Abstract
The sex determination process in cucurbits involves the control of stamen or carpel development during the specification of male or female flowers from a bisexual floral meristem, a function coordinated by ethylene. A gain-of-function mutation in the miR164-binding site of CpCUC2B, ortholog of the Arabidopsis transcription factor gene CUC2, not only produced ectopic floral meristems and organs, but also suppressed the development of carpels and promoted the development of stamens. The cuc2b mutation induced the transcription of CpCUC2B in the apical shoots of plants after female flowering but repressed other CUC genes regulated by miR164, suggesting a conserved functional redundancy of these genes in the development of squash flowers. The synergistic androecious phenotype of the double mutant between cuc2b and etr2b, an ethylene-insensitive mutation that enhances the production of male flowers, demonstrated that CpCUC2B arrests the development of carpels independently of ethylene and CpWIP1B. The transcriptional regulation of CpCUC1, CpCUC2, and ethylene genes in cuc2b and ethylene mutants also confirms this conclusion. However, the epistasis of cuc2b over aco1a, a mutation that suppresses stamen arrest in female flowers, and the down-regulation of CpACS27A in cuc2b female apical shoots, indicated that CpCUC2B promotes stamen development by suppressing the late ethylene production.
Keywords: Boundary specification, CUC2, Cucurbita pepo, ethylene, female flowering, flower meristem
The CpCUC2B boundary gene plays an essential role in squash sex determination. The gene prevents carpel formation, independently of ethylene, and promotes stamen development by inhibiting the ethylene biosynthesis gene CpACS27A.
Introduction
The zucchini morphotype of Cucurbita pepo is one of the most economically significant cucurbit crops. The species is monoecious, carrying unisexual male and female flowers on the same individual plant, but the number of female and male flowers per plant varies between the different cultivars. Unisexual flowers are individually developed at each node in the main and secondary shoots, with the plant exhibiting three sequential sexual developmental phases: an initial male phase; a mixed phase characterized by alternating production of female and male flowers; and a predominantly or exclusively female phase (Peñaranda et al., 2007; Manzano et al., 2010, 2013). These phases are regulated by a combination of environmental, hormonal, and genetic factors (Martínez and Jamilena, 2021).
Ethylene has been established as the primary regulator of sex determination in cucurbits (Manzano et al., 2010; Pannell, 2017). Elevated levels of this hormone promote female flower development and facilitate the maturation of floral organs (Manzano et al., 2013), whereas suppression of ethylene production or impairment of its perception leads to the conversion of females into male or bisexual flowers, changing monoecy into androecy or andromonoecy (Martínez et al., 2014; García et al., 2020a, b). Several genes associated with this developmental process have been identified in cucurbits, including ACS and ACO genes, such as CsACS2 in cucumber, CmACS7 in melon, CitACS4 in watermelon, and CpACS27A and CpACO1 in zucchini. Mutations in these genes promote the conversion of female into bisexual or hermaphrodite flowers, indicating their key role in stamen arrest during development of female flowers (Boualem et al., 2008, 2009; Martínez et al., 2014; Manzano et al., 2016; Cebrián et al., 2022). In contrast, loss-of-function mutation in other ethylene biosynthesis genes, including cucumber and melon ACS11 (Boualem et al., 2015) and cucumber CsACO2 (Chen et al., 2016), converts female into male flowers and thus monoecy into androecy, indicating that they function as promoters of carpel development at early stages of flower development (Martínez and Jamilena, 2021). The sex phenotypes of four ethylene-insensitive mutants in zucchini have also demonstrated that the different ethylene receptors cooperate in the female flower bud to promote the development of carpels and the arrest of stamens during female flower development. Single and double ethylene-insensitive mutants are, in fact, andromonoecious or androecious depending on the dosage and strength of mutant alleles in each genotype (García et al., 2020a, b).
In addition to ethylene biosynthesis and perception genes, various transcription factor genes have been identified in cucurbits that control sex determination. The ethylene-responsive transcription factor genes ERF110 and ERF31 not only show an ethylene-sensitive expression but also regulate the production of ethylene by transcriptional activation of the ethylene biosynthesis genes ACS11 and ACS2, respectively (Pan et al., 2018; Tao et al., 2018). The WIP1 gene of both cucumber and melon encodes a zinc finger transcription factor with a key role in carpel abortion during the development of male flowers and is transcriptionally regulated by ACS11 and ACO2 (Boualem et al., 2015; Chen et al., 2016). Mutations in WIP1 lead to ginoecy in melon, cucumber, and watermelon (Martin et al., 2009; Hu et al., 2017; Zhang et al., 2020). The WIP1 transcription factor regulates the development of male flowers by repressing the transcription of the carpel identity gene CRABS CLAW (CRC) through a TOPLESS-mediated histone deacetylation mechanism, and therefore CRC mutants are androecious (Zhang et al., 2022).
This study introduces a new player in the sex determination gene network of the cucurbits, the C. pepo transcription factor gene CUP-SHAPED COTYLEDON 2B (CpCUC2B). We show that an miR164-resistant mutation in the CpCUC2B boundary specification gene of zucchini not only increased the number of floral meristems, the number of floral organs, and the serration of leaves, but also promoted the conversion of female into male flowers, leading to a nearly androecious phenotype. Therefore, the gene mediates the arrest of carpels during the development of male flowers.
Materials and methods
Plant material for mutant isolation
An ethyl methanesulfonate (EMS) mutant collection composed of 3751 families of C. pepo was generated at the University of Almeria in the zucchini genetic background MU-CU-16 (García et al., 2018). A high-throughput screening was performed looking for alterations in vegetative and reproductive development in 123 M2 families, grown to maturity under standard greenhouse conditions during the spring–summer 2020 season in Almeria, Spain. The phenotypic analysis resulted in the identification of a mutant line (cuc2b) with a reduced proportion of female flowers, as well as alterations in the development of the floral organs.
The cuc2b locus was mapped by crossing mutant M2 plants with the scallop line UPV-196. The F1 plants were then self-pollinated and the resulting F2 segregating population was used to perform bulk segregant analysis sequencing (BSA-seq). In parallel, mutant M2 plants were crossed twice with MU-CU-16 to generate the segregating populations BC1S1 and BC2S1 in the MU-CU-16 background. These segregating plants were evaluated to validate the causal mutation of the mutant phenotype.
Phenotypic evaluation of the cuc2b mutant line
More than 600 BC1S1 and BC2S1 plants were phenotyped for sex expression in the autumn–winter 2021 season and in the spring–summer 2022 season. The sex phenotype of each plant was determined by scoring the presence of pistillate or staminate flowers at the first 60 nodes of the plants. This evaluation led to the classification of plants into three groups: wild type (wt/wt), characterized by a proportion of female flowers comparable with the genetic background (30%); mutants (cuc2b/cuc2b) with a reduced percentage of female flowers and alterations in the development of floral organ development; and heterozygotes (wt/cuc2b), which show an intermediate phenotype of sex expression.
These plants were also analyzed for the presence of alterations in vegetative and reproductive organs: number of flowers per node, number of floral organs in male and female flowers, as well as alterations in leaf morphology.
Identification of the cuc2b mutation by BSA-seq coupled with whole-genome sequencing
To identify the causal mutation of the cuc2b phenotype, wild-type (WT) and mutant plants derived from F2 segregating populations were subjected to whole-genome sequencing (WGS). Genomic DNA from 15 WT and 15 cuc2b plants was extracted from young leaves using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987). The concentration of all samples was adjusted for an equal representation of each plant DNA. The WT DNA samples were pooled to create the WT-bulk, and the mutant DNA samples were pooled to create the MUT-bulk. DNA was also sequenced from 10 plants from the scallop line UPV-196.
DNA from each bulk was fragmented to an average size of 350 bp and was end-repaired, A-tailed, and ligated with Illumina adapters. The Illumina NovaSeq 6000 platform was used to perform genome DNA sequencing by Novogene Co., Ltd, generating 150 bp paired-end reads. Sequencing data were aligned with the reference C. pepo genome v4.1 using BWA software (Li and Durbin, 2009) (parameters: mem -t 4 -k 32 -M), and single nucleotide polymorphism (SNP) variations were detected using SAMtools (Li, 2011) and BCFtools (Danecek et al., 2021) with the following parameter: mpileup -m 2 -F 0.002'. The resulting SNPs were annotated with the ANNOVAR software (Wang et al., 2010).
The resulting VCF file was used for quantitative trait locus sequencing (QTL-seq) analysis using the QTLseqr package of R (Mansfeld and Grumet, 2018). The SNPs from the two bulks present in the file were filtered using the ‘filterSNPs’ function with the following parameters: total depth between 30 and 200, reference allele frequency 0.3, and genotype quality >50. To detect putative QTLs, the SNP index and ΔSNP index were obtained using the function ‘runQTLseqAnalysis’ (Takagi et al., 2013). Identification of candidate QTL regions was performed using a 1 Mb sliding window, and the confidence intervals (90, 95, and 99%) for the ΔSNP indices were determined using 10 000 simulations for each bulk.
To establish candidate mutations, the SNPs present in the putative QTL were filtered according to the following parameters: alternative allele frequency (AF) in the WT-bulk <0.35, AF>0.65 in the mutant-bulk, genotype quality ≥30, and read depth ≥7. Common variants between this and other mutant lines already sequenced in the laboratory were filtered out, as they are variants that were fixed in the genetic background line MU-CU-16 after de novo sequencing and assembly of the reference genome. Those variants that were specific for the scallop line UPV-196 were also discarded. Positions with canonical EMS changes (G>A or C>T transitions) were selected as candidate mutations and used to conduct a fine mapping study to identify the causal mutation of the phenotype.
Validation of the cuc2b mutation by high-throughput genotyping of individual segregating plants
A total of 356 BC1S1 and 280 BC2S1 plants in the genetic background MU-CU-16 were subjected to high-throughput genotyping using competitive allele-specific PCR (KASP) technology according to the manufacturer’s instructions. The primers for the putative SNPs were synthesized by LGC Genomics®, and the KASP assay was performed on the CFX96 Touch real-time PCR detection system (Bio-Rad®) using the LGC KASP genotyping protocol. PCR cycling conditions were analyzed using CFX Maestro™ software (Bio-Rad®) to determine the genotype of each individual plant for the SNP candidates analyzed.
Bioinformatic analysis: phylogeny, protein stability, and regulatory networks.
Homologous protein sequences and putative miR164-binding sites were identified using the NCBI BLAST tool (https://blast.ncbi.nlm.nih.gov). Multiple sequence alignments of CUC2 transcription factors from other species and miR164-binding sites were performed with Clustal Omega (Sievers et al., 2011). The secondary structure of Pri-miR164 was predicted by miRNA-fold (Tav et al., 2016) and the graphic representation was performed using the FORNA online tool (Kerpedjiev et al., 2015).
