Abstract
At least eight conserved motifs are visible in the totivirus RNA-dependent RNA polymerase (RDRP). We have systematically altered each of these in the Saccharomyces cerevisiae double-stranded RNA virus ScVL1 by substituting the conserved motifs from a giardiavirus. The results help define the conserved regions of the RDRP involved in polymerase function and those essential for other reasons.
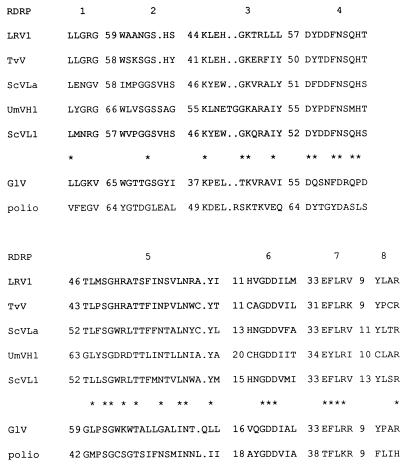
Among the known RNA-dependent RNA polymerase (RDRP) sequences of double-stranded RNA (dsRNA) viruses, only the RDRPs of the noninfectious dsRNA viruses of simple eukaryotes (most being members of the family Totiviridae) form a well-defined group with significant sequence similarities throughout (2). In addition to the “universally” conserved polymerase residues such as the GDD motif (10), this group of five RDRPs shares a set of conserved motifs that extend beyond those present in other viral RDRPs. Several more putative members of the family Totiviridae have been added to this alignment, and the eight conserved motifs remain (3). One other dsRNA virus whose RDRP displays significant similarity throughout to the RDRPs of noninfectious dsRNA viruses of simple eukaryotes is GlV, the Giardia lamblia virus. GlV differs from the dsRNA viruses of lower eukaryotes in that it is infectious to its host. The eight conserved motifs of the RDRPs of members of the family Totiviridae are shown in Fig. 1, compared to GlV and poliovirus RDRPs. Mutations in motifs 4, 5, and 6 have demonstrated their importance for polymerase function (15, 16).
FIG. 1.
Conserved motifs in RDRPs of members of the family Totiviridae compared to those of poliovirus. Spacing between motifs (in amino acids) is indicated for each RDRP. Asterisks indicate residues identical in all Totiviridae RDRPs. Periods indicate insertions made to maximize alignment. Sequences are those previously aligned (2) with the addition of poliovirus (11, 13). LRV1, Leishmania RNA virus 1; TvV, Trichomonas vaginales virus; ScVLa and ScVL1, S. cerevisiae virus; UmVH1, Ustilago maydis virus; GlV, G. lamblia virus.
The Saccharomyces cerevisiae virus L1, or ScVL1 (also known as ScVL-A), is the prototypical member of the family Totiviridae, having two open reading frames (cap and pol) encoding a capsid protein (Cap) and an RNA-dependent RNA polymerase (Pol). The 4.6-kb dsRNA genome is encapsidated into a 39-nm icosahedral particle comprised of 60 dimers of capsid protein (4). The RDRP is expressed as a Cap-Pol fusion protein by −1 ribosomal frameshifting (5, 9). ScVM1 is also encapsidated in ScVL1-derived particles, so that ScVM1 is a satellite of ScVL1 (6). ScVM1 encodes the k1 killer toxin, as well as immunity to the toxin (1, 17).
Motifs 5 and 6 of the ScVL1 RDRP have been examined by scanning-alanine mutagenesis and domain swaps (14, 15). In spite of the high degree of conservation in these regions, these domains are not interchangeable between ScV and Sindbis virus, reovirus, Middleburg virus, or poliovirus (15).
