Abstract
Sequence analysis identified significant variation in the second exon of equine infectious anemia virus (EIAV) rev. Functional analysis indicated that limited amino acid variation in Rev significantly altered the export activity of the protein but did not affect Rev-dependent alternative splicing. EIAV Rev can mediate export through two independent cis-acting Rev-responsive elements (RREs), and differences among Rev variants were more pronounced when both RREs were present. Variation in Rev may be an important mechanism for regulation of virus replication in vivo and may contribute to changes in clinical disease.
Equine infectious anemia virus (EIAV) is a member of the lentivirus subfamily of retroviruses and possesses many of the characteristic features of that subfamily including a complex genome organization, tropism for cells of the monocyte/macrophage lineage, and establishment of a persistent, lifelong infection. Whereas lentivirus infections are typically characterized by a slow, chronic disease, EIAV can induce a rapid, variable disease course in horses. Horses which survive early clinical episodes carry a lifelong, persistent infection with low viral load. The rapid changes between clinical stages of disease which occur during EIAV infection provide for an excellent model for analyzing factors which contribute to lentivirus pathogenesis and persistence. One factor important in EIAV persistence and pathogenesis is genetic and antigenic variation. Genetic mutations in the viral env gene are associated with the occurrence of antigenic-variant viruses, and the role of antigenic variation in EIAV persistence has been extensively studied (14, 20, 29, 32, 35). Additional clusters of genetic variation are found in the virus long terminal repeat and in the region of gp45/Rev overlap (1, 4, 21, 27). The biological significance of variation in these regions is not clear; however, genetic changes which alter levels of viral gene expression may be important factors in viral pathogenesis in vivo. EIAV replicates in cells of the monocyte/macrophage lineage (28), and the severity of clinical and pathological signs of disease is closely associated with levels of viral replication in these cells (3). Therefore, variation in viral regulatory elements may modulate overall levels of virus replication and contribute to changes in clinical disease course.
Lentiviruses utilize complex mechanisms to regulate virus replication. The regulatory protein Rev functions to direct the nuclear export of incompletely spliced viral RNAs encoding viral structural proteins during the late phase of virus replication. Numerous lentiviruses utilize Rev-dependent RNA export pathways (reviewed in reference 9), and the human immunodeficiency virus type 1 (HIV-1) Rev-mediated RNA export pathway is the best-characterized pathway. HIV-1 Rev binds a secondary structure in the viral pre-mRNA called the Rev-responsive element (RRE) (8, 24, 42), multimerizes (22, 23, 34, 41), and then utilizes a non-mRNA nuclear export pathway to redirect movement of incompletely spliced RNA from the nucleus (2, 10, 11, 40). Discrete functional domains within the protein mediate the interactions of Rev with cellular proteins and viral RNA required for nuclear localization, RNA binding, multimerization, and nuclear export.
EIAV Rev is a 165-amino-acid protein translated from a bicistronic four-exon mRNA coding for both Tat and Rev (7, 39). The nuclear export signal (NES) of EIAV Rev has been mapped to amino acids 31 to 55 (12), and domain swapping experiments have shown that the EIAV Rev NES can substitute for the HIV-1 or visna virus NES (2, 25, 30). Other functional domains of EIAV Rev have not yet been identified. EIAV Rev also has an additional, apparently unique function among complex retrovirus export proteins. Whereas HIV-1 Rev and human T-cell leukemia virus type 1 Rex inhibit the expression of both Tat- and Rev-specific mRNAs in facilitating the export of incompletely spliced mRNAs, EIAV Rev specifically down-regulates its own production, independent of Tat, by promoting exon 3 skipping of the bicistronic mRNA (Fig. 1) (13, 26). This mechanism allows for continuous production of Tat, while Rev synthesis is limited. Although the mechanism by which the alternative splicing occurs has yet to be completely delineated, it has been proposed that binding of EIAV Rev to an RRE overlapping exon 3 interferes with SR protein-RNA or SR protein-snRNP interactions (13). The disruption of SR protein binding is thought to result in exon 3 exclusion (13). This multifunctional nature of EIAV Rev highlights its importance in regulation of virus gene expression and replication. As such, genetic variation in Rev may have added significance in vivo.
FIG. 1.
