Abstract
In apparent contrast to earlier work on Epstein-Barr virus (EBV) carriage in the general Caucasian population, in vitro virus isolations from human immunodeficiency virus (HIV)-positive male homosexual cohorts have shown frequent examples of multiple EBV infection and an overall prevalence of type 2 EBV strains exceeding 30%. Here we ask to what extent these findings might hold true in another T-cell-immunocompromised cohort, HIV-positive hemophilic patients. Resident EBV strains were rescued within lymphoblastoid cell lines derived from the blood and throat washings of 39 such individuals, using the same in vitro protocols of virus isolation as for the homosexual cohort. A mean of 19 independent cell lines was made per patient, and in each case the resident virus was characterized by PCR-based viral genomic analysis and by immunoblotting to reveal the viral “EBNAprint.” By these criteria a significant proportion (14 of 39) of the hemophilic cohort carried more than one EBV strain, suggesting that T-cell impairment does indeed sensitize virus carriers to reinfection with new strains of exogenously transmitted virus. However, the overall incidence of type 2 EBV infection was 10%, which is close to that observed in the earlier work with healthy carriers and substantially lower than that seen in HIV-positive homosexuals. We infer that type 2 EBV is relatively rare in the general Caucasian population but has become endemic in the homosexual community.
Epstein-Barr virus (EBV), a gammaherpesvirus widespread in human populations, is usually carried as a lifelong asymptomatic infection yet is etiologically linked to a number of different malignancies (reviewed in reference 37). It was the study of one such malignancy, the endemic (African) form of Burkitt’s lymphoma (BL), which first identified two distinct types of EBV isolates (1, 9), originally called A and B and now designated types 1 and 2. These are essentially homologous across large stretches of the genome but are distinguished by linked polymorphisms in the latent genes encoding the nuclear antigens EBNA2, -3A, -3B, and -3C (1, 9, 41). The virus type can therefore be determined at the DNA level by PCR amplification across these polymorphic loci (2, 41, 47) and at the protein level by using monoclonal antibodies (MAbs) or human sera with type-specific reactivities against these antigens (40, 45). Furthermore, within each broad type, individual virus strains can be distinguished by screening across other defined polymorphisms in the viral genome (4, 30, 34, 35) and by determining the strain-specific “EBNAprint,” i.e., the precise sizes of the virus-coded EBNA1, -2, -3A, -3B, and -3C proteins as visualized in immunoblots (19, 53). Studies on endemic BL tumors and subsequently on virus isolates rescued as in vitro-transformed lymphoblastoid cell lines (LCLs) from healthy donors suggested that type 1 and type 2 strains were of roughly equal prevalence in at least some parts of equatorial Africa (57). By contrast, the much rarer cases of EBV-positive sporadic BL occurring in children in the Western world almost all carried a type 1 virus (14, 58). This again appeared to reflect the situation within the general population in these areas, where, based on virus isolations, type 1 strains were prevalent and <10% of individuals carried type 2 virus (20, 53). The balance of evidence from such work also strongly suggested that healthy virus carriers harbored a single virus strain; multiple infection either with different virus types or with different strains of the same type appeared to be extremely rare (20, 53; reviewed in reference 17).
The generality of these conclusions has subsequently been challenged by observations of AIDS patients in Western communities. Such patients show an unusually high risk both of Burkitt-like lymphoma, arising relatively early in the course of AIDS and (like sporadic BL) EBV genome positive in roughly 30% of cases (22, 48), and of immunoblastic lymphoma arising in late-stage AIDS and (like posttransplant lymphoma) EBV genome positive in the majority if not all cases (22, 32). Unexpectedly, both of these AIDS-associated lymphomas and also a third EBV-associated malignancy seen in this patient group, Hodgkin’s disease, were found to carry a type 2 EBV strain in 25 to 50% of EBV genome-positive cases (6, 10, 14, 21, 36, 46). These observations, and an earlier report detecting coresident EBV genotypes in an AIDS lymphoma patient (26), prompted several studies of EBV infection in HIV-positive patients as a whole, analyzing the types of virus that were detectable in the oropharynx and circulating B-cell pool either directly by PCR analysis or by virus rescue in vitro (7, 29, 31, 44, 50, 51, 54, 55). This work clearly showed that the prevalence of type 2 virus infection in such patients was at least 30%, substantially higher than had been apparent from most studies on the general population from which these patients were drawn. Interestingly, detailed analysis of such in vitro isolates revealed that most of the HIV-positive patients with detectable type 2 EBV also carried a coresident type 1 strain, while another 25% of the patients carried multiple viruses, all of type 1 (54).
The full biological significance of these findings is still not known. One interpretation is that the EBV carrier state in HIV-positive patients faithfully magnifies that which exists generally in the immunocompetent population and that the full range of resident strains are simply easier to detect in this immunocompromised setting because of the higher overall viral load. If that were the case, earlier studies (20, 53) must have seriously underestimated the true prevalence of type 2 virus in the general Caucasian population (3, 47); this is possible in that type 2 strains may be subdominant in vivo and are rescued less efficiently than type 1 strains in in vitro transformation assays (38). In seeking to address these issues, we noted at the outset that almost all of the work to date on HIV-positive patients, including our own recent analysis (54, 55), had been confined to male homosexual cohorts. We reasoned that if the above-described arguments were true, then a similar pattern of virus isolations should be obtained from other T-cell-immunocompromised patient groups. In the present work we have used the same methods as employed with the HIV-positive homosexual cohort (54) to screen an equally large cohort of hemophilic patients who, over a decade earlier, had acquired HIV iatrogenically from contaminated Factor VIII concentrates.
MATERIALS AND METHODS
Patients.
The study group comprised 48 HIV-positive hemophilic patients. These included 31 adult males (mean age, 39 years; range, 22 to 71) attending outpatient clinics at the Queen Elizabeth Hospital, Birmingham (designated QEH), or at Churchill Hospital, Oxford (OX), and 17 adolescent males (mean age, 16; range, 13 to 20) attending outpatient clinics at the Birmingham Children’s Hospital (BCH). All patients had been infected with HIV in the early 1980s from contaminated batches of factor VIII concentrate; they were studied between 1993 and 1996, some 10 to 15 years following HIV infection, at a time when almost all were significantly immunocompromised and showed depressed CD4+ T-cell counts (median count, 150/mm3; range, 0 to 820). A control group consisted of 38 HIV-negative adolescent hemophilic patients, again sampled via outpatient clinics at Birmingham Children’s Hospital and spanning the same age range as the 17 HIV-infected adolescent patients described above. Heparinized blood (20 to 30 ml) and throat washings were taken and processed as described previously (54), with most patients being sampled on two separate occasions over the 3-year study period. Blood samples provided plasma for serological studies and mononuclear cells for virus isolation and in vitro regression assays, while throat washings provided cell-free filtrates for virus isolation. Data thus obtained from the HIV-positive and HIV-negative hemophilic patient groups are compared with corresponding results from 40 HIV-positive male homosexuals studied in parallel (54) and from 56 HIV-negative adult control donors analyzed in an earlier study (53).
Immunological assays.
Plasma preparations were first heat inactivated at 56°C for 30 min and then stored at −20°C until testing for immunoglobulin G antibody titers to the EBV capsid antigen (VCA) by standard immunofluorescence assay (53). When yields of peripheral blood mononuclear cells (PBM) from EBV-seropositive patients were sufficiently high, an aliquot of cells was screened for in vitro regression of B95.8 strain EBV-induced transformation to provide a measure of EBV-specific cytotoxic T-lymphocyte (CTL) memory function; results were expressed as the minimum seeding of PBM per 0.3-ml microtest plate well required to observe a 50% incidence of regression in replicate wells, as described previously (35). To allow comparison between patient groups, individual regression titer end points were classified as being within the normal range for most healthy EBV carriers (<4.5 × 105 PBM/well), weak compared to those for most healthy carriers (4.5 × 105 to 6 × 105 PBM/well), or undetectable (>6 × 105 PBM/well).
Virus isolation.
The methods for isolation of virus from blood and from throat washings were exactly those used in the parallel study of HIV-positive male homosexuals (54). Briefly, PBM were cultured in 0.3-ml microtest plate wells at a range of cell dilutions (six replicates/seeding) in RPMI 1640 medium and 10% (vol/vol) fetal calf serum, initially supplemented with 0.1 μg of cyclosporin A per ml to inhibit T-cell activation, and individual wells developing spontaneously transformed foci of outgrowth were expanded to provide LCLs carrying the patient’s endogenous EBV isolate(s). Throat washings were similarly assayed for transforming virus by using PBM from cord blood or from adult EBV-seronegative donors as a source of indicator cells. Note that these transformation assays were each conducted over a 12-week period; all wells which developed microscopically visible foci within that time were expanded irrespective of growth rate; hence, the time from initial seeding to LCL cryostorage could be up to 6 months. Occasionally wells developed transformed foci which failed to expand, and in such cases the culture was used to provide a DNA sample only.
