Abstract
TFAP2C is a well-established regulator of the trophoblast lineage in mice and humans, but a handful of studies indicate that TFAP2C may play an important role in pluripotency. Here we hypothesize and provide new evidence that TFAP2C functions as an activator of trophoblast and pluripotency genes during preimplantation embryo development.
Keywords: TFAP2C, Preimplantation embryo, First cell-fate decision, Pluripotency, Hippo signaling
Preimplantation embryo development comprises a series of molecular and cellular events that transform a totipotent zygote into a multi-lineage blastocyst. The first cell-fate decision occurs during the morula-to-blastocyst transition and involves the formation of the multipotent trophoblast and pluripotent inner cell mass (ICM) lineages (Karasek et al. 2020). Importantly, the underlying transcriptional mechanism(s) that govern the switch from embryonic totipotency to multipotency and pluripotency cellular states are not well established. The elucidation of these molecular mechanisms is of great importance to the fields of stem cell biology and reproductive biology.
Transcription factor AP2 gamma (TFAP2C) represents a well-established regulator of the trophoblast lineage in mammalian embryos. Most studies on TFAP2C in early embryos and during pregnancy indicate that it functions as a pro-trophoblast lineage transcription factor (TF). For example, earlier studies using knockout mice demonstrated that TFAP2C is required for placental development (Werling and Schorle, 2002). Conversely, forced expression of TFAP2C in mouse embryonic stem (ES) cells induces a trophoblast cell-fate (Kuckenberg et al. 2010). Altogether, these studies show that TFAP2C is crucial for trophoblast lineage development.
Remarkably, a handful of studies in mouse preimplantation embryos and human ES cells indicate that TFAP2C may play a more diverse role in lineage formation and/or the establishment of pluripotency. In this Point of View article, we hypothesize and provide new experimental evidence that TFAP2C governs the first cell-fate decision in mouse preimplantation embryos by positively regulating both pluripotency and trophoblast lineage genes. We postulate that TFAP2C accomplishes this by forming interactions with distinct regulatory proteins in the outer and inner blastomeres of morulae.
The molecular mechanism by which TFAP2C regulates trophoblast lineage formation was revealed by two landmark studies published by our laboratory over the last decade. These studies revealed that TFAP2C functions as a master regulator of genes required for apical cell polarity, tight junction biogenesis, and blastocoele formation (Choi et al. 2012). Furthermore, loss-of-function and gain-of-function studies revealed that TFAP2C promotes trophoblast lineage specification by activation of Caudal-type homeobox 2 (Cdx2) (Cao et al. 2015). TFAP2C can positively regulate Cdx2 expression via two mechanisms. The first mechanism involves transcriptional activation of Cdx2 by binding to an enhancer (Cao et al. 2015). The second mechanism involves negative regulation of the HIPPO signaling pathway in the outer cells of morulae by activation of cell polarity and Rho-associated protein kinase (Rock) genes (Cao et al. 2015). A more recent study by another group provided evidence that TFAP2C may cooperate with Tea domain transcription factor 4 (TEAD4) and Ras homolog family member (RHOA) to regulate the onset of cell polarization in mouse preimplantation embryos (Zhu et al. 2020). Altogether, these studies highlight the importance of TFAP2C in cellular processes that underlie trophoblast lineage formation in preimplantation embryos.
Despite the preponderance of evidence supporting a role for TFAP2C in different aspects of trophoblast biology, we postulate that TFAP2C plays an important function in the establishment of pluripotency. Evidence in support of this comes from three intriguing reports. An earlier publication from our laboratory demonstrated that TFAP2C is not required for Oct4 repression during trophoblast lineage formation in mouse preimplantation embryos, but instead is required for Oct4 activation and/or maintenance (Choi et al. 2013). Chromatin immunoprecipitation (ChIP) experiments in mouse ES cells revealed that TFAP2C binding was enriched at an OCT4/SOX2 binding motif located within the Oct4 distal enhancer (Choi et al. 2013). In support of this finding, a proximity ligation assay (PLA) revealed that OCT4 and TFAP2C proteins physically interact in the nuclei of preimplantation embryos (Choi et al. 2013). Two recent studies in human naïve and primed ES cells revealed a vital role for TFAP2C in controlling OCT4 expression by binding to an OCT4 enhancer and promoting open chromatin (Pastor et al. 2018, Chen et al. 2018). Collectively, these studies indicate that TFAP2C may play a pivotal role in the establishment and/or maintenance of pluripotency gene expression in mice and humans.
