Highlights
-
•
Development of safe and effective ASFV vaccines remains an essential need.
-
•
Summary of the research on the gene/protein-based ASFV vaccines.
-
•
Detailed description of 11 major structural proteins.
-
•
Emphasis on the roles of the viral gene/proteins in stimulation of the specific immunity.
-
•
Discussion on the other factors that may influence vaccine development.
Keywords: African swine fever virus, Vaccine, Immunization regimen, Structural proteins, Specific immunity
Abstract
African swine fever (ASF) is a highly contagious acute hemorrhagic viral disease, with the mortality rate of up to 100 % in domestic pigs. In recent years, ASF outbreaks have caused huge economic losses in numerous countries and regions, especially in Asia. Therefore, there is a pressing need to develop safe and effective vaccines against infection of the causative pathogen, African swine fever virus (ASFV). ASFV contains a large genome composed of double-stranded DNA with a size of 170–194 kb, which encodes nearly 200 viral proteins. Understanding the function of these complex genes/proteins and their roles in the generation of protective immunity will help in the development of ASFV vaccines. In this article, the gene/protein-based vaccine candidate are summarized, and the structural proteins which have been previously reported to protect animals from the virus challenge were emphatically described.
Graphical abstract
1. ASF prevalence
The African swine fever is an acute, virulent and hemorrhagic infectious disease, which can cause very high mortality (up to 100 %) in domestic pigs and wild boars. The disease poses a constant threat to the global swine industry. Infected pigs, Ornithodoros ticks and contaminated materials can all contribute to the rapid spread of ASFV (Dixon et al., 2020; Plowright et al., 1969). Interestingly, ASFV appears to establish a long-term persistent/latent infection in pigs that were cured previously from ASFV infection (Chambaro et al., 2020). Studies have shown that the virulence or route of infection do not affect the ability of the ASFV infected animals to shed virus to the surrounding environment for at least 70 days after infection (de Carvalho Ferreira et al., 2012). The large-scale quarantine and culling of infected animals are widely used as the standard procedures for the prevention and control of ASFV epidemic. Additionally, biosecurity measures are implemented to protect the feeding environment (De Lorenzi et al., 2020).
ASF was first identified in Kenya in 1921, and since then it has been prevalent in Africa, affecting more than 30 African countries (Penrith et al., 2019; Sánchez-Vizcaíno et al., 2015). ASFV was initially introduced in Portugal in 1957. However, it re-emerged in Lisbon, Portugal in 1960, and then rapidly spread to Spain, Sardinia, and other western European countries. Until the mid-1990s, except in Sardinia, where it was still endemic, it was eradicated in the western Europe (Cwynar et al., 2019). The first reported outbreak of ASFV in Russia was in 1977. In 2007, ASFV was introduced to Georgia from the Caucasus region. From then on, it spread to many eastern European countries including the Russian Federation and Ukraine (Dixon et al., 2020). China experienced a large-scale outbreak of ASF in 2018, which was the first occurrence of ASF in Asia. The isolated strain showed a high degree of homology with the Georgia strain (Ge et al., 2018; Zhao et al., 2019). Since then, the virus further spread to other Asian countries, including Mongolia, North Korea, Cambodia, Laos, the Philippines, Timor-Leste, and Papua New Guinea (Tran et al., 2021). In 2021, ASF broke out in the Dominican Republic (Gonzales et al., 2021), marking the first infection detected in the Western hemisphere since 1984. This incident brought the attention of the North American pig industry, which then began to be vigilant and concerned about the invasion of ASF (https://www.avma.org/javma-news/2021-09-15/african-swine-fever-reaches-caribbean) (Goonewardene et al., 2022). In January 2022, another two countries reported ASFV infection: North Macedonia and Thailand (https://www.woah.org/en/disease/african-swine-fever/#ui-id-2) (Zhao et al., 2023). To date, ASF has been detected and reported in 74 countries since 2005 (https://www.woah.org/en/disease/african-swine-fever/#ui-id-2).
2. ASFV and ASFV vaccine
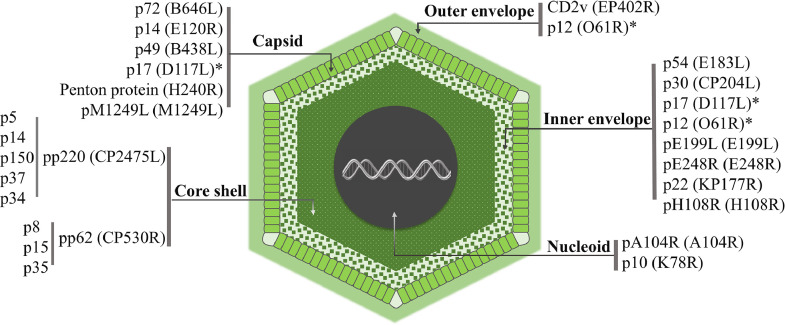
ASFV is a double-stranded large DNA arbovirus, which is the only member of the Asfarviridae family. ASFV has an icosahedral envelope structure with an average diameter of 200 nm and consists of five parts: Outer envelope, Capsid, Inner envelope, Core shell, and Nucleoid (Fig. 1). Its genome ranges in length from about 170 to 194 kb with the open reading frames (ORFs) length of 150–167 kb. Based on the variable 3′-end of the B646L gene, that encodes a main capsid protein, p72, (Boshoff et al., 2007), ASFV is defined as 24 genotypes (I–XXIV) (Njau et al., 2021). The genome can encode 150–200 proteins, including 68 structural proteins and more than 100 non-structural proteins (Alejo et al., 2018; Wang et al., 2021b). The large number of proteins indicates that the structure of ASFV is rather complex and that they may play different roles during viral invasion and proliferation. The functions of most of the proteins are still unclear.
Fig. 1.
Schematic diagram of the ASFV structure and location of some structural proteins. ASFV is composed of an outer envelope, capsid, inner membrane, inner capsid, and nucleoid. The proteins in each layer play different roles at different stages of ASFV infection. * Indicates that the protein can be detected in different parts of the virion.
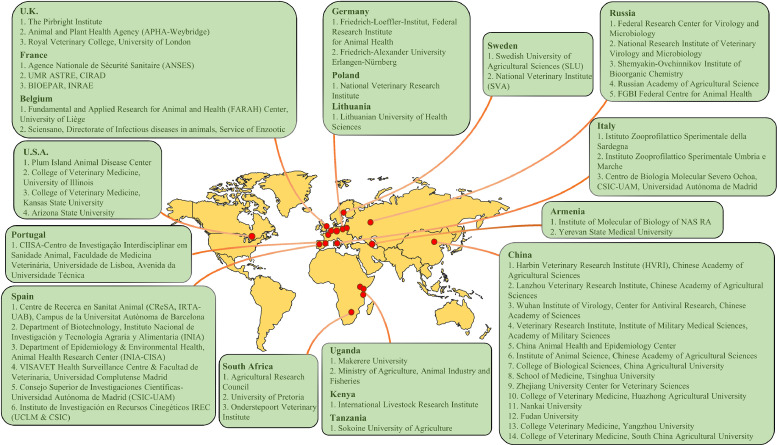
Laboratories around the world are now racing to accelerate the development of ASFV vaccines (Fig. 2). With the joint efforts of scientists, the attenuated vaccine has made a great progress. Most of the live attenuated vaccines candidate for ASFV have demonstrated 100 % protection experimentally (Liu et al., 2021a). An attenuated vaccine strain, ASFV-G-ΔI177L, developed by the Plum Island Animal Disease Center of the United States (PIADC-USDA) has been proven to be safe and highly efficacious in the challenge studies using parental ASFV-G (Borca et al., 2021). The safety of the ASFV-G-∆I177L was further evaluated by NAVETCO, a company of the Ministry of Agriculture and Rural Development in Vietnam (Tran et al., 2022). The results demonstrated that the pigs who received high viral loads of the attenuated virus didn't develop any clinical symptoms associated with ASF up to 90 or 180 days after vaccination. In addition, the results of necropsies conducted at the end of the experiment also confirmed that no pathological changes related to ASF had occurred (Borca et al., 2023). In February 2023, AVAC Vietnam and PIADC-USDA set out to promote another ASFV vaccine, which contained six genes knocked out strain (ASFV-G-△MGF): MGF505-1R, MGF360-12L, MGF360-13L, MGF360-14L, MGF505-2R and MGF50-3R (O'Donnell et al., 2015). This vaccine has been reported to provide excellent protection against the Georgia strain (ASFV-G) in animals after a single intramuscular injection (O'Donnell et al., 2015). The vaccine candidate was passaged five times in domestic pigs according to VICH (International Cooperation on Harmonization of Technical Requirements for Registration of Veterinary Medicinal Products) guideline 41, and there was no significant virulence reversion observed (Deutschmann et al., 2023). Attenuated live vaccine candidate HLJ/18-7GD from China had the following seven genes deleted: MGF505-1R, MGF505-2R, MGF505-3R, MGF360-12L, MGF360-13L, MGF360-14L, and EP402R (Chen et al., 2020). It provided protection against challenges with a prevalent highly virulent genotype II HLJ/18-like variant and Georgia07-like strain within the 28 days observation (Wang et al., 2024). However, HLJ/18-7GD did not induce solid cross-protection against genotype I low virulent ASFV challenge in pigs (Wang et al., 2024). Recently, the research team of Harbin Veterinary Research Institute in China isolated three recombinant strains of ASFV genotype I and genotype II, which are reported to produce high lethality and high transmission rates, as well as resistance towards the existing ASFV vaccine, which was derived from the genotype II (Zhao et al., 2023). The emergence of recombinant ASFV strains has imposed serious challenges to the prevention and control of ASF epidemics.
Fig. 2.
Major laboratories worldwide conducting research on ASFV vaccine.
Currently, biological control and culling remain the primary methods for controlling spread of the ASFV. Therefore, development of an effective ASFV vaccine is urgent and necessary. The ASFV vaccines developed so far have failed to provide complete protection against both homologous and heterologous viral strains infections. Inactivated virus vaccines are considered to be safe, though previous experience suggests that inactivated ASFV vaccine may not only fail to provide complete immune protection for domestic pigs (Stone and Hess, 1967), but also even produce antibody-dependent enhancement (ADE) effect to further exacerbating the symptoms of infected pigs (Yoon et al., 1996). The attenuated vaccines have been shown to induce neutralizing antibodies in the vaccinated animals (Silva et al., 2022) to provide excellent protection for the animals. However, there are still several issues that need to be addressed, such as differentiating infected individuals from vaccinated individuals (DIVA), persistent infection in pigs leading to immunosuppression, low viral symptoms, poor cross-protection, reassortment with wild-type strain, and reemergence of virulence. Therefore, gene engineered vaccines such as nucleic acid vaccines and protein subunit vaccines against ASFV have attracted the attention of scholars. These types of vaccines can stimulate both humoral and cellular immunity, leading to production of the protective antibodies. For the development of such vaccines, it is crucial to select appropriate and effective antigens or antigen combinations. Additionally, appropriate antigen expression vectors and vaccine adjuvants are also indispensable factors for the vaccine development. An effective ASFV vaccine should be able to induce humoral immunity to block the entry of ASFV into its target cells and/or produce antibody-dependent cell-mediated cytotoxicity (ADCC) effects. Vaccination should also induce the production of cytotoxic T lymphocytes (CTLs) that can specifically recognize infected cells. T cells, especially CD8+ T cells, can activate cellular immunity to lyse target cells and play an important role in protective immunity against ASFV (Oura et al., 2005). Therefore, it is necessary to screen antigens or epitopes that can induce specific cellular immunity. The effectiveness of combining these antigens or epitopes still requires extensive experimental verification. Furthermore, there are inconsistencies in the literature regarding whether certain antigens produce positive immune effects.