Identification of virus isolates.
The methods of genomic and EBNAprint analyses were essentially as described in earlier work (28, 53, 54). Briefly, DNA from LCL preparations was subjected to PCR amplification with primer-probe combinations across type-specific polymorphisms within the EBNA2 and EBNA3C genes (41), across the 33-bp repeat region of the latent membrane protein 1 (LMP1) gene (34), and across a locus in LMP1 which in some virus strains displays a 30-bp deletion relative to the B95.8 prototype (34). Reference EBV strains included in the genomic analysis were B95.8 (type 1, 4.5 copies of the 33-bp LMP1 repeat, undeleted at the 30-bp LMP1 locus) and AG876 (type 2, 4 copies of the 33-bp LMP1 repeat, deleted at the 30-bp LMP1 locus). Note that although it is possible for heterogeneity across the 33-bp repeat region to be generated during replication of a single EBV strain (49), in practice we found that the great majority of LCL clones established by B-cell transformation with a reference laboratory strain retained the same number of repeats as shown by the cell lines from which the reference virus was prepared (53a).
EBNAprints of LCL protein extracts were produced by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separation and immunoblotting as described previously (54). Reference cell extracts were provided by the EBV-negative B lymphoma line BJAB, the type 1 EBV-transformed LCL C2+ OBA, and the type 2 EBV-transformed LCL C2+ BL16. Antibody preparations used in immunoblotting were the EBNA1-specific MAb IH4 (15), the EBNA2-specific MAb PE2 (56), the type 1 EBNA3C-specific MAb E3C-A10 (33), and human sera AM (preferentially reactive against EBNA1), MS (preferentially reactive against type 1 EBNA3C), and NZ (preferentially reactive against type 2 EBNA3C) (54). On some occasions, immunoblots were also probed with the LMP1-specific MAb pool CS1-4 (39). Blots were developed as described previously (54).
On occasions where the above-described genomic and EBNAprint analyses left some doubt about the relationship between coresident virus isolates, a naturally polymorphic region of the EBNA1 gene (4) was amplified by using the primers 5′ GAAAAGAGGCCCAGGAGTCCCAGTAGTCAG 3′ (B95.8 coordinates 109081 to 109110) and 5′ AACAGCACGCATGATGTCTACTGGGGATTT 3′ (B95.8 coordinates 109969-109940). The cycle for amplification was 35 cycles of 94°C for 60 s, 62°C for 90 s, and 72°C for 240 s. The PCR product was run out on a 1% agarose gel and purified with the Qiaquick gel extraction kit (Qiagen) according to the manufacturer’s instructions. Purified DNA was eluted in a 50-μl volume, of which 15 μl was then incorporated into the sequencing reaction. Sequencing was carried out according to the manufacturer’s instructions with an Amplicycle sequencing kit (Perkin-Elmer) and a 32P-end-labelled internal sequencing primer, 5′ AGAAGGCCCAAGCACTGGAC 3′ (B95.8 coordinates 109278 to 109297), to determine the nucleotide sequence of EBNA1 codons 470 to 510.
RESULTS
EBV immune status of hemophilic patients.
We first set out to determine the status of the HIV-positive hemophilic patients with respect to prior EBV infection, by using anti-VCA antibodies as a serological indicator, and to measure the prevailing level of EBV-specific T-cell immunity in EBV-infected patients, by using the in vitro regression assay. For this part of the study, the 17 adolescent patients are considered separately from the rest of the cohort so as to allow their direct comparison with an adolescent control group of 38 HIV-negative hemophilic patients. As shown in Table 1, roughly two-thirds of these adolescent patients had a history of EBV infection, both in the HIV-positive cohort and in the HIV-negative control group. However anti-VCA titers were fivefold higher in HIV-positive patients than in HIV-negative patients, an elevation consistent with that reported for other T-cell-immunocompromised cohorts and probably reflecting increased EBV antigenic load in vivo (52). Furthermore, the level of EBV-specific T-cell immunity was markedly reduced in the HIV-positive patients, with six of the seven individuals analyzed failing to register an end point in the in vitro regression assay; by contrast, regression end points for EBV-infected hemophilic patients in the HIV-negative control group were mostly within the normal range.
TABLE 1.
Immunological status of study cohorts
| Age groupa | Cohort | Anti-VCA immunoglobulin G responses
|
No. with the following EBV-specific CTL response/totalb:
|
|||
|---|---|---|---|---|---|---|
| No. seropositive/total (% seropositive) | Mean titer | Normal | Weak | Undetectable | ||
| Adolescent | HIV-positive hemophilic | 11/17 (65) | 1:1,970 | 1/7 | 0/7 | 6/7 |
| HIV-negative hemophilic | 25/38 (66) | 1:389 | 9/14 | 3/14 | 2/14 | |
| Adult | HIV-positive hemophilic | 28/31 (90) | 1:2,430 | 1/12 | 1/12 | 10/12 |
| HIV-negative healthy control | 50/56 (89) | 1:428 | 20/24 | 4/24 | 0/24 | |
| HIV-positive homosexual | 40/40 (100) | 1:1,347 | 4/16 | 3/16 | 9/16 | |
Patients classified as adolescent were 13 to 20 years old at the time of study; those classified as adult were >20 years old.
EBV-specific CTL responses in selected EBV-seropositive patients were quantified by using the in vitro regression assay, and results were expressed as the number of PBM seeded per 0.3-ml microtest plate well for a 50% incidence of regression; responses were classified as being normal (<4.5 × 105 PBM/well), weak (4.5 × 105 to 6 × 105 PBM/well), or undetectable (>6 × 105 PBM/well).
The results obtained from the 31 adult HIV-positive hemophilic patients are also presented in Table 1. As might be expected, a larger proportion (90%) of these individuals had serological evidence of an existing EBV infection, matching the incidence seen in our previous screening of a healthy adult control group (53). Once again, however, the mean anti-VCA titer was at least fivefold higher in the HIV-positive patients than in controls, while EBV-specific T-cell immunity as measured in the regression assay was very significantly impaired. Table 1 also includes for comparison the corresponding values obtained in a parallel study of 40 HIV-positive male homosexuals, every one of whom proved to be EBV infected (54). These patients also showed elevated anti-VCA titers and impaired T-cell immunity compared to control values, although the differences were not quite as marked as with the HIV-positive hemophilic patients.
EBV isolations from HIV-positive hemophilic patients.
Virus isolates were prepared from the blood and/or throat washings of all 39 HIV-positive hemophilic patients (11 adolescent and 28 adult) who had serological evidence of prior EBV infection. A mean of 19 independent EBV isolates (range, 4 to 39) per patient was obtained. The different patterns of results are described below.
(i) Patients with a single detectable EBV strain.
