Abstract
Aims
Heart failure (HF) has shared genetic architecture with its risk factors: atrial fibrillation (AF), body mass index (BMI), coronary heart disease (CHD), systolic blood pressure (SBP), and type 2 diabetes (T2D). We aim to assess the association and risk prediction performance of risk‐factor polygenic risk scores (PRSs) for incident HF and its subtypes in bi‐racial populations.
Methods and results
Five PRSs were constructed for AF, BMI, CHD, SBP, and T2D in White participants of the Atherosclerosis Risk in Communities (ARIC) study. The associations between PRSs and incident HF and its subtypes were assessed using Cox models, and the risk prediction performance of PRSs was assessed using C statistics. Replication was performed in the ARIC study Black and Cardiovascular Health Study (CHS) White participants. In 8624 ARIC study Whites, 1922 (31% cumulative incidence) HF cases developed over 30 years of follow‐up. PRSs of AF, BMI, and CHD were associated with incident HF (P < 0.001), where PRSAF showed the strongest association [hazard ratio (HR): 1.47, 95% confidence interval (CI): 1.41–1.53]. Only the addition of PRSAF to the ARIC study HF risk equation improved C statistics for 10 year risk prediction from 0.812 to 0.829 (∆C: 0.017, 95% CI: 0.009–0.026). The PRSAF was associated with both incident HF with reduced ejection fraction (HR: 1.43, 95% CI: 1.27–1.60) and incident HF with preserved ejection fraction (HR: 1.46, 95% CI: 1.33–1.62). The associations between PRSAF and incident HF and its subtypes, as well as the improved risk prediction, were replicated in the ARIC study Blacks and the CHS Whites (P < 0.050). Protein analyses revealed that N‐terminal pro‐brain natriuretic peptide and other 98 proteins were associated with PRSAF.
Conclusions
The PRSAF was associated with incident HF and its subtypes and had significant incremental value over an established HF risk prediction equation.
Keywords: Polygenic risk score, Heart failure, Risk prediction, Cohort
Introduction
Heart failure (HF) is a complex disorder with known comorbidities and lifestyle risk factors that affects more than 6 million American adults, and its prevalence continues to grow. 1 The genetic heritability (h2) of HF, defined as the amount of inherited genetic variation associated with HF, was characterized by a Swedish adoption study at 26%, suggesting that individuals may have a genetic predisposition to developing HF. 2 A genome‐wide association study (GWAS) of 26 studies from the Heart Failure Molecular Epidemiology for Therapeutic Targets (HERMES) Consortium identified 12 independent single nucleotide polymorphisms (SNPs) significantly associated with HF. Of these SNPs, nine were also associated with HF risk factors, including atrial fibrillation (AF), coronary heart disease (CHD), low‐density lipoprotein cholesterol, type 2 diabetes (T2D), body mass index (BMI), systolic blood pressure (SBP), and diastolic blood pressure. 3 The latest GWAS of HF, which integrated HERMES and biobank resources with more than 1.5 million participants, identified 47 risk loci, confirming the shared genetic aetiology between HF and its risk factors. 4 However, the relations between genetic variants and HF incidence remain elusive.
Risk prediction equations have been developed that integrate clinical risk factors to facilitate identification of individuals at high risk of developing HF so that primary prevention can be more effectively administered. Polygenic risk scores (PRSs), summarizing the estimated genetic effects of SNPs from GWAS, may offer an opportunity to enhance disease risk prediction so at‐risk individuals can be identified earlier with the goal to improve disease surveillance and better inform treatment plans, according to a statement from the American Heart Association. 5 PRSs have been successfully implemented in predicting disease outcomes, such as CHD and stroke, demonstrating improvement beyond clinical risk factors. 6 , 7 , 8 Evidence has also suggested that PRSs based on clinical risk factors may have predictive power on clinical outcomes. 8 , 9 Currently, there are no genome‐wide PRSs of HF that have become available, and few studies have been conducted focusing on HF. Therefore, we sought to investigate the added value of PRSs, derived from clinical risk factors, on HF risk prediction over an established prediction equation in the Atherosclerosis Risk in Communities (ARIC) study, 10 a longitudinal cohort with 30 year follow‐up of HF, and to replicate our findings in the Cardiovascular Health Study (CHS). We further utilized proteome data from the ARIC study to unravel the underlying pathways associated with the PRSs that improved HF risk prediction.
Methods
Study population
The ARIC study is a prospective cohort study comprised of 15 792 men and women, mostly Blacks and Whites, to investigate the aetiology of atherosclerosis and cardiovascular disease (CVD). Initial enrolment (1987–89) occurred in four US communities: Forsyth County, North Carolina; Jackson, Mississippi; suburban Minneapolis, Minnesota; and Washington County, Maryland, followed by nine clinical visits. The CHS is a longitudinal study comprised of men and women aged 65 years and older to investigate the importance of risk factors related to CHD and stroke. Initial enrolment (1989–90) occurred in four communities: Forsyth County, North Carolina; Sacramento County, California; Washington County, Maryland; and Pittsburgh, Pennsylvania, and follow‐up visits occurred until 1999. The design of two studies has previously been published. 11 , 12
Present analyses included the ARIC study Blacks and Whites from baseline visit (1987–89), Visit 3 (1993–95), and Visit 4 (1996–98) and the CHS Whites from baseline visit (1989–90). Participants with prevalent HF or those who did not have genetic information, clinical risk factors at the respective visit, or HF follow‐up were excluded (Supporting Information, Figure S1 ). All study participants provided informed consent, and study protocols were approved by institutional review boards at all participating institutions.
HF outcome and clinical risk factors
In the ARIC study, incident HF was defined using hospitalization or death records with discharge code International Classification of Diseases (ICD)‐9428, in any position, or ICD‐10150. Starting in 2005, adjudication of all hospitalizations with potential HF‐related ICD discharge codes was implemented, with retrieval and abstraction of eligible medical records and subsequent review for evidence of HF signs and symptoms. 13 Participants with ejection fraction ≥50% with HF symptoms were further defined as HF with preserved ejection fraction (HFpEF), and participants with ejection fraction <50% were defined as HF with reduced ejection fraction (HFrEF), which included both HFrEF (ejection fraction <40%) and HF with mid‐range ejection fraction (HFmrEF) (ejection fraction between 40% and 49%) per 2016 European Society of Cardiology (ESC) HF subtype definition. 14 , 15 In the current analysis, our primary outcome was incident HF occurring after Visit 1 until 31 December 2018, and secondary outcomes included incident HFpEF and HFrEF adjudicated after 2005 until 31 December 2018. Participants at the Jackson, Mississippi, field centre included incident HF cases occurring after Visit 1 until 31 December 2017.