In the present work, rather than introducing radical mutations into the ScVL1 RDRP, we chose to swap analogous conserved motifs from the GlV RDRP into the ScVL1 RDRP. In this manner, we hoped to obtain subtle phenotypic changes in the ScVL1 RDRP, as opposed to the lethal phenotypes observed in some cases during previous engineering of mutations into the ScVL1 RDRP (15). We reasoned that swapping conserved domains would be more likely to conserve secondary and tertiary structure than would alanine substitutions. In this way, we hoped to separate the functions of the Pol domain. Our results show that while all chimeric ScVL1 RDRPs retained the ability to package viral RNA, there were marked differences in the ability of chimeric RDRPs to cure ScVL1 and to support the ScVM1 satellite virus in the absence of ScVL1.
Motif swaps in regions 1 to 8 were constructed by oligonucleotide-mediated site-directed mutagenesis (12) of appropriate restriction fragments in vectors from which single-stranded DNA could be produced such that the GIV sequences of Fig. 1 replaced those of ScVL1. After sequence verification, altered restriction fragments were used to replace the wild-type sequences in the L1 cDNA expression vector. Details are available upon request.
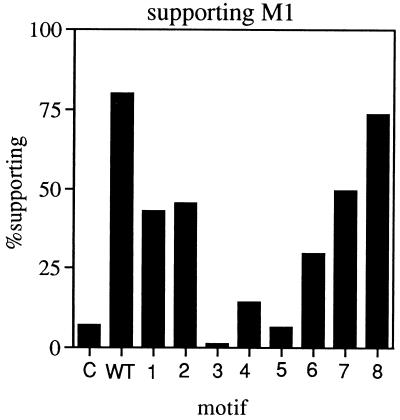
A subset of ScVL1 chimeras are capable of supporting the M1 satellite virus.
We tested the ability of the swapped Cap-Pol fusions to rescue the M1 satellite virus in the absence of the L1 helper virus. Expression of L1 gene products launched from a cDNA has been shown to be capable of rescuing the M1 satellite (6, 19). In this assay, the nucleus of a strain incapable of supporting ScVL1 due to a mak mutation (strain T120, a mak10 mutant, in this case), but producing Cap and Cap-Pol from an L1 cDNA copy, is combined with cytoplasm containing ScVL1 and ScVM1 (6, 19). Cytoductants in which expression of Cap and Cap-Pol or Cap and mutant Cap-Pol successfully rescues the M1 satellite will retain the killer phenotype associated with the M1 satellite.
As expected, expression of wild-type Cap-Pol (pG3L1) successfully maintains the M1 satellite in the absence of ScVL1 (Fig. 2, lane 2) to a level of 80% ± 10%. The M1 satellite is lost in strains which do not express the L1 cDNA (pG3; Fig. 2, lane 1). Cap-Pols carrying swaps in motifs 3, 4, 5, and 6 are significantly impaired in the support of ScVM1, while swaps in motifs 1, 2, 7, and 8 have milder effects or no effects (motif 8). Loss of M1 in cytoductions has been verified by Northern blotting in some cases (data not shown).
FIG. 2.
Ability of mutant Cap-Pols to support ScVM1. Cytoduction assays were performed as described in Materials and Methods to assay replicase and transcriptase activities of mutant versions of the ScV Cap-Pol. About 100 cytoductants derived from each mutant were tested for the killer phenotype. C, negative control (vector without cDNA insert); WT, wild-type positive control (unmodified ScV Cap-Pol). 1 to 8, mutants carrying GlV-ScV motif swaps in regions 1 to 8.
Curing of ScVL1.
Several interfering phenomena have been described for the ScV system. One type of interference results from overproduction of Cap and Cap-Pol from a cDNA (18). In this case, curing (exclusion) of ScVL1 is ascribed to titration of limiting host factors by cDNA-encoded Cap-Pol. The cDNA used in these experiments was unable to launch an infectious cycle since it was missing 8 nucleotides at the 3′ end of the L1 sequence and contained an additional vector sequence at its 5′ end. Mutations which abolished production of Cap-Pol also abolished curing. By comparing Cap-Pol carrying mutations in one or two amino acids in conserved regions 5 and 6, it was found that, in general, mutants with impaired RDRP activity also failed to exclude (cure) ScVL1. Exclusion, in this case, was a stochastic process; multiple rounds of curing resulted in gradual, complete exclusion of ScVL1 (and, consequently, ScVM1) from cells (18).