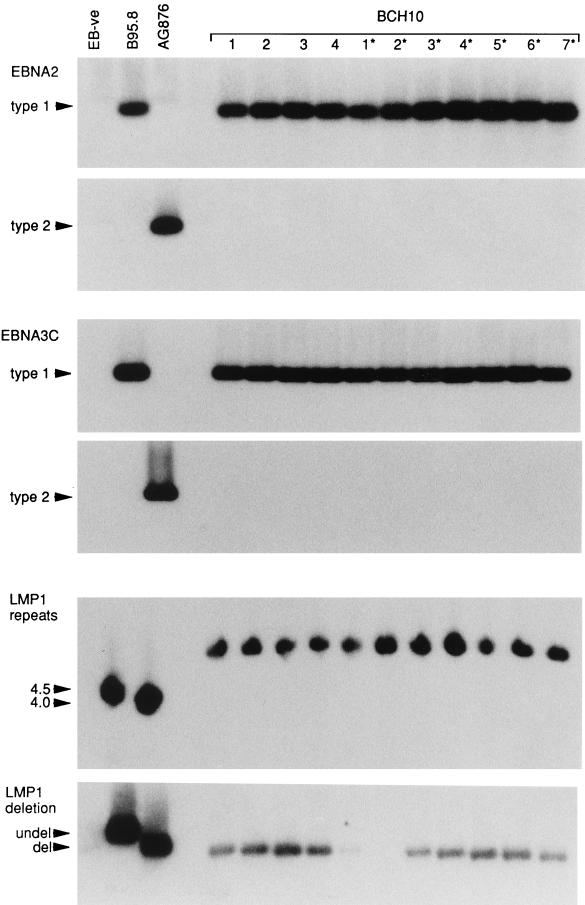
In 25 of the 39 patients studied, all EBV isolates rescued from a single individual were of a single unique strain. These findings are illustrated here with reference to BCH10, a patient from whom a total of 34 independent EBV isolates (14 from blood and 20 from throat washings) were rescued in vitro from two occasions of testing 2 years apart. Figure 1 presents the results of EBV genomic analysis of four blood-derived isolates (isolates 1 to 4) and seven throat washing-derived isolates (isolates 1* to 7*); these are representative of the data from all 34 isolates from this patient. Every virus isolate gave a type 1-specific PCR amplification product at both the EBNA2 and EBNA3C polymorphic loci, and there was no evidence of any coresident type 2 virus. Furthermore, every one of these type 1 isolates was identical at two well-characterized polymorphisms (the 33-bp repeat and 30-bp deletion loci) within the LMP1 gene sequence; thus, as shown in Fig. 1, each isolate had six copies of the 33-bp repeat and displayed a 30-bp deletion. Note that this and all ensuing PCR assays included as reference virus isolates the prototype 1 B95.8 strain (4.5 copies of the 33-bp repeat and undeleted at the 30-bp locus) and the prototype 2 AG876 strain (4 copies of the 33-bp repeat and deleted at the 30-bp locus) (34).
FIG. 1.
PCR analysis of EBV strains carried by blood-derived LCLs 1 to 4 and throat washing-derived LCLs 1* to 7* from patient BCH10. For virus typing, EBNA2 gene amplification was carried out with a common 5′ primer and type-specific 3′ primers, and EBNA3 gene amplification was carried out with common 5′ and 3′ primers; in each case, the products were probed separately with type-specific probes. For strain detection, LMP1 gene amplifications were carried out across the 33-bp repeat and 30-bp deletion loci. Assays included reference DNA samples from the type 1 EBV-carrying B95.8 cell line (4.5 copies of the 33-bp repeat, undeleted [undel] at the 30-bp locus), the type 2 EBV-carrying AG876 cell line (4 copies of the 33-bp repeat, deleted [del] at the 30-bp locus), and the EBV-negative (EB-ve) BJAB cell line. All BCH10-derived isolates were type 1 and were identical by strain-specific marker analysis. (Longer exposures of the LMP1 deletion gel showed that isolate 2* was deleted at the 30-bp locus.)
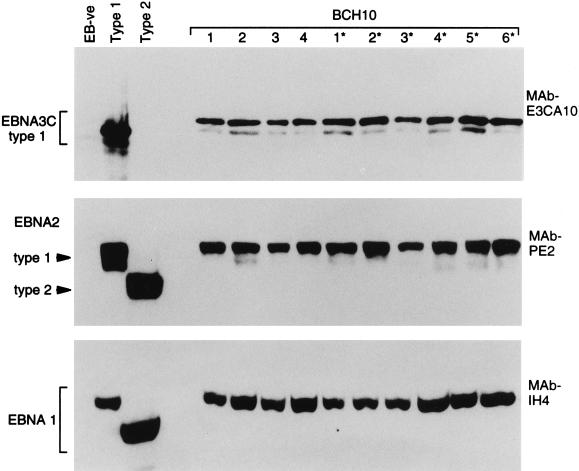
The implication from these data, that patient BCH10 carried a single type 1 virus strain, was further supported by the immunoblotting data. Here, protein extracts from the BCH10-derived LCLs were separated by SDS-PAGE, and the blots were probed with MAbs to EBNA3C, EBNA2, and EBNA1; on the same gels were included reference extracts from the EBV-negative B-cell line BJAB, from a type 1 virus-transformed LCL, and from a type 2 virus-transformed LCL. All 34 lines from patient BCH10 produced the same EBNAprint; representative blots for blood-derived LCLs 1 to 4 and throat washing-derived LCLs 1* to 6* are shown in Fig. 2. The sizes of the EBNA3C, EBNA2, and EBNA1 proteins are uniform in every lane, strongly suggesting that each line carries the identical viral strain. Note also that this virus can be classified as a type 1 strain from the EBNAprint: first the EBNA3C protein was detectable by using the type 1 EBNA3C-specific MAb E3CA10, and second, the EBNA2 protein lay within the size range (80 to 90 kDa) typical of a type 1 allelic product and well above the 75-kDa position characteristic of type 2 EBNA2.
FIG. 2.
EBNAprint analysis of EBV strains carried by blood-derived LCLs 1 to 4 and throat washing-derived LCLs 1* to 6* from patient BCH10. Protein extracts were separated by SDS-PAGE, and the immunoblots were then probed with the type 1 EBNA3-specific MAb E3CA10, the EBNA2-specific MAb PE2, and the EBNA1-specific MAb IH4. Immunoblots included reference protein samples from the type 1 EBV-transformed LCL C2+ OBA, the type 2 EBV-transformed LCL C2+ BL16, and the EBV-negative (EB-ve) BJAB cell line. All BCH10-derived isolates gave identical EBNAprints.
The individual data for all 25 patients with a single detectable EBV strain are presented in Table 2 (see below); in 24 cases the resident strain was of type 1, and in a single case it was of type 2.
TABLE 2.
EBV infections in HIV-positive hemophilic patients
| Patient | No. of virus isolatesa from:
|
No. and type of resident EBV strains | |
|---|---|---|---|
| Blood | Throat | ||
| BCH1 | 16 | 23 | Single type 1 strain |
| BCH2 | 10 | 9 | Single type 1 strain |
| BCH10 | 14 | 20 | Single type 1 strain |
| BCH12 | 10 | 25 | Single type 1 strain |
| BCH14 | 9 | 11 | Single type 1 strain |
| BCH16 | 17 | 10 | Single type 1 strain |
| QEH2 | 9 | 11 | Single type 1 strain |
| QEH3 | 2 | 13 | Single type 1 strain |
| QEH6 | 15 | 7 | Single type 1 strain |
| QEH8 | 15 | 14 | Single type 1 strain |
| QEH10 | 13 | 14 | Single type 1 strain |
| QEH11 | 2 | 19 | Single type 1 strain |
| QEH13 | 12 | Single type 1 strain | |
| QEH14 | 8 | 20 | Single type 1 strain |
| QEH17 | 12 | 16 | Single type 1 strain |
| QEH19 | 11 | 12 | Single type 1 strain |
| QEH21 | 8 | 2 | Single type 1 strain |
| QEH23 | 6 | 10 | Single type 1 strain |
| QEH24 | 9 | 9 | Single type 1 strain |
| QEH26 | 7 | Single type 1 strain | |
| QEH27 | 4 | 12 | Single type 1 strain |
| OX4 | 9 | 16 | Single type 1 strain |
| OX7 | 4 | Single type 1 strain | |
| OX12 | 12 | Single type 1 strain | |
| OX1 | 10 | Single type 2 strain | |
| BCH3 | 2 | 3 | Multiple type 1 strains |
| BCH6 | 11 | 9 | Multiple type 1 strains |
| BCH8 | 12 | 19 | Multiple type 1 strains |
| BCH9 | 2 | 18 | Multiple type 1 strains |
| QEH4 | 12 | Multiple type 1 strains | |
| QEH5 | 8 | 9 | Multiple type 1 strains |
| QEH9 | 4 | 18 | Multiple type 1 strains |
| QEH18 | 2 | 13 | Multiple type 1 strains |
| QEH25 | 10 | Multiple type 1 strains | |
| OX14 | 3 | 8 | Multiple type 1 strains |
| OX17 | 8 | Multiple type 1 strains | |
| BCH7 | 12 | 9 | Type 1 and type 2 strains |
| QEH15 | 12 | 11 | Type 1 and type 2 strains |
| OX2 | 2 | 11 | Type 1 and type 2 strains |
The mean number of virus isolates per patient was 19 overall, 20.7 for patients with a single detectable strain, and 16.0 for patients with multiple detectable strains. Boldface indicates the sites at which different coresident EBV strains could be detected.
(ii) Patients with multiple EBV strains.