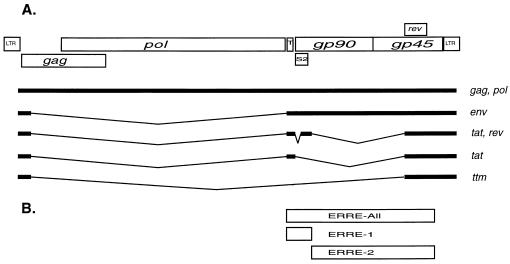
Organization of the EIAV genome. (A) Known ORFs and predominant mRNAs isolated from virus-infected tissue culture cells (19). LTR, long terminal repeat. (B) Location of EIAV regions inserted into pDM138 CAT constructs (17). pERRE-All contains nucleotides 5280 to 7534, pERRE-1 contains a short 5′ RRE sequence overlapping the first Rev exon (nucleotides 5280 to 5834), and pERRE-2 contains a major portion of the remaining downstream EIAV sequence present in pERRE-All (nucleotides 5837 to 7534) (15). All numbering of nucleotides in the present report is based on that of Kawakami et al. (19).
Rev is absolutely required for expression of lentivirus structural genes and production of new virus. Therefore, factors which modulate Rev activity and, consequently, alter levels of viral gene expression may be important in regulating virus replication in vivo. Rev-attenuated phenotypes have been identified during asymptomatic stages of HIV-1 infection, suggesting that variation in Rev could alter virus replication in vivo and contribute to the clinical outcome of infection (16, 18). It would be expected that viral phenotypes that included highly competent Rev phenotypes would be present during periods of rapid virus replication, whereas the production of “attenuated” or latent virus may be important for evasion of an active host immune response during periods of long-term persistence. Indeed, using multiple assays Hua et al. (16) showed that HIV-1 Rev clones obtained from asymptomatic infections were less functional than wild-type Rev. Restricted expression of viral structural genes is a common strategy of persistent viruses (33), and these findings suggest that variation in Rev may be an important factor in lentivirus pathogenesis. We had previously identified extensive nucleotide substitutions in the Rev open reading frame (ORF) from virus obtained from a horse experimentally infected with EIAV (1). The coexistence of putative Rev-competent and Rev-deficient phenotypes suggested that variation in EIAV Rev may contribute to virus persistence through regulation of structural gene expression. The goal of the present study was to further characterize genetic variation in EIAV Rev and to determine if variation in EIAV Rev altered biological activity.
Genetic variation in EIAV Rev.
To further explore the potential role of Rev variation in EIAV pathogenesis, we analyzed Rev cDNAs obtained from cells inoculated with either the Th-1, Th-6, or MA-1 virus isolate (Fig. 2A and B), as well as additional EIAV Rev sequences available in GenBank (Fig. 2C). Th-1 and Th-6 are field-derived virus isolates of EIAV recovered during the first and sixth febrile cycles, respectively, of a horse experimentally inoculated with whole blood from an EIAV-seropositive, naturally infected horse (1, 6). MA-1 is a cell culture-adapted, avirulent virus derived from Th-1 by in vitro passage in equine dermal (ED) cells (5, 6). The analysis indicated a high degree of genetic variation in Rev exon 2. The sequences represent those from a variety of isolates, including virulent (Wyoming) and avirulent (MA-1) EIAV as well as in vivo- (Th-1, Th-6) and in vitro-adapted (MA-1) virus. Some of the sequences were derived from a single proviral clone isolated by limiting dilution and thus are representative of a predominant virus (i.e., P3.21), whereas other sequences represent quasispecies obtained following PCR amplification and cloning of viral cDNA or proviral DNA (Th-1.51). Numerous amino acid substitutions were found in the NES and in a 71-amino-acid region encoded by the center portion of the exon. The changes included deletions as well as amino acid substitutions, some of which were associated with the appearance of premature stop codons. In many cases, identical substitutions were found to reoccur or to occur at specific amino acids, regardless of the virus isolate. Examples include valine/alanine at amino acid 105 and isoleucine/arginine/asparagine at amino acid 113. In other cases, a single change was diagnostic of a particular virus isolate. For example, all of the MA-1 quasispecies contained a glycine at amino acid 39, whereas cDNA clones from the related Th-1 virus or the unrelated Wyoming strain of virus encode an aspartic acid at that location. Also, the glutamine and serine amino acids present at positions 134 and 135, respectively, were more frequent in MA-1 quasispecies, whereas the glycine-to-aspartic acid change at amino acid 115 was found only in the virulent Wyoming strain of EIAV. Within the NES, 80% of the variation occurred at amino acids reported to be necessary for Rev activity (25). While it is possible that the observed variation in Rev is merely a consequence of random variation or that it reflects selection for changes in the overlapping gp45 reading frame, recent findings suggest a role for Rev variation in the biology of EIAV in vivo. During successive febrile periods in a pony experimentally inoculated with EIAV, nucleotide and amino acid variation in EIAV Rev accumulated at approximately the same rate as that observed in gp90 and more frequently than was observed in gp45 or in the long terminal repeat (31). Taken together, these findings support the hypothesis that variation in EIAV Rev is biologically significant.