Some of the vaccine studies that used nucleic acid or protein subunits are summarized in the table below (Table 1) with an intention to provide references for those who are interested in the selection of appropriate immunogens for vaccine development since development of effective gene engineered vaccines would require selection of the ASFV genes and proteins that can stimulate both humoral and cellular immunity.
Table 1.
Gene engineered subunit vaccine candidates and their effectiveness.
| Gene (Protein) name | Vaccine type | Challenge | Specific antibodies | Neutralizing antibodies | T cell response | Clinical outcome (n = protected animals/ total animals) | Reference |
|---|---|---|---|---|---|---|---|
| A151R, B119L, B602L, EP402RΔPRR, B438R, K205R-A104R | Adenovirus | /a | Yb | / | Y | / | (Lokhandwala et al., 2017) |
| CP204L (p32), E183L (p54), CP530R (pp62), B646L (p72), | Adenovirus | / | Y | / | Y | / | (Lokhandwala et al., 2016) |
| B646L (p72), E183L (p54), O61R (p12), EP402R (CD2v) | Vaccinia virus | / | Y | / | Partial | / | (Lopera-Madrid et al., 2017) |
| A151R, B119L, B602L, EP402RΔPRR, B438L, K205R, A104R, CP530R (pp62), B646L (p72) | Adenovirus | Y | Y | / | Y | No protection | (Lokhandwala et al., 2019) |
| EP153R, K78R (p10), CP530R (p15), CP80R, I329L, H108R, K196R, CP312R, F334L, NP419L, NP868R, B66L, H339R, R298L, K145R, B385R, F165R, F778R, S273R, MGF100-1L, A224L, MGF505-6R, B175L | Adenovirus | Y | / | / | / | No protection | (Cadenas-Fernández et al., 2020) |
| 42 replication-deficient adenovirus-Vectored multicistronic expression cassettes encoding ASFV antigens | Adenovirus | Y | Y | / | / | No protection | (Zajac et al., 2023) |
| CP2475L (p220) | Protein | / | Y | / | Y | / | (Zajac et al., 2022) |
| B646L (p72), CP204L (p30), E183L (p54), E199L, EP153R, EP364R, F317L, I329L, MGF360, MGF505 | Protein | Y | Y | / | Y | Full protection (n = 6/6); Viremia was significantly reduced | (Goatley et al., 2020) |
| CP204L (p30), E183L (p54), B646L (p72), KP177R (p22) | Protein | Y | Y | Y | / | No protection; Slight delay of clinical disease and viremia | (Neilan et al., 2004) |
| EP402R (CD2v) | Protein | Y | Y | / | / | Full protection (n = 3/3) | (Ruiz-Gonzalvo et al., 1996) |
| CP204L (p30), E183L (p54) | Protein | Y | Y | Y | / | Partial protection (n = 3/6) | (Gómez-Puertas et al., 1998) |
| CP204L (p30), E183L (p54) | Protein | Y | Y | Y | / | Full protection (n = 2/2) | (Barderas et al., 2001) |
| M448R, MGF505–7R | DNA | Y | Y | / | Y | Partial protection (n = 3/5) | (Bosch-Camós et al., 2021) |
| CP204L (p32), E183L (p54), EP402R (CD2v) | DNA | Y | Y | / | Y | Partial protection (n = 4/6) | (Argilaguet et al., 2013a) |
| ASFV DNA library | DNA | Y | Y | Y | Y | Partial protection (n = 6/10) | (Lacasta et al., 2014) |
| CP204L (p30), E183L (p54), EP402R (CD2v) | DNA | Y | Y | / | Y | Partial protection (n = 2/6); The onset time of clinical symptoms was delayed | (Argilaguet et al., 2012) |
| B646L (p72), MGF110-5L, CP204L, CP530R (pp62), I73R, I215L, A151R, C129R, E146L, L8L, M448R, MGF110-4L | DNA | Y | Y | / | Y | No protection; reduced viral load in some tissues | (Netherton et al., 2019) |
| CP204L (p32), E183L (p54), EP402R (CD2v), B646L (p72), D117L (p17), CP530R (p15, p35) | DNA–Protein | Y | Y | Nc | N | No protection; Accelerated onset of clinical symptoms, viremia, and death | (Sunwoo et al., 2019) |
This experiment or statistic was not performed.
Yes.
No.
ASFV gene or protein-based vaccines include protein subunit vaccine, DNA vaccine, virus vectored vaccine and DNA-protein combined vaccine. Among them, virus vectored vaccines did not show satisfactory results in the clinical studies (Table 1). While the DNA or protein vaccines usually select vaccine components from the genes or proteins of CP204L (p30), E183L (p54), B646L (p72), CP530R (pp62), CP2475L (p220), or EP402R (CD2v), etc. The vaccines are designed by the different study groups and contained different ASFV gene/protein combinations, which were tested with different protocols performed by the different laboratories from different countries. Thus, it is hard to estimate that (1) which viral protein(s) induced the neutralizing antibodies efficiently, (2) whether there are any potential synergistic effects or inhibitions between the different viral proteins, or for a single protein, (3) which part produces ADE effect, and (4) which part possesses neutralizing epitope. Therefore, it remains inconclusive to choose the best vaccine candidate for further study.
On the other hand, the infection of pigs with the ASFV viral particles is independent of the viral outer membrane protein, CD2v (Borca et al., 1998), which further increase the complexity of the vaccine design. For example, anti-ASFV sera collected from ASF survivors cannot inhibit ASFV replication in vitro (Walczak et al., 2022). The pigs which are immunized with CD2v protein alone expressed by baculovirus did not produce neutralizing antibodies but survived from the attack with the homologous strain E75 (Ruiz-Gonzalvo et al., 1996). Tandem fusion of p30, p54 and CD2v as a DNA vaccine can induce strong humoral and cellular immunity (Argilaguet et al., 2012). When the DNA vector was modified by replacement of pCMV with pBacPH-GFP PolyA, the immune protection effect was greatly improved (Argilaguet et al., 2013). However, a combined DNA-protein vaccine containing immunogenic antigens CD2v, p32, p54, p72, p17, p15, and p35 displayed the opposite effect that the vaccination accelerated clinical symptoms and death in pigs (Sunwoo et al., 2019).
In the face of the fact that the viral protein functions and the pathogenic mechanism of ASFV are not well understood, increasing the number of antigens in vaccines may not enhance the efficacy of vaccines. The reasons might be that the irrelevant antibodies interfere with the neutralization of the virus, or ADE effects assist the virus infection. Furthermore, failure in the cross-protection between the heterologous strains is also a reason for reduced efficacy.
3. The major structural proteins in ASFV vaccine development
With the efforts of scientists worldwide over the years, several ASFV proteins that provide immune protection were identified. These proteins bring hope for the development of effective gene engineered vaccine formulations. The structures and functions of these ASFV proteins that are reported to produce neutralizing antibodies or show protective effect in the animal studies are reviewed below.
3.1. CD2v
The outer envelope is the layer acquired by ASFV from the host cell membrane during budding process. Some pEP402R (CD2v) proteins have been detected in the outer layer of budding virions from ASFV, regardless of viral virulence (Borca et al., 2020a). CD2v is a type I transmembrane protein encoded by EP402R and is located on the outer envelope of ASFV. It has a signal peptide (SP), a transmembrane region (TM), and two immunoglobulin-like domains (IG) (Rodríguez et al., 1993) and can be highly glycosylated, which is important for the viral release, cellular infection and immune evasion of ASFV (Watanabe et al., 2019). CD2v can be cleaved into N-terminal glycosylated fragments and C-terminal unglycosylated fragments. This cleavage process occurs in the endoplasmic reticulum or Golgi region. The C-terminal unglycosylated fragment and the full-length protein are mainly concentrated in the membrane region surrounding the virus factory. The C-terminal proline-rich repeat of CD2v can interact with SHP37 and AP-1, which facilitate intracellular virion movement, protein transport, and endocytosis (Kay-Jackson et al., 2004; Pérez-Núñez et al., 2015). The N-terminal glycosylated fragments were detected in the more dispersed manner in the cytoplasm (Goatley and Dixon, 2011). The effect of CD2v on the ability of ASFV to infect appears to depend on the strain or virulence of the virus (Borca et al., 1998; Gladue et al., 2020; Monteagudo et al., 2017). As well, a linear epitope at amino acids 28–51 of the extracellular domain of CD2v has been reported (Ren et al., 2022). Transcription of EP402R that encodes CD2v occurs late in viral replication and is not necessary for viral replication. The name, CD2v, was given as its N-terminal extracellular domain resembles with the T lymphocyte surface adhesion receptor CD2. ASFV CD2v interacts with CSF2RA to activate the JAK2-STAT3 pathway and inhibit apoptosis, and thus maintains the survival of infected cells to promote viral replication (Gao et al., 2023). Both CD2v and host CD2 can induce hemadsorption phenomenon (HAD) around the cells expressing the protein, resembling a rosette of red blood cells. This phenomenon is necessary for the binding of extracellular virus to erythrocytes, promoting the spread of the virus in vivo and regulating the immune response (Borca et al., 1998; Chaulagain et al., 2021; Rodríguez et al., 1993). However, there was no HAD phenomenon in ASFV genotype I. The hemadsorption of African swine fever virus is determined by signal peptide and N-glycosylation of N-terminal-CD2v (Pérez-Núñez et al., 2023). The candidate attenuated vaccine GΔDKE-CmutQ96R/K108D mutates the amino acid of CD2v, resulting in no binding to red blood cells in vitro. In the immunized pigs, strong early ASFV-specific antibody and cellular responses were detected along with reduced levels of virus in blood. The protection rate against antigen-challenge was more than 80 % (Rathakrishnan et al., 2023). The ASFV-infected cells cannot exhibit the HAD phenomenon when the gene EP153R is deleted, indicating that EP153R is likely playing a role in the interaction between CD2v and the corresponding cellular receptor (Galindo et al., 2000). One research group managed to generate expression vectors of CD2v from the HAD+ strain and explored the effect of single viral protein on rosette formation. Results showed that the expression of CD2v was enough to induce rosette formation, but not the EP153R indicating that the expression of CD2v is necessary and sufficient for HAD formation, independently of the expression of any other ASFV protein and ruled out a role of EP153R in HAD (Pérez-Núñez et al., 2023). The estimated surface density of most envelope proteins is very low and the abundance of CD2v is one of the lowest among the ASFV structural proteins detected so far. This characteristic is of relevance since the surface density of an antigen plays a key role in antigenicity and immunogenicity (Zhu, 2022). The low surface density reduces the affinity of antibodies for virions and also reduces the immunogenicity of protective antigens (Zhu, 2022). It has been noted that the low surface density of CD2v on extracellular virions may be a result of intracellular localization of majority of the expressed CD2v rather than on the cell surface (Goatley and Dixon, 2011; Lu et al., 2023).
