Abstract
The nonstructural proteins NS1 and NS2 are thought to be expressed from the p7 promoter of Aedes densonucleosis virus (AeDNV). To study gene expression from the p7 promoter, eight different plasmids were constructed by fusing β-galactosidase or β-glucuronidase into the genome so that the reporter gene was in different open reading frames and under the transcriptional control of the p7 promoter. After transfection into C6/36 Aedes albopictus cells, constructs generated comparable amounts of RNA, but only the NS1 and NS2 fusion constructs produced appreciable levels of active enzyme. NS1 and NS2 fusion constructs contained wild-type AeDNV sequences from the p7 promoter downstream to nucleotide 458. The remaining constructs, with the exception of p7GUS.rf3, lacked some or all of these necessary sequences and inefficiently produced protein. These data suggest that sequences downstream of the p7 promoter play a role in translational regulation of gene expression from the p7 promoter of AeDNV.
Densonucleosis viruses are parvoviruses of arthropods. Three densoviruses that differ substantially in their genomic organization have been well characterized. In the Junonia coenia densovirus (7), the genes for the structural and nonstructural proteins are encoded on opposite strands. In the Aedes aegypti densovirus (AeDNV) (1, 2) and the closely related Aedes albopictus parvovirus (AaPV) (5), the genes for the nonstructural and structural proteins are encoded on the same strand, similar to those of mammalian parvoviruses. Gene expression from the viral promoters is transactivated by the viral NS1 protein (3, 9), but little else is known regarding the control of gene expression of densoviruses. Gene expression in the mammalian parvoviruses is controlled at several different levels, including transcription initiation, RNA splicing, translation initiation, and protein processing (4, 18–20, 26). Gene expression of the densoviruses is likely to be similarly complex.
The genome of AeDNV is a negative-sense DNA molecule 4,009 nucleotides in length (2). Preliminary data suggest that the genome codes for two polyadenylated transcripts (unpublished data). One is a full-length transcript approximately 3,500 nucleotides in length that originates from a yet-unidentified position near the p7 promoter and presumably terminates at the polyadenylation signal located at 92 map units. The second transcript, which is approximately 1,200 nucleotides in length, originates at an unidentified position near the p61 promoter and is also believed to be terminated at the polyadenylation signal at 92 map units. There are three open reading frames (ORFs), which encompass nearly the entire genome. The left ORF, 2,262 nucleotides in length, encodes the nonstructural protein NS1. The middle ORF, which lies entirely within the left ORF and is 1,158 nucleotides in length, presumably encodes the nonstructural protein NS2. The right ORF encodes the structural proteins VP1 and VP2. It is thought that a portion of the amino terminus of VP1 is proteolytically cleaved to produce VP2 (2).
The AeDNV genome does not have any apparent consensus splice sequences, and thus, the transcript that arises from the p7 promoter is not believed to be spliced. Both the left and middle ORFs must therefore be translated from the p7 transcript, but in the absence of any splicing event, expression of both ORFs from one transcript is unusual. Generally, the first AUG from the 5′ end of the mRNA is most efficiently utilized for translation, and downstream AUGs are infrequently used as the start of translation (15).
Here we report that, under basal expression conditions, both the NS1 and NS2 AUGs are efficiently used. In addition, sequences that are required for gene expression are located within the first 100 nucleotides downstream of the p7 promoter, and these sequences are likely involved in a translational regulatory mechanism affecting gene expression from the p7 promoter.
MATERIALS AND METHODS
Construction of plasmids. (i) Reporter gene expression plasmids.
Four nonfusion constructs in which the reporter gene’s wild-type ATG and downstream sequences were preserved during subcloning, and the reporter gene’s ATG was used for expression, were made with β-glucuronidase (GUS). The plasmid p7GUS was made by subcloning the KpnI/EcoRI fragment (the left end of the virus) from pUCA (3) into the KpnI and EcoRI sites of pBluescript KS− (Stratagene, La Jolla, Calif.). The XbaI/SstI fragment from pBI101 (Clontech, Palo Alto, Calif.) containing the GUS gene cassette was then subcloned into the XbaI and SstI sites of the pBluescript construct, thereby placing the GUS gene out of frame with the viral NS1 ATG. The plasmid pUCA.GUS was made by subcloning the Eco47III/HincII fragment from pUCA (the right end of the AeDNV genome) into the Ecl136 site of p7GUS, supplying a functional polyadenylation signal. The plasmid pUCA.GUSSma− was made by digesting pUCA.GUS with SmaI and religating. This resulted in a deletion of 29 nucleotides, thereby shifting the GUS gene cassette in frame with the viral NS1 ATG. The plasmid p7GUS.rf3 was made by subcloning the SmaI/NsiI fragment containing the GUS gene cassette from pUCA.GUS into the MscI and NsiI sites of pUCA, thereby placing the GUS gene cassette out of frame with both NS1 and NS2 ATGs.
The β-galactosidase (β-Gal) gene was used to generate two NS1 gene fusion constructs. The plasmid p7βgalNS1 was made by first digesting pMC1871 (Pharmacia, Piscataway, N.J.) with XmaI. The ends of the DNA were filled in with Klenow fragment, and the DNA was subsequently digested with PstI to yield the 3,100-nucleotide β-Gal gene. The β-Gal gene was then subcloned into the MscI and NsiI sites of pUCA. The plasmid pGAL1 was made by subcloning the SmaI/PstI fragment from pMC1871 into the MscI and NsiI sites of pUCA. This plasmid had an additional single-base deletion of a guanine residue at the MscI/SmaI fusion site.