FIG. 2.
Amino acid sequence alignments of the product of Rev exon 2 (amino acids 31 to 165). (A) Amino acid sequence of the product of exon 2 of MA-1 Rev (4) and amino acid sequences encoded by cDNAs isolated from MA-1-infected ED cells (ME) and MA-1-infected horse macrophage cultures (HMC) (MM). (B) Amino acid sequences of products of cDNAs isolated from Th-1-infected HMC (A22, B11, F22, H21) and viral DNAs isolated from an EIAV-positive horse at the first and sixth febrile cycles (Th-1 and Th-6, respectively) (1). Missing sequences are due to the use of an internal 5′ primer for PCR amplification. (C) Amino acid sequences deduced from in vivo Rev exon 2 sequences obtained from GenBank (accession no. X63059, X16988, M14855, M18385, M18386, M18387, M18388, M87580, and M93674).
Two independent RREs can mediate EIAV Rev-dependent export.
To assess the functional activity of Rev variants, we developed an in vitro nuclear export assay similar to that widely used in functional assays of other lentivirus Rev proteins (17, 25, 37). In other complex retroviruses, transactivation of the Rev/Rex RNA export pathway occurs through an interaction with a single RRE (reviewed in reference 9). For the majority of lentiviruses, the RRE is located near the surface transmembrane envelope region; two exceptions are feline immunodeficiency virus and human T-cell leukemia virus type 1, for which the RREs have been mapped near the 3′ end of the genome (9, 36, 38). Surprisingly, previous studies have identified two cis-acting regions in EIAV which are able to act as RREs (15, 26). However, the specific binding of Rev with only one element overlapping the 3′ end of the first rev exon has been shown (13). The exact location of the second RRE has not been identified, and initial studies were performed to confirm the presence of two RREs. A pDM138-derived reporter plasmid, pERRE-All, was constructed which has the chloramphenicol acetyltransferase (CAT) gene and a region containing all of the putative EIAV RRE sequences within an intron flanked by HIV-1 splice sites (15, 17). Additional reporter plasmids containing EIAV regions previously shown to be able to act as RREs were constructed (26): pERRE-1 contains a short 5′ RRE sequence overlapping the first Rev exon, and pERRE-2 contains a major portion of the remaining downstream EIAV sequence present in pERRE-All (15). The locations of the EIAV sequence present in the reporter constructs are shown in Fig. 1B. For functional assays, 293 cells were seeded in triplicate at 1 × 105 to 5 × 105 cells/well in six-well tissue culture dishes. The next day cells were transfected with 0.2 μg of reporter plasmid, 0.2 μg of beta-galactosidase reporter plasmid pCH110 (Pharmacia, Uppsala, Sweden) or pSV–beta-galactosidase (Promega, Madison, Wis.), and 1 μg of an MA-1 Rev expression plasmid or empty vector. pUC19 DNA was added to bring the total amount of DNA transfected in each well to 2 μg. Cells were transfected by calcium phosphate coprecipitation, and the medium was changed the next day. Two days posttransfection cells were harvested in phosphate-buffered saline containing 5 mM EDTA, pelleted, resuspended in 300 μl of 0.25 M Tris, pH 7.5, and lysed by three rounds of freezing and thawing. Fifty microliters of lysate was assayed for beta-galactosidase activity, and these values were used to normalize lysates for CAT assays. Reaction mixtures for CAT assays were equalized with 0.25 M Tris, pH 7.5, to a final volume of 92 μl and incubated at 37°C with 5 μl of 20 mM acetyl coenzyme A and 3 μl of [14C]chloramphenicol (50 mCi/mmol). Unacetylated and acetylated forms were separated by thin-layer chromatography and quantified with a Molecular Dynamics phosphorImager (Sunnyvale, Calif.). The percentage of acetylation was calculated for each transfection, and the data represents the average acetylation and standard error of the mean for all experiments.