In the CHS, incident HF was defined by a physician diagnosis of symptomatic HF plus supporting evidence of medical treatment of HF or supportive clinical findings on echocardiography, contrast ventriculography, or chest radiography. HF was further subtyped into HFpEF and HFrEF on the basis of findings from echocardiography and cardiac catheterization reports. 16 Primary outcome was all‐cause incident HF occurring after baseline exam until 2015.
Clinical exams were administered at each study visit, and the current analyses used risk factors collected from the ARIC study Visits 1 and 4, and the CHS Visit 1, unless stated otherwise. Details about demographic and clinical information collection are provided in Supporting Information, Methods section.
Proteomic measurements
At the ARIC study Visit 3, blood samples were collected and stored at −80°C, and the relative concentrations of plasma proteins or protein complexes were measured using an aptamer‐based approach 17 by SomaLogic Inc. (Boulder, CO, USA). Details of the ARIC study proteomic measures and quality control were described previously. 18 A total of 4877 aptamers measuring 4697 unique proteins or protein complexes passed quality control and were included in the current proteomic analyses.
Statistical analysis
Five PRSs for HF risk factors (AF, BMI, CHD, SBP, and T2D) were constructed in the ARIC study Whites for primary analyses, while PRSAF was constructed in the ARIC study Blacks and the CHS Whites for replication analyses. PRSs were calculated using pre‐defined allele weights developed by Khera et al. 7 , 19 for AF, BMI, CHD, and T2D and Vaura et al. 20 for SBP (Supporting Information, Methods section). PRSs were further standardized with a mean of 0 and standard deviation (SD) of 1 prior to analyses. More than 90% of SNPs reported in Khera et al. 7 , 19 and Vaura et al. 20 were included in the ARIC study Whites PRS calculations. Details are explained in Supporting Information, Methods section and Table S1 .
The cumulative incidence of HF was calculated using Kaplan–Meier curves to account for censored participants. Primary analyses were conducted in the ARIC study Whites at Visit 1 because the five PRSs originally developed were based on European ancestry. 7 , 19 , 20 Cox models were used to test the association between incident HF and each PRS. Models were adjusted for age, sex, BMI, SBP, heart rate, smoking status, prevalent CHD, prevalent T2D, blood pressure‐lowering medication use, as defined in the ARIC study HF prediction equation, 10 and study centre, prevalent AF, and the first 10 genetic principal components. Proportional hazard assumptions for PRSs were assessed through visual inspection of Schoenfeld residual plots, and no violation of proportionality was observed. An additional Cox model was fit including all nominal significant PRSs (P < 0.050) identified in individual PRS analyses. We evaluated the PRS risk reclassification and prediction performance by computing C statistics calculated for 10, 20, and 30 years. A censoring‐adjusted C statistic proposed by Uno et al. 21 was used, which has an equivalent interpretation as Harrell's C for censored survival data. The average 95% confidence intervals (CIs) of the C statistics were calculated via 100 iterations of perturbation resampling. We further conducted stratified analysis by sex and age groups to examine potential effect modifications of each PRS, and finally, we tested the association between PRS quintiles and incident HF. Statistical significance was determined at P < 0.010 to account for five PRSs analysed. We performed secondary analyses to estimate the effect of PRSAF on HF subtypes, HFrEF and HFpEF, using data collected from participants at the ARIC study Visit 4 and the CHS baseline visit. Cox models were conducted for HFrEF and HFpEF, respectively: Model 1 adjusted for risk factors in the ARIC study HF prediction equation, 10 study centre, prevalent AF, and first 10 genetic principal components, and Model 2 additionally adjusted for natural log‐transformed N‐terminal pro‐brain natriuretic peptide (NT‐proBNP) levels. Replication analyses were performed in the ARIC study Blacks and the CHS Whites using the aforementioned models.
Protein pathway analysis
To explore the biological pathways underlying HF‐associated PRSs, we related PRSAF with proteomic measures in the ARIC study Whites at Visit 3. Linear regressions were performed to determine associations between inverse‐normal transformed protein values and PRSAF adjusting for age, sex, study centre, and the first 10 principal components. Proteins significantly associated with PRSAF [false discovery rate (FDR) <0.050] were further considered in the pathway analysis. A sensitivity analysis was performed on PRSAF by further adjusting for estimated glomerular filtration rate the use of anti‐coagulates at Visit 3. The functional annotations of those significantly associated proteins were determined using Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes pathway analysis. A protein–protein interaction network was further created using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING). Detailed methods are explained in Supporting Information, Methods section.
Analyses were performed using RStudio v3.6.2 in conjunction with the survival, survminer, and survC1 packages. Two‐sided P < 0.050 was considered statistically significant, unless specified otherwise.
Results
Study population characteristics
Baseline characteristics of the ARIC study and CHS participants are shown in Table 1 . Primary analyses consisted of 8624 ARIC study Whites at Visit 1 (52% female) with a mean age of 54.2 years. During a median of 27.2 year follow‐up (inter‐quartile range, 18.12–30.12), 1922 (31% cumulative incidence) participants developed HF. HF cases tended to have a higher prevalence of comorbidities and unfavourable clinical risk factors such as current smokers and higher blood pressure levels. Replication analyses were performed in 2525 ARIC study Blacks (61% female, mean age 53.3 years) and 3156 CHS Whites (61% female, mean age 72.3 years). In the ARIC study Blacks, during a median of 24.9 year follow‐up (inter‐quartile range, 15.62–28.94), 735 (37% cumulative incidence) participants developed HF. In the CHS Whites, 1077 (57% cumulative incidence) participants developed HF during a median of 13.28 year follow‐up (inter‐quartile range, 7.93–19.41). PRSs were approximately normally distributed in the ARIC study and the CHS, and HF cases tended to have a higher median standardized PRS than those who remained free of HF during follow‐up (Figure 1 ).
Table 1.
