Abstract
This letter demonstrates the potential of novel cryptic proteins resulting from TAR DNA-binding protein 43 (TDP-43) dysfunction as markers of TDP-43 pathology in neurodegenerative diseases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13024-024-00718-8.
To the editor
Aberrant accumulation of the RNA binding protein TDP-43 in the cytoplasm and its depletion from the nucleus are pathological hallmarks of several neurodegenerative diseases including subsets of frontotemporal lobar degeneration (FTLD-TDP), amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (AD-TDP). Loss of TDP-43 from the nucleus impairs its ability to repress cryptic exon (CE) inclusion during RNA splicing [1]. Consequently, CEs are anomalously included in critical transcripts such as STMN2 and UNC13A [2–9], which can produce truncated or destabilized RNAs and lead to a loss of their function. Recently, we and others demonstrated that some transcripts with in-frame CEs produce stable CE-containing novel proteins detectable in cerebrospinal fluid (CSF) from patients with FTLD-TDP or ALS [10, 11]. One notable example is a cryptic protein derived from the gene hepatoma-derived growth factor-like protein 2 (HDGFL2-CE), a histone-binding protein expressed throughout the brain. Accordingly, CSF HDGFL2-CE could potentially serve as a sensitive biomarker of TDP-43 pathology, illuminating the contribution of pathological TDP-43 to the clinical variability in TDP-43 proteinopathies [11]. But testing whether CSF HDGFL2-CE is indicative of TDP-43 pathology is hampered by the lack of robust methods to measure pathological TDP-43 in biofluids. To determine if HDGFL2-CE abundance can be used as a readout of the presence of TDP-43 pathology, we probed whether HDGFL2-CE is preferentially expressed in affected neuroanatomical regions with TDP-43 proteinopathy in a cohort of well-characterized post-mortem tissues from FTLD-TDP and AD-TDP cases, and whether HDGFL2-CE abundance associates with pathological TDP-43 burden.
Based on the predicted structure of HDGFL2-CE protein (Fig. S1A), we had generated a previously described rabbit polyclonal HDGFL2-CE antibody (Mayo-LP) [10] that specifically detects HDGFL2-CE but not wild-type HDGFL2 (HDGFL2-WT) proteins in lysates from TDP-43-depleted human induced pluripotent stem cells (iPSC) (Fig. S1B). Using a commercial C-terminal HDGFL2-WT antibody as the capture antibody, and our Mayo-LP HDGFL2-CE antibody as the detection antibody, we then developed a Meso Scale Discovery (MSD) immunoassay that dose-dependently detected endogenous HDGFL2-CE protein in 500‒8000 ng of total protein lysate from TDP-43-depleted iPSC-derived neurons [10]. We have since further optimized the assay by biotinylating the capture antibody, using streptavidin MSD plates, and testing different diluents (Fig. S1C−E). When using MSD Diluent 35, our modified assay detected HDGFL2-CE but not HDGFL2-WT in 16 ng of total protein in lysates from HEK293T cells overexpressing these proteins (Fig. S1D). To determine whether our assay is sufficiently sensitive to detect endogenous HDGFL2-CE, we used lysates from control and TDP-43-depleted iPSCs. Compared to Diluent 35, Diluent 100 provided a better signal to noise ratio detecting endogenous HDGFL2-CE in as little as 125 ng of total protein from TDP-43-depleted iPSC lysates (Fig. S1E).