Both β-Gal and GUS genes were used to construct gene fusions with the viral NS2 ORF. The plasmid p7βgalNS2 was made by first partially digesting pGAL1 with BamHI. Linear full-length fragments were then isolated, and the ends of the linear fragments were filled in with Klenow fragment (addition of 4 nucleotides). Linear fragments were circularized by ligation, and clones that had altered BamHI sites nearest the reporter gene fusion site were isolated. The plasmid p7GUSNS2 was made by first digesting pUCA.GUS with XmaI. The ends of the linear DNA were filled in with Klenow fragment, and then linear DNA was subsequently digested with NsiI. The small fragment, approximately 2 kb in length and containing the GUS gene, was subcloned into the MscI and NsiI sites of pUCA.
A DraIII deletion mutation was introduced into both NS1 and NS2 β-Gal gene fusion constructs. The NS1 gene fusion DraIII deletion mutant, p7βgalNS1.D3−, was made by digesting pGAL1 with EcoRV (the EcoRV fragment contained an alternate DraIII site that was removed to facilitate mutation of the DraIII site near the p7 promoter). The large fragment was isolated and ligated to form pGAL1.erv. pGAL1.erv was digested with DraIII, the overhanging nucleotides were removed from the linear fragments with T4 DNA polymerase, and the fragments were religated. pGAL1.erv clones that lacked a DraIII site were isolated. The EcoRV fragment from p7βgalNS1, which contained the remainder of the β-Gal gene, was then subcloned into the EcoRV site of pGAL1.erv. Sequencing revealed that the DraIII mutation was a deletion of 6 nucleotides (Fig. 1) rather than the expected 3-base deletion. The NS2 gene fusion DraIII deletion mutant, p7βgalNS2.D3−, was made by using p7βgalNS1.D3− as the starting construct to preserve the identical DraIII mutation. The reading frameshift was accomplished by first partially digesting p7βgalNS1.D3− with BamHI. Full-length linear fragments were isolated, and the ends of the linear fragments were filled in with Klenow fragment (addition of 4 nucleotides). Linear fragments were circularized by ligation. Clones that had altered BamHI sites nearest the reporter gene fusion site were isolated.
FIG. 1.
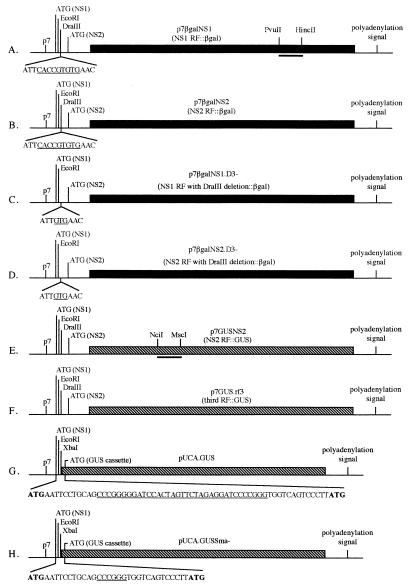
Schematic representation of the eight constructs used to transfect C6/36 cells. Dark and hatched boxes represent β-Gal and GUS genes, respectively. (A and B) Gene fusions into the NS1 and NS2 reading frames (RF), respectively. The underlined sequence indicates the DraIII restriction site. The bar between the PvuII and HincII sites indicates where the riboprobe used in the RNase protection assay protects the RNA from all β-Gal-containing constructs. (C and D) NS1 and NS2 gene fusions with the 6-nucleotide deletion at the DraIII site. The underlined sequence indicates what remains of the DraIII site. (E) An NS2 reading frame gene fusion. The bar between the NciI and MscI sites indicates where the riboprobe protects RNA from all GUS-containing constructs. (F) GUS gene cassette fused into the third reading frame. (G) GUS gene cassette fused immediately downstream of the p7 promoter. The viral NS1 ATG and the wild-type GUS ATG are shown in boldface and are the first and second ATGs downstream of the p7 promoter, respectively. The underlined sequence indicates nucleotides between the two SmaI sites (CCC/GGG). (H) The underlined sequence indicates nucleotides that remained between the SmaI sites after a deletion mutation was introduced into pUCA.GUS.
In all expression plasmids, regions near the p7 promoter were sequenced to confirm cloning sites and to confirm that mutation sites were altered as expected. Sequencing was done on an automated sequencer at MacroMolecular Resources, Colorado State University.
(ii) Runoff transcription plasmids.
Internal fragments of both reporter genes were subcloned into vectors that contained bacteriophage RNA polymerase promoters for use in transcribing labeled riboprobes. pBluescript KS−, which contains both T7 and T3 RNA polymerase promoter sequences, was the final vector for both reporter genes. The plasmid pBGUS was made by digesting p7GUS with SstI and MscI. The 3,800-bp fragment was isolated, and overhanging ends were removed with T4 DNA polymerase and then circularized by ligation with T4 DNA ligase. The plasmid p3ZGAL was made by subcloning the SstI/HincII (nucleotides 1962 to 2900) lacZ fragment from pMC1871 into the SstI and HincII sites of the pGEM3Z (Promega, Madison, Wis.) vector. The plasmid p7lacZ was made by subcloning the SstI/PstI fragment from p3ZGAL into the SstI and PstI sites of p7GUS.
Cell culture.
C6/36 A. albopictus cells (10) were maintained in L15 medium (Gibco BRL, Gaithersburg, Md.) containing 10% fetal bovine serum and 1% penicillin-streptomycin in 25-cm2 flasks at 28°C.
Transfections.