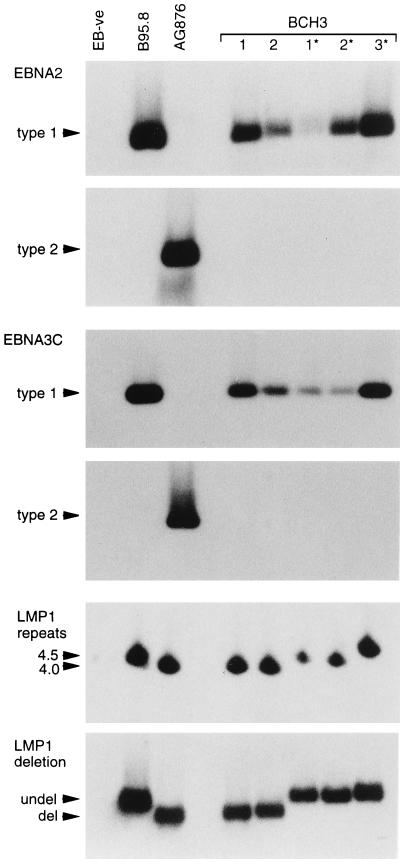
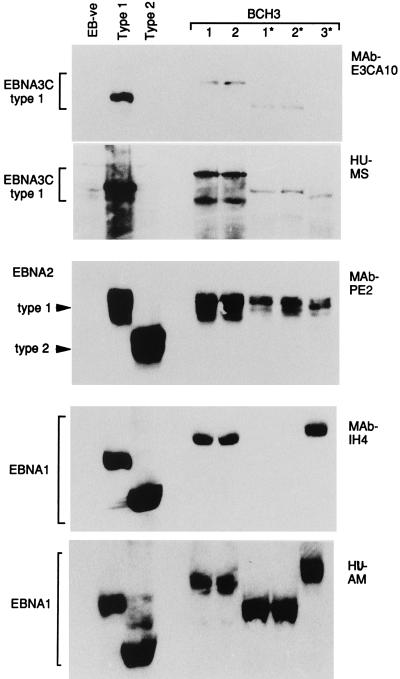
The remaining 14 patients gave evidence of infection with more than one EBV strain. For example, patient BCH3, who could be studied on a single occasion only and from whom only five viral isolations were made, was interesting in that one type 1 strain was detected in the blood and two other distinct type 1 strains were detected in the throat. Figure 3 presents the PCR analysis of genomic markers, from which it is clear that all five isolates were type 1 at both the EBNA2 and EBNA3C loci. However, three different patterns (shown by blood isolates 1 and 2, throat isolates 1* and 2*, and throat isolate 3*) were observed from analysis of the polymorphic LMP1 loci. These different viral identities were fully confirmed by the immunoblot analysis illustrated in Fig. 4. Although all isolates encoded EBNA2 proteins which migrated to similar positions within the type 1 size range, the EBNA1 and EBNA3C immunoblots revealed three different EBNAprints. In the case of EBNA1, the three resident viral strains could be distinguished by EBNA1 size and by the fact that the EBNA1 encoded by throat isolates 1* and 2* lacked the MAb IH4 epitope and had to be detected by using a polyvalent human serum, AM, with strong anti-EBNA1 reactivity. In the case of EBNA3C, the strains were again distinguishable on the basis of their different-sized EBNA3C proteins and by the absence of the MAb E3CA10 epitope in the protein encoded by throat isolate 3*.
FIG. 3.
PCR analysis of EBV strains carried by blood-derived isolates 1 and 2 and throat washing-derived isolates 1* to 3* from patient BCH3. Assays were conducted with controls as described for Fig. 1. All BCH3-derived isolates were type 1 but could be distinguished into three groups (1, 2 v 1*, 2* v 3*) by strain-specific marker analysis.
FIG. 4.
EBNAprint analysis of EBV strains carried by blood-derived isolates 1 and 2 and throat washing-derived isolates 1* to 3* from patient BCH3. Immunoblots were conducted with controls as described for Fig. 2; additional blots were probed with polyclonal human (HU) sera from a type 1 virus-infected donor, MS (known to be reactive to type 1 EBNA3C), and from donor AM (known to be selectively reactive to EBNA1). The BCH3-derived isolates could be classified into three groups (1, 2 v 1*, 2* v 3*) from their distinct EBNAprints.
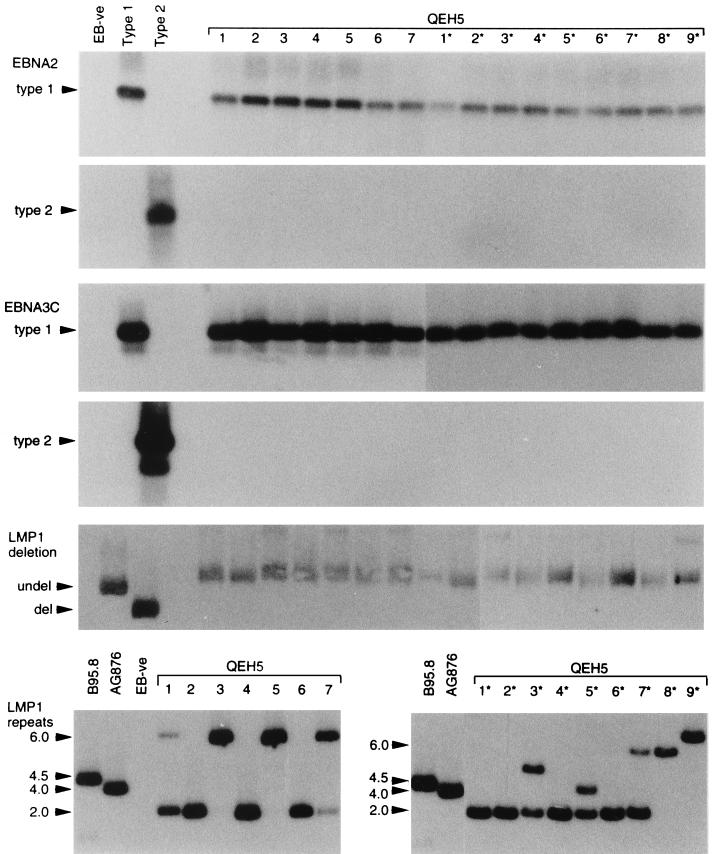
A second example of coinfection with distinct type 1 EBV strains comes from patient QEH5. Figure 5 shows the PCR analysis of viral genome markers for blood-derived isolates 1 to 7 and for throat washing-derived isolates 1* to 9*. All isolates were exclusively type 1 at the EBNA2 and EBNA3C type-specific loci, and all were undeleted at the LMP1 locus. However, when the analysis was extended to the LMP1 33-bp repeat region, there was evidence of viruses with either two or six repeats in the blood and of both these viruses plus additional repeat size variants in the throat; note that some LCLs gave two signals in the LMP1 repeat assay, suggesting that these are mixed populations of cells carrying different virus isolates. On their own, such findings are not diagnostic of coinfection with independent viral strains, because LMP1 repeat size heterogeneity might be generated by nonhomologous recombination during replication of a single parental strain (49). However, the subsequent EBNAprint analysis strongly suggested that isolates with two repeats (e.g., LCLs 2, 4, and 2*) and isolates with six repeats (e.g., LCLs 3, 5, and 8*) represented distinct strains. Thus, as shown in Fig. 6, these sets of isolates encoded slightly different-sized EBNA3C, EBNA2, and EBNA1 proteins. Interestingly, when we extended the protein analysis by immunoblotting with the LMP1-specific MAb pool CS1-4, we noted that differences in LMP1 size appeared to reflect the different numbers of 33-bp repeats; such a correlation had not been observed in earlier studies comparing the LMP1 proteins encoded by EBV strains known to be independent of one another (28). Therefore, as a final check on the identity of the isolates from patient QEH5, we went on to amplify and sequence across a naturally polymorphic region of the EBNA1 gene (codons 470 to 510) from a number of QEH5-derived LCLs. The sets of isolates first distinguished on the basis of LMP1 repeat numbers had clearly different sequence changes vis-à-vis B95.8 in this region and therefore must represent independent viral strains. Both strains were rescued from this patient on three successive occasions of testing.
FIG. 5.
PCR analysis of EBV strains carried by blood-derived isolates 1 to 7 and throat washing-derived isolates 1* to 9* from patient QEH5. Assays were conducted with controls as described for Fig. 1. All QEH5-derived isolates were type 1 and undeleted at the LMP1 30-bp locus but showed different numbers of LMP1 33-bp repeats. Note that some lanes display more than one PCR product in the LMP1 repeat analysis, implying that in such cases the LCL is a mixture of cells carrying different virus strains.