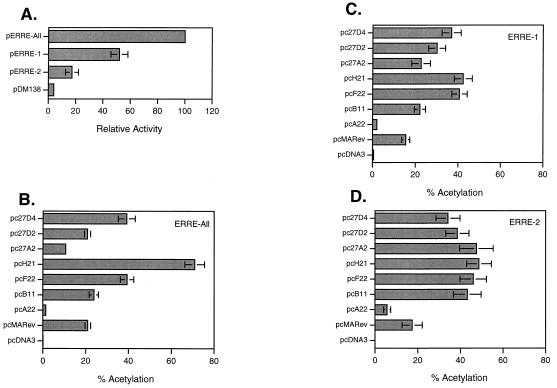
All three ERRE reporter plasmids were found to contain cis-acting elements able to mediate Rev-dependent RNA export (Fig. 3A). For purposes of comparison, results are presented as percentages of activity found with pERRE-All, which is shown as 100%. pERRE-1, containing the RNA element shown by Gontarek and Derse to interact with glutathione S-transferase–Rev in vitro (13), produced the majority (52%) of activity found with pERRE-All. In contrast, assays with pERRE-2 resulted in only 17% of the activity seen with the pERRE-All vector. There was also a low level of transactivation of the background vector, pDM138. These findings confirm previous studies indicating that EIAV Rev can mediate nuclear export through two separate RREs (26). In addition, they provide quantitative results which indicate that the primary RRE is contained within ERRE-1 and encompasses Rev exon 1. Although both RREs appear to be required for maximum efficiency of the Rev-dependent export pathway, the significance of ERRE-2 as an important mediator of RNA export is questionable. The results presented here suggest that ERRE-2 functions primarily as an enhancer of ERRE-1 rather than as an independent mediator of RNA export. Further studies are needed to more clearly ascertain the mechanism of the EIAV dual-RRE export pathway.
FIG. 3.
In vitro assays of EIAV Rev activity. Transfection experiments were performed as described in the text. Two days posttransfection cells were harvested and lysates were normalized for the CAT reactions by a beta-galactosidase assay. Individual experiments included triplicate wells, and the data shown represents the means of at least three separate experiments. Error bars denote the standard errors of the means for all experiments. (A) Rev can trans activate through two discrete regions of EIAV. pcMARev transactivation of CAT reporter plasmids pERRE-All, pERRE-1, pERRE-2, and pDM138 is shown. Each reporter plasmid contains the EIAV sequences shown in Fig. 1B; pDM138 is the background reporter plasmid. Transfections and CAT assays were performed as described in the text, except that lysates from wells with pERRE-All were diluted fivefold to allow for the comparison. (B) Transactivation of pDM138 CAT reporter plasmid pERRE-All by EIAV Rev variants showing that amino acid variation in Rev alters the biological activity of the protein. (C and D) Transactivation of pDM138 CAT reporter plasmids pERRE-1 (C) and pERRE-2 (D) by EIAV Rev variants indicating that the full effects of variation require both RRE regions. CAT assays were performed for each experiment under conditions that resulted in approximately 20% acetylation for the pcMARev lysates. Therefore, although the activities of the reporter plasmids differ, all experiments appear on the same scale.
Variation in Rev alters biological activity.
