SUMMARY
The World Health Organization has established a fungal priority pathogens list that includes species critical or highly important to human health. Among them is the order Mucorales, a fungal group comprising at least 39 species responsible for the life-threatening infection known as mucormycosis. Despite the continuous rise in cases and the poor prognosis due to innate resistance to most antifungal drugs used in the clinic, Mucorales has received limited attention, partly because of the difficulties in performing genetic manipulations. The COVID-19 pandemic has further escalated cases, with some patients experiencing the COVID-19-associated mucormycosis, highlighting the urgent need to increase knowledge about these fungi. This review addresses significant challenges in treating the disease, including delayed and poor diagnosis, the lack of accurate global incidence estimation, and the limited treatment options. Furthermore, it focuses on the most recent discoveries regarding the mechanisms and genes involved in the development of the disease, antifungal resistance, and the host defense response. Substantial advancements have been made in identifying key fungal genes responsible for invasion and tissue damage, host receptors exploited by the fungus to invade tissues, and mechanisms of antifungal resistance. This knowledge is expected to pave the way for the development of new antifungals to combat mucormycosis. In addition, we anticipate significant progress in characterizing Mucorales biology, particularly the mechanisms involved in pathogenesis and antifungal resistance, with the possibilities offered by CRISPR-Cas9 technology for genetic manipulation of the previously intractable Mucorales species.
KEYWORDS: CotH, mucoricin, host responses, RNAi, iron uptake, dimorphism, genome duplication, Rhizopus, Mucor, invasins
INTRODUCTION
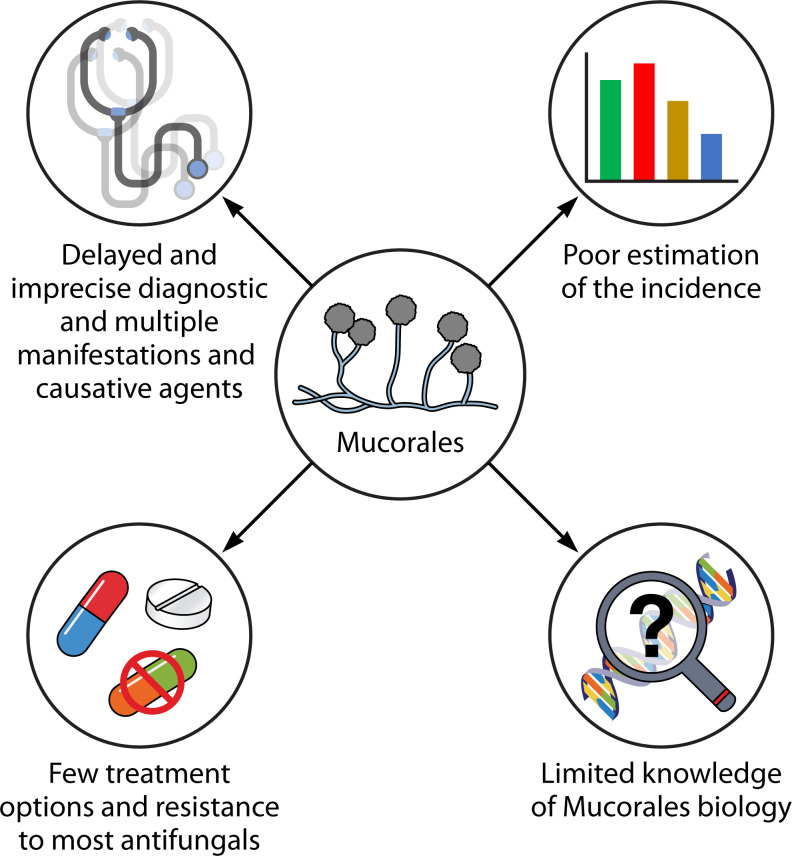
Mucormycosis is an invasive fungal infection caused by various species of fungi belonging to the order Mucorales. It primarily affects individuals with compromised immune systems, such as those with uncontrolled diabetes, organ transplant recipients, or patients with hematological malignancies (HMs). However, it can also impact immunocompetent individuals with skin breaks due to trauma or burns (1). The number of mucormycosis cases has been continuously increasing, with a significant surge observed in patients recovering from coronavirus disease 2019 (COVID-19). As a response to this growing concern, the World Health Organization (WHO) has included Mucorales in the high group of importance in its first fungal priority pathogens list. This recognition underscores the need to comprehend and address the challenges posed by mucormycosis (2). The WHO report emphasizes the necessity for heightened surveillance, research, development, and innovation, as well as public health interventions to improve the overall response to the priority fungal pathogens (2, 3). In the specific case of Mucorales, the report highlights the absence of accurate global incidence, limited information about standardized diagnosis and evidence-based treatments, and inadequate determination of antifungal resistance (Fig. 1) (2).
Fig 1.
Problems associated with the management of mucormycosis.
Mucormycosis stands out among pathogenic fungi due to its tendency to cause disproportionately severe infections with remarkably high morbidity and mortality rates, even in relatively immunocompetent individuals (4). Despite aggressive surgical interventions and intensive antifungal treatments, mucormycosis exhibits a distressingly high mortality rate, ranging from 50% to nearly 100%, depending on the specific manifestation (4, 5). In contrast, mortality rates for candidiasis and aspergillosis range from 20% to 50% and 35% to 45%, respectively (6, 7). Despite the introduction of newer antifungals, particularly triazoles with activity against Mucorales, such as posaconazole and isavuconazole, the high mortality rate for mucormycosis has not significantly improved in recent years (8, 9). A recent systematic review found that 90-day mortality (41.0%) remained comparable to that reported previously (44%–46%) (9). This excessively high mortality rate might be attributed to a multitude of factors, including late diagnosis, the innate antifungal resistance of Mucorales, limited treatment options, and the scarce understanding of the infective process associated with each causative agent (Fig. 1).
Phylogenetically distant from Ascomycetes and Basidiomycetes, the limited availability of specific and effective antifungal agents against Mucorales can be largely attributed to the general lack of comprehensive understanding of its biology. This knowledge gap is exacerbated by the intrinsic difficulty of genetic manipulation in Mucorales, historically prompting researchers to prefer alternative and more genetically tractable model systems (10). The overall lack of understanding of the Mucorales order has resulted in categorizing all infections caused by these fungi under the broad term “mucormycosis,” disregarding the significant heterogeneity among the genera and species that can cause infection and exhibit distinct behaviors. The specific knowledge of each species and their mechanisms of infection and virulence is even more limited.
The recent years have witnessed significant progress in knowing aspects of the pathogenesis of Mucorales compared to previous decades. This progress can be primarily attributed to the perseverance of a reduced number of investigators, who have developed novel study models and advanced methodologies for genetic manipulation. The objective of this review is to comprehensively describe and analyze these recent advancements, with a particular focus on the infection-related mechanisms, virulence factors, host molecular responses to infection, and antifungal resistance mechanisms.
EPIDEMIOLOGY AND INCIDENCE OF MUCORMYCOSIS, AN EMERGING DISEASE
Although the precise incidence of the mucormycosis is unknown, it is rising globally, becoming a significant public health concern (11–13). A systematic review covering a broad timeframe before COVID-19 pandemic (2000–2017) revealed that Europe exhibited the majority of cases (34%), followed by Asia (31%), and North or South America (28%), while lower rates were reported in Africa, Australia, and New Zealand (3%) (14). However, more recent estimated burden of mucormycosis per country consistently supports a greater prevalence (about 80 times) of mucormycosis cases in India and Pakistan than in developed countries before the COVID-19 pandemic (13, 15), contradicting the previous study that attributed the highest number of cases to Europe. This disparity can potentially be attributed to under-reporting that may have occurred during the study period in Asian countries, thereby highlighting that the precise diagnosis of mucormycosis continues to be one of the primary challenges in healthcare facilities. Although a simple potassium hydroxide (KOH) wet mount can detect the typical mucoralean hyphae in tissue samples, the requirement of trained staff and the low sensitivity of culture may be responsible for these variations. Additionally, diagnosing mucormycosis shows a scarcity of commercial and standardized methods (1, 8) across different countries, which can further vary during different data collection periods. The sum of these factors could explain why global epidemiological data on mucormycosis can be contradictory and not fully reliable. Despite the discrepancies in the incidence of mucormycosis across different countries, timeline comparisons of epidemiological data consistently demonstrate a steady rise in case numbers before the COVID-19 pandemic (13, 15, 16). This surge in mucormycosis incidence over recent years is what prompted the scientific community to designate it as an emerging disease (4, 13).
The incidence of predisposing conditions for mucormycosis, such as diabetes mellitus, HMs, transplantation, prolonged neutropenia, iron overload, corticosteroids, and trauma, varies significantly across different geographical regions (15). Diabetes mellitus is the predominant underlying disease in mucormycosis patients globally, with a significant rise in diabetes prevalence observed worldwide (4, 15). Uncontrolled type II diabetes is the most common type in diabetic patients with mucormycosis, and diabetes has been identified as a major risk factor in a high percentage of cases, particularly in India, Mexico, Middle Eastern, and North African countries (17–20). Furthermore, diabetic ketoacidosis (DKA) is a well-characterized predisposing factor for mucormycosis, primarily due to the increased availability of iron in the serum (21).
HMs and hematopoietic stem cell transplantation (HSCT) are frequently observed as the predominant underlying conditions in mucormycosis cases across Europe, the USA, and Australia (13). Acute myeloid leukemia is the most common type of HM associated with mucormycosis, comprising a significant percentage of cases in various studies (13, 22). Prolonged neutropenia and HSCT are important risk factors for mucormycosis, with varying incidence rates reported in different regions, ranging from 0.29% for HSCT recipients in the USA to 1%–2% in developing countries such as India, Iran, and South America (13). Solid organ malignancies and solid organ transplantation (SOT) are also significant risk factors for mucormycosis, with varying reported prevalence rates in different countries (13). The incidence of mucormycosis in different organ transplant recipients varies, with renal, liver, heart, and lung recipients experiencing distinct rates (23). Calcineurin inhibitors, such as tacrolimus, show a potential association with a lower risk, while renal failure, diabetes mellitus, and prior antifungal drug use are linked to a higher risk (13, 24).
Chronic administration of corticosteroids and other immunosuppressive agents has long been recognized as an important risk factor for mucormycosis, even before the COVID-19 pandemic (25, 26). However, the widespread use of corticosteroids in COVID-19 patients, particularly in India, has been associated with the alarming surge in mucormycosis cases (27, 28). This increase highlights the profound impact of the pandemic on mucormycosis incidence, underscoring the urgent need for heightened awareness and vigilance in managing fungal infections.
Immunocompetent individuals may also be affected when the fungal sporangiospores are inoculated directly into the skin as a result of trauma or burns, leading to cutaneous manifestations. Various studies have found that nearly 20% of patients with mucormycosis have no underlying medical condition. Trauma can range from minor incidents resulting in small open wounds to major events, including natural disasters, surgical procedures, and combat-related injuries (13, 29, 30).
The increase in COVID-19-associated mucormycosis (CAM) cases in India marks the most important outbreak of mucormycosis to date (11, 12, 27), with the staggering number of approximately 41,000 cases reported by the end of June 2021, resulting in around 3,200 deaths (31). Referral sites witnessed a corresponding increase in mucormycosis cases, with one study observing a rise from 0.03% to 2.5% of all hospital admissions between January and May 2021, indicating a significant impact of the pandemic on mucormycosis incidence (32). Studies about the etiology of the localized epidemic in India suggest both host and environmental factors contributed to the outbreak (28, 33, 34). Host factors include diabetes mellitus, diabetic ketoacidosis during COVID-19, and renal transplantation (28). In addition, there is strong evidence that glucocorticoid therapy, including cumulative dose, and zinc supplementation were associated with CAM. Additionally, rural residence was significantly linked with CAM, which may be due to higher levels of sporangiospores in the rural environment (28). In fact, a high burden of Mucorales sporangiospores both in hospital and outdoor environments has been reported in India during and even before the CAM epidemic (35, 36). In the same line, the sporangiospore count in the air in bedrooms of non-hospitalized CAM patients was significantly higher than in the air in other rooms in their homes (34). Interestingly, a strong uptick in environmental sporangiospore counts, likely due to favorable weather conditions (warm, high evaporation, and low humidity) for the dispersal of fungal propagules, preceded the CAM outbreak (33). Additionally, whole-genome sequencing (WGS) of 15 isolates of different species (7 of Rhizopus delemar, 4 of Rhizopus arrhizus, 2 Rhizopus microsporus, and 2 Lichtheimia ornata) revealed significant genomic differences among the isolates of the same species, suggesting that they are not clonal and the absence of nosocomial transmission (33). Similar situations have been observed in smaller outbreaks outside India in which WGS has been applied to the isolates, such as one in a burn unit in France and another in Joplin after a tornado. In the first case, the infections were caused by multiple strains of Mucor circinelloides that seemed patient specific, suggesting that the patients were more likely infected from a pool of diverse strains from the environment rather than from direct transmission among them (37). Similarly, the Apophysomyces trapeziformis strains causing the outbreak in Joplin were not clonal (38), supporting the high genomic heterogeneity of the mucormycosis-causative agents. The general idea is the existence of remarkable genetic diversity in mucormycosis-causing pathogens that extends beyond the species level, and it is difficult to genetically link them to a definitive environmental source (39).
DIAGNOSIS OF MUCORMYCOSIS
It is crucial to recognize that mucormycosis is a serious condition that requires early diagnosis and prompt initiation of appropriate treatment. Delayed diagnosis and management can have devastating consequences, including tissue loss, disseminated infection, and, in severe cases, death. However, achieving an accurate diagnosis of mucormycosis remains one of the foremost challenges in the management of this disease. Currently, most hospitals lack reliable serological, PCR-based, or skin tests to diagnose mucormycosis, which cannot be excluded solely based on sterile culture results. Histopathology, direct microscopy, and culture of diverse clinical specimens, supported with imaging techniques, are the classical and primary diagnostic approaches for mucormycosis (1, 5). In this sense, the diagnosis of mucormycosis necessitates specialized personnel and techniques that are not always readily available in clinical settings, especially in developing countries. As a result, the use of routine diagnostic techniques rather than those specifically tailored for mucormycosis leads to misdiagnosis and confusion of mucormycosis with other filamentous fungal infections, such as aspergillosis, resulting in inappropriate treatment from the outset.
Direct microscopy of KOH wet mounts with fluorescent brighteners such as blankophor and calcofluor white, along with a fluorescence microscope, allows for a rapid presumptive diagnosis of mucormycosis (1). This method is recommended by the European Confederation of Medical Mycology and Mycoses Study Group Education and Research Consortium in conjunction with histopathology to define surgical margins during invasive fungal infection surgery. Immunohistochemistry with monoclonal antibodies against R. arrhizus (syn. R. oryzae) also helps to differentiate mucormycosis from aspergillosis when cultures are negative (1).
Culture is critical to the diagnosis of mucormycosis as it allows for genus and species identification and antifungal susceptibility testing. Medically significant Mucorales are thermotolerant and grow rapidly at 37°C on various carbohydrate substrates. Colony formation typically occurs within 24–48 hours, and identification is based on colony and microscopic morphology and growth temperature (40). Positive cultures from sterile sites confirm the diagnosis, whereas those from non-sterile sites require correlation with clinical and radiological data. However, the low sensitivity of the cultures, up to 50% false negatives, necessitates optimal specimen handling and collaboration between clinicians and microbiology laboratories (41).
A definitive diagnosis of mucormycosis relies on identifying characteristic fungal hyphae of Mucorales in tissue biopsies or bronchoalveolar lavage (BAL) samples from patients with pulmonary mucormycosis. Histopathology plays a crucial role in distinguishing the presence of the pathogenic fungus in the specimen from a culture contaminant, and it is essential in determining whether there is an invasion of blood vessels (42). Additionally, histopathology can detect co-infections with other molds. Mucorales fungi typically produce wide (5–20 µm), thin-walled, ribbon-like hyphae with minimal septations (pauciseptate) and right-angle branching, in contrast to the narrower (3–5 µm), septate hyphae with acute-angle branching observed in Aspergillus species and other hyaline molds. Routine hematoxylin and eosin stains may only reveal the cell wall without internal structures or occasionally degenerate hyphae. Stains such as Grocott methenamine-silver (GMS) and periodic acid-Schiff (PAS) can aid in highlighting the fungal wall, with PAS providing better visualization of the surrounding tissue compared to GMS (42).
In addition to traditional diagnostic techniques for mucormycosis, other more recent molecular methods are slowly being implemented in the clinical setting, with PCR standing out as the most reliable and user-friendly approach. Quantitative PCR (qPCR) methods for serum, tissue, and BAL offer rapid results (around 3 hours) and earlier diagnosis of mucormycosis compared to conventional methods and imaging (43, 44). These qPCR assays are mainly based on detecting ITS1 or ITS2 ribosomal regions or the 18S region of rDNA for several species of Mucorales (45, 46). Besides ribosomal RNA, more specific targets are proposed, such as the sporangiospore coating protein homolog cotH genes as promising targets unique to Mucorales, showing favorable results in a mouse model (47). Serum is an ideal specimen for the detection of Mucorales because sampling is noninvasive, and angioinvasiveness of Mucorales results in high levels of fungal DNA in serum (48). They have been proven to be suitable for screening and monitoring high-risk patients, including follow-up after treatment initiation. Interestingly, recent multicenter studies with a significant number of patients (MODIMUCOR study) showed good reproducibility and performance of qPCR assays in serum samples, supporting the inclusion of this technique as part of the diagnostic strategy for mucormycosis (43, 44). In addition to in-house qPCR methods, there are at least three commercial qPCR kits for Mucorales (MucorGenius from PathoNostics, MycoGENIE from Ademtech, and Fungiplex from Bruker) that facilitate the testing for mucormycosis in serum, BAL, or biopsies (49). MucorGenius is the most evaluated kit with a sensitivity of 75% in serum (50) and 90% in respiratory samples, where a specificity of 95% was observed (51). MycoGENIE, which is able to identify both Aspergillus and Mucorales, showed a sensitivity of 80% and a specificity of 95% in serum (52).
Although qPCR is most likely to be adopted in clinical settings, other strategies are being explored. Matrix-assisted laser desorption ionization time-of-flight has demonstrated reliability and speed as a method for species-level identification. However, its effectiveness depends on the availability of in-house databases (53). In addition, breath volatile metabolite profiles were tested as a novel diagnostic method in a murine model of invasive mucormycosis. Distinct profiles of the metabolite sesquiterpene were identified in three Mucorales species, offering the potential for non-invasive diagnosis and therapy monitoring, but further evaluation is required to validate its efficacy (54).
MUCORMYCOSIS SHOWS DIFFERENT CLINICAL MANIFESTATIONS
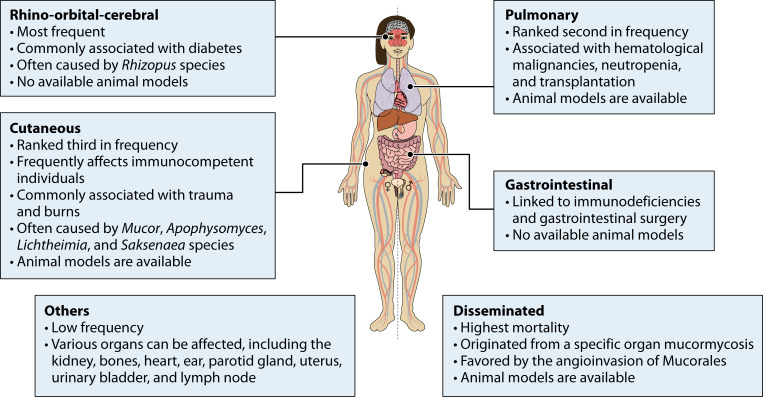
Mucormycosis is an infection that can manifest in various forms and affect different organs, resulting in a wide range of clinical presentations (Fig. 2) (14, 15, 55). The variability of its manifestations makes mucormycosis a complex and challenging disease to diagnose and treat. The most common forms of mucormycosis include rhino-orbital-cerebral (ROC), pulmonary, cutaneous, gastrointestinal, and disseminated infections. Other less common clinical forms of mucormycosis involve the kidney, bone, heart, ear, parotid gland, uterus, urinary bladder, and lymph nodes (14, 15, 55). Each form exhibits distinct characteristics based on the site of infection and the route of fungal entry into the body (55).
Fig 2.
Manifestations of mucormycosis. Most remarkable features of the different types of infections caused by Mucorales are indicated.
Rhino-orbital-cerebral
ROC mucormycosis (Fig. 2) is the most common manifestation of this fungal infection, and it is mainly associated with diabetic ketoacidosis or with uncontrolled diabetes mellitus. Other risk factors include SOT, corticosteroid therapy, chronic kidney disease, and intravenous drug use (14, 15). This is a severe manifestation primarily affecting the nasal and orbital regions, with the potential to invade cerebral tissue and lead to life-threatening complications. Patients often present with symptoms such as nasal congestion, facial pain, proptosis (forward displacement of the eye), chemosis (swelling of the conjunctiva), and visual disturbances. The infection can rapidly progress to involve adjacent structures, including the orbit, paranasal sinuses, and brain (55, 56). One of the distinguishing features of ROC mucormycosis is the presence of black eschar, which is a necrotic, black-colored tissue resulting from fungal invasion and subsequent tissue necrosis (55, 57). The presence of a black eschar in CAM patients led to the entirely incorrect assumption that it was caused by black fungi, which are phaeoid fungi unrelated to Mucorales. Cranial computed tomographic (CT) or magnetic resonance imaging is strongly recommended for the diagnosis of ROC, followed by endoscopic biopsy if sinusitis is present (1). Treatment of ROC mucormycosis requires a multidisciplinary approach involving ophthalmologists, otolaryngologists, and infectious disease specialists. The mainstay of treatment is aggressive surgical debridement to remove necrotic tissue and control the fungal invasion. Antifungal therapy with agents such as amphotericin B or posaconazole is administered intravenously to target the fungal infection systemically (26, 58, 59). The prognosis for rhino-orbital mucormycosis remains challenging, with a high mortality rate despite appropriate treatment, due to the aggressive nature of the infection, delay in diagnosis, and underlying immunocompromised state of the patient (4).
Pulmonary
The pulmonary manifestation (Fig. 2) is the second most prevalent type of mucormycosis and predominantly affects patients with hematological disorders and transplant recipients (15). The clinical features of this manifestation are characterized by a wide spectrum of respiratory symptoms. Patients may present with fever, cough, shortness of breath, chest pain, and hemoptysis (coughing up blood). These symptoms can be nonspecific and similar to other respiratory infections, making early diagnosis difficult (40). One of the distinguishing features of pulmonary mucormycosis is the rapid progression and potential for dissemination of the infection, which has been reported in 40% of the cases (4). Diagnosis of pulmonary mucormycosis involves a combination of clinical evaluation, radiological imaging, and microbiological examination. Chest imaging, CT scans, may reveal characteristic findings, including the reverse halo sign, an area of ground-glass opacity surrounded by a ring of consolidation, nodules, cavitation, or infarction (1). Because imaging studies can be nonspecific, microscopic examination of respiratory specimens, such as bronchoalveolar lavage fluid or lung biopsy specimens, can help demonstrate the presence of fungal hyphae invading the lung tissue (25, 40, 60). Prompt and aggressive treatment is essential in managing the pulmonary mucormycosis. This usually involves a combination of antifungal therapy and surgery (25, 40, 61). The prognosis for pulmonary mucormycosis remains poor, primarily due to the underlying immunosuppressed state of affected individuals and the aggressive nature of the infection (4, 60).