The phylogenetic relationship of CpCUC2 with other homologous proteins (Supplementary Table S1) was studied using MEGA X software (Kumar et al., 2018) with MUSCLE alignment (Edgar, 2004) and the Maximum Likelihood method based on the Poisson correction mode (Zuckerkandl and Pauling, 1965), with 2000 bootstrap replicates. The analysis of conserved motifs was done with the MEME tool (https://meme-suite.org/meme/tools/meme; Bailey et al., 2015) and motif identification in the PFAM database (Mistry et al., 2021). The gene structure was determined using the GSDS 2 tool (Hu et al., 2015). Cis-regulatory elements were analyzed on the PlantCare website (Rombauts et al., 1999). Schematic representations were performed with TBtools (Chen et al., 2020).
The regulatory network of CpCUC genes was inferred using the ARACNE algorithm (Margolin et al., 2006) and visualized using Cytoscape software (Shannon et al., 2003). The inputs for the software were an expression matrix derived from the expression data of different plant organs and the C. pepo transcription factors of the PlantTFDB (Jin et al., 2017). Minimum mutual information (MI), the value used to measure the dependence between two random variables, was obtained, and interactions with P-values <1e–4 were selected after 100 bootstrap iterations.
Gene expression analysis
Three approaches were followed to perform gene expression analysis: (i) RNA-seq of samples from different plant organs of the genetic background MU-CU-16; (ii) RNA-seq of samples from different organs of the WT and ethylene-insensitive mutants etr2b, etr1b, and etr1a-1 (García et al., 2020a, b); and (iii) quantitative real-time PCR (qRT-PCR) from apical shoots of WT, cuc2b plants, and the ethylene-deficient and insensitive mutants aco1a and etr2b (García et al., 2020b; Cebrián et al., 2022), respectively.
The following plant organs were analyzed by RNA-seq in MU-CU-16: apical shoots of plants in the developmental stages of three nodes (male apical shoots, before female flowering) and 12 nodes (female apical shoots, after female flowering), male and female flowers with a corolla length of 30 mm, leaves of 10 mm, and roots from seedlings 17–21 d after germination. For each sample, three biological replicates have been taken from at least three plants per replicate. For RNA isolation, the Omega Biotek EZNA® plant RNA kit (R6827-01) was used following the manufacturer’s protocol. The RNA was then eluted in nuclease-free water and immediately prepared for sequencing. Samples were sequenced by BGI Genomics using the DNBseq platform, generating 150 bp paired-end reads and 6 Gb of raw data per sample.
The transcriptomes of the WT and ethylene mutants etr1b and etr1a-1 (García et al., 2020a) were analyzed by RNA-seq in apical shoots of plants with either three or 12 nodes. Library construction and sequencing of ethylene perception mutant samples were performed by Novogene. The sequencing platform used was the Illumina NovaSeq 6000 Sequencing System, generating 150 bp paired-end reads and obtaining 6 Gb of raw data per sample. Fragments per kilobase of transcript per million mapped reads (FPKM) values were obtained for the analysis of results using the BALLGOWN package in R (Frazee et al., 2014, Preprint), and these data were used to create heatmaps with TBtools.
The analysis of gene expression by qRT-PCR was carried out in samples of WT and cuc2b, aco1a (Cebrián et al., 2022), and etr2b (García et al., 2020b) plants grown in a greenhouse during the spring–summer season. Apical shoots from adult plants were collected in triplicate for each genotype, sampling at least five plants per replicate. Total RNA was isolated with the Omega Biotek EZNA® plant RNA Kit (R6827-01). RNA was converted to cDNA with the ADNc RevertAidTM kit (Thermo Fisher Scientific®). qPCR was carried out in a 10 μl total volume with 1× Top Green qPCR Super Mix (Bio-Rad®) on the thermocycler of the CFX-96 Touch Real-Time PCR Detection System (Bio-Rad®). Gene expression values were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001). Two constitutive genes were used as internal reference, CpEF1α and CpACT. Primers for gene expression analysis are shown in Supplementary Table S2. To validate the miR164-binding and cutting site of CpCUC2B, two pairs of primers were used for qPCR, one designed in the 3' region of the transcript, and the other flanking the predicted binding site of miR164 (Supplementary Table S2).
Statistical analysis
Data were subjected to an ANOVA using the statistical software Statgraphic Centurion XVIII. Differences between samples were separated by least significant difference at a significance level of P≤0.05.
Results
The mutant cuc2b shows a nearly androecious sex phenotype
The sex phenotype of the monoecious species C. pepo is characterized by two different developmental phases in the main shoot, a first male phase in which plants produce only male flowers, and a second female phase in which plants alternate the production of male and female flowers (Fig. 1A). The transition between the two phases, that is, the node at which the plants start to produce female flowers, is called female flowering transition.
Fig. 1.
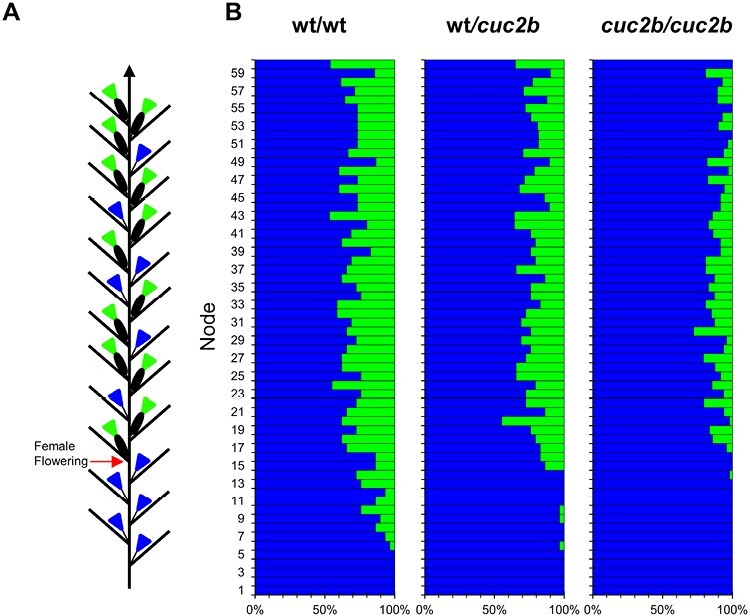
Sex phenotypes of the WT and cuc2b mutant. (A) Schematic representation of the distribution of males and females in the monoecious squash plant. Blue=male flower; green=female flower. The red arrow indicates the female flowering transition (FFT), the node at which the first female flower is developed. (B) Distribution of male and female flowers in the first 60 nodes of the wt/wt, wt/cuc2b, and cuc2b/cuc2b mutant plants (total plants=636). In each node, blue and green bars represent the percentage of male and female flowers of the total plants analyzed, respectively.
The phenotypic screening of an EMS mutant collection of C. pepo allowed the identification of a novel mutant called cuc2b which is altered in the female flowering transition and the determination of sex. For phenotyping analysis, the mutation was introgressed in the genetic background of MU-CU-16 by two successive backcrosses, and sex phenotyping was performed in 185 plants from the BC2S1 segregating population. In homozygous wt/wt plants, the female flowering transition occurred around node 11, and plants produced an average of 23.7% of female flowers per plant (Table 1; Fig. 1B). Mutant plants showed a very late female flowering transition and a nearly androecious phenotype, with a very reduced number of female flowers per plant (Table 1). Homozygous cuc2b/cuc2b plants started female flowering around node 30 and produced an average of only 7.6% female flowers per plant (Table 1). Heterozygous wt/cuc2b plants showed an intermediate sex phenotype (Table 1), indicating that the mutation is semi-dominant.
Table 1.
Sex expression of WT and cuc2b mutant plants
| Genotype | Female flowering transition | Female flowers per plant (%) |
|---|---|---|
| wt/wt | 11.8 ± 0.6 a | 23.7 ± 1.2 a |
| wt/cuc2b | 15.8 ± 0.4 b | 19.0 ± 0.7 b |
| cuc2b/cuc2b | 30.4 ± 2.7 c | 7.6 ± 1.1 c |
Female flowering transition indicates the node at which the first female flower emerges. Different letters in the same column indicate significant differences between phenotypes (ANOVA, P≤0.05).
The cuc2b flowers are impaired in the specification of the floral meristem and the separation of floral organs
The cuc2b mutation altered the development of the floral meristem and the primordia of the lateral organs (Fig. 2). Cucurbita pepo plants only develop a single male or female flower in each leaf axil, and the WT of the population studied did so. However, in cuc2b plants, 37.4% of male nodes and 17.6% or female nodes developed two flowers (Fig. 2A; Table 2). Some of these double flowers fused along the pedicels (male flowers) or along the pedicels and ovaries (female flowers) (Fig. 2A), indicating accessory flowers derived from a very early division of the axillary floral meristem.
Fig. 2.
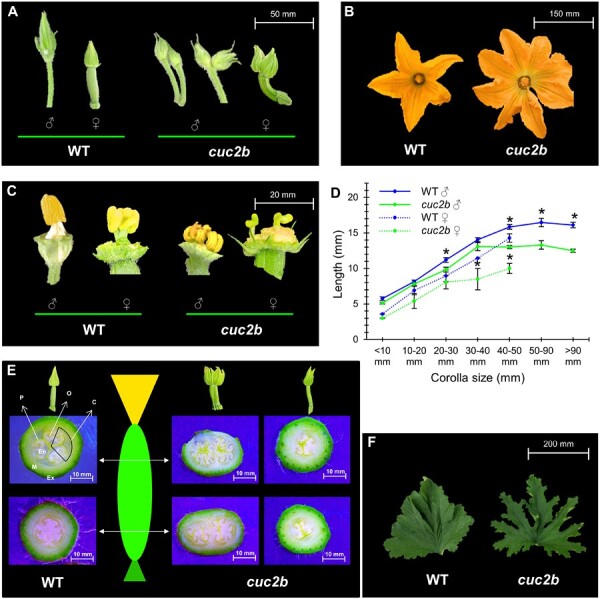
Effects of the cuc2b mutation on flower and leaf development. (A) Double male and female flowers in cuc2b nodes. (B) Increase in the number of petals in cuc2b flowers. (C) Alterations in the number and development of stamens and carpels. (D) Length of reproductive organs (stamen in male flowers and pistils in female flowers) relative to corolla length. Error bars represent the SE. Asterisks (*) indicate significant differences between genotypes at a specific corolla development stage (ANOVA, P≤0.05). (E) Transverse sections of WT and cuc2b ovaries in single and double flowers. Both distal sections (close to the corolla) and proximal sections (close to the pedicel) are shown. P, placenta; O, ovule; C, carpel; En, endocarp; M, mesocarp; Ex, exocarp. (F) WT and cuc2b leaf serration.
Table 2.
Effect of the cuc2b mutation on the number of flowers per node and the number of floral organs
| Node | Double flowers (%) | No. of sepals | No. of petals | No. of stamens | No. of carpels | Bisexual flowers (%) a | |
|---|---|---|---|---|---|---|---|
| WT | ♀ | 5.03 ± 0.003 b | 5.03 ± 0.003 a | 3.1 ± 0.06 b | 0 | ||
| ♂ | 5 ± 0.0 b | 5 ± 0.0 a | 3 ± 0.0 a | ||||
| cuc2b single flowers | ♀ | 5.76 ± 0.16 c | 5.81 ± 0.22 b | 2.71 ± 0.26 ab | 2.4 | ||
| ♂ | 5.53 ± 0.08 c | 5.60 ± 0.12 b | 4.22 ± 0.11 b | ||||
| cuc2b double flowers | ♀ | 17.6 | 3.86 ± 0.31 a | 5.18 ± 0.35 ab | 2.05 ± 0.30 a | 2.4 | |
| ♂ | 37.4 | 4.37 ± 0.16 a | 5.09 ± 0.31 a | 4.10 ± 0.28 b |
a Ovary-bearing flowers with anthers but without a style or stigma from the total of flowers analyzed of each genotype. n=250 male flowers and 90 female flowers. Different letters in the same column indicate significant differences between genotypes (ANOVA, P≤0.05).