Lipofectin reagent (Gibco BRL), was used to transfect C6/36 cells according to the manufacturer’s recommendations. Fifty microliters of Lipofectin reagent and 20 μg of supercoiled plasmid DNA (10 μg of each of two plasmids) were added to 6 ml of L15 medium to make the Lipofectin mixture. The mixture was homogenized by vortexing and allowed to complex at room temperature for at least 15 min prior to application to cells. C6/36 cells were grown to a 50 to 70% confluent monolayer in a 75-cm2 flask and washed once with sterile phosphate-buffered saline (PBS), and 6 ml of Lipofectin mixture was added. Cells were incubated with Lipofectin mixture for 6 h at room temperature. After incubation, the Lipofectin mixture was removed, cells were washed once with sterile PBS, and 15 ml of fresh L15 medium containing 10% fetal bovine serum and 1% penicillin-streptomycin was added. Cells were then incubated at 28°C for 48 h.
Total protein (TP) assays.
Protein standards that ranged from 100 to 1,000 μg/ml were prepared with bovine serum albumin (Pierce Chemical Company, Rockford, Ill.). Aliquots of each standard were mixed with 1 ml of Coomassie blue reagent (Pierce Chemical Company), and standards were measured in a Beckman DU 640 spectrophotometer (Beckman Instruments, Inc., Fullerton, Calif.) at 595 nm. Appropriate volumes of cell lysates (prepared as described below) were mixed with 1 ml of Coomassie blue reagent, and the absorbance at 595 nm was measured for each sample. Sample concentrations were automatically calculated by the spectrophotometer based on the standard concentrations.
Protein expression assays.
Expression assays were designed such that two plasmids were used for each transfection and were performed with the following pairs of plasmids: pUCA.GUS-p7βgalNS1, pUCA.GUSSma−-p7βgalNS1, p7βgalNS1-p7GUSNS2, p7βgalNS1.D3−-p7GUSNS2, p7βgalNS2-p7GUSNS2, p7βgalNS2.D3−-p7GUSNS2, and p7GUS.rf3-p7βgalNS1. One plasmid contained GUS, and the second contained β-Gal. In each transfection, one plasmid, either p7GUSNS2 or p7βgalNS1, was known to express protein and was used as an internal standard to control for variation in efficiency of transfection. After cotransfection of C6/36 cells, β-Gal activity, GUS activity, and TP were measured. Thus, β-Gal activity per microgram of TP and GUS activity per microgram of TP were determined for each cotransfection and for untransfected C6/36 cells (negative controls). Negative control values (β-Gal per microgram of TP and GUS per microgram of TP) were subtracted from each corresponding transfection value. By using β-gal per microgram of TP and GUS per microgram of TP, ratios of β-Gal to GUS activity and/or GUS to β-Gal activity were then determined for each transfection. For cotransfections that contained p7βgalNS1, a ratio of GUS to β-Gal activity was calculated, and for cotransfections that contained p7GUSNS2, a ratio of β-Gal to GUS activity was determined.
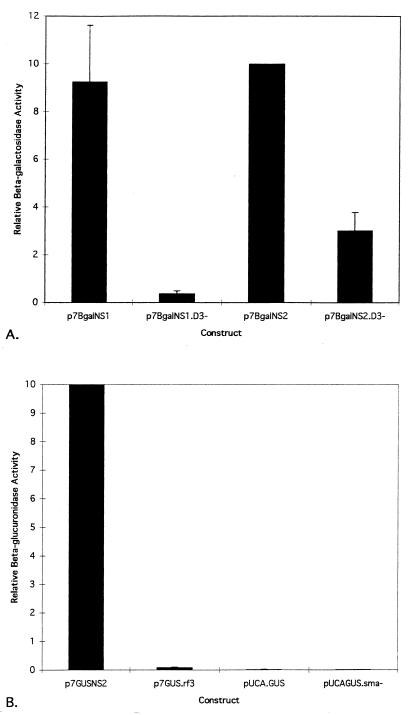
Within each replicate of cotransfections for which p7GUSNS2 was the standardizing plasmid, the p7βgalNS2/p7GUSNS2 ratio was arbitrarily given a value of 10. The normalization factor (N) was determined by dividing 10 by the measured value of the p7βgalNS2/p7GUSNS2 ratio, and each β-Gal/GUS ratio was then multiplied by N. Similarly, within each replicate for which p7βgalNS1 was the standardizing plasmid, the p7GUSNS2/p7βgalNS1 ratio was arbitrarily given a value of 10. N was determined by dividing 10 by the measured value of the p7GUSNS2/p7βgalNS1 ratio, and each GUS/β-Gal ratio was then multiplied by N. The averages and standard deviations were then determined for three replicate transfections.
After transfection, cells were dislodged by vigorously shaking the flask. A 1-ml aliquot of the cell suspension was transferred to a 1.7-ml microcentrifuge tube for the protein assay, and the remainder of cells were used for RNA isolation. Cells were pelleted in the 1.7-ml microcentrifuge tube by centrifugation (2,000 × g, 23°C, 5 min). Growth medium was removed, and cells were resuspended in 500 μl of PBS. Cells were again pelleted, and PBS was removed. Cells were then lysed in 250 μl of lysis solution (100 mM sodium phosphate [pH 7.8], 0.2% Triton X-100, 1 mM dithiothreitol), and lysates were analyzed for TP and for enzyme activity with luminometry-based Galacto-Light and GUS-Light kits (Tropix, Bedford, Mass.) according to the manufacturer’s recommendations. Appropriate sample volumes were used to ensure that the measurement of relative light units was in the linear range of the luminometer. Sample aliquots were pipetted into Turner Luminometer disposable cuvettes (Turner Designs, Sunnyvale, Calif.). At 1-min intervals, 180 (GUS-Light) or 200 (Galacto-Light) μl of reaction buffer (100 mM potassium phosphate [pH 8.0], 1 mM magnesium chloride, 1× Glucuron or 1× Galacton chemiluminescent substrate [Tropix]) was sequentially added to each cuvette. After each sample had been incubated at room temperature for 60 min, 300 μl of light emission accelerator was added, and the sample was immediately measured in a Turner TD-20e Luminometer (3-s delay, 15-s integration period; Turner Designs).