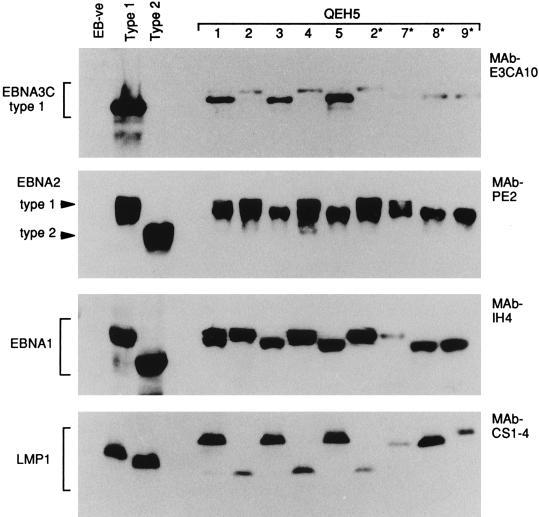
FIG. 6.
EBNAprint analysis of EBV strains carried by blood-derived isolates 1 to 5 and throat washing-derived isolates 2* and 7* to 9* from patient QEH5. Immunoblots were conducted with controls as described for Fig. 2; an additional blot was probed with the LMP1-specific MAb pool CS1-4. The two main EBNAprint patterns are illustrated by isolates 1, 3, 5, and 8* and by isolates 2, 4, and 2*, respectively.
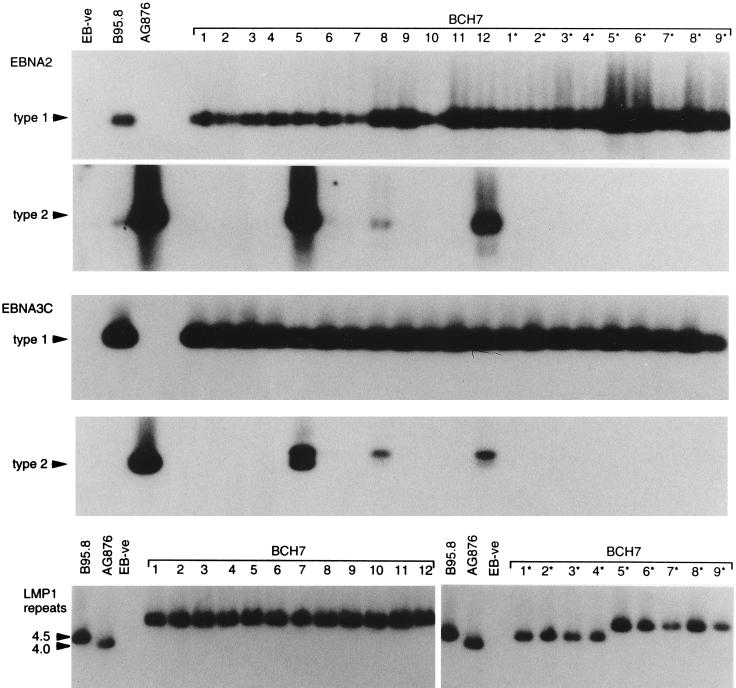
In most of the cases (11 of 14) where multiple EBV infections were identified, the coresident viral strains were all of type 1; only three individuals were found to carry both type 1 and type 2 viruses. These are exemplified by patient BCH7, from whom 12 blood-derived (1 to 12) and nine throat washing-derived (1* to 9*) LCLs were rescued in vitro. As shown by the PCR amplification data in Fig. 7, all of these gave type 1-specific signals at both EBNA2 and EBNA3C polymorphic loci. However, three of the blood-derived LCLs 5, 8, and 12 also gave type 2-specific signals at both loci. This result was confirmed on several occasions of testing, including when the initial freezings of these particular cell lines were resuscitated and reanalyzed. The type 2 virus-carrying cells appeared to be a minor subpopulation in these LCLs, however, since their presence was barely detectable on immunoblots (data not shown). This type 2 virus was not detectable in the throat washing isolates from BCH7; instead, we found two type 1 viruses in the throat, one which had the same PCR profile as the dominant virus in the blood, with 5.5 copies of the 33-bp repeat and undeleted at the 30-bp locus, and a different type 1 strain, with 4.5 copies of the 33-bp repeat and undeleted at the 30-bp locus (Fig. 7 and data not shown). Of the two other individuals giving evidence of type 1 plus type 2 coinfection, patient OX1 resembled BCH7 in that both virus types were detectable in the blood but only type 1 virus was detectable in the throat. By contrast, patient QEH15 was positive for both virus types in the blood and in the throat, a result which was observed on two independent occasions of testing 2 years apart. The individual data for all 11 patients carrying multiple type 1 EBV strains and for the three patients carrying both type 1 and type 2 strains are presented in Table 2.
FIG. 7.
PCR analysis of EBV strains carried by blood isolates 1 to 12 and throat washing-derived isolates 1* to 9* from patient BCH7. Assays were conducted with controls as described for Fig. 1. All LCLs carried type 1 virus, but LCLs 5, 8, and 12 also contained cells carrying a type 2 virus; among the type 1 isolates, one was dominant in the blood, but a second type 1 virus, distinguishable by LMP1 repeat size, was detectable in the throat (isolates 1* to 4*).
DISCUSSION
This work set out to resolve a number of apparently contradictory findings in the literature regarding the number and type of EBV strains present within virus carriers. On the one hand, the analysis of healthy EBV-infected individuals in Western countries suggests that most if not all carry a single virus strain only and that this is of type 1 in at least 90% of cases (reviewed in reference 17). Such conclusions are based on detailed virus isolation studies (20, 53), including prospective sampling of individual donors over several years (53), and on the fact that familial transmission of particular virus strains can be traced retrospectively between generations (19). On the other hand, virus isolation studies of HIV-positive patients, largely involving male homosexual cohorts, show that at least half of such patients carry multiple EBV strains and that the incidence of type 2 virus infection exceeds 30% (7, 31, 44, 54, 55). This prompts a number of questions. In particular, do the data for T-cell-immunocompromised patients magnify what is occurring (albeit undetected) in healthy virus carriers, or are they specific to the immunocompromised patient group? If the latter, to what extent are the atypical features of EBV carriage in HIV-positive homosexuals a consequence of immune impairment per se and to what extent do they reflect the increased risk of hematogeneous viral transmissions associated with a homosexual lifestyle? We sought to address these questions by applying exactly the same EBV isolation protocols to a second HIV-positive cohort, namely, hemophilic patients who had become iatrogenically infected with HIV over a decade earlier from contaminated batches of Factor VIII concentrate.
The preliminary assays of anti-VCA antibody titers and of EBV-specific T-cell immunity in these hemophilic patients (Table 1) were important for the following reasons. First, they showed that approximately one-third of adolescent hemophilic patients, whether from the HIV-positive study group or HIV-negative controls, had no serological evidence of prior EBV infection, a value in line with that expected for this age group in the general population of the United Kingdom (25). Likewise, adult HIV-positive hemophilic patients had a 90% incidence of EBV seropositivity, again mirroring that seen in the general adult population (53). Hence, one can discount the possibility that Factor VIII preparations can act as a source of EBV infection; it is most likely, therefore, that hemophilic patients acquire EBV via the natural oral route. Second, using elevated anti-VCA titers and depressed virus-specific T-cell responses as surrogate markers of immune T-cell impairment (52), we found that the HIV-positive hemophilic patients as a group were at least as immunocompromised as the HIV-positive homosexuals examined in a parallel study (54). This is further supported by comparing general criteria of immune status, such as CD4+ T-cell counts and the proportion of patients with symptoms of AIDS (see footnotes a and b of Table 3). Hence, any differences between the two groups in their range of resident EBV strains cannot be ascribed to differences in host immune competence. The two studies were also equally rigorous in that here we screened 745 independent virus isolates from 39 hemophilic patients, compared to 560 isolates from 35 homosexual patients in the parallel work (54). Given these numbers, any differences in results are also unlikely to have arisen through sampling error alone.
TABLE 3.
EBV infections in HIV-positive study cohorts
| No. and type of resident EBV strains | No. positive/total (% positive) in:
|
|
|---|---|---|
| HIV-positive hemophilic cohorta | HIV-positive homosexual cohortb | |
| Single type 1 strain | 24/39 (62) | 17/35 (49) |
| Multiple type 1 strains | 11/39 (28) | 7/35 (20) |
| Single type 2 strain | 1/39 (2.5) | 1/35 (3) |
| Multiple type 2 strains | 1/35 (3) | |
| Type 1 + type 2 coinfection | 3/39 (7.7) | 9/35 (26) |
| Overall type 2 EBV infectionc | 4/39 (10) | 11/35 (31) |
Data are based on the present analysis of 726 EBV isolates from 39 HIV-positive hemophilic patients. These patients had a median CD4+ T-cell count of 150/mm3 (range, 0 to 820). At the time of study, 13 of 39 had clinical symptoms of AIDS but none had either oral hairy leukoplakia or an AIDS-associated malignancy.