Baseline characteristics of participants from the ARIC study and the CHS enrolled at baseline visits
| ARIC study Whites | ARIC study Blacks | CHS study Whites | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Full population | HF cases | HF‐free controls | Full population | HF cases | HF‐free controls | Full population | HF cases | HF‐free controls | |
| N | 8624 | 1922 | 6702 | 2525 | 735 | 1790 | 3156 | 1077 | 2079 |
| Age (years) | 54.22 (5.68) | 56.34 (5.36) | 53.61 (5.63) | 53.29 (5.75) | 54.65 (5.63) | 52.73 (5.71) | 72.30 (5.36) | 72.59 (5.27) | 72.16 (5.40) |
| Sex (female) | 4520 (52) | 879 (46) | 3641 (54) | 1548 (61) | 440 (60) | 1108 (62) | 1919 (61) | 636 (59) | 1283 (62) |
| BMI (kg/m2) | 26.86 (4.71) | 28.16 (5.17) | 26.49 (4.50) | 29.50 (5.97) | 31.19 (6.92) | 28.81 (5.38) | 26.28 (4.43) | 26.83 (4.58) | 26.00 (4.32) |
| SBP (mmHg) | 118.18 (16.86) | 123.69 (17.90) | 116.60 (16.21) | 128.55 (21.08) | 133.29 (22.81) | 126.60 (20.02) | 136.29 (21.06) | 139.30 (22.28) | 134.73 (20.23) |
| Heart rate (b.p.m.) | 66.44 (9.92) | 67.14 (10.65) | 66.24 (9.69) | 66.61 (10.81) | 67.99 (11.71) | 66.04 (10.37) | 64.84 (11.00) | 65.03 (11.07) | 64.74 (10.96) |
| BP‐lowering drug users | 1525 (18) | 526 (27) | 999 (15) | 954 (38) | 369 (50) | 585 (33) | 1118 (35) | 457 (42) | 661 (43) |
| Smoking status | |||||||||
| Current | 2129 (25) | 591 (31) | 1538 (23) | 734 (29) | 229 (31) | 505 (28) | 365 (12) | 117 (11) | 248 (12) |
| Former | 3044 (35) | 728 (38) | 2316 (35) | 615 (24) | 187 (25) | 428 (24) | 1287 (41) | 456 (42) | 831 (40) |
| Never | 3451 (40) | 603 (31) | 2848 (42) | 1176 (47) | 319 (43) | 857 (48) | 1504 (48) | 504 (47) | 1000 (48) |
| Prevalent CHD | 379 (4) | 198 (10) | 181 (3) | 79 (3) | 42 (6) | 37 (2) | 0 (0) | 0 (0) | 0 (0) |
| Prevalent AF | 98 (1) | 32 (2) | 66 (1) | 56 (2) | 18 (2) | 38 (2) | 64 (2) | 31 (3) | 33 (2) |
| Prevalent diabetes | 698 (8) | 281 (15) | 417 (6) | 469 (19) | 240 (33) | 229 (13) | 375 (12) | 158 (5) | 217 (10) |
| Follow‐up time (years) | 27.20 (18.12, 30.12) | 19.08 (12.31, 25.05) | 29.20 (21.31, 30.43) | 24.93 (15.62, 28.94) | 17.45 (10.77, 23.40) | 28.17 (19.95, 29.42) | 13.28 (7.93, 19.41) | 11.29 (6.82, 16.44) | 14.36 (8.57, 20.99) |
AF, atrial fibrillation; ARIC, Atherosclerosis Risk in Communities; BMI, body mass index; BP, blood pressure; CHD, coronary heart disease; CHS, Cardiovascular Health Study; HF, heart failure; SBP, systolic blood pressure.
Follow‐up time is reported as median and percentiles (25th, 75th), all other continuous measures are reported as mean and standard deviation, and categorical measures are reported as frequency and percentage.
Figure 1.
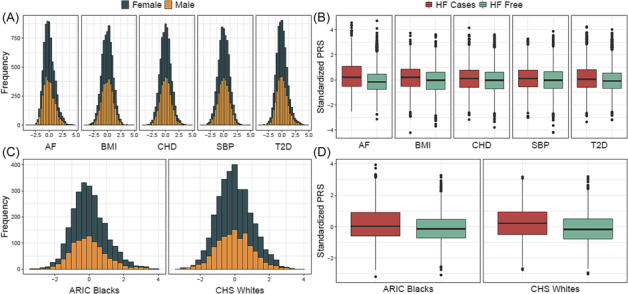
Distribution of polygenic risk scores (PRSs) for atrial fibrillation (AF), body mass index (BMI), coronary heart disease (CHD), systolic blood pressure (SBP), and type 2 diabetes (T2D) in the Atherosclerosis Risk in Communities (ARIC) study Whites by (A) sex and (B) heart failure status. Distribution of PRSAF in the ARIC study Blacks and the Cardiovascular Health Study (CHS) Whites by (C) sex and (D) heart failure status.
Polygenic risk scores with HF association and risk prediction
PRSAF, PRSBMI, and PRSCHD were significantly associated with incident HF in the ARIC study Whites (P < 0.001, Table 2 ). Per SD increase of PRSAF, PRSBMI, and PRSCHD was associated with 47%, 11%, and 12% higher risk of HF after accounting for HF risk factors. PRSSBP and PRST2D were not significantly associated with incident HF. Sex‐ and age‐stratified analyses yielded similar results between men and women and between early and late middle‐aged (age <55 vs. ≥55 years) adults. The association was further assessed by regressing incident HF on PRSAF, PRSBMI, and PRSCHD simultaneously. Each PRS remained significant (P < 0.050), and their effects did not attenuate materially (Supporting Information, Table S2 ). We investigated the relationship between incident HF and PRS quintiles for AF, BMI, and CHD. Kaplan–Meier curves graphically indicated a graded effect of quintiles on the risk of HF for PRSAF, and a markedly increased risk was observed in the fifth quintile (Figure 2A ). Slightly less graded effects were observed from PRSBMI (Figure 2B ) and PRSCHD (Figure 2C ) quintiles on the risk of HF. Log‐rank tests suggested statistical differences across the PRS quintiles for AF, BMI, and CHD (P < 0.0001). PRSAF quintiles had the greatest magnitude of effect on HF risk compared with PRSBMI and PRSCHD quintiles (Supporting Information, Table S2 ). Participants with a PRSAF in the top 20% had a more than three‐fold risk for HF compared with those in the bottom 20% [hazard ratio (HR): 3.02, 95% CI: 2.62–3.48].
Table 2.
Hazard ratios and 95% confidence intervals of polygenic risk score on incident heart failure in Whites from the ARIC study at Visit 1 (1987–89)
| N (HF events) | AF PRS | BMI PRS | CHD PRS | SBP PRS | T2D PRS | |
|---|---|---|---|---|---|---|
| All | 8624 (1992) | 1.47 (1.41–1.53) | 1.11 (1.05–1.16) | 1.12 (1.07–1.17) | 1.05 (1.00–1.10) | 1.03 (0.98–1.09) |
| Women | 4520 (879) | 1.45 (1.37–1.54) | 1.14 (1.05–1.23) | 1.07 (1.01–1.15) | 1.04 (0.96–1.12) | 1.04 (0.96–1.12) |
| Men | 4104 (1043) | 1.49 (1.40–1.57) | 1.07 (1.00–1.15) | 1.17 (1.10–1.24) | 1.06 (0.99–1.13) | 1.03 (0.95–1.11) |
| Age <55 | 4501 (683) | 1.41 (1.31–1.51) | 1.19 (1.08–1.30) | 1.05 (0.97–1.13) | 1.10 (1.01–1.20) | 1.01 (0.92–1.11) |
| Age ≥55 | 4123 (1239) | 1.49 (1.41–1.57) | 1.06 (1.00–1.13) | 1.14 (1.08–1.21) | 0.99 (0.93–1.05) | 1.04 (0.97–1.11) |
AF, atrial fibrillation; ARIC, Atherosclerosis Risk in Communities; BMI, body mass index; CHD, coronary heart disease; HF, heart failure; PRS, polygenic risk score; SBP, systolic blood pressure; T2D, type 2 diabetes.