To further investigate the role of TFAP2C in pluripotency gene expression, we conducted a series of loss-of-function and gain-of-function experiments in mouse preimplantation embryos (Fig. 1). For these experiments, we focused on Oct4 and Nanog. Oct4 is both maternally and zygotically expressed (Jaenisch & Young 2008), whereas Nanog is only zygotically expressed after the 2-cell stage (Miyanari & Torres-Padilla 2012). Recently, we conducted an extensive series of experiments examining the role of TFAP2C in Sox2 regulation, and we have a manuscript in preparation supporting a direct role of TFAP2C in Sox2 expression and localization. In the first set of experiments, we microinjected either 100 μM Tfap2c or control siRNA into fertilized 1-cell embryos and cultured them to the 8-cell and morula stages. Quantitative real-time PCR (qRT-PCR) analysis revealed that Oct4, Nanog, and Tfap2c transcripts were significantly reduced at the 8-cell and morula stages in Tfap2c siRNA injected embryos (Fig. 1a). TFAP2C protein was undetectable by immunofluorescence (data not shown), like we previously showed (Choi et al. 2012, Cao et al. 2015).
Figure 1.
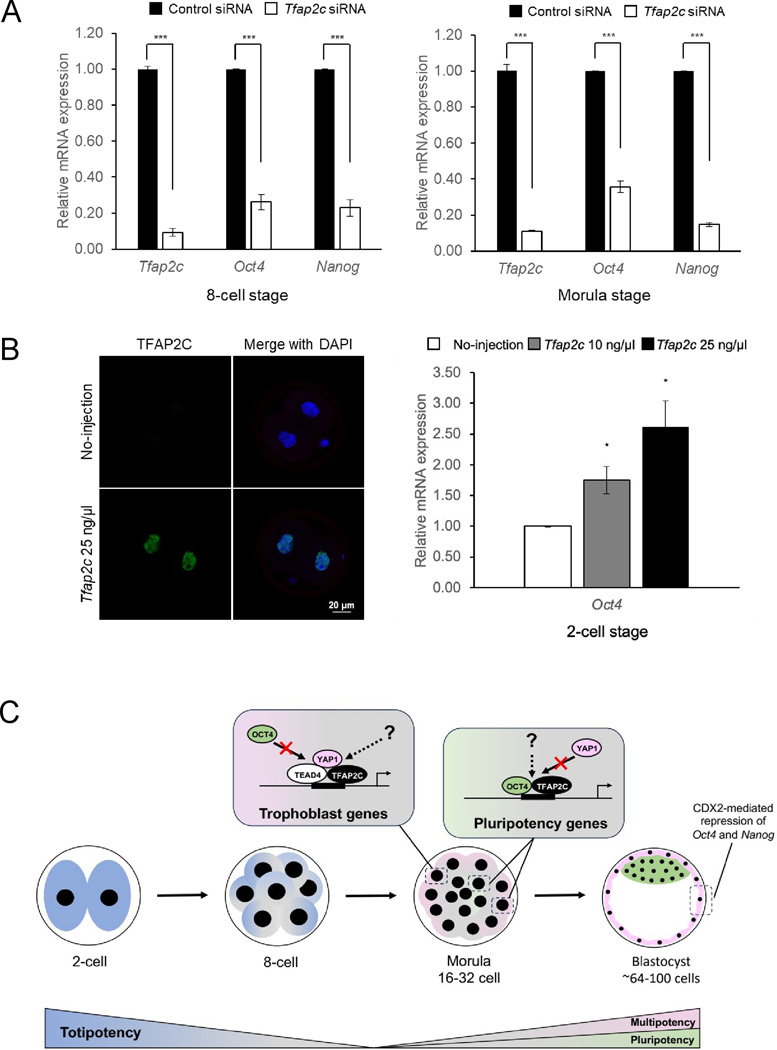
Additional evidence supporting a role for TFAP2C in positive regulation of pluripotency genes and a working model proposing how TFAP2C regulates conflicting cell-fates. (A) qRT-PCR analysis of Tfap2c, Oct4, and Nanog transcripts in TFAP2C knockdown embryos at the 8-cell and morula stages. Ubtf was used as an endogenous control. Values are means ± SD. *** P<0.001 (data shown are from n=3 biological replicates; Student’s t-test). (B) Immunofluorescence staining of TFAP2C in control embryos and embryos microinjected with Tfap2c cRNA. Embryos were counterstained with DAPI. Scale bar = 20μm. qRT-PCR analysis of Oct4 transcripts in 2-cell embryos overexpressing TFAP2C. Ubtf was used as an endogenous control. Values are means ± SD. * P<0.05 (data shown are from n=3 biological replicates, Student’s t-test). (C) Working model illustrating how TFAP2C regulates pluripotency and trophoblast genes in preimplantation embryos. Between the 2-cell and 8-cell stages, there is a decrease in totipotency, and then during the 8-cell and morula stages, there is a switch to pluripotent and multipotent cellular states. Between the 8-cell and morula stages, TFAP2C activates or upregulates the expression of trophoblast genes such as Cdx2 by forming a transcriptional complex with YAP1 and TEAD4 in the outer blastomeres of morulae. During the same stages, TFAP2C forms alternate transcriptional complexes with OCT4 and/or other regulatory proteins that favor pluripotency gene expression. During the morula-to-blastocyst transition, Oct4 and Nanog are downregulated in the multipotent trophoblast lineage via high levels of CDX2 induced by TFAP2C and YAP1/TEAD4.