RNA isolation.
C6/36 cells were transfected under conditions identical to those for the protein assays. Forty-eight hours posttransfection, approximately 108 cells were used for total RNA isolation (14).
Antisense riboprobes.
Riboprobes to the β-Gal gene and GUS gene were transcribed from PvuII-linearized p7lacZ and NciI-linearized pBGUS, respectively. After templates p7lacZ and pBGUS were linearized, they were extracted once with phenol-chloroform-isoamyl alcohol (50:49:1) that was equilibrated to pH 7.4 with Tris-Cl, ethanol precipitated, and redissolved in diethylpyrocarbonate-treated water. T3 and T7 RNA polymerases were then used to transcribe [α-32P]CTP-labeled (800 Ci/mmol, 10 mCi/ml; New England Nuclear) riboprobes from p7lacZ and pBGUS templates, respectively, with the Maxiscript kit (Ambion, Austin, Tex.). Probes were labeled to a specific activity of 109 cpm/μg. Probes were gel purified according to the manufacturer’s instructions, and after elution, probes were ethanol precipitated in the presence of yeast tRNA and redissolved in 200 μl of hybridization buffer {80% formamide, 40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 6.4), 0.4 M NaCl, 1 mM EDTA}. A portion of the 566-nucleotide β-Gal probe hybridized to lacZ RNA corresponding to nucleotides 6284 to 6512 of pMC1871 and resulted in a protected fragment of 229 nucleotides in the RNase protection assay. Most of the 203-nucleotide GUS probe hybridized to GUS RNA corresponding to nucleotides 2986 to 3173 of pBI101 and resulted in a protected fragment of 188 nucleotides in the RNase protection assay.
RNase protection assay.
RNA from C6/36 cells transfected as described for the protein assays was analyzed by RNase protection assay (8). To ensure that the probe was present in vast excess over target RNA in the RNase protection assay, serial dilutions of sample RNA were hybridized to a constant amount of probe (1.5 × 105 cpm of each probe) to demonstrate that increasing signal intensity corresponded to increasing sample input. Thirty micrograms of total RNA from each transfection was lyophilized in a SpeedVac vacuum desiccator (Savant Instruments, Holbrook, N.Y.) and redissolved in 30 μl of hybridization buffer (80% formamide, 40 mM PIPES [pH 6.4], 0.4 M NaCl, 1 mM EDTA). The RNase protection assay was optimal when 1.5 × 105 cpm of each probe was used, hybridizations were done at 45°C, and RNase digestions were done with 350 μl of RNase digestion solution {5 μl of RNase cocktail (500 U of RNase A per ml, 20,000 U of RNase T1 [Ambion] per ml), 35 μl of RNase digestion buffer (10 mM Tris-Cl [pH 7.5], 300 mM NaCl, 5 mM EDTA), 310 μl of diethylpyrocarbonate-treated water}. Protected fragments were fractionated by electrophoresis on a Tris-borate-EDTA–8 M urea–5% polyacrylamide gel (200 by 160 by 0.7 mm). Samples were electrophoresed at 200 V until the bromophenol blue dye front was near the bottom of the gel. The gel was then transferred and dried onto chromatography paper (grade 1514A; Micro Filtration Systems). The dried gel was exposed to a PhosphorImaging screen (Molecular Dynamics, Sunnyvale, Calif.) overnight, after which the screen was scanned, yielding a 16-bit gel image. The image was then imported into NIH Image version 1.6, and values of intensity of lacZ- and GUS-protected fragments in each sample were determined. Untransfected C6/36 cells (negative controls) were also probed for β-Gal and GUS RNA. Within each transfection replicate, negative control β-Gal RNA and GUS RNA values were subtracted from each corresponding sample value.
Within each replicate of transfections, β-Gal RNA/GUS RNA and/or GUS RNA/β-Gal RNA ratios were determined for each sample. For transfections that contained p7βgalNS1, GUS RNA/β-Gal RNA ratios were determined, and for transfections that contained p7GUSNS2, β-Gal RNA/GUS RNA ratios were determined. For transfections in which p7GUSNS2 was the standardizing plasmid, the p7βgalNS2/p7GUSNS2 RNA ratio was arbitrarily assigned a value of 10. N was determined by dividing 10 by the measured value of the p7βgalNS2 RNA/p7GUSNS2 RNA ratio, and each β-Gal RNA/GUS RNA ratio was subsequently multiplied by N. For transfections in which p7βgalNS1 was the standardizing plasmid, the p7GUSNS2/p7βgalNS1 RNA ratio was arbitrarily assigned a value of 10. Here N was determined by dividing 10 by the measured value of the p7GUSNS2/p7βgalNS1 RNA ratio, and each GUS/β-Gal RNA ratio was then multiplied by N. The averages and standard deviations were then determined for three replicate transfections.
