Abstract
Nucleocapsid protein (NCp7) of human immunodeficiency virus type 1 is found covering the genomic RNA in the interior of the viral particle. It is a highly basic protein with two zinc fingers of the form CX2CX4HX4C which exhibit strong affinity for a zinc cation. To study the structure-function relationship of the N-terminal zinc finger of NCp7, this domain was either deleted or changed to CX2CX4CX4C. We examined virus formation and structure as well as proviral DNA synthesis. Our data show that these two NC mutations result in the formation of particles with an abnormal core morphology and impair the end of proviral DNA synthesis, leading to noninfectious viruses.
The nucleocapsid protein (NCp7) of human immunodeficiency virus type 1 (HIV-1) is a highly basic protein which is tightly associated with the genomic RNA dimer in mature viral particles to form the ribonucleoprotein complex (14). NCp7 is derived from the C terminus of the Pr55gag precursor following proteolytic cleavage (18, 19, 29). NCp7 contains two zinc fingers of the form CX2CX4HX4C (49) with high affinity for a zinc cation (23, 45), and which are close to each other as shown by 1H nuclear magnetic resonance spectroscopy analyses and molecular modelling (36–38).
The NCp7 protein is involved in essential steps of genome replication since it promotes annealing of the tRNA3Lys primer to the genomic primer binding site (3, 4, 16) and minus-strand DNA transfer during proviral DNA synthesis (2, 13, 17, 28, 42). In addition, NCp7 appears to abolish nonspecific reverse transcription due to self-priming that can take place either at the 3′ end or at nicks in the genomic RNA (28, 32, 34) and to enhance efficiency and processivity of the reverse transcriptase (RT) enzyme (30, 40, 42, 46, 51). These functional properties of NCp7 seem to be related to the nucleic acid annealing activity of the protein in vitro (31). In fact, NCp7 promotes rapid and extensive hybridization of two complementary nucleic acid sequences by destabilizing intramolecular duplexes and by favoring formation of the most stable intermolecular duplex (33, 47).
During virion formation, NC protein, as part of Pr55gag and/or as mature NCp7, is thought to bind to the viral RNA (11, 12), resulting in genomic RNA dimerization and packaging (14, 16) and in the formation of the virion nucleocapsid structure (8, 39, 41). Moreover, NC protein is able to stabilize dimeric RNA, converting it from the immature to the mature, stable form (22, 24).
Extensive mutational analyses of HIV-1 NCp7 have shown that substitutions of highly conserved residues thought to modify the overall conformation of the protein result in the production of viral particles defective in replication (12, 39, 41, 43). Analysis of the NC zinc finger mutant virus shows a strong defect in genomic RNA packaging (1, 20, 27, 35). Although both fingers are required for encapsidation of viral RNA and for infectivity, they are not functionally equivalent and their respective positions cannot be exchanged (26). On the other hand, substituting basic residues for neutral amino acids reduces genomic RNA packaging and results in the attenuation of NC mutant viruses (5).
In an attempt to study the structure-function relationships of the N-terminal zinc finger of HIV-1 NC protein during different steps of the viral replication cycle, the first zinc finger was either deleted (to create mutant ΔD1) or changed to a CCCC motif (to create mutant H23C). Substituting His23 for Cys causes structural modifications in the N-terminal zinc finger which disrupt the proximity of the two zinc fingers and result in a misfolded protein (15). The H23C substitution does not, however, interfere with the strong affinity of the mutated zinc finger for the zinc cation. This is in contrast to other mutations, such as the substitution of Cys for Ser or His for Ala, which prevent zinc coordination (1, 20, 27, 35).
H23C and ΔD1 were obtained by site-directed mutagenesis performed on the pNL4-3 HIV-1 molecular clone as previously described (39) with the oligonucleotides 5′-GCAAAGAAGGGTGCATAGCC-3′ (for H23C) and 5′-GAAAGACTGTTAAGGGTGGCAGGGCCCC-3′ (for ΔD1). As previously reported, both mutants were completely defective in replication in SupT1 and HeLa P4 cells (15, 27).
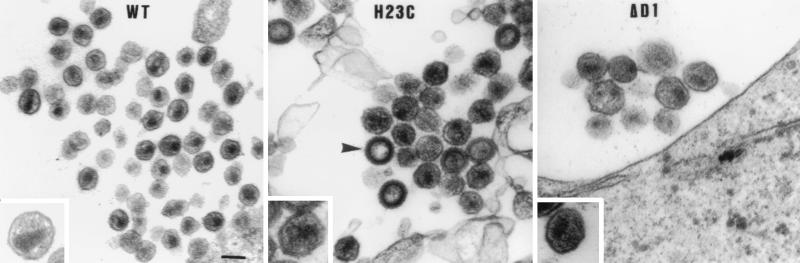
To analyze the morphology of the NC mutant viruses, HeLa P4 cells (10) were transfected by the calcium phosphate precipitation method (44) with wild-type (wt) or mutant pNL4-3 and processed for thin-layer electron microscopy (Fig. 1). Cells transfected with the wt provirus showed numerous viral particles budding at the plasma membrane and extracellular mature virions, with morphology typical of HIV, including central electron-dense material corresponding to the core (Fig. 1, inset). However, many particles produced by cells transfected with both mutants had a mature but abnormal morphology (Fig. 1). Under higher magnification (Fig. 1, insets), a majority of mutant virions were observed to be characterized by (i) a strong electron density of the whole particle and (ii) an abnormal core structure which did not correspond to the typical cone-shaped core of the mature wt particles, probably due to a nucleocapsid with a modified ultrastructure. Moreover, 28 and 12% of the H23C and ΔD1 released particles, respectively, exhibited a typical immature morphology characterized by a doughnut or ring shape corresponding to a thick electron-dense outer shell and an electron-lucent center where no typical cone-shaped core was detectable. Also, NC mutant virions had a mean diameter of 134 nm, which is larger than that of wt particles (106 nm). Moreover, we observed that after ultracentrifugation, the amount of CAp24 detected by enzyme-linked immunosorbent assay was five times lower for NC mutant particles than for wt particles (data not shown), indicating that particles with an abnormal core morphology are probably unstable. These observations suggest that the first zinc finger structure of NC protein is probably critical for a stable core structure.
FIG. 1.
Electron microscopy of wt and NC mutant virions. Particles derived from the wt, H23C, and ΔD1 transfected HeLa P4 cells were postfixed with 1% osmium tetroxide and embedded in epon 72 h posttransfection. Grids were counterstained with uranyl acetate and lead citrate. The arrowhead shows an immature particle with a thick electron-dense outer shell and an electron-lucent center lacking a typical cone-shaped core. The bar represents 100 nm. (Insets) higher magnification of either a normal mature particle for the wt virus (with a central electron-dense material corresponding to the cone-shaped core) or a representative example of mature particles with a typical abnormal morphology for H23C and ΔD1 virions.
To determine the densities of the NC mutant virions, viral supernatants were concentrated and separated on 20 to 50% sucrose density gradients as previously described (5). The H23C and ΔD1 mutant viruses had densities of 1.179 and 1.171 g/ml, respectively, which is very close to that of the wt virus (1.18 g/ml).
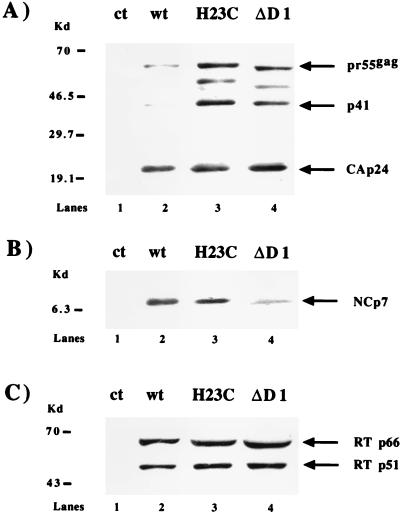
Three days after transfection, virions released during a period of 24 h were pelleted through a 20% sucrose cushion and viral proteins were analyzed by immunoblotting with anti-CAp24, anti-NCp7, and anti-RTp66/p51 antibodies (Fig. 2). We observed that Pr55gag processing was affected in the H23C and ΔD1 mutants, as judged by an increase in the prominence of (i) the Pr55gag precursor itself and (ii) the p41-processed intermediate known to contain the MAp17 and CAp24/25 sequences, as well as another intermediate containing the NC region (Fig. 2A). The ratios of the precursors to mature CAp24 were approximately 10% for the wt, 50% for ΔD1, and 70% for H23C, as determined by scanning densitometry. Mature NCp7 was detected for both mutants (Fig. 2B). The use of antibodies directed against RTp66/p51 allowed us to determine no significant difference in the amount of RT protein present in NC mutant virions compared to the wt (Fig. 2C). In conclusion, the gag structural proteins and RT are present in mutant virions, although a minor defect in Pr55gag processing was observed for both mutants.