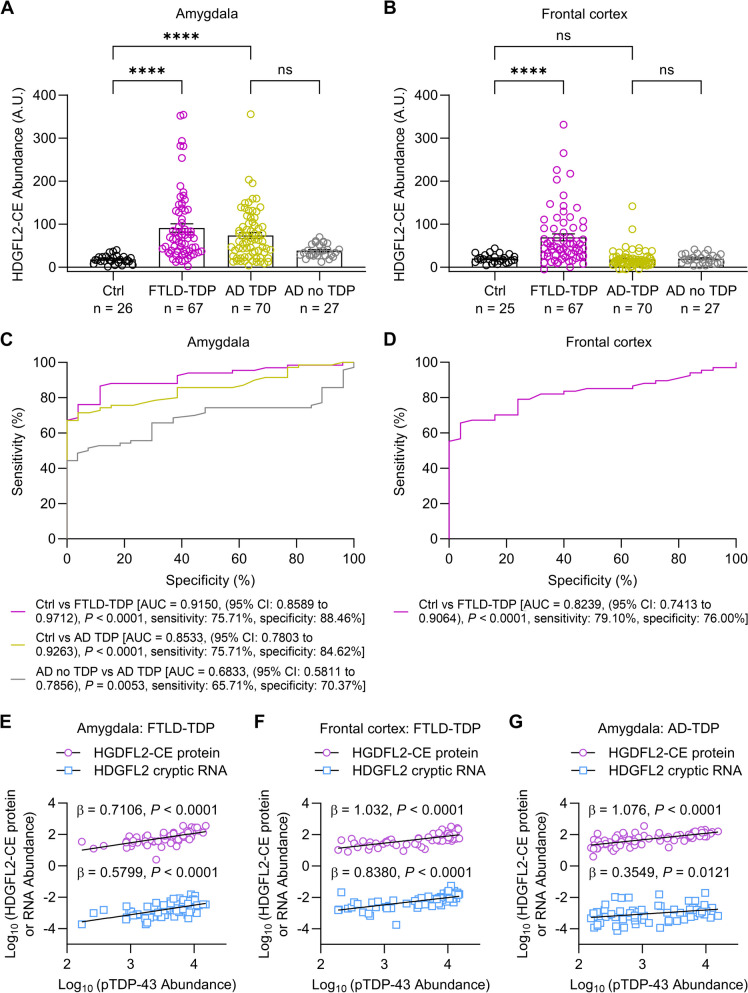
Next, we tested if our optimized assay could detect HDGFL2-CE in brain regions of FTLD-TDP (amygdala and frontal cortex) and AD-TDP (amygdala) characterized by TDP-43 pathology [7]. Compared to cognitively normal controls (controls), HDGFL2-CE was significantly increased in the amygdala of FTLD-TDP and AD-TDP cases in unadjusted analysis and when adjusting for age at death and sex (Fig. 1A, Table S1). In contrast, frontal cortex HDGFL2-CE was significantly increased only in FTLD-TDP cases when compared to controls (Fig. 1B, Table S1). When comparing AD cases without TDP-43 pathology (AD no TDP) to AD-TDP cases, HDGFL2-CE was significantly increased in the amygdala, but not the frontal cortex, in AD-TDP (Fig. 1A, Table S1) – an expected result given the paucity of TDP-43 pathology in the frontal cortex of AD-TDP cases.
Fig. 1.
HDGFL2-CE proteins are increased in brain regions with TDP-43 pathology in FTLD-TDP and AD-TDP, and distinguish these TDP-43 proteinopathies from non-TDP-43 controls. Immunoassay quantification of HDGFL2-CE proteins in the amygdala (A) and frontal cortex (B) of cognitively normal controls (Ctrl, n = 27, n = 26 amygdala and n = 25 frontal cortex), FTLD-TDP (n = 67), AD-TDP (n = 70) and AD no TDP (n = 27). Data are presented as mean ± s.e.m. * P < 0.05 and **** P < 0.0001, ns: not significant. (C, D) Area under the receiver operating characteristic curves (AUC) showing the discriminatory capability of HDGFL2-CE in the amygdala or frontal cortex to distinguish FTLD-TDP from Ctrl (pink), AD-TDP from Ctrl (gold), and AD-TDP from AD no TDP (black). AUC values are shown. (E–G) Scatterplots of HDGFL2-CE protein and RNA abundance with pTDP-43 abundance in the amygdala (E) and frontal cortex (F) of FTLD-TDP patients, as well as in the amygdala of AD-TDP patients (G). Regression coefficients (β) and P values from linear regression analysis of pTDP-43 with HDGFL2-CE protein and RNA adjusting for age and sex are shown
The presence of HDGFL2-CE differentiated individuals with and without TDP-43 pathology in the amygdala: HDGFL2-CE distinguished between controls and individuals with TDP-43 pathology with an area under the receiver operating characteristic curve (AUC) of 0.85 and 0.92 for AD-TDP and FTLD-TDP, respectively, indicating good to excellent discriminatory ability (Fig. 1C). When assessing whether amygdala HDGFL2-CE protein distinguishes AD no TDP from AD-TDP, we found moderate discriminatory ability (AUC of 0.68, Fig. 1C). In the frontal cortex, HDGFL2-CE differentiated controls and FTLD-TDP with an AUC of 0.82, indicating good discriminatory ability (Fig. 1D).
Finally, phosphorylated TDP-43 (pTDP-43) burden in the amygdala and frontal cortex significantly associated with HDGFL2-CE protein and HDGFL2-CE RNA abundance in FTLD-TDP in both unadjusted analysis and in analysis adjusting for age at death, sex, and RNA integrity number (RIN), the latter for analysis of HDGFL2-CE RNA only (Fig. 1E, F, Table S2). In the amygdala of AD-TDP cases, pTDP-43 burden also associated with HDGFL2-CE protein and RNA in unadjusted and adjusted analyses (Fig. 1G, Table S2). However, estimated β coefficients where higher for HDGFL2-CE protein than HDGFL2-CE RNA indicating that HDGFL2-CE protein serves as a more accurate indicator of TDP-43 dysfunction.