RESULTS
The presumed initiation codons for the NS1 and NS2 ORFs are the first and second ATGs following the TATA box defining the p7 promoter. These initiation codons are separated by 73 nucleotides (Fig. 2). To investigate whether one or both of these start codons were utilized, two β-Gal fusion constructs were made. Plasmids p7βgalNS1 and p7βgalNS2 contained translational fusions of β-Gal with two viral ORFs, ORF1 (NS1) and ORF2 (NS2) (Fig. 1). Both NS1 and NS2 fusion proteins were expressed at similar levels in C6/36 cells (Fig. 3). Since only one RNA is likely transcribed from the p7 promoter (unpublished data) and consensus splice junctions are not evident between the AUGs, it seems likely that both AUGs are recognized on the p7 transcript with similar efficiencies. To determine if a third AUG could be utilized in the third reading frame on the p7 transcript, the plasmid p7GUS.rf3 was constructed (Fig. 1). The third ATG was not originally in the viral sequence but was from the GUS gene, and the restriction site used for cloning into the viral genome was the same as that used to make the NS1 and NS2 gene fusion plasmids. Comparison of p7GUS.rf3 with p7GUSNS2 (the GUS analog to p7βgalNS2) (Fig. 3) showed that an AUG located in the third reading frame was not recognized with an efficiency comparable to that of the NS2 AUG. Thus, only the first two AUGs in the transcript, which correspond to the NS1 ORF and NS2 ORF, were used.
FIG. 2.
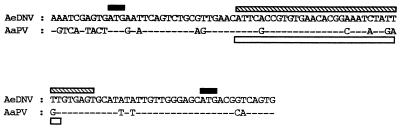
Sequence comparison of nucleotides downstream of the nonstructural promoter between AeDNV (290 to 388) and AaPV (325 to 423). The solid boxes indicate NS1 and NS2 ATGs of AeDNV, respectively. Regions of AeDNV and AaPV sequence that correspond to the secondary structures shown in Fig. 4 are indicated by the hatched and open boxes, respectively.
FIG. 3.
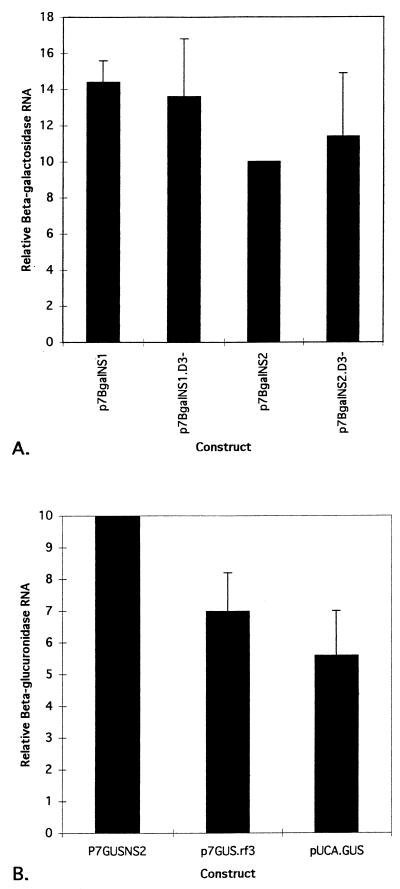
Comparison of relative enzyme production among β-Gal expression plasmids and among GUS expression plasmids. Both β-Gal and GUS genes were used to generate fusions into the viral NS2 reading frame, and therefore, protein production from p7βgalNS2 and p7GUSNS2 represents the same expression level from the AeDNV genome. Accordingly, the normalization of β-Gal/GUS and GUS/β-Gal ratios was designed such that p7βgalNS2/p7GUSNS2 and p7GUSNS2/p7βgalNS1 ratios were arbitrarily assigned a value of 10. Thus, p7βgalNS2 and p7GUSNS2 values shown do not have standard deviations. (A) Relative β-Gal activity. The determination of β-Gal/GUS activity ratios for each transfection included negative control values as described in Materials and Methods. p7GUSNS2 was used in each transfection as the internal standard to control for variation in transfection efficiency. Means and standard deviations were determined for three replicate transfections. (B) Relative GUS activity. The determination of GUS/β-Gal activity ratios for each transfection included negative control values as described in Materials and Methods. Here, p7βgalNS1 was used in each transfection as the internal standard to control for variation in transfection efficiency. Means and standard deviations were determined for three replicate transfections (pUCA.GUSSma− does not have a standard deviation because it was analyzed only once).
To determine how close to the putative p7 promoter sequence a gene of interest could be inserted and expressed, the plasmid pUCA.GUS was constructed by insertion of the GUS gene and several restriction sites from the plasmid pBI101 into the EcoRI site immediately downstream of the NS1 initiation codon of AeDNV (Fig. 1). This plasmid retained the NS1 initiation codon, but it was out of frame with the GUS gene. In order for GUS to be expressed, translation would have to begin at the second AUG codon, which originated from the GUS gene and was 59 nucleotides downstream of the NS1 AUG. This construct did not express the GUS reporter gene (Fig. 3). In an attempt to alleviate the lack of expression exhibited by pUCA.GUS, it was modified to make pUCA.GUSSma−, in which 29 nucleotides were removed from the restriction site array between the two ATGs, thereby placing the GUS ATG in frame with the viral NS1 ATG (Fig. 1). pUCA.GUSSma− also failed to produce GUS activity (Fig. 3). However, as described below, pUCA.GUS produced RNA, which suggested that transcription was not severely affected; rather, translation was not initiated at either AUG in this construct.
p7βgalNS1 and p7βgalNS2 contained nearly 200 nucleotides of viral sequence downstream of the p7 promoter and were capable of expressing the reporter gene. In contrast, pUCA.GUS and pUCA.GUSSma− had little viral sequence downstream of the promoter and were unable to express the reporter gene. This suggested that sequences downstream of the p7 promoter were necessary for gene expression.