FIG. 2.
Western blot analysis of virion proteins. HeLa P4 cells were transfected with wt pNL4-3, H23C, or ΔD1. Twenty-four-hour cell-free virus was collected 72 h posttransfection and pelleted through a 20% sucrose cushion. Samples (adjusted for equal amounts of mature CAp24 by prior Western blotting with anti-CAp24 monoclonal antibodies) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a gradient of 5 to 20% polyacrylamide and analyzed by Western blotting with anti-CAp24 monoclonal antibodies (A) and anti-NCp7 (B) and anti-RT (C) polyclonal antibodies. The relative positions of the HIV-1 proteins are indicated on the right. Note the presence of a maturation intermediate containing the NC region (as implied by the faster migration of this product for the ΔD1 mutant; see panel A, lane 4). Note also the presence of Gag intermediates at 41 and 49 kDa not seen in the wt virions. Molecular mass markers are shown on the left. ct, mock-transfected cells.
We examined the virion genomic RNA content of the NC mutants by slot blot hybridization, as described previously (39), by probing with a randomly 32P-labeled 5.3-kb SacI-SalI fragment of the pNL4-3 plasmid corresponding to the gag and pol sequences. Genomic RNA packaging was approximately 10% of the wt level for both mutants, which is similar to levels found in other studies involving mutations of the Zn2+-chelating residues (data not shown) (27, 35).
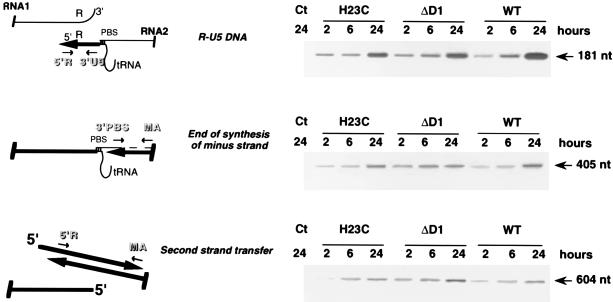
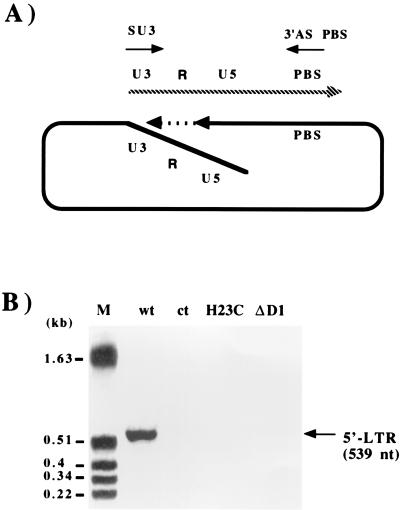
The implications of NCp7 in the reverse transcription process in vitro prompted us to use a PCR-based system to analyze the major steps of proviral DNA synthesis in vivo. Infection of SupT1 cells was performed by addition of 24-h cell-free virus produced 3 days after transfection in the presence of 3 U of RQ1 DNase (Promega) per ml and concentrated 10 times in a Biomax OSI column. After extracting DNA 2, 6, and 24 h postinfection, we used PCR and a corresponding set of primers to detect R-U5 DNA (which consists mainly of strong-stop cDNA), the end of minus-strand DNA synthesis, and second-strand transfer (Fig. 3). The absence of plasmid pNL4-3 was confirmed with primers specific for the pUC vector (data not shown). Our results show that the levels of R-U5 DNA were similar for the wt and the NC mutant viruses, as were the extents of minus-strand DNA and second-strand transfer (Fig. 3). To detect possible defects at the very end of proviral DNA synthesis, we used a primer localized at the 5′ end of the long terminal repeat (LTR) U3 sequence (Fig. 4) (6). This revealed an amplified fragment of the appropriate size in cells infected with wt virus, but no product was detectable for either NC mutant, suggesting that although reverse transcription of the viral genome was complete for the mutants (as indicated by observable second-strand transfer) (Fig. 3), proviral DNA synthesis leading to the formation of the 5′ LTR was incomplete.