In a second set of experiments, we tested whether overexpression of TFAP2C could augment Oct4 expression at the 2-cell stage. Notably, in human naïve ES cells and ES cells undergoing differentiation, TFAP2C can function as a pioneer transcription factor by opening chromatin and inducing gene expression (Pastor et al. 2018, Li et al. 2019). For this experiment, we focused exclusively on Oct4 because Nanog is normally expressed at very low levels at the 2-cell stage and is almost below the level of detection by real-time PCR for relative quantification (data not shown). One-cell embryos were injected with 10 or 25 ng/μl of Tfap2c complimentary RNA (cRNA). Embryos were then cultured to the 2-cell stage. Microinjection of Tfap2c cRNA markedly increased TFAP2C protein in 2-cell embryos (Fig. 1b). Furthermore, qRT-PCR analysis revealed that overexpression of TFAP2C significantly induced Oct4 expression in a dose-dependent manner (Fig. 1b). Collectively, these experiments provide additional evidence that TFAP2C can positively regulate Oct4 and Nanog expression during early embryogenesis in mice. Future research studies are necessary to show whether TFAP2C functions as an inducer or a modulator of Oct4 and Nanog.
So, what are the molecular mechanisms TFAP2C utilizes to regulate gene expression in the trophoblast lineage versus the pluripotent ICM? It is well established in mice that the trophoblast and pluripotent ICM lineages represent opposing cell-fates that negatively regulate the formation of one another. For example, during the morula-to-blastocyst transition, OCT4 and NANOG repress Cdx2 transcription to promote ICM development, while CDX2 negatively regulates Oct4 and Nanog to support trophoblast lineage development (Strumpf et al. 2005, Karasek et al. 2020). Thus, how does TFAP2C regulate gene expression in these conflicting cell-fates? We first must consider that TFAP2C is ubiquitously expressed from the 1-cell stage up until the late morula stage before it becomes downregulated in the ICM (Kuckenberg et al. 2010, Choi et al. 2012). During this period, pluripotency genes such as Oct4 and Nanog and trophoblast genes such as Cdx2 are upregulated around the 8-cell stage in preparation for the first cell-fate decision. Based on this overlapping expression pattern, we postulate that TFAP2C forms transcriptional complexes with distinct regulatory proteins in the outer and inner blastomeres during the 8-cell-to-morula and morula-to-blastocyst transitions.