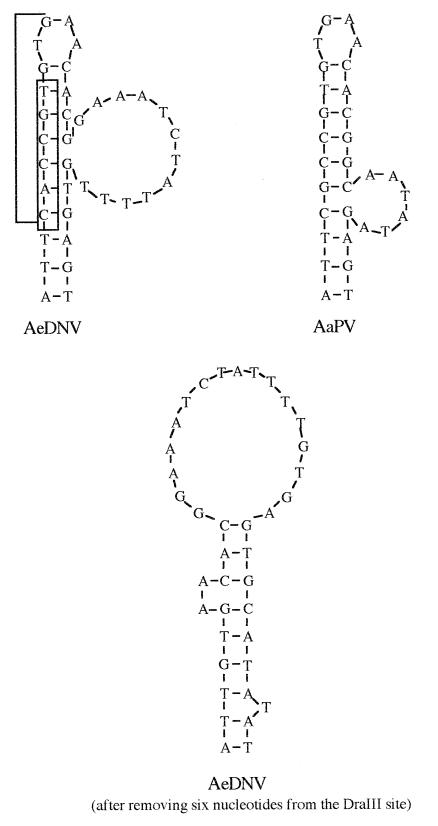
To identify regions of potential importance, AeDNV sequences downstream of the p7 promoter in the vicinity of the NS1 and NS2 ATGs (nucleotides 290 to 388) were compared to those downstream of the nonstructural promoter of AaPV (nucleotides 325 to 423) (5), a closely related mosquito densovirus (Fig. 2). These sequences were analyzed for potential RNA secondary structures and were both predicted to contain stem-loop structures (Fig. 4). To explore the possible role of this putative secondary structure in expression from the p7 promoter of AeDNV, p7βgalNS1.D3− and p7βgalNS2.D3− were constructed by removal of six nucleotides from the secondary structure at the DraIII site to alter its form (Fig. 4) and potentially its function without altering the reading frame. Indeed, both p7βgalNS1.D3− and p7βgalNS2.D3− resulted in lower levels of β-Gal activity (Fig. 3). p7βgalNS1.D3− showed approximately a 20-fold reduction and p7βgalNS2.D3− showed approximately a threefold reduction in protein production. These data show that the six nucleotides removed are necessary for efficient gene expression from the p7 promoter.
FIG. 4.
Predicted secondary structures for AeDNV and AaPV conserved regions. The bracketed nucleotides in AeDNV indicate the DraIII recognition sequence, and the boxed nucleotides are those that were removed at the DraIII site. Sequences were analyzed for RNA secondary structures with MacDNAsisPro (Hitachi Software Engineering Co., Ltd.).
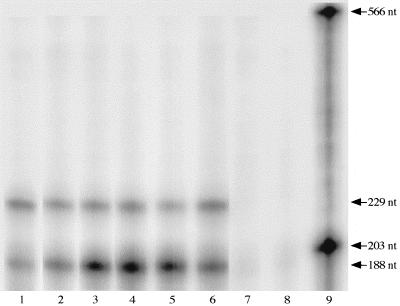
To determine if the sequences between the NS1 and NS2 AUG codons influenced transcription or translation, RNase protection was used to measure RNA production from each construct (Fig. 5). Densitometric analysis of the gel image revealed that all GUS constructs produced comparable levels of RNA, as did all β-Gal constructs (Fig. 6). The amount of RNA from pUCA.GUS, which produced no protein, and the amount of RNA from p7GUSNS2, which produced high levels of protein, differed by less than a factor of 2. Likewise, p7βgalNS1.D3− and p7βgalNS2.D3− produced lower levels of protein than p7βgalNS1, but all three constructs produced similar levels of RNA. Since all constructs were capable of producing RNA, those that did not produce protein were likely deficient in translation.
FIG. 5.
RNase protection assay. The 566- and 203-nucleotide (nt) fragments are full-length β-Gal probe and GUS probe, respectively. The 229- and 188-nucleotide fragments are the β-Gal- and GUS-protected fragments, respectively. RNA samples in each lane are from C6/36 cells cotransfected with expression plasmids. Lane 1, pUCA.GUS-p7βgalNS1. Lane 2, p7βgalNS1-p7GUSNS2. Lane 3, p7βgalNS1.D3−-p7GUSNS2. Lane 4, p7βgalNS2-p7GUSNS2. Lane 5, p7βgalNS2.D3−-p7GUSNS2. Lane 6, p7GUS.rf3-p7βgalNS1. Lane 7, RNA from untransfected C6/36 cells. Lane 8, β-Gal and GUS riboprobes digested with RNase. Lane 9, β-Gal and GUS riboprobes not digested with RNase.
FIG. 6.
Quantitative analysis of the protected fragments from the RNase protection assay. Protected fragments were visualized by exposing the RNase protection gel to a PhosphorImaging screen (Molecular Dynamics). The screen was scanned, which converted the gel image to a digital format. NIH Image version 1.6 was then used to densitometrically analyze the gel image, resulting in values of intensity for each protected fragment. Levels of RNA produced from constructs in each transfection were then used to determine β-Gal/GUS RNA and GUS/β-Gal RNA ratios as described in Materials and Methods. As with the enzyme activity ratios, normalization of RNA ratios was designed such that p7βgalNS2/p7GUSNS2 and p7GUSNS2/p7βgalNS1 RNA ratios within each replicate were assigned a value of 10. Hence, p7βgalNS2 and p7GUSNS2 RNA values shown do not have standard deviations. (A) Relative β-Gal RNA production. Negative control values were included in the determination of β-Gal/GUS RNA ratios as described in Materials and Methods. p7GUSNS2 was used in each transfection as an internal standard to control for variation in transfection efficiency. Means and standard deviations were determined for three replicate transfections. (B) Relative GUS RNA production. Negative control values were included in the determination of GUS/β-Gal RNA ratios as described in Materials and Methods. p7βgalNS1 was used as the internal standard. RNA production from pUCA.GUSSma− was not analyzed. Means and standard deviations were determined for three replicate transfections.