To determine whether amino acid variation in Rev resulted in differences in biological activity, eight variant Rev cDNAs, four from MA-1-infected ED cells and four from Th-1-infected macrophages, were cloned into the eukaryotic expression vector pCR3 (Invitrogen, Carlsbad, Calif.) for functional analysis. Western blot analysis with Rev-specific polyclonal antibodies confirmed that all variants were expressed at similar levels (data not shown). Variant cDNAs were assayed in transient-expression assays by cotransfection with Rev reporter plasmids containing ERRE-All, ERRE-1, or ERRE-2 as described above. The results indicated that amino acid changes encoded by the Rev ORF significantly altered biological activity (P < 0.0001) when both RREs were present (Fig. 3B). Variants ranged from being inactive (A22) to producing activities greater than threefold that of MA-1 Rev (H21). EIAV Rev variants F22, H21, and 27D4 were significantly more active than all other variants. In general, the range of biological variation was greater in the Th-1-derived Revs, consistent with the greater degree of sequence diversity of those clones compared to that of the MA-1-derived clones. The results are consistent with studies analyzing variation in HIV-1 Rev (16) and support the hypothesis that biological changes in Rev activity may have significance in vivo. In addition, our results demonstrate that variation in regions outside the NES can also alter nuclear export activity. Because all assays were done with a single RRE sequence, we cannot rule out the possibility that compensatory mutations in the RRE minimize the biological effects of Rev variation in vivo. Such analysis requires further mapping of the EIAV RREs.
Given the possibility of a synergistic interaction contributing to EIAV Rev-dependent export, we further characterized the effects of variation with the individual RREs by assaying the nine variants with both pERRE-1 and pERRE-2. Since the functional activities of the reporter plasmids differ (Fig. 3A), the experiments with the separate reporter plasmids were performed under CAT assay conditions in which the acetylation of pcMARev lysates was approximately 20%. An analysis of the individual Rev variants with the pERRE-1 reporter plasmid produced a pattern of activity relative to MA-1 Rev similar to that observed with ERRE-All (Fig. 3B and C). However, with the exception of A22, the differences among the variants were less pronounced than those observed with the pERRE-All reporter plasmid. H21 was only twofold more active than MA-1 Rev, while 27A2 and 27D2 were slightly more active than MA-1 Rev. Surprisingly, the pattern of variation seen with pERRE-All was abolished when the variants were assayed with the pERRE-2 reporter plasmid (Fig. 3D). A22 was still inactive, but all other variants were more active than MA-1 Rev and not significantly different from each other. Together, these results indicate that genetic variation in Rev alters biological activity and that the effects of Rev variation are enhanced in the presence of both RREs. The mechanism(s) by which EIAV Rev utilizes two separate RREs is unknown, and it is not clear why the differences among the variants are decreased when only one RRE is present. As suggested above, the downstream RRE may function primarily as an enhancer element of nuclear export mediated by RRE-1. If so, the effects of variation in regions of Rev important for interaction between RRE-1 and RRE-2 may require the presence of both elements for observable differences in biological activity.
Rev-dependent alternative splicing is not affected by variation.
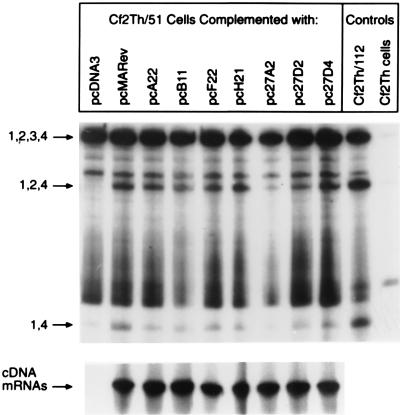
The current model of EIAV Rev-dependent alternative splicing proposes that the binding of Rev to ERRE-1 interferes with SR protein-RNA or SR protein-snRNP interactions to promote exon 3 skipping (13). The significance of this phenomenon in terms of virus replication is not known, although alternative splicing may play an important role in regulation of virus replication. Therefore, studies were undertaken to determine if the Rev sequence variants differed in splice site utilization during EIAV mRNA processing. To analyze Rev-mediated alternative splicing patterns, we developed Cf2Th cell lines stably transfected with a Rev-defective (Cf2th/51) or Rev-competent (Cf2th/112) provirus by G418 selection (data not shown). Cf2Th/51 cells were then trans-complemented with the variant cDNAs. Cells were seeded at 2 × 105 cells/well