Abstract
Simple Summary
The human gastrointestinal tract harbors a complex and dynamic population of microorganisms (mostly bacteria and viruses) that are called the gut microbiota. This microbial community is known to exert a marked influence on the host health. Among the different bacterial families present in the gut microbiota, Enterobacteriaceae represents only a small fraction of total bacteria in healthy individuals, whereas it can abnormally proliferate in the gut of individuals affected by intestinal or extra-intestinal diseases such as obesity, inflammatory bowel disease or metabolic disorders. Our review explores the recent knowledge discussing the dynamics of Enterobacteriaceae in the human gut in healthy or disease-associated conditions. We also describe emerging mechanistic studies that could explain how Enterobacteriaceae proliferate in cases of gut microbiota imbalance in patients as well as the consequences such expansions could have on host health.
Abstract
The human gut microbiota plays a crucial role in maintaining host health. Our review explores the prevalence and dynamics of Enterobacteriaceae, a bacterial family within the Proteobacteria phylum, in the human gut which represents a small fraction of the gut microbiota in healthy conditions. Even though their roles are not yet fully understood, Enterobacteriaceae and especially Escherichia coli (E. coli) play a part in creating an anaerobic environment, producing vitamins and protecting against pathogenic infections. The composition and residency of E. coli strains in the gut fluctuate among individuals and is influenced by many factors such as geography, diet and health. Dysbiosis, characterized by alterations in the microbial composition of the gut microbiota, is associated with various diseases, including obesity, inflammatory bowel diseases and metabolic disorders. A consistent pattern in dysbiosis is the expansion of Proteobacteria, particularly Enterobacteriaceae, which has been proposed as a potential marker for intestinal and extra-intestinal inflammatory diseases. Here we develop the potential mechanisms contributing to Enterobacteriaceae proliferation during dysbiosis, including changes in oxygen levels, alterations in mucosal substrates and dietary factors. Better knowledge of these mechanisms is important for developing strategies to restore a balanced gut microbiota and reduce the negative consequences of the Enterobacteriaceae bloom.
Keywords: human gut microbiota, Enterobacteriaceae, Escherichia coli, gut dysbiosis
1. Proteobacteria/Enterobacteriaceae in the Healthy Human Gut
1.1. Taxonomy, Diversity and Abundance
The human gut microbiota harbors approximately 1014 microorganisms, comprising more than 1000 bacterial species belonging to only few bacterial phyla [1]. Firmicutes and Bacteroidota, comprising 90% of the overall phylum composition, stand out as the predominant microbial phyla in the gut. Actinobacteria, Proteobacteria, Fusobacteria and Verrucomicrobia are also found as part of the normal gut microbiota but are represented to a lesser extent [2]. The phylum Proteobacteria is systematically present in the gut of healthy humans with a low abundance (less than 5% of total microbial abundance) [3]. Based on 16S rDNA sequence analysis, the Proteobacteria phylum is divided into six classes: Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Deltaproteobacteria, Epsilonproteobacteria and Zetaproteobacteria. In the human gut, Gammaproteobacteria and Deltaproteobacteria are the most commonly found, Gammaproteobacteria being mainly represented by the Enterobacteriaceae family. The Enterobacteriaceae family comprises both commensals and opportunistic disease-causing pathogens. However, Enterobacteriaceae usually constitutes less than 1% of the healthy gut microbiota. This family includes a diversity of bacterial genera including Escherichia, Enterobacter and Klebsiella. The species Escherichia coli was shown to be, by far, the most dominant Enterobacteriaceae in healthy humans [4]. For these reasons, most data available in the literature regarding Enterobacteriaceae in the gut microbiota frequently concern E. coli, even if other minor bacterial species could have important functions as well.
Cross-sectional studies of human adults demonstrated that E. coli is a member of the intestinal microbiome of over 90% of individuals [5]. The number of distinct E. coli strains per host was also assessed in individuals and the average number was found to be between 1.1 and 3.5 E. coli strains per individual [6]. Based on molecular analysis, the different E. coli strains can be divided into nine defined phylogenetic groups (A, B1, B2, C, D, E, F, G and H). The prevalence of E. coli phylogenetic groups in the gut of healthy individuals varies between studies and appears to depend on different factors such as geographic location and diet. While phylogroup A was dominant in industrialized countries in the 1980s [5], the proportion of phylogroup B2 strongly increased over the last 30 years and is now as prevalent as phylogroup A in healthy individuals [7]. Also, the presence of a dominant strain from phylogroups E and F appears to tolerate the co-occurrence of other minor E. coli strains, whereas the establishment of a dominant strain from phylogroup B2 seems to limit gut colonization by other E. coli populations [8].