FIG. 3.
Analysis of early and late steps of viral DNA synthesis by PCR. Infection of HeLa P4 cells was performed with wt and mutant viruses adjusted to the same level of viral RNA. Viral DNA synthesis was analyzed by PCR with primer pairs as follows: 5′R (5′-GGTCTCTCTGGTTAGACCA-3′) and 3′ U5 (5′-CTGCTAGAGATTTTCCACAC-3′) to generate R-U5 DNA, 3′PBS (5′-ACTTGAAAGCGAAAGTAAAGC-3′) and MA (5′-TGATGCACACAAAGAGGAC-3′) to detect the end of synthesis of minus-strand cDNA, and 5′R and MA to study second-strand transfer. DNA was extracted from either total cellular extract (R-U5 DNA and synthesis of the minus-strand DNA) or nuclear fraction (second-strand transfer) as previously described (5). The localization of each primer pair relative to the viral genome is represented on the left. The absence of plasmid pNL4-3 was confirmed by using primers localized within the flanking DNA (5′ check [5′-AGGCAGTCTAGTCCCCAG-3′]) and pUC (3′ check [5′-TCGTCACATGTTCTTTCCTG-3′]). The cycling times were 5 min at 94°C for denaturation, 30 s at 58°C for annealing, and 30 s at 72°C for extension. Thirty cycles were performed. Ct, mock-transfected cells; PBS, primer binding site.
FIG. 4.
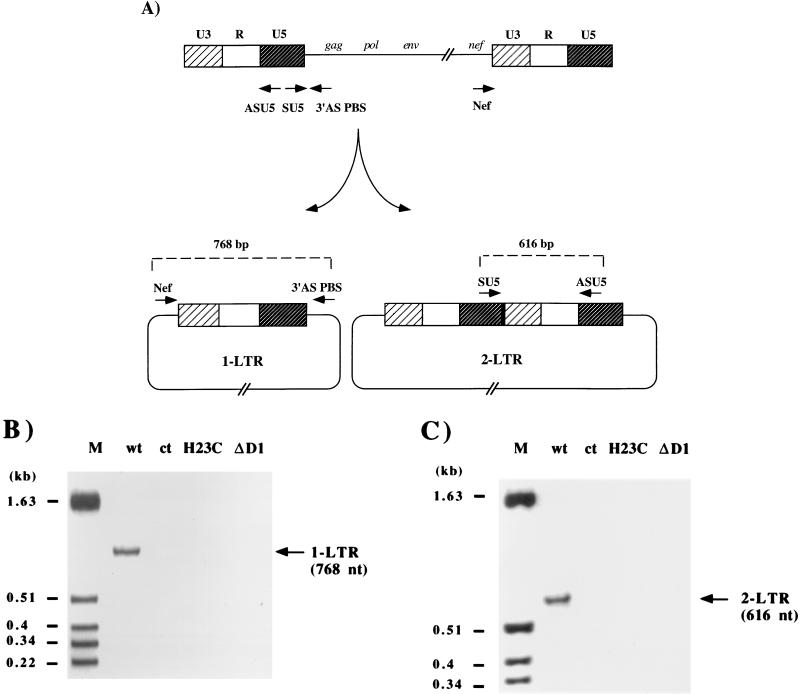
PCR analysis of the end of proviral DNA synthesis. (A) Schematic representation of 5′ LTR synthesis and localization of the sense primer SU3 (5′-GCACCATCCAAAGGTCAGTGG-3′) and the antisense primer ASPBS (5′-CTCCTCTGGCTTTACTTTCGC-3′). The dashed line indicates the DNA strand displacement necessary for 5′ LTR synthesis. (B) DNA extracted from infected cells (5) was analyzed by PCR as described in the legend for Fig. 3. Molecular size markers are indicated on the left.