DISCUSSION
We have shown that both the NS1 ORF and, as previously demonstrated (3), the NS2 ORF are expressed and that there are sequences downstream of the p7 promoter that are essential for expression of both ORFs in C6/36 cells. Removal of these sequences abolished protein production. Furthermore, removing as few as six nucleotides from within the DraIII site, which altered the predicted secondary structure in the RNA (Fig. 4), resulted in a drastic decrease in protein production. Variations in rates of transcription initiation, elongation, and/or termination should have resulted in altered RNA levels and would have been measurable by the RNase protection assay. However, the RNase protection assay showed that all constructs produced similar amounts of RNA, and thus, transcription does not seem to be severely affected.
The assembly of the 40S ribosomal subunit and cellular initiation factors onto the mRNA is the first step of translation. This complex then scans the RNA to find the primary translational start codon, usually the first AUG downstream of the 5′ end of the transcript. The remaining components of the translational machinery then associate with the complex, and synthesis of a new protein begins. The first AUG downstream of the p7 TATA box of AeDNV is that of the NS1 ORF. This AUG was shown to be utilized for gene expression and thus must be contained in the p7 transcript. Although not yet determined, it is likely that the 5′ end of the p7 transcript will map to a position downstream of the p7 TATA box but upstream from the NS1 AUG. Based on this assumption, the NS1 AUG is expected to be the primary translational start codon on the p7 transcript. However, both NS1 and NS2 ORFs are expressed at about equal efficiencies, which suggests that alternative and/or additional mechanisms are involved in expression from the p7 transcript.
In eukaryotic and many viral systems, processing of the primary transcript allows expression of more than one protein from a single transcription unit. In the AeDNV p7 transcript, two scenarios that could explain the expression of both NS1 and NS2 ORFs can be imagined. For example, a portion of the transcript could be cleaved so that the NS1 AUG is removed, thereby leaving the NS2 AUG as the primary start codon. If the DraIII deletion interfered with this type of RNA processing (e.g., hindered the removal of the 5′ end of the primary transcript), expression of the NS2 ORF should be affected much more than that of the NS1 ORF. Fewer transcripts that used the NS2 AUG as the start codon would be generated. Alternatively, the transcript could be spliced to place the NS2 ORF in frame with the NS1 AUG. If the DraIII deletion interfered with a splicing event between the two ORFs, expression of the NS2 ORF should again be affected more than that of the NS1 ORF. In contrast, our data showed that expression of the NS1 ORF is affected to a greater degree than expression of the NS2 ORF, suggesting that neither type of RNA processing occurs and further supporting the belief that AeDNV transcripts are not differentially spliced.
Translation initiation can be influenced by the cap structure (11, 25, 27) and the poly(A) tail (11, 22, 24, 25), but these would be expected to be identical among our constructs and unlikely to explain the observed differences in expression. The sequence context in which a given AUG codon occurs can affect the efficiency at which it is recognized by a ribosome, and evidence suggests that the optimal nucleotide context in eukaryotic systems is (A/G)CCAUGG (16). The contexts of the NS1 and NS2 AUGs of AeDNV are not identical to the proposed optimal sequence, but they do have similarities to it. The NS1 AUG context, GUGAUGG, conforms to the proposed optimal sequence only by having a purine located at the −3 position (three nucleotides upstream from the AUG). However, in determination of the optimal AUG context, only 25% of examined sequences had a G at the −3 position (16). In contrast, the NS2 AUG context, AGCAUGA, matches the optimal sequence at the −3 and −1 positions. Furthermore, the −3 residue of the NS2 AUG context is an A, which was the most commonly observed purine residue at the −3 position (16). Thus, the NS2 AUG appears to be in a more optimal nucleotide context than the NS1 AUG. Since neither the NS1 AUG nor the NS2 AUG context perfectly conforms to the proposed optimal sequence, both are predicted to be in a suboptimal context. However, since the proposed optimal sequence context was derived primarily from higher eukaryotic mRNAs, these predictions may not be accurate, because the optimal sequence may be different for mosquito systems.
Insertion of a secondary structure downstream of an AUG in a poor sequence context has been shown to increase the efficiency at which that start codon is used, and the enhancement is most efficient when the secondary structure is 14 nucleotides downstream of the AUG, the distance between the leading edge of a ribosome and its AUG recognition site (17). In AeDNV, the predicted secondary structure is located approximately 20 nucleotides downstream of the NS1 AUG. In pUCA.GUS and pUCA.GUSSma−, the secondary structure has been removed, and neither the NS1 AUG nor the GUS AUG was used at all, indicating that these start codons may in fact be in a suboptimal nucleotide context (the nucleotide context of the GUS gene AUG, CUUAUGU, has a higher degree of divergence from the proposed optimal context than either the NS1 or the NS2 AUG). Furthermore, the putative RNA secondary structure was altered in p7βgalNS1.D3−, which resulted in a drastic decrease in the efficiency at which the NS1 AUG was used and is in good agreement with Kozak’s data (17). However, the NS2 AUG is downstream of the putative secondary structure but is also affected by alteration of the RNA structure. Based on this observation, it is unclear whether the important feature of the secondary structure is stability or three-dimensional shape. If stability is more important, the primary function of the RNA structure might be to enhance translation initiation from the NS1 AUG. If shape of the structure is more important, the structure might have the potential for modulating expression from both NS1 and NS2 AUGs. Since RNA secondary structures are also known to sometimes constitute internal ribosome entry sites (6, 12, 13, 21, 23), the role of the putative RNA secondary structure of AeDNV as an internal ribosome entry site cannot be excluded.