3.2. p12
The p12 structural protein of ASFV is encoded by the O61R gene, which is associated with viral membrane precursors, assembled particles, and intracellular mature virus (Salas and Andrés, 2013). The p12 has three molecular forms with the molecular weight of 10, 12 and 17 kDa respectively, only the 17 kDa-form presents in the viral particles (Angulo et al., 1993). The viral attachment protein p12 is localized in the outer membrane, and can interact with cellular protein receptors to promote the attachment of ASFV to host cells (Galindo et al., 1997). However, the antisera of p12 could not neutralize the virus and inhibit virus binding to the host cells (Angulo et al., 1993). Immunofluorescence microscopy of ASFV-infected cells showed that p12 is located at the inner envelope of the ASFV structure (Salas and Andrés, 2013). And upon ASFV entry into the cell, p12 is found at the perinuclear virus factories and along with virus particles. p12 spreads throughout the cytoplasm and to the cell surface at later timepoints post-infection. This localization pattern is nearly identical to that of viral protein p17 (Salas and Andrés, 2013).
3.3. p72
The p72 structural protein of ASFV is encoded by gene B646L and has a relative molecular weight of 73.2 kDa. It plays a role in the formation of the viral capsid in the late period of viral replication and serve as the main component of the viral icosahedron. It accounts for about 32 % of the total weight of viral particles (Neilan et al., 2004). The monomer p72 has a double jelly-roll structure, which is a feature shared by many other icosahedral viruses (San Martín and van Raaij, 2018). The p72 protein forms a trimeric structure within the viral icosahedron. Each p72 contains two p72 tandem roll domains, which is comprised of eight β-sheets. The trimeric structure of p72 contains six roll barrels, which combine to form a pseudohexamer. The bottom of trimeric structure is anchored to the inner membrane of the virus, while the top propeller structure extends outside of the virus outer envelope, forming an exposed area (Liu et al., 2019a; Wang et al., 2019). The exposed region along with the N-terminal domain reveal specific differences between virus types. Based on the analysis of the spatial structure, the position of this region may contribute to the formation of conformational epitopes, which are receptor-binding regions on the cell surface (Wang et al., 2019). pB602L is an important non-structural protein of ASFV. It acts as a molecular chaperone that assists in proper folding of p72 to form a trimer (Epifano et al., 2006), which is necessary for the formation of the viral icosahedral capsid as well as increased trypsin-resistance (Liu et al., 2019a). Expression of p72 alone in HEK293F cells results in the formation of soluble aggregates, since correctly folded and assembled p72 can only be obtained when p72 and pB602L are co-expressed (Liu et al., 2019a). Newly synthesized p72 molecules are distributed approximately evenly between the cytoplasm and endoplasmic reticulum (ER) membrane. It appears that p72 is incorporated into capsid and/or matrix precursors on the ER membrane during virus assembly (Cobbold and Wileman, 1998). p72 is the major antigen detected in the blood of virus-infected pigs. Currently, the most sensitive technique for ASFV detection on the market is the p72-based detection assay. Based on the differences in the 3′ -terminus of B646L that encodes p72, 24 different p72 genotypes have been identified for currently isolated ASFV. However, the current method of distinguishing p72 genotypes may not detect the difference in p72 epitopes (Wang et al., 2021a). In vitro, monoclonal antibody 135D4 can recognize the region of p72 from amino acid residue 400–404 (GVINE) and can immunoprecipitate p72 (Borca et al., 1994). Monoclonal antibodies prepared by expression of p72 protein using baculovirus can recognize four groups of linear epitopes, which are aa156–165, aa265–280, aa280–294 and aa290–303. These four groups were all located in the highly conserved region of p72 (Heimerman et al., 2018). So far, no major linear or conformational epitopes have been identified in the exposed region of p72 (Heimerman et al., 2018; Phillips, 2016). When purified p72 was treated at 96 °C for 10 min, native PAGE gel analysis showed that most of the trimer spikes were still intact and stable, indicating high thermal stability of the trimer (Liu et al., 2019a). The p72 trimer spike outer surface contains numerous charged residues, and due to its high thermal stability, no obvious glycosylation sites were observed in the Cryo-EM structure (Wang et al., 2021a). Animals infected with ASFV rapidly produced large amounts of p72 antibodies. However, it is generally believed that these antibodies don't have neutralizing activity and can't provide immune protection or even produce ADE effects (Neilan et al., 2004) likely due to no relative epitopes are present in the exposed region. p72 is commonly used for virus detection by ELISA, due to its high amount and prolonged presence in the blood circulation.
3.4. p17
The p17 structural protein of ASFV is encoded by the gene D117L, which exhibits high genetic stability (Yáñez et al., 1995). p17 is a major transmembrane protein of ASFV and is located in the capsid and inner envelope (Andrés et al., 2020). It has been identified as a structural phosphoprotein that is mainly expressed during late stages of virus replication (Muñoz and Tabarés, 2022). This essential and highly abundant protein is required for capsid assembly and icosahedral morphogenesis (Suárez et al., 2010a). In the viral capsid, p17 is located just below the capsid shell and binds tightly to the base domain of the major capsid protein p72, firmly anchoring the p72 capsid to the inner membrane (Liu et al., 2019b; Wang et al., 2019). Inhibiting p17 by the specific rabbit polyclonal sera prevents the proteolytic processing of polyproteins pp220 and pp62, and in turn, hinders the interaction between capsid proteins and viral membrane precursors. Thus, inhibition of p17 prevents the construction of viral helical precursors (Suárez et al., 2010a). p17 promotes mitophagy by facilitating the interaction between cellular proteins translocase of outer mitochondrial membrane 70 (TOMM70) and the mitophagy receptor SQSTM1 and inhibits the innate immune response by degrading mitochondrial antiviral-signaling protein (MAVS). This suggests a role of p17 in immune escape by ASFV (Hu et al., 2023). p17 can also inhibit cell proliferation through ER stress and induce reactive oxygen species (ROS)-mediated cell cycle arrest, suggesting that p17 may be involved in the pathogenesis of ASFV (Xia et al., 2020). p17 is a specific antigen in the immunoreaction of pig sera with neutralizing antibodies (Muñoz and Tabarés, 2022). Protein Immunization with a mixture of 44 antigens followed by ELISA analysis demonstrated that p17 was an immunogenic protein of ASFV (Jancovich et al., 2018).
3.5. p30 and p54
Both p54 and p30 are the structural proteins involved in viral entry, and both are localized to the inner envelope of ASFV. The antigenic properties of these structural proteins during ASFV infection (Sánchez et al., 2013). Oviedo et al. found that recombinant p30 is more efficient than p54 for antibody detection by ELISA, and p54 presents better reactivity than p30 in Western blot (Cubillos et al., 2013; Oviedo et al., 1997). The combined use of these two proteins for ASFV serological diagnosis can improve the sensitivity of the assay (Oviedo et al., 1997).
The p30 protein is encoded by the CP204L gene with a relative molecular weight of 30 kDa and is involved in viral internalization process (Hernáez et al., 2016). Its expression usually begins within 2–4 h after infection and persists throughout the infection cycle (Lithgow et al., 2014). Studies have shown that the interaction of p30 with the host cellular proteins (RPSA, DAB2, CAPG, and ARPC5) might be involved in the viral internalization process mediated by clathrin-mediated endocytosis and macropinocytosis. In addition, p30 may also regulate innate immunity by interacting with the innate immune regulators such as DAB2, PARP9, RPSA, OAS1, and VBP1 (Chen et al., 2022b). p30 exhibits excellent antigenicity. Anti-p30 antibody can be detected 8–12 days after vaccination, making it an important target for early diagnosis of the virus infection (Li et al., 2022). The C-terminus of p30 has been identified as an immunodominant region (Murgia et al., 2019). In one study, the ASFV Georgia strain was selected to explore the antigenic region of p30 protein, and the results demonstrated that three of the four linear epitopes were highly conserved. Among these, the linear epitopes 116–125 and 146–160 in the C-terminal antigenic region were found to be more immunodominant (Wu et al., 2020). Recombinant poxviruses with p30 have been utilized in the development of vaccines against ASFV. One of the most advanced poxvirus vaccine vector is Modified Vaccinia Virus Ankara (MVA) (Sánchez-Sampedro et al., 2015), and its promoter for antigen expression plays a crucial role in determining whether the replicative viral vector can activate durable humoral and cellular immunity (Brun et al., 2008). Previous studies have shown that repeated vaccination with the recombinant vaccine virus MVA-prMVA13.5L-p30 constructed using the natural promoter of MVA, can induce high levels of anti-p30 antibodies (Lopera-Madrid et al., 2021). p54 is encoded by the E183L gene and has a relative molecular mass of 25 kDa. It is associated with virus attachment (Gómez-Puertas et al., 1998) and can induce apoptosis of infected cells in the early stage of ASFV infection (Hernáez et al., 2004). It contains a transmembrane domain, a Gly-Gly-X motif, as well as a recognition sequence for processing several ASFV structural proteins. The sequence of Gly-Gly-X is believed to be a proteolytic site in the viral or cellular protein precursors (López-Otín et al., 1989). Immunoelectron microscopy showed that p54 is localized in the viral factories of infected cells (Rodriguez et al., 1996). During viral infection, p54 in the microtubule organizing center binds to the light chain of cytoplasmic dynein LC8, which is essential for the recruitment of envelope precursors to assembly sites (Rodríguez et al., 2004). This phenomenon suggests a possible mechanism by which ASFV particles are transported to the perinuclear factory region through directional movement along the minus end of microtubules (Alonso et al., 2001). In one study, recombinant fusion proteins formed by the major outer membrane lipoprotein I (Oprl) of Pseudomonas aeruginosa, combined with p30 or p54, were used to immunize mice, and the serum collected was able to neutralize more than 86 % of ASFV in vitro (Zhang et al., 2022). In another study, researchers used the fusion proteins ZPM (Z12-P30-modified p54) and ZPMT (Z12-p30-modified p54-T cell epitope) to immunize mice. These fusion proteins were formed by fusion of p30 and p54/p54 T cell epitopes with a novel cell-penetrating peptide Z12. The serum from immunized mice neutralized more than 85 % of ASFV in vitro. Among the two fusion proteins, ZPMT induced the most potent neutralizing antibody responses and cellular immunity (Zhang et al., 2021).