in six-well tissue culture plates and transfected with 9 μg of Rev variant plasmid or empty-vector DNA by liposome-mediated transfection (Boehringer Manneheim, Indianapolis, Ind.). Two days posttransfection, RNA was isolated and cDNAs were amplified by reverse transcription-PCR (RT-PCR) using primers which spanned all EIAV splice donor and splice acceptor sites (5′ primer: CGCAGACCCTACCTGTTG [nucleotide 354]; 3′ primer: TCTTCAGGTAACGACTGCC [nucleotide 7301]). To allow for sensitive visualization of the splicing products, the 5′ primer was end labeled with 32P. RT-PCR was performed as described by the manufacturer (Perkin-Elmer, Foster City, Calif.). PCR was performed at an annealing temperature of 55°C and run for 25 cycles. DNA from the RT-PCR reactions was phenol-chloroform extracted, ethanol precipitated, and resuspended in 40 μl of 0.1× Tris-EDTA buffer. Ten microliters of each reaction mixture was electrophoresed through a denaturing (7 M urea) 5% polyacrylamide gel. Gels were fixed, dried, and then exposed to film for visualization. As a control, Rev mRNAs were amplified from transfected plasmids with pCR3-specific primers (5′ primer: ATACGACTCACTATAGGG; 3′ primer: ATTTAGGTGACACTATAG). The results indicated that both the 1,2,3,4 exon mRNA and the alternately spliced 1,2,4 exon mRNA were present in Cf2th/112 cells and in all Cf2th/51 cells trans-complemented with variant-Rev cDNAs, including that of the functionally inactive A22 (Fig. 4). In contrast, the 1,2,4 exon mRNA was not detected in Cf2Th/51 cells alone or in cells trans-complemented with vector DNA. The failure to detect differences in alternative splicing patterns suggests that variation present in the cDNAs we examined is not important for Rev-mediated alternative splicing. The finding that A22 was similar to the products of other variant cDNAs in splice site utilization indicates that the nuclear export function of Rev is independent of exon 3 skipping. Therefore, these two functions most likely occupy separate domains in the protein, although both functions may require RRE binding.
FIG. 4.
Amino acid variation does not alter Rev-dependent alternative splicing. Rev-defective cells were transfected with 9 μg of variant-Rev plasmids. Total RNA was isolated and reverse transcribed with random hexamer primers. cDNA was amplified by PCR with EIAV primers specific for exon 1 and exon 4 by using a 5′ primer that was end-labeled with 32P. PCR products were isolated and separated by electrophoreses through a denaturing 5% polyacrylamide gel. The locations of EIAV splicing products are shown. As a control, mRNAs from transfected plasmids were amplified with pCR3-specific primers flanking the Rev insert.
Overall, our findings demonstrate that variation can enhance or attenuate the EIAV Rev phenotype. Previous studies with HIV-1 have shown that variation within the HIV-1 Rev NES can alter the biological phenotype and that the observed changes in function were consistent in both in vitro assays and studies of virus replication (16). We have demonstrated that variation in regions other than the NES can also modulate Rev activity in vitro. Given the extent of Rev variation we have observed, our results suggest that variation in Rev may be an important mechanism for modulating levels of virus replication during the course of clinical disease. Indeed, Leroux et al. reported rapid variation in Rev during sequential febrile cycles of a pony experimentally inoculated with EIAV (21). Functional Rev is absolutely required for production of infectious virus, and it might be expected that Rev-defective or Rev-attenuated genotypes would be rapidly selected against during replication in vivo. However, factors which decrease Rev activity and decrease levels of viral gene expression may have a selective advantage and allow virus to persist in the face of an active host immune response. Further structural and functional analysis of in vivo-derived variants at different stages of the clinical disease course are needed to delineate the role of Rev in lentivirus pathogenesis.
Acknowledgments
We thank Yvonne Wannemuehler, Teresa A. Smith, and Mary Jane Long for technical assistance, Eric Vaughn for helpful discussions, and Wendy Maury and C. Martin Stoltzfus for critical review of this manuscript.
This work was supported in part by USDA grant 96-02102 (S.C.) and PHS grants AI30025 (S.C.) and AI35477 (T.J.H.). M.E.H. is supported by a National Science Foundation Graduate Research Fellowship.