Retention of CEs in mRNAs owing to TDP-43 dysfunction is well-documented in FTLD-TDP, ALS and AD-TDP [2–8], but the identification that certain in-frame CEs generate stable cryptic proteins is new [8, 10, 11]. Here, we explored the recently identified HDGFL2-CE protein. By developing a sensitive and specific immunoassay to detect HDGFL2-CE proteins, we observed that HDGFL2-CE is significantly increased in brain regions with TDP-43 pathology in FTLD-TDP and AD-TDP, associates with pTDP-43 burden, and can distinguish individuals with TDP-43 pathology from those without. In line with our findings, Irwin et al. used their HDGFL2-CE antibody to perform immunofluorescent staining of motor cortex and hippocampus tissues from patients with ALS-FTD demonstrating that HDGFL2-CE proteins accumulate in cells exhibiting pTDP-43 pathology [11]. They additionally found that, compared to controls, CSF HDGFL2-CE was statistically significantly higher in individuals likely to have TDP-43 pathology, namely presymptomatic or symptomatic C9orf72 repeat expansion carriers and patients with sporadic ALS [11].
Collectively, these findings show that the presence of HDGFL2-CE in the brain is a sensitive reporter of TDP-43 pathology in neurodegenerative diseases. These findings empower CSF HDGFL2-CE as a surrogate marker of TDP-43 pathology and dysfunction, which in turn would inform the selection of ideal participants for clinical trials of potential TDP-43-based therapeutics, and potentially enable precision medicine strategies for pathological subtypes of FTLD and AD.
Supplementary Information
Acknowledgements
We thank the patients and their families for their contributions to this study.
Authors’ contributions
L.P., Y.-J.Z., M.E.W., M.P., and T.F.G. conceptualized and designed the study. A.C., L.M.D., P.C.O, M.Y., K.J.W. and M.P. carried-out the investigation. N.N.I. and T.C. generated 3D structure of cryptic HDGFL2 proteins. B.R., M.D., C.C., N.R.G., B.F.B., D.S.K., R.C.P., K.A.J., B.O., A.D.G., D.W.D., M.P., M.E.W., Y.-J.Z. and L.P. provided resources. A.C., T.F.G., M.P., M.E.W., Y.-J.Z. and L.P performed the data analysis. E.A.A., A.C., A.D.G, T.F.G, M.P., M.E.W., Y.-J.Z. and L.P. wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Target ALS Foundation (M.P., M.E.W., Y.-J.Z. and L.P.,), Robert Packard Center for ALS Research at Johns Hopkins (L.P.), the National Institutes of Health/National Institute on Aging [(5P30AG0062677: L.P.), (ALLFTD U19AG063911: N.R.G., B.F.B., D.S.K., R.C.P. D.W.D. and L.P.), (R01AG037491: K.A.J.)], the National Institutes of Health/National Institute of Neurological Disorders and Stroke [(U54NS123743: A.D.G., M.P. and L.P.); (R35NS097273: L.P.); (R35NS097263: A.D.G.); (P01NS084974: D.W.D., T.F.G., L.P. and Y.-J.Z); (RF1NS120992: K.A.J and M.P.); (R01NS117461: T.F.G. and Y.-J.Z.); and (R21NS127331: Y.-J.Z.)].
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files, or available from the corresponding authors upon reasonable request.
Declarations
Ethics approval and consent to participate
Human postmortem brain tissues from amygdala and frontal cortex were provided by the Mayo Clinic Florida Brain Bank. Diagnosis was independently ascertained by trained neurologists and neuropathologists upon neurological and pathological examinations, respectively. All participants, or the next of kin, provided written informed consent, and all protocols were reviewed and approved by the Mayo Clinic Institutional Review Board and Ethics Committee.
Consent for publication
All authors have reviewed the final manuscript and consent to publication.