3.6. p22
The p22 structural transmembrane protein is encoded by the ASFV gene KP177R, and is transcribed early during virus replication (Camacho and Viñuela, 1991). It was first detected in the cell membrane of host cells in the late stages of infection (Camacho and Viñuela, 1991). p22 is localized on the inner membrane of the virus and the surface of infected cells (Alejo et al., 2018). However, the function of p22 remains unclear. Deletion of KP177R did not affect viral replication, and the virus yield was similar to that of the parental strain used in the study (high-virulence field isolate Georgia2010). This suggests that p22 does not appear to be involved in virus replication or virulence (Vuono et al., 2021). The high-throughput methods identified p22 interacting host proteins, which were related to biological processes such as cell binding, cell structure, signal transduction, and cell adhesion (Zhu et al., 2021). The C-terminal portion of p22 (p22Ct; amino acids 42–189) was produced in E. coli expression system and was subsequently used to immunize mice. A high antibody titer was detected in these mice, indicating that the p22 C-terminal fragment is highly immunogenic (Díaz et al., 2022).
3.7. pp220 and pp62
The pp220 and pp62 are two multiprotein precursors of ASFV proteins. pp220 is encoded by the gene CP2475L with a relative molecular mass of 281.5 kDa and expressed late during virus replication. It can be cleaved by the virus-encoded Small Ubiquitin-related Modifier (SUMO)-like protease pS273R to produce mature viral proteins p5, p14, p34, p37 and p150, which also locate in the virions (Andrés et al., 2001; Simón-Mateo et al., 1993). Infected cells contain various processed forms of pp220, but only the structural proteins p34 and p150 can be detected in mature virions. pp62, encoded by CP530R with a relative molecular weight of 60.5 kDa, is also proteolytically hydrolyzed by pS273R to mature virion proteins p8, p15 and p35. These decomposed virion proteins constitute the main components of the virion core and capsid, accounting for about 32 % of the total viral protein. The hydrolysis process of pp220 and pp62 occurs simultaneously during the process of virus assembly (Andrés et al., 2002). pp220 is located directly underneath the inner capsule. The correct binding of pp220 to the cell membrane is essential for regulating the correct processing of pp220 and the correct packaging of virions (Heath et al., 2003). Complexes of the C-terminus of p150 and other protein hydrolysates of pp220 may mediate interaction between capsid proteins (Li et al., 2023). pp62 is required for proper assembly and maturation of the core of the viral particle (Suárez et al., 2010b), and pp62 processing requires the expression of pp220. Processing of pp220 and pp62 also requires the expression of the major capsid protein p72. The correct processing of pp220 and pp62 is a crucial characteristic of mature ASFV virions. pp220 polyprotein contains at least four T cell epitopes, which are located at position 161–169, 859–867, 1363–1371 and 1463–1471. Strong IFN-γ responses can be induced in domestic pigs by immunization with pp220 (Zajac et al., 2022). Previous studies have predicted that epitopes IADAINQEF, QIYKTLLEY and SLYPTQFDY (located in pp220) were highly conserved T cell epitopes in ASFV genome and could therefore be a promising candidate for vaccine development (Herrera and Bisa, 2021). p35, formed by pp62 hydrolysis, is a completely new folded structure that consists of two helical bundle lobes: a large "head domain" and a relatively small "tail domain" (Li et al., 2020). In solution, p35 mainly exists in the monomeric state, while its dimeric state is observed at low pH. This suggests that the conformation of p35 may change during virus entry and uncoating. There are two charged patches on the surface of p35: a continuous positively charged bulge composed of lysine and arginine residues and a negatively charged patch composed of aspartic acid and glutamic acid residues (Li et al., 2020). Positively charged veneers play an important role in lipid binding activity and can promote core-shell binding to the inner membrane. The function of the negative charge on the other side may be associated with interaction with various structural proteins. During viral assembly, membrane-associated proteins such as p35 serve as docking scaffolds to recruit other components in the host membrane and core-shell, further forming stable and mature virions (Li et al., 2020). One study identified antibodies from 23 ASFV-positive pig serum samples, and the results showed that all the samples contained antibodies against p30, p54 and pp62. These results indicate the strong immunogenic properties of p30, p54 and pp62 (Luong et al., 2022).
3.8. pA104R
pA104R encoded by A104R is a histone-like protein that is expressed late in the ASFV replication cycle. It is essential for ASFV genome packaging and replication (Frouco et al., 2017)Particularly, pA104R is located in the nucleoid to facilitate nucleoid assembly (Alejo et al., 2018) by binding to dsDNA with a higher affinity (Liu et al., 2020). pA104R works with ASFV topoisomerase II (pP1192R) to regulate DNA supercoiling of ASFV (Frouco et al., 2017). The binding of pA104R to DNA is disrupted by stilbene derivatives SD1 and SD4 and this disruption inhibits ASFV replication in porcine macrophages (Liu et al., 2020). Deletion of the pA104R gene in ASFV was found to lead to an apparent decrease in virulence (Ramirez-Medina et al., 2022). pA104R is also known as an antagonist of type I interferon signaling, which exerts its function through interfering with multiple signaling pathways (Chen et al., 2023). After infecting pigs with nonpathogenic ASFV, pA104R could elicit a strong antibody response, which was higher in asymptomatic pigs than in chronically infected pigs (Reis et al., 2007). Therefore, the role of the pA104R antibody in the protective response should be further investigated.
3.9. p10
p10 is encoded by the K78R gene of ASFV. It has a nuclear import capacity and accumulates in the nucleus during viral replication (Nunes-Correia et al., 2008). The prokaryotic expression of p10 protein showed a strong DNA binding activity with similar affinities for both double-stranded and single-stranded DNA (Muñoz et al., 1993). The C-terminal region of p10 contains a helix-turn-helix motif and this structural arrangement promotes a stable overall interaction configuration with the dsDNA. The Lys-enriched C-terminal helix tends to interact with the minor groove of dsDNA, while the N-terminal helix rich in serine residues forms highly stable interaction with the major groove of dsDNA. These two misaligned helices are connected with a flexible loop (Istrate et al., 2022). Antibodies against p10 were identified among the antibodies produced by pigs that survived ASFV infection, but the strength of the antibody response varied (Kollnberger et al., 2002; Reis et al., 2007).
3.10. Other ASFV protein
Based on the two studies reported, it is highly possible that the viral neutralizing epitopes are also located in other ASFV proteins such as B602L, K78R, and K205R (Arias et al., 2017; Urbano and Ferreira, 2022). Additional studies are required to confirm efficacy of these epitopes.
4. The cellular and humoral immunity in ASFV infection
Macrophages (Mφ) and dendritic cells (DC) are phagocytes as part of the innate immunity. In adaptive immunity, these cells belong to a group of professional antigen-presenting cells (APCs), who bridge innate immunity and adaptive immunity. ASFV targets monocytes/macrophages severely affects their phagocytic property, and antigen processing and presenting abilities. ASFV was reported to be able to recruit several elements of the macrophage translation machinery to the viral factory, enabling efficient replication and translation of ASFV in cells (Alonso et al., 2001; Franzoni et al., 2018). Additionally, multiple type-I IFN suppressing genes were activated in ASFV-infected macrophages within the multigene families MGF360 and MGF530/505 (Zhang et al., 2022). This would induce an immuno-suppressive state in the host, resulting in severe sickness and death, before the immune system could respond adequately, of most animals infected with the virulent strain of ASFV (Schäfer et al., 2022).
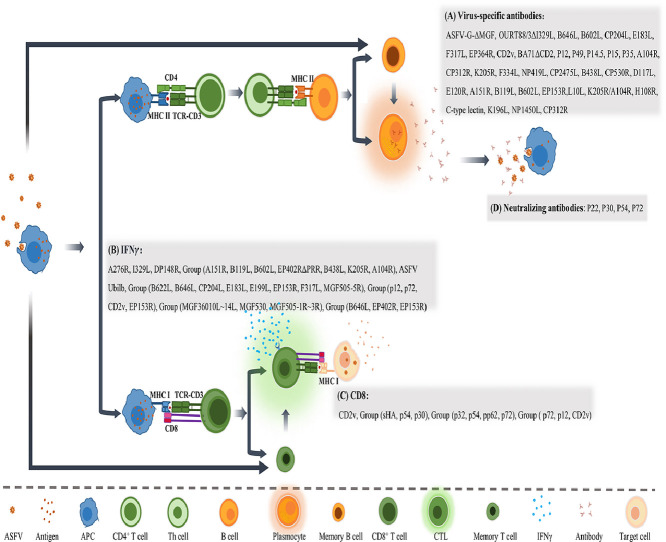
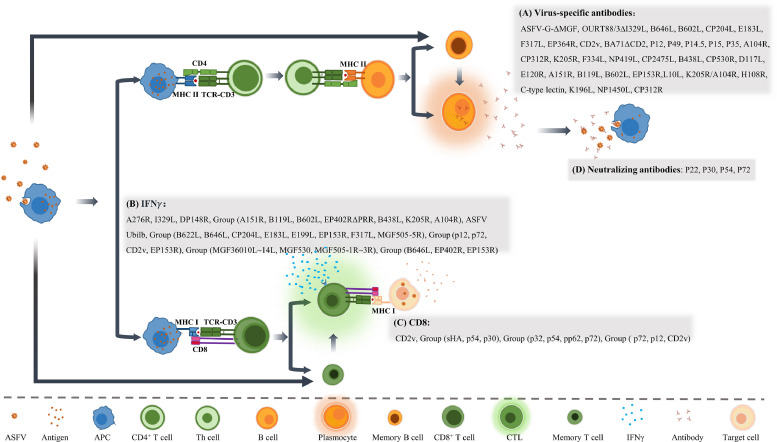
Adaptive immunity includes humoral immunity and cellular immunity. Humoral immunity is facilitated mainly through the activation of B cells capable of producing functional antibodies, which can specifically recognize and bind to foreign antigens and neutralize them through various mechanisms (Fig. 3). The development of effective protein subunit vaccines requires correct identification of viral antigens that could generate potent neutralizing antibodies. Currently, there are some controversies regarding strength of the antibody to neutralize ASFV (Escribano et al., 2013; Franzoni et al., 2018). There is no direct correlation between the level of neutralizing antibodies and protection. A study indicated that non-neutralizing antibodies can inhibit Porcine epidemic diarrhea virus (PEDV) infection in vivo via indirect mechanisms dependent on the Fc receptor (Abreu-Mota et al., 2018; Jegaskanda et al., 2013). However, we are convinced that high levels of neutralizing antibodies are essential for complete protection of the pig population, based on the following observations. Virus-neutralizing activity was observed in nearly 100 % of animals vaccinated with live attenuated vaccine candidates (Zhao et al., 2023). Feeding newborn piglets with breast milk containing ASFV antibody or anti-venom serum significantly reduced clinical symptoms compared with the untreated group (Schlafer et al., 1984). Immunization with p30, p54, p72 and p22 proteins expressed by baculovirus produced neutralizing antibodies but did not show an obvious protective effect, but the clinical course was delayed (Neilan et al., 2004). These data suggested that humoral immunity and the generation of neutralizing antibodies partially attenuate or delay the clinical course of ASFV-infected animals. However, attempts on identifying a suitable immunogen or epitope combination to stimulate production of more potent neutralizing antibodies have not yet been successful. Therefore, further research for a comprehensive understanding of immunogenic proteins of ASFV is urgently needed.