At present, these data suggest that expression from both NS1 and NS2 ORFs is regulated at the level of translation. It also appears that the NS1 and NS2 AUGs have different potentials for being used as start codons based on the proposed optimal nucleotide context surrounding start codons (16). The NS1 AUG is in a less optimal sequence context and likely requires the help of an RNA secondary structure to allow the ribosomal complex to initiate there. The NS2 AUG is also in a suboptimal sequence context but in a better context than the NS1 AUG. Therefore, the NS2 AUG might be more likely to function as a start codon independently of the putative RNA secondary structure, but use of the NS2 AUG as a start codon is still dependent upon upstream sequences in an unknown manner.
Further characterization of these regulatory sequences within the AeDNV genome will be necessary to reveal mechanisms as well as possible cofactors involved in the control of gene expression of AeDNV. Such information will facilitate the use of AeDNV as a genetic shuttle and as a cloning vector. Further experiments to determine the exact role of AeDNV sequence positions 290 to 388 are likely to identify a new mechanism of gene regulation within the Parvoviridae.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (NIAID AI25629 and AI28781) and the John D. and Catherine T. MacArthur Foundation.
REFERENCES
- 1.Afanasiev B N, Galev E E, Buchatsky L P, Kozlov Y V, Baev A A. Organization of the densovirus genome, with particular reference to the mosquito densovirus. Dokl Biol Sci. 1990;311:275–278. [Google Scholar]
- 2.Afanasiev B N, Galyov E E, Buchatsky L P, Kozlov Y V. Nucleotide sequence and genomic organization of Aedes densonucleosis virus. Virology. 1991;185:323–336. doi: 10.1016/0042-6822(91)90780-f. [DOI] [PubMed] [Google Scholar]
- 3.Afanasiev B N, Kozlov Y V, Carlson J O, Beaty B J. Densovirus of Aedes aegypti as an expression vector in mosquito cells. Exp Parasitol. 1994;79:322–339. doi: 10.1006/expr.1994.1095. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Asher E, Aloni Y. Transcription of minute virus of mice, an autonomous parvovirus, may be regulated by attenuation. J Virol. 1984;52:266–276. doi: 10.1128/jvi.52.1.266-276.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boublik Y, Jousset F X, Bergoin M. Complete nucleotide sequence and genomic organization of the Aedes albopictus parvovirus (AaPV) pathogenic for Aedes aegypti larvae. Virology. 1994;200:752–763. doi: 10.1006/viro.1994.1239. [DOI] [PubMed] [Google Scholar]
- 6.Brown E A, Day S P, Jansen R W, Lemon S M. The 5′ nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J Virol. 1991;65:5828–5838. doi: 10.1128/jvi.65.11.5828-5838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumas B, Jourdan M, Pascaud A M, Bergoin M. Complete nucleotide sequence of the cloned infectious genome of Junonia coenia densovirus reveals an organization unique among parvoviruses. Virology. 1992;191:202–222. doi: 10.1016/0042-6822(92)90182-o. [DOI] [PubMed] [Google Scholar]
- 8.Gilman M. Ribonuclease protection assay. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 4.7.1–4.7.8. [Google Scholar]
- 9.Giraud C, Devauchelle G, Bergoin M. The densovirus of Junonia coenia (Jc DNV) as an insect cell expression vector. Virology. 1992;186:207–218. doi: 10.1016/0042-6822(92)90075-z. [DOI] [PubMed] [Google Scholar]
- 10.Igarashi A. Isolation of a Singh’s Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J Gen Virol. 1978;40:531–544. doi: 10.1099/0022-1317-40-3-531. [DOI] [PubMed] [Google Scholar]
- 11.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 13.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kingston R E, Chomczynski P, Sacchi N. Guanidinium methods for total RNA preparation. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 4.2.1–4.2.8. [Google Scholar]
- 15.Kozak M. Evaluation of the “scanning model” for initiation of protein synthesis in eucaryotes. Cell. 1980;22:7–8. doi: 10.1016/0092-8674(80)90148-8. [DOI] [PubMed] [Google Scholar]
- 16.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krauskopf A, Aloni Y. A cellular repressor regulates transcription initiation from the minute virus of mice P38 promoter. Nucleic Acids Res. 1994;22:828–834. doi: 10.1093/nar/22.5.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krauskopf A, Bengal E, Aloni Y. The block to transcription elongation at the minute virus of mice attenuator is regulated by cellular elongation factors. Mol Cell Biol. 1991;11:3515–3521. doi: 10.1128/mcb.11.7.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krauskopf A, Resnekov O, Aloni Y. A cis downstream element participates in regulation of in vitro transcription initiation from the P38 promoter of minute virus of mice. J Virol. 1990;64:354–360. doi: 10.1128/jvi.64.1.354-360.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 22.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rijnbrand R C, Abbink T E, Haasnoot P C, Spaan W J, Bredenbeek P J. The influence of AUG codons in the hepatitis C virus 5′ nontranslated region on translation and mapping of the translation initiation window. Virology. 1996;226:47–56. doi: 10.1006/viro.1996.0626. [DOI] [PubMed] [Google Scholar]
- 24.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 25.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 26.Schoborg R V, Pintel D J. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology. 1991;181:22–34. doi: 10.1016/0042-6822(91)90466-o. [DOI] [PubMed] [Google Scholar]
- 27.Shatkin A J. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]