The number and separation of floral organs differed between WT and mutant male and female flowers (Fig. 2B, C; Table 2). In single flowers, the number of sepals and petals increased from 5 in WT to 5.53 and 5.60 in the male flower of cuc2b and to 5.76 and 5.81 in the female flowers of cuc2b, respectively. The number of stamens also increased from 3 in the WT to 4.22, but the number of carpels was reduced from 3 to 2.71 carpels in the cuc2b mutant flowers (Table 2). In the double cuc2b flowers, the number of floral organs per flower varied with respect to those of the single cuc2b flowers (Table 2). The number of sepals and petals was reduced compared with that of the single flowers, but the number of stamens and carpels was similar in the single and double flowers of cuc2b (Table 2), suggesting that the division of the axillary floral meristem probably occurred after the initiation of the petal primordia, but before the initiation of stamens and carpels. Furthermore, compared with the WT, cuc2b flowers appeared to have a reduced stamen and pistil length, indicating that the mutation also altered the growth of sexual organs, especially during the later stages of flower development (Fig. 2D). Moreover, 2.4% of total cuc2b flowers were bisexual and developed both carpels and stamens, (Table 2).
The cross-sections of the single and double mutant ovaries indicated that exocarp and mesocarp develop normally, but the development of the endocarp and placenta is partially or completely abolished (Fig. 2E). In bisexual flowers, the endocarp and placenta tissue was also completely abolished. The WT pistil comprises three fused styles and stigmas, and an inferior ovary with three carpels, each developing two rows of placentas and ovules along the ovary (Fig. 2E). Although the phenotype was variable among flowers, most cuc2b pistils comprise fewer than three carpels at the distal end close to the flower corolla, with an average number of 2.71 and 2.05 styles, stigmas, and placentas in single and double cuc2b flowers, respectively. As we move towards the proximal end, the endocarp of the cuc2b ovary was replaced by mesocarp tissue and the number of placentas was further reduced (Fig. 2E). These flower defects caused partial female fertility, with a very reduced number of seeds compared with WT flowers (Table 3). However, the male flower of cuc2b was fully fertile, producing a similar number of seeds when used as a pollinator of the WT flowers (Table 3).
Table 3.
Female and male fertility of WT, heterozygous, and homozygous cuc2b mutant plants
| ♀ Parent | ♂ Parent | No. of fruits | No. of seeds | No. of seeds/fruit |
|---|---|---|---|---|
| wt/wt | wt/wt | 30 | 10 000 | 333 |
| wt/wt | cuc2b/cuc2b | 7 | 1490 | 213 |
| wt/wt | wt/cuc2b | 1 | 300 | 300 |
| wt/cuc2b | wt/wt | 2 | 420 | 210 |
| wt/cuc2b | wt/cuc2b | 3 | 600 | 200 |
| cuc2b/cuc2b | wt/wt | 6 | 168 | 28 |
| cuc2b/cuc2b | cuc2b/cuc2b | 2 | 65 | 33 |
In contrast to the defects found in flower meristem specification, the apical and axillary vegetative meristems of cuc2b plants developed normally (Supplementary Fig. S1). The cuc2b leaf, however, also had defective development patterns, with the mutation enhancing the serration of the leaf margin (Fig. 2F), similar to what occurs for miR164 mutants and miR164-resistant alleles of CUC2 in Arabidopsis (Nikovics et al., 2006).
cuc2b disrupts the NAC-like gene CpCUC2B on C. pepo chromosome 6
To elucidate the causative mutation of the cuc2b phenotype, the mutant was crossed twice with the zucchini cultivar MU-CU-16 (background genotype of the EMS population), and the obtained BC2 generation was selfed to generate a BC2S1 segregating population. The cuc2b mutant was also crossed with the scallop cultivar UPV-196 and then selfed to generate an F2 segregating population. The BC2S1 population on the MU-CU-16 background was used for the phenotyping of mutants, as the two backcrosses reduced the number of EMS mutations other than cuc2b. On the other hand, the F2 population was used for BSA-seq. A total of 15 WT and 15 cuc2b F2 plants were separated in two DNA bulks (WT and cuc2b) and used for WGS. More than 80 million reads were obtained that covered >97% of the reference genome with an average depth of 40 and 37.21 in the WT- and cuc2b-bulk, respectively (Table 4). The 1.51 and 1.49 million SNPs in the WT- and mutant-bulk were used to run the QTLseqr package, calculating and plotting the SNP index and ΔSNP index against the position of the genome. This allowed the identification of a single region on chromosome 6 where the average SNP index was higher in the WT-bulk than in the mutant-bulk (Fig. 3A). The ΔSNP index values in this region of chromosome 6 (position 2 982 470 to 5 702 905) differed significantly from 0, indicating this region harbors a major QTL (QTL6) that is probably responsible for the cuc2b phenotype (Fig. 3A).
Table 4.
Summary of sequencing data and filtering for SNPs in WT and cuc2b DNA bulks
| Whole-genome sequencing data | WT | cuc2b | ||||
|---|---|---|---|---|---|---|
| No. of reads | 90 123 030 | 83 373 064 | ||||
| Mapped reads (%) | 97.87 | 97.42 | ||||
| Average depth | 40.00 | 37.21 | ||||
| Coverage at least 4× (%) | 95.65 | 94.99 | ||||
| SNPs | ||||||
| No. of SNPs | 1 513 525 | |||||
| EMS SNPs (G>A/C>T) | 141 960 | |||||
| AF (WT-bulk) <0.35; AF (cuc2b-bulk) >0.65 | 2888 | |||||
| Exonic SNPs | 263 | |||||
| Non-common SNPs | 97 | |||||
| EMS SNPs (GQ >90; DP >10) | 39 | |||||
| Non-synonymous SNPs | 16 | |||||
| Non-synonymous SNPs on QTL6 | 2 | |||||
| Candidate SNPs | ||||||
| Chr | Position | Ref | Alt | Gene ID | Effect | Annotation |
|---|---|---|---|---|---|---|
| 6 | 4168895 | C | T | Cp4.1LG06g06610 | S230F | NAC domain protein |
| 6 | 4671155 | G | A | Cp4.1LG06g08960 | V884M | Autophagy-related protein 18f-like |
AF, allelic frequency; GQ, genotype quality; DP, read depth.
Fig. 3.
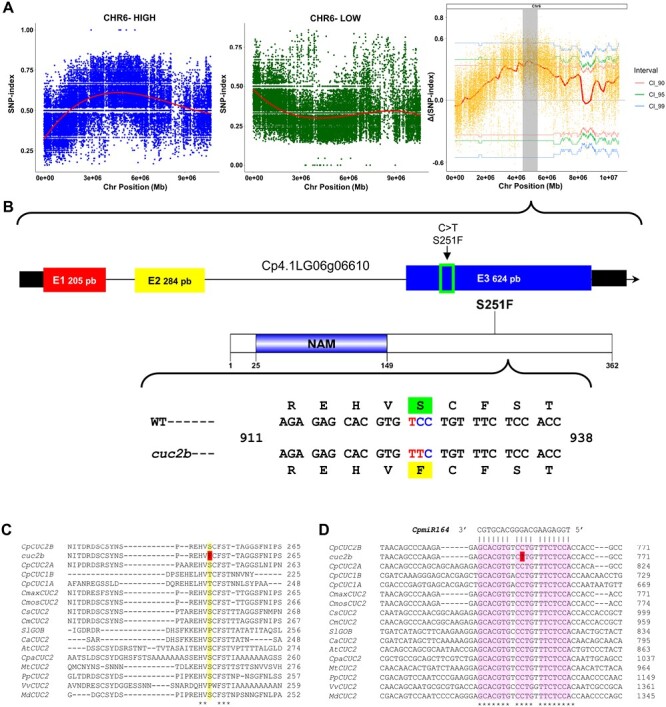
Identification of the cuc2b causal mutation by BSA-seq. (A) SNP index plots of WT and mutant bulks, and the ΔSNP index plot on chromosome 6. The fine mapping of this region allows the identification of an SNP in the candidate region of chromosome 6 as the causal mutation of the cuc2b phenotype. (B) Genomic location of the cuc2b mutation in exon 3 of a NAC-like gene (Cp4.1LG06g06610) in Cucurbita pepo subgenome B. The gene was called CpCUC2B because it is coding for a protein with a high identity to Arabidopsis thaliana transcription factor CUC2. (C) The cuc2b mutation causes a change of serine to phenylalanine in residue 251 of CpCUC2B. (D) Multiple alignment of the miR164-binding site in the mRNA sequence of the CUC2 genes of different organisms (Cp, Cucurbita pepo; Cmax, Cucurbita maxima; Cmos, Cucurbita moschata; Cs, Cucumis sativus; Cm, Cucumis melo; Ca, Capsicum annuum; At, Arabidopsis thaliana; Cpa, Carica papaya; Mt, Medicago truncatula; Pp, Prunus persica; Vv, Vitis vinifera; Md, Malus domestica). The cuc2b mutation hits a very conserved nucleotide of the miR164-binding site of CpCUC2B.
Although the two bulks were made up of the most extreme phenotypes, given that the cuc2b mutant was semi-dominant, its causal mutation should have a minimum SNP index of 0.66 in the mutant-bulk and a maximum SNP index of 0.33 in the WT-bulk. Therefore, the >141 000 canonical EMS mutations (C>T or G>A) identified were filtered by their allele frequency (AF>0.65 in the cuc2b-bulk and AF<0.35 in the WT-bulk), which resulted in 2888 mutations. Only 97 of the filtered mutations were exonic and absent from other lines in the mutant collection, 16 were non-synonymous, and only two of them were located in QTL6 (SNP1: Cp4.1LG06:4168895 and SNP2: Cp4.1LG06:4671155). SNP1 was a C>T transition in the gene Cp4.1LG06g06610, which encodes a transcription factor containing the NAC domain (Table 3), while SNP2 was a C>T transition in the gene Cp4.1LG06g08960, which encodes the 18f-like protein related to autophagy (Table 4).
To confirm the causal mutation of the cuc2b phenotype, a total of 177 plants from a BC1S1 population were genotyped for both SNP1 and SNP2 in QTL6. Only SNP1 co-segregated perfectly with the cuc2b locus, but 13% recombination was found between cuc2b and SNP2 (Supplementary Dataset S1). Furthermore, we genotyped two additional segregating populations, 179 plants from a BC1S1 generation and 280 plants from a BC2S1 generation, finding a perfect co-segregation between SNP1 and the cuc2b phenotype (Supplementary Dataset S1). Based on the co-segregation between SNP1 and the mutant phenotype in 636 plants, we concluded that the CUC2B locus is probably encoding the NAC transcription factor gene CpCUC2B.