Cutaneous
This is the third most common type of mucormycosis, often affecting immunocompetent patients, and includes not only skin infection but also subcutaneous tissue involvement (Fig. 2) (15). Several skin injuries, ranging from bites of small animals to trauma resulting from combat injuries or natural catastrophes, expose internal tissues to environmental sources of fungal sporangiospores, increasing their susceptibility to mucormycosis (15, 29, 30). The clinical features of cutaneous mucormycosis are characterized by the presence of necrotic skin lesions, often accompanied by pain, swelling, and erythema (29). These lesions may progress rapidly, leading to tissue destruction and possible dissemination to deeper structures. The characteristic black eschar may also be observed at the site of the initial skin lesion. Diagnosis is often made by both histopathology and culture (62). Similar to rhino-orbital mucormycosis, cutaneous mucormycosis requires a multidisciplinary treatment approach involving dermatologists, surgeons, and infectious disease specialists (4, 63). The prognosis for cutaneous mucormycosis depends on several factors, including the extent of tissue involvement, underlying comorbidities, and timeliness of diagnosis and treatment. Early detection and intervention are critical to prevent the progression of the infection and minimize the risk of complications, such as tissue loss, dissemination to vital organs, and mortality (4, 64).
Gastrointestinal
Gastrointestinal mucormycosis (Fig. 2) affects the gastrointestinal tract, particularly in individuals with compromised immune systems or those who have undergone gastrointestinal surgery. It is also the most common manifestation of mucormycosis in neonates, with a high associated mortality (65). It can cause symptoms such as abdominal pain, gastrointestinal bleeding, and intestinal perforation (55, 59). Early endoscopic biopsy and histopathological examination are the gold standard for its diagnosis (66). A recent outbreak of gastrointestinal and disseminated mucormycosis was linked to contaminated batches of yogurt, highlighting the potential for foodborne transmission of this fungal infection (67).
Disseminated
Disseminated mucormycosis (Fig. 2) refers to a widespread infection involving multiple organs and occurs when any of the aforementioned manifestations cannot be controlled by the immune system or clinical treatment, and the angioinvasive nature of the Mucorales fungi causes the disease to spread throughout the body. These cases have the highest mortality rate, reaching 100% in patients with severe underlying risk factors (4, 14).
CAUSATIVE AGENTS OF MUCORMYCOSIS
Mucorales, like most human pathogenic fungi, are saprophytes that have evolved to thrive in natural environments different from the human body. Therefore, they have developed mechanisms to obtain nutrients and avoid predation under these conditions, independent of human infection. Some species may have serendipitously evolved physiologies that allow them to infect weakened humans and behave as opportunistic pathogens (68). The order Mucorales comprises 14 families, 56 genera, and more than 330 species (69). Of these, at least 11 genera and 39 species have been identified as causative agents of mucormycosis (16). The most important characteristics of the major genera causing mucormycosis are described below.
Rhizopus spp.
Rhizopus is by far the most common genus associated with mucormycosis, accounting for approximately half of the diagnosed cases worldwide, including CAM (14, 70, 71). In order to clarify the nomenclature, the current most employed species names are used in this review: R. arrhizus as synonymous with R. oryzae and R. delemar as synonymous with R. arrhizus var. delemar (72, 73). Most epidemiological studies include cases caused by either species as caused by R. arrhizus, even though these are two different species. Most infections identified at the species level are caused by R. arrhizus, followed by R. microsporus, with sporadic cases caused by R. homothallicus, R. schipperae, and R. stolonifer, although it should be noted that a significant number of isolates are identified only at the genus level (14, 70). In India, there are indications that the infections due to R. microsporus and R. homothallicus are increasing (74).
Rhizopus spp. can cause any mucormycosis manifestations, but they predominantly cause ROC, followed by pulmonary and cutaneous manifestations. They are also the major causative agent in all continents (14). Genome duplication seems to have played an important role in the evolution of Rhizopus spp., resulting in the duplication of genes that may contribute to antifungal resistance and pathogenesis with the gain of specific functions (75–78). Phylogenomic analyses of more than 220 sequenced genomes of Rhizopus spp., mostly of R. arrhizus, suggest that they are part of a monophyletic group (79). The genomes of Rhizopus spp., like those of most Mucorales, are in the draft stage and contain many scaffolds, making it difficult to perform a comprehensive characterization of the genome structure and function (79). The genetic diversity of Rhizopus spp. extends beyond the species level, as evidenced by WGS of 70 Rhizopus isolates (45 R. arrhizus, 19 R. delemar, and 6 R. microsporus) from patients with mucormycosis, and hospital and regional environments, which revealed that hospitals and surrounding communities are home to genomically diverse Rhizopus (39).
Rhizopus spp. show high resistance to most available antifungal drugs, with wide variation in susceptibility within specific species to antifungal agents with significant anti-Mucorales activity (80), which is a common feature of Mucorales. The MIC50 (The Clinical and Laboratory Standards Institute, CLSI) of amphotericin B is 0.03–4 µg/mL for R. arrhizus and 0.06–4 μg/mL for R. microsporus, the MIC50 of posaconazole is 0.03–32 µg/mL for R. arrhizus and 0.06–16 μg/mL for R. microsporus, and the MIC50 of isavuconazole is 0.125–2 μg/mL for R. arrhizus and 0.125–1 μg/mL for R. microsporus (81, 82). Epidemiological cut-off value (ECV) of posaconazole is the same for both species (2 µg/mL), whereas the ECV of amphotericin B is higher for R. arrhizus (4 µg/mL) than for R. microsporus (2 µg/mL) (82).
Mucor spp.
Mucor is the second most common genus in terms of mucormycosis cases, followed closely by Lichtheimia (14). This genus is incredibly diverse and probably polyphyletic (79), and is by far the largest genus in the Mucorales, currently comprising 76 recognized species, 13 of which are known to be involved in infections (70). Mucor spp. can cause many manifestations of mucormycosis, although cutaneous infection is the most common, followed by disseminated and pulmonary (14). Due to its relevance, more than 150 genomes have been sequenced, with the genome of M. lusitanicus having the highest quality genome sequence within the early-diverging fungi (EDF), as it is represented in a very few numbers of scaffolds that are close to the number of chromosomes, with the centromeres identified (83). In addition, procedures for genetic manipulation of this species are well established, including gene deletion and tagging (83–85). Mucor spp. are particularly amenable to genetic manipulation, and procedures have also been developed for M. circinelloides (86). Some Mucor species that cause mucormycosis (M. racemosus, M. lusitanicus, M. circinelloides, M. subtilissimus, and M. fragilis) exhibit dimorphism in that they can grow either as a yeast or as a mycelium. The known stimuli and pathways involved in the dimorphism of Mucorales will be described in a separate section as they play a role in the virulence of the dimorphic species, especially in Mucor spp. This dimorphism has also been observed in species of other genera such as Rhizomucor miehei, Rhizomucor pusillus, Mycotypha microspore, and Cokeromyces recurvatus (87–89).
The most clinically relevant Mucor species, M. circinelloides, has MIC50 (CLSI) of amphotericin B, posaconazole, and isavuconazole of 0.03–4, 0.06–16, and 0.125–>16 µg/mL, respectively (81, 82), while the ECVs of amphotericin B and posaconazole are 2 and 4 µg/mL, respectively (82).
Lichtheimia spp.
The number of infections caused by Lichtheimia spp. is similar worldwide to that caused by Mucor spp., although in Europe and Africa, it far exceeds the number of cases produced by Mucor and is the second most commonly reported mucormycosis-causing agent after Rhizopus spp. (14). In contrast, the number of cases in America is rather low (14, 90). Interestingly, the phagocytic susceptibility of Lichtheimia species depends on their geographical origin (91).
The only three species (L. corymbifera, L. ramosa, and L. ornata) that have been found to cause human infections (70) produce multiple manifestations with a predominance of cutaneous followed by ROC and pulmonary infections (14). The MIC50 (CLSI) of amphotericin B, posaconazole, and isavuconazole for L. corymbifera is 0.06–16, 0.06–4, and 1–2 μg/mL, respectively (81, 82), while the ECVs of amphotericin B and posaconazole are 2 µg/mL (82).
Other genera
Rhizopus, Mucor, and Lichtheimia together account for 75% of all mucormycosis cases worldwide, but there are other genera that are important locally or by the type of manifestation that they produce (14). For example, Apophysomyces spp., especially A. variabilis, are the second most common cause of mucormycosis in India (74). Together with Saksenaea spp., they cause mainly cutaneous infections (14), while Cunninghamella spp. predominantly cause pulmonary disease with an associated mortality higher than that caused by other Mucorales (14).
EXPERIMENTAL MODELS TO STUDY MUCORMYCOSIS
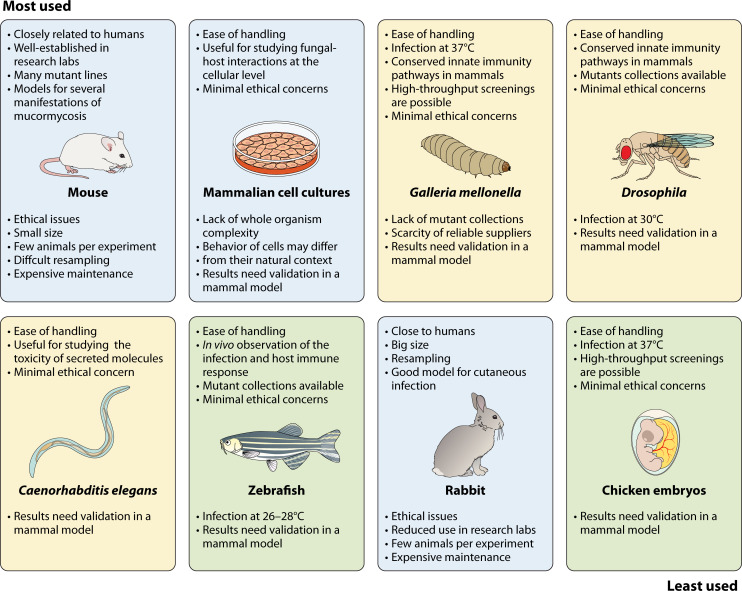
The small number of cases makes it difficult to study mucormycosis in humans and to conduct clinical trials prior to therapy approval. Therefore, the use of experimental models is even more necessary in mucormycosis than in other infections. However, the complex nature of mucormycosis, with its diverse clinical presentations and underlying predisposing factors, has led to the use of multiple animal models to comprehensively study the disease (Fig. 3). Mammalian species are generally considered the gold standard due to their anatomical, immunological, and physiological similarity to humans, and they are the preferred models for characterizing disease progression, host response, and therapeutic treatments. However, ethical, social, and logistical constraints limit the widespread use of mammalian models for large-scale screening. As a result, alternative models such as zebrafish, insects, and worms have been developed to elucidate the molecular mechanisms that govern pathogenicity and resistance to antifungal drugs, as well as to evaluate the efficacy of novel antifungal agents and therapeutic approaches. Therefore, the choice of the animal model depends on the specific research question to be addressed. In addition, some fundamental questions about the interaction between the fungus and the host can be addressed using mammalian cell cultures. It is important to note that results obtained from alternative animal models or cell cultures must be validated in at least one mammalian model. In this section, we will critically evaluate the advantages and limitations of the most commonly used animal models for the study of mucormycosis. Specific details on these models of mucormycosis can be found in comprehensive reviews (92, 93).
Fig 3.
Experimental models to study mucormycosis. The models are displayed in order of frequency of use, with the most used in the top left and the least used in the bottom right. Text above and below each image describes the advantages and limitations of each model, respectively. Mammals are shown in light blue boxes, other vertebrates in green boxes, and invertebrates in yellow boxes.
Mammalian models
Mammalian models provide a closer approximation to humans, although differences in anatomy, physiology, and immune response may affect the extrapolation of results. Several mammals (Fig. 3) have been used to study mucormycosis, ranging from typical laboratory species to more exotic species such as bank voles or Asian water buffalo calves. The most commonly used species are rabbits and mice, which have established models for pulmonary, cutaneous/subcutaneous, and disseminated mucormycosis (92, 93). These models also include procedures to chemically induce immunosuppression and DKA, mimicking the most common human predisposing conditions to infection. In mice and rabbits, two types of molecules, cytostatics and corticosteroids, are employed to induce immunosuppression. Cytostatics, particularly cyclophosphamide in mice, target replicating cells, including those in the bone marrow, while leaving resident immune cells, including tissue macrophages, relatively unaffected (94). On the other hand, corticosteroids, mainly cortisone acetate in mice, impair the antifungal function of neutrophils without reducing their number in the blood (95). Because of their different effects on the immune response, some studies combine both molecules to achieve complete immunosuppression in animals. DKA is induced by the application of streptozotocin in mice (96) and alloxan in rabbits (97), resulting in the ablation of insulin-producing beta cells in the pancreas.
The study of mucormycosis in rabbits has the advantage of their large size, which facilitates observation of symptoms and allows for repeated sampling. However, the majority of recent studies have been conducted in mice, primarily because of the smaller space requirements for rearing and maintaining mouse colonies (98). Mice are more integrated into laboratory research, making them more readily available and manageable. In addition, the ability to generate genetically modified strains, particularly with the advent of CRISPR/Cas technology, further contributes to their popularity.
Both rabbits and mice can be infected through different routes, depending on the specific research question. In humans, pulmonary mucormycosis often occurs in immunosuppressed individuals by the inhalation of sporangiospores (99). However, infecting animals with aerosols containing sporangiospores is technically challenging and has not been used in mucormycosis research. Nevertheless, experimental models of pulmonary mucormycosis have been developed by direct application of sporangiospore solution into the nostrils (intranasal) or into the trachea (intratracheal/endotracheal). Although the manner in which sporangiospores land in the airways may not exactly mimic human conditions, these experimental pulmonary models closely resemble human infection because sporangiospores are exposed to all host defense mechanisms. They must overcome the array of host defense molecules secreted by the epithelium and the mucociliary clearance, adhere to and disrupt the airway epithelium to initiate infection (100, 101). Additionally, similar to humans, infection in animals occurs only under predisposing conditions such as immunosuppression and DKA (92).
Disseminated infection, the most dangerous manifestation of mucormycosis, often originates from a specific organ mucormycosis, such as pulmonary or cutaneous mucormycosis (4), when the fungus enters the bloodstream and infects other organs. This entire process has been replicated in mice (102, 103) and rabbits (104). However, the limited number of studies and associated technical challenges have resulted in a lack of standardized models for disseminated mucormycosis initiated from focal infections (93). To address these challenges, an increasing number of studies have employed intravenous infection with sporangiospores. This method offers advantages such as ease of implementation, high reproducibility, and the ability to induce a fatal disease even in animals lacking predisposing factors (105). Consequently, it has been used extensively to investigate the pathogenesis and efficacy of therapeutic approaches, especially antifungal drugs. However, it faces the problem of important differences compared to naturally disseminated infection in humans. The method bypasses the establishment of the initial infections in the primary organ, and instead, sporangiospores rather than hyphal fragments travel in the bloodstream and provoke the disseminated infection (106). These differences must be considered when analyzing the pathophysiology, the role of fungal factors, and the virulence among species and strains.
Models of cutaneous mucormycosis have also been established by subcutaneous inoculation of sporangiospores in diabetic rabbits and rats (107, 108), and in neutropenic mice treated with cyclophosphamide and cortisone acetate to impair tissue macrophage clearance of sporangiospores (109). However, these models have received limited attention and require further research to be well established. The situation is even more challenging for other forms of mucormycosis, such as ROC and gastrointestinal infections, because reliable animal models for studying these manifestations are currently lacking (92, 93).
Alternative animal models
The need for alternatives to mammalian models arises from the requirement for large numbers of animals for certain studies, which conflicts with ethical considerations and the high costs associated with the use of mammalian models. The continued completion of Mucorales genomes (79), along with significant progress in establishing protocols for manipulating recalcitrant species (110), is expected to catalyze a surge in functional genomics, which would require high-throughput screening strategies capable of determining the role of individual genes in virulence. Although alternative models can be utilized to study many mechanisms underlying human fungal infections, their use is also limited by the evolutionary distance between these models and mammals (Fig. 3). Therefore, discoveries made in these models need to be validated in mammalian infection models.
Two major groups of models have been developed (Fig. 3): one based on vertebrates and the other on invertebrates. In the vertebrate models, zebrafish and chicken embryos have been utilized, although their use has been limited to a few studies. Models of mucormycosis have been developed in both larval and adult zebrafish (111, 112), but the larval model is particularly interesting because its transparency allows for the observation of the infection and the corresponding host immune response (111). Although zebrafish models have contributed to our understanding of the disease, they face the problem that the fish are maintained at 26°C–28°C, lacking the temperature protection that humans have against infection. On the other hand, chicken embryos can be easily maintained at 37°C and have been used for high-throughput screening to evaluate the virulence of mucormycosis-causing fungi (113). However, like zebrafish models, chicken embryo models have not been extensively studied in the context of mucormycosis (93).
Invertebrates are increasingly being used as an alternative model for mucormycosis due to the low ethical concerns and the ability to study large numbers of individuals at a reduced cost. Species used to study mucormycosis include Drosophila melanogaster, Caenorhabditis elegans, and Galleria mellonella (Fig. 3). C. elegans is a recent addition that has been used to identify secreted molecules that may play a role in the virulence of Mucorales. The lethality observed in this nematode correlates well with the virulence observed in DKA mice infected intraperitoneally (114–116). D. melanogaster possesses many defense mechanisms found in mammals, including signaling pathways that are critical regulators of immune responses and the ability to fight against many microbes primarily through the production of conserved antimicrobial peptides and phagocytosis by plasmatocytes, which are comparable to mammalian macrophages (117). Its genetics are well understood, with large mutant collections available, and it is easy to grow, manipulate, and analyze in large numbers (118). These advantages have led to the establishment of a model to dissect the immunopathogenesis of mucormycosis (119). The use of this model has provided insights into the factors that mediate host-pathogen interactions, factors that modify fungal virulence, virulence determinants, and the efficacy of novel antifungal strategies (93, 120, 121).
One of the major limitations of Drosophila is that infection experiments are conducted at temperatures below 30°C (122). In contrast, infection experiments with G. mellonella larvae can be performed at 37°C, although their maintenance temperature is lower, making them easier to handle (123, 124). In addition, the larvae are inexpensive and readily available as they are commonly traded as fish bait or food for exotic animals. Their large size compared to other invertebrate models facilitates the infection process, which is performed by injection into the hind prolegs with a syringe (123). Furthermore, similar to Drosophila, G. mellonella larvae possess several innate immune pathways that are conserved in mammals (125). Therefore, this model has been adapted for the study of Mucorales, especially for the analysis of virulence and the evaluation of antifungal treatments (124). Interestingly, studies comparing the virulence of different strains of the same species, which is necessary when analyzing the gene function through the generation of knockout mutants, have revealed a strong correlation between virulence in G. mellonella and in mice (121, 126). However, one of the major limitations of the G. mellonella model is the lack of mutant collections and the scarcity of reliable companies that provide larvae under optimal conditions (127).
Mammalian cell cultures
The cultures of tumor or primary mammalian cells from different epithelia and phagocytes have been used to address specific questions regarding the interaction between the fungus and mammalian cells (Fig. 3). Their use has led to significant advances in the understanding of infection, including the identification of fungal invasins (76, 128) and toxins (129), as well as host receptors bound by invasins (76, 130), and the analysis of fungal invasion and cytotoxicity on epithelial cells. In addition, they have been instrumental in characterizing the transcriptomic responses of both the fungus and the host during these interactions (77, 130, 131). The most commonly used mammalian cells include the A549 tumor cell line derived from type II alveolar cells (76, 77, 129, 132), primary alveolar epithelial cells (76, 129), tumor nasal epithelial cells (RPMI 2650) (76), human umbilical vein endothelial cells (HUVECs) (77, 129, 133–135), bone marrow-derived macrophages (BMDMs) (130), primary polymorphonuclear neutrophils (130), and the murine macrophage cell line J774A.1 (131, 136).
The use of mammalian cell cultures has some disadvantages due to the lack of complexity compared to a whole organism, where multiple immune cells and signaling pathways are involved in the response to the fungus. In addition, the behavior of cells in culture may not fully represent the characteristics of the same cells within their natural tissue environment. Therefore, validation in a mammalian model is strictly necessary.
HOST RESPONSE TO MUCORALES
The innate immune system plays a predominant role in the defense against fungal infections, including mucormycosis, because constant contact with spores in daily life does not lead to infection in individuals without predisposing factors or epithelial damage. Consequently, fungal infections often exhibit opportunistic behavior (68). Epithelia play an essential role as physical barriers, secreting antimicrobial peptides and recruiting innate immune cells through the production of cytokines and chemokines (100, 101). Host responses have been extensively studied in the fungi that cause the most common infections, such as Candida albicans (137) and Aspergillus fumigatus (138). Many of the host defense mechanisms against these species are expected to be common to Mucorales infections, although some differences may exist due to differences in cell wall composition (139–141).
During the development of mucormycosis, sporangiospores come into contact with different types of epithelia depending on the manifestation of the mucormycosis (Fig. 4). However, the response of the epithelium is largely unknown, with the response of the airway epithelium being the most studied. In healthy individuals, pulmonary epithelial cells play a central role in antimicrobial immunity by clearing spores from the airways via the mucociliary escalator and by producing antimicrobial peptides and inflammatory mediators that direct early decisions about innate and adaptive responses. However, little is known about how these cells promote antifungal immunity (100, 101). In vitro studies with Mucorales have shown that sporangiospores are endocytosed by the epithelial cell lines A549 and RPMI 2650 (76, 132). Similar studies with Aspergillus conidia showed that A549 efficiently kills the phagocytized conidia, but this has not been proved in Mucorales (142). Surprisingly, internalization of sporangiospores by respiratory epithelial cells was not observed in mice infected with L. corymbifera, contradicting the in vitro results with cell lines (143). Unfortunately, equivalent experiments with other Mucorales species have not been carried out to confirm the in vivo results with L. corymbifera. Therefore, further in vivo analyses are needed to understand the role of epithelial cells in the response to Mucorales sporangiospores.