Previous work indicated that one- and two-LTR circle forms are generated within the nucleus, probably by host activities since incubation of deproteinized linear HIV-1 cDNA with cell extracts leads to the formation of both forms, thus excluding NC protein from this process (7, 21, 48, 49, 50). We used these findings to further examine proviral DNA synthesis. The quantity of DNA used for PCR was adjusted for equal amounts of plus-strand DNA in infected cells. The nuclear fraction was analyzed by PCR 20 h after infection, using primers which amplify the one- or two-LTR DNA circles (Fig. 5A). No circle forms were detected after amplification in cells infected with either the H23C or ΔD1 mutant virus, while they were detected for wt HIV-1 (Fig. 5B and C). This experiment suggests that mutant proviral DNA is unable to form DNA circles because of defective ends and/or inefficient translocation in the nucleus.
FIG. 5.
PCR analysis of one- and two-LTR proviral DNA forms in infected HeLa P4 cells. (A) Schematic representation of the various forms of unintegrated circular HIV-1 viral DNA and the positions of the primer pairs used for PCR. Viral DNA nuclear import was analyzed by PCR with primers Nef (5′-GTTTTCCAGTCACACCTCAGG-3′) and 3′ASPBS (5′-CTCCTCTGGCTTTACTTTCGC-3′) to detect the single-LTR circular form (1-LTR) (B) and primers SU5 (5′-GACCCTTTTAGTCAGTGTGG-3′) and ASU5 (5′-CCAGAGTCACACAACAGACG-3′) to detect the double-LTR circular form (2-LTR) (C). Molecular size markers are indicated on each panel.
In the present study, the importance of the correct spatial arrangement of the proximal CCHC zinc finger has been assessed with respect to the biological functions of HIV-1 NC protein. Electron micrographs reveal that the majority of the NC mutant particles had either an immature or an abnormal core morphology compared to that of the wt virus, although no significant variation was detected in their respective densities. This impairment of the viral core structure may be related to defects in polyprotein precursor processing observed for both NC mutants but not observed in the wt. We assume that a misfolded NC domain within Pr55gag could negatively influence the conformation of the precursor and/or the stability of gag and gag-pol oligomers. This could be related to a defect in viral assembly.
Early reverse transcripts, such as R-U5 DNA, were detected by PCR for both NC mutants in infected cells, indicating that neither virus entry into cells nor the beginning of reverse transcription was affected. Moreover, viral DNA synthesis seemed to be complete as far as the second-strand transfer for H23C and ΔD1 mutants, providing evidence for a fully functional involvement of the mutated NC proteins during reverse transcription in vivo. Interestingly, we observed that the final step of viral DNA synthesis leading to synthesis of the 5′ LTR did not proceed correctly for either the H23C or the ΔD1 variant. This is interesting in light of the observation that neither of the one- and two-LTR circle forms could be detected for these mutants; due to defective ends, these molecules were unable to generate circle forms or to integrate. We assume that the H23C and ΔD1 mutations affected the stability of the reverse transcription complex and led to a defect in DNA strand displacement necessary for 5′ LTR synthesis, which has been reported to be slow and inefficient (25), and/or the loss of protection of viral DNA against exonucleases.
Moreover, correct integration may require functional cooperation between the NC and integrase (IN) proteins, as observed in vitro (9). Similarly, putative interactions between NC protein and cellular proteins involved in this process could also be affected. Further investigations are needed to determine whether NC mutant proteins and/or viral DNA is present within the nuclei of infected cells.
Taken together, these results show that the conformation of the NC protein is critical not only for virus assembly but also for complete proviral DNA synthesis and/or integration. This suggests that the NC protein acts as a chaperone protein during the course of the viral life cycle, mediated by its multimeric organization and enabling the production of infectious particles.
Acknowledgments
Thanks are due to Biomérieux for providing the anti-CAp24 monoclonal antibody and to Gérard Morel for critical assistance in analysis of electron microscopy data. We are grateful to Michael Rau for critical reading of the manuscript.