The mutation cuc2b affected the third exon of the gene, producing a C>T transition at the 753 nucleotide position of the CpCUC2B transcript, and a serine to phenylalanine substitution at position 251 of the protein (S251F) (Fig. 3B). When homologous protein sequences from different plants were aligned, it was found that S251 was not a very conserved residue between plants, and bioinformatic analysis with the SNAP2 tool predicted a reduced effect of S251F substitution on protein function (Fig. 3C; Supplementary Figs S2, S3). However, the 21 nucleotide sequence around the CpCUC2B mutation was highly conserved in plants. This conserved region of mRNA corresponds to the miR164-binding site (Fig. 3D), a negative post-transcriptional regulator of these NAC-like transcription factors in different plant species (Mallory et al., 2004; Adam et al., 2011) (Supplementary Fig. S4). The mutation affects nucleotide 10 of the binding site in mRNA, which is part of the miR164 slicing site (Voinnet, 2009).
Phylogeny and molecular structure of CpCUC2B
An NCBI BLASTn and BLASTp search with nucleotide and amino acid sequences of Cp4.1LG06g06610 demonstrated a high homology with Arabidopsis CUP-SHAPED COTYLEDON clade genes CUC1, CUC2, and CUC3, three transcription factor genes of the NAC family. We searched for other CUC genes in the C. pepo genome. Since the genome of C. pepo is duplicated (Sun et al., 2017; Montero-Pau et al., 2018), we found seven CUC homologs. The clustal analysis performed with the three Arabidopsis and the seven squash CUC proteins indicated that Cp4.1LG06g06610 probably corresponded to Arabidopsis CUC2 (Fig. 4A) and, since it is located in the B subgenome, the gene was called CpCUC2B. Its paralog in subgenome A, CpCUC2A (Cp4.1LG02g01950), was found in a syntenic block between chromosomes 6 and 2 (Cucurbit Genomic Database, http://cucurbitgenomics.org/). The two paralogs shared >80% homology. The other five squash CUC genes were clustered with Arabidopsis CUC1 or CUC3 (Fig. 4A), suggesting that all these gene functions are conserved between Arabidopsis and squash.
Fig. 4.
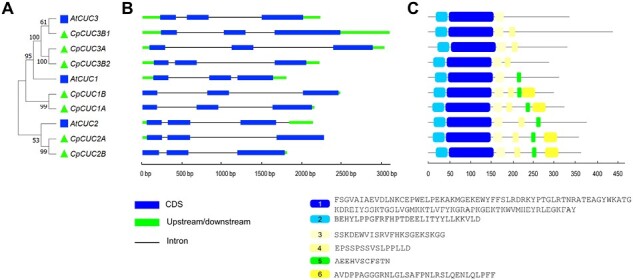
Phylogenetic relationships, gene structure, and architecture of conserved protein motifs in CUC genes from Cucurbita pepo and Arabidopsis thaliana. (A). Phylogenetic tree constructed based on the CUC full-length protein sequences. (B) Exon–intron structure of CUC genes. (C) Schematic representation of the conserved motifs in CUC proteins. Motifs 1 and 2 comprise the NAC domain of this family of transcription factors. Motifs 4, 5, and 6 correspond to motifs I, II, and III in Adams et al. (2011).
The CpCUC2B and CpCUC2A paralogs, but also the other CUC-like squash genes, were found to have the same molecular structure as those of Arabidopsis and other species, with three exons and two introns (Fig. 4B), as well as the same domain architecture as CUC-like transcription factors (Fig. 4C). The miR164-binding site was only found in the CpCUC1 and CpCUC2B transcripts, indicating a common post-transcriptional regulation of the CUC1 and CUC2 genes in Arabidopsis and squash.
The analysis of conserved motifs using the MEME tool (https://meme-suite.org/meme/tools/meme) allowed the identification of six different motifs, the same domain architecture as CUC-like transcription factors (Fig. 4C). Motif 1 and motif 2 correspond to the conserved non-apical-meristem (NAM) domain, characteristic of NAC genes in this category. Motif 4 corresponds to motif I in Adam et al. (2011) and comprised the L motif in Larsson et al. (2012). Motifs 5 and 6 were specific to CpCUC1 and CpCUC2 and were absent in CpCUC3. Motif 5, defined as motif II in Adam et al. (2011) or motif V in Larsson et al. (2012), corresponds to the miR164-binding site in the CUC mRNA sequence, while the function of motif 6, defined as motif III in Adam et al. (2011), has not been determined yet (Maugarny et al., 2016).
Tissular regulation of the C. pepo miR164 and CUC genes
Since CUC1 and CUC2 expression is known to be regulated by miR164 in Arabidopsis and other species, a search was conducted to identify potential loci transcribing this specific miRNA. Six miR164 loci were identified in the C. pepo genome, located on chromosomes 3, 8, 4, 10, 18, and 19, respectively (Supplementary Fig. S5). The MIR164C1 transcript (LOC111803168 on chromosome 10) and the MIR164C2 transcript (LOC111782191 on chromosome 19) were previously annotated as non-coding RNAs (ncRNAs) consisting of 410 and 1088 nucleotides, respectively, in the NCBI database (Supplementary Fig. S5). The loci MIR164A1, MIR164A2, MIR164B1, and MIR164B2 were also identified in our C. pepo RNA-seq projects, which included transcripts of 421, 399, 410, and 210 bp, respectively (Supplementary Fig. S5).
RNA-seq data from different plant organs revealed that the highest expression of MIR164 genes was in the apical shoots and in female and male flowers, including the ovary (Fig. 5A). MIR164C1 was detected in all plant organs, with higher expression levels in male and female flowers (flowers with a 30 mm length corolla). MIR164C2 exhibited higher expression in leaves and in female and male flowers, but appeared to have reduced expression in other organs. MIR164B1 showed predominant expression in the apical shoots, but reduced expression in other organs, and the same was true for MIR164B2, but with a lower expression level. MIR164B1 showed higher expression in the apical shoots of plants after female flowering (female apical shoots) than in the apical shoots of plants before female flowering (male apical shoots), suggesting the involvement of this miR164b1 in the regulation of the female flowering transition (Fig. 5A). MIR164A2 did not show detectable expression in the tissues analyzed, while MIR164A1 showed reduced expression compared with the other MIR164 loci (Fig. 5A).
Fig. 5.
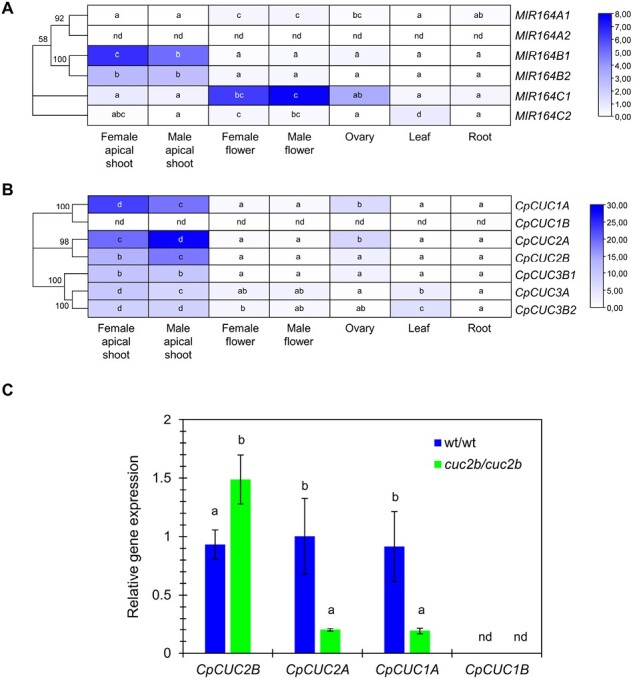
Expression patterns of (A) MIR164 and (B) CpCUC genes in different organs of WT plants: apical shoot at the three (male apical shoot before female flowering) and 12 nodes stage (female apical shoot after female flowering), male and female flowers with a corolla length of 30 mm, ovaries from flowers at the same developmental stage, and leaves of 10 mm and roots from plantlets 17–21 d after germination. FPKM values were used to generate the heatmap with hierarchical clustering analysis. The scale represents the relative signal intensity of the FPKM values. (C) qRT-PCR of CpCUC genes in the apical shoots of WT and cuc2b plants after female flowering. PCR primers for CpCUC genes were designed in the flanking regions of the predicted miR164-binding sites. Different letters indicate significant differences between samples for each gene (ANOVA, P≤0.05); nd, no detectable expression.
The same RNA-seq data also revealed that, in general, the transcription of CUC genes was negatively correlated with MIR164 expression (Fig. 5B). The CUC genes were expressed mainly in the apical shoots of plants producing male and female flowers (Fig. 5B). They were also found to be expressed in the ovary and the leaf, but showed reduced expression in female and male flowers (Fig. 5B). CpCUC2A and CpCUC2B showed a high expression in the apical shoots producing both lateral male flowers (male apical shoots) or female flowers (female apical shoots), with their expression negatively correlated with the abundance of MIR164B1 transcripts, namely higher in the apical shoots of plants before female flowering, and therefore producing only male flowers (Fig. 5B). Female flowering was then related to an up-regulation of MIR164B1 and a down-regulation of CpCUC2A and CpCUC2B. The ovaries also exhibited high expression of the genes CpCUC1A, CpCUC2A, and CpCUC2B, although the values were lower than in the apical shoots. In male and female flowers, leaves, and roots, CUC genes showed very low expression levels (Fig. 5B). The CpCUC1B transcript was not detectable in the different tissues analyzed (Fig. 5B).
Given the remarkable expression of CpCUC genes in the apical shoot, qRT-PCR was used to compare the relative expression of CpCUC2B, but also CpCUC2A, CpCUC1A, and CpCUC1B, in the apical shoots of WT and cuc2b plants after the female flowering transition (Fig. 5C). Given that these genes could be regulated by miR164, primers for gene expression were designed in the flanking regions of the predicted miR164-binding site, so detecting only the non-degraded mRNA. It was confirmed that CpCUC1B is not expressed in the apical shoots. The CpCUC2B gene was found to be overexpressed in the apical shoots of cuc2b plants relative to the WT (Fig. 5C). These results indicate that cuc2b is likely to be a miR164-resistant allelic variant that increases CpCUC2B transcription, which explains the mutant gain-of-function phenotype. To validate the predicted binding and cutting site of miR164, relative gene expression was also assessed using primers designed in the 3' region of the transcript, outside the miR164-binding site (Supplementary Fig. S6). With the use of the latter primers, the level of CpCUC2B transcripts was the same in WT and cuc2b plants (Supplementary Fig. S6). Therefore, the differences in expression between WT and cuc2b with the primers flanking the miR164-binding site would indicate a reduced integrity of the WT transcript in that region, confirming the miR164 binding and cutting site. On the other hand, the cuc2b mutation diminished the expression of CpCUC2A and CpCUC1A in the apical shoot of plants, suggesting a possible increased competitive action of miR164 on these two genes (Fig. 5B).