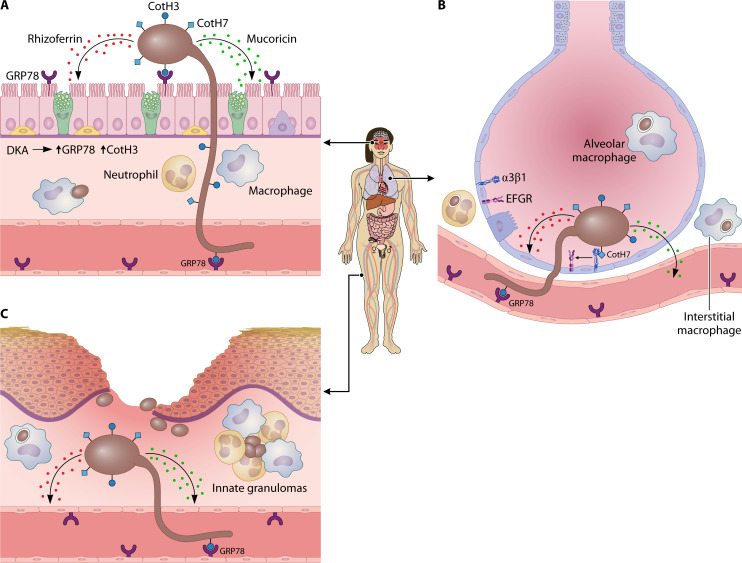
Fig 4.
Fungal invasion and cellular response of the host. The most significant routes of infection are illustrated: rhino-orbital, pulmonary, and skin breaches. (A) Rhino-orbital infections primarily occur in individuals with DKA, likely because associated features induce the expression of both GRP78 and Rhizopus CotH3. Rhizopus CotH3 binds to GRP78, facilitating invasion of both respiratory epithelium and vascular tissue, respectively. Attachment to epithelial cells is possible only for germlings (enlarged for better visualization), but the entry of sporangiospores (brown spheres) after breaching epithelial integrity could also be a potential route. Epithelial damage is mediated by the secretion of mucoricin. Other secreted molecules, such as rhizoferrin, may also contribute to tissue damage. Hyphae cannot be phagocytosed by phagocytes, but they are sensitive to neutrophil cationic peptides. (B) Pulmonary infections are frequently associated with a deficient immune system. A similar host response to nasal infection occurs, except for the presence of alveolar macrophages, which play a primary role in sporangiospore phagocytosis. Despite a significant influx of neutrophils, inflammatory monocytes, and dendritic cells in the lungs of immunocompetent mice, most sporangiospores are phagocytosed by alveolar macrophages, with only a few by interstitial macrophages and neutrophils. Failure in the immune response reduces this reaction, potentially resulting in fungal invasion, mediated by fungal CotH7, which interacts with integrin β1 on alveolar epithelial cells, leading to the activation of epidermal growth factor receptor (EGFR). (C) Skin damage due to trauma or burns can result in mucormycosis in immunocompetent individuals. Breaches in the skin epithelium facilitate the entry of sporangiospores, which readily adhere to laminin or type IV collagen of the basement membrane. This entry triggers a robust immune response, recruiting various types of immune cells that either phagocytose the sporangiospores or restrict their dispersion and germination by forming early innate granulomas, in which the sporangiospores keep alive for long time. Similar structures have been observed in other types of mucormycosis in animal models and humans. Eventually, the fungus may reach the vascular system and disseminate.
Similar to other fungal infections, phagocytic cells, including alveolar macrophages, in the respiratory tract play an important role in protection against pulmonary mucormycosis (Fig. 4). Experimental pulmonary infection of immunocompetent mice with sporangiospores of R. arrhizus and R. delemar revealed that clearance was slower than with A. fumigatus conidia, and a significant proportion of the Rhizopus sporangiospores remained viable in the lung for up to 10 days after infection (130). In these experiments, the Rhizopus sporangiospores were predominantly phagocytosed by alveolar macrophages, but a previous experiment showed that Lichtheimia sporangiospores were also phagocytosed in the bronchi by mononuclear phagocytes, in both cases resulting in the inhibition of sporangiospore swelling (130, 143). Furthermore, in the case of Rhizopus, the absence of swelling inside the alveolar macrophages is likely due to iron starvation (Fig. 5A), as has been observed in BMDMs (130). Therefore, this result links the role of iron in preventing germination with classical predisposing conditions that lead to an increase in free iron such as ketoacidosis and the use of deferoxamine in dialysis (144, 145). Similarly, predisposing factors such as hyperglycemia, acidosis, and steroids are thought to promote mucormycosis by impairing the phagocytic function of alveolar macrophages (146–148). Another mechanism of inhibition of Rhizopus sporangiospore germination has been described in in vitro cultures of rat alveolar macrophages, where the generation of nitric oxide (NO) blocked germination (149). NO serves as a central mediator in macrophage effector responses due to its potent antimicrobial properties and its role in signaling through various immune pathways, showing antimicrobial activity against a wide variety of fungi (150–152). However, the role of NO in mucormycosis requires further investigation because the blockage is not observed in a human macrophage line (149), and several Mucorales spp. inhibit NO production in vitro (153).
Fig 5.
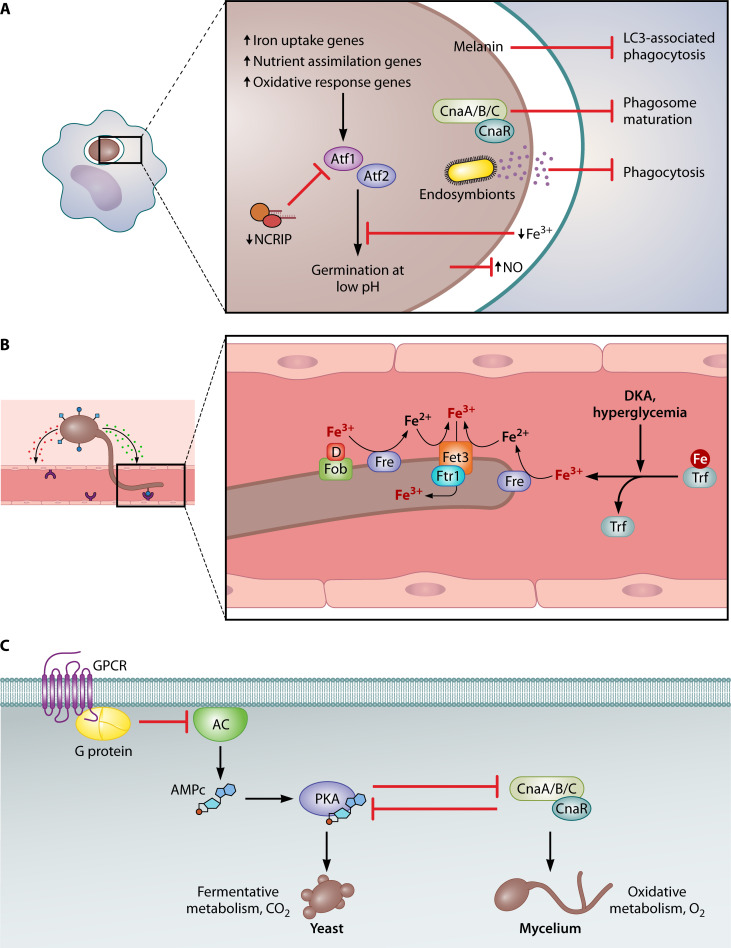
Mechanisms involved in the fungal colonization of the host. (A) Phagocytosis represents one of the primary host responses to prevent fungal infection. Mucorales species have evolved mechanisms to hinder phagosome maturation, with melanin- and calcineurin-dependent mechanisms observed in Rhizopus and Mucor, respectively. They also employ strategies to evade phagocytosis, such as the endosymbiont-mediated secretion of antiphagocytic factors in certain R. microsporus isolates. Furthermore, phagocytosis triggers a significant remodeling of fungal gene expression in response to reactive oxygen species and nutritional immunity, including iron deficiency, which inhibits spore germination. In Mucor, the non-canonical RNAi pathway (NCRIP) plays a significant role in this transcriptomic response. This includes the involvement of transcription factors Atf1 and Atf2, which are essential for germination under acidic conditions. (B) Iron plays a crucial role in Mucorales infectivity, as an increase in free iron resulting from conditions like diabetic ketoacidosis, hyperglycemia, and other forms of acidosis can enhance their virulence. These fungi employ a high-affinity reductive system for iron uptake, comprising a ferric reductase (Fre), multicopper ferroxidase (Fet3), and high-affinity iron permease (Ftr1). Additionally, at least Rhizopus can acquire iron bound to xenosiderophores, such as deferoxamine (D), utilizing the Fob1 and Fob2 proteins. Trf refers to transferrin. (C) Mucor and other Mucorales species exhibit growth dimorphism. In Mucor, mycelial growth is associated with virulence. The transition from yeast to hyphae is regulated by oxygen levels, CO2 levels, and the carbon source. The calcineurin and PKA pathways play opposing roles in dimorphism. G-proteins repress yeast growth under low oxygen levels by inhibiting adenylate cyclase (AC).
In addition to macrophages, neutrophils are also important in preventing infection, as neutropenia is a major risk factor for mucormycosis (154, 155). Experimental pulmonary infection with Rhizopus sporangiospores results in a significant influx of neutrophils and inflammatory monocytes into the lung (Fig. 4) (130, 148). Neutrophil chemotaxis is induced by the initiation of germination (155). These neutrophils phagocytose few sporangiospores, but they surround and confine them to the point of the infection. These structures look like the initiation of cell clusters of phagocytes around the sporangiospores observed in the zebrafish model and in rabbits, resembling early innate granulomas (Fig. 4C) (107, 111, 156). Similar structures have been observed in human disseminated mucormycosis (130). Interestingly, sporangiospores in these clusters remain alive for a long time, at least 15 days in rabbits, and immunosuppression or alloxan-induced diabetes results in germination and fungal growth (107, 156). In contrast to the high resistance of sporangiospores to neutrophil killing in vitro, swollen sporangiospores and hyphae are sensitive to neutrophil cationic peptides (157). Also, in vitro studies suggest that interferon-γ and granulocyte-macrophage colony-stimulating factor increase the activity of polymorphonuclear leukocytes against Mucorales (158). Indeed, sporangiospores in rabbit granulomas are eventually killed, indicating the ability of neutrophils and probably other innate immune cells to kill the fungus (107).
The role of adaptive immunity in mucormycosis has received less attention than innate immunity, but it likely plays a role because lymphopenia is associated with rapid disease progression and mortality in pulmonary mucormycosis and mucormycosis in patients with hematological malignancies (159, 160). Dendritic cells recognize β-glucan exposed after Rhizopus germination through the C-type lectin receptor dectin-1, resulting in the production of high levels of interleukin 23 (IL-23), which drives T helper 17 cells (TH-17) responses (161). Interestingly, mice deficient in caspase recruitment domain-containing protein 9 (Card9), an essential adaptor molecule downstream of C-type lectin receptors, show increased susceptibility to cutaneous mucormycosis caused by Mucor irregularis, which is associated with impairments in TH cells, including TH-17, and neutrophil responses (162). In addition, IL-17 receptor genetically deficient mice have lower survival rates than wild-type strains following intravenous infection with Rhizopus sporangiospores, supporting a role for IL-17 signaling in the response to infection (163). Similarly, experimental pulmonary mucormycosis induced in IFN-γ-deficient mice has revealed that IFN-γ signaling is not required for survival but improves fungal clearance (164). It is expected that increased use of mutants in key genes of the mammalian immune response will contribute to the understanding of the host response to mucormycosis.
MECHANISMS INVOLVED IN THE FUNGAL COLONIZATION OF THE HOST
Response to macrophages
Mucorales must adapt their metabolism and physiology in order to grow and obtain nutrients in the host environment while at the same time repelling host defenses. As mentioned earlier, in the case of pulmonary mucormycosis, the first line of defense will be the phagocytes in the respiratory tract. However, mucoralean sporangiospores can survive phagocytosis (130, 135, 165, 166). Two mechanisms have been described that prevent sporangiospore killing after phagocytosis by arresting phagosome maturation (Fig. 5A). In Rhizopus sporangiospores, melanin induces the phagosome maturation arrest by targeting LC3-associated phagocytosis, an important antifungal pathway that regulates early events in A. fumigatus phagosome biogenesis (130). In Mucor sporangiospores, the blockage of phagosome maturation in a murine macrophage cell line and BMDMs depends on the calcineurin signaling pathway (135, 166). Calcineurin is a Ca2+- and calmodulin-dependent serine-threonine phosphatase composed of a catalytic subunit with phosphatase activity and a regulatory subunit that interacts with Ca2+-calmodulin. Calcineurin acts by dephosphorylating transcription factors and promoting their nuclear localization, resulting in the regulation of specific transcriptional responses (167). In the case of M. lusitanicus, there are three catalytic calcineurin subunits (designated CnaA, CnaB, and CnaC) and one regulatory subunit, CnaR (168). Mutants with a deletion of cnaR grow in a yeast-like form and are unable to inhibit phagosome maturation in in vitro cultures of murine macrophages. This suggests that the ability to grow as mycelium is necessary to arrest phagosome maturation (166). However, the isolation of mutants in bycA (bypass of calcineurin) that suppress the effect of cnaR deletion on morphology, but not on phagosome maturation, suggests that the inhibition of phagosome maturation is independent of morphology and relies on an as yet uncharacterized mechanism (135).
In addition to blocking phagosome maturation, Mucor sporangiospores are able to germinate inside the phagosome and kill macrophages in vitro (165, 166) and induce apoptotic cell death in macrophages, but not neutrophils, in an adult zebrafish model of infection (112). The ability of some Mucor strains to germinate inside the macrophage depends on their size. M. lusitanicus shows sporangiospore size dimorphism, with some strains producing large multinucleate and others small mononucleate sporangiospores. Only large sporangiospores are able to germinate inside the macrophages and are virulent in G. mellonella (165). The size of the sporangiospores is controlled by the calcineurin pathway and G protein signaling pathways. Deletion mutants of either cnaA or the gpa11 and gpa12, which encode heterotrimeric G-alpha subunits, result in larger sporangiospores (168, 169). These two regulatory mechanisms appear to be linked because Gpa11 and Gpa12 activate cnaA expression at the mRNA level, but the relationship is not fully understood because mutants for gpa11 and gpa12 are avirulent, whereas cnaA mutants are more virulent than the wild-type strain in DKA mice infected intraperitoneally (169). The presence of these mechanisms in other Mucorales species causing mucormycosis has not been investigated.
A third mechanism that may contribute to the inability of macrophages to kill Mucorales sporangiospores is based on the suppression of NO production (153). The ability to block NO production is widespread in Mucorales in vitro in murine macrophage cell lines, and it is not associated with a reduction in mRNA or protein levels of the macrophage nitric oxide synthase gene. In R. delemar, suppressive activity requires fungal to operate by two different uncharacterized mechanisms: one that removes NO from the media and another that requires direct contact with macrophages (153).
The transcriptomic response of both Rhizopus and Mucor to phagocytosis has been studied in vitro to gain insight into the fungal response to phagocytosis. The study of the interaction of R. delemar with BMDMs revealed that genes involved in iron acquisition were upregulated after phagocytosis (Fig. 5A) (130). The transcriptomic response of Mucor sporangiospores to phagocytosis by a murine cell line of macrophages uncovered a profound remodeling of the gene expression with more than 9% of the genes changing the expression. The enrichment of genes involved in nutrient assimilation and metabolism suggests that the fungus alters its metabolic pathways to utilize alternative nutrient sources for germination inside the phagosome. Additionally, genes associated with the response to oxidative stress were also enriched, possibly explaining the survival of sporangiospores against macrophage attack (131). Deletion of highly induced fungal genes in response to the interaction revealed that the transcription factors Atf1, Atf2, and Gcn4, the extracellular proteins Chi1 and Pps1, and the aquaporin Aqp1 play key roles in germination within the phagosome and virulence in immunosuppressed mice infected intravenously. Further transcriptomic analysis of the mutants in atf1 and atf2 (Fig. 5A; Table 1) suggests the existence of an Atf-mediated pathway that responds to oxidative stress, macronutrient metabolism, and germination at low pH levels (131).
TABLE 1.
Genes proved to be involved in pathogenesis in Mucorales
| Gene | Product | Role in virulence | Species in which function has been proved | References |
|---|---|---|---|---|
| cotH3a | Extracellular protein CotH3 | Host cell invasion of nasal and endothelial epithelia | R. delemar | (76, 128) |
| cotH7 | Extracellular protein CotH7 | Host cell invasion of lung epithelium | R. delemar | (76) |
| cotH3a, cotH4 | Extracellular protein CotH3, CotH4 | Unknown | M. lusitanicus | (121) |
| RO3G_06568 | Mucoricin | Host cell damage | R. delemar | (129) |
| ftr1 | High-affinity iron permease | High-affinity reductive system for iron uptake | R. delemar | (170) |
| fet3a, fet3b, fet3c | Ferroxidases | High-affinity reductive system for iron uptake | M. lusitanicus | (171) |
| fob1, fob2 | Ferrioxamine receptors | Siderophore uptake | R. delemar | (172) |
| cnbR | Calcineurin regulatory B subunit | Dimorphism, phagosome maturation arrest | M. lusitanicus | (166, 168) |
| cnaA | Calcineurin catalytic A subunit | Dimorphism, spore size | M. lusitanicus | (168, 169) |
| atf1, atf2 | Regulatory factors | Response to macrophage phagocytosis | M. lusitanicus | (131) |
| r3b2 | Atypical RNase III of the non-canonical RNAi pathway | Response to macrophage phagocytosis | M. lusitanicus | (173) |
| pkaR1 | Regulatory subunit of cAMP-dependent protein kinase A | Dimorphism | M. lusitanicus | (174) |
| rfs | Rhizoferrin | Iron uptake? | M. lusitanicus | (114) |
| gpa11, gpa12 | G-protein alpha subunits | Sporangiospore size | M. lusitanicus | (169) |
| Gpb1 | Heterotrimeric G-protein beta subunits | Dimorphism | M. lusitanicus | (174) |
| mcplD | Phospholipase D | Signaling? | M. lusitanicus | (126) |
| mcmyo5 | Myosin V | Intracellular transport, dimorphism | M. lusitanicus | (126) |
| wex1 | Exonuclease | Unknown | M. lusitanicus | (175) |
| mcwc-1a | White-collar 1 regulatory protein | Unknown | M. lusitanicus | (176) |
CotH3 proteins from M. lusitanicus and R. delemar belong to different CotH protein clades.
Gene expression is regulated by RNA interference (RNAi) pathways, and the response to phagocytosis and virulence is not an exception (173). One such RNAi pathway, the non-canonical RNAi pathway (NCRIP), represses the expression of most phagocytosis-responsive genes, including atf1 and atf2, under saprophytic conditions, and this repression is released by phagocytosis (Fig. 5A). Therefore, mutations in r3b2 (atypical RNaseIII) or rdrp-1 (RNA-dependent RNA polymerase-1) (Table 1), key genes of this pathway, induce a strong misregulation of genes responding to phagocytosis in vitro, including oxidative stress response genes, and reduced virulence in disseminated infection of immunosuppressed mice (173).
Mechanisms of cell adhesion, invasion, and tissue damage
As mentioned above, epithelial cells constitute a defensive barrier against external threats. During the invasion process, Rhizopus sporangiospores attach to epithelial cells via CotH proteins. These proteins are distinct kinases that constitute structural components of the spore coat of some organisms (177, 178). CotH proteins were originally discovered as a structural component of the dormant spores of Bacillus subtilis, where they are required for normal germination (178, 179). Widely distributed in bacteria, CotH proteins are also found in green algae, some protists, and EDF (78, 177). The R. delemar genome encodes eight CotH proteins, three of which act as invasins in interactions with different epithelial cells (76, 77, 128). Thus, the CotH3 interacts with the glucose-regulated protein 78 (GRP78) to invade and damage the nasal epithelial cells, whereas CotH7 interacts with the integrin α3β1 of the alveolar epithelial cells, leading to the activation of the epidermal growth factor receptor (EGFR) and subsequent host cell invasion (Fig. 4) (76). GRP78, also known as BiP or HSPA5, plays a crucial role as a major chaperone in the endoplasmic reticulum (ER), contributing significantly to the folding and maturation of nascent polypeptides within the ER compartment (180). Notably, the expression of both GRP78 on nasal epithelial cells and CotH3 in R. delemar is induced by high glucose, iron, and ketone body levels, which are hallmarks of DKA, the major predisposing factor for ROC mucormycosis (76). The overexpression of GRP78 may result in cell surface localization, likely due to the saturation of the retrieval system that maintains the protein within the ER (181). Consistent with the role of these proteins in mucormycosis, knockdown GRP78 or CotH3 function, either by shRNA or by specific antibody inhibition, reduces invasion and epithelial damage and protects DKA mice from mucormycosis (128, 133, 182). Remarkably, inhibition of GRP78 does not affect the infective process of Candida or Aspergillus, suggesting a different and specific mechanism for host cell invasion by Mucorales (133). Similarly, an antibody against the integrin β1 inhibits invasion of alveolar epithelial cells by R. delemar and protects immunosuppressed mice from pulmonary mucormycosis (76).
A specific feature of mucormycosis is the presence of extensive angioinvasion via penetration and damage of the endothelial cells lining of the blood vessels (5). Therefore, the interaction of the fungus with endothelial cells is an important step in mucormycosis, and the resulting vascular thrombosis and necrosis can impede the arrival of defense cells at the point of infection and the proper delivery of antifungal agents. For this invasion process to occur, the fungus must come into contact with endothelial cells, which are separated from endothelial cells by basement membranes containing laminin and collagen IV (Fig. 4C) (183). Tissue damage induced by either external forces or wounds resulting from diabetes or chemotherapy exposes these extracellular proteins, which serve as adhesion points for fungal spores but not hyphae (184, 185). Some fungal pathogens, such as Candida, use surface proteins to invade host cells through the interaction of agglutinin-like sequences (Als), which are glycoproteins that interact with cadherins (186, 187). Similarly, Aspergillus depends on the thaumatin-like protein CalA to interact with laminin and to invade lung epithelial and endothelial cells through the α5β1 integrin (188, 189). In the case of mucormycosis, early studies revealed that R. delemar germlings, but not sporangiospores, use the highly similar CotH2 and CotH3 to interact with GRP78, allowing fungal adhesion and invasion of endothelial cells in vitro (133). Transcriptomic analysis of endothelial cells interacting with Rhizopus or Mucor showed that the platelet-derived growth factor receptor B (PDGFRB) is upregulated and that its inhibition reduces the cell damage caused by Mucorales (77). However, the implications of PDGFRB as a receptor or coreceptor with the abovementioned proteins remain to be further characterized.
The number of CotH genes varies among the different lineages of Mucoromycotina, Mortierellomycotina, and Neocallimastigomycota lineages (77, 78). In M. lusitanicus, 17 genes encoding CotH proteins have been identified (121). Five of these CotH-encoding genes were disrupted using CRISPR/Cas9, and it was found that CotH3 and CotH4 were found to be important for sporangiospore wall formation and composition, with mutants in CotH3 or CotH4 (Table 1) exhibiting reduced virulence in insects (D. melanogaster and G. mellonella) and DKA mice (121). The importance of CotH proteins in the infection process and their almost specific distribution in mucormycosis-causing fungi in the eukaryotic kingdom motivated the development and testing of polyclonal antibodies raised against peptides of CotH3, which protected DKA and neutropenic mice from mucormycosis (182).