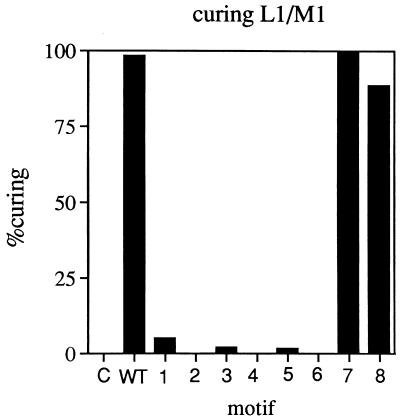
Yeast expression vectors containing the full-length L1 cDNA or an L1 cDNA with a swap in one of the conserved motifs were transformed into a strain in which ScVL1 and ScVM1 were initially present. After selection for cells which harbored the expression vector (Trp+ cells), 100 independent transformants were streaked for single colonies on nonselective medium (YPD) (8) for two rounds of growth. Loss of the vector before assaying of transformants for the killer phenotype is necessary since ScVM1 may be supported by ScVL1 or by the L1 cDNA (18). The loss of the vector after growth on nonselective medium was detected by replica plating of colonies on SC lacking Trp (8). The killer phenotype (K+/K−) of each transformant was assayed by streaking it on a killer plate containing a lawn of toxin-sensitive S7 cells. The presence of ScVM1 (and hence ScVL1) was ascertained by examining killer plates for a region of clearing surrounding the colony tested. The curing efficiency is determined by the ratio of the number of Trp− K− colonies divided by the number of Trp− colonies after plasmid loss. Expression of wild-type Cap and Cap-Pol (Fig. 3, WT) cures ScVL1, as noted previously (18). That curing is dependent on expression of the L1 cDNA is seen by the lack of curing in cells which initially harbor a plasmid that is without the L1 cDNA (Fig. 3C). Swaps in conserved motifs 1 through 6 (Fig. 3, motifs 1 to 6) abolish curing of ScVL1. Cap-Pol with a swap in conserved motif 7 or 8 (Fig. 3, motifs 7 and 8) retains the ability to cure ScVL1.
FIG. 3.
Curing of ScVM1 by expression of Cap and Cap-Pol from expression vectors. Curing experiments were performed as described in Materials and Methods, with about 100 clones of each mutant tested for the killer phenotype. Labeling is as in Fig. 2.
RDRP activity and curing are separable.
These experiments demonstrate that support of M1 requires motifs 3 to 6, while curing of L1 requires motifs 1 and 2 in addition (motifs 1 to 6 are residues 944 to 1229 of Cap-Pol). This is consistent with previous results showing that mutants A766D and S855P are fully functional for M1 rescue and packaging but defective in curing (20) and that mutations in motifs 5 and 6 affect both M1 rescue and L1 curing (15). Specific domains are necessary for support of M1, but curing requires all but the C terminus of Pol (beyond residue 1262, domain 7).
Packaging activity of mutant Cap-Pol.
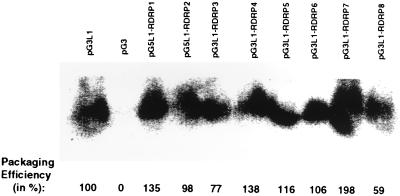
An N-terminal region of Cap-Pol up to approximately residue 858 is responsible for the recognition and packaging of plus-stranded RNAs into nascent viral particles (7, 20). None of the conserved motifs in the RDRPs of members of the family Totiviridae are included in this region; the first conserved motif begins at residue 944. Consequently, we expected all of the GlV-ScV motif swap mutants of Cap-Pol to have approximately normal packaging abilities. Packaging was assayed by production of an artificial transcript with the packaging signal from M1 (21) in cells expressing Cap and Cap-Pol from L1 cDNA expression vectors constructed from the wild type and motif 1 to 8 GlV-ScV substitutions.