Regulatory network of CpCUC genes
To elucidate the different elements that may regulate CUC genes in response to various stresses or hormones, cis-regulatory elements (CREs) in the promoter region of Arabidopsis and squash CUC genes were identified by searching in a 1000 bp region upstream of the ATG start codon (Supplementary Fig. S7A). The presence of regulatory elements that respond to abiotic stresses was notably observed, highlighting the importance of CUC genes in environmental stress responses. Moreover, CUC genes appear to respond to MYB-, MYC-, and WRKY-like transcription factors, all of them involved in stress response. Elements responsive to jasmonic acid (JA) and gibberellins (GAs) were found in the CpCUC2B promoter, while its paralog CpCUC2A was found to be responsive to ethylene, JA, and abscisic acid (ABA) (Supplementary Fig. S7A).
The transcriptomic data of C. pepo obtained by our group in different RNA sequencing projects was used to predict the potential regulatory network of CpCUC1 and CpCUC2, the miR164-regulated CUC genes. We found that there are 99 genes regulated by CpCUC2A, 90 by CpCUC2B, and 48 by CpCUC1A (Supplementary Dataset S2). CpCUC2B, CpCUC2A, and CpCUC1A appear to regulate several meristem-determining genes such as CpSOL2/CRN, CpCLAVATA2, and CpSQN, whose mutants in Arabidopsis exhibit alterations in the number of carpels (Müller et al., 2008). They also regulate certain homeodomain protein genes (CpAH22, CpPDF2, and CpARID1) that affect embryonic and gamete development (Tan and Irish, 2006; Ogawa et al., 2015; Li et al., 2017). The expression of CUC genes was also related to hormone transduction pathways, particularly those involved in ABA or auxin signaling. Regarding organ boundary maintenance, the presence of two MYB-like transcription factor genes, CpMYB105 and CpMYB117, with significant effects on organ patterning in Arabidopsis is important (Lee et al., 2009; Gomez et al., 2011). CpCUC2B appears to also regulate CpWIP1B, encoding a key transcription factor orchestrating the development of male flowers in cucurbits (Supplementary Fig. S7B).
Crosstalk between CpCUC2B and ethylene in flower development and sex determination
Given that ethylene is the main regulator of sex determination in cucurbits, we studied the crosstalk between CpCUC2B and ethylene in the control of sex determination. Three different approaches were used for this purpose: (i) determining the phenotype of double mutants between cuc2b and aco1a and etr2b, two mutations affecting ethylene biosynthesis and signaling, respectively; (ii) comparing the expression of ethylene genes in WT and cuc2b; and (iii) comparing the expression of CUC and MIR164 genes in the WT and the ethylene-deficient and insensitive mutants aco1a, etr1a-1, etr1b, and etr2b.
The flower phenotypes of single and double mutants cuc2b/aco1a and cuc2b/etr2b are shown in Table 5 and Supplementary Fig. S8. Both aco1a and etr2b exhibited a partial andromonoecious phenotype (García et al., 2020b; Cebrián et al., 2022), with 25.9% and 2.5% of total flowers converted into bisexual or hermaphrodite flowers in aco1a and etr2b, respectively (Table 5). Therefore, both CpACO1A and CpETR2B are involved in the arrest of stamens during the development of female flowers. Like cuc2b, the single etr2b also showed a decrease in the number of pistillate flowers per plant (7.6%), indicating that etr2b directly converted a large number of female flowers into male flowers (Table 5). The ethylene response mediated by CpETR2B is therefore involved not only in the arrest of stamen but also in the promotion of carpel development in female flowers. This is contrary to the function of CpCUC2B, which, as demonstrated by the maleness phenotype of the gain-of-function cuc2b, acts as a repressor of carpel development for the specification of male flowers. However, single aco1a and etr2b mutants did not show the ectopic floral meristem or ectopic floral organs of cuc2b, producing only one flower per node and five sepals and petals, and three stamens or carpels per male of female flower, respectively (Table 5).
Table 5.
Phenotypic evaluation of single and double mutants between cuc2b and the ethylene-deficient and ethylene-insensitive mutants aco1a and etr2b
| Node | Double flowers (%) | Flowers with changes in the number of floral organs with respect to the WT (%) | Bisexual flowers (%)a | Pistillate flowers (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Sepals | Petals | Stamens | Carpels | |||||
| wt/wt | ♀ | 0 | 0 | 0 | 0 | 0 | 23.7 ± 1.2 | |
| ♂ | 0 | 0 | 0 | 0 | ||||
| cuc2b | ♀ | 17.6 | 76 | 63 | 72 | 2.4 | 7.6 ± 1.1 | |
| ♂ | 37.4 | 69 | 60 | 81 | ||||
| aco1a | ♀ | 0 | 0 | 0 | 0 | 25.9 | 35.01 ± 3.4 | |
| ♂ | 0 | 0 | 0 | 0 | ||||
| etr2b | ♀ | 0 | 0 | 0 | 0 | 2.5 | 7.84 ± 1.35 | |
| ♂ | 0 | 0 | 0 | 0 | ||||
| cuc2b/aco1a | ♀ | 0 | 67 | 33 | 12 | 2.6 | 6.35 ± 2.15 | |
| ♀ | 17 | 32 | 65 | 39 | – | |||
| cuc2b/etr2b | ♀ | – | – | – | – | 0.0 ± 0.0 | ||
| ♀ | 26 | 65 | 40 | 56 | ||||
a Ovary-bearing flowers with anthers from the total of flowers analyzed of each genotype.
The homozygous double mutant cuc2b/aco1a showed a similar reduction in the percentage of pistillate flowers per plant (6.35%) to cuc2b (7.6%) (Table 5), and very different from the one shown by aco1a (35.01%), indicating that cuc2b is epistatic over aco1a in the control of sex determination. A total of 2.6% of the total flowers in the double mutant were bisexual (Supplementary Fig. S8), similar to the cuc2b single mutant, indicating a slight lack of stamen arrest in those floral meristems determined as female flowers, but the reduced number of female flowers in the double mutant cuc2b/aco1a and the environmental dependence of the stamen development phenotype made it difficult to establish a solid correlation between the effect of both mutations in the phenotype. The specific phenotypic defects of cuc2b were more pronounced in male flowers than in female double mutant flowers. Therefore, the percentage of affected flowers was lower in cuc2b/aco1a than in cuc2b (Table 5). Thus, double flowers occurred in male but not in female nodes of the double mutant, and changes in the number of floral organs affected male flowers more than female flowers (Table 5).
On the other hand, the homozygous double mutant cuc2b/etr2b was completely androecious and produced no pistillate flowers at all. The higher severity of the cuc2b/etr2b phenotype compared with that of the single mutants, with complete suppression of carpel development and complete conversion of female into male flowers, indicates a synergistic effect of cuc2b and etr2b on sex determination. Male flowers of cuc2b/etr2b showed the specific defects of the single mutant cuc2b, with double flowers and a higher number of sepals, petals, and stamens than in the WT (Table 5).
In Arabidopsis, ethylene is known to regulate the expression of the miR164 and CUC genes (Li et al., 2013). To investigate whether ethylene can also modulate the expression of the CUC/miR164 module in the apical shoot of squash, its expression was comparatively studied in WT and ethylene-deficient (aco1a) and ethylene-insensitive (etr1a-1, etr1b, and etr2b) mutants. Both etr1b and etr1a-1 confer an androecious phenotype, and plants only produced male flowers (García et al., 2020a). RNA-seq data from apical shoots of plants in the male and female stages of development confirmed that the MIR164B1 or MIR164B2 loci (highly expressed in the apical shoots of both WT and mutant plants) were up-regulated in the apical shoots of plants producing female flowers, but ethylene-insensitive mutations etr1b and etr1a-1 did not produce a significant change in their expression in comparison with the WT at the same stage of development (Fig. 6A).
Fig. 6.
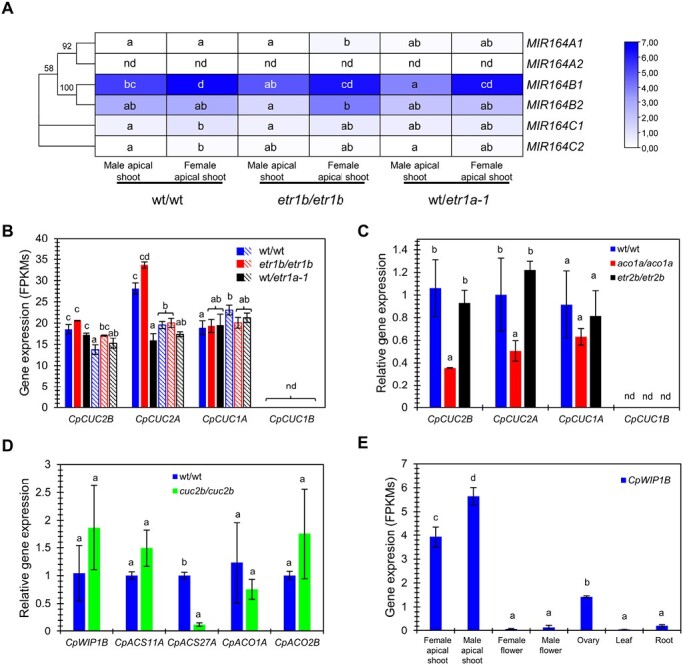
Crosstalk between ethylene and CpCUC genes in the regulation of sex determination in C. pepo. Gene expression of miR164 and CpCUC genes in ethylene-deficient and insensitive mutants, and of ethylene and other sex-regulating genes in the cuc2b mutant is shown. (A) Expression patterns of MIR164 loci and (B) CpCUC genes regulated by miR164 in the apical shoots of the ethylene-insensitive androecious mutants etr1b and etr1a-1 plants at the three nodes (male apical shoot, solid color) and 12 nodes stage (female apical shoot, striped pattern color). FPKM values were used to generate the heatmaps with hierarchical clustering analysis. The scale represents the relative signal intensity of the FPKM values. (C) Relative gene expression obtained by qRT-PCR of CpCUC genes regulated by miR164 in the apical shoots of WT and ethylene-deficient (aco1a) and ethylene-insensitive (etr2b) mutants. (D) Relative gene expression obtained by qRT-PCR of ethylene biosynthesis genes and the transcription factor gene CpWIP1B in the apical shoots of WT and cuc2b plants. (E) Expression patterns of CpWIP1B in different organs of WT plants: apical shoot at the three (male apical shoot, before female flowering) and 12 nodes stage (female apical shoot, after female flowering), male and female flowers with a corolla length of 30 mm, ovaries from flowers at the same developmental stage, and leaves of 10 mm and roots from plantlets of 17–21 d after germination. FPKM values were used to generate the graph. Different letters indicate significant differences between samples in each gene (ANOVA, P≤0.05). nd, no detectable expression.