Data are based on previous analysis of 560 EBV isolates from 35 HIV-positive homosexual patients. These patients had a median CD4+ T-cell count of 155/mm3 (range, 0 to 650). At the time of study, 14 of 35 had clinical symptoms of AIDS but none had either oral hairy leukoplakia or an AIDS-associated malignancy.
The values for type 2 virus prevalence in the two cohorts are significantly different (P < 0.05).
Table 3 summarizes the overall data from the two studies in terms of the numbers of individuals with single or with multiple EBV strains and the numbers with type 1 and/or type 2 virus infection. These allow us to draw conclusions on two major issues. The first issue is whether overt multiple infections are a feature of all T-cell-immunocompromised groups. We were concerned that the frequency of multiple infection previously seen in the HIV-positive homosexual patients might reflect the atypical acquisition of additional EBV strains via a hematogeneous route from sexual contacts. Likewise, allograft recipients, another immunocompromised group for whom multiple EBV infections have been reported (18, 20), might also have acquired virus by an atypical route, namely, from B cells in the allograft itself (16, 23). In fact, we found that a substantial proportion (36%) of HIV-positive hemophilic patients also carry two or more detectable EBV strains, and, as argued earlier, these almost certainly have been acquired by the natural route. As an additional control to ask whether hemophilia per se is a predisposing factor, we also screened several HIV-negative hemophilic patients without finding evidence of coresident EBV strains (55a). The data therefore suggest very strongly that immune T-cell impairment is the major determinant of the overt multiple EBV infections seen in HIV-positive individuals. It is also interesting that both here (Table 2) and in the homosexual cohort (54), multiple infections with virus strains of the same type are more frequently found in the throat than in the blood; possible reasons for this are discussed elsewhere (54).
It remains a moot point, however, whether the acquisition of multiple EBV strains is genuinely restricted to immunocompromised individuals or is merely easier to detect in such cases because of the elevated virus load in vivo. In this context, some studies based on the direct PCR amplification of ex vivo samples, either across polymorphisms in the EBNA1 gene (4) or in the BamHI-F region of the genome (30), suggest that different strains coexist in the blood and/or throat of healthy immunocompetent carriers. On the other hand, in vitro isolation studies indicate the dominance of a single transforming strain in the great majority of such carriers, with any observed heterogeneity among isolates from a single individual being the result of intrastrain recombination at repeat sequences in the viral genome (20, 53). Final resolution of this issue will require a combination of these two approaches and must take into account the possible involvement of transformation-defective viruses that would not be rescued in vitro. Nevertheless, the power of in vitro isolation methods should not be underestimated. In the present work the detection of coresident EBV strains in HIV-positive hemophilic patients was not dependent on establishing unusually large numbers of virus isolates per patient; the mean numbers of isolates from individuals with overt multiple infection were actually lower than those for individuals with only one detectable strain (see Table 2, footnote a). Furthermore, in most cases where coinfection occurred, the different resident strains were each represented several times in the panel of LCLs derived from that individual, and so their presence was not difficult to detect. The uniformity of virus strains observed among independent virus isolates from healthy carriers (20, 53) is therefore more striking when viewed in this light.
The second issue being addressed by this study is whether type 2 virus strains are prevalent in all T-cell-immunocompromised groups. Our data strongly suggest that this is not the case. Thus, we found that just 10% of HIV-positive hemophiliacs had detectable type 2 EBV infection, and in only one patient was a type 2 virus the dominant strain. This is significantly different (P < 0.05) from what was seen in HIV-positive homosexuals, where >30% of patients carried a type 2 virus, often as the dominant or equidominant strain rescuable in vitro (54). Note that although the slower growth of type 2 virus-transformed LCLs can bias against type 2 virus detection in in vitro isolation assays (38), our protocols are specifically designed to minimize this problem. Whatever residual bias exists, however, must apply equally to the studies of both HIV-positive cohorts. We conclude, therefore, that there is a real difference in type 2 virus prevalence between the hemophilic and homosexual patient groups. It is well established that homosexual groups can harbor a greater range of infectious agents than are endemic in the general population. By contrast, apart from iatrogenic infections from Factor VIII preparations, hemophilic patients tend to mirror the general population in their viral flora. The example of human herpesvirus 8, a sexually transmitted gammaherpesvirus, is especially pertinent here; the incidence of human herpesvirus 8 infection in HIV-positive homosexuals is markedly higher than that seen in HIV-positive hemophilic patients or in the general population (12, 27).
We interpret our findings as showing that type 2 strains of EBV, a type most commonly seen in equatorial Africa, have become endemic in homosexual communities in the West, whereas they remain relatively rare in the general population as a whole. The previously reported high incidence of type 2 virus infection (7, 29, 31, 44, 50, 51, 54, 55) and of type 2 virus-positive malignancies (6, 10, 14, 21, 36, 46) among HIV-positive patients almost certainly reflects the predominance of male homosexuals in these patient cohorts, especially during the first 10 years of the AIDS epidemic (5). It is also necessary to reexamine the notion, to which these earlier findings gave rise, that type 2 viruses enjoy some kind of selective advantage over type 1 strains in the immunologically compromised host. There is no a priori reason for this, and it seems to us more likely that both viruses are able to take equal advantage of impaired T-cell surveillance. The implication of our findings is that the relative prevalence rates of type 1 and type 2 viruses, at least in the general population in the United Kingdom, are in line with those first suggested from in vitro isolation studies of healthy individuals, where the great majority carried type 1 virus and only 5 to 10% carried type 2 (53). Interestingly, an extensive survey of virus isolates from Dutch bone marrow transplant recipients led to similar conclusions regarding type 2 prevalence (20). Such observations are also in line with the European data from three EBV-associated tumors, posttransplant lymphoma, sporadic BL, and Hodgkin’s disease, arising in HIV-negative patients; these tumors were found to carry a type 1 strain in the great majority of EBV genome-positive cases (8a, 10, 13, 24, 42, 58). This preponderance of type 1 viruses cannot be due to nonpathogenicity of type 2 EBV, since type 2 strains are well represented in these same categories of malignancy arising in (predominantly homosexual) AIDS cohorts (6, 10, 14, 21, 36, 46). The question of type 2 virus prevalence in other Western societies remains to be resolved and will require in vitro isolation work to complement the results of direct PCR amplification assays. It is possible that in North America, where a greater proportion of the population is of African descent, type 2 prevalence may be higher (47). Even here, however, the data from EBV-associated tumors arising in HIV-negative patients strongly suggest that type 1 viruses are predominant (8, 11, 14, 29a). It will be important to map the epidemiology of type 1 and type 2 virus infections more accurately on a worldwide basis so that we can begin to understand how EBV coevolution with the human species led to the emergence of these distinct virus types.
ACKNOWLEDGMENTS
This work was supported by the Cancer Research Campaign, United Kingdom, and by the Endowment Fund of the University Hospital Birmingham.
We thank Susan Williams for photography and Deborah Williams for excellent secretarial assistance.
REFERENCES
- 1.Adldinger H K, Delius H, Freese U K, Clarke J, Bornkamm G W. A putative transforming gene of Jijoye virus differs from that of Epstein-Barr virus prototypes. Virology. 1985;141:221–234. doi: 10.1016/0042-6822(85)90253-3. [DOI] [PubMed] [Google Scholar]
- 2.Aitken C, Sengupta S K, Aedes C, Moss D J, Sculley T B. Heterogeneity within the Epstein-Barr virus nuclear antigen 2 gene in different strains of Epstein-Barr virus. J Gen Virol. 1994;75:95–100. doi: 10.1099/0022-1317-75-1-95. [DOI] [PubMed] [Google Scholar]
- 3.Appolloni A, Sculley T B. Detection of A-type and B-type Epstein-Barr virus in throat washings and lymphocytes. Virology. 1994;202:978–981. doi: 10.1006/viro.1994.1422. [DOI] [PubMed] [Google Scholar]
- 4.Bhatia K, Raj A, Guttierez M I, Judde J-G, Spangler G, Venkatesh H, Magrath I T. Variation in the sequence of Epstein-Barr virus nuclear antigen 1 in normal peripheral blood lymphocytes and in Burkitt’s lymphomas. Oncogene. 1996;13:177–181. [PubMed] [Google Scholar]
- 5.Biggar R J, Rabkin C S. The epidemiology of AIDS-related neoplasms. Haematol/Oncol Clin N Am. 1996;10:997–1010. doi: 10.1016/s0889-8588(05)70380-4. [DOI] [PubMed] [Google Scholar]
- 6.Boyle M J, Sewell W A, Sculley T B, Apolloni A, Turner J J, Swanson C E, Penny R, Cooper D A. Subtypes of Epstein-Barr virus in human immunodeficiency virus-associated non-Hodgkin lymphomas. Blood. 1991;78:3004–3001. [PubMed] [Google Scholar]
- 7.Buisson M, Morand P, Genoulaz O, Bourgeat M-J, Micoud M, Seigneurin J-M. Changes in the dominant Epstein-Barr virus type during human immunodeficiency virus infection. J Gen Virol. 1994;75:431–437. doi: 10.1099/0022-1317-75-2-431. [DOI] [PubMed] [Google Scholar]
- 8.Cen H, Williams P A, McWilliams H P, Breinig M C, Ho M, McKnight J L C. Evidence for restricted Epstein-Barr virus latent gene expression and anti-EBNA antibody response in solid organ transplant recipients with post-transplant lymphoproliferative disorders. Blood. 1993;81:1393–1403. [PubMed] [Google Scholar]
- 8a.Crawford, D. H. Personal communication.