Fig. 3.
Specific immune response induced by ASFV genes and/or its encoded proteins. ASFV infected antigen-presenting cells, such as macrophages, stimulate both humoral and cellular immune response through MHC II and MHC I pathway respectively, to generate virus specific antibodies and CTLs, as well as adaptive immune memory. The gene-engineered ASFVs, or viral genes/proteins are listed that could produce (A) Virus-specific antibodies, (B) IFNγ, (C) CD8+ T cells, and (D) Neutralizing antibodies.
Cellular immunity is employed to eliminate pathogens by targeting intracellular microorganisms and directly killing infected cells. The detailed relationship between ASFV infection and cellular immunity has been reviewed and analyzed extensively by Schäfer and collegues (Schäfer et al., 2022). Briefly, CD8+ T cells are primarily activated through MHC I to secrete IFN-γ which lyses the target cells (Fig. 3). Therefore, IFN-γ and CD8+ T cells are generally used as detection indicators to assess the protective potential of cellular immunity. In comparison to antigen-based subunit vaccines, DNA vaccines can induce both cellular and humoral immunity, making it a promising vaccine type against ASFV (Borca et al., 2020b). Anna Lacasta et al. constructed a plasmid library of ASFV to immunize pigs, and the results showed that it could stimulate both cellular and humoral immunity, and provide 60 % protection against E75, a highly virulent genotype I strain (Lacasta et al., 2014). Lynnette et al. used rAd5 and MVA as vectors to express eight ASFV genes (B602L, B646L (p72), CP204L (p30), E183L (p54), E199L, EP153R (C-type lectin), F317L and MGF505-5R) to immunize pigs. The results showed that this combination could activate cellular immunity and achieve 100 % lethal protection against another highly virulent genotype I strain, OUR T88/1 (Goatley et al., 2020).
5. Several key points that need to be considered
The ASFV virion contains five parts, from which four parts can be present in virus-infected cells, and include the core-shell, inner envelope, capsid, and outer envelope. Theoretically, all four components could stimulate humoral or cellular immune responses. Based on the research findings so far, several major ASFV proteins such as CD2v, p30, p54, p72, pp220 and pp62 are the most immunogenic proteins for the development of nucleic acid and protein subunit vaccines. These proteins play important roles in the process of ASFV infection or replication. Additional ASFV proteins can also inhibit apoptosis of the infected host cells as well as inhibit MHC class I expression. These mechanisms likely disrupt the virus capture by antibodies and NK and/or antigen-specific T cells, and undermine the efficacy of vaccine-induced immunity (Escribano et al., 2013). Additional studies are required to determine specific ASFV protein or protein group capable of inducing complete protection.
The results from the multi-antigen combination DNA (EP402R, B646L, CP204L, D117L) and protein (p15, p35, p54, p17) vaccines showed that they accelerated the occurrence of viremia and death (Sunwoo et al., 2019). On the other hand, immunization with baculovirus expressing p54 and p30 proteins provided 50 % protection rate (Gómez-Puertas et al., 1998). Interestingly, immunization with the same baculovirus expressing p54, p30, p72 and p22 proteins did not have any protective effect and only delayed clinical symptoms (Neilan et al., 2004). Therefore, we suggested the next step would be to analyze the ASFV structural proteins from the perspective of protein location and function, and to screen for more effective antigen combinations. In addition to structural proteins, some non-structural proteins or enzymes also play an important role in the process of viral replication and immune evasion. For example, p72 can be correctly folded and assembled only when co-expressed with pB602L. EP153R is involved in the interaction between CD2v and its corresponding cell receptors. Hence, these proteins should also be taken into consideration in the vaccine design.
Epitopes are crucial in the process of antigen recognition, which subsequently induces antibody production and cell-mediated immune response against pathogens. Therefore, identifying the antigenic epitopes of antibodies is essential for developing epitope-based vaccines as well as diagnostics. Antigenic epitopes always contain approximately 5–15aa and can be divided into linear and spatial epitopes (Berger and Lapthorn, 2016). The majority of the antigenic epitopes are spatial epitopes. Linear epitopes are continuous fragments of protein in the primary structure. In contrast, spatial epitopes are neighboring amino acid residues on the surface of protein, but these amino acids are discontinuous in the primary protein structure. For example, p35 has a conformational change from monomer to dimer, which can lead to better display of spatial epitopes. The baculovirus expression vector is currently used to express bioactive proteins. However, it remains unclear whether the ASFV proteins will be fully functional when expressed by baculovirus expression system.
Different immune adjuvants have different mechanisms of action and effects on vaccines. Appropriate immune adjuvants can nonspecifically enhance or modify the body's adaptive immune response to the corresponding antigen, thereby boosting the immunogenicity of the vaccine. The Ad5-ASFV adenovirus vector vaccine prepared with BioMize (VaxLiant, NE, USA) and ISA-201™ (Seppic, NJ, USA) adjuvants was used to immunize piglets with a cocktail therapy, and the results showed that the pp62-specific antibody were significantly higher in the ISA-201™ group compared to the BioMize adjuvant group (Zajac et al., 2023). Chen et al. used the new Porcine reproductive and respiratory syndrome virus (PRRSV)-specific IgM monoclonal antibody (Mab)-PR5nf1 as a vaccine adjuvant to study the efficacy of in enhancing the protection (Chen et al., 2022a). The results showed that the addition of PRRSV-specific IgM to the PRRSV-KIV vaccine could significantly improve the overall survival rate (SOR) and enhance the cell-mediated immunity (CMI). When evaluating a vaccine for safety and effectiveness, FDA considers adjuvants as a component of the vaccine rather than approved separately. Common adjuvants in existing vaccines include aluminum adjuvant, such as AS01 and CpG 1018 (https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/common-ingredients-fda-approved-vaccines) and Matrix-M, which has been used in COVID-19 vaccines (https://www.fda.gov/media/173097/download). Nonetheless, it is still necessary to develop new adjuvants or combinations of adjuvants that can effectively enhance antigen-specific immune responses.
Lastly, different vectors may have different effects on antigen expression and immune effect (Šimčíková et al., 2015; Yuan et al., 1999). In addition, it is difficult to compare the results of immune protection in pigs due to variations in immunization routes and challenge ways.
6. Summary and outlook
At present, all pig farms have established protection programs such as large-scale quarantine and culling against ASFV attacks, which can cause huge economic losses (De Lorenzi et al., 2020; Liu et al., 2021b; Tian et al., 2021). Therefore, vaccine development remains the best choice to combat ASF as a preventative measure. Selecting ASFV proteins that can induce humoral and cellular immune responses in pigs and produce improved levels of neutralizing antibodies is crucial for the development of ASFV subunit vaccines. In the long term, gene engineered vaccine has shown promising potential in a fight against ASFV.
With the current understanding of ASFV proteins, we propose that one should focus on investigating viral structural proteins that are involved in the virus adsorption, internalization, and release, as well as the immune evasion mechanisms to generate gene/protein-based vaccine candidates. Additionally, more research should be conducted to optimize the different proteins combinations, vaccine efficacy, adjuvants, and immunization schedules to elicit robust immune protection against different strains of ASFV.
CRediT authorship contribution statement
Ning Wang: Methodology, Formal analysis, Investigation, Visualization, Writing – original draft. Pan Huang: Investigation, Visualization, Writing – original draft. Jun Zhang: Methodology, Formal analysis, Investigation, Visualization, Writing – original draft. Minqi Lin: Methodology, Formal analysis, Investigation, Visualization, Writing – original draft. Xiaoru Lai: Investigation, Visualization, Writing – original draft. Jianwen Chen: Investigation, Visualization, Writing – original draft. Chungen Pan: Conceptualization, Supervision, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare that there is no potential conflict of interest in this article.
Acknowledgments
This study was supported by Science and Technology Planning Project of Guangzhou (2024B03J1362).
Data availability
No data was used for the research described in the article.
References
- Abreu-Mota T., Hagen K.R., Cooper K., et al. Non-neutralizing antibodies elicited by recombinant Lassa-Rabies vaccine are critical for protection against Lassa fever. Nat. Commun. 2018;9(1):4223. doi: 10.1038/s41467-018-06741-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejo A., Matamoros T., Guerra M., et al. A proteomic atlas of the African swine fever virus particle. J. Virol. 2018;92(23) doi: 10.1128/jvi.01293-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C., Miskin J., Hernáez B., et al. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 2001;75(20):9819–9827. doi: 10.1128/jvi.75.20.9819-9827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés G., Alejo A., Salas J., et al. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. J. Virol. 2002;76(24):12473–12482. doi: 10.1128/jvi.76.24.12473-12482.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés G., Alejo A., Simón-Mateo C., et al. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 2001;276(1):780–787. doi: 10.1074/jbc.M006844200. [DOI] [PubMed] [Google Scholar]
- Andrés G., Charro D., Matamoros T., et al. The cryo-EM structure of African swine fever virus unravels a unique architecture comprising two icosahedral protein capsids and two lipoprotein membranes. J. Biol. Chem. 2020;295(1):1–12. doi: 10.1074/jbc.AC119.011196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo A., Viñuela E., Alcamí A. Inhibition of African swine fever virus binding and infectivity by purified recombinant virus attachment protein p12. J. Virol. 1993;67(9):5463–5471. doi: 10.1128/jvi.67.9.5463-5471.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argilaguet J.M., Pérez-Martín E., López S., et al. BacMam immunization partially protects pigs against sublethal challenge with African swine fever virus. Antiviral Res. 2013;98(1):61–65. doi: 10.1016/j.antiviral.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Argilaguet J.M., Perez-Martin E., Nofrarias M., et al. DNA vaccination partially protects against African swine fever virus lethal challenge in the absence of antibodies. PLoS ONE. 2012;7(9):e40942. doi: 10.1371/journal.pone.0040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias M., De La Torre A., Dixon L., et al. Approaches and perspectives for development of African swine fever virus vaccines. Vaccines (Basel) 2017;5(4) doi: 10.3390/vaccines5040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barderas M.G., Rodríguez F., Gómez-Puertas P., et al. Antigenic and immunogenic properties of a chimera of two immunodominant African swine fever virus proteins. Arch Virol. 2001;146(9):1681–1691.doi. doi: 10.1007/s007050170056. [DOI] [PubMed] [Google Scholar]
- Berger P., Lapthorn A.J. The molecular relationship between antigenic domains and epitopes on hCG. Mol. Immunol. 2016;76:134–145. doi: 10.1016/j.molimm.2016.06.015. [DOI] [PubMed] [Google Scholar]
- Borca M.V., Carrillo C., Zsak L., et al. Deletion of a CD2-like gene, 8-DR, from African swine fever virus affects viral infection in domestic swine. J. Virol. 1998;72(4):2881–2889. doi: 10.1128/jvi.72.4.2881-2889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borca M.V., Irusta P., Carrillo C., et al. African swine fever virus structural protein p72 contains a conformational neutralizing epitope. Virology. 1994;201(2):413–418. doi: 10.1006/viro.1994.1311. [DOI] [PubMed] [Google Scholar]
- Borca M.V., O'donnell V., Holinka L.G., et al. Deletion of CD2-like gene from the genome of African swine fever virus strain Georgia does not attenuate virulence in swine. Sci. Rep. 2020;10(1):494. doi: 10.1038/s41598-020-57455-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borca M.V., Ramirez-Medina E., Silva E., et al. ASF vaccine candidate ASFV-G-∆I177L does not exhibit residual virulence in long-term clinical studies. Pathogens. 2023;12(6) doi: 10.3390/pathogens12060805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borca M.V., Ramirez-Medina E., Silva E., et al. ASFV-G-∆I177L as an effective oral nasal vaccine against the Eurasia strain of Africa swine fever. Viruses. 2021;13(5) doi: 10.3390/v13050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borca M.V., Ramirez-Medina E., Silva E., et al. Development of a highly effective African swine fever virus vaccine by deletion of the I177L gene results in sterile immunity against the current epidemic Eurasia strain. J. Virol. 2020;94(7) doi: 10.1128/jvi.02017-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Camós L., López E., Collado J., et al. M448R and MGF505-7R: Two African Swine Fever Virus Antigens Commonly Recognized by ASFV-Specific T-Cells and with Protective Potential. Vaccines (Basel) 2021;9(5):508. doi: 10.3390/vaccines9050508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff C.I., Bastos A.D., Gerber L.J., et al. Genetic characterisation of African swine fever viruses from outbreaks in southern Africa (1973-1999) Vet. Microbiol. 2007;121(1–2):45–55. doi: 10.1016/j.vetmic.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Brun A., Albina E., Barret T., et al. Antigen delivery systems for veterinary vaccine development. Viral-vector based delivery systems. Vaccine. 2008;26(51):6508–6528. doi: 10.1016/j.vaccine.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas-Fernández E., Sánchez-Vizcaíno J.M., Kosowska A., et al. Adenovirus-vectored African swine fever virus antigens cocktail is not protective against virulent arm07 isolate in Eurasian wild boar. Pathogens. 2020;9(3) doi: 10.3390/pathogens9030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho A., Viñuela E. Protein p22 of African swine fever virus: an early structural protein that is incorporated into the membrane of infected cells. Virology. 1991;181(1):251–257. doi: 10.1016/0042-6822(91)90490-3. [DOI] [PubMed] [Google Scholar]
- Chambaro H.M., Sasaki M., Sinkala Y., et al. Evidence for exposure of asymptomatic domestic pigs to African swine fever virus during an inter-epidemic period in Zambia. Transbound. Emerg. Dis. 2020;67(6):2741–2752. doi: 10.1111/tbed.13630. [DOI] [PubMed] [Google Scholar]
- Chaulagain S., Delhon G.A., Khatiwada S., et al. African swine fever virus CD2v protein induces β-interferon expression and apoptosis in swine peripheral blood mononuclear cells. Viruses. 2021;13(8) doi: 10.3390/v13081480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Li L., Guo S., et al. African swine fever virus pA104R protein acts as a suppressor of type I interferon signaling. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1169699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Liu B., Zhang X., et al. A porcine reproductive and respiratory syndrome virus (PRRSV)-specific IgM as a novel adjuvant for an inactivated PRRSV vaccine improves protection efficiency and enhances cell-mediated immunity against heterologous PRRSV challenge. Vet. Res. 2022;53(1):65. doi: 10.1186/s13567-022-01082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zhao D., He X., et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci. China Life Sci. 2020;63(5):623–634. doi: 10.1007/s11427-020-1657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Chen X., Liang Y., et al. Interaction network of African swine fever virus structural protein p30 with host proteins. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.971888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold C., Wileman T. The major structural protein of African swine fever virus, p73, is packaged into large structures, indicative of viral capsid or matrix precursors, on the endoplasmic reticulum. J. Virol. 1998;72(6):5215–5223. doi: 10.1128/jvi.72.6.5215-5223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos C., Gómez-Sebastian S., Moreno N., et al. African swine fever virus serodiagnosis: a general review with a focus on the analyses of African serum samples. Virus Res. 2013;173(1):159–167. doi: 10.1016/j.virusres.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Cwynar P., Stojkov J., Wlazlak K. African swine fever status in Europe. Viruses. 2019;11(4) doi: 10.3390/v11040310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho Ferreira H.C., Weesendorp E., Elbers A.R., et al. African swine fever virus excretion patterns in persistently infected animals: a quantitative approach. Vet. Microbiol. 2012;160(3–4):327–340. doi: 10.1016/j.vetmic.2012.06.025. [DOI] [PubMed] [Google Scholar]
- De Lorenzi G., Borella L., Alborali G.L., et al. African swine fever: a review of cleaning and disinfection procedures in commercial pig holdings. Res. Vet. Sci. 2020;132:262–267. doi: 10.1016/j.rvsc.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Deutschmann P., Forth J.H., Sehl-Ewert J., et al. Assessment of African swine fever vaccine candidate ASFV-G-∆MGF in a reversion to virulence study. NPJ Vaccines. 2023;8(1):78. doi: 10.1038/s41541-023-00669-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz C., Salát J., Břínek Kolařová D., et al. Examination of immunogenic properties of recombinant antigens based on p22 protein from African swine fever virus. J. Vet. Res. 2022;66(3):297–304. doi: 10.2478/jvetres-2022-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.K., Stahl K., Jori F., et al. African swine fever epidemiology and control. Annu Rev. Anim. Biosci. 2020;8:221–246. doi: 10.1146/annurev-animal-021419-083741. [DOI] [PubMed] [Google Scholar]
- Epifano C., Krijnse-Locker J., Salas M.L., et al. The African swine fever virus nonstructural protein pB602L is required for formation of the icosahedral capsid of the virus particle. J. Virol. 2006;80(24):12260–12270. doi: 10.1128/jvi.01323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano J.M., Galindo I., Alonso C. Antibody-mediated neutralization of African swine fever virus: myths and facts. Virus Res. 2013;173(1):101–109. doi: 10.1016/j.virusres.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Franzoni G., Dei Giudici S., Oggiano A. Infection, modulation and responses of antigen-presenting cells to African swine fever viruses. Virus Res. 2018;258:73–80. doi: 10.1016/j.virusres.2018.10.007. [DOI] [PubMed] [Google Scholar]
- Frouco G., Freitas F.B., Coelho J., et al. DNA-binding properties of African swine fever virus pA104R, a histone-like protein involved in viral replication and transcription. J. Virol. 2017;91(12) doi: 10.1128/jvi.02498-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo I., Almazán F., Bustos M.J., et al. African swine fever virus EP153R open reading frame encodes a glycoprotein involved in the hemadsorption of infected cells. Virology. 2000;266(2):340–351. doi: 10.1006/viro.1999.0080. [DOI] [PubMed] [Google Scholar]
- Galindo I., Viñuela E., Carrascosa A.L. Protein cell receptors mediate the saturable interaction of African swine fever virus attachment protein p12 with the surface of permissive cells. Virus Res. 1997;49(2):193–204. doi: 10.1016/s0168-1702(97)00037-3. [DOI] [PubMed] [Google Scholar]
- Gao Q., Yang Y., Luo Y., et al. African swine fever virus envelope glycoprotein CD2v interacts with host CSF2RA to regulate the JAK2-STAT3 pathway and inhibit apoptosis to facilitate virus replication. J. Virol. 2023;97(4) doi: 10.1128/jvi.01889-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S., Li J., Fan X., et al. Molecular characterization of African swine fever virus, China, 2018. Emerg. Infect. Dis. 2018;24(11):2131–2133. doi: 10.3201/eid2411.181274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladue D.P., O'donnell V., Ramirez-Medina E., et al. Deletion of CD2-like (CD2v) and C-type lectin-like (EP153R) genes from African swine fever virus Georgia-∆9GL abrogates its effectiveness as an experimental vaccine. Viruses. 2020;12(10) doi: 10.3390/v12101185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goatley L.C., Dixon L.K. Processing and localization of the African swine fever virus CD2v transmembrane protein. J. Virol. 2011;85(7):3294–3305. doi: 10.1128/jvi.01994-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goatley L.C., Reis A.L., Portugal R., et al. A pool of eight virally vectored African swine fever antigens protect pigs against fatal disease. Vaccines (Basel) 2020;8(2) doi: 10.3390/vaccines8020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Puertas P., Rodríguez F., Oviedo J.M., et al. The African swine fever virus proteins p54 and p30 are involved in two distinct steps of virus attachment and both contribute to the antibody-mediated protective immune response. Virology. 1998;243(2):461–471. doi: 10.1006/viro.1998.9068. [DOI] [PubMed] [Google Scholar]
- Gonzales W., Moreno C., Duran U., et al. African swine fever in the Dominican Republic. Transbound. Emerg. Dis. 2021;68(6):3018–3019. doi: 10.1111/tbed.14341. [DOI] [PubMed] [Google Scholar]
- Goonewardene K.B., Onyilagha C., Goolia M., et al. Superficial inguinal lymph nodes for screening dead pigs for African swine fever. Viruses. 2022;14(1) doi: 10.3390/v14010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath C.M., Windsor M., Wileman T. Membrane association facilitates the correct processing of pp220 during production of the major matrix proteins of African swine fever virus. J. Virol. 2003;77(3):1682–1690. doi: 10.1128/jvi.77.3.1682-1690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimerman M.E., Murgia M.V., Wu P., et al. Linear epitopes in African swine fever virus p72 recognized by monoclonal antibodies prepared against baculovirus-expressed antigen. J. Vet. Diagn. Invest. 2018;30(3):406–412. doi: 10.1177/1040638717753966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernáez B., Díaz-Gil G., García-Gallo M., et al. The African swine fever virus dynein-binding protein p54 induces infected cell apoptosis. FEBS Lett. 2004;569(1–3):224–228. doi: 10.1016/j.febslet.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Hernáez B., Guerra M., Salas M.L., et al. African swine fever virus undergoes outer envelope disruption, capsid disassembly and inner envelope fusion before core release from multivesicular endosomes. PLoS Pathog. 2016;12(4) doi: 10.1371/journal.ppat.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera L.R.M., Bisa E.P. In silico analysis of highly conserved cytotoxic T-cell epitopes in the structural proteins of African swine fever virus. Vet. World. 2021;14(10):2625–2633. doi: 10.14202/vetworld.2021.2625-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Zhong G., Ding S., et al. African swine fever virus protein p17 promotes mitophagy by facilitating the interaction of SQSTM1 with TOMM70. Virulence. 2023;14(1) doi: 10.1080/21505594.2023.2232707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istrate C., Marques J., Bule P., et al. In silico characterization of African swine fever virus nucleoprotein p10 interaction with DNA. Viruses. 2022;14(11) doi: 10.3390/v14112348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancovich J.K., Chapman D., Hansen D.T., et al. Immunization of pigs by DNA prime and recombinant vaccinia virus boost to identify and rank African swine fever virus immunogenic and protective proteins. J. Virol. 2018;92(8) doi: 10.1128/jvi.02219-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegaskanda S., Job E.R., Kramski M., et al. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J. Immunol. 2013;190(4):1837–1848. doi: 10.4049/jimmunol.1201574. [DOI] [PubMed] [Google Scholar]
- Kay-Jackson P.C., Goatley L.C., Cox L., et al. The CD2v protein of African swine fever virus interacts with the actin-binding adaptor protein SH3P7. J. Gen. Virol. 2004;85(Pt 1):119–130. doi: 10.1099/vir.0.19435-0. [DOI] [PubMed] [Google Scholar]
- Kollnberger S.D., Gutierrez-Castañeda B., Foster-Cuevas M., et al. Identification of the principal serological immunodeterminants of African swine fever virus by screening a virus cDNA library with antibody. J. Gen. Virol. 2002;83(Pt 6):1331–1342. doi: 10.1099/0022-1317-83-6-1331. [DOI] [PubMed] [Google Scholar]
- Lacasta A., Ballester M., Monteagudo P.L., et al. Expression library immunization can confer protection against lethal challenge with African swine fever virus. J. Virol. 2014;88(22):13322–13332. doi: 10.1128/JVI.01893-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhang Q., Liu Y., et al. Indirect ELISA using multi-antigenic dominants of p30, p54 and p72 recombinant proteins to detect antibodies against African swine fever virus in pigs. Viruses. 2022;14(12) doi: 10.3390/v14122660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Fu D., Zhang G., et al. Crystal structure of the African swine fever virus structural protein p35 reveals its role for core shell assembly. Protein Cell. 2020;11(8):600–605. doi: 10.1007/s13238-020-00730-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu Q., Shao L., et al. Structural insights into the assembly of the African swine fever virus inner capsid. J. Virol. 2023;97(6) doi: 10.1128/jvi.00268-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow P., Takamatsu H., Werling D., et al. Correlation of cell surface marker expression with African swine fever virus infection. Vet. Microbiol. 2014;168(2–4):413–419. doi: 10.1016/j.vetmic.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang X., Mao R., et al. Research progress on live attenuated vaccine against African swine fever virus. Microb. Pathog. 2021;158 doi: 10.1016/j.micpath.2021.105024. [DOI] [PubMed] [Google Scholar]
- Liu Q., Ma B., Qian N., et al. Structure of the African swine fever virus major capsid protein p72. Cell Res. 2019;29(11):953–955. doi: 10.1038/s41422-019-0232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Sun Y., Chai Y., et al. The structural basis of African swine fever virus pA104R binding to DNA and its inhibition by stilbene derivatives. Proc. Natl. Acad. Sci. U. S. A. 2020;117(20):11000–11009. doi: 10.1073/pnas.1922523117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Luo Y., Wang Y., et al. Cryo-EM structure of the African swine fever virus. Cell Host Microbe. 2019;26(6):836–843.e833. doi: 10.1016/j.chom.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang X., Qi W., et al. Prevention and control strategies of African swine fever and progress on pig farm repopulation in China. Viruses. 2021;13(12) doi: 10.3390/v13122552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokhandwala S., Petrovan V., Popescu L., et al. Adenovirus-vectored African Swine Fever Virus antigen cocktails are immunogenic but not protective against intranasal challenge with Georgia 2007/1 isolate. Vet Microbiol. 2019;235:10–20. doi: 10.1016/j.vetmic.2019.06.006. [DOI] [PubMed] [Google Scholar]
- Lokhandwala S., Waghela S.D., Bray J., et al. Induction of robust immune responses in swine by using a cocktail of adenovirus-vectored African swine fever virus antigens. Clin Vaccine Immunol. 2016;23(11):888–900. doi: 10.1128/cvi.00395-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokhandwala S., Waghela S.D., Bray J., et al. Adenovirus-vectored novel African Swine Fever Virus antigens elicit robust immune responses in swine. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopera-Madrid J., Medina-Magües L.G., Gladue D.P., et al. Optimization in the expression of ASFV proteins for the development of subunit vaccines using poxviruses as delivery vectors. Sci. Rep. 2021;11(1):23476. doi: 10.1038/s41598-021-02949-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopera-Madrid J., Osorio J.E., He Y., et al. Safety and immunogenicity of mammalian cell derived and Modified Vaccinia Ankara vectored African swine fever subunit antigens in swine. Vet Immunol Immunopathol. 2017;185:20–33. doi: 10.1016/j.vetimm.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C., Simón-Mateo C., Martínez L., et al. Gly-Gly-X, a novel consensus sequence for the proteolytic processing of viral and cellular proteins. J. Biol. Chem. 1989;264(16):9107–9110. [PubMed] [Google Scholar]
- Lu W., Bai Y., Zhang S., et al. An intracellular epitope of ASFV CD2v protein elicits humoral and cellular immune responses. Animals (Basel) 2023;13(12) doi: 10.3390/ani13121967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong H.Q., Lai H.T., Do L.D., et al. Differential antibody responses in sows and finishing pigs naturally infected with African swine fever virus under field conditions. Virus Res. 2022;307 doi: 10.1016/j.virusres.2021.198621. [DOI] [PubMed] [Google Scholar]
- Monteagudo P.L., Lacasta A., López E., et al. BA71ΔCD2: a new recombinant live attenuated African swine fever virus with cross-protective capabilities. J. Virol. 2017;91(21) doi: 10.1128/jvi.01058-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz A.L., Tabarés E. Characteristics of the major structural proteins of African swine fever virus: role as antigens in the induction of neutralizing antibodies. A review. Virology. 2022;571:46–51. doi: 10.1016/j.virol.2022.04.001. [DOI] [PubMed] [Google Scholar]
- Muñoz M., Freije J.M., Salas M.L., et al. Structure and expression in E. coli of the gene coding for protein p10 of African swine fever virus. Arch. Virol. 1993;130(1–2):93–107. doi: 10.1007/bf01318999. [DOI] [PubMed] [Google Scholar]
- Murgia M.V., Mogler M., Certoma A., et al. Evaluation of an African swine fever (ASF) vaccine strategy incorporating priming with an alphavirus-expressed antigen followed by boosting with attenuated ASF virus. Arch. Virol. 2019;164(2):359–370. doi: 10.1007/s00705-018-4071-8. [DOI] [PubMed] [Google Scholar]
- Neilan J.G., Zsak L., Lu Z., et al. Neutralizing antibodies to African swine fever virus proteins p30, p54, and p72 are not sufficient for antibody-mediated protection. Virology. 2004;319(2):337–342. doi: 10.1016/j.virol.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Netherton C.L., Goatley L.C., Reis A.L., et al. Identification and immunogenicity of African swine fever virus antigens. Front Immunol. 2019;10:1318. doi: 10.3389/fimmu.2019.01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njau E.P., Machuka E.M., Cleaveland S., et al. African swine fever virus (ASFV): biology, genomics and genotypes circulating in Sub-Saharan Africa. Viruses. 2021;13(11) doi: 10.3390/v13112285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Correia I., Rodríguez J.M., Eulálio A., et al. African swine fever virus p10 protein exhibits nuclear import capacity and accumulates in the nucleus during viral infection. Vet. Microbiol. 2008;130(1–2):47–59. doi: 10.1016/j.vetmic.2007.12.010. [DOI] [PubMed] [Google Scholar]
- O'donnell V., Holinka L.G., Gladue D.P., et al. African swine fever virus Georgia isolate harboring deletions of MGF360 and MGF505 genes is attenuated in swine and confers protection against challenge with virulent parental virus. J. Virol. 2015;89(11):6048–6056. doi: 10.1128/jvi.00554-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oura C.a.L., Denyer M.S., Takamatsu H., et al. In vivo depletion of CD8+ T lymphocytes abrogates protective immunity to African swine fever virus. J. Gen. Virol. 2005;86(Pt 9):2445–2450. doi: 10.1099/vir.0.81038-0. [DOI] [PubMed] [Google Scholar]
- Oviedo J.M., Rodríguez F., Gómez-Puertas P., et al. High level expression of the major antigenic African swine fever virus proteins p54 and p30 in baculovirus and their potential use as diagnostic reagents. J. Virol. Methods. 1997;64(1):27–35. doi: 10.1016/s0166-0934(96)02140-4. [DOI] [PubMed] [Google Scholar]
- Penrith M.L., Bastos A.D., Etter E.M.C., et al. Epidemiology of African swine fever in Africa today: sylvatic cycle versus socio-economic imperatives. Transbound. Emerg. Dis. 2019;66(2):672–686. doi: 10.1111/tbed.13117. [DOI] [PubMed] [Google Scholar]
- Pérez-Núñez D., García-Belmonte R., Riera E., et al. Signal peptide and N-glycosylation of N-terminal-CD2v determine the hemadsorption of African swine fever virus. J. Virol. 2023 doi: 10.1128/jvi.01030-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Núñez D., García-Urdiales E., Martínez-Bonet M., et al. CD2v interacts with adaptor protein AP-1 during African swine fever infection. PLoS ONE. 2015;10(4) doi: 10.1371/journal.pone.0123714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.E. Kansas State University; 2016. Epitope Mapping of African Swine Fever Virus p72 Capsid Protein Using Polyclonal Swine Sera and Monoclonal Antibodies. [Google Scholar]
- Plowright W., Parker J., Peirce M.A. African swine fever virus in ticks (Ornithodoros moubata, murray) collected from animal burrows in Tanzania. Nature. 1969;221(5185):1071–1073. doi: 10.1038/2211071a0. [DOI] [PubMed] [Google Scholar]
- Ramirez-Medina E., Vuono E.A., Pruitt S., et al. Deletion of African swine fever virus histone-like protein, A104R from the Georgia isolate drastically reduces virus virulence in domestic pigs. Viruses. 2022;14(5) doi: 10.3390/v14051112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathakrishnan A., Reis A.L., Petrovan V., et al. A protective multiple gene-deleted African swine fever virus genotype II, Georgia 2007/1, expressing a modified non-haemadsorbing CD2v protein. Emerg. Microbes Infect. 2023;12(2) doi: 10.1080/22221751.2023.2265661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis A.L., Parkhouse R.M.E., Penedos A.R., et al. Systematic analysis of longitudinal serological responses of pigs infected experimentally with African swine fever virus. J. Gen. Virol. 2007;88(Pt 9):2426–2434. doi: 10.1099/vir.0.82857-0. [DOI] [PubMed] [Google Scholar]
- Ren D., Ding P., Liu S., et al. Development and characterization of recombinant ASFV CD2v protein nanoparticle-induced monoclonal antibody. Int. J. Biol. Macromol. 2022;209(Pt A):533–541. doi: 10.1016/j.ijbiomac.2022.03.069. [DOI] [PubMed] [Google Scholar]
- Rodriguez F., Ley V., Gómez-Puertas P., et al. The structural protein p54 is essential for African swine fever virus viability. Virus Res. 1996;40(2):161–167. doi: 10.1016/0168-1702(95)01268-0. [DOI] [PubMed] [Google Scholar]
- Rodríguez J.M., García-Escudero R., Salas M.L., et al. African swine fever virus structural protein p54 is essential for the recruitment of envelope precursors to assembly sites. J. Virol. 2004;78(8):4299. doi: 10.1128/jvi.78.8.4299-4313.2004. -1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez J.M., Yáñez R.J., Almazán F., et al. African swine fever virus encodes a CD2 homolog responsible for the adhesion of erythrocytes to infected cells. J. Virol. 1993;67(9):5312–5320. doi: 10.1128/jvi.67.9.5312-5320.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gonzalvo F., Rodríguez F., Escribano J.M. Functional and immunological properties of the baculovirus-expressed hemagglutinin of African swine fever virus. Virology. 1996;218(1):285–289. doi: 10.1006/viro.1996.0193. [DOI] [PubMed] [Google Scholar]
- Salas M.L., Andrés G. African swine fever virus morphogenesis. Virus Res. 2013;173(1):29–41. doi: 10.1016/j.virusres.2012.09.016. [DOI] [PubMed] [Google Scholar]
- San Martín C., Van Raaij M.J. The so far farthest reaches of the double jelly roll capsid protein fold. Virol. J. 2018;15(1):181. doi: 10.1186/s12985-018-1097-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Sampedro L., Perdiguero B., Mejías-Pérez E., et al. The evolution of poxvirus vaccines. Viruses. 2015;7(4):1726–1803. doi: 10.3390/v7041726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Vizcaíno J.M., Mur L., Gomez-Villamandos J.C., et al. An update on the epidemiology and pathology of African swine fever. J. Comp. Pathol. 2015;152(1):9–21. doi: 10.1016/j.jcpa.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Sánchez E.G., Quintas A., Nogal M., et al. African swine fever virus controls the host transcription and cellular machinery of protein synthesis. Virus Res. 2013;173(1):58–75. doi: 10.1016/j.virusres.2012.10.025. [DOI] [PubMed] [Google Scholar]
- Schäfer A., Franzoni G., Netherton C.L., et al. Adaptive cellular immunity against African swine fever virus infections. Pathogens. 2022;11(2) doi: 10.3390/pathogens11020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlafer D.H., Mebus C.A., Mcvicar J.W. African swine fever in neonatal pigs: passively acquired protection from colostrum or serum of recovered pigs. Am. J. Vet. Res. 1984;45(7):1367–1372. [PubMed] [Google Scholar]
- Silva E.B., Krug P.W., Ramirez-Medina E., et al. The presence of virus neutralizing antibodies is highly associated with protection against virulent challenge in domestic pigs immunized with ASFV live attenuated vaccine candidates. Pathogens. 2022;11(11) doi: 10.3390/pathogens11111311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimčíková M., Prather K.L., Prazeres D.M., et al. Towards effective non-viral gene delivery vector. Biotechnol. Genet. Eng. Rev. 2015;31(1–2):82–107. doi: 10.1080/02648725.2016.1178011. [DOI] [PubMed] [Google Scholar]
- Simón-Mateo C., Andrés G., Viñuela E. Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus. EMBO J. 1993;12(7):2977–2987. doi: 10.1002/j.1460-2075.1993.tb05960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.S., Hess W.R. Antibody response to inactivated preparations of African swine fever virus in pigs. Am. J. Vet. Res. 1967;28(123):475–481. [PubMed] [Google Scholar]
- Suárez C., Gutiérrez-Berzal J., Andrés G., et al. African swine fever virus protein p17 is essential for the progression of viral membrane precursors toward icosahedral intermediates. J. Virol. 2010;84(15):7484–7499. doi: 10.1128/jvi.00600-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez C., Salas M.L., Rodríguez J.M. African swine fever virus polyprotein pp62 is essential for viral core development. J. Virol. 2010;84(1):176–187. doi: 10.1128/jvi.01858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo S.Y., Perez-Nunez D., Morozov I., et al. DNA-protein vaccination strategy does not protect from challenge with African swine fever virus Armenia 2007 strain. Vaccines (Basel) 2019;7(1) doi: 10.3390/vaccines7010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Luo Y., Wen T., et al. A quadruple protection procedure for resuming pig production in small-scale ASFV-positive farms in China. Curr. Res. Microb. Sci. 2021;2 doi: 10.1016/j.crmicr.2020.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H.T.T., Truong A.D., Dang A.K., et al. Genetic characterization of African swine fever viruses circulating in North Central region of Vietnam. Transbound. Emerg. Dis. 2021;68(3):1697–1699. doi: 10.1111/tbed.13835. [DOI] [PubMed] [Google Scholar]
- Tran X.H., Phuong L.T.T., Huy N.Q., et al. Evaluation of the safety profile of the ASFV vaccine candidate ASFV-G-ΔI177L. Viruses. 2022;14(5) doi: 10.3390/v14050896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbano A.C., Ferreira F. African swine fever control and prevention: an update on vaccine development. Emerg. Microbes Infect. 2022;11(1):2021–2033. doi: 10.1080/22221751.2022.2108342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuono E.A., Ramirez-Medina E., Pruitt S., et al. Evaluation of the function of the ASFV KP177R gene, encoding for structural protein p22, in the process of virus replication and in swine virulence. Viruses. 2021;13(6) doi: 10.3390/v13060986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak M., Juszkiewicz M., Szymankiewicz K., et al. ASF -survivors' sera do not inhibit African swine fever virus replication in vitro. J. Vet. Res. 2022;66(1):21–27. doi: 10.2478/jvetres-2022-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Xie M., Wu W., et al. Structures and functional diversities of ASFV proteins. Viruses. 2021;13(11) doi: 10.3390/v13112124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhao D., Wang J., et al. Architecture of African swine fever virus and implications for viral assembly. Science. 2019;366(6465):640–644. doi: 10.1126/science.aaz1439. [DOI] [PubMed] [Google Scholar]
- Wang Y., Kang W., Yang W., et al. Structure of African swine fever virus and associated molecular mechanisms underlying infection and immunosuppression: a review. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.715582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhang J., Li F., et al. The attenuated African swine fever vaccine HLJ/18-7GD provides protection against emerging prevalent genotype II variants in China. Emerg. Microbes Infect. 2024;13(1) doi: 10.1080/22221751.2023.2300464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Bowden T.A., Wilson I.A., et al. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 2019;1863(10):1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Lowe A.D., Rodríguez Y.Y., et al. Antigenic regions of African swine fever virus phosphoprotein P30. Transbound. Emerg. Dis. 2020 doi: 10.1111/tbed.13533. [DOI] [PubMed] [Google Scholar]
- Xia N., Wang H., Liu X., et al. African swine fever virus structural protein p17 inhibits cell proliferation through ER stress-ROS mediated cell cycle arrest. Viruses. 2020;13(1) doi: 10.3390/v13010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez R.J., Rodríguez J.M., Nogal M.L., et al. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208(1):249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- Yoon K.J., Wu L.L., Zimmerman J.J., et al. Antibody-dependent enhancement (ADE) of porcine reproductive and respiratory syndrome virus (PRRSV) infection in pigs. Viral Immunol. 1996;9(1):51–63. doi: 10.1089/vim.1996.9.51. [DOI] [PubMed] [Google Scholar]
- Yuan Z.H., Fang X., Zheng L.J., et al. HBV DNA vaccine immunization the effects of DNA vector constructs and routes of gene delivery on the induction of antiviral immunity. Chin. J. Microbiol. Immunol. 1999;19(01):29–32. doi: 10.3760/j:issn:0254-5101.1999.01.011. [DOI] [Google Scholar]
- Zajac M.D., Sangewar N., Lokhandwala S., et al. Adenovirus-vectored African swine fever virus pp220 Induces robust antibody, IFN-gamma, and CTL responses in pigs. Front. Vet. Sci. 2022;9 doi: 10.3389/fvets.2022.921481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac M.D., Trujillo J.D., Yao J., et al. Immunization of pigs with replication-incompetent adenovirus-vectored African swine fever virus multi-antigens induced humoral immune responses but no protection following contact challenge. Front. Vet. Sci. 2023;10 doi: 10.3389/fvets.2023.1208275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Liu W., Gao Z., et al. Antigenic and immunogenic properties of recombinant proteins consisting of two immunodominant African swine fever virus proteins fused with bacterial lipoprotein OprI. Virol. J. 2022;19(1):16. doi: 10.1186/s12985-022-01747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Liu W., Gao Z., et al. Antigenicity and immunogenicity of recombinant proteins comprising African swine fever virus proteins p30 and p54 fused to a cell-penetrating peptide. Int. Immunopharmacol. 2021;101(Pt A) doi: 10.1016/j.intimp.2021.108251. [DOI] [PubMed] [Google Scholar]
- Zhao D., Liu R., Zhang X., et al. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg. Microbes. Infect. 2019;8(1):438–447. doi: 10.1080/22221751.2019.1590128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Sun E., Huang L., et al. Highly lethal genotype I and II recombinant African swine fever viruses detected in pigs. Nat. Commun. 2023;14(1):3096. doi: 10.1038/s41467-023-38868-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.J. African swine fever vaccinology: the biological challenges from immunological perspectives. Viruses. 2022;14(9) doi: 10.3390/v14092021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Fan B., Zhou J., et al. A high-throughput method to analyze the interaction proteins with p22 protein of African swine fever virus in vitro. Front. Vet. Sci. 2021;8 doi: 10.3389/fvets.2021.719859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.