The CpCUC1A and CpCUC1B genes were not regulated by female flowering, nor by ethylene-insensitive mutations etr1a-1 and etr1b (Fig. 6B). However, CpCUC2A and CpCUC2B were down-regulated in the apical shoots of plants producing female flowers relative to those in earlier stages, when the plant is only producing male flowers (Fig. 2B). This regulation could be mediated by miR164, as the MIR164B1 gene is induced in the apical shoot upon female flowering. Only etr1a-1 was found to down-regulate CpCUC2A in the male phase of development, validating the specific ethylene regulation site predicted by ARACNE in CpCUC2A (Fig. 6B). However, this regulation does not appear to have anything to do with sex expression, as at this stage both WT and etr1a-1 were producing male flowers. No transcriptional changes were found for CpCUC genes in the apical shoots of ethylene-insensitive mutants at the female phase of development (Fig. 6B), when the WT were producing female flowers, but the ethylene mutants were still producing male flowers.
The qPCR expression analysis with primers at the miR164-binding site confirmed that the CpCUC transcripts accumulated similarly in the apical shoots of both the WT and the ethylene-insensitive mutant etr2b (Fig. 6C). However, the ethylene mutation aco1a reduced the accumulation of CpCUC2A and CpCUC2B transcripts in the apical shoots of the plant in the female phase of development (Fig. 6C). The mutations aco1a and etr2b produce an andromonoecious sex phenotype, consistent with a reduction in ethylene production or ethylene sensitivity in the lateral floral meristems of the apical shoot, and the suppression of stamen arrest in female flowers (García et al., 2020b; Cebrián et al., 2022). Therefore, the down-regulation of CpCUC2A and CpCUC2B in aco1a must occur at later stages of flower development, when the floral meristem is already determined as a female flower.
The transcription of ethylene biosynthesis sex-determining genes was finally compared in the apical shoots of WT and cuc2b plants at the female stage of development (Fig. 6D). Only the ethylene biosynthesis gene CpACS27A showed a significant down-regulation in cuc2b compared with the WT. CpACS27A is the partner enzyme of CpACO1A in the production of late ethylene required for stamen arrest in female flowers, which explains the development of bisexual flowers in cuc2b. None of the ethylene biosynthesis genes involved the promotion of carpels at earlier stages of female flower development (CpACS11A and CpACO2B) changed its expression in cuc2b (Fig. 6D), indicating that CpCUC2B suppresses carpel development on a pathway other than that of ethylene. Therefore, the phenotypes of the double mutants cuc2b/etr2b and cuc2b/aco1a and the gene expression data indicate that CpCUC2B and ethylene act on two independent pathways controlling the development of carpels, but interact within the same pathway for the regulation of stamen development.
The function to CpCUC2B is similar to that of the transcription factor WIP1, which aborts the development of carpels for the specification of male flowers. However, CpCUC2B is not negatively regulated by ethylene that promotes carpel development (Fig. 6B, C), as occurs for WIP1 of melon and cucumber. The squash CpWIP1B is not regulated by CpCUC2B (Fig. 6D), indicating that CpCUC2B performs its function independently of CpWIP1B. RNA-seq data showed that CpWIP1B is not specifically expressed in male flowers, but is also expressed in female flowers and in other plant organs (Fig. 6E). However, CpWIP1B was found to have a higher expression in the apical shoots of plants producing male flowers than in the apical shoots of plants producing female flowers (Fig. 6E), which may indicate a conserved role in the determination of squash male flowers.
Discussion
cuc2b is an miR164-resistant gain-of-function mutation in CpCUC2B
The squash mutant cuc2b has been isolated from a direct screening of an EMS population of squash (García et al., 2018). Both QTL-seq analysis and fine mapping have shown that cuc2b is a canonical EMS transition C>T that produces an S251F change in the transcription factor CpCUC2B. So, the mutation was also called cuc2b. The gene CpCUC2B and its paralog CpCUC2A share high homology with another five genes in the duplicated genome of squash (CpCUC1A, CpCUC1B, CpCUC3A, CpCUC3B1, and CpCUC3B2), all of them coding for NAC-domain transcription factors, represented by the petunia NO APICAL MERISTEM (NAM), Antirrhinum CUP, and the three Arabidopsis CUC1, CUC2, and CUC3 genes (Souer et al., 1996; Aida et al., 1997; Weir et al., 2004). Based on the sequence of the NAC domain, but also on their miR164 post-transcriptional regulation, different plant NACs are divided into two clades (Maugarny et al., 2016). The first clade, represented by NAM, CUC1, and CUC2, along with squash CpCUC1A/B and CpCUC2A/B, displays a binding site for miR164. The second clade is represented by CUC3 and, like the three squash CpCUC3 genes, is not regulated by miR164. The tissular expression patterns of the six squash MIR164 genes and the seven CpCUC genes found in the genome of C. pepo confirmed that CUC/miR164 is a conserved developmental regulation module in both angiosperms and gymnosperms (Adam et al., 2011; Vialette-Guiraud et al., 2011). The negative regulation of squash CpCUC1 and CpCUC2 by miR164 was evident in the apical shoots of plants during the female flowering transition, where MIR164 (especially locus MIR164B1) was found to be up-regulated and CpCUC2A and CpCUC2B down-regulated (Fig. 5). The expression of the CpCUC1 and CpCUC2 genes was also negatively correlated with the expression of the MIR164 loci in different plant organs (Fig. 5), demonstrating that these squash genes are not only those showing the highest sequence identity with Arabidopsis CUC1 and CUC2, but are also regulated by miR164.
Different pieces of evidence indicate that squash cuc2b is an miR164-resistant and gain-of-function mutation in CpCUC2B. First, the change produced by the cuc2b mutation affects a non-conserved residue of the protein sequence but, at a nucleotide level, the transition C>T affects position 10 of the binding site of miR164, which is a known cutting site position for the post-transcriptional regulation of target mRNA (Voinnet, 2009). Second, the effects of this mutation, which leads to accessory floral meristems and floral organs, suppressed carpel development, and increased leaf serration, resembled those of mutants at MIR164 loci or miR164-binding sites of CUC1 and CUC2 in Arabidopsis mutants (Laufs et al., 2004; Mallory et al., 2004; Baker et al., 2005; Larue et al., 2009) as well as the tomato Gob4-d (Berger et al., 2009). Third, in the apical shoot, where axillary floral meristems are determined as male or female flowers, and floral organs and leaves are defining their boundaries, the complete transcripts of CpCUC2B were accumulated more strongly in cuc2b than in the WT. Although amino acid substitution appears to have little effect on the protein stability (Supplementary Fig. S3), a potential impact on protein function and mutant phenotype cannot be excluded. The reduced expression of the other CpCUC1 and CpCUC2 genes (Fig. 5) in the same apical shoots suggests a competition between the different CUC genes for binding miR164, but clearly demonstrates a major function for CpCUC2B in the developmental functions defined by cuc2b. Despite this, given the functional redundancy of Arabidopsis CUC genes, with cuc1 and cuc2 single mutants having a nearly normal phenotype (Takada et al., 2001), it is likely that the partial penetrance of cuc2b in different plants and flowers may depend on the combined effect of CpCUC1 and CpCUC2 genes, up-regulating CpCUC2B, but down-regulating CpCUC2A and CpCUC1A in overlapping tissues. Therefore, it cannot be ruled out that CpCUC1A and CpCUC2A can complement the defects produced by cuc2b in certain developmental processes.
CpCUC2B has multiple functions in the formation of the axillary floral meristem and the development of the lateral organs
The phenotype of cuc2b has shown that CpCUC2B plays a relevant role in defining the boundaries of the floral organs and the leaf. The role of CUC genes in boundary specification and lateral organ separation is conserved in different plant species, including the dicots Petunia, Arabidopsis, tomato, and Medicago (Souer et al., 1996; Aida et al., 1997; Weir et al., 2004; Berger et al., 2009; Cheng et al., 2012), and monocots such as rice and sugarcane (Chang et al., 2021; Aslam et al., 2022). For this reason, this family of genes are known as boundary genes. The inferred regulatory network of the CpCUC2A/B gene also supports the involvement of these genes in organ patterning and in reproductive development.
The loss of function of cuc mutations hinders the formation of boundaries in the floral meristem and the separation of floral organs, leading to flowers with fused and a reduced number of floral organs (Aida et al., 1997; Vroemen et al., 2003; Hibara et al., 2006). During leaf development, loss-of-function mutations result in fused or a low number of leaflets of compound leaves and reduced serration of Arabidopsis simple leaves (Blein et al., 2008; Berger et al., 2009; Cheng et al., 2012; Wang and Dane, 2013). In contrast, the squash cuc2b gain-of-function mutation leads to an increase in the number of sepals, petals, and stamens, and to a deeper and larger leaf serration, a phenotype also exhibited by overexpressed CUC mutants resulting from a defective miR164 regulation in Arabidopsis (Laufs et al., 2004; Baker et al., 2005; Nikovics et al., 2006) and the miR164-resistant variant of tomato SlGOB (Berger et al., 2009).
In squash, solitary male or female flowers are formed on the axil of each leaf. The development of double male and female flowers in the squash cuc2b mutant therefore shows the crucial role of CpCUC2B in the formation of the floral meristem, a function that has not been previously reported for the CUC genes. However, the cuc2b mutation does not appear to be very severe since only 17.6 and 37.4 of the cuc2b nodes were occupied by double flowers, and the rest of the nodes develop single flowers. Double flowers are likely to be due to the formation of an accessory boundary in the floral meristem that eventually results in two separated or fused male or female flowers (Fig. 2). Division of the floral meristem must occur after the initiation of the primordia of sepals and petals, because in these double flowers the number of sepals and petals is reduced, while in single flowers the number of sepals and petals increases compared with the WT (Table 2). Although floral meristems do not appear to be affected by cuc mutations in Arabidopsis, the cuc2b double axillary flowers resemble those of the axillary vegetative meristem in Arabidopsis cuc gain-of-function mutants, which develop accessory vegetative buds in leaf axils (Raman et al., 2008). Therefore, it is likely that CUC genes are required for the formation of axillary meristems, be they axillary flowers or secondary axillary shoots.