- 9.Dambaugh T, Hennessy K, Chamnankit L, Kieff E. U2 region of Epstein-Barr virus DNA may encoded Epstein-Barr nuclear antigen 2. Proc Natl Acad Sci USA. 1984;81:7632–7636. doi: 10.1073/pnas.81.23.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Re V, Boiocchi M, De Vita S, Dolcetti R, Gloghini A, Uccini S, Baroni C, Scarpa A, Cattoretti G, Carbone A. Subtypes of Epstein-Barr virus in HIV-1-associated and HIV-1-unrelated Hodgkin’s Disease cases. Int J Cancer. 1993;54:895–898. doi: 10.1002/ijc.2910540603. [DOI] [PubMed] [Google Scholar]
- 11.Frank D, Cesarman E, Liu Y F, Michler R E, Knowles D M. Post-transplantation lymphoproliferative disorders frequently contain type A but not type B Epstein-Barr virus. Blood. 1995;85:1396–1403. [PubMed] [Google Scholar]
- 12.Gao S-J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with or without Kaposi’s sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 13.Gledhill S, Gallagher A, Jones D B, Krajewski A S, Alexander F E, Klee E, Wright D H, O’Brien C, Onions D E, Jarrett R F. Viral involvement in Hodgkin’s Disease—detection of clonal type A Epstein-Barr virus genomes in tumour samples. Br J Cancer. 1991;64:227–232. doi: 10.1038/bjc.1991.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldschmidts W L, Bhatia K, Franklin-Johnson J, Akar N, Guttierez M I, Shibata D, Carolan M, Levine A, Magrath I T. Epstein-Barr virus genotypes in AIDS-associated lymphomas are similar to those in endemic Burkitt’s lymphoma. Leukaemia. 1992;6:875–878. [PubMed] [Google Scholar]
- 15.Grasser F A, Murray P G, Kremmer E, Klein K, Remberger K, Feiden W, Reynolds G, Niedobitek G, Young L S, Muller-Lantzsch N. Monoclonal antibodies directed against the Epstein-Barr virus-encoded nuclear antigen 1: immunohistochemical detection of EBNA1 in the malignant cells of Hodgkin’s disease. Blood. 1994;84:3792–3799. [PubMed] [Google Scholar]
- 16.Gratama J W. Epstein-Barr virus infections of bone marrow transplantation recipients. In: Forman S J, Blume K G, Thomas E D, editors. Bone marrow transplantation. New York, N.Y: Blackwell Scientific Publications; 1994. pp. 429–442. [Google Scholar]
- 17.Gratama J W, Ernberg I. Molecular epidemiology of Epstein-Barr virus infection. Adv Cancer Res. 1995;67:197–255. [PubMed] [Google Scholar]
- 18.Gratama J W, Lennette E T, Lonnqvist B, Oosterveer M A P, Klein G, Ringden O, Ernberg I. Detection of multiple Epstein-Barr viral strains in allogeneic bone marrow transplant recipients. J Med Virol. 1992;37:39–47. doi: 10.1002/jmv.1890370107. [DOI] [PubMed] [Google Scholar]
- 19.Gratama J W, Oosterveer M A P, Klein G, Ernberg I. EBNA size polymorphism can be used to trace Epstein-Barr virus spread within families. J Virol. 1990;64:4703–4708. doi: 10.1128/jvi.64.10.4703-4708.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gratama J W, Oosterveer M A P, Weimar W, Sintnicolaas K, Sizoo W, Bolhuis R L H, Ernberg I. Detection of multiple ‘Ebnotypes’ in individual Epstein-Barr virus carriers following lymphocyte transformation by virus derived from peripheral blood and oropharynx. J Gen Virol. 1994;75:85–94. doi: 10.1099/0022-1317-75-1-85. [DOI] [PubMed] [Google Scholar]
- 21.Gunthel C J, Ng V, McGrath M, Herndier B, Shiramizu B. Association of Epstein-Barr virus types 1 and 2 with acquired immunodeficiency syndrome-related primary central nervous system lymphomas. Blood. 1994;83:618–619. [PubMed] [Google Scholar]
- 22.Hamilton-Dutoit S J, Raphael M, Ardouin J, Diebold J, Lisse I, Pederson C, Oksenhendler E, Marelle L, Pallesen G. In situ demonstration of Epstein-Barr virus small RNAs (EBER1) in acquired immunodeficiency syndrome-related lymphomas: correlation with tumour morphology and primary site. Blood. 1993;82:619–624. [PubMed] [Google Scholar]
- 23.Haque T J, Crawford D H. Transmission of Epstein-Barr virus during transplantation. Rev Med Virol. 1996;6:77–84. doi: 10.1002/(SICI)1099-1654(199606)6:2<77::AID-RMV166>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 24.Haque T, Thomas J A, Falk K I, Parratt R, Hunt B J, Yacoub M, Crawford D H. Transmission of donor Epstein-Barr virus (EBV) in transplanted organs causes lymphoproliferative disease in EBV-seronegative recipients. J Gen Virol. 1996;77:1169–1172. doi: 10.1099/0022-1317-77-6-1169. [DOI] [PubMed] [Google Scholar]
- 25.Henle W, Henle G. Seroepidemiology of the virus. In: Epstein M A, Achong B G, editors. The Epstein-Barr virus. Berlin, Germany: Springer-Verlag; 1979. pp. 61–78. [Google Scholar]
- 26.Katz B Z, Andiman W A, Eastman R, Martin K, Miller G. Infection with two genotypes of Epstein-Barr virus in an infant with AIDS and lymphoma of the central nervous system. J Infect Dis. 1986;153:601–604. doi: 10.1093/infdis/153.3.601. [DOI] [PubMed] [Google Scholar]
- 27.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi’s sarcoma- associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 28.Khanim F, Yao Q-Y, Niedobitek G, Sihota S, Rickinson A B, Young L S. Analysis of Epstein-Barr virus gene polymorphisms in normal donors and in virus-associated tumours from different geographic locations. Blood. 1996;88:3491–3501. [PubMed] [Google Scholar]
- 29.Kyaw M T, Hurren L, Evans L, Moss D J, Cooper D A, Benson E, Esmore E, Sculley T B. Expression of B-type Epstein-Barr virus in HIV-infected patients and cardiac transplant recipients. AIDS Res Hum Retroviruses. 1992;8:1869–1874. doi: 10.1089/aid.1992.8.1869. [DOI] [PubMed] [Google Scholar]
- 29a.Liebowitz, D. Personal communication.