Mucorales adhesion and subsequent cell invasion are associated with extensive host cell damage and necrosis (5). In vitro damage to A549 cells and HUVECs by multiple Mucorales species supports this is as a characteristic feature of mucormycosis-causing Mucorales. This common pattern of infection led to the discovery of the mucormycosis-associated toxin known as mucoricin (129). This hyphal-secreted/shed toxin shares structural and functional features with the plant toxin ricin and induces necrosis and apoptosis and alters permeability in mammalian cells in vitro, as well as inhibits protein synthesis (129). Damage to host cells likely explains the symptoms observed in toxin-inoculated mice, which show enhanced angioinvasion, inflammation, and tissue destruction. Interestingly, anti-mucoricin antibodies protect mice from the inflammation and tissue damage associated with mucormycosis (129).
Mucoricin is not the only protein secreted by Mucorales to facilitate the infectious process (Fig. 4). Secreted R. delemar peptides have been screened, and three peptides (CesT, Colicin, and a Ca2+/calmodulin-dependent protein kinase/ligand) with immunomodulatory activity have been detected (190). Additionally, some Mucoromycota species establish symbiotic interactions with bacteria (191, 192). One of the best characterized is the mutualistic interaction between R. microsporus and Mycetohabitans rhizoxinica (193–196). Rhizoxin, a toxin produced by the bacteria-colonizing R. microsporus, inhibits plant defenses and allows both organisms to infect the plant, causing the rice seedling disease (196–198). However, no differences in Rhizopus virulence have been found between bacteria-containing isolates and endosymbiont-free strains (199, 200). Nevertheless, a recent clinical isolate of R. microsporus requires its bacterial endosymbiont, Ralstonia picketii, for virulence, antiphagocytic activity, and inhibition of phagosomal maturation (201). Sporangiospores harboring bacteria were more resistant to clearance by macrophages, and in vivo experiments validated that Ralstonia was crucial for immune system evasion and the progress of the infection (Fig. 5A) (201). Importantly, the nature of the substance(s) secreted during this symbiosis remains to be further characterized. The identification of this isolate suggests that other clinical isolates may harbor bacteria involved in virulence, although a large number is not expected as only few cases have been described.
Iron uptake
Iron is an essential element for almost all living organisms, including human pathogens. An important component of the innate immune system is nutritional immunity, a group of sophisticated strategies that limit the access to trace minerals, including iron (202, 203). In healthy individuals, the amount of iron in the bloodstream is low and bound to iron-binding proteins such as transferrin, ferritin, or lactoferrin. However, certain pathologies can elevate serum-free iron levels, making individuals susceptible to infections, especially Mucorales (204). Hyperglycemia, DKA, and other forms of acidosis impair the ability of transferrin to chelate iron, making it available for pathogen growth (Fig. 5B). Serum iron can also increase in cases of patients undergoing dialysis or multiple transfusions (21, 144).
Fungi have developed strategies to obtain iron in conditions where it is limited, and those that are pathogens must acquire it from their hosts either from transferrin or from the most abundant source of iron, which is the heme group of hemoglobin. Fungi have three basic mechanisms for iron uptake: a reductive mechanism that can be of high or low affinity, the uptake of iron bound to siderophores, and the uptake of iron from the heme group (205). In Mucorales, a high-affinity reductive mechanism and the use of siderophores have been implicated in iron acquisition during the infection (170, 171, 206).
Reductive mechanisms
This mechanism consists of three main steps: (i) the extracellular reduction of ferric to ferrous iron performed by a high-affinity NADPH-dependent ferric reductase encoded by the fre genes, (ii) the reoxidation back to ferric iron by an O2-dependent multicopper ferroxidase encoded by the fet3 genes, and (iii) the transport of this ferric iron into the cell interior by a high-affinity iron permease encoded by the ftr1 genes (Fig. 5B) (207, 208). Low iron availability induces the expression of this system in R. delemar (170), M. lusitanicus (171), and L. corymbifera (209). RNAi silencing of R. delemar ftr1 and deletion of the three M. lusitanicus fet3 genes (Table 1) have revealed that this system is required for iron uptake and virulence in DKA and immunosuppressed mice, respectively (170, 171). Three ferroxidase genes, fet3a, fet3b, and fet3c, have been identified in M. lusitanicus, and fet3c is the key virulence factor, although there is partial redundancy with the other two paralogs. An interesting observation is that the expression of these paralogous genes depends on the dimorphic state. The fet3a gene is expressed in the yeast form of the fungus, while the other two are specifically expressed in the mycelium. Thus, the high-affinity iron uptake mechanism is linked to dimorphism, linking these two processes involved in the virulence of the fungus (171).
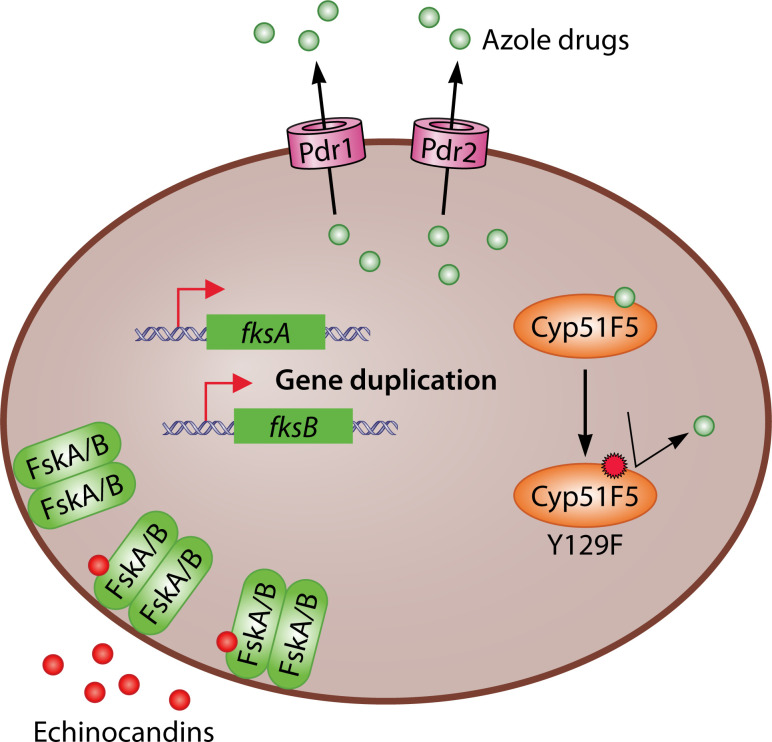
Siderophores
Siderophores are small organic molecules secreted by bacteria and fungi that exhibit exceptionally high affinities and specificities for binding extracellular ferric iron. Some Mucorales produce rhizoferrin, a siderophore belonging to the polycarboxylate family that is synthesized by a non-ribosomal peptide synthetase encoded by the rfs gene (Table 1) (114, 206, 210–212). The expression of this gene is induced by low iron concentrations, macrophage interaction, and infection of mice (114). While data from R. microscopus indicate that this siderophore is inefficient at chelating iron from serum (213), the deletion of this gene in M. lusitanicus reduces virulence in intraperitoneally infected DKA mice, suggesting a role for rhizoferrin in virulence (114). In addition, the ability of Mucorales to uptake xenosiderophores, siderophores produced by other microorganisms, such as bacterial deferoxamine used in hemodialysis patients to reduce iron overload toxicity, has long been recognized. The iron-rich form of deferoxamine (ferrioxamine) induces the expression of Rhizopus fob1 and fob2, which encode two surface receptors that bind this siderophore and allow iron uptake by the high-affinity reductive mechanism (Fig. 5B). Homologs for these two genes have been identified in all genomes of the analyzed Mucorales species (172).
Growth dimorphism in Mucorales species
Some mucormycosis-causing species of Mucorales can grow as yeast or mycelium depending on the environmental conditions. The study of dimorphism has provided a remarkable understanding of the molecular mechanisms and the spatiotemporal evolution of mucormycosis. Initially thought to be the result of species transmutation from Saccharomyces cerevisiae (214), Mucor dimorphism was first described in the 19th century by the research of Louis Pasteur, who not only noted the dimorphic growth of M. racemosus but also the importance of O2 concentration as a determinant of yeast or hyphal growth (215). Although there are some variations among Mucor species (216), the presence of fermentable hexoses and the absence of oxygen have been proposed as the two main determinants for yeast growth induction, whereas aerobic growth is linked to hyphal growth (216, 217). Related to the previously mentioned factors, CO2 concentration has also been described as an important factor in the induction of yeast growth (Fig. 5C) (218–220). Chemical inhibitors of mitochondrial function (such as β-phenylethyl alcohol, antimycin A, potassium cyanide, oligomycin) and the inhibition of the synthesis of essential mitochondrial components by chloramphenicol result in yeast growth (221–223). Taken together, these observations highlight the importance of the oxidative/fermentative metabolism in the regulation of dimorphism. In addition, nitrogen metabolism has also been implicated in the dimorphic transition. The enzyme ornithine decarboxylase (ODC), which catalyzes the conversion of ornithine to putrescine, is active during the air-induced yeast-to-hyphae transition in several Mucor species. In addition, inhibition of ODC by diamino butanone blocks this transition (224–227). In combination with all of the above factors, high levels of cyclic AMP (cAMP) promote yeast growth, and it has also been reported to participate in the inhibition of ODC synthesis in M. racemosus (216, 228).
In terms of its relationship to pathogenesis, both yeast and hyphal forms and the dynamic transition between them have been reported to be important in fungal infective processes. One of the best studied systems regarding the relevance of dimorphism is the primary human pathogenic fungus, Candida. In this fungus, both morphological forms are important for virulence, as each form is involved in different stages of the infective process (229). Yeast growth is critical for bloodstream dissemination, whereas hyphal growth is important for cell adhesion and penetration of epithelial barriers (230–233). In addition, the dimorphic transition is critical in other fungal pathogens such as Penicillium marneffei, Blastomyces dermatitidis, Coccidioides spp., Histoplasma capsulatum, Sporothrix schenckii, and Paracoccidioides spp., where the transition from hyphae to yeast enables host colonization (234). In Mucorales, most of the studies focused on the pathogenic relevance of dimorphism have been performed in M. lusitanicus due to the availability of genetic and molecular tools (10) that have allowed the dissection of several mechanisms underlying pathogenesis-related dimorphism. Detailed analyses of the calcineurin signaling pathway have revealed its importance in the regulation of dimorphism in M. lusitanicus (Fig. 5C) and its potential as an antifungal drug target. Knockout mutation of the regulatory subunit cnaR results in a yeast-locked phenotype with reduced virulence in G. mellonella (168). Conversely, gain-of-function mutations in either cnaA or cnaR result in hyphal phenotype (168). Remarkably, these mutants revealed the different host responses to the yeast or hyphal forms. Only when growing as hyphae, M. lusitanicus induces the production of proinflammatory cytokines and the production of the proangiogenic growth factor FGF-2. In contrast, the yeast form of M. lusitanicus undergoes phagosome maturation, whereas sporangiospores and germinated sporangiospores are better able to evade the host immune system (166).
In addition to its role in ODC regulation, cAMP is a secondary messenger that activates the protein kinase A (PKA) signaling pathway to promote yeast growth (217, 235). Interestingly, calcineurin and the PKA pathway are interlocked in the control of dimorphisms, as its activity is increased when yeast growth is forced by deletion of cnaR (Table 1) or treatment with the calcineurin inhibitor FK506 (168). PKA is a heterotetrameric holoenzyme composed of two regulatory subunits (PKAR) and two catalytic subunits (PKAC). Activation of G protein-coupled receptors leads to the GDP-GTP exchange on the G proteins, which can regulate the activity of the adenylate cyclase and control cAMP levels in the cell (Fig. 5C). cAMP is bound by the PKAR subunits, which provoke the release and activation of the PKAC subunits. Three of the four PKAR subunit genes encoded by the M. lusitanicus genome are directly involved in dimorphism. A gain-of-function mutation of the pkaR1 gene (Table 1) favors hyphal growth, and its deletion causes defects in the yeast-to-hyphal transition. Knockout of pkaR2 forces M. lusitanicus into its hyphal form, and in the case of pkaR4, the only subunit found to be essential, its knockout (in heterokaryosis) leads to abnormal hyphal emission during the transition from an anaerobic environment to aerobic conditions (235).
The PKA pathway is interlocked with the G proteins in the control of Mucor dimorphism and virulence. The G protein beta subunit 1 (Gpb1) regulates pkaR1 (Fig. 5C; Table 1), and its deletion resulted in the same phenotype described for pkaR1 mutants and reduced virulence (174). Interestingly, and given its relationship to oxidative/fermentative metabolism, a mutant in M. lusitanicus alcohol dehydrogenase 1 (adh1), which is unable to perform fermentative metabolism, showed a higher virulence and increased resistance to phagocytosis (115). It is also worth mentioning the implications of a phospholipase D (mcpID) in the regulation of dimorphism regulation. Mutation of the mcpID gene induces yeast-like-locked phenotype and reduces virulence in mice (126), although the molecular link between this phospholipase D and dimorphism regulation remains to be elucidated. In addition, other molecular mechanisms have been linked to dimorphism and virulence, including mechanisms related to substance secretion. ADP-ribosylation factors (Arf), which are important for vesicular trafficking, have been linked to growth morphology and, thus, to virulence. In M. lusitanicus, mutations in either Arf1 or Arf2 result in increased virulence, with Arf2 being essential for proper yeast development and Arf1 mutation causing defects in hyphal formation (116). The role of two Arf-like (Arl) proteins has also been studied in M. lusitanicus, showing that Arl1 is highly induced during the yeast-to-hyphae transition and that mutations in Arl2 cause defects in vesicular trafficking, increased antifungal susceptibility, and higher virulence of the cell-free medium (236).
Other secreted molecules with a role in virulence
In addition to mucoricin, secretion of other molecules has also been reported to be a determinant of virulence in Mucorales. One of the most concerning mucormycosis outcomes is the trauma-related necrotizing myocutaneous mucormycosis due to its high morbidity and mortality rates (237, 238). This has been associated with extreme mechanical forces, as in the case of the Joplin tornado in Missouri (29). Interestingly, tornado shear challenge (TSC) by magnetic stirring induces transient and increased virulence in several Mucorales species due to the release of soluble factors (120). Unfortunately, the molecular mechanisms underlying this phenomenon remain unknown, although the inhibition of the calcineurin/Hsp90 pathway in R. arrhizus and deletion of two calcineurin catalytic A subunits (cnaA and cnaB) and the regulatory B subunit (cnbR) of M. lusitanicus resulted in increased virulence of sporangiospores following TSC, suggesting a key regulatory role of the calcineurin/Hsp90 pathway in this mechanism of hypervirulence (120).
Other genes involved in the virulence of Mucorales
Among the many mechanisms involved in virulence, a complete network of gene expression regulation under the control of a white-collar photoreceptor has been reported to be important for the infective process. M. lusitanicus has three different white-collar 1 photoreceptors (designated Mcwc-1a, Mcwc-1b, and Mcwc-1c) that are specialized for different responses to light (239). Remarkably, although they have been associated with virulence in other fungi (240–242), their role in mucormycosis was unknown. A recent study has showed that Mcwc-1a (Table 1) was required for full virulence in M. lusitanicus and that several genes putatively involved in virulence are regulated by Mcwc-1a during the infection (176). Finally, a comparative analysis in terms of RNAi regulation between a virulent and an avirulent strain of M. lusitanicus served to identify the importance of Wex1, a putative DEDDy exonuclease with RNase domains (Table 1), as an essential virulence factor (175). Taken together, these findings reveal a complex machinery involved in the pathogenesis of Mucorales and highlight the importance and need to deepen and broaden the studies on the molecular mechanisms of this disease.
MECHANISMS OF ANTIFUNGAL RESISTANCE
The high morbidity and mortality of mucormycosis are explained not only by the complex mechanisms involved in host invasion and evasion of host defenses but also by the characteristic antifungal drug resistance of Mucorales. Understanding the mechanisms underlying antifungal resistance in Mucorales is crucial for the effective treatment of this disease.
As previously noted, Mucorales naturally possess intrinsic resistance to commonly used antifungal agents, resulting in limited treatment options (80), and clinically relevant members of the Mucorales displayed distinct patterns of antifungal drug resistance in vitro (243). Notably, amphotericin B emerges as the most potent antifungal agent against all Mucorales species tested, as indicated by MIC values falling within the range associated with susceptibility for A. fumigatus. Conversely, voriconazole exhibited elevated MICs against all species examined, exceeding the threshold recognized for clinical efficacy against A. fumigatus. Interestingly, isavuconazole and posaconazole have demonstrated varying degrees of in vitro activity against selected members of the Mucorales, with efficacy dependent on the specific species involved (243). These findings underscore the importance of accurately identifying the causative species in order to select an appropriate antifungal agent. The clinical use of antifungal agents aligns with the in vitro findings, with the lipid formulation of amphotericin B serving as the first-line therapy in most cases (80). However, this long-standing antifungal agent is burdened with numerous side effects, particularly increased toxicity in patients with renal insufficiency (244). In addition, amphotericin B exhibits limited efficacy in severe cases of mucormycosis and disseminated infections, leaving clinicians with limited treatment options (4).
Azoles are heterocyclic compounds that are among the most frequently used drugs to eradicate fungal infections (245, 246). Triazoles target cell wall synthesis through inhibition of ergosterol synthesis by blocking the activity of cytochrome P450 (CYP)-dependent lanosterol 14α-demethylase (Erg11) (247). Mucorales show species-specific differences in susceptibility to medium- (e.g., isavuconazole) and long-tailed azoles (e.g., posaconazole) in vitro and in vivo (80, 81, 103, 248–250) and therefore limited to salvage therapies (251). Characteristically, Mucorales are intrinsically resistant to short-tailed azoles (fluconazole and voriconazole) (252–255). Similar resistances have been linked to point mutations for the two major pathogenic fungi, Candida and Aspergillus. The first reported mutation was an arginine to lysine substitution (R467K) in a C. albicans isolate, and several other resistance mutations have been reported over the years (256, 257). In the case of Aspergillus, it is a naturally occurring T301I substitution confers intrinsic short-tailed resistance (258, 259). Mucorales possess two paralogs of the P450 (CYP)-dependent lanosterol 14α-demethylase, designated F1 and F5 (260). Analysis of both proteins revealed that a naturally occurring tyrosine substitution for a phenylalanine residue (Y129F) (Fig. 6) explains the intrinsic resistance of Mucorales to short-tailed azoles (260). A structural analysis based on the known structure of the cytochrome P450 (CYP)-dependent lanosterol 14α-demethylases revealed a mechanism for this mutation based on the disruption of a water-mediated hydrogen bond (261). Furthermore, mimicking this mutation in the homologous protein of S. cerevisiae rendered the yeast resistant to short-tailed azoles (261). The occurrence of other point mutations in genes involved in ergosterol biosynthesis has also been reported in Mucorales (262), raising concerns about the consequences of emerging resistance to the current antifungal drugs. In addition, functional analysis of the three M. lusitanicus erg6 genes encoding the sterol 24-C-methyltransferase suggests that one of the three genes, erg6b, plays a critical role in the azole resistance of M. lusitanicus. Analysis of the sterol composition of the erg6b knockout mutant suggests the existence of four alternative pathways for the biosynthesis of sterols that produce ergosterol and other alternative, non-toxic sterol products. Switching between these pathways in response to azole treatment may contribute to azole resistance (263).
Fig 6.
Mechanism of innate antifungal drugs resistance in Mucorales. Resistance of Mucorales to antifungals is linked to gene duplication of genes encoding antifungal efflux pumps (pdr), targets of echinocandins (fks), and proteins involved in biosynthesis of azoles (erg6 and erg11). In addition, the mutation Y129F (spiked red ball) in one of the Erg11 proteins (Cyp51F5) causes resistance to short-tailed azoles.
Echinocandins, such as micafungin, are another antifungal drug that target cell wall synthesis. These drugs target the β-(1,3)-d-glucan synthase (Fks) (264). A study in M. lusitanicus has revealed that Mucorales are resistant to micafungin by a dual mechanism. This fungus has three copies of the fks genes (fksA, fksB, and fksC), and interestingly, this increase in gene copy number is key to micafungin resistance as the knockout of either fksA or fksB results in increased susceptibility to micafungin (Fig. 6) (265). Additionally, to the duplication of the fks genes, calcineurin also regulates the expression of fksA and fksB, and its deletion results in a decreased expression of these genes and an increased susceptibility to micafungin, supporting the involvement of the calcineurin pathway in micafungin resistance (265).
Another example of gene duplication in Mucorales as a strategy for antifungal resistance is the high gene copy number of the antifungal efflux pump paralogs. ATP-binding cassette (ABC) transporters have been frequently linked to antifungal drug resistance in Candida, Aspergillus, and Cryptococcus (266–269), and their role in antifungal drug resistance in Mucorales has recently been characterized (270). The genome of M. lusitanicus encodes eight ABC transporters, of which pdr1 is overexpressed in the presence of azoles, and its deletion causes increased susceptibility to posaconazole, ravuconazole, and isavuconazole (Fig. 6) (270). Interestingly, pdr1 deletion also induces an increase in the expression of two other ABC transporter copies, pdr2 and pdr6, suggesting a compensatory effect of the different efflux pumps and highlighting that an intricate network regulates the ability of Mucorales to expel antifungal drugs.
In addition to intrinsic resistance mechanisms to antifungal drugs, Mucorales also has an acquired resistance mechanism based on the RNAi machinery, which has been extensively characterized in M. lusitanicus. This mechanism involves alterations in the expression of specific genes through the RNAi machinery, leading to reduced susceptibility to antifungal agents. As mentioned above, exposure of M. lusitanicus to FK506 forces non-virulent yeast growth. However, the observation of FK506-forced yeast colonies in M. lusitanicus led to the discovery of revertant sectors that exhibited resistant hyphal growth (271). Approximately 70% of the isolates harbored mutations in either the fkbA (which encodes the FKBP12 that complexes with FK506) or the calcineurin subunits CnaA and CnaB, but the remaining 30% lacked mutations in these genes (271). Interestingly, these isolates without mutations in any of the analyzed genes displayed a transient resistance that reverted to the wild-type growth (yeast growth in the presence of FK506) after a few cycles in drug-free media. Further characterization of these isolates revealed that in the presence of FK506, these resistant isolates showed decreased levels of fkbA/FKPB12 and increased levels of sRNAs targeting the fkbA gene (271). Because of transient phenotype, these isolates were termed epimutants. Characterization of the corresponding M. lusitanicus RNAi-mediated epimutation machinery revealed that it employs most of the components of the canonical RNAi pathway, with some peculiarities such as a more relevant role for the Dicer-1 enzyme (Dcl-1) (271, 272). Remarkably, deletion of key genes of the NCRP increases the generation of epimutants, establishing for the first time a negative regulatory function of this pathway over the epimutational pathway (272). In addition to FK506 resistance, the RNAi-mediated epimutational pathway has also proven to be an important mechanism for generating resistance to other antifungal compounds. This pathway is involved in the generation of resistance to 5-fluoroorotic acid (5-FOA), a compound that is converted to a toxic derivative through the uracil synthesis pathway (273). Isolated 5-FOA epimutants showed increased levels of sRNAs targeting both the orotate phosphoribosyl transferase (pyrF) and orotidine-5′-monophosphate decarboxylase (pyrG), which convert 5-FOA to its toxic derivative (274). This highlights the adaptive nature of this mechanism and raises concerns about the advantages that this pathway confers to Mucorales to develop antifungal resistance. Importantly, the function of this pathway has also been validated in vivo in murine models (275). FK506 epimutants were proven to be stable and resistant to antifungal administration in several infected organs (275), and furthermore, sporangiospores recovered from the brain showed a higher in vitro frequency of epimutation when exposed to FK506 than those extracted from other organs and sporangiospores that had not undergone infection (275). Although not yet proven, it is possible that this mechanism also operates to generate transient resistance to antifungal drugs used in the clinic. Ultimately, a comprehensive understanding of the mechanisms driving antifungal resistance in Mucorales is and will be fundamental to guide clinicians and researchers in optimizing treatment strategies and improving patient outcomes in mucormycosis.