CpCUC2B controls squash sex determination by arresting the development of carpels and suppressing the arrest of stamens
The role of CpCUC2B in sex determination is more specific for squash and perhaps other cucurbits. The formation of female and male unisexual flowers in monoecious and dioecious plants results from the arrest of either stamens or carpels in a floral meristem that is initiated as hermaphrodite. In cucurbits, ethylene is the hormone that determines sex, arresting stamen development and promoting carpel development for the determination of female flowers. A reduction in ethylene at very early stages of flower development leads to the activation of the WIP1 zinc finger transcription factor, which arrests the development of carpels during the determination of male flowers (Martin et al., 2009; Hu et al., 2017; Zhang et al., 2020). The role of CpCUC2B is similar to that described for WIP1 and opposite to that of ethylene. The increased number of male flowers in cuc2b plants and the reduced number of carpels in cuc2b pistillate flowers clearly show that the gene is required to arrest carpel development for the determination of male flowers. Comparison of the transcriptomes of male and hermaphrodite flowers in the androdioecious tree Tapiscia sinensis also suggested that CUC genes may promote the formation of male flowers by suppressing carpel development in hermaphrodite floral meristems (Xin et al., 2019). Furthermore, the fact that 2.4% of the cuc2b pistillate flowers were classified as bisexual, developing an ovary and stamens, but without a pistil or stigma, also indicates that CpCUC2B is also able to promote the development of stamens in a floral meristem that was already determined as female. Down-regulation of CpACS27A in cuc2b female apical shoots also supports the conclusion that CpCUC2B suppresses stamen arrest in female flowers. This function was described for WIP1 in melon and cucumber, which is negatively regulated by ethylene biosynthesis genes such as ACS11 and ACO2 at very early stages of flower development (Boualem et al., 2015; Chen et al., 2016), but later suppresses the activation of ethylene biosynthesis genes that are involved in stamen abortion (CmACS7, CsACS2, CitACS4, and CpACS27 orthologs), which leads to the floral meristem being determined as a male flower (Martínez and Jamilena, 2021). The squash CpWIP1B is highly expressed in the apical shoots of plants at the male phase of development (Fig. 6E), suggesting that it cooperates with CpCUC2B, and perhaps other CUC-like genes, in the specification of male flowers. The function of the CUC genes, as repressors of growth and organ outgrowth to define boundaries and organ separation (Aida and Tasaka, 2006), could have been exploited to promote carpel abortion during the evolutionary emergence of unisexual flowers in monoecious cucurbits. The WIP1 transcription factor has also been described as an inhibitor of growth and development by activating genes involved in programmed cell death and repressing genes involved in meristem growth (Roldan et al., 2020). In the few pistillate flowers of cuc2b, the ovary phenotype is similar to that of Arabidopsis mutants that express miR164-resistant forms of CUC1 and CUC2 (Sieber et al., 2007; Kamiuchi et al., 2014), and indicates that CpCUC2B conserves its role in placenta and ovule development.
How is CpCUC2B integrated in the genetic framework that controls cucurbit sex determination? Most of the known cucurbit sex-determining genes are ethylene biosynthesis and signaling genes. So, we studied the genetic interactions between cuc2b and the already known mutations in ethylene biosynthesis and signaling pathways. It was known that squash ethylene receptors CpETR1A, CpETR1B, and CpETR2B perceive not only the early ethylene produced by ACS11 and ACO2, promoting in this way development of carpels by repressing WIP1, but also the late ethylene that is produced by the enzymes CpACS27A and CpACO1A and that triggers stamen abortion. The phenotypes of single and double mutants, and the expression of CpCUC2B, CpWIP1B, and ethylene sex-determining genes in the ethylene and cuc2b mutants, permitted the development of the model shown in Fig. 7. The ethylene-insensitive mutations (etr1b, etr1a, etr1a-1, and etr2b) convert female flowers into male or hermaphrodite flowers, thus generating a sexual phenotype that goes from andromonoecy to androecy, depending on the severity of each mutation and the level of ethylene insensitivity of each mutant (García et al., 2020a, b). However, the aco1a mutation only converts female flowers into hermaphrodite flowers but does not produce any change in the number of male and pistillate flowers per plant (Cebrián et al., 2022). The sex phenotype of the double mutant cuc2b/aco1a demonstrated that the cuc2b mutation is epistatic over aco1a and, given that the CpACS27A gene is significantly down-regulated in cuc2b, it is likely that CpCUC2B promotes stamen development by suppressing CpACS27A, the key enzyme involved in late ethylene production and stamen abortion in female flowers (Fig. 7). Down-regulation of CpCUC2A and CpCUC2B in the apical shoots of aco1a may indicate that this late ethylene may finally activate the CUC genes required for the specification and separation of female floral organs (Fig. 7). This conclusion is also supported by the reduced boundary-specific cuc2b phenotypes in the female flowers compared with male flowers of the double mutant cuc2b/aco1a (Table 5).
Fig. 7.
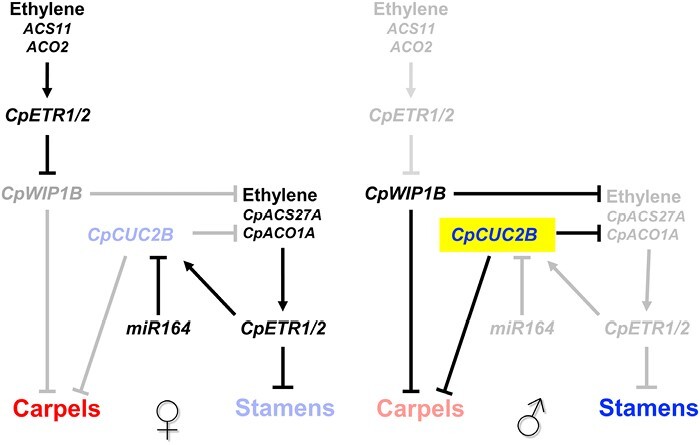
Role of CpCUC2B and miR164 in the gene network model regulating the arrest of stamens and carpels for female and male flower determination, respectively. The function of CpCUC2B in the arrest of carpel development for the development of male flowers is independent of ethylene produced by ACS11 and ACO2, and of the transcription factor CpWIP1B. However, CpCUC2B inhibits the ethylene produced by CpACS27A and CpACO1A, the enzymes involved in the arrest of stamen during the development of female flowers. Genes that are active in male or female flower development are highlighted in black or a more intense color, while those that are repressed are in gray or a paler color.
On the other hand, the androecious phenotype of the double mutant cuc2b/etr2b, where all female flowers were converted into male flowers, demonstrated a synergistic effect of the cuc2b and etr2b mutations on male flower production. The role of CpCUC2B in carpel abortion during the determination of a male flower is therefore independent of ethylene (Fig. 7), a regulation that differs from that of WIP1, whose expression is negatively regulated by ACS11 and ACO2 in female floral meristems of melon and cucumber (Boualem et al., 2015; Chen et al., 2016). The fact that the study of gene expression did not show transcriptional regulation between the CpCUC and CpETR genes (Fig. 6) also supports the independent action of CpCUC2B and ethylene in the suppression or promotion of carpel development in squash flowers. A more in-depth study would be required to determine whether the role of ethylene in the promotion of carpel development could be mediated by the down-regulation of CpWIP1B in squash. The present data, however, indicate that CpWIP1B may be involved in male flower specification, since the gene is highly expressed in the apical shoots of plants at the early male phase of development (Fig. 6E).
Supplementary data
The following supplementary data are available at JXB online.
Dataset S1. Validation of the cuc2b mutation.
Dataset S2. Regulatory network of CUC genes inferred by the ARACNE algorithm.
Table S1. List of the CUC genes and proteins used in alignments and phylogenetic analysis.
Table S2. Primers used for gene expression and mRNA degradation analysis by qPCR.
Fig. S1. Effects of the cuc2b mutation on vegetative development.
Fig. S2. Multiple alignment of CpCUC2 amino acid sequences with homologous CUC2 proteins from diverse species.
Fig. S3. Effect of cuc2b mutation on protein stability.
Fig. S4. Multiple alignment of CpmiR164 mature sequence with homologous miR164 from diverse plant species.
Fig. S5. Loci encoding miR164 in C. pepo.
Fig. S6. qRT-PCR of the CpCUC2B gene.
Fig. S7. CUC gene regulation network.
Fig. S8. Phenotypes of single and double mutants of etr2b, aco1a, and cuc2b.
Contributor Information
María Segura, Department of Biology and Geology. Agri-food Campus of International Excellence (CeiA3) and Research Center CIAIMBITAL, University of Almería, 04120 Almería, Spain.
Alicia García, Department of Biology and Geology. Agri-food Campus of International Excellence (CeiA3) and Research Center CIAIMBITAL, University of Almería, 04120 Almería, Spain.
Germán Gamarra, Department of Biology and Geology. Agri-food Campus of International Excellence (CeiA3) and Research Center CIAIMBITAL, University of Almería, 04120 Almería, Spain.
Álvaro Benítez, Department of Biology and Geology. Agri-food Campus of International Excellence (CeiA3) and Research Center CIAIMBITAL, University of Almería, 04120 Almería, Spain.
Jessica Iglesias-Moya, Department of Biology and Geology. Agri-food Campus of International Excellence (CeiA3) and Research Center CIAIMBITAL, University of Almería, 04120 Almería, Spain.
Cecilia Martínez, Department of Biology and Geology. Agri-food Campus of International Excellence (CeiA3) and Research Center CIAIMBITAL, University of Almería, 04120 Almería, Spain.
Manuel Jamilena, Department of Biology and Geology. Agri-food Campus of International Excellence (CeiA3) and Research Center CIAIMBITAL, University of Almería, 04120 Almería, Spain.
Rainer Melzer, University College Dublin, Ireland.
Author contributions
MJ and CM: design and coordination of the research; MS: conducting most of the experiments and data analysis; AG, AB, GG, and JI-M: collaboration in data analysis; MJ and MS: writing and revision. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare no conflict of interest regarding this publication.
Funding
This work was supported by grant PID2020-118080RB-C21, funded by ‘Ministerio de Ciencia e Innovación’ together with EU FEDER funds. MS acknowledges the D.I scholarship program (DIN2018-010127) from ‘Ministerio de Ciencia, Innovación y Universidades’ with the company Green Breeding Biotech SL; JI-M acknowledges the FPI scholarship (PRE2018-085149) from ‘Ministerio de Ciencia, Innovación y Universidades’; and AG received a Margarita Salas postdoctoral fellowship (RR_A_2022_05) of ‘Ministerio de Universidades’ funded by NextGenerationEU program.
Data availability
All relevant data can be found within the manuscript and its supporting materials. All the raw reads generated in this study have been deposited in the public database of the National Center of Biotechnology under BioProject PRJNA1042934.