- 30.Lung M L, Lam W P, Chan K H, Li S, Sham J, Choy D. Direct detection of Epstein-Barr virus in peripheral blood and comparison of Epstein-Barr virus genotypes present in direct specimens and lymphoblastoid cell lines established from nasopharyngeal carcinoma patients and healthy carriers in Hong Kong. Int J Cancer. 1992;52:174–177. doi: 10.1002/ijc.2910520203. [DOI] [PubMed] [Google Scholar]
- 31.Luxton J C, Williams A, Weller I, Crawford D H. Epstein-Barr virus infection of HIV-seropositive individuals is transiently suppressed by high dose acyclovir treatment. AIDS. 1993;7:1337–1343. doi: 10.1097/00002030-199310000-00006. [DOI] [PubMed] [Google Scholar]
- 32.MacMahon E M, Glass J D, Hayward D, Mann R B, Becker P S, Charache P, McArthur J C, Ambinder R F. Epstein-Barr virus in AIDS-related primary central nervous system lymphoma. Lancet. 1991;338:969–973. doi: 10.1016/0140-6736(91)91837-k. [DOI] [PubMed] [Google Scholar]
- 33.Maunders M J, Petti L, Rowe M. Precipitation of the Epstein-Barr virus protein EBNA2 by an EBNA3C-specific monoclonal antibody. J Gen Virol. 1994;75:769–778. doi: 10.1099/0022-1317-75-4-769. [DOI] [PubMed] [Google Scholar]
- 34.Miller W E, Edwards R H, Walling D M, Taab-Traub N. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J Gen Virol. 1994;75:2729–2740. doi: 10.1099/0022-1317-75-10-2729. [DOI] [PubMed] [Google Scholar]
- 35.Moss D J, Rickinson A B, Pope J H. Long-term T cell-mediated immunity to Epstein-Barr virus in man. I. Complete regression of virus-induced transformation in cultures of seropositive donor leukocytes. Int J Cancer. 1978;22:662–668. doi: 10.1002/ijc.2910220604. [DOI] [PubMed] [Google Scholar]
- 36.Ometto L, Menin C, Masiero S, Bonaldi L, del Mistro A, Cattelan A M, D’Andrea E, de Rossi A, Chieco-Bianci L. Molecular profile of Epstein-Barr virus in human immunodeficiency virus type 1-related lymphadenopathies and lymphomas. Blood. 1997;90:313–322. [PubMed] [Google Scholar]
- 37.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 38.Rickinson A B, Young L S, Rowe M. Influence of the Epstein-Barr virus nuclear antigen EBNA2 on the growth phenotype of virus-transformed B cells. J Virol. 1987;61:1310–1317. doi: 10.1128/jvi.61.5.1310-1317.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rowe M, Evans H S, Young L S, Hennessy K, Kieff E, Rickinson A B. Monoclonal antibodies to the latent membrane protein of Epstein-Barr virus reveal heterogeneity of the protein and inducible expression in virus-transformed cells. J Gen Virol. 1987;68:1575–1586. doi: 10.1099/0022-1317-68-6-1575. [DOI] [PubMed] [Google Scholar]
- 40.Rowe M, Young L S, Cadwallader K, Petti L, Kieff E, Rickinson A B. Distinction between Epstein-Barr virus type A (EBNA2A) and type B (EBNA2B) isolates extends to the EBNA3 family of nuclear proteins. J Virol. 1989;63:1031–1039. doi: 10.1128/jvi.63.3.1031-1039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A, Kieff E. Epstein-Barr virus type 1 (EBV-1) and 2 (EBV-2) differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandvej K, Peh S C, Storstein Andresen B, Pallesen G. Identification of potential hot spots in the carboxy-terminal part of the Epstein-Barr virus (EBV) BNLF-1 gene in both malignant and benign EBV-associated diseases: high frequency of a 30 bp deletion in Malaysian and Danish peripheral T cell lymphomas. Blood. 1994;84:4053–4060. [PubMed] [Google Scholar]
- 43.Schuster V, Ott G, Seidenspinner S, Kreth H W. Common Epstein-Barr virus (EBV) type 1 variant strains in both malignant and benign EBV-associated disorders. Blood. 1996;87:1579–1585. [PubMed] [Google Scholar]
- 44.Sculley T B, Apolloni A, Hurren L, Moss D J, Cooper D A. Coinfection with A- and B-type Epstein-Barr virus in human immunodeficiency virus-positive subjects. J Infect Dis. 1990;162:643–648. doi: 10.1093/infdis/162.3.642. [DOI] [PubMed] [Google Scholar]
- 45.Sculley T B, Apolloni A, Moss D J, Mueller-Lantsch N, Misko I S, Cooper D A. Expression of Epstein-Barr virus nuclear antigens 3, 4 and 6 are altered in cell lines containing B type virus. Virology. 1989;171:401–408. doi: 10.1016/0042-6822(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 46.Shibata D, Weiss L M, Hernandez A M, Nathwani B N, Bernstein L, Levine A M. Epstein-Barr virus-associated non-Hodgkin’s lymphoma in patients infected with the human immunodeficiency virus. Blood. 1993;81:2102–2109. [PubMed] [Google Scholar]
- 47.Sixbey J W, Shirley P, Chesney P J, Buntin D M, Resnick L. Detection of a second widespread strain of Epstein-Barr virus. Lancet. 1989;ii:761–765. doi: 10.1016/s0140-6736(89)90829-5. [DOI] [PubMed] [Google Scholar]
- 48.Subar M, Neri A, Inghirami G, Knowles D M, Dalla-Favera R. Frequent c-myc oncogene activation and infrequent presence of Epstein-Barr virus genome in AIDS-associated lymphoma. Blood. 1988;72:667–671. [PubMed] [Google Scholar]
- 49.Walling D M, Clark N M, Markovitz D M, Frank T S, Brawn D K, Eisenberg E, Krutchkoff D J, Felix D H, Raab-Traub N. Epstein-Barr virus co-infection and recombination in non-human-immunodeficiency virus-associated oral hairy leukoplakia. J Infect Dis. 1995;171:1122–1130. doi: 10.1093/infdis/171.5.1122. [DOI] [PubMed] [Google Scholar]
- 50.Walling D M, Edmistan S N, Sixbey J W, Abdel-Hamid M, Resnick L, Raab-Traub N. Co-infection of multiple strains of the Epstein-Barr virus in human immunodeficiency virus-associated hairy leukoplakia. Proc Natl Acad Sci USA. 1992;89:6560–6564. doi: 10.1073/pnas.89.14.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walling D M, Perkins A G, Webster-Cyriaque J, Resnick L, Raab-Traub N. The Epstein-Barr virus EBNA-2 gene in oral hairy leukoplakia: strain variation, genetic recombination, and transcriptional expression. J Virol. 1994;68:7918–7926. doi: 10.1128/jvi.68.12.7918-7926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao Q Y, Rickinson A B, Gaston J S H, Epstein M A. In vitro analysis of the Epstein-Barr virus:host balance in long-term renal allograft recipients. Int J Cancer. 1985;35:43–49. doi: 10.1002/ijc.2910350108. [DOI] [PubMed] [Google Scholar]
- 53.Yao Q Y, Rowe M, Martin B, Young L S, Rickinson A B. The Epstein-Barr virus carrier state: dominance of a single growth-transforming isolate in the blood and in the oropharynx of healthy virus carriers. J Gen Virol. 1991;72:1579–1590. doi: 10.1099/0022-1317-72-7-1579. [DOI] [PubMed] [Google Scholar]
- 53a.Yao, Q. Y., and R. J. Tierney. Unpublished results.
- 54.Yao Q Y, Tierney R J, Croom-Carter D, Dukers D, Cooper G M, Ellis C J, Rowe M, Rickinson A B. Frequency of multiple EBV infections in T-cell-immunocompromised individuals. J Virol. 1996;70:4884–4894. doi: 10.1128/jvi.70.8.4884-4894.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao Q Y, Tierney R J, Croom-Carter D, Cooper J, Ellis C, Rowe M, Rickinson A B. Isolation of intertypic recombinants of Epstein-Barr virus from T-cell-immunocompromised individuals. J Virol. 1996;70:4895–4903. doi: 10.1128/jvi.70.8.4895-4903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55a.Yao, Q. Y., et al. Unpublished results.
- 56.Young L S, Alfieri C, Hennessy K, Evans H, O’Hara C, Anderson K C, Ritz J, Shapiro R S, Rickinson A, Kieff E, Cohen J I. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med. 1989;321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- 57.Young L S, Yao Q Y, Rooney C M, Sculley T B, Moss D J, Rupani H, Laux G, Bornkamm G W, Rickinson A B. New type B isolates of Epstein-Barr virus from Burkitt’s lymphoma and from normal individuals in endemic areas. J Gen Virol. 1987;68:2853–2862. doi: 10.1099/0022-1317-68-11-2853. [DOI] [PubMed] [Google Scholar]
- 58.Zimber U, Adldinger H K, Lenoir G M, Vuillaume M, Knebel-Doeberitz M V, Laux G, Desgranges C, Wittman P, Freese U K, Schneider U, Bornkamm G W. Geographical prevalence of two Epstein-Barr virus types. Virology. 1986;154:56–66. doi: 10.1016/0042-6822(86)90429-0. [DOI] [PubMed] [Google Scholar]