CONCLUSIONS
Mucormycosis is a highly complex disease caused by at least 39 species of Mucorales, resulting in diverse manifestations that affect various organs. The knowledge of the biology of Mucorales, including the mechanisms causing the disease and resistance to antifungal drugs, is currently very limited due to the genetic intractability of these fungi. However, significant advances have been made in this century, such as the discovery that these fungi, particularly Rhizopus, utilize different CotH proteins to attach to and invade epithelial tissues through the recognition of tissue-specific host receptors. Additionally, mucoricin has been identified as one of the toxins secreted by the fungi that causes tissue damage. Several reports suggest that Mucorales species secrete other molecules important for the infection, including a molecule synthesized by bacterial endosymbionts. In addition, two iron uptake systems have been shown to play a critical role in the infection, and several other genes involved in fungal virulence have been identified through the generation of targeted mutants in M. lusitanicus. The use of M. lusitanicus has also allowed the identification of an RNAi-mediated mechanism that generates transient resistance to antifungal drugs. Despite these achievements, a disturbing conclusion arising from these studies is the lack of validation of results obtained with one causative agent in others. Most of the knowledge comes from Rhizopus, Mucor, and Lichtheimia, which makes sense as they are the more common mucormycosis-causative agents. However, the role of many mechanisms in infection and fungal resistance needs to be validated among them. Additionally, in some countries, like India, the frequency of infections caused by other species, such as Apophysomyces, is notably high. Therefore, more research efforts should be made to extend the observations made in one species. Also, much work remains to be done to improve diagnosis and to develop new antifungal agents against Mucorales.
ACKNOWLEDGMENTS
The researchers are funded by Fundación Séneca-Agencia de Ciencia y Tecnología de la Región de Murcia, Spain (21969/PI/22) to V.G. and grants PGC2018-097452-B-I00 to V.G. and PID2021-124674NB-I00 to F.E.N. funded by MCIN/AEI/ 10.13039/501100011033 by “ERDF A way of making Europe,” by the “European Union."
Biographies

Carlos Lax is a Ph.D. student in the Genetics and Microbiology department of the University of Murcia. His current research focuses on unraveling the molecular mechanisms of virulence of Mucorales, developing genetics tools for the study of basal fungi and their genomic and epigenetic characterization. He is interested in expanding the molecular resources available for basal fungi to provide, combined with comprehensive genomic studies, the most detailed information about the pathogenesis, ecology, and evolution of these organisms and, more importantly, improving himself so his interests and ambitions do not exceed his skills and talents.

Francisco E. Nicolás is a distinguished biologist with a rich academic journey. Graduating from the University of Murcia (Spain) in 2001 with a degree in Biology, he went on to earn his PhD in Genetics from the same institution in 2006. Dr. Molina's career includes a tenure as a senior researcher at the University of East Anglia (UK) for three and a half years before returning to the University of Murcia on a prestigious "Ramón y Cajal" contract. Since 2021, he has held the position of Assistant Professor at the University of Murcia. With a keen interest in genetics and gene regulation mechanisms, Dr. Molina has dedicated 23 years to exploring the fascinating world of fungi, particularly focusing on the neglected group, Mucorales, as a promising model for groundbreaking discoveries.

Eusebio Navarro obtained his Ph.D. in Biological Sciences at the University of Murcia, Spain, by studying the light regulation of cellular processes in the Mucorales fungus Mucor lusitanicus. During his postdoctoral research at the Center for Edaphology and Applied Biology of Segura (Spain), he shifted his focus to studying salt stress processes in pea and pepper plants. Since obtaining an assistant professor position at the University of Murcia, he has dedicated over 25 years to studying Mucorales genetics, with a primary focus on the regulation by light and the functioning of the non-canonical pathway of gene silencing (NCRIP). More recently, his research has evolved to concentrate on opportunistic infections, specifically mucormycosis, caused by this group of fungi.

Victoriano Garre is a Professor of Genetics at the University of Murcia, Spain. He earned his PhD from the same institution and has maintained a close association with it throughout his career, holding various positions, except for a postdoctoral period at the Institute of Botany in Münster, Germany. With a profound interest in the genetic and epigenetic regulation of gene expression in Mucorales fungi, he has dedicated over 30 years to advancing and promoting their study. Driven by the conviction that a comprehensive understanding of life on the planet necessitates the study of neglected fungal groups like Mucorales, his current focus revolves around unraveling the molecular mechanisms enabling Mucorales to cause mucormycosis, a life-threatening infection with limited treatment options. Simultaneously, he endeavors to unveil the role of DNA adenine methylation in the regulation of gene expression, as it is an epigenetic mark abundant in early-diverging fungi, including Mucorales.
Contributor Information
Victoriano Garre, Email: vgarre@um.es.
Mark D. Rose, Georgetown University, Washington, DC, USA
REFERENCES
- 1. Cornely OA, Alastruey-Izquierdo A, Arenz D, Chen SCA, Dannaoui E, Hochhegger B, Hoenigl M, Jensen HE, Lagrou K, Lewis RE, et al. 2019. Global guideline for the diagnosis and management of mucormycosis: an initiative of the European confederation of medical mycology in cooperation with the mycoses study group education and research consortium. Lancet Infect Dis 19:e405–e421. doi: 10.1016/S1473-3099(19)30312-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. WHO fungal priority pathogens list to guide research, development and public health action. World Health Organization (WHO). Available from: https://www.who.int/publications/ i/item/9789240060241 [Google Scholar]
- 3. Fisher MC, Denning DW. 2023. The WHO fungal priority pathogens list as a game-changer. Nat Rev Microbiol 21:211–212. doi: 10.1038/s41579-023-00861-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roden MM, Zaoutis TE, Buchanan WL, Knudsen TA, Sarkisova TA, Schaufele RL, Sein M, Sein T, Chiou CC, Chu JH, Kontoyiannis DP, Walsh TJ. 2005. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin Infect Dis 41:634–653. doi: 10.1086/432579 [DOI] [PubMed] [Google Scholar]
- 5. Spellberg B, Edwards J, Ibrahim A. 2005. Novel perspectives on mucormycosis: pathophysiology, presentation, and management. Clin Microbiol Rev 18:556–569. doi: 10.1128/CMR.18.3.556-569.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, Marr KA, Pfaller MA, Chang C-H, Webster KM. 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis 48:1695–1703. doi: 10.1086/599039 [DOI] [PubMed] [Google Scholar]
- 7. Steinbach WJ, Marr KA, Anaissie EJ, Azie N, Quan SP, Meier-Kriesche HU, Apewokin S, Horn DL. 2012. Clinical epidemiology of 960 patients with invasive aspergillosis from the PATH alliance registry. J Infect 65:453–464. doi: 10.1016/j.jinf.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 8. Lamoth F. 2023. Novel approaches in the management of mucormycosis. Curr Fungal Infect Rep 17:98–107. doi: 10.1007/s12281-023-00463-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jeong W, Keighley C, Wolfe R, Lee WL, Slavin MA, Chen SCA, Kong DCM. 2019. Contemporary management and clinical outcomes of mucormycosis: a systematic review and meta-analysis of case reports. Int J Antimicrob Agents 53:589–597. doi: 10.1016/j.ijantimicag.2019.01.002 [DOI] [PubMed] [Google Scholar]
- 10. Lax C, Cánovas-Márquez JT, Tahiri G, Navarro E, Garre V, Nicolás FE. 2022. Genetic manipulation in Mucorales and new developments to study mucormycosis. Int J Mol Sci 23:3454. doi: 10.3390/ijms23073454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tayabali K, Pothiwalla H, Narayanan S. 2023. Epidemiology of COVID-19–associated mucormycosis. Curr Fungal Infect Rep 17:1–20. doi: 10.1007/s12281-023-00464-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pasquier G. 2023. COVID-19-associated mucormycosis in India: why such an outbreak? J Mycol Med 33:101393. doi: 10.1016/j.mycmed.2023.101393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skiada A, Pavleas I, Drogari-Apiranthitou M. 2020. Epidemiology and diagnosis of mucormycosis: an update. J Fungi 6:265. doi: 10.3390/jof6040265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jeong W, Keighley C, Wolfe R, Lee WL, Slavin MA, Kong DCM, Chen S-A. 2019. The epidemiology and clinical manifestations of mucormycosis: a systematic review and meta-analysis of case reports. Clin Microbiol Infect 25:26–34. doi: 10.1016/j.cmi.2018.07.011 [DOI] [PubMed] [Google Scholar]
- 15. Prakash H, Chakrabarti A. 2019. Global epidemiology of mucormycosis. J Fungi 5:26. doi: 10.3390/jof5010026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pham D, Howard-Jones AR, Sparks R, Stefani M, Sivalingam V, Halliday CL, Beardsley J, Chen S-A. 2023. Epidemiology, modern diagnostics, and the management of Mucorales infections. J Fungi 9:659. doi: 10.3390/jof9060659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Corzo-León DE, Chora-Hernández LD, Rodríguez-Zulueta AP, Walsh TJ. 2018. Diabetes mellitus as the major risk factor for mucormycosis in Mexico: epidemiology, diagnosis, and outcomes of reported cases. Med Mycol 56:29–43. doi: 10.1093/mmy/myx017 [DOI] [PubMed] [Google Scholar]
- 18. Patel A, Kaur H, Xess I, Michael JS, Savio J, Rudramurthy S, Singh R, Shastri P, Umabala P, Sardana R, Kindo A, Capoor MR, Mohan S, Muthu V, Agarwal R, Chakrabarti A. 2020. A multicentre observational study on the epidemiology, risk factors, management and outcomes of mucormycosis in India. Clin Microbiol Infect 26:944. doi: 10.1016/j.cmi.2019.11.021 [DOI] [PubMed] [Google Scholar]
- 19. Lanternier F, Dannaoui E, Morizot G, Elie C, Garcia-Hermoso D, Huerre M, Bitar D, Dromer F, Lortholary O, French Mycosis Study Group . 2012. A global analysis of mucormycosis in France: the RetroZygo study (2005-2007). Clin Infect Dis 54 Suppl 1:S35–S43. doi: 10.1093/cid/cir880 [DOI] [PubMed] [Google Scholar]
- 20. Stemler J, Hamed K, Salmanton-García J, Rezaei-Matehkolaei A, Gräfe SK, Sal E, Zarrouk M, Seidel D, Abdelaziz Khedr R, Ben-Ami R, Ben-Chetrit E, Roth Y, Cornely OA. 2020. Mucormycosis in the Middle East and North Africa: analysis of the Fungiscope registry and cases from the literature. Mycoses 63:1060–1068. doi: 10.1111/myc.13123 [DOI] [PubMed] [Google Scholar]
- 21. Ibrahim AS, Spellberg B, Edwards J. 2008. Iron acquisition: a novel perspective on mucormycosis pathogenesis and treatment. Curr Opin Infect Dis 21:620–625. doi: 10.1097/QCO.0b013e3283165fd1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skiada A, Pagano L, Groll A, Zimmerli S, Dupont B, Lagrou K, Lass-Florl C, Bouza E, Klimko N, Gaustad P, Richardson M, Hamal P, Akova M, Meis JF, Rodriguez-Tudela J-L, Roilides E, Mitrousia-Ziouva A, Petrikkos G, European Confederation of Medical Mycology Working Group on Zygomycosis . 2011. Zygomycosis in Europe: analysis of 230 cases accrued by the registry of the European confederation of medical mycology (ECMM) working group on zygomycosis between 2005 and 2007. Clin Microbiol Infect 17:1859–1867. doi: 10.1111/j.1469-0691.2010.03456.x [DOI] [PubMed] [Google Scholar]
- 23. Almyroudis NG, Sutton DA, Linden P, Rinaldi MG, Fung J, Kusne S. 2006. Zygomycosis in solid organ transplant recipients in a tertiary transplant center and review of the literature. Am J Transplant 6:2365–2374. doi: 10.1111/j.1600-6143.2006.01496.x [DOI] [PubMed] [Google Scholar]
- 24. Singh N, Aguado JM, Bonatti H, Forrest G, Gupta KL, Safdar N, John GT, Pursell KJ, Muñoz P, Patel R, et al. 2009. Zygomycosis in solid organ transplant recipients: a prospective, matched case-control study to assess risks for disease and outcome. J Infect Dis 200:1002–1011. doi: 10.1086/605445 [DOI] [PubMed] [Google Scholar]
- 25. Hoang K, Abdo T, Reinersman JM, Lu R, Higuita NIA. 2020. A case of invasive pulmonary mucormycosis resulting from short courses of corticosteroids in a well-controlled diabetic patient. Med Mycol Case Rep 29:22–24. doi: 10.1016/j.mmcr.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kontoyiannis DP, Lewis RE. 2011. How I treat mucormycosis. Blood 118:1216–1224. doi: 10.1182/blood-2011-03-316430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh AK, Singh R, Joshi SR, Misra A. 2021. Mucormycosis in COVID-19: a systematic review of cases reported worldwide and in India. Diabetes Metab Syndr 15:102146. doi: 10.1016/j.dsx.2021.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muthu V, Agarwal R, Rudramurthy SM, Thangaraju D, Shevkani MR, Patel AK, Shastri PS, Tayade A, Bhandari S, Gella V, et al. 2023. Multicenter case–control study of COVID-19–associated mucormycosis outbreak, India. Emerg Infect Dis 29:8–19. doi: 10.3201/eid2901.220926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neblett Fanfair R, Benedict K, Bos J, Bennett SD, Lo Y-C, Adebanjo T, Etienne K, Deak E, Derado G, Shieh W-J, Drew C, Zaki S, Sugerman D, Gade L, Thompson EH, Sutton DA, Engelthaler DM, Schupp JM, Brandt ME, Harris JR, Lockhart SR, Turabelidze G, Park BJ. 2012. Necrotizing cutaneous mucormycosis after a tornado in Joplin, Missouri, in 2011. N Engl J Med 367:2214–2225. doi: 10.1056/NEJMoa1204781 [DOI] [PubMed] [Google Scholar]
- 30. Walsh TJ, Hospenthal DR, Petraitis V, Kontoyiannis DP. 2019. Necrotizing mucormycosis of wounds following combat injuries, natural disasters, burns, and other trauma. J Fungi 5:57. doi: 10.3390/jof5030057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. National Centre for Disease Control . 2021. CD alert: Covid-19 associated mucormycosis. Available from: https://ncdc.mohfw.gov.in/WriteReadData/l892s/20590398661627904360.pdf
- 32. Anand T, Mukherjee A, Satija A, Velamuri PS, Singh KJ, Das M, Josten K, Yadav PD, Sahay RR, Keche AY, et al. 2022. A case control investigation of COVID-19 associated mucormycosis in India. BMC Infect Dis 22:856. doi: 10.1186/s12879-022-07844-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chowdhary A, Gupta N, Wurster S, Kumar R, Mohabir JT, Tatavarthy S, Mittal V, Rani P, Barman P, Sachdeva N, Singh A, Sharma B, Jiang Y, Cuomo CA, Kontoyiannis DP. 2023. Multimodal analysis of the COVID‐19‐associated mucormycosis outbreak in Delhi, India indicates the convergence of clinical and environmental risk factors. Mycoses 66:515–526. doi: 10.1111/myc.13578 [DOI] [PubMed] [Google Scholar]
- 34. Ghosh AK, Singh R, Reddy S, Singh S, Rudramurthy SM, Kaur H, Choudhary H, Chakrabarti A. 2022. Evaluation of environmental Mucorales contamination in and around the residence of COVID-19-associated mucormycosis patients. Front Cell Infect Microbiol 12:953750. doi: 10.3389/fcimb.2022.953750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Biswal M, Gupta P, Kanaujia R, Kaur K, Kaur H, Vyas A, Hallur V, Behera B, Padaki P, Savio J, Nagaraj S, Chunchanur SK, Shwetha JV, Ambica R, Nagdeo N, Khuraijam R, Priyolakshmi N, Patel K, Thamke D, Dash L, Jadhav D, Bharmal R, Bhattacharya S, Rudramurthy SM, Chakrabarti A. 2022. Evaluation of hospital environment for presence of Mucorales during COVID-19-associated mucormycosis outbreak in India – a multi-centre study. J Hosp Infect 122:173–179. doi: 10.1016/j.jhin.2022.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prakash H, Singh S, Rudramurthy SM, Singh P, Mehta N, Shaw D, Ghosh AK. 2020. An aero mycological analysis of Mucormycetes in indoor and outdoor environments of northern India. Med Mycol 58:118–123. doi: 10.1093/mmy/myz031 [DOI] [PubMed] [Google Scholar]
- 37. Garcia-Hermoso D, Criscuolo A, Lee SC, Legrand M, Chaouat M, Denis B, Lafaurie M, Rouveau M, Soler C, Schaal J-V, Mimoun M, Mebazaa A, Heitman J, Dromer F, Brisse S, Bretagne S, Alanio A. 2018. Outbreak of invasive wound mucormycosis in a burn unit due to multiple strains of Mucor circinelloides f. circinelloides resolved by whole-genome sequencing. mBio 9:e00573-18. doi: 10.1128/mBio.00573-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Etienne KA, Gillece J, Hilsabeck R, Schupp JM, Colman R, Lockhart SR, Gade L, Thompson EH, Sutton DA, Neblett-Fanfair R, Park BJ, Turabelidze G, Keim P, Brandt ME, Deak E, Engelthaler DM. 2012. Whole genome sequence typing to investigate the Apophysomyces outbreak following a tornado in Joplin, Missouri, 2011. PLoS One 7:e49989. doi: 10.1371/journal.pone.0049989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nguyen MH, Kaul D, Muto C, Cheng SJ, Richter RA, Bruno VM, Liu G, Beyhan S, Sundermann AJ, Mounaud S, Pasculle AW, Nierman WC, Driscoll E, Cumbie R, Clancy CJ, Dupont CL. 2020. Genetic diversity of clinical and environmental Mucorales isolates obtained from an investigation of mucormycosis cases among solid organ transplant recipients. Microb Genom 6:mgen000473. doi: 10.1099/mgen.0.000473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walsh TJ, Gamaletsou MN, McGinnis MR, Hayden RT, Kontoyiannis DP. 2012. Early clinical and laboratory diagnosis of invasive pulmonary, extrapulmonary, and disseminated mucormycosis (zygomycosis). Clin Infect Dis 54 Suppl 1:S55–S60. doi: 10.1093/cid/cir868 [DOI] [PubMed] [Google Scholar]
- 41. Lackner M, Caramalho R, Lass-Flörl C. 2014. Laboratory diagnosis of mucormycosis: current status and future perspectives. Future Microbiol 9:683–695. doi: 10.2217/fmb.14.23 [DOI] [PubMed] [Google Scholar]
- 42. Guarner J, Brandt ME. 2011. Histopathologic diagnosis of fungal infections in the 21st century. Clin Microbiol Rev 24:247–280. doi: 10.1128/CMR.00053-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Millon L, Caillot D, Berceanu A, Bretagne S, Lanternier F, Morio F, Letscher-Bru V, Dalle F, Denis B, Alanio A, Boutoille D, Bougnoux ME, Botterel F, Chouaki T, Charbonnier A, Ader F, Dupont D, Bellanger AP, Rocchi S, Scherer E, Gbaguidi-Haore H, Herbrecht R. 2022. Evaluation of serum Mucorales polymerase chain reaction (PCR) for the diagnosis of mucormycoses: the MODIMUCOR prospective trial. Clin Infect Dis 75:777–785. doi: 10.1093/cid/ciab1066 [DOI] [PubMed] [Google Scholar]
- 44. Rocchi S, Scherer E, Mengoli C, Alanio A, Botterel F, Bougnoux ME, Bretagne S, Cogliati M, Cornu M, Dalle F, et al. 2021. Interlaboratory evaluation of Mucorales PCR assays for testing serum specimens: a study by the fungal PCR initiative and the Modimucor study group. Med Mycol 59:126–138. doi: 10.1093/mmy/myaa036 [DOI] [PubMed] [Google Scholar]
- 45. Bernal-Martínez L, Buitrago MJ, Castelli MV, Rodriguez-Tudela JL, Cuenca-Estrella M. 2013. Development of a single tube multiplex real-time PCR to detect the most clinically relevant Mucormycetes species. Clin Microbiol Infect 19:E1–E7. doi: 10.1111/j.1469-0691.2012.03976.x [DOI] [PubMed] [Google Scholar]
- 46. Zaman K, Rudramurthy SM, Das A, Panda N, Honnavar P, Kaur H, Chakrabarti A. 2017. Molecular diagnosis of rhino-orbito-cerebral mucormycosis from fresh tissue samples. J Med Microbiol 66:1124–1129. doi: 10.1099/jmm.0.000560 [DOI] [PubMed] [Google Scholar]
- 47. Baldin C, Soliman SSM, Jeon HH, Alkhazraji S, Gebremariam T, Gu Y, Bruno VM, Cornely OA, Leather HL, Sugrue MW, Wingard JR, Stevens DA, Edwards JE, Ibrahim AS. 2018. PCR-based approach targeting Mucorales-specific gene gamily for diagnosis of mucormycosis. J Clin Microbiol 56:e00746-18. doi: 10.1128/JCM.00746-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Millon L, Herbrecht R, Grenouillet F, Morio F, Alanio A, Letscher-Bru V, Cassaing S, Chouaki T, Kauffmann-Lacroix C, Poirier P, Toubas D, Augereau O, Rocchi S, Garcia-Hermoso D, Bretagne S, French Mycosis Study Group . 2016. Early diagnosis and monitoring of mucormycosis by detection of circulating DNA in serum: retrospective analysis of 44 cases collected through the French surveillance network of invasive fungal infections (RESSIF). Clin Microbiol Infect 22:810. doi: 10.1016/j.cmi.2015.12.006 [DOI] [PubMed] [Google Scholar]
- 49. Dannaoui E. 2022. Recent developments in the diagnosis of mucormycosis. J Fungi 8:457. doi: 10.3390/jof8050457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mercier T, Reynders M, Beuselinck K, Guldentops E, Maertens J, Lagrou K. 2019. Serial detection of circulating Mucorales DNA in invasive mucormycosis: a retrospective multicenter evaluation. J Fungi (Basel) 5:113. doi: 10.3390/jof5040113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guegan H, Iriart X, Bougnoux ME, Berry A, Robert-Gangneux F, Gangneux JP. 2020. Evaluation of MucorGenius Mucorales PCR assay for the diagnosis of pulmonary mucormycosis. J Infect 81:311–317. doi: 10.1016/j.jinf.2020.05.051 [DOI] [PubMed] [Google Scholar]
- 52. Imbert S, Portejoie L, Pfister E, Tauzin B, Revers M, Uthurriague J, Hernandez-Grande M, Lafon ME, Jubert C, Issa N, Dumas PY, Delhaes L. 2023. A multiplex PCR and DNA-sequencing workflow on serum for the diagnosis and species identification for invasive aspergillosis and mucormycosis. J Clin Microbiol 61:e0140922. doi: 10.1128/jcm.01409-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schwarz P, Guedouar H, Laouiti F, Grenouillet F, Dannaoui E. 2019. Identification of Mucorales by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J Fungi 5:56. doi: 10.3390/jof5030056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koshy S, Ismail N, Astudillo CL, Haeger CM, Aloum O, Acharige MT, Farmakiotis D, Baden LR, Marty FM, Kontoyiannis DP, Fredenburgh L, Koo S. 2017. Breath-based diagnosis of invasive mucormycosis (IM). Open Forum Infect Dis 4:S53–S54. doi: 10.1093/ofid/ofx162.124 [DOI] [Google Scholar]
- 55. Petrikkos G, Skiada A, Lortholary O, Roilides E, Walsh TJ, Kontoyiannis DP. 2012. Epidemiology and clinical manifestations of mucormycosis. Clin Infect Dis 54 Suppl 1:S23–S34. doi: 10.1093/cid/cir866 [DOI] [PubMed] [Google Scholar]
- 56. Ribes JA, Vanover-Sams CL, Baker DJ. 2000. Zygomycetes in human disease. Clin Microbiol Rev 13:236–301. doi: 10.1128/CMR.13.2.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Veisi A, Bagheri A, Eshaghi M, Rikhtehgar MH, Rezaei Kanavi M, Farjad R. 2022. Rhino-orbital mucormycosis during steroid therapy in COVID-19 patients: a case report. Eur J Ophthalmol 32:NP11–NP16. doi: 10.1177/11206721211009450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pagano L, Cornely OA, Busca A, Caira M, Cesaro S, Gasbarrino C, Girmenia C, Heinz WJ, Herbrecht R, Lass-Flörl C, Nosari A, Potenza L, Racil Z, Rickerts V, Sheppard DC, Simon A, Ullmann AJ, Valentini CG, Vehreschild JJ, Candoni A, Vehreschild MJGT. 2013. Combined antifungal approach for the treatment of invasive mucormycosis in patients with hematologic diseases: a report from the SEIFEM and FUNGISCOPE registries. Haematologica 98:e127–e130. doi: 10.3324/haematol.2012.083063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rammaert B, Lanternier F, Zahar JR, Dannaoui E, Bougnoux ME, Lecuit M, Lortholary O. 2012. Healthcare-associated mucormycosis. Clin Infect Dis 54 Suppl 1:S44–S54. doi: 10.1093/cid/cir867 [DOI] [PubMed] [Google Scholar]
- 60. Marras TK, Daley CL. 2002. Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin Chest Med 23:553–567. doi: 10.1016/s0272-5231(02)00019-9 [DOI] [PubMed] [Google Scholar]
- 61. Cesaro S, Pillon M, Calore E, Alaggio R, Gamba P, Bergamo S, Mainardi C, Toffolutti T, Pegoraro A, Messina C. 2011. Combination antifungal therapy and surgery for the treatment of invasive pulmonary aspergillosis after hematopoietic stem cell transplantation. Pediatr Rep 3:e18. doi: 10.4081/pr.2011.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Skiada A, Drogari-Apiranthitou M, Pavleas I, Daikou E, Petrikkos G. 2022. Global cutaneous mucormycosis: a systematic review. J Fungi 8:194. doi: 10.3390/jof8020194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sheldon WH, Bauer H. 1959. The development of the acute inflammatory response to experimental cutaneous mucormycosis in normal and diabetic rabbits. J Exp Med 110:845–852. doi: 10.1084/jem.110.6.845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kennedy KJ, Daveson K, Slavin MA, van Hal SJ, Sorrell TC, Lee A, Marriott DJ, Chapman B, Halliday CL, Hajkowicz K, Athan E, Bak N, Cheong E, Heath CH, Morrissey CO, Kidd S, Beresford R, Blyth C, Korman TM, Robinson JO, Meyer W, Chen SC-A, Australia and New Zealand Mycoses Interest Group of the Australasian Society for Infectious Diseases . 2016. Mucormycosis in Australia: contemporary epidemiology and outcomes. Clin Microbiol Infect 22:775–781. doi: 10.1016/j.cmi.2016.01.005 [DOI] [PubMed] [Google Scholar]
- 65. Roilides E, Zaoutis TE, Katragkou A, Benjamin DK, Walsh TJ. 2009. Zygomycosis in neonates: an uncommon but life-threatening infection. Am J Perinatol 26:565–573. doi: 10.1055/s-0029-1220775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Addasi Y, Nguyen AH, Sabri A, Ahmad F, Rangray R, Velagapudi M. 2023. Gastrointestinal mucormycosis: a clinical review. Gastroenterology Res 16:249–253. doi: 10.14740/gr1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lee SC, Billmyre RB, Li A, Carson S, Sykes SM, Huh EY, Mieczkowski P, Ko DC, Cuomo CA, Heitman J. 2014. Analysis of a food-borne fungal pathogen outbreak: virulence and genome of a Mucor circinelloides isolate from yogurt. mBio 5:e01390-14. doi: 10.1128/mBio.01390-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rokas A. 2022. Evolution of the human pathogenic lifestyle in fungi. Nat Microbiol 7:607–619. doi: 10.1038/s41564-022-01112-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Naranjo-Ortiz MA, Gabaldón T. 2019. Fungal evolution: diversity, taxonomy and phylogeny of the fungi. Biol Rev Camb Philos Soc 94:2101–2137. doi: 10.1111/brv.12550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Walther G, Wagner L, Kurzai O. 2019. Updates on the taxonomy of Mucorales with an emphasis on clinically important taxa. J Fungi 5:106. doi: 10.3390/jof5040106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hoenigl M, Seidel D, Sprute R, Cunha C, Oliverio M, Goldman GH, Ibrahim AS, Carvalho A. 2022. COVID-19-associated fungal infections. Nat Microbiol 7:1127–1140. doi: 10.1038/s41564-022-01172-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gryganskyi AP, Golan J, Dolatabadi S, Mondo S, Robb S, Idnurm A, Muszewska A, Steczkiewicz K, Masonjones S, Liao H-L, Gajdeczka MT, Anike F, Vuek A, Anishchenko IM, Voigt K, de Hoog GS, Smith ME, Heitman J, Vilgalys R, Stajich JE. 2018. Phylogenetic and phylogenomic definition of Rhizopus species. G3 (Bethesda) 8:2007–2018. doi: 10.1534/g3.118.200235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dolatabadi S, de Hoog GS, Meis JF, Walther G. 2014. Species boundaries and nomenclature of Rhizopus arrhizus (syn. R. oryzae). Mycoses 57 Suppl 3:108–127. doi: 10.1111/myc.12228 [DOI] [PubMed] [Google Scholar]
- 74. Prakash H, Chakrabarti A. 2021. Epidemiology of mucormycosis in India. Microorganisms 9:523. doi: 10.3390/microorganisms9030523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ma L-J, Ibrahim AS, Skory C, Grabherr MG, Burger G, Butler M, Elias M, Idnurm A, Lang BF, Sone T, et al. 2009. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet 5:e1000549. doi: 10.1371/journal.pgen.1000549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Alqarihi A, Gebremariam T, Gu Y, Swidergall M, Alkhazraji S, Soliman SSM, Bruno VM, Edwards JE, Filler SG, Uppuluri P, Ibrahim AS. 2020. GRP78 and integrins play different roles in host cell invasion during mucormycosis. mBio 11:e01087-20. doi: 10.1128/mBio.01087-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chibucos MC, Soliman S, Gebremariam T, Lee H, Daugherty S, Orvis J, Shetty AC, Crabtree J, Hazen TH, Etienne KA, Kumari P, O’Connor TD, Rasko DA, Filler SG, Fraser CM, Lockhart SR, Skory CD, Ibrahim AS, Bruno VM. 2016. An integrated genomic and transcriptomic survey of mucormycosis-causing fungi. Nat Commun 7:12218. doi: 10.1038/ncomms12218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang Y, Chang Y, Ortañez J, Peña JF, Carter-House D, Reynolds NK, Smith ME, Benny G, Mondo SJ, Salamov A, Lipzen A, Pangilinan J, Guo J, LaButti K, Andreopolous W, Tritt A, Keymanesh K, Yan M, Barry K, Grigoriev IV, Spatafora JW, Stajich JE. 2023. Divergent evolution of early terrestrial fungi reveals the evolution of mucormycosis pathogenicity factors. Genome Biol Evol 15:evad046. doi: 10.1093/gbe/evad046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gryganskyi AP, Golan J, Muszewska A, Idnurm A, Dolatabadi S, Mondo SJ, Kutovenko VB, Kutovenko VO, Gajdeczka MT, Anishchenko IM, et al. 2023. Sequencing the genomes of the first terrestrial fungal lineages: what have we learned? Microorganisms 11:1830. doi: 10.3390/microorganisms11071830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dannaoui E. 2017. Antifungal resistance in Mucorales. Int J Antimicrob Agents 50:617–621. doi: 10.1016/j.ijantimicag.2017.08.010 [DOI] [PubMed] [Google Scholar]
- 81. Arendrup MC, Jensen RH, Meletiadis J. 2015. In vitro activity of isavuconazole and comparators against clinical isolates of the Mucorales order. Antimicrob Agents Chemother 59:7735–7742. doi: 10.1128/AAC.01919-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Espinel-Ingroff A, Chakrabarti A, Chowdhary A, Cordoba S, Dannaoui E, Dufresne P, Fothergill A, Ghannoum M, Gonzalez GM, Guarro J, Kidd S, Lass-Flörl C, Meis JF, Pelaez T, Tortorano AM, Turnidge J. 2015. Multicenter evaluation of MIC distributions for epidemiologic cutoff value definition to detect amphotericin B, posaconazole, and itraconazole resistance among the most clinically relevant species of Mucorales. Antimicrob Agents Chemother 59:1745–1750. doi: 10.1128/AAC.04435-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Navarro-Mendoza MI, Pérez-Arques C, Panchal S, Nicolás FE, Mondo SJ, Ganguly P, Pangilinan J, Grigoriev IV, Heitman J, Sanyal K, Garre V. 2019. Early diverging fungus Mucor circinelloides lacks centromeric histone CENP-A and displays a mosaic of point and regional centromeres. Curr Biol 29:3791–3802. doi: 10.1016/j.cub.2019.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nagy G, Szebenyi C, Csernetics Á, Vaz AG, Tóth EJ, Vágvölgyi C, Papp T. 2017. Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides. Sci Rep 7:16800. doi: 10.1038/s41598-017-17118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nicolás FE, Navarro-Mendoza MI, Pérez-Arques C, López-García S, Navarro E, Torres-Martínez S, Garre V. 2018. Molecular tools for carotenogenesis analysis in the mucoral Mucor circinelloides, p 221–237. In Methods in molecular biology. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 86. Yang J, Cánovas-Márquez JT, Li P, Li S, Niu J, Wang X, Nazir Y, López-García S, Garre V, Song Y. 2021. Deletion of plasma membrane malate transporters increased lipid accumulation in the oleaginous fungus Mucor circinelloides WJ11. J Agric Food Chem 69:9632–9641. doi: 10.1021/acs.jafc.1c03307 [DOI] [PubMed] [Google Scholar]
- 87. Sharma N, Schwartzman JD, Gutmann EJ, Marotti JD, Liu X. 2018. Cokeromyces recurvatus in a Papanicolaou test: an exceedingly rare finding that can be mistaken for Paracoccidioides brasiliensis. Cytojournal 15:5. doi: 10.4103/cytojournal.cytojournal_35_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ghormade V, Pathan E, Deshpande MV. 2012. Yeast-hypha dimorphism in zygomycetous fungi, p 87–104. In Ruiz-Herrera J (ed), Dimorphic fungi: their importance as models for differentiation and fungal pathogenesis. Bentham Science Publishers Ltd. [Google Scholar]
- 89. Trachuk P, Szymczak WA, Muscarella P, Sarwar UN. 2018. A case of invasive gastrointestinal Mycotypha infection in a patient with neutropenia. Case Rep Infect Dis 2018:5864175. doi: 10.1155/2018/5864175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Badali H, Cañete-Gibas C, McCarthy D, Patterson H, Sanders C, David MP, Mele J, Fan H, Wiederhold NP. 2021. Epidemiology and antifungal susceptibilities of mucoralean fungi in clinical samples from the United States. J Clin Microbiol 59:e0123021. doi: 10.1128/JCM.01230-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hassan MIA, Cseresnyes Z, Al-Zaben N, Dahse H-M, Vilela de Oliveira RJ, Walther G, Voigt K, Figge MT. 2019. The geographical region of origin determines the phagocytic vulnerability of Lichtheimia strains. Environ Microbiol 21:4563–4581. doi: 10.1111/1462-2920.14752 [DOI] [PubMed] [Google Scholar]
- 92. Kamei K. 2001. Animal models of zygomycosis - Absidia, Rhizopus, Rhizomucor, and Cunninghamella. Mycopathologia 152:5–13. doi: 10.1023/a:1011900630987 [DOI] [PubMed] [Google Scholar]
- 93. Jacobsen I. 2019. Animal models to study mucormycosis. J Fungi 5:27. doi: 10.3390/jof5020027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kamei A, Gao G, Neale G, Loh LN, Vogel P, Thomas PG, Tuomanen EI, Murray PJ. 2016. Exogenous remodeling of lung resident macrophages protects against infectious consequences of bone marrow-suppressive chemotherapy. Proc Natl Acad Sci U S A 113:E6153–E6161. doi: 10.1073/pnas.1607787113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Philippe B, Ibrahim-Granet O, Prévost MC, Gougerot-Pocidalo MA, Sanchez Perez M, Van der Meeren A, Latgé JP. 2003. Killing of Aspergillus fumigatus by alveolar macrophages is mediated by reactive oxidant intermediates. Infect Immun 71:3034–3042. doi: 10.1128/IAI.71.6.3034-3042.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Goyal SN, Reddy NM, Patil KR, Nakhate KT, Ojha S, Patil CR, Agrawal YO. 2016. Challenges and issues with streptozotocin-induced diabetes – a clinically relevant animal model to understand the diabetes pathogenesis and evaluate therapeutics. Chem Biol Interact 244:49–63. doi: 10.1016/j.cbi.2015.11.032 [DOI] [PubMed] [Google Scholar]
- 97. Bauer H, Flanagan JF, Sheldon WH. 1955. Experimental cerebral mucormycosis in rabbits with alloxan diabetes. Yale J Biol Med 28:29–36. [PMC free article] [PubMed] [Google Scholar]
- 98. Jacobsen ID. 2023. Mouse models of mucormycosis. Methods Mol Biol 2667:181–196. doi: 10.1007/978-1-0716-3199-7_14 [DOI] [PubMed] [Google Scholar]
- 99. Danion F, Coste A, Le Hyaric C, Melenotte C, Lamoth F, Calandra T, Garcia-Hermoso D, Aimanianda V, Lanternier F, Lortholary O. 2023. What is new in pulmonary mucormycosis? J Fungi (Basel) 9:307. doi: 10.3390/jof9030307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Whitsett JA, Alenghat T. 2015. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol 16:27–35. doi: 10.1038/ni.3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hernández-Santos N, Wiesner DL, Fites JS, McDermott AJ, Warner T, Wüthrich M, Klein BS. 2018. Lung epithelial cells coordinate innate lymphocytes and immunity against pulmonary fungal infection. Cell Host Microbe 23:511–522. doi: 10.1016/j.chom.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schulze B, Rambach G, Schwartze VU, Voigt K, Schubert K, Speth C, Jacobsen ID. 2017. Ketoacidosis alone does not predispose to mucormycosis by Lichtheimia in a murine pulmonary infection model. Virulence 8:1657–1667. doi: 10.1080/21505594.2017.1360460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Luo G, Gebremariam T, Lee H, French SW, Wiederhold NP, Patterson TF, Filler SG, Ibrahim AS. 2013. Efficacy of liposomal amphotericin B and posaconazole in intratracheal models of murine mucormycosis. Antimicrob Agents Chemother 57:3340–3347. doi: 10.1128/AAC.00313-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Reinhardt DJ, Licata I, Kaplan W, Ajello L, Chandler FW, Ellis JJ. 1981. Experimental cerebral zygomycosis in alloxan-diabetic rabbits: variation in virulence among zygomycetes. Med Mycol 19:245–255. doi: 10.1080/00362178185380421 [DOI] [PubMed] [Google Scholar]
- 105. Smith JMB, Jones RH. 1973. Localization and fate of Absidia ramosa spores after intravenous inoculation of mice. J Comp Pathol 83:49–55. doi: 10.1016/0021-9975(73)90026-1 [DOI] [PubMed] [Google Scholar]
- 106. Ben-Ami R, Luna M, Lewis RE, Walsh TJ, Kontoyiannis DP. 2009. A clinicopathological study of pulmonary mucormycosis in cancer patients: extensive angioinvasion but limited inflammatory response. J Infect 59:134–138. doi: 10.1016/j.jinf.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sheldon WH, Bauer H. 1958. Activation of quiescent mucormycotic granulomata in rabbits by induction of acute alloxan diabetes. J Exp Med 108:171–178. doi: 10.1084/jem.108.1.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sheldon WH, Bauer H. 1960. Tissue mast cells and acute inflammation in experimental cutaneous mucormycosis of normal, 48/80-treated, and diabetic rats. J Exp Med 112:1069–1084. doi: 10.1084/jem.112.6.1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Lewis RE, Ben-Ami R, Best L, Albert N, Walsh TJ, Kontoyiannis DP. 2013. Tacrolimus enhances the potency of posaconazole against Rhizopus oryzae in vitro and in an experimental model of mucormycosis. J Infect Dis 207:834–841. doi: 10.1093/infdis/jis767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lax C, Navarro-Mendoza MI, Pérez-Arques C, Navarro E, Nicolás FE, Garre V. 2021. Stable and reproducible homologous recombination enables CRISPR-based engineering in the fungus Rhizopus microsporus Cell Rep Methods 1:100124. doi: 10.1016/j.crmeth.2021.100124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Voelz K, Gratacap RL, Wheeler RT. 2015. A zebrafish larval model reveals early tissue-specific innate immune responses to Mucor circinelloides. Dis Model Mech 8:1375–1388. doi: 10.1242/dmm.019992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. López-Muñoz A, Nicolás FE, García-Moreno D, Pérez-Oliva AB, Navarro-Mendoza MI, Hernández-Oñate MA, Herrera-Estrella A, Torres-Martínez S, Ruiz-Vázquez RM, Garre V, Mulero V. 2018. An adult zebrafish model reveals that mucormycosis induces apoptosis of infected macrophages. Sci Rep 8:12802. doi: 10.1038/s41598-018-30754-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Schwartze VU, Hoffmann K, Nyilasi I, Papp T, Vágvölgyi C, de Hoog S, Voigt K, Jacobsen ID. 2012. Lichtheimia species exhibit differences in virulence potential. PLoS One 7:e40908. doi: 10.1371/journal.pone.0040908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Alejandre-Castañeda V, Patiño-Medina JA, Valle-Maldonado MI, Nuñez-Anita RE, Santoyo G, Castro-Cerritos KV, Ortiz-Alvarado R, Corrales-Escobosa AR, Ramírez-Díaz MI, Gutiérrez-Corona JF, López-Torres A, Garre V, Meza-Carmen V. 2022. Secretion of the siderophore rhizoferrin is regulated by the cAMP-PKA pathway and is involved in the virulence of Mucor lusitanicus. Sci Rep 12:10649. doi: 10.1038/s41598-022-14515-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Díaz-Pérez SP, Patiño-Medina JA, Valle-Maldonado MI, López-Torres A, Jácome-Galarza IE, Anaya-Martínez V, Gómez-Ruiz V, Campos-García J, Nuñez-Anita RE, Ortiz-Alvarado R, Ramírez-Díaz MI, Gutiérrez-Corona JF, Meza-Carmen V. 2020. Alteration of fermentative metabolism enhances Mucor circinelloides virulence. Infect Immun 88:e00434-19. doi: 10.1128/IAI.00434-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Patiño-Medina JA, Maldonado-Herrera G, Pérez-Arques C, Alejandre-Castañeda V, Reyes-Mares NY, Valle-Maldonado MI, Campos-García J, Ortiz-Alvarado R, Jácome-Galarza IE, Ramírez-Díaz MI, Garre V, Meza-Carmen V. 2018. Control of morphology and virulence by ADP-ribosylation factors (Arf) in Mucor circinelloides. Curr Genet 64:853–869. doi: 10.1007/s00294-017-0798-0 [DOI] [PubMed] [Google Scholar]
- 117. Lemaitre B, Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu Rev Immunol 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615 [DOI] [PubMed] [Google Scholar]
- 118. Lionakis MS, Kontoyiannis DP. 2012. Drosophila melanogaster as a model organism for invasive aspergillosis, p 455–468. In Brand AC, MacCallum DM (ed), Host-fungus interactions. Humana Press, Totowa, NJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chamilos G, Lewis RE, Hu J, Xiao L, Zal T, Gilliet M, Halder G, Kontoyiannis DP. 2008. Drosophila melanogaster as a model host to dissect the immunopathogenesis of zygomycosis. Proc Natl Acad Sci U S A 105:9367–9372. doi: 10.1073/pnas.0709578105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wurster S, Tatara AM, Albert ND, Ibrahim AS, Heitman J, Lee SC, Shetty AC, McCracken C, Graf KT, Mikos AG, Bruno VM, Kontoyiannis DP. 2020. Tornadic shear stress induces a transient, calcineurin-dependent hypervirulent phenotype in Mucorales molds. mBio 11:1–15. doi: 10.1128/mBio.01414-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Szebenyi C, Gu Y, Gebremariam T, Kocsubé S, Kiss-Vetráb S, Jáger O, Patai R, Spisák K, Sinka R, Binder U, Homa M, Vágvölgyi C, Ibrahim AS, Nagy G, Papp T. 2023. cotH genes are necessary for normal spore formation and virulence in Mucor lusitanicus. mBio 14:e0338622. doi: 10.1128/mbio.03386-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chamilos G, Lionakis MS, Lewis RE, Kontoyiannis DP. 2007. Role of mini-host models in the study of medically important fungi. Lancet Infect Dis 7:42–55. doi: 10.1016/S1473-3099(06)70686-7 [DOI] [PubMed] [Google Scholar]
- 123. Singulani JL, Scorzoni L, de Oliveira HC, Marcos CM, Assato PA, Fusco-Almeida A, Mendes-Giannini M. 2018. Applications of invertebrate animal models to dimorphic fungal infections. J Fungi 4:118. doi: 10.3390/jof4040118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Maurer E, Hörtnagl C, Lackner M, Grässle D, Naschberger V, Moser P, Segal E, Semis M, Lass-Flörl C, Binder U. 2019. Galleria mellonella as a model system to study virulence potential of mucormycetes and evaluation of antifungal treatment. Med Mycol 57:351–362. doi: 10.1093/mmy/myy042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pereira T, de Barros P, Fugisaki L, Rossoni R, Ribeiro F, de Menezes R, Junqueira J, Scorzoni L. 2018. Recent advances in the use of Galleria mellonella model to study immune responses against human pathogens. J Fungi 4:128. doi: 10.3390/jof4040128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Trieu TA, Navarro-Mendoza MI, Pérez-Arques C, Sanchis M, Capilla J, Navarro-Rodriguez P, Lopez-Fernandez L, Torres-Martínez S, Garre V, Ruiz-Vázquez RM, Nicolás FE. 2017. RNAi-based functional genomics identifies new virulence determinants in mucormycosis. PLoS Pathog 13:e1006150. doi: 10.1371/journal.ppat.1006150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Trevijano-Contador N, Zaragoza O. 2019. Immune response of Galleria mellonella against human fungal pathogens. J Fungi 5:3. doi: 10.3390/jof5010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Gebremariam T, Liu M, Luo G, Bruno V, Phan QT, Waring AJ, Edwards JE, Filler SG, Yeaman MR, Ibrahim AS. 2014. CotH3 mediates fungal invasion of host cells during mucormycosis. J Clin Invest 124:237–250. doi: 10.1172/JCI71349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Soliman SSM, Baldin C, Gu Y, Singh S, Gebremariam T, Swidergall M, Alqarihi A, Youssef EG, Alkhazraji S, Pikoulas A, Perske C, Venkataramani V, Rich A, Bruno VM, Hotopp JD, Mantis NJ, Edwards JE, Filler SG, Chamilos G, Vitetta ES, Ibrahim AS. 2021. Mucoricin is a ricin-like toxin that is critical for the pathogenesis of mucormycosis. Nat Microbiol 6:313–326. doi: 10.1038/s41564-020-00837-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Andrianaki AM, Kyrmizi I, Thanopoulou K, Baldin C, Drakos E, Soliman SSM, Shetty AC, McCracken C, Akoumianaki T, Stylianou K, Ioannou P, Pontikoglou C, Papadaki HA, Tzardi M, Belle V, Etienne E, Beauvais A, Samonis G, Kontoyiannis DP, Andreakos E, Bruno VM, Ibrahim AS, Chamilos G. 2018. Iron restriction inside macrophages regulates pulmonary host defense against Rhizopus species. Nat Commun 9:3333. doi: 10.1038/s41467-018-05820-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Pérez-Arques C, Navarro-Mendoza MI, Murcia L, Lax C, Martínez-García P, Heitman J, Nicolás FE, Garre V. 2019. Mucor circinelloides thrives inside the phagosome through an ATF-mediated germination pathway. mBio 10:e02765-18. doi: 10.1128/mBio.02765-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Watkins TN, Gebremariam T, Swidergall M, Shetty AC, Graf KT, Alqarihi A, Alkhazraji S, Alsaadi AI, Edwards VL, Filler SG, Ibrahim AS, Bruno VM. 2018. Inhibition of EGFR signaling protects from mucormycosis. mBio 9:e01384-18. doi: 10.1128/mBio.01384-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Liu M, Spellberg B, Phan QT, Fu Y, Fu Y, Lee AS, Edwards JE, Filler SG, Ibrahim AS. 2010. The endothelial cell receptor GRP78 is required for mucormycosis pathogenesis in diabetic mice. J Clin Invest 120:1914–1924. doi: 10.1172/JCI42164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ibrahim AS, Spellberg B, Avanessian V, Fu Y, Edwards JE. 2005. Rhizopus oryzae adheres to, is phagocytosed by, and damages endothelial cells in vitro. Infect Immun 73:778–783. doi: 10.1128/IAI.73.2.778-783.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Vellanki S, Billmyre RB, Lorenzen A, Campbell M, Turner B, Huh EY, Heitman J, Lee SC. 2020. A novel resistance pathway for calcineurin inhibitors in the human-pathogenic Mucorales Mucor circinelloides. mBio 11:e02949-19. doi: 10.1128/mBio.02949-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Osorio-Concepción M, Lax C, Navarro E, Nicolás FE, Garre V. 2021. DNA methylation on N6‐adenine regulates the hyphal development during dimorphism in the early‐diverging fungus Mucor lusitanicus. J Fungi 7:738. doi: 10.3390/jof7090738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. 2018. Invasive candidiasis. Nat Rev Dis Primers 4:18026. doi: 10.1038/nrdp.2018.26 [DOI] [PubMed] [Google Scholar]
- 138. Latgé J-P, Chamilos G. 2019. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev 33:e00140-18. doi: 10.1128/CMR.00140-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Mélida H, Sain D, Stajich JE, Bulone V. 2015. Deciphering the uniqueness of Mucoromycotina cell walls by combining biochemical and phylogenomic approaches. Environ Microbiol 17:1649–1662. doi: 10.1111/1462-2920.12601 [DOI] [PubMed] [Google Scholar]
- 140. Bartnicki-Garcia S. 1968. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu Rev Microbiol 22:87–108. doi: 10.1146/annurev.mi.22.100168.000511 [DOI] [PubMed] [Google Scholar]
- 141. Lecointe K, Cornu M, Leroy J, Coulon P, Sendid B. 2019. Polysaccharides cell wall architecture of Mucorales. Front Microbiol 10:469. doi: 10.3389/fmicb.2019.00469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Wasylnka JA, Moore MM. 2003. Aspergillus fumigatus conidia survive and germinate in acidic organelles of A549 epithelial cells. J Cell Sci 116:1579–1587. doi: 10.1242/jcs.00329 [DOI] [PubMed] [Google Scholar]
- 143. Rammaert B, Jouvion G, de Chaumont F, Garcia-Hermoso D, Szczepaniak C, Renaudat C, Olivo-Marin J-C, Chrétien F, Dromer F, Bretagne S. 2015. Absence of fungal spore internalization by bronchial epithelium in mouse models evidenced by a new bioimaging approach and transmission electronic microscopy. Am J Pathol 185:2421–2430. doi: 10.1016/j.ajpath.2015.04.027 [DOI] [PubMed] [Google Scholar]
- 144. Artis WM, Fountain JA, Delcher HK, Jones HE. 1982. A mechanism of susceptibility to mucormycosis in diabetic ketoacidosis transferrin and iron availability. Diabetes 31:1109–1114. doi: 10.2337/diacare.31.12.1109 [DOI] [PubMed] [Google Scholar]
- 145. Boelaert JR, Van Cutsem J, de Locht M, Schneider Y-J, Crichton RR. 1994. Deferoxamine augments growth and pathogenicity of Rhizopus, while hydroxypyridinone chelators have no effect. Kidney Int 45:667–671. doi: 10.1038/ki.1994.89 [DOI] [PubMed] [Google Scholar]
- 146. Brunet K, Arrivé F, Martellosio J-P, Lamarche I, Marchand S, Rammaert B. 2021. Corticosteroids alter alveolar macrophage control of Lichtheimia corymbifera spores in an ex vivo mouse model. Med Mycol 59:694–700. doi: 10.1093/mmy/myaa104 [DOI] [PubMed] [Google Scholar]
- 147. Chinn RY, Diamond RD. 1982. Generation of chemotactic factors by Rhizopus oryzae in the presence and absence of serum: relationship to hyphal damage mediated by human neutrophils and effects of hyperglycemia and ketoacidosis. Infect Immun 38:1123–1129. doi: 10.1128/iai.38.3.1123-1129.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Waldorf AR, Ruderman N, Diamond RD. 1984. Specific susceptibility to mucormycosis in murine diabetes and bronchoalveolar macrophage defense against Rhizopus. J Clin Invest 74:150–160. doi: 10.1172/JCI111395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Jorens PG, Boelaert JR, Halloy V, Zamora R, Schneider YJ, Herman AG. 1995. Human and rat macrophages mediate fungistatic activity against Rhizopus species differently: in vitro and ex vivo studies. Infect Immun 63:4489–4494. doi: 10.1128/iai.63.11.4489-4494.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Bath PM, Coleman CM, Gordon AL, Lim WS, Webb AJ. 2021. Nitric oxide for the prevention and treatment of viral, bacterial, protozoal and fungal infections. F1000Res 10:536. doi: 10.12688/f1000research.51270.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bogdan C. 2015. Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol 36:161–178. doi: 10.1016/j.it.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 152. Lundberg JO, Weitzberg E. 2022. Nitric oxide signaling in health and disease. Cell 185:2853–2878. doi: 10.1016/j.cell.2022.06.010 [DOI] [PubMed] [Google Scholar]
- 153. Soare AY, Bruno VM. 2024. Mucorales fungi suppress nitric oxide production by macrophages. mBio 15:e0284823. doi: 10.1128/mbio.02848-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Pagano L, Ricci P, Tonso A, Nosari A, Cudillo L, Montillo M, Cenacchi A, Pacilli L, Fabbiano F, Del Favero A, GIMEMA Infection Program . 1997. Mucormycosis in patients with haematological malignancies: a retrospective clinical study of 37 cases. Br J Haematol 99:331–336. doi: 10.1046/j.1365-2141.1997.3983214.x [DOI] [PubMed] [Google Scholar]
- 155. Waldorf AR, Diamond RD. 1985. Neutrophil chemotactic responses induced by fresh and swollen Rhizopus oryzae spores and Aspergillus fumigatus conidia. Infect Immun 48:458–463. doi: 10.1128/iai.48.2.458-463.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Inglesfield S, Jasiulewicz A, Hopwood M, Tyrrell J, Youlden G, Mazon-Moya M, Millington OR, Mostowy S, Jabbari S, Voelz K. 2018. Robust phagocyte recruitment controls the opportunistic fungal pathogen Mucor circinelloides in innate granulomas in vivo. mBio 9:e02010-17. doi: 10.1128/mBio.02010-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Levitz SM, Selsted ME, Ganz T, Lehrer RI, Diamond RD, Levitz SM, Selsted ME, Ganz T, Lehrer RI, Diamond RD. 1986. In vitro killing of spores and hyphae of Aspergillus fumigatus and Rhizopus oryzae by rabbit neutrophil cationic peptides and bronchoalveolar macrophages. J Infect Dis 154:483–489. doi: 10.1093/infdis/154.3.483 [DOI] [PubMed] [Google Scholar]
- 158. Gil-Lamaignere C, Simitsopoulou M, Roilides E, Maloukou A, Winn RM, Walsh TJ. 2005. Interferon‐γ and granulocyte‐macrophage colony‐stimulating factor augment the activity of polymorphonuclear leukocytes against medically important zygomycetes. J Infect Dis 191:1180–1187. doi: 10.1086/428503 [DOI] [PubMed] [Google Scholar]
- 159. Lewis RE, Georgiadou SP, Sampsonas F, Chamilos G, Kontoyiannis DP. 2014. Risk factors for early mortality in haematological malignancy patients with pulmonary mucormycosis. Mycoses 57:49–55. doi: 10.1111/myc.12101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kyvernitakis A, Torres HA, Jiang Y, Chamilos G, Lewis RE, Kontoyiannis DP. 2016. Initial use of combination treatment does not impact survival of 106 patients with haematologic malignancies and mucormycosis: a propensity score analysis. Clin Microbiol Infect 22:811. doi: 10.1016/j.cmi.2016.03.029 [DOI] [PubMed] [Google Scholar]
- 161. Chamilos G, Ganguly D, Lande R, Gregorio J, Meller S, Goldman WE, Gilliet M, Kontoyiannis DP. 2010. Generation of IL-23 producing dendritic cells (DCs) by airborne fungi regulates fungal pathogenicity via the induction of TH-17 responses. PLoS One 5:e12955. doi: 10.1371/journal.pone.0012955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Sun L, Zhang S, Wan Z, Li R, Yu J. 2021. In vivo and in vitro impairments in T helper cell and neutrophil responses against Mucor irregularis in Card9 knockout mice. Infect Immun 89:e00040-21. doi: 10.1128/IAI.00040-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Dos Santos AR, Fraga-Silva TF, Almeida D de F, Dos Santos RF, Finato AC, Amorim BC, Andrade MI, Soares CT, Lara VS, Almeida NL, de Arruda OS, de Arruda MS, Venturini J. 2020. Rhizopus-host interplay of disseminated mucormycosis in immunocompetent mice. Future Microbiol 15:739–752. doi: 10.2217/fmb-2019-0246 [DOI] [PubMed] [Google Scholar]
- 164. Dos Santos AR, Fraga-Silva TF, de Fátima Almeida-Donanzam D, Dos Santos RF, Finato AC, Soares CT, Lara VS, Almeida NLM, Andrade MI, de Arruda OS, de Arruda MSP, Venturini J. 2022. IFN-γ mediated signaling improves fungal clearance in experimental pulmonary mucormycosis. Mycopathologia 187:15–30. doi: 10.1007/s11046-021-00598-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Li CH, Cervantes M, Springer DJ, Boekhout T, Ruiz-Vazquez RM, Torres-Martinez SR, Heitman J, Lee SC. 2011. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog 7:e1002086. doi: 10.1371/journal.ppat.1002086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Lee SC, Li A, Calo S, Inoue M, Tonthat NK, Bain JM, Louw J, Shinohara ML, Erwig LP, Schumacher MA, Ko DC, Heitman J. 2015. Calcineurin orchestrates dimorphic transitions, antifungal drug responses and host-pathogen interactions of the pathogenic mucoralean fungus Mucor circinelloides. Mol Microbiol 97:844–865. doi: 10.1111/mmi.13071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Sassone-Corsi P. 2012. The cyclic AMP pathway. Cold Spring Harb Perspect Biol 4:a011148. doi: 10.1101/cshperspect.a011148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Lee SC, Li A, Calo S, Heitman J. 2013. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog 9:e1003625. doi: 10.1371/journal.ppat.1003625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Patiño-Medina JA, Reyes-Mares NY, Valle-Maldonado MI, Jácome-Galarza IE, Pérez-Arques C, Nuñez-Anita RE, Campos-García J, Anaya-Martínez V, Ortiz-Alvarado R, Ramírez-Díaz MI, Chan Lee S, Garre V, Meza-Carmen V. 2019. Heterotrimeric G-alpha subunits Gpa11 and Gpa12 define a transduction pathway that control spore size and virulence in Mucor circinelloides. PLoS One 14:e0226682. doi: 10.1371/journal.pone.0226682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Ibrahim AS, Gebremariam T, Lin L, Luo G, Husseiny MI, Skory CD, Fu Y, French SW, Edwards JE, Spellberg B. 2010. The high affinity iron permease is a key virulence factor required for Rhizopus oryzae pathogenesis. Mol Microbiol 77:587–604. doi: 10.1111/j.1365-2958.2010.07234.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Navarro-Mendoza MI, Pérez-Arques C, Murcia L, Martínez-García P, Lax C, Sanchis M, Capilla J, Nicolás FE, Garre V. 2018. Components of a new gene family of ferroxidases involved in virulence are functionally specialized in fungal dimorphism. Sci Rep 8:7660. doi: 10.1038/s41598-018-26051-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Liu M, Lin L, Gebremariam T, Luo G, Skory CD, French SW, Chou TF, Edwards JE, Ibrahim AS. 2015. Fob1 and Fob2 proteins are virulence determinants of Rhizopus oryzae via facilitating iron uptake from ferrioxamine. PLoS Pathog 11:e1004842. doi: 10.1371/journal.ppat.1004842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Pérez-Arques C, Navarro-Mendoza MI, Murcia L, Navarro E, Garre V, Nicolás FE. 2020. A non-canonical RNAi pathway controls virulence and genome stability in Mucorales. PLoS Genet 16:e1008611. doi: 10.1371/journal.pgen.1008611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Valle-Maldonado MI, Patiño-Medina JA, Pérez-Arques C, Reyes-Mares NY, Jácome-Galarza IE, Ortíz-Alvarado R, Vellanki S, Ramírez-Díaz MI, Lee SC, Garre V, Meza-Carmen V. 2020. The heterotrimeric G‐protein beta subunit Gpb1 controls hyphal growth under low oxygen conditions through the protein kinase A pathway and is essential for virulence in the fungus Mucor circinelloides. Cell Microbiol 22:e13236. doi: 10.1111/cmi.13236 [DOI] [PubMed] [Google Scholar]
- 175. Pérez-Arques C, Navarro-Mendoza MI, Murcia L, Navarro E, Garre V, Nicolás FE. 2021. The RNAi mechanism regulates a new exonuclease gene involved in the virulence of Mucorales. Int J Mol Sci 22:2282. doi: 10.3390/ijms22052282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Pérez-Arques C, Navarro-Mendoza MI, Murcia L, Lax C, Sanchis M, Capilla J, Navarro E, Garre V, Nicolás FE. 2021. A mucoralean white collar-1 photoreceptor controls virulence by regulating an intricate gene network during host interactions. Microorganisms 9:459. doi: 10.3390/microorganisms9020459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Nguyen KB, Sreelatha A, Durrant ES, Lopez-Garrido J, Muszewska A, Dudkiewicz M, Grynberg M, Yee S, Pogliano K, Tomchick DR, Pawłowski K, Dixon JE, Tagliabracci VS. 2016. Phosphorylation of spore coat proteins by a family of atypical protein kinases. Proc Natl Acad Sci U S A 113:E3482–E3491. doi: 10.1073/pnas.1605917113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. McKenney PT, Driks A, Eichenberger P. 2013. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol 11:33–44. doi: 10.1038/nrmicro2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Freitas C, Plannic J, Isticato R, Pelosi A, Zilhão R, Serrano M, Baccigalupi L, Ricca E, Elsholz AKW, Losick R, O Henriques A. 2020. A protein phosphorylation module patterns the Bacillus subtilis spore outer coat. Mol Microbiol 114:934–951. doi: 10.1111/mmi.14562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Ibrahim IM, Abdelmalek DH, Elfiky AA. 2019. GRP78: a cell’s response to stress. Life Sci 226:156–163. doi: 10.1016/j.lfs.2019.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Zhang Y, Liu R, Ni M, Gill P, Lee AS. 2010. Cell surface relocalization of the endoplasmic reticulum chaperone and unfolded protein response regulator GRP78/BiP. J Biol Chem 285:15065–15075. doi: 10.1074/jbc.M109.087445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Gebremariam T, Alkhazraji S, Soliman SSM, Gu Y, Jeon HH, Zhang L, French SW, Stevens DA, Edwards JE, Filler SG, Uppuluri P, Ibrahim AS. 2019. Anti-CotH3 antibodies protect mice from mucormycosis by prevention of invasion and augmenting opsonophagocytosis. Sci Adv 5:eaaw1327. doi: 10.1126/sciadv.aaw1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Mak KM, Mei R. 2017. Basement membrane type IV collagen and laminin: an overview of their biology and value as fibrosis biomarkers of liver disease. Anat Rec 300:1371–1390. doi: 10.1002/ar.23567 [DOI] [PubMed] [Google Scholar]
- 184. Bouchara JP, Oumeziane NA, Lissitzky JC, Larcher G, Tronchin G, Chabasse D. 1996. Attachment of spores of the human pathogenic fungus Rhizopus oryzae to extracellular matrix components. Eur J Cell Biol 70:76–83. [PubMed] [Google Scholar]
- 185. Huang Y, Kyriakides TR. 2020. The role of extracellular matrix in the pathophysiology of diabetic wounds. Matrix Biol Plus 6–7:100037. doi: 10.1016/j.mbplus.2020.100037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wächtler B, Citiulo F, Jablonowski N, Förster S, Dalle F, Schaller M, Wilson D, Hube B. 2012. Candida albicans-epithelial interactions: dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PLoS One 7:e36952. doi: 10.1371/journal.pone.0036952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Zhu W, Phan QT, Boontheung P, Solis NV, Loo JA, Filler SG. 2012. EGFR and HER2 receptor kinase signaling mediate epithelial cell invasion by Candida albicans during oropharyngeal infection. Proc Natl Acad Sci U S A 109:14194–14199. doi: 10.1073/pnas.1117676109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Liu H, Lee MJ, Solis NV, Phan QT, Swidergall M, Ralph B, Ibrahim AS, Sheppard DC, Filler SG. 2016. Aspergillus fumigatus CalA binds to integrin α5β1 and mediates host cell invasion. Nat Microbiol 2:16211. doi: 10.1038/nmicrobiol.2016.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Upadhyay SK, Mahajan L, Ramjee S, Singh Y, Basir SF, Madan T. 2009. Identification and characterization of a laminin-binding protein of Aspergillus fumigatus: extracellular thaumatin domain protein (AfCalAp). J Med Microbiol 58:714–722. doi: 10.1099/jmm.0.005991-0 [DOI] [PubMed] [Google Scholar]
- 190. Soliman SSM, El-Labbad EM, Abu-Qiyas A, Fayed B, Hamoda AM, Al-Rawi AM, Dakalbab S, El-Shorbagi ANA, Hamad M, Ibrahim AS, Mohammad MG. 2022. Novel secreted peptides from Rhizopus arrhizus var. delemar with immunomodulatory effects that enhance fungal pathogenesis. Front Microbiol 13:863133. doi: 10.3389/fmicb.2022.863133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Kobayashi DY, Crouch JA. 2009. Bacterial/fungal interactions: from pathogens to mutualistic endosymbionts. Annu Rev Phytopathol 47:63–82. doi: 10.1146/annurev-phyto-080508-081729 [DOI] [PubMed] [Google Scholar]
- 192. Pawlowska TE, Gaspar ML, Lastovetsky OA, Mondo SJ, Real-Ramirez I, Shakya E, Bonfante P. 2018. Biology of fungi and their bacterial endosymbionts. Annu Rev Phytopathol 56:289–309. doi: 10.1146/annurev-phyto-080417-045914 [DOI] [PubMed] [Google Scholar]
- 193. Carter ME, Carpenter SCD, Dubrow ZE, Sabol MR, Rinaldi FC, Lastovetsky OA, Mondo SJ, Pawlowska TE, Bogdanove AJ. 2020. A TAL effector-like protein of an endofungal bacterium increases the stress tolerance and alters the transcriptome of the host. Proc Natl Acad Sci U S A 117:17122–17129. doi: 10.1073/pnas.2003857117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Mondo SJ, Lastovetsky OA, Gaspar ML, Schwardt NH, Barber CC, Riley R, Sun H, Grigoriev IV, Pawlowska TE. 2017. Bacterial endosymbionts influence host sexuality and reveal reproductive genes of early divergent fungi. Nat Commun 8:1843. doi: 10.1038/s41467-017-02052-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Lackner G, Moebius N, Hertweck C. 2011. Endofungal bacterium controls its host by an HRP type III secretion system. ISME J 5:252–261. doi: 10.1038/ismej.2010.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Partida-Martinez L.P, Hertweck C. 2005. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437:884–888. doi: 10.1038/nature03997 [DOI] [PubMed] [Google Scholar]
- 197. Scherlach K, Partida-Martinez LP, Dahse H-M, Hertweck C. 2006. Antimitotic rhizoxin derivatives from a cultured bacterial endosymbiont of the rice pathogenic fungus Rhizopus microsporus. J Am Chem Soc 128:11529–11536. doi: 10.1021/ja062953o [DOI] [PubMed] [Google Scholar]
- 198. Partida-Martinez LP, de Looss CF, Ishida K, Ishida M, Roth M, Buder K, Hertweck C. 2007. Rhizonin, the first mycotoxin isolated from the zygomycota, is not a fungal metabolite but is produced by bacterial endosymbionts. Appl Environ Microbiol 73:793–797. doi: 10.1128/AEM.01784-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Ibrahim AS, Gebremariam T, Liu M, Chamilos G, Kontoyiannis D, Mink R, Kwon-Chung KJ, Fu Y, Skory CD, Edwards JE, Spellberg B. 2008. Bacterial endosymbiosis is widely present among zygomycetes but does not contribute to the pathogenesis of mucormycosis. J Infect Dis 198:1083–1090. doi: 10.1086/591461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Partida-Martinez LP, Bandemer S, Rüchel R, Dannaoui E, Hertweck C. 2008. Lack of evidence of endosymbiotic toxin‐producing bacteria in clinical Rhizopus isolates. Mycoses 51:266–269. doi: 10.1111/j.1439-0507.2007.01477.x [DOI] [PubMed] [Google Scholar]
- 201. Itabangi H, Sephton-Clark PCS, Tamayo DP, Zhou X, Starling GP, Mahamoud Z, Insua I, Probert M, Correia J, Moynihan PJ, Gebremariam T, Gu Y, Ibrahim AS, Brown GD, King JS, Ballou ER, Voelz K. 2022. A bacterial endosymbiont of the fungus Rhizopus microsporus drives phagocyte evasion and opportunistic virulence. Curr Biol 32:1115–1130. doi: 10.1016/j.cub.2022.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Kehl-Fie TE, Skaar EP. 2010. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol 14:218–224. doi: 10.1016/j.cbpa.2009.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen–host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Gebremariam T, Lin L, Liu M, Kontoyiannis DP, French S, Edwards JE, Filler SG, Ibrahim AS. 2016. Bicarbonate correction of ketoacidosis alters host-pathogen interactions and alleviates mucormycosis. J Clin Invest 126:2280–2294. doi: 10.1172/JCI82744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Philpott CC. 2006. Iron uptake in fungi: a system for every source. Biochim Biophys Acta 1763:636–645. doi: 10.1016/j.bbamcr.2006.05.008 [DOI] [PubMed] [Google Scholar]
- 206. Carroll CS, Grieve CL, Murugathasan I, Bennet AJ, Czekster CM, Liu H, Naismith J, Moore MM. 2017. The rhizoferrin biosynthetic gene in the fungal pathogen Rhizopus delemar is a novel member of the NIS gene family. Int J Biochem Cell Biol 89:136–146. doi: 10.1016/j.biocel.2017.06.005 [DOI] [PubMed] [Google Scholar]
- 207. Lindahl PA. 2019. A comprehensive mechanistic model of iron metabolism in Saccharomyces cerevisiae. Metallomics 11:1779–1799. doi: 10.1039/c9mt00199a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Hassett RF, Romeo AM, Kosman DJ. 1998. Regulation of high affinity iron uptake in the yeast Saccharomyces cerevisiae. J Biol Chem 273:7628–7636. doi: 10.1074/jbc.273.13.7628 [DOI] [PubMed] [Google Scholar]
- 209. Schwartze VU, Winter S, Shelest E, Marcet-Houben M, Horn F, Wehner S, Linde J, Valiante V, Sammeth M, Riege K, Nowrousian M, Kaerger K, Jacobsen ID, Marz M, Brakhage AA, Gabaldón T, Böcker S, Voigt K. 2014. Gene expansion shapes genome architecture in the human pathogen Lichtheimia corymbifera: an evolutionary genomics analysis in the ancient terrestrial Mucorales (Mucoromycotina). PLoS Genet 10:e1004496. doi: 10.1371/journal.pgen.1004496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Thieken A, Winkelmann G. 1992. Rhizoferrin: a complexone type siderophore of the Mucorales and Entomophthorales (zygomycetes). FEMS Microbiol Lett 73:37–41. doi: 10.1016/0378-1097(92)90579-d [DOI] [PubMed] [Google Scholar]
- 211. Škríba A, Patil RH, Hubáček P, Dobiáš R, Palyzová A, Marešová H, Pluháček T, Havlíček V. 2020. Rhizoferrin glycosylation in Rhizopus microsporus. J Fungi (Basel) 6:89. doi: 10.3390/jof6020089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. de Locht M, Boelaert JR, Schneider YJ. 1994. Iron uptake from ferrioxamine and from ferrirhizoferrin by germinating spores of Rhizopus microsporus. Biochem Pharmacol 47:1843–1850. doi: 10.1016/0006-2952(94)90314-x [DOI] [PubMed] [Google Scholar]
- 213. Boelaert JR, de Locht M, Van Cutsem J, Kerrels V, Cantinieaux B, Verdonck A, Van Landuyt HW, Schneider YJ. 1993. Mucormycosis during deferoxamine therapy is a siderophore-mediated infection. In vitro and in vivo animal studies. J Clin Invest 91:1979–1986. doi: 10.1172/JCI116419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Bartnicki-Garcia S. 1963. Symposium on biochemical bases of morphogenesis in fungi. III. Mold-yeast dimorphism of Mucor. Bacteriol Rev 27:293–304. doi: 10.1128/br.27.3.293-304.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Pasteur L. 1876. Etudes sur la biere [Google Scholar]
- 216. Orlowski M. 1991. Mucor dimorphism. Microbiol Rev 55:234–258. doi: 10.1128/mr.55.2.234-258.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Wolff AM, Appel KF, Petersen JB, Poulsen U, Arnau J. 2002. Identification and analysis of genes involved in the control of dimorphism in Mucor circinelloides (syn. racemosus). FEMS Yeast Res 2:203–213. doi: 10.1111/j.1567-1364.2002.tb00085.x [DOI] [PubMed] [Google Scholar]
- 218. Bartnicki-García S, Nickerson WJ. 1962. Nutrition, growth, and morphogenesis of Mucor rouxii. J Bacteriol 84:841–858. doi: 10.1128/jb.84.4.841-858.1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Bartnicki-Garcia S, Nickerson WJ. 1962. Induction of yeast-like development in Mucor by carbon dioxide. J Bacteriol 84:829–840. doi: 10.1128/jb.84.4.829-840.1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Bartnicki-Garcia S, Nickerson WJ. 1962. Assimilation of carbon dioxide and morphogenesis of Mucor rouxii. Biochim Biophys Acta 64:548–551. doi: 10.1016/0006-3002(62)90314-1 [DOI] [PubMed] [Google Scholar]
- 221. Friedenthal M, Epstein A, Passeron S. 1974. Effect of potassium cyanide, glucose and anaerobiosis on morphogenesis of Mucor rouxii. Microbiology 82:15–24. doi: 10.1099/00221287-82-1-15 [DOI] [Google Scholar]
- 222. Schulz BE, Kraepelin G, Hinkelmann W. 1974. Factors affecting dimorphism in Mycotypha (Mucorales): a correlation with the fermentation/respiration equilibrium. J Gen Microbiol 82:1–13. doi: 10.1099/00221287-82-1-1 [DOI] [PubMed] [Google Scholar]
- 223. Clark-Walker GD. 1973. Relationship between dimorphology and respiration in Mucor genevensis studied with chloramphenicol. J Bacteriol 116:972–980. doi: 10.1128/jb.116.2.972-980.1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Inderlied CB, Cihlar RL, Sypherd PS. 1980. Regulation of ornithine decarboxylase during morphogenesis of Mucor racemosus. J Bacteriol 141:699–706. doi: 10.1128/jb.141.2.699-706.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Martinez-Pacheco M, Rodriguez G, Reyna G, Calvo-Mendez C, Ruiz-Herrera J. 1989. Inhibition of the yeast-mycelial transition and the phorogenesis of Mucorales by diamino butanone. Arch Microbiol 151:10–14. doi: 10.1007/BF00444661 [DOI] [PubMed] [Google Scholar]
- 226. Ruiz-Herrera J, Calvo-Mendez C. 1987. Effect of ornithine decarboxylase inhibitors on the germination of sporangiospores of Mucorales. Exp Mycol 11:287–296. doi: 10.1016/0147-5975(87)90017-X [DOI] [Google Scholar]
- 227. Calvo-Mendez C, Martinez-Pacheco M, Ruiz-Herrera J. 1987. Regulation of ornithine decarboxylase activity in Mucor bacilliformis and Mucor rouxii. Exp Mycol 11:270–277. doi: 10.1016/0147-5975(87)90015-6 [DOI] [Google Scholar]
- 228. Ito ET, Cihlar RL, Inderlied CB. 1982. Lipid synthesis during morphogenesis of Mucor racemosus. J Bacteriol 152:880–887. doi: 10.1128/jb.152.2.880-887.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Jacobsen ID, Wilson D, Wächtler B, Brunke S, Naglik JR, Hube B. 2012. Candida albicans dimorphism as a therapeutic target . Expert Rev Anti Infect Ther 10:85–93. doi: 10.1586/eri.11.152 [DOI] [PubMed] [Google Scholar]
- 230. Fradin C, Kretschmar M, Nichterlein T, Gaillardin C, d’Enfert C, Hube B. 2003. Stage‐specific gene expression of Candida albicans in human blood. Mol Microbiol 47:1523–1543. doi: 10.1046/j.1365-2958.2003.03396.x [DOI] [PubMed] [Google Scholar]
- 231. Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Köhler JR, Kadosh D, Lopez-Ribot JL. 2010. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog 6:e1000828. doi: 10.1371/journal.ppat.1000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Phan QT, Belanger PH, Filler SG. 2000. Role of hyphal formation in interactions of Candida albicans with endothelial cells. Infect Immun 68:3485–3490. doi: 10.1128/IAI.68.6.3485-3490.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Wächtler B, Wilson D, Haedicke K, Dalle F, Hube B. 2011. From attachment to damage: defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS One 6:e17046. doi: 10.1371/journal.pone.0017046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Boyce KJ, Andrianopoulos A. 2015. Fungal dimorphism: the switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. FEMS Microbiol Rev 39:797–811. doi: 10.1093/femsre/fuv035 [DOI] [PubMed] [Google Scholar]
- 235. Ocampo J, Fernandez Nuñez L, Silva F, Pereyra E, Moreno S, Garre V, Rossi S. 2009. A subunit of protein kinase a regulates growth and differentiation in the fungus Mucor circinelloides. Eukaryot Cell 8:933–944. doi: 10.1128/EC.00026-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Patiño-Medina JA, Valle-Maldonado MI, Maldonado-Herrera G, Pérez-Arques C, Jácome-Galarza IE, Díaz-Pérez C, Díaz-Pérez AL, Araiza-Cervantes CA, Villagomez-Castro JC, Campos-García J, Ramírez-Díaz MI, Garre V, Meza-Carmen V. 2019. Role of Arf-like proteins (Arl1 and Arl2) of Mucor circinelloides in virulence and antifungal susceptibility. Fungal Genet Biol 129:40–51. doi: 10.1016/j.fgb.2019.04.011 [DOI] [PubMed] [Google Scholar]
- 237. Warkentien TE, Shaikh F, Weintrob AC, Rodriguez CJ, Murray CK, Lloyd BA, Ganesan A, Aggarwal D, Carson ML, Tribble DR, Infectious Disease Clinical Research Program Trauma Infectious Disease Outcomes Study Group . 2015. Impact of Mucorales and other invasive molds on clinical outcomes of polymicrobial traumatic wound infections. J Clin Microbiol 53:2262–2270. doi: 10.1128/JCM.00835-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Chander J, Stchigel AM, Alastruey-Izquierdo A, Jayant M, Bala K, Rani H, Handa U, Punia RS, Dalal U, Attri AK, Monzon A, Cano-Lira JF, Guarro J. 2015. Fungal necrotizing fasciitis, an emerging infectious disease caused by Apophysomyces (Mucorales). Rev Iberoam Micol 32:93–98. doi: 10.1016/j.riam.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 239. Silva F, Torres-Martínez S, Garre V. 2006. Distinct white collar-1 genes control specific light responses in Mucor circinelloides. Mol Microbiol 61:1023–1037. doi: 10.1111/j.1365-2958.2006.05291.x [DOI] [PubMed] [Google Scholar]
- 240. Ruiz-Roldán MC, Garre V, Guarro J, Mariné M, Roncero MIG. 2008. Role of the white collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum. Eukaryot Cell 7:1227–1230. doi: 10.1128/EC.00072-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Canessa P, Schumacher J, Hevia MA, Tudzynski P, Larrondo LF. 2013. Assessing the effects of light on differentiation and virulence of the plant pathogen Botrytis cinerea: characterization of the white collar complex. PLoS ONE 8:e84223. doi: 10.1371/journal.pone.0084223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Zhu P, Idnurm A. 2018. The contribution of the white collar complex to Cryptococcus neoformans virulence is independent of its light-sensing capabilities. Fungal Genet Biol 121:56–64. doi: 10.1016/j.fgb.2018.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Borman AM, Fraser M, Patterson Z, Palmer MD, Johnson EM. 2021. In vitro antifungal drug resistance profiles of clinically relevant members of the Mucorales (Mucoromycota) especially with the newer triazoles. J Fungi 7:271. doi: 10.3390/jof7040271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Sabra R, Branch RA. 1990. Amphotericin B nephrotoxicity. Drug Saf 5:94–108. doi: 10.2165/00002018-199005020-00003 [DOI] [PubMed] [Google Scholar]
- 245. Shafiei M, Peyton L, Hashemzadeh M, Foroumadi A. 2020. History of the development of antifungal azoles: a review on structures, SAR, and mechanism of action. Bioorg Chem 104:104240. doi: 10.1016/j.bioorg.2020.104240 [DOI] [PubMed] [Google Scholar]
- 246. Lass-Flörl C. 2011. Triazole antifungal agents in invasive fungal infections: a comparative review. Drugs 71:2405–2419. doi: 10.2165/11596540-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 247. Watson PF, Rose ME, Ellis SW, England H, Kelly SL. 1989. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem Biophys Res Commun 164:1170–1175. doi: 10.1016/0006-291x(89)91792-0 [DOI] [PubMed] [Google Scholar]
- 248. Lamoth F, Lewis RE, Kontoyiannis DP. 2022. Investigational antifungal agents for invasive mycoses: a clinical perspective. Clin Infect Dis 75:534–544. doi: 10.1093/cid/ciab1070 [DOI] [PubMed] [Google Scholar]
- 249. Chowdhary A, Singh PK, Kathuria S, Hagen F, Meis JF. 2015. Comparison of the EUCAST and CLSI broth microdilution methods for testing isavuconazole, posaconazole, and amphotericin B against molecularly identified Mucorales species. Antimicrob Agents Chemother 59:7882–7887. doi: 10.1128/AAC.02107-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Luo G, Gebremariam T, Lee H, Edwards JE, Kovanda L, Ibrahim AS. 2014. Isavuconazole therapy protects immunosuppressed mice from mucormycosis. Antimicrob Agents Chemother 58:2450–2453. doi: 10.1128/AAC.02301-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Cornely OA, Arikan-Akdagli S, Dannaoui E, Groll AH, Lagrou K, Chakrabarti A, Lanternier F, Pagano L, Skiada A, Akova M, et al. 2014. ESCMID and ECMM joint clinical guidelines for the diagnosis and management of mucormycosis 2013. Clin Microbiol Infect 20 Suppl 3:5–26. doi: 10.1111/1469-0691.12371 [DOI] [PubMed] [Google Scholar]
- 252. Vitale RG, de Hoog GS, Schwarz P, Dannaoui E, Deng S, Machouart M, Voigt K, van de Sande WWJ, Dolatabadi S, Meis JF, Walther G. 2012. Antifungal susceptibility and phylogeny of opportunistic members of the order Mucorales. J Clin Microbiol 50:66–75. doi: 10.1128/JCM.06133-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Alastruey-Izquierdo A, Castelli MV, Cuesta I, Zaragoza O, Monzón A, Mellado E, Rodríguez-Tudela JL. 2009. In vitro activity of antifungals against zygomycetes. Clin Microbiol Infect 15:71–76. doi: 10.1111/j.1469-0691.2009.02984.x [DOI] [PubMed] [Google Scholar]
- 254. Almyroudis NG, Sutton DA, Fothergill AW, Rinaldi MG, Kusne S. 2007. In vitro susceptibilities of 217 clinical isolates of zygomycetes to conventional and new antifungal agents. Antimicrob Agents Chemother 51:2587–2590. doi: 10.1128/AAC.00452-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Ritz N, Ammann RA, Aebischer CC, Gugger M, Jaton K, Schmid RA, Aebi C. 2005. Failure of voriconazole to cure disseminated zygomycosis in an immunocompromised child. Eur J Pediatr 164:231–235. doi: 10.1007/s00431-004-1606-7 [DOI] [PubMed] [Google Scholar]
- 256. White TC. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob Agents Chemother 41:1488–1494. doi: 10.1128/AAC.41.7.1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Sanglard D, Ischer F, Koymans L, Bille J. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α- demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother 42:241–253. doi: 10.1128/AAC.42.2.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Leonardelli F, Macedo D, Dudiuk C, Cabeza MS, Gamarra S, Garcia-Effron G. 2016. Aspergillus fumigatus intrinsic fluconazole resistance is due to the naturally occurring T301I substitution in Cyp51Ap. Antimicrob Agents Chemother 60:5420–5426. doi: 10.1128/AAC.00905-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Hagiwara D, Watanabe A, Kamei K, Goldman GH. 2016. Epidemiological and genomic landscape of azole resistance mechanisms in Aspergillus fungi. Front Microbiol 7:1382. doi: 10.3389/fmicb.2016.01382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Caramalho R, Tyndall JDA, Monk BC, Larentis T, Lass-Flörl C, Lackner M. 2017. Intrinsic short-tailed azole resistance in mucormycetes is due to an evolutionary conserved aminoacid substitution of the lanosterol 14α-demethylase. Sci Rep 7:15898. doi: 10.1038/s41598-017-16123-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Sagatova AA, Keniya MV, Wilson RK, Sabherwal M, Tyndall JDA, Monk BC. 2016. Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14α-demethylase. Sci Rep 6:26213. doi: 10.1038/srep26213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Idnurm A, Xu M. 2022. Identification of the ergC gene involved in polyene drug sensitivity in the Mucorales species Phycomyces blakesleeanus. Mol Biol Rep 49:981–987. doi: 10.1007/s11033-021-06917-6 [DOI] [PubMed] [Google Scholar]
- 263. Bauer K, Rafael B, Vágó B, Kiss-Vetráb S, Molnár A, Szebenyi C, Varga M, Szekeres A, Vágvölgyi C, Papp T, Nagy G. 2023. Characterization of the sterol 24-C-methyltransferase genes reveals a network of alternative sterol biosynthetic pathways in Mucor lusitanicus Microbiol Spectr 11:e0031523. doi: 10.1128/spectrum.00315-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Denning DW. 2002. Echinocandins: a new class of antifungal. J Antimicrob Chemother 49:889–891. doi: 10.1093/jac/dkf045 [DOI] [PubMed] [Google Scholar]
- 265. Garcia A, Huh EY, Lee SC. 2023. Serine/threonine phosphatase calcineurin orchestrates the intrinsic resistance to micafungin in the human-pathogenic fungus Mucor circinelloides. Antimicrob Agents Chemother 67:e0068622. doi: 10.1128/aac.00686-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Fraczek MG, Bromley M, Buied A, Moore CB, Rajendran R, Rautemaa R, Ramage G, Denning DW, Bowyer P. 2013. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J Antimicrob Chemother 68:1486–1496. doi: 10.1093/jac/dkt075 [DOI] [PubMed] [Google Scholar]
- 267. Holmes AR, Cardno TS, Strouse JJ, Ivnitski-Steele I, Keniya MV, Lackovic K, Monk BC, Sklar LA, Cannon RD. 2016. Targeting efflux pumps to overcome antifungal drug resistance. Future Med Chem 8:1485–1501. doi: 10.4155/fmc-2016-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Rocha MFG, Bandeira SP, de Alencar LP, Melo LM, Sales JA, Paiva M de AN, Teixeira CEC, Castelo-Branco D de SCM, Pereira-Neto W de A, Cordeiro R de A, Sidrim JJC, Brilhante RSN. 2017. Azole resistance in Candida albicans from animals: highlights on efflux pump activity and gene overexpression. Mycoses 60:462–468. doi: 10.1111/myc.12611 [DOI] [PubMed] [Google Scholar]
- 269. Chang M, Sionov E, Khanal Lamichhane A, Kwon-Chung KJ, Chang YC. 2018. Roles of three Cryptococcus neoformans and Cryptococcus gattii efflux pump-coding genes in response to drug treatment. Antimicrob Agents Chemother 62:e01751-17. doi: 10.1128/AAC.01751-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Nagy G, Kiss S, Varghese R, Bauer K, Szebenyi C, Kocsubé S, Homa M, Bodai L, Zsindely N, Nagy G, Vágvölgyi C, Papp T. 2021. Characterization of three pleiotropic drug resistance transporter genes and their participation in the azole resistance of Mucor circinelloides. Front Cell Infect Microbiol 11:660347. doi: 10.3389/fcimb.2021.660347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Calo S, Shertz-Wall C, Lee SC, Bastidas RJ, Nicolás FE, Granek JA, Mieczkowski P, Torres-Martínez S, Ruiz-Vázquez RM, Cardenas ME, Heitman J. 2014. Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature 513:555–558. doi: 10.1038/nature13575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Calo S, Nicolás FE, Lee SC, Vila A, Cervantes M, Torres-Martinez S, Ruiz-Vazquez RM, Cardenas ME, Heitman J. 2017. A non-canonical RNA degradation pathway suppresses RNAi-dependent epimutations in the human fungal pathogen Mucor circinelloides. PLoS Genet 13:e1006686. doi: 10.1371/journal.pgen.1006686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Boeke JD, Trueheart J, Natsoulis G, Fink GR. 1987. 5-fluoroorotic acid as a selective agent in yeast molecular genetics, p 164–175. In Methods in enzymology [DOI] [PubMed] [Google Scholar]
- 274. Chang Z, Billmyre RB, Lee SC, Heitman J. 2019. Broad antifungal resistance mediated by RNAi-dependent epimutation in the basal human fungal pathogen Mucor circinelloides. PLoS Genet 15:e1007957. doi: 10.1371/journal.pgen.1007957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Chang Z, Heitman J. 2019. Drug-resistant epimutants exhibit organ-specific stability and induction during murine infections caused by the human fungal pathogen Mucor circinelloides. mBio 10:e02579-19. doi: 10.1128/mBio.02579-19 [DOI] [PMC free article] [PubMed] [Google Scholar]