The test transcript is packaged in particles by all of the mutant Cap-Pols created by GlV-ScV motif substitution (Fig. 4). Packaging is detectable over a range of about 300-fold (20), but none of the mutants tested showed nearly this much variance from the wild type; all were within a twofold variation of the wild type (Fig. 4), which is within the range of experimental error of the method (21). We conclude that none of the mutational changes tested affects packaging efficiency and that probably all of the mutant Cap-Pol proteins are made in approximately normal quantities.
FIG. 4.
Northern blot of RNAs extracted from peak fractions of CsCl gradients containing viral particles derived from the two-plasmid packaging assay. RNAs were extracted from equal amounts of fractions and run on a 1.4% nondenaturing agarose gel, and a Northern blot was performed as previously described (8). The minus-strand RNA probe was specific for vector portions of the test transcript. Relative packaging efficiencies were calculated from the PhosphorImager scan of the gel normalized to the amount of cells harvested for particles in each case.
Curing of ScVL1 requires domains in Cap-Pol not required for polymerase activity, as well as those required for the RDRP. Motifs 1 to 6 are all required for curing (that is, in the region between 944 and 1229). In addition, mutants A766D and S855P (20) and PD1175AA, TK1177AA, and KL1207AA (18) all have reduced or no curing activity. None of these point mutations fall in any of the conserved motifs of the RDRP (and do not affect RDRP activity), but all are consistent with a requirement for overall sequence integrity in all but the carboxy-terminal domain (after residue 1262) for the curing activity of Cap-Pol.
This work is the first to explore the functions of motifs 1, 2, 4, 7, and 8 in members of the family Totiviridae. Of these, motif 4, which is highly conserved in this group, is clearly of functional importance for the polymerase, in agreement with results obtained with encephalomyocarditis virus (16). Motifs 1, 2, and 8 do not appear to be part of the active site of the polymerase, although motifs 1 and 2 play a role in the curing of ScVL1. This corresponds well to the inability to find decent homologs of motifs 1, 2, and 8 in the poliovirus RDRP (Fig. 1).
Interestingly, more drastic substitutions in motif 8 (MMIK, rather than YPAR, for YSLR) resulted in complete loss of both curing and polymerase activities for the ScV Cap-Pol (0% curing and 6% support of M1). Since this motif appears to be neither part of the active site of the enzyme nor crucial for curing, the highly conserved arginine in this motif may play an important role in the tertiary-structure formation of the RDRP of members of the family Totiviridae. This arginine is conserved in three additional RDRPs from putative members of the family Totiviridae (3).
Acknowledgments
We thank our colleagues Michael Holland, Tzy-Hwa Tzeng, C. C. Wang, Reed Wickner, and Wensheng Yao for plasmids.
We thank the National Institutes of Health for support (grant GM22200).
REFERENCES
- 1.Bostian K A, Jayachandran S, Tipper D J. A glycosylated protoxin in killer yeast: models for its structure and maturation. Cell. 1983;32:169–180. doi: 10.1016/0092-8674(83)90507-x. [DOI] [PubMed] [Google Scholar]
- 2.Bruenn J A. A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acids Res. 1993;21:5667–5669. doi: 10.1093/nar/21.24.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruenn, J. A. Viruses of fungi and protozoans: is everyone sick? In C. J. Hurst (ed.), Viral ecology, in press. Academic Press, Inc., New York, N.Y.