References
- Adam H, Marguerettaz M, Qadri R, et al. 2011. Divergent expression patterns of miR164 and CUP-SHAPED COTYLEDON genes in palms and other monocots: implication for the evolution of meristem function in angiosperms. Molecular Biology and Evolution 28, 1439–1454. [DOI] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M.. 1997. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. The Plant Cell 9, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Tasaka M.. 2006. Genetic control of shoot organ boundaries. Current Opinion in Plant Biology 9, 72–77. [DOI] [PubMed] [Google Scholar]
- Aslam M, She Z, Jakada BH, Fakher B, Greaves JG, Yan M, Chen Y, Zheng P, Cheng Y, Qin Y.. 2022. Interspecific complementation restoration of phenotype in Arabidopsis cuc2cuc3 mutant by sugarcane CUC2 gene. BMC Plant Biology 22, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, Noble WS.. 2015. The MEME suite. Nucleic Acids Research 43, W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CC, Sieber P, Wellmer F, Meyerowitz EM.. 2005. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Current Biology 15, 303–315. [DOI] [PubMed] [Google Scholar]
- Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, Zinder M, Samach A, Eshed Y, Ori N.. 2009. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136, 823–832. [DOI] [PubMed] [Google Scholar]
- Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P.. 2008. A conserved molecular framework for compound leaf development. Science 322, 1835–1839. [DOI] [PubMed] [Google Scholar]
- Boualem A, Fergany M, Fernandez R, et al. 2008. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 321, 836–838. [DOI] [PubMed] [Google Scholar]
- Boualem A, Troadec C, Camps C, et al. 2015. A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science 350, 688–691. [DOI] [PubMed] [Google Scholar]
- Boualem A, Troadec C, Kovalski I, Sari MA, Perl-Treves R, Bendahmane A.. 2009. A conserved ethylene biosynthesis enzyme leads to andromonoecy in two Cucumis species. PLoS One 4, e6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrián G, Iglesias-Moya J, Romero J, Martínez C, Garrido D, Jamilena M.. 2022. The ethylene biosynthesis gene CpACO1A: a new player in the regulation of sex determination and female flower development in Cucurbita pepo. Frontiers in Plant Science 12, 817922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, Xu R, Xun Q, Liu J, Zhong T, Ding Y, Ding C.. 2021. OsmiR164-targeted OsNAM, a boundary gene, plays important roles in rice leaf and panicle development. The Plant Journal 106, 41–55. [DOI] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R.. 2020. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant 13, 1194–1202. [DOI] [PubMed] [Google Scholar]
- Chen H, Sun J, Li S, et al. 2016. An ACC oxidase gene essential for cucumber carpel development. Molecular Plant 9, 1315–1327. [DOI] [PubMed] [Google Scholar]
- Cheng X, Peng J, Ma J, Tang Y, Chen R, Mysore KS, Wen J.. 2012. NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula. New Phytologist 195, 71–84. [DOI] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, et al. 2021. Twelve years of SAMtools and BCFtools. GigaScience 10, giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19, 11–15. [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT.. 2014. Flexible isoform-level differential expression analysis with Ballgown. bioRxiv 003665. [Preprint]. [Google Scholar]
- García A, Aguado E, Garrido D, Martínez C, Jamilena M.. 2020a. Two androecious mutations reveal the crucial role of ethylene receptors in the initiation of female flower development in Cucurbita pepo. The Plant Journal 103, 1548–1560. [DOI] [PubMed] [Google Scholar]
- García A, Aguado E, Martínez C, Loska D, Beltrán S, Valenzuela JL, Garrido D, Jamilena M.. 2020b. The ethylene receptors CpETR1A and CpETR2B cooperate in the control of sex determination in Cucurbita pepo. Journal of Experimental Botany 71, 154–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A, Aguado E, Parra G, et al. 2018. Phenomic and genomic characterization of a mutant platform in Cucurbita pepo. Frontiers in Plant Science 9, 1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez MD, Urbez C, Perez-Amador MA, Carbonell J.. 2011. Characterization of constricted fruit (ctf) mutant uncovers a role for AtMYB117/LOF1 in ovule and fruit development in Arabidopsis thaliana. PLoS One 6, e18760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M.. 2006. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. The Plant Cell 18, 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G.. 2015. GSDS 20: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Li D, Liu X, Qi J, Gao D, Zhao S, Huang S, Sun J, Yang L.. 2017. Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Molecular Plant 10, 1575–1578. [DOI] [PubMed] [Google Scholar]
- Jin J, Tian F, Yang D-C, Meng Y-Q, Kong L, Luo J, Gao G.. 2017. PlantTFDB 40: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Research 45, D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiuchi Y, Yamamoto K, Furutani M, Tasaka M, Aida M.. 2014. The CUC1 and CUC2 genes promote carpel margin meristem formation during Arabidopsis gynoecium development. Frontiers in Plant Science 5, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P, Hammer S, Hofacker IL.. 2015. Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics 31, 3377–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35, 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Sundström JF, Sitbon F, von Arnold S.. 2012. Expression of PaNAC01, a Picea abies CUP-SHAPED COTYLEDON orthologue, is regulated by polar auxin transport and associated with differentiation of the shoot apical meristem and formation of separated cotyledons. Annals of Botany 110, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue CT, Wen J, Walker JC.. 2009. A microRNA–transcription factor module regulates lateral organ size and patterning in Arabidopsis. The Plant Journal 58, 450–463. [DOI] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J.. 2004. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. [DOI] [PubMed] [Google Scholar]
- Lee D-K, Geisler M, Springer PS.. 2009. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 136, 2423–2432. [DOI] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics (Oxford, England) 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wu W, Zhao Y, Zheng B.. 2017. A reciprocal inhibition between ARID1 and MET1 in male and female gametes in Arabidopsis. Journal of Integrative Plant Biology 59, 657–668. [DOI] [PubMed] [Google Scholar]
- Li Z, Peng J, Wen X, Guo H.. 2013. ETHYLENE-INSENSITIVE3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. The Plant Cell 25, 3311–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B.. 2004. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Current Biology 14, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Mansfeld BN, Grumet R.. 2018. QTLseqr: an R package for bulk segregant analysis with next-generation sequencing. The Plant Genome 11, 180006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano S, Aguado E, Martínez C, Megías Z, García A, Jamilena M.. 2016. The ethylene biosynthesis gene CitACS4 regulates monoecy/andromonoecy in watermelon (Citrullus lanatus). PLoS One 11, 154362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano S, Martínez C, Domínguez V, Avalos E, Garrido D, Gómez P, Jamilena M.. 2010. A major gene conferring reduced ethylene sensitivity and maleness in Cucurbita pepo. Journal of Plant Growth Regulation 29, 73–80. [Google Scholar]
- Manzano S, Martínez C, Megías Z, Garrido D, Jamilena M.. 2013. Involvement of ethylene biosynthesis and signalling in the transition from male to female flowering in the monoecious Cucurbita pepo. Journal of Plant Growth Regulation 32, 789–798. [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Favera RD, Califano A.. 2006. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Troadec C, Boualem A, Rajab M, Fernandez R, Morin H, Pitrat M, Dogimont C, Bendahmane A.. 2009. A transposon-induced epigenetic change leads to sex determination in melon. Nature 461, 1135–1138. [DOI] [PubMed] [Google Scholar]
- Martínez C, Jamilena M.. 2021. To be a male or a female flower, a question of ethylene in cucurbits. Current Opinion in Plant Biology 59, 101981. [DOI] [PubMed] [Google Scholar]
- Martínez C, Manzano S, Megías Z, Barrera A, Boualem A, Garrido D, Bendahmane A, Jamilena M.. 2014. Molecular and functional characterization of CpACS27A gene reveals its involvement in monoecy instability and other associated traits in squash (Cucurbita pepo L). Planta 239, 1201–1215. [DOI] [PubMed] [Google Scholar]
- Maugarny A, Gonçalves B, Arnaud N, Laufs P.. 2016. CUC transcription factors: to the meristem and beyond. In: Gonzalez DH, ed. Plant transcription factors. Evolutionary, structural and functional aspects. Academic Press, 229–247. [Google Scholar]
- Mistry J, Chuguransky S, Williams L, et al. 2021. Pfam: the protein families database in 2021. Nucleic Acids Research 49, D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Pau J, Blanca J, Bombarely A, et al. 2018. De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnology Journal 16, 1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Bleckmann A, Simon R.. 2008. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. The Plant Cell 20, 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P.. 2006. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. The Plant Cell 18, 2929–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa E, Yamada Y, Sezaki N, Kosaka S, Kondo H, Kamata N, Abe M, Komeda Y, Takahashi T.. 2015. ATML1 and PDF2 play a redundant and essential role in Arabidopsis embryo development. Plant and Cell Physiology 56, 1183–1192. [DOI] [PubMed] [Google Scholar]
- Pan J, Wang G, Wen H, Du H, Lian H, He H, Pan J, Cai R.. 2018. Differential gene expression caused by the F and M loci provides insight into ethylene-mediated female flower differentiation in cucumber. Frontiers in Plant Science 9, 1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannell JR. 2017. Plant sex determination. Current Biology 27, R191–R197. [DOI] [PubMed] [Google Scholar]
- Peñaranda A, Payan MC, Garrido D, Gómez P, Jamilena M.. 2007. Production of fruits with attached flowers in zucchini squash is correlated with the arrest of maturation of female flowers. Jpurnal of Horticultural Science and Biotechnology 82, 579–584. [Google Scholar]
- Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K.. 2008. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. The Plant Journal 55, 65–76. [DOI] [PubMed] [Google Scholar]
- Roldan MVG, Izhaq F, Verdenaud M, et al. 2020. Integrative genome-wide analysis reveals the role of WIP proteins in inhibition of growth and development. Communications Biology 3, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombauts S, Déhais P, Van Montagu M, Rouzé P.. 1999. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Research 27, 295–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM.. 2007. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R.. 1996. The No Apical Meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. [DOI] [PubMed] [Google Scholar]
- Sun H, Wu S, Zhang G, et al. 2017. Karyotype stability and unbiased fractionation in the paleo-allotetraploid cucurbita genomes. Molecular Plant 10, 1293–1306. [DOI] [PubMed] [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M.. 2001. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Takagi H, Abe A, Yoshida K, et al. 2013. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. The Plant Journal 74, 174–183. [DOI] [PubMed] [Google Scholar]
- Tan QK-G, Irish VF.. 2006. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiology 140, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Niu H, Wang Z, Zhang W, Wang H, Wang S, Zhang X, Li Z.. 2018. Ethylene responsive factor ERF110 mediates ethylene-regulated transcription of a sex determination-related orthologous gene in two Cucumis species. Journal of Experimental Botany 69, 2953–2965. [DOI] [PubMed] [Google Scholar]
- Tav C, Tempel S, Poligny L, Tahi F.. 2016. miRNAFold: a web server for fast miRNA precursor prediction in genomes. Nucleic Acids Research 44, W181–W184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialette-Guiraud ACM, Adam H, Finet C, Jasinski S, Jouannic S, Scutt CP.. 2011. Insights from ANA-grade angiosperms into the early evolution of CUP-SHAPED COTYLEDON genes. Annals of Botany 107, 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. [DOI] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MACJ, de Vries SC.. 2003. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in arabidopsis. The Plant Cell 15, 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H.. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research 38, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Dane F.. 2013. NAC (NAM/ATAF/CUC) transcription factors in different stresses and their signaling pathway. Acta Physiologiae Plantarum 35, 1397–1408. [Google Scholar]
- Weir I, Lu J, Cook H, Causier B, Schwarz-Sommer Z, Davies B.. 2004. Cupuliformis establishes lateral organ boundaries in Antirrhinum. Development 131, 915–922. [DOI] [PubMed] [Google Scholar]
- Xin G-L, Liu J-Q, Liu J, Ren X-L, Du X-M, Liu W-Z.. 2019. Anatomy and RNA-Seq reveal important gene pathways regulating sex differentiation in a functionally androdioecious tree, Tapiscia sinensis. BMC Plant Biology 19, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Guo S, Ji G, et al. 2020. A unique chromosome translocation disrupting ClWIP1 leads to gynoecy in watermelon. The Plant Journal 101, 265–277. [DOI] [PubMed] [Google Scholar]
- Zhang S, Tan F-Q, Chung C-H, et al. 2022. The control of carpel determinacy pathway leads to sex determination in cucurbits. Science 378, 543–549. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L.. 1965. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, eds. Evolving genes and proteins. Academic Press, 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials. All the raw reads generated in this study have been deposited in the public database of the National Center of Biotechnology under BioProject PRJNA1042934.