This work was supported by ANRS, SIDACTION, MGEN (Mutuelle Générale de l’Education Nationale), and European Community grant CT 96-0675.
REFERENCES
- 1.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J L. Transactivation of the minus-strand transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barat C, Schatz O, Le Grice S, Darlix J L. Analysis of the interaction of HIV-1 replication primer tRNALys,3 with nucleocapsid protein and reverse transcriptase. J Mol Biol. 1993;231:185–190. doi: 10.1006/jmbi.1993.1273. [DOI] [PubMed] [Google Scholar]
- 4.Barat C, Lullien V, Schatz O, Keith G, Nugeyre M-T, Grüninger Leitch F, Barré-Sinoussi F, LeGrice S, Darlix J-L. HIV-1 reverse transcriptase specifically interacts with the anti-codon domain of its cognate primer tRNA. EMBO J. 1989;8:3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthoux L, Péchoux C, Ottmann M, Morel G, Darlix J-L. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J Virol. 1997;71:6973–6981. doi: 10.1128/jvi.71.9.6973-6981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 8.Carrière C, Gay B, Chazal N, Morin N, Boulanger P. Sequence requirements for encapsidation of deletion mutants and chimeras of human immunodeficiency virus type 1 Gag precursor into retrovirus-like particles. J Virol. 1995;69:2366–2377. doi: 10.1128/jvi.69.4.2366-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carteau S, Batson S C, Poljak L, Mouscadet J-F, de Rocquigny H, Darlix J-L, Roques B P, Käs E, Auclair C. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J Virol. 1997;71:6225–6229. doi: 10.1128/jvi.71.8.6225-6229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clavel F, Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol. 1994;68:1179–1185. doi: 10.1128/jvi.68.2.1179-1185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darlix J L, Vincent A, Gabus C, de Rocquigny H, Roques B. Transactivation of the 5′ to 3′ viral DNA strand-transfer by the nucleocapsid protein during reverse transcription of HIV-1 RNA. C R Acad Sci Ser III Sci Vie. 1993;316:763–771. [PubMed] [Google Scholar]
- 14.Darlix J-L, Gabus C, Nugeyre M-T, Clavel F, Barré-Sinoussi F. Cis elements and trans acting factors involved in the RNA dimerization of HIV-1. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 15.Deméné H, Dong C Z, Ottmann M, Rouyez M C, Jullian N, Morellet N, Mély Y, Darlix J L, Fouyrnié-Zaluski M C, Saragosti S, Roques B P. 1H NMR structure and biological studies of the His23→Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critical for viral infectivity. Biochemistry. 1994;33:11707–11716. doi: 10.1021/bi00205a006. [DOI] [PubMed] [Google Scholar]
- 16.de Rocquigny H, Gabus C, Vincent A, Fournié-Zaluski M C, Roques B, Darlix J L. Viral RNA annealing activities of HIV-1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Stefano J J. Interaction of human immunodeficiency virus nucleocapsid protein with a structure mimicking a replication intermediate. J Biol Chem. 1996;271:16350–16356. [PubMed] [Google Scholar]
- 18.Di Marzo Veronese F, Copeland T D, Oroszlan S, Gallo R C, Sarngadharan M G. Biochemical and immunological analysis of human immunodeficiency virus gag gene products p17 and p24. J Virol. 1988;62:795–801. doi: 10.1128/jvi.62.3.795-801.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Marzo Veronese F, Rahman R, Copeland T, Oroszlan S, Gallo R C, Sarngadharan M G. Immunological and chemical analysis of p6, the carboxyl-terminal fragment of HIV p15. AIDS Res Hum Retroviruses. 1987;3:253–264. doi: 10.1089/aid.1987.3.253. [DOI] [PubMed] [Google Scholar]
- 20.Dorfman T, Luban J, Goff S P, Haseltine W A, Göttlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farnet C M, Haseltine W A. Circularization of human immunodeficiency virus type 1 DNA in vitro. J Virol. 1991;65:6942–6952. doi: 10.1128/jvi.65.12.6942-6952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Y X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzgerald D W, Coleman J E. Physicochemical properties of cloned nucleocapsid protein from HIV-1. Interactions with metal ions. Biochemistry. 1991;30:5195–5201. doi: 10.1021/bi00235a012. [DOI] [PubMed] [Google Scholar]