REFERENCES
- 1.Alexandersen S, Carpenter S. Characterization of variable regions in the envelope and S3 open reading frame of equine infectious anemia virus. J Virol. 1991;65:4255–4262. doi: 10.1128/jvi.65.8.4255-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogerd H, Greene W C. Dominant negative mutants of human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev fail to multimerize in vivo. J Virol. 1993;67:2496–2502. doi: 10.1128/jvi.67.5.2496-2502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter S, Alexandersen S. Pathogenesis of equine infectious anemia virus infection. Semin Virol. 1992;3:157–166. [Google Scholar]
- 4.Carpenter S, Alexandersen S, Long M J, Perryman S, Chesebro B. Identification of a hypervariable region in the long terminal repeat of equine infectious anemia virus. J Virol. 1991;65:1605–1610. doi: 10.1128/jvi.65.3.1605-1610.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter S, Chesebro B. Change in host cell tropism associated with in vitro replication of equine infectious anemia virus. J Virol. 1989;63:2492–2496. doi: 10.1128/jvi.63.6.2492-2496.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter S, Evans L H, Sevoian M, Chesebro B. Role of the host immune response in selection of equine infectious anemia virus variants. J Virol. 1987;61:3783–3789. doi: 10.1128/jvi.61.12.3783-3789.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll R, Derse D. Translation of equine infectious anemia virus bicistronic tat-rev mRNA requires leaky ribosome scanning of the tat CTG initiation codon. J Virol. 1993;67:1433–1440. doi: 10.1128/jvi.67.3.1433-1440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cochrane A W, Chen C-H, Rosen C A. Specific interaction of the human immunodeficiency virus rev protein with a structured region in the env mRNA. Proc Natl Acad Sci USA. 1990;87:1198–1202. doi: 10.1073/pnas.87.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen B R. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol Rev. 1992;56:375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer U, Huber J, Boelens W C, Mattal I W, Luhrmann R. The HIV-1 rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 11.Fridell R A, Bogerd H P, Cullen B R. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci USA. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fridell R A, Partin K M, Carpenter S, Cullen B R. Identification of the activation domain of equine infectious anemia virus Rev. J Virol. 1993;67:7317–7323. doi: 10.1128/jvi.67.12.7317-7323.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gontarek R R, Derse D. Interactions among SR proteins, an exonic splicing enhancer, and a lentivirus Rev protein regulate alternative splicing. Mol Cell Biol. 1996;16:2325–2331. doi: 10.1128/mcb.16.5.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond S A, Cook S J, Lichtenstein D L, Issel C J, Montelaro R C. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J Virol. 1997;71:3840–3852. doi: 10.1128/jvi.71.5.3840-3852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris, M. E., G. J. Smith III, F. J. Kim, and T. J. Hope. 1997. Unpublished data.
- 16.Hua J, Caffrey J J, Cullen B R. Functional consequences of natural sequence variation in the activation domain of HIV-1 Rev. Virology. 1996;222:423–429. doi: 10.1006/viro.1996.0439. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, Hope T J, Bond B L, McDonald D, Grahl K, Parslow T G. Minimal Rev-response element for type 1 human immunodeficiency virus. J Virol. 1991;65:2131–2134. doi: 10.1128/jvi.65.4.2131-2134.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iversen A K N, Shpaer E G, Rodrigo A G, Hirsch M S, Walker B D, Sheppard H W, Merigan T C, Mullins J I. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J Virol. 1995;69:5743–5753. doi: 10.1128/jvi.69.9.5743-5753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawakami T, Sherman L, Dahlbert J, Gazit A, Yaniv A, Tronick S R, Aaronson S A. Nucleotide sequence analysis of equine infectious anemia proviral DNA. Virology. 1987;158:300–312. doi: 10.1016/0042-6822(87)90202-9. [DOI] [PubMed] [Google Scholar]
- 20.Kono Y. Recurrences of equine infectious anemia. In: Bryans J T, Gerber H, editors. Proceedings of the Conference on Equine Infectious Diseases. S. Basel, Switzerland: Karger; 1972. pp. 175–186. [Google Scholar]
- 21.Leroux C, Issel C J, Montelaro R C. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J Virol. 1997;71:9627–9639. doi: 10.1128/jvi.71.12.9627-9639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madore S J, Tiley L S, Malim M H, Cullen B R. Sequence requirements for rev multimerization in vivo. Virology. 1994;202:186–194. doi: 10.1006/viro.1994.1334. [DOI] [PubMed] [Google Scholar]