Hazard ratios (95% confidence intervals) were derived from Cox hazard proportional regressions adjusting for age, sex, centre, body mass index, smoking status, systolic blood pressure, blood pressure‐lowering medication use, heart rate, prevalent diabetes, prevalent coronary heart disease, prevalent atrial fibrillation, and the first 10 principal components.
Figure 2.
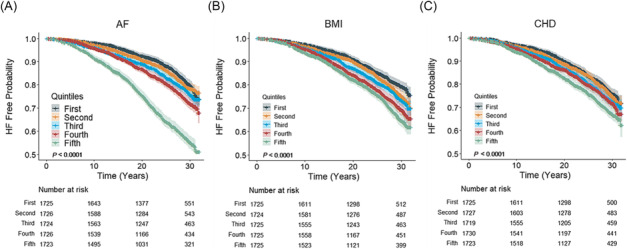
Kaplan–Meier survival curves for incident heart failure by quintiles of polygenic risk scores for (A) atrial fibrillation (AF), (B) body mass index (BMI), and (C) coronary heart disease (CHD) in Whites from the Atherosclerosis Risk in Communities study. Shaded areas represent 95% confidence intervals, and P values are obtained from log‐rank tests.
When evaluating discrimination of risk prediction models, we observed that the addition of PRSAF significantly improved prediction of HF lifetime risk (from 10 to 30 years) over the ARIC study HF prediction equation with further adjustment of prevalent AF. Individually adding PRSBMI and PRSCHD did not improve the risk prediction, and the addition of PRSAF, PRSBMI, and PRSCHD simultaneously compared with only PRSAF did not significantly improve the discrimination for HF risk either. With 10 years of follow‐up in the ARIC study Whites, the C statistic increased from 0.812 (95% CI: 0.792–0.831) to 0.829 (95% CI: 0.810–0.848) with the addition of PRSAF to the ARIC study HF prediction equation (∆C: 0.017, 95% CI: 0.009–0.026). The incremental value of PRSAF over the ARIC study HF prediction equation was similar when observing 20 and 30 year follow‐up, though the overall predictability weakened over time (Supporting Information, Table S3 ).
To understand the relationship between PRSAF and HF subtypes, we examined their associations among 5964 (54% female) ARIC study Whites at Visit 4. There were 280 (37%) HFrEF and 357 (48%) HFpEF cases ascertained, among a total of 750 incident HF cases, with a median of 14.0 year follow‐up (Supporting Information, Table S4 ). Compared with Visit 1, Visit 4 participants were older and had higher prevalence of HF risk factors, such as CHD and diabetes. PRSAF showed a strong association with incident all‐cause HF (HR: 1.43, 95% CI: 1.34–1.54), as well as both HFrEF (HR: 1.43, 95% CI: 1.27–1.60) and HFpEF (HR: 1.46, 95% CI: 1.33–1.62), and the additional adjustment of NT‐proBNP, a diagnostic biomarker of HF, did not alter the associations (Table 3 ).
Table 3.
Association between polygenic risk score of atrial fibrillation and incident heart failure and its subtypes in participants from the ARIC study and the CHS
| N (events) | Model 1 | Model 2 | ||||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |||
| ARIC study Whites | All HF | 5964 (750) | 1.43 (1.34–1.54) | 7.01 × 10−24 | 1.40 (1.30–1.50) | 5.07 × 10−21 |
| HFpEF | 5571 (357) | 1.46 (1.33–1.62) | 7.08 × 10−14 | 1.44 (1.30–1.59) | 1.13 × 10−12 | |
| HFrEF | 5494 (280) | 1.43 (1.27–1.60) | 7.73 × 10−10 | 1.38 (1.24–1.55) | 1.87 × 10−8 | |
| ARIC study Blacks | All‐cause | 1398 (201) | 1.30 (1.13–1.49) | 0.0002 | 1.28 (1.11–1.47) | 0.0005 |
| HFpEF | 1287 (90) | 1.38 (1.12–1.69) | 0.0024 | 1.34 (1.09–1.65) | 0.0049 | |
| HFrEF | 1289 (92) | 1.28 (1.04–1.57) | 0.0201 | 1.26 (1.03–1.55) | 0.0249 | |
| CHS Whites | All‐cause | 2200 (779) | 1.47 (1.37–1.57) | 7.68 × 10−29 | 1.44 (1.34–1.54) | 2.66 × 10−25 |
| HFpEF | 1677 (256) | 1.60 (1.42–1.79) | 1.86 × 10−15 | 1.57 (1.40–1.76) | 2.99 × 10−14 | |
| HFrEF | 1620 (199) | 1.52 (1.33–1.73) | 4.48 × 10−10 | 1.47 (1.29–1.68) | 1.20 × 10−8 | |
ARIC, Atherosclerosis Risk in Communities; CHS, Cardiovascular Health Study; CI, confidence interval; HF, heart failure; HFpEF, heart failure with preserved ejection fraction; HFrEF, heart failure with reduced ejection fraction; HR, hazard ratio; NT‐proBNP, N‐terminal pro‐brain natriuretic peptide.
Model 1 adjusted for age, sex, study centre, body mass index, systolic blood pressure, blood pressure‐lowering medication use, heart rate, smoking status, prevalent coronary heart disease, prevalent diabetes, prevalent atrial fibrillation (not adjusted in Blacks due to small number of cases), and the first 10 principal components. Model 2 further adjusted for log‐transformed NT‐proBNP. The CHS models do not include adjustment of prevalent coronary heart disease or the first 10 principal components.
We further analysed incident AF as a time‐dependent covariate in the ARIC study Whites to assess the influence that the clinical manifestation of AF has on the risk of HF. From baseline until end of follow‐up in 2018, there were 2097 (24%) participants that developed AF and 999 had a PRSAF in the top 20% (Supporting Information, Table S5 ). There was a 28% higher risk of HF per SD increase in PRSAF (P = 1.32 × 10−26, 95% CI: 1.22–1.34) after adjusting for risk factors in the ARIC study HF prediction equation, 10 the first 10 genetic principal components, and AF diagnosis. Though the risk of HF was slightly attenuated, the PRSAF remained significantly associated with incident HF after accounting for incident AF from over 30 years of follow‐up.
Replication of PRSAF association and risk prediction
We next constructed PRSs in the ARIC study Blacks and the CHS Whites using the aforementioned allele weights and performed replication analyses to test the association of PRSAF with incident HF and the prediction performance. In the ARIC study Blacks, PRSAF showed significant association with incident HF (HR: 1.29, 95% CI: 1.20–1.39). The addition of PRSAF into the ARIC study HF prediction equation showed a significant improvement in the discrimination of HF risk with 10 years of follow‐up: C statistic increased to 0.810 from 0.795 (∆C: 0.015, 95% CI: 0.004–0.026). In the CHS, PRSAF showed a significant association with incident HF (HR: 1.46, 95% CI: 1.38–1.55). With 10 years of follow‐up, the addition of PRSAF into the HF equation showed a significant improvement in the discrimination of HF: C statistic increased to 0.734 from 0.719 (∆C: 0.015, 95% CI: 0.004–0.025).