- 24.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuentes G M, Rodriguez-Rodriguez L, Palaniappan C, Fay P J, Bambara R A. Strand displacement synthesis of the long terminal repeats by HIV-1 reverse transcriptase. J Biol Chem. 1996;271:1966–1971. doi: 10.1074/jbc.271.4.1966. [DOI] [PubMed] [Google Scholar]
- 26.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorelick R J, Nigida S M, Jr, Bess J W, Jr, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson L E, Bowers M A, Sowder II R C, Serabyn S A, Johnson D G, Bess J W, Jr, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji X, Klarman G J, Preston B D. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry. 1996;35:132–143. doi: 10.1021/bi951707e. [DOI] [PubMed] [Google Scholar]
- 31.Khan R, Chang H O, Kaluarachchi K, Giedroc D P. Interaction of retroviral nucleocapsid proteins with transfer RNAPhe: a lead ribosyme and 1H NMR study. Nucleic Acids Res. 1996;24:3568–3575. doi: 10.1093/nar/24.18.3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lapadat-Tapolsky M, Gabus C, Rau M, Darlix J L. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and its variability. J Mol Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- 33.Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix J L. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Quan Y, Arts E J, Li Z, Preston B D, de Rocquigny H, Roques B P, Darlix J-L, Kleiman L, Parniak M A, Wainberg M A. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA3Lys in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuno A, Ido E, Goto T, Kuwata T, Nakai M, Hayami M. Mutational analysis of two zinc finger motifs in HIV type 1 nucleocapsid proteins: effect on proteolytic processing of gag precursors and particle formation. AIDS Res Hum Retroviruses. 1996;12:793–800. doi: 10.1089/aid.1996.12.793. [DOI] [PubMed] [Google Scholar]
- 36.Morellet N, de Rocquigny H, Mély Y, Jullian N, Déméné H, Ottmann M, Gérard D, Darlix J L, Fournie-Zaluski M C, Roques B P. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J Mol Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- 37.Morellet N, Jullian N, de Rocquigny H, Maigret B, Darlix J L, Roques B. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ominchinski J G, Clore G M, Sakaguchi K, Appella E, Gronenborn A M. Structural characterization of a 39 residue synthetic peptide containing the two zinc binding domains from the HIV-1 p7 nucleocapsid protein by CD and NMR spectroscopy. FEBS Lett. 1991;292:25–30. doi: 10.1016/0014-5793(91)80825-n. [DOI] [PubMed] [Google Scholar]
- 39.Ottmann M, Gabus C, Darlix J-L. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peliska J A, Balasubramanian S, Giedroc D P, Benkovic S J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNaseH activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 41.Poon D T K, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez-Rodriguez L, Tsuchihashi Z, Fuentez G M, Bambara R A, Fay P J. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J Biol Chem. 1995;270:15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 43.Schmalzbauer E, Strack B, Dannull J, Guehmann S, Moelling K. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J Virol. 1996;70:771–777. doi: 10.1128/jvi.70.2.771-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song W, Lahiri D K. Efficient transfection of DNA by mixing cells in suspension with calcium phosphate. Nucleic Acids Res. 1995;23:3609–3611. doi: 10.1093/nar/23.17.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.South T L, Blake P R, Hare D R, Summers M F. C-terminal retroviral-type zinc finger domain from the HIV-1 nucleocapsid protein is structurally similar to the N-terminal zinc finger domain. Biochemistry. 1991;30:6342–6349. doi: 10.1021/bi00239a036. [DOI] [PubMed] [Google Scholar]
- 46.Tanchou V, Gabus C, Rogemond V, Darlix J L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J Mol Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- 47.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varmus H, Brown P. Retroviruses. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 53–108. [Google Scholar]
- 49.Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985;40:9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- 50.Whitcomb J M, Kumar R, Hughes S H. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J Virol. 1990;64:4903–4906. doi: 10.1128/jvi.64.10.4903-4906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]