- 23.Malim M H, Cullen B R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 24.Malim M H, Hauber J, Le S-Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–256. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 25.Mancuso V A, Hope T J, Zhu L, Derse D, Phillips T, Parslow T G. Posttranscriptional effector domains in the Rev proteins of feline immunodeficiency virus and equine infectious anemia virus. J Virol. 1994;68:1998–2001. doi: 10.1128/jvi.68.3.1998-2001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martarano L, Stephens R, Rice N, Derse D. Equine infectious anemia virus trans-regulatory protein Rev controls viral mRNA stability, accumulation, and alternative splicing. J Virol. 1994;68:3102–3111. doi: 10.1128/jvi.68.5.3102-3111.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maury W, Perryman S, Oaks J L, Seid B K, Crawford T, McGuire T, Carpenter S. Localized sequence heterogeneity in the long terminal repeats of in vivo isolates of equine infectious anemia virus. J Virol. 1997;71:4929–4937. doi: 10.1128/jvi.71.7.4929-4937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGuire T C, Crawford T B, Henson J B. Immunofluorescent localization of equine infectious anemia virus in tissue. Am J Pathol. 1971;62:283–294. [PMC free article] [PubMed] [Google Scholar]
- 29.McGuire T C, Tumas D B, Byrne K M, Hines M T, Leib S R, Brassfield A L, O’Rourke K I, Perryman L E. Major histocompatibility complex-restricted CD8+ cytotoxic T lymphocytes from horses with equine infectious anemia virus recognize Env and Gag/PR proteins. J Virol. 1994;68:1459–1467. doi: 10.1128/jvi.68.3.1459-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer B E, Meinkoth J L, Malim M H. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J Virol. 1996;70:2350–2359. doi: 10.1128/jvi.70.4.2350-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michael N L, d’Arcy L, Ehrenberg P K, Redfield R R. Naturally occurring genotypes of the human immunodeficiency virus type 1 long terminal repeat display a wide range of basal and Tat-induced transcriptional activities. J Virol. 1994;68:3163–3174. doi: 10.1128/jvi.68.5.3163-3174.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montelaro R C, Parekh B, Orrego A, Issel C J. Antigenic variation during persistent infection by equine infectious anemia virus, a retrovirus. J Biol Chem. 1984;259:10539–10544. [PubMed] [Google Scholar]
- 33.Oldstone M B A. Viral persistence. Cell. 1989;56:517–520. doi: 10.1016/0092-8674(89)90573-4. [DOI] [PubMed] [Google Scholar]
- 34.Olsen H, Cochrane A, Dillon P, Nalin C, Rosen C. Interaction of the human immunodeficiency virus type 1 rev protein with a structured region in env mRNA is dependent on multimer formation mediated through a basic stretch of amino acids. Genes Dev. 1990;4:1357–1364. doi: 10.1101/gad.4.8.1357. [DOI] [PubMed] [Google Scholar]
- 35.Payne S L, Fang F-D, Liu C-P, Dhruva B R, Rwambo P, Issel C J, Montelaro R C. Antigenic variation and lentivirus persistence: variations in envelope gene sequences during EIAV infection resemble changes reported for sequential isolates of HIV. Virology. 1987;161:321–331. doi: 10.1016/0042-6822(87)90124-3. [DOI] [PubMed] [Google Scholar]
- 36.Phillips T R, Lamont C, Konings D A M, Shacklett B L, Hamson C A, Luciw P A, Elder J H. Identification of the Rev transactivation and Rev-responsive elements of feline immunodeficiency virus. J Virol. 1992;66:5464–5471. doi: 10.1128/jvi.66.9.5464-5471.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth J, Dobbelstein M. Export of hepatitis B virus RNA on a Rev-like pathway: inhibition by the regenerating liver inhibitory factor IκBα. J Virol. 1997;71:8933–8939. doi: 10.1128/jvi.71.11.8933-8939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saltarelli M J, Schoborg R, Pavlakis G N, Clements J E. Identification of the caprine arthritis encephalitis virus rev protein and its cis-acting rev-responsive element. Virology. 1994;199:47–55. doi: 10.1006/viro.1994.1096. [DOI] [PubMed] [Google Scholar]
- 39.Stephens R M, Derse D, Rice N R. Cloning and characterization of cDNAs encoding equine infectious anemia Tat and putative Rev proteins. J Virol. 1990;64:3716–3725. doi: 10.1128/jvi.64.8.3716-3725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 41.Zapp M, Hope T, Parslow T, Green M. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus rev protein: a dual function for an arginine-rich motif. Proc Natl Acad Sci USA. 1988;88:7734–7738. doi: 10.1073/pnas.88.17.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zapp M L, Green M R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]