- 4.Cheng R H, Caston J R, Wang G-J, Gu R, Smith T J, Baker T S, Bozarth R F, Trus B L, Cheng N, Wickner R B, Steven A C. Fungal virus capsids: cytoplasmic compartments for the replication of double-stranded RNA formed as icosahedral shells of asymmetric Gag dimers. J Mol Biol. 1994;244:255–258. doi: 10.1006/jmbi.1994.1726. [DOI] [PubMed] [Google Scholar]
- 5.Diamond M E, Dowhanick J J, Nemeroff M E, Pietras D F, Tu C, Bruenn J A. Overlapping genes in a yeast double-stranded RNA virus. J Virol. 1989;63:3983–3990. doi: 10.1128/jvi.63.9.3983-3990.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimura T, Esteban R, Esteban L M, Wickner R B. Portable encapsidation signal of the L-A double-stranded RNA virus of S. cerevisiae. Cell. 1990;62:819–828. doi: 10.1016/0092-8674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- 7.Fujimura T, Ribas J C, Makhov A M, Wickner R B. Pol of gag-pol fusion protein required for encapsidation of viral RNA of yeast L-A virus. Nature. 1992;359:746–749. doi: 10.1038/359746a0. [DOI] [PubMed] [Google Scholar]
- 8.Huan B-F, Shen Y, Bruenn J A. In vivo mapping of a sequence required for interference with the yeast killer virus. Proc Natl Acad Sci USA. 1991;88:1271–1275. doi: 10.1073/pnas.88.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Icho T, Wickner R B. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J Biol Chem. 1989;264:6716–6723. [PubMed] [Google Scholar]
- 10.Jablonski S A, Luo M, Morrow C D. Enzymatic activity of poliovirus RNA polymerase mutants with single amino acid changes in the conserved YGDD amino acid motif. J Virol. 1993;65:4565–4572. doi: 10.1128/jvi.65.9.4565-4572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitamura N, Semler B L, Rothberg P G, Larsen G R, Adler C J, Dorner A J, Emini E A, Hanecak R, Lee J J, van der Werf S, Anderson C W, Wimmer E. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981;291:547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- 12.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Racaniello V R, Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci USA. 1981;78:4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribas J C, Fujimura T, Wickner R B. A cryptic RNA-binding domain in the Pol region of the L-A double-stranded RNA virus Gag-Pol fusion protein. J Virol. 1994;68:6014–6020. doi: 10.1128/jvi.68.9.6014-6020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribas J C, Wickner R B. RNA-dependent RNA polymerase consensus sequence of the L-A double-stranded RNA virus: definition of essential domains. Proc Natl Acad Sci USA. 1992;89:2185–2189. doi: 10.1073/pnas.89.6.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankar S, Porter A G. Point mutations which drastically affect the polymerization activity of encephalomyocarditis virus RNA-dependent RNA polymerase correspond to the active site of Escherichia coli DNA polymerase I. J Biol Chem. 1992;267:10168–10176. [PubMed] [Google Scholar]
- 17.Skipper N, Thomas D Y, Lau P C. Cloning and sequencing of the preprotoxin-coding region of the yeast M1 double-stranded RNA. EMBO J. 1984;3:107–111. doi: 10.1002/j.1460-2075.1984.tb01769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valle R P, Wickner R B. Elimination of L-A double-stranded RNA virus of Saccharomyces cerevisiae by expression of gag and gag-pol from L-A cDNA clone. J Virol. 1993;67:2764–2771. doi: 10.1128/jvi.67.5.2764-2771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickner R B, Icho T, Fujimura T, Widner W R. Expression of yeast L-A double-stranded RNA virus proteins produces derepressed replication: a ski− phenocopy. J Virol. 1991;65:155–161. doi: 10.1128/jvi.65.1.155-161.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao W, Muqtadir K, Bruenn J A. Packaging in a yeast double-stranded RNA virus. J Virol. 1995;69:1917–1919. doi: 10.1128/jvi.69.3.1917-1919.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao W-S, Adelman K, Bruenn J A. In vitro selection of packaging sites in a double-stranded RNA virus. J Virol. 1997;71:2157–2162. doi: 10.1128/jvi.71.3.2157-2162.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]