We propose that in the emerging trophoblast lineage, TFAP2C forms a transcriptional complex with the HIPPO signaling pathway effector Yes-associated protein 1 (YAP1) and TEAD4 to positively regulate trophoblast genes (Fig. 1c). We and others demonstrated that TFAP2C establishes interactions with YAP1 and TEAD4 in the outer blastomeres of morulae (Karasek et al. 2020, Chi et al. 2020). Notably, the HIPPO pathway plays a crucial role in the first cell-fate decision (Karasek et al. 2020). For instance, in the outer blastomeres of morulae, the HIPPO signaling pathway is inactivated allowing YAP1 to localize to the nuclei and regulate trophoblast lineage genes such as Cdx2 (Karasek et al. 2020). Conversely, on the inside of the embryo, the HIPPO pathway is active, and YAP1 becomes phosphorylated by LATS Kinase, forcing it to stay in the cytoplasm, allowing the expression of Sox2 (Karasek et al. 2020). In support of our above observations, unpublished qRT-PCR data generated from RNA interference studies in our laboratory revealed that TFAP2C and YAP1 regulate similar sets of genes required for trophoblast lineage development (data not shown). Thus, it is conceivable that TFAP2C converges with YAP1 and TEAD4 in the outer blastomeres to positively regulate trophoblast genes. Further research using a combination of genetic approaches and RNA sequencing will be necessary to establish the genetic relationship between TFAP2C, YAP1, and TEAD4.
So how does TFAP2C positively regulate pluripotency genes such as Oct4 and Nanog? These genes are initially ubiquitously expressed before they become restricted to the ICM. We postulate that TFAP2C forms unique transcriptional complexes with regulatory proteins that favor pluripotency gene expression (Fig. 1c). One potential candidate is OCT4 protein itself. It is well known that OCT4 represents one of several core pluripotency TFs that can autoregulate itself as well as positively regulate other pluripotency genes (Jaenisch & Young 2008). Based on our published preliminary data discussed earlier (Choi et al. 2013), we reason that TFAP2C may interact with OCT4 protein to positively regulate Oct4 and Nanog on the inside and outside blastomeres. Other lineage-specific TFs and epigenetic regulators may serve as strong candidates for targeting TFAP2C to the promoters and enhancers of Oct4, Nanog, and other pluripotency genes.
In conclusion, we are just starting to understand how TFAP2C can regulate the formation of two conflicting cell-fates in early mouse embryos. Experimental data indicate that TFAP2C is an important regulator of both trophoblast and pluripotency genes. Emerging data in the field indicate that TFAP2C may regulate trophoblast genes in collaboration with YAP1 and TEAD4. The mechanisms by which TFAP2C activates and/or maintains the expression of pluripotency genes required for ICM development are largely unknown. TFAP2C likely forms regulatory complexes with distinct regulatory proteins to promote the expression of pluripotency genes. More research is necessary to establish the molecular mechanism(s) by which TFAP2C co-regulates trophoblast lineage and pluripotency gene expression during preimplantation development.
Acknowledgements
Animal use protocols were approved by Michigan State University’s Institutional Animal Care and Use Committee. The authors thank Ms. Katie Wilson for insightful comments and suggestions on the manuscript. The authors apologize to researchers in the field whose work we were unable to cite due to space limitations. We thank the anonymous reviewer and associate editor for excellent suggestions.
Funding
This work was supported by a grant from the National Institutes of Child Health and Development (NICHD) to Jason G. Knott (HD095371). Chad S. Driscoll and Mohamed Ashry were supported by T32 doctoral and post-doctoral fellowships, respectively, from the NICHD (HD087166).
Footnotes
Declaration of interests
Jason G. Knott is an associate editor of Reproduction. Jason was not involved in the review or editorial process for this paper, on which he is listed as an author The authors declare there is no conflict of interest that could be perceived as prejudicing the impartiality of this point of view.
References
- Cao Z, Carey TS, Ganguly A, Wilson CA, Paul S & Knott JG 2015. Transcription factor AP-2γ induces early Cdx2 expression and represses HIPPO signaling to specify the trophectoderm lineage. Development 142 1606–1615. ( 10.1242/dev.120238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Liu W, Zimmerman J, Pastor WA, Kim R, Hosohama L, Ho J, Aslanyan M, Gell JJ, Jacobsen SE & Clark AT 2018. The TFAP2C-Regulated OCT4 Naive Enhancer Is Involved in Human Germline Formation. Cell Rep 25 3591–3602. ( 10.1016/j.celrep.2018.12.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi F, Sharpley MS, Nagaraj R, Roy SS & Banerjee U 2020. Glycolysis-Independent Glucose Metabolism Distinguishes TE from ICM Fate during Mammalian Embryogenesis. Dev Cell 53 9–26 ( 10.1016/j.devcel.2020.02.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I, Carey TS, Wilson CA & Knott JG 2012. Transcription factor AP-2γ is a core regulator of tight junction biogenesis and cavity formation during mouse early embryogenesis. Development 139 4623-4632. ( 10.1242/dev.086645) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I, Carey TS, Wilson CA & Knott JG 2013. Evidence that transcription factor AP-2γ is not required for Oct4 repression in mouse blastocysts. PLoS One 8 e65771. 10.1371/journal.pone.0065771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R & Young R 2008. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132 567–582. ( 10.1016/j.cell.2008.01.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasek C, Ashry M, Driscoll CS & Knott JG 2020. A tale of two cell-fates: role of the Hippo signaling pathway and transcription factors in early lineage formation in mouse preimplantation embryos. Mol Hum Reprod 26 653–664. ( 10.1093/molehr/gaaa052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuckenberg P, Buhl S, Woynecki T, van Fürden B, Tolkunova E, Seiffe F, Moser M, Tomlin A, Winterhager E & Schorle H 2010. The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol Cell Biol 30 3310–3320. ( 10.1128/MCB.01215-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang Y, Torkelson JL, Shankar G, Pattison JM, Zhen HH, Fang F, Duren Z, Xin J, Gaddam S, Melo SP, Piekos SN, Li J, Liaw EJ, Chen L, Wernig M, Wong WH, Chang HY & Oro AE 2019. TFAP2C- and p63-dependent networks sequentially rearrange chromatin landscapes to drive human epidermal lineage commitment. Cell Stem Cell 24 271–284. ( 10.1016/j.stem.2018.12.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y& Torres-Padilla ME 2012. Control of ground-state pluripotency by allelic regulation of Nanog. Nature 483 470–473. ( 10.1038/nature10807) [DOI] [PubMed] [Google Scholar]
- Pastor WA, Liu W, Chen D, Ho J, Kim R, Hunt TJ, Lukianchikov A, Liu X, Polo JM, Jacobsen SE & Clark AT 2018. TFAP2C regulates transcription in human naive pluripotency by opening enhancers. Nat Cell Biol 20 553–564. ( 10.1038/s41556-018-0089-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F & Rossant J 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132 2093–2102. ( 10.1242/dev.01801) [DOI] [PubMed] [Google Scholar]
- Werling U & Schorle H 2002. Transcription factor gene AP-2 gamma essential for early murine development. Mol Cell Biol 22 3149–3156. ( 10.1128/MCB.22.9.3149-3156.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Cornwall-Scoones J, Wang P, Handford CE, Na J, Thomson M & Zernicka-Goetz M. 2020. Developmental clock and mechanism of de novo polarization of the mouse embryo. Science 370 6522. ( 10.1126/science.abd2703) [DOI] [PMC free article] [PubMed] [Google Scholar]