Competing interests
B.F.B. receives institutional research grant support from Alector, Biogen, Transposon, Cognition Therapeutics, and GE Healthcare. B.F.B. receives an honorarium for SAB activities for the Tau Consortium. L.P. is a consultant for Expansion Therapeutics. D.S.K. serves on a Data Safety Monitoring Board for the Dominantly Inherited Alzheimer Network Treatment Unit study. D.S.K. served on a Data Safety monitoring Board for a tau therapeutic for Biogen (until 2021) but received no personal compensation. D.S.K. is an investigator in clinical trials sponsored by Biogen, Lilly Pharmaceuticals, and the University of Southern California. D.S.K. has served as a consultant for Roche, Samus Therapeutics, Magellan Health, Biovie and Alzeca Biosciences but receives no personal compensation. D.S.K. attended an Eisai advisory board meeting for lecanemab. A.D.G. is a scientific founder of Maze Therapeutics.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Anna Calliari and Lillian M. Daughrity contributed equally to this work.
Change history
7/27/2024
A Correction to this paper has been published: 10.1186/s13024-024-00744-6
Contributor Information
Michael E. Ward, Email: wardme@nih.gov
Yong-Jie Zhang, Email: zhang.yongjie@mayo.edu.
Leonard Petrucelli, Email: petrucelli.leonard@mayo.edu.
References
- 1.Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349:650–5. 10.1126/science.aab0983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown AL, Wilkins OG, Keuss MJ, Hill SE, Zanovello M, Lee WC, Bampton A, Lee FCY, Masino L, Qi YA, et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature. 2022;603:131–7. 10.1038/s41586-022-04436-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma XR, Prudencio M, Koike Y, Vatsavayai SC, Kim G, Harbinski F, Briner A, Rodriguez CM, Guo C, Akiyama T, et al. TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature. 2022;603:124–30. 10.1038/s41586-022-04424-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melamed Z, Lopez-Erauskin J, Baughn MW, Zhang O, Drenner K, Sun Y, Freyermuth F, McMahon MA, Beccari MS, Artates JW, et al. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat Neurosci. 2019;22:180–90. 10.1038/s41593-018-0293-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klim JR, Williams LA, Limone F, Guerra San Juan I, Davis-Dusenbery BN, Mordes DA, Burberry A, Steinbaugh MJ, Gamage KK, Kirchner R, et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci. 2019;22:167–79. 10.1038/s41593-018-0300-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prudencio M, Humphrey J, Pickles S, Brown AL, Hill SE, Kachergus JM, Shi J, Heckman MG, Spiegel MR, Cook C, et al. Truncated stathmin-2 is a marker of TDP-43 pathology in frontotemporal dementia. J Clin Invest. 2020;130:6080–92. 10.1172/JCI139741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estades Ayuso V, Pickles S, Todd T, Yue M, Jansen-West K, Song Y, Gonzalez Bejarano J, Rawlinson B, DeTure M, Graff-Radford NR, et al. TDP-43-regulated cryptic RNAs accumulate in Alzheimer’s disease brains. Mol Neurodegener. 2023;18:57. 10.1186/s13024-023-00646-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang K, Ling JP, Redding-Ochoa J, An Y, Li L, Dean SA, Blanchard TG, Pylyukh T, Barrett A, Irwin KE, et al. Loss of TDP-43 splicing repression occurs early in the aging population and is associated with Alzheimer’s disease neuropathologic changes and cognitive decline. Acta Neuropathol. 2023;147:4. 10.1007/s00401-023-02653-2 [DOI] [PubMed] [Google Scholar]
- 9.Chung M, Carter EK, Veire AM, Dammer EB, Chang J, Duong DM, Raj N, Bassell GJ, Glass JD, Gendron TF, et al. Cryptic exon inclusion is a molecular signature of LATE-NC in aging brains. Acta Neuropathol. 2024;147:29. 10.1007/s00401-023-02671-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seddighi S, Qi YA, Brown AL, Wilkins OG, Bereda C, Belair C, Zhang YJ, Prudencio M, Keuss MJ, Khandeshi A, et al. Mis-spliced transcripts generate de novo proteins in TDP-43-related ALS/FTD. Sci Transl Med. 2024;16:eadg7162. 10.1126/scitranslmed.adg7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irwin KE, Jasin P, Braunstein KE, Sinha IR, Garret MA, Bowden KD, Chang K, Troncoso JC, Moghekar A, Oh ES, et al. A fluid biomarker reveals loss of TDP-43 splicing repression in presymptomatic ALS-FTD. Nat Med. 2024;30:382. 10.1038/s41591-023-02788-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coban MA, Morrison J, Maharjan S, Hernandez Medina DH, Li W, Zhang YS, Freeman WD, Radisky ES, Le Roch KG, Weisend CM, et al. Attacking COVID-19 progression using multi-drug therapy for synergetic target engagement. Biomolecules. 2021;11:787. 10.3390/biom11060787 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files, or available from the corresponding authors upon reasonable request.