We also performed replication analyses to test the association of PRSAF with HF subtypes in the ARIC study Blacks and the CHS Whites. Similar to the findings from the ARIC study Whites, PRSAF showed a strong association with incident all‐cause HF, as well as both HFrEF and HFpEF, in the ARIC study Blacks and the CHS Whites, and the additional adjustment of NT‐proBNP did not significantly alter the associations (Table 3 ).
PRSAF with proteome
To investigate the potential biological pathways underlying PRSAF, we related PRSAF with 4877 proteins measured by SomaScan from 7339 ARIC study Whites at Visit 3. We found 99 proteins that were significantly associated with PRSAF (FDR < 0.050; Supporting Information, Table S6 ). Protein NT‐proBNP showed the strongest positive association (β = 0.090, P = 2.23 × 10–12), and amyloid‐like protein 1 showed the strongest negative association (β = −0.072, P = 4.18 × 10–7; Figure 3A ). Using GO enrichment, those 99 proteins were significantly enriched in one biological process, cell adhesion, and 10 cell components such as extracellular region and extracellular space, and one molecular function, calcium ion binding (Figure 3B ). Sensitivity analysis adjusting for estimated glomerular filtration rate and use of anti‐coagulates yielded similar results (data not shown).
Figure 3.
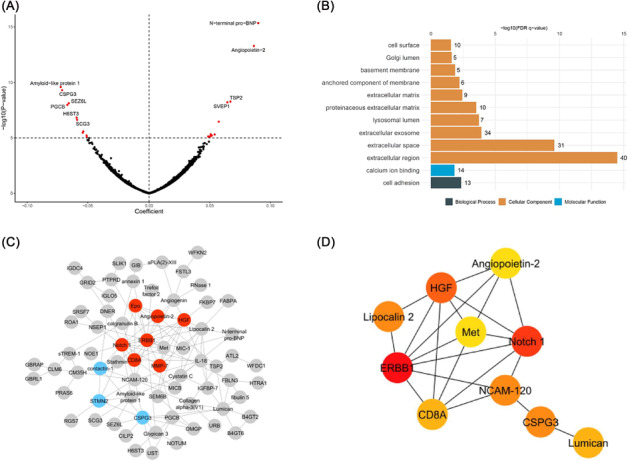
Protein pathway analysis in Whites from the Atherosclerosis Risk in Communities study. (A) Volcano plot of 4877 plasma proteins on the polygenic risk score for atrial fibrillation. Significant proteins [false discovery rate (FDR) P value] are shown in red. (B) Significantly enriched (FDR P value <0.05) functional annotations of 99 proteins. (C) Protein–protein interaction (PPI) network of 99 proteins. Colours represent clusters identified in network: Cluster 1 is red, and Cluster 2 is blue. (D) Top 10 hub genes identified in PPI network for having the greatest degree of network connections where colours indicate the protein's connectivity degree ranking in the entire PPI network: red is high, orange is moderate, and yellow is low.
Among those 99 proteins, a protein–protein interaction network with a total of 97 nodes and 140 edges was created using the STRING database with the top 2 clusters highlighted in Figure 3C and the enriched pathways listed in Supporting Information, Table S7 . The proteins involved in the first cluster included angiopoietin‐2, erythropoietin (Epo), T‐cell surface glycoprotein CD8 alpha chain (CD8A), epidermal growth factor receptor (ERBB1), hepatocyte growth factor (HGF), matrix metallopeptidase 7 (MMP‐7), and neurogenic locus notch homologue protein 1 (Notch 1). Furthermore, the top 10 nodes with the greatest degree of network connections were identified, and a subnetwork was configured (Figure 3D ). Among the top 10 hub proteins, ERBB1 had the greatest connectivity degree of 18.
Discussion
In a longitudinal cohort, we observed that PRSAF was prospectively associated with incident HF, HFpEF and HFrEF, and improved 10 year risk prediction over an established HF risk equation in Whites, and the findings were replicated among independent samples of Whites and Blacks. The effects were consistent across sex and age groups, and the increased risk for HF in the high genetic risk group (top PRS quintile) against the low genetic risk group (bottom PRS quintile) was three‐fold after accounting for clinical risk factors. Proteome analyses identified a few proteins associated with PRSAF, revealing potential underlying biological pathways. Our findings suggest the potential value of incorporating PRSs into HF risk prediction.
PRSs have recently been applied to multiple CVD outcomes such as CHD, AF, and stroke, 8 , 22 , 23 with the goal to optimize the screening and prevention of the disease. While genetically predicted risk factors, including CHD and AF, have been shown to be associated with HF, 24 few studies have tested the relationship between incident HF and its subtypes with PRSs for HF risk factors. We have identified and replicated PRSAF that relates to increased risk of HF and its subtypes, HFrEF and HFpEF. We demonstrated that PRSAF explained a reasonable amount of the variance for AF, and individuals with high genetic susceptibility had increased risk of developing HF, independent of clinical risk factors. Therefore, PRSAF could be considered as a new risk factor reflecting lifetime risk of AF, which offers an additional opportunity to identify individuals at risk of HF, beyond the clinical risk factors. Future studies are warranted to assess the prognostic value of PRSAF in HF patients to inform management decision, as studies have shown that clinical AF negatively influences HF prognosis. 25
One of the utilities of PRSs is to aid in prediction of individuals developing disease later in life. The additive value of a PRS for coronary artery disease over an established pooled cohort equation 26 has been shown in multiple settings to increase 10 year C statistics by 2%, 6 , 22 , 27 , 28 which is consistent with our current analysis: ~2% increase of C statistics for PRSAF of 10 year prediction of HF. AF and HF are known to be closely associated, with each predisposing to the other, 29 and recent studies demonstrate a plausible causal relationship between AF and HF. 30 However, AF is not considered in the ARIC study HF risk prediction equation, 10 Pooled Cohort equations to Prevent HF (PCP‐HF), 31 or a machine learning‐based race‐specific prediction model for HF, 32 which were developed primarily in middle‐aged populations. This is possibly due to the low prevalence of AF in the middle‐aged population, as the overall prevalence of AF in the United States is 1–2%, which in part limits the predictability of prevalent AF. The risk of AF has a rapid incline after age 65, 33 while the PRSs of AF capture the lifetime genetic susceptibility of AF, making it a powerful predictor of incident HF. We observed that PRSAF still remained significantly associated with incident HF after accounting for incident AF as a time‐dependent covariate, suggesting that genetic susceptibility of AF influences the risk of HF. We also observed that the additive value of prevalent AF over the ARIC study HF risk prediction equation only increased C statistics by 0.1% for 10 year prediction of HF. Though PRSAF is derived to represent the genetic risk of AF, we have shown that PRSAF is significantly associated with incident HF, with or without clinical manifestation of AF, and that the addition of PRSAF into the ARIC study HF risk prediction equation improved the discrimination of incident HF. The ARIC study HF risk prediction equation includes a parsimonious set of clinical risk factors, which yields a decent prediction performance for HF. Although PCP‐HF and/or machine learning‐based race‐specific HF prediction model improves the accuracy of HF prediction over the ARIC study HF risk prediction equation, they require additional clinical variables, as well as biomarker information, which are not typically assessed in routine exams. Given the widely available personal genetic information, that is, biobanks, adding a PRSAF into a parsimonious HF risk prediction equation, which accounts for people at risk for AF in late life, may aid in identifying individuals at risk for HF early. Of note, most studies have looked at 10 year risk prediction while our current analyses have showed that PRSs can improve up to 30 year risk prediction of HF, further supporting the use of genetically predicted AF to improve risk prediction.
NT‐proBNP is a well‐known biomarker of HF and is used in HF diagnosis and prognosis. 34 As expected, results from our proteome analysis showed that NT‐proBNP had the strongest positive effect with PRSAF, and importantly, the adjustment of NT‐proBNP in our ARIC study HF risk prediction did not attenuate the effect of PRSAF, which suggests that the PRSAF captures additional biological function. In the proteome analysis, the top hub protein, ERBB1, was negatively associated with PRSAF. Evidence is not yet available to support an association between ERBB1 and HF; however, an association between ERBB1 and the progression of AF has been previously reported. 35 Furthermore, protein angiopoietin‐2 is the second strongest positive association with PRSAF and is identified in Cluster 1 as well as in the top 10 hub proteins due to a high degree of interactions. Previous studies have shown angiopoietin‐2 to be associated with various CVDs including HF, 36 and the performance as a biomarker for HF is comparable with NT‐proBNP in adults. 37 A recent study reported a direct association between increased levels of angiopoietin‐2 and increased risk of HF in a middle‐aged ethnically diverse population and argued that the increased angiopoietin‐2 levels precede heart disease and may contribute to the disease pathogenesis. 38 Additional investigation of angiopoietin‐2 on HF is warranted as well as in younger age groups.
Some notable strengths of our study include being the first to test the association between PRSs and HF prediction, inclusion of two race groups, replication in an independent cohort, and incorporating proteomic measures to identify PRS‐related pathways. Furthermore, we have assessed and identified an association between PRSAF and HF subtypes, HFrEF and HFpEF. The predictive performance of PRSs is considered poor in African Americans, as most human genetics research studies are characterized by a large proportion of European ancestry. 39 Despite data limitations, a recent study found that the PRSs derived from women of European ancestry for breast cancer risk generalized well and were significantly associated with breast cancer risk for women with European, Latinx, and African ancestries. 40 In our current study, we found the findings of PRSAF in Whites generalized well in Blacks. This is likely benefitted from a small proportion of Blacks included in the original AF GWAS that was used to derive PRSAF by Khera et al. 7 , 41 Future studies are needed to derive multi‐ancestry PRSs and to examine the associations with HF in ancestries other than European.
Our study has some limitations also: first, though the proportion was low, the ARIC study and the CHS were included as contributing cohorts in the original GWAS for AF used to derive PRSs, which could lead to an overestimation of PRS performance. Importantly, our primary outcome was incident HF, which is independent of the original GWAS for HF risk factors. Second, the current analyses only focused on five established risk factors of HF. The prediction of HF remains to be evaluated by incorporating PRSs for other risk factors, such as low‐density lipoprotein cholesterol or diastolic blood pressure, and HF itself, by incorporating a comprehensive HF GWAS summary. Third, addition of PRSAF had a 1.7% increase in 10 year C statistics and is consistent with previous studies that examined PRS predictability of CVD; however, the clinical utility still remains to be evaluated. Fourth, initial enrolment in the ARIC study began 30 years ago in 1989, and the HF definitions, diagnosis, therapies, and prognoses have since been updated. However, end of follow‐up was until 2018, and the longer length of follow‐up time may offer more insight on the aetiology of HF. Lastly, our analyses were performed in the ARIC study and the CHS, and prediction performance was compared with the ARIC study HF risk equation. It is unclear what performance is achievable in other HF prediction equations or in individuals of different ancestries and cohorts.
In conclusion, we demonstrated that PRSAF was associated with incident HF and its subtypes and improved 10 year risk prediction of HF beyond an established HF risk equation in the ARIC study Whites, and the results were replicated in the ARIC study Blacks and the CHS Whites. Our findings suggest that a PRSAF may be useful in identifying individuals with high risk of HF. Future application of PRS for AF in large prospective studies of HF to validate its prediction performance is warranted.
Conflict of interest
A.M.S. has received research support from Novartis and Philips Ultrasound through Brigham and Women's Hospital and consulting fees from Philips Ultrasound and Edwards Lifesciences. R.C.H. has received research support from Denka Seiken through Baylor College of Medicine and consulting fees from Denka Seiken. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. All other authors declare no conflicts of interest.
Funding
The Atherosclerosis Risk in Communities (ARIC) study has been funded in whole or in part with federal funds from the National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH), Department of Health and Human Services, under Contract Nos. HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I, and HHSN268201700005I. Funding was also supported by NHLBI R01HL087641, R01HL059367, and R01HL086694; National Human Genome Research Institute Contract U01HG004402; and NIH Contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research. SomaLogic Inc. conducted the SomaScan assays in exchange for use of the ARIC study data. This work was supported in part by NIH/NHLBI Grants R01HL105756, R01HL134320, R01HL141824, R01HL148218, and R01HL160793. A.M.S. was supported by NIH/NHLBI Grants R01HL135008, R01HL143224, R01HL150342, R01HL148218, and K24HL152008. The Cardiovascular Health Study (CHS) was supported by Contracts HHSN268201200036C, HHSN268200800007C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and 75N92021D00006 and Grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, and U01HL130114 from the NHLBI, with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided by R01AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS‐NHLBI.org. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Supporting information
Table S1. Polygenic risk scores performance in participants from the Atherosclerosis Risk in Communities (ARIC) study at visit 1 (1987–1989) and the Cardiovascular Health Study (CHS) at visit 1 (1989–1990).
Table S2. Additional cox proportional hazard models in Whites from the Atherosclerosis Risk in Communities (ARIC) study.
Table S3. C‐statistics and delta C of polygenic risk scores over heart failure risk prediction model in Whites from the Atherosclerosis Risk in Communities (ARIC) Study.
Table S4. Characteristics of participants from the Atherosclerosis Risk in Communities (ARIC) study at Visit 4 (1996–1999) and Cardiovascular Health Study (CHS) at Visit 1 (1989–1990) included for analysis of incident heart failure subtypes.
Table S5. Further evaluation of atrial fibrillation in Whites (n = 8,624) from the Atherosclerosis Risk in Communities (ARIC) study.
Table S6. Proteins significantly associated with polygenic risk score for atrial fibrillation in Whites from the Atherosclerosis Risk in Communities (ARIC) study at visit 3.
Table S7. Protein pathway analysis top clusters identified and their enriched pathways.
Figure S1. Flow chart of study design and sample selection.
Acknowledgements
The authors thank the staff and participants of the ARIC study and the CHS for their important contributions.
Alkis, T. , Luo, X. , Wall, K. , Brody, J. , Bartz, T. , Chang, P. P. , Norby, F. L. , Hoogeveen, R. C. , Morrison, A. C. , Ballantyne, C. M. , Coresh, J. , Boerwinkle, E. , Psaty, B. M. , Shah, A. M. , and Yu, B. (2024) A polygenic risk score of atrial fibrillation improves prediction of lifetime risk for heart failure. ESC Heart Failure, 11: 1086–1096. 10.1002/ehf2.14665.
References
- 1. Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, et al. Heart disease and stroke statistics—2019 update: A report from the American Heart Association. Circulation 2019;139:e56‐e528. doi: 10.1161/CIR.0000000000000659 [DOI] [PubMed] [Google Scholar]
- 2. Lindgren MP, PirouziFard MN, Smith JG, Sundquist J, Sundquist K, Zöller B. A Swedish nationwide adoption study of the heritability of heart failure. JAMA Cardiol 2018;3:703‐710. doi: 10.1001/jamacardio.2018.1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shah S, Henry A, Roselli C, Lin H, Sveinbjörnsson G, Fatemifar G, et al. Genome‐wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun 2020;11:163. doi: 10.1038/s41467-019-13690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Levin MG, Tsao NL, Singhal P, Liu C, Vy HMT, Paranjpe I, et al. Genome‐wide association and multi‐trait analyses characterize the common genetic architecture of heart failure. Nat Commun 2022;13:6914. doi: 10.1038/s41467-022-34216-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O'Sullivan JW, Raghavan S, Marquez‐Luna C, Luzum JA, Damrauer SM, Ashley EA, et al. Polygenic risk scores for cardiovascular disease: A scientific statement from the American Heart Association. Circulation 2022;146:e93‐e118. doi: 10.1161/CIR.0000000000001077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elliott J, Bodinier B, Bond TA, Chadeau‐Hyam M, Evangelou E, Moons KGM, et al. Predictive accuracy of a polygenic risk score‐enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA 2020;323:636‐645. doi: 10.1001/jama.2019.22241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome‐wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219‐1224. doi: 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abraham G, Malik R, Yonova‐Doing E, Salim A, Wang T, Danesh J, et al. Genomic risk score offers predictive performance comparable to clinical risk factors for ischaemic stroke. Nat Commun 2019;10:5819. doi: 10.1038/s41467-019-13848-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krapohl E, Patel H, Newhouse S, Curtis CJ, von Stumm S, Dale PS, et al. Multi‐polygenic score approach to trait prediction. Mol Psychiatry 2018;23:1368‐1374. doi: 10.1038/mp.2017.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Agarwal SK, Chambless LE, Ballantyne CM, Astor B, Bertoni AG, Chang PP, et al. Prediction of incident heart failure in general practice: The Atherosclerosis Risk in Communities (ARIC) study. Circ Heart Fail 2012;5:422‐429. doi: 10.1161/CIRCHEARTFAILURE.111.964841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wright JD, Folsom AR, Coresh J, Sharrett AR, Couper D, Wagenknecht LE, et al. The ARIC (Atherosclerosis Risk in Communities) study: JACC Focus Seminar 3/8. J Am Coll Cardiol 2021;77:2939‐2959. doi: 10.1016/j.jacc.2021.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fried LP, Borhani NO., Enright P, et al. (1991). The cardiovascular health study: Design and rationale. Annals of Epidemiology, 1(3), 263–276. doi: 10.1016/1047-2797(91)90005-w [DOI] [PubMed] [Google Scholar]
- 13. Rosamond WD, Chang PP, Baggett C, Johnson A, Bertoni AG, Shahar E, et al. Classification of heart failure in the Atherosclerosis Risk in Communities (ARIC) study: A comparison of diagnostic criteria. Circ Heart Fail 2012;5:152‐159. doi: 10.1161/CIRCHEARTFAILURE.111.963199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelly JP, Mentz RJ, Mebazaa A, Voors AA, Butler J, Roessig L, et al. Patient selection in heart failure with preserved ejection fraction clinical trials. J Am Coll Cardiol 2015;65:1668‐1682. doi: 10.1016/j.jacc.2015.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JGF, Coats AJS, et al. 2016 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J 2016;37:2129‐2200. doi: 10.1093/eurheartj/ehw128 [DOI] [PubMed] [Google Scholar]
- 16. Lemaitre RN, Jensen PN, Hoofnagle A, McKnight B, Fretts AM, King IB, et al. Plasma ceramides and sphingomyelins in relation to heart failure risk: The Cardiovascular Health Study. Circ Heart Fail 2019;12:e005708. doi: 10.1161/CIRCHEARTFAILURE.118.005708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parrinello CM, Grams ME, Couper D, Ballantyne CM, Hoogeveen RC, Eckfeldt JH, et al. Recalibration of blood analytes over 25 years in the Atherosclerosis Risk in Communities study: Impact of recalibration on chronic kidney disease prevalence and incidence. Clin Chem 2015;61:938‐947. doi: 10.1373/clinchem.2015.238873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walker KA, Chen J, Zhang J, Fornage M, Yang Y, Zhou L, et al. Large‐scale plasma proteomic analysis identifies proteins and pathways associated with dementia risk. Nat Aging 2021;1:473‐489. doi: 10.1038/s43587-021-00064-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khera AV, Chaffin M, Wade KH, Zahid S, Brancale J, Xia R, et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell 2019;177:587‐596 e589. doi: 10.1016/j.cell.2019.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vaura F, Kauko A, Suvila K, Havulinna AS, Mars N, Salomaa V, et al. Polygenic risk scores predict hypertension onset and cardiovascular risk. Hypertension 2021;77:1119‐1127. doi: 10.1161/HYPERTENSIONAHA.120.16471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uno H, Cai T, Pencina MJ, D'Agostino RB, Wei LJ. On the C‐statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med 2011;30:1105‐1117. doi: 10.1002/sim.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mosley JD, Gupta DK, Tan J, Yao J, Wells QS, Shaffer CM, et al. Predictive accuracy of a polygenic risk score compared with a clinical risk score for incident coronary heart disease. JAMA 2020;323:627‐635. doi: 10.1001/jama.2019.21782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weng LC, Preis SR, Hulme OL, Larson MG, Choi SH, Wang B, et al. Genetic predisposition, clinical risk factor burden, and lifetime risk of atrial fibrillation. Circulation 2018;137:1027‐1038. doi: 10.1161/CIRCULATIONAHA.117.031431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kwok MK, Schooling CM. Mendelian randomization study on atrial fibrillation and cardiovascular disease subtypes. Sci Rep 2021;11:18682. doi: 10.1038/s41598-021-98058-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dries DL, Exner DV, Gersh BJ, Domanski MJ, Waclawiw MA, Stevenson LW. Atrial fibrillation is associated with an increased risk for mortality and heart failure progression in patients with asymptomatic and symptomatic left ventricular systolic dysfunction: A retrospective analysis of the SOLVD trials. J Am Coll Cardiol 1998;32:695‐703. doi: 10.1016/s0735-1097(98)00297-6 [DOI] [PubMed] [Google Scholar]
- 26. Goff DC Jr, Lloyd‐Jones DM, Bennett G, Coady S, D'Agostino RB, Gibbons R, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 2014;129:S49‐S73. doi: 10.1161/01.cir.0000437741.48606.98 [DOI] [PubMed] [Google Scholar]
- 27. Hindy G, Aragam KG, Ng K, Chaffin M, Lotta LA, Baras A, et al. Genome‐wide polygenic score, clinical risk factors, and long‐term trajectories of coronary artery disease. Arterioscler Thromb Vasc Biol 2020;40:2738‐2746. doi: 10.1161/ATVBAHA.120.314856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun L, Pennells L, Kaptoge S, Nelson CP, Ritchie SC, Abraham G, et al. Polygenic risk scores in cardiovascular risk prediction: A cohort study and modelling analyses. PLoS Med 2021;18:e1003498. doi: 10.1371/journal.pmed.1003498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. al‐Khatib SM, Benjamin EJ, Albert CM, Alonso A, Chauhan C, Chen PS, et al. Advancing research on the complex interrelations between atrial fibrillation and heart failure: A report from a US National Heart, Lung, and Blood Institute Virtual Workshop. Circulation 2020;141:1915‐1926. doi: 10.1161/CIRCULATIONAHA.119.045204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu M, Tan J, Yang J, Gao X, Yang Y. Use of Mendelian randomization to evaluate the effect of atrial fibrillation on cardiovascular diseases and cardiac death. ESC Heart Fail 2023;10:628‐636. doi: 10.1002/ehf2.14237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khan SS, Ning H, Shah SJ, Yancy CW, Carnethon M, Berry JD, et al. 10‐year risk equations for incident heart failure in the general population. J Am Coll Cardiol 2019;73:2388‐2397. doi: 10.1016/j.jacc.2019.02.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Segar MW, Jaeger BC, Patel KV, Nambi V, Ndumele CE, Correa A, et al. Development and validation of machine learning‐based race‐specific models to predict 10‐year risk of heart failure: A multicohort analysis. Circulation 2021;143:2370‐2383. doi: 10.1161/CIRCULATIONAHA.120.053134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kornej J, Borschel CS, Benjamin EJ, Schnabel RB. Epidemiology of atrial fibrillation in the 21st century: Novel methods and new insights. Circ Res 2020;127:4‐20. doi: 10.1161/CIRCRESAHA.120.316340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maries L, Manitiu I. Diagnostic and prognostic values of B‐type natriuretic peptides (BNP) and N‐terminal fragment brain natriuretic peptides (NT‐pro‐BNP). Cardiovasc J Afr 2013;24:286‐289. doi: 10.5830/CVJA-2013-055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Büttner P, Ueberham L, Shoemaker MB, Roden DM, Dinov B, Hindricks G, et al. Identification of central regulators of calcium signaling and ECM–receptor interaction genetically associated with the progression and recurrence of atrial fibrillation. Front Genet 2018;9:162. doi: 10.3389/fgene.2018.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chong AY, Caine GJ, Freestone B, Blann AD, Lip GY. Plasma angiopoietin‐1, angiopoietin‐2, and angiopoietin receptor tie‐2 levels in congestive heart failure. J Am Coll Cardiol 2004;43:423‐428. doi: 10.1016/j.jacc.2003.08.042 [DOI] [PubMed] [Google Scholar]
- 37. Lukasz A, Beutel G, Kümpers P, Denecke A, Westhoff‐Bleck M, Schieffer B, et al. Angiopoietin‐2 in adults with congenital heart disease and heart failure. PLoS ONE 2013;8:e66861. doi: 10.1371/journal.pone.0066861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peplinski BS, Houston BA, Bluemke DA, Kawut SM, Kolb TM, Kronmal RA, et al. Associations of angiopoietins with heart failure incidence and severity. J Card Fail 2021;27:786‐795. doi: 10.1016/j.cardfail.2021.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duncan L, Shen H, Gelaye B, Meijsen J, Ressler K, Feldman M, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun 2019;10:3328. doi: 10.1038/s41467-019-11112-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu C, Zeinomar N, Chung WK, Kiryluk K, Gharavi AG, Hripcsak G, et al. Generalizability of polygenic risk scores for breast cancer among women with European, African, and Latinx ancestry. JAMA Netw Open 2021;4:e2119084. doi: 10.1001/jamanetworkopen.2021.19084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.METASTROKE Consortium of the ISGCChristophersen IE, Rienstra M, Roselli C, Yin X, Geelhoed B, Barnard J, et al. Large‐scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet 2017;49:946‐952. doi: 10.1038/ng.3843Neurology Working Group of the CHARGE Consortiumthe AFGen Consortium [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Polygenic risk scores performance in participants from the Atherosclerosis Risk in Communities (ARIC) study at visit 1 (1987–1989) and the Cardiovascular Health Study (CHS) at visit 1 (1989–1990).
Table S2. Additional cox proportional hazard models in Whites from the Atherosclerosis Risk in Communities (ARIC) study.
Table S3. C‐statistics and delta C of polygenic risk scores over heart failure risk prediction model in Whites from the Atherosclerosis Risk in Communities (ARIC) Study.
Table S4. Characteristics of participants from the Atherosclerosis Risk in Communities (ARIC) study at Visit 4 (1996–1999) and Cardiovascular Health Study (CHS) at Visit 1 (1989–1990) included for analysis of incident heart failure subtypes.
Table S5. Further evaluation of atrial fibrillation in Whites (n = 8,624) from the Atherosclerosis Risk in Communities (ARIC) study.
Table S6. Proteins significantly associated with polygenic risk score for atrial fibrillation in Whites from the Atherosclerosis Risk in Communities (ARIC) study at visit 3.
Table S7. Protein pathway analysis top clusters identified and their enriched pathways.
Figure S1. Flow chart of study design and sample selection.


