhibit HIV-1 gene expression. New Biol. 1991;3:759–768. [PubMed] [Google Scholar]
- 47.Niederman T M J, Garcia J V, Hastings W R, Luria S, Ratner L. Human immunodeficiency virus type 1 Nef protein inhibits NF-kB induction in human T cells. J Virol. 1992;66:6213–6219. doi: 10.1128/jvi.66.10.6213-6219.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandori M W, Fitch N J S, Craig H M, Richman D D, Spina C A, Guatelli J C. Producer-cell modification of human immunodeficiency virus type 1: Nef is a virion protein. J Virol. 1996;70:4283–4290. doi: 10.1128/jvi.70.7.4283-4290.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porter D C, Melsen L R, Compans R W, Morrow C D. Release of virus-like particles from cells infected with poliovirus replicons which express human immunodeficiency virus type 1 Gag. J Virol. 1996;70:2643–2649. doi: 10.1128/jvi.70.4.2643-2649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell D F J, Martin M A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rein A, Schultz A. Different recombinant murine leukemia viruses use different cell surface receptors. Virology. 1984;136:144–152. doi: 10.1016/0042-6822(84)90255-1. [DOI] [PubMed] [Google Scholar]
- 52.Rossi G B, Verani P, Macchi B, Federico M, Orecchia A, Nicoletti L, Buttò S, Lazzarin A, Mariani G, Ippolito G, Manzari V. Recovery of HIV-related retroviruses from Italian patients with AIDS or AIDS-related complex and from asymptomatic at-risk individuals. Ann NY Acad Sci. 1987;511:390–400. doi: 10.1111/j.1749-6632.1987.tb36268.x. [DOI] [PubMed] [Google Scholar]
- 53.Rsuche J R, Lynn D L, Robert-Guroff M, Langlosis A J, Lyerly H K, Carson H, Krohn K, Ranki A, Gallo R C, Bolognesi D P, Putney S D, Matthews T J. Humoral immune response to the entire human immunodeficiency virus envelope glycoprotein made in insect cells. Proc Natl Acad Sci USA. 1987;84:6924–6928. doi: 10.1073/pnas.84.19.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakai H, Shibata R, Sakuragi J I, Sakuragi S, Kawamura M, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 Vif protein for maturation of virus particles. J Virol. 1993;67:1663–1667. doi: 10.1128/jvi.67.3.1663-1666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 56.Schubert U, Clouse K A, Strebel K. Augmentation of virus secretion by the human immunodeficiency virus type 1 Vpu protein is cell type independent and occurs in cultured human primary macrophages and lymphocytes. J Virol. 1995;69:7699–7711. doi: 10.1128/jvi.69.12.7699-7711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simm M, Shahabuddin M, Chao W, Allan J S, Volsky D J. Aberrant Gag protein composition of a human immunodeficiency virus type 1 vif mutant produced in primary lymphocytes. J Virol. 1995;69:4582–4586. doi: 10.1128/jvi.69.7.4582-4586.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith A J, Cho M-I, Hammarskjold M L, Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into virus-like particles. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sova P, Volsky D J. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spina C A, Kwoh T J, Chowers M Y, Guatelli J C, Richman D D. The importance of Nef in the induction of human immunodeficiency virus type 1 replication for primary quiescent CD4 lymphocytes. J Exp Med. 1994;179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steck F, Rubin H. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. I. Establishment of interference. Virology. 1966;29:628–641. doi: 10.1016/0042-6822(66)90287-x. [DOI] [PubMed] [Google Scholar]
- 62.Steck F, Rubin H. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. II. Early steps of infection by RSV or cells under conditions of interference. Virology. 1966;29:642–653. doi: 10.1016/0042-6822(66)90288-1. [DOI] [PubMed] [Google Scholar]
- 63.Steffy K R, Wong-Staal F. Tansdominant inhibition of wild-type human immunodeficiency virus type 2 replication by an envelope deletion mutant. J Virol. 1993;67:1854–1859. doi: 10.1128/jvi.67.4.1854-1859.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stevenson M, Meier C, Mann A M, Chapman N, Waslak A. Envelope glycoprotein of HIV induces interference and cytolysis resistance in CD4+ cells: mechanism for persistence in AIDS. Cell. 1989;83:483–486. doi: 10.1016/0092-8674(88)90168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swingler S, Gallay G, Camaur D, Song J, Abo A, Trono D. The Nef protein of human immunodeficiency virus type 1 enhances serine phosphorylation of the viral matrix. J Virol. 1997;71:4372–4377. doi: 10.1128/jvi.71.6.4372-4377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taddeo B, Federico M, Titti F, Rossi G B, Verani P. Homologous superinfection of both producer and nonproducer HIV-infected cells is blocked at a late retrotranscription step. Virology. 1993;194:441–452. doi: 10.1006/viro.1993.1283. [DOI] [PubMed] [Google Scholar]
- 67.Trono D, Feinberg M B, Baltimore D. HIV-1 Gag mutants can dominantly interfere with the replication of the wild-type virus. Cell. 1989;59:113–120. doi: 10.1016/0092-8674(89)90874-x. [DOI] [PubMed] [Google Scholar]
- 67a.Vicenzi, E. Personal communication.
- 68.Vogt P K, Ishizaki R. Reciprocal patterns of genetic resistance to avian tumor viruses in two lines of chicken. Virology. 1965;26:664–672. doi: 10.1016/0042-6822(65)90329-6. [DOI] [PubMed] [Google Scholar]
- 69.Volsky D J, Simm M, Shahabuddin M, Li G, Chao W, Potash M J. Interference to human immunodeficiency virus type 1 infection in the absence of downmodulation of the principal virus receptor, CD4. J Virol. 1996;70:3823–3833. doi: 10.1128/jvi.70.6.3823-3833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wigler M, Sweet R, Sim G K, Wold B, Pellicer A, Lacy E, Maniatis T, Silverstein S, Awel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979;16:758–777. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]