1.2. Strain Residency in the Gut
Longitudinal studies in healthy individuals have reported that the gut microbiota composition is relatively stable at high taxonomic levels. Nevertheless, recent studies present evidence suggesting a significant turnover rate at the strain level, at least within populations of Enterobacteriaceae [4,9,10]. In one of these studies, strain residency was evaluated in eight participants for an average time of 500 days [4]. Only 31% of Enterobacteriaceae clones were considered as long-term residential strains, whereas the others were defined as short-term transient strains. It is crucial to emphasize that the permanence of strains can vary significantly among individuals. While some individuals may host several resident strains for extended periods, others may experience the successive emergence and disappearance of clonal populations over time [4]. Thus, it appears that Enterobacteriaceae residency is a dynamic process and resident clones turn over at different rates in individuals. E. coli strains are not all equal with respect to their ability to colonize and reside in the adult human gut. Depending on their properties, ingested E. coli strains are able to establish in the gut and become resident for a long period or are quickly lost at the rate of gut transit. Several factors are thought to influence E. coli residency in the gut such as socio-economic factors, diet, health, travel or exposure to antibiotics. However, the rules governing E. coli residency in the gut are far from being fully understood. Nonetheless, it has been shown that long-lived resident E. coli are phylogenetically distinct from short-lived transients and belong preferentially to phylogenetic groups A, B2 and F [4]. Gene association studies are now warranted to better define residency-associated traits and understand this important aspect of microbial ecology.
1.3. Functional Roles of Enterobacteriaceae in the Human Gut
Despite the fact that Enterobacteriaceae are detected in nearly all individuals, the ecological roles of this bacterial family in the human gut are poorly understood. As of now, the functions assigned to Enterobacteriaceae include the following:
Maintenance of an efficient anaerobic environment in the gut
As facultative anaerobes, most Enterobacteriaceae, and in particular E. coli, deplete oxygen diffusing from the mucosal surface and therefore participate to the establishment of a perfect environment for strict anaerobes which constitute almost all members of the gut microbiota. Several works suggest that E. coli develops a relationship with the anaerobic members of the gut microbiota. Within the gastrointestinal tract, anaerobic bacteria break down complex polysaccharides, releasing simple carbohydrates that E. coli metabolizes for its growth. In return, E. coli helps create an anaerobic environment by scavenging oxygen [11]. This function is important not only in adults but also in the neonates. Indeed, E. coli is one of the first bacteria to colonize the gut of neonates at birth. This newly established and rapidly growing E. coli population then changes the structure and function of the epithelial cells in ways that appear crucial for healthy microbiome development [12]. In particular, oxygen consumption by E. coli and other facultative anaerobes creates hospitable conditions for strict anaerobes to colonize and become dominant inhabitants of the gut [13].
Production of vitamins
In humans and animals, vitamin K, also referred as menaquinone, has multiple roles and one of them is to be a co-factor for many important enzymes. E. coli is known to produce menaquinone during anaerobic growth and uses it as an electron carrier in the respiratory chain [14]. With other members of the gut microbiota, E. coli modulates menaquinone concentrations in the human gut and contributes to supplying the host with its vitamin K requirement [15]. E. coli is also able to synthesize vitamin B12 through modifications of corrinoids [16]. Vitamin B12, also known as cyanocobalamin, is atypical among vitamins because it is not produced by plants or the majority of vertebrates. Instead, it is exclusively synthesized by bacteria and archaea. The essential nature of these pathways makes many eukaryotes, including humans, dependent on microbial corrinoids. Nevertheless, the direct impact of gut microbial communities on the levels of cobalamin (vitamin B12) in the host is a matter of debate. Indeed, cobalamin produced in the colon, where microbial numbers are highest, is probably not bioavailable for the host because the receptors necessary for absorbing the vitamin are mostly found in the small intestine. In humans, diet is instead more likely to constitute the primary source of cobalamin [17]. Nevertheless, the biological significance of vitamin production by Enterobacteriaceae and contribution to host requirement is probably very low because (i) they represent only a small population in the gut microbiota and (ii) vitamin synthesis in the gut relies on multiple bacteria from diverse phyla and results from complex microbial–microbial cooperation [18].
Protection against gut pathogen infections
The best characterized role of Enterobacteriaceae in gut homeostasis concerns its contribution to resistance against colonization by exogenous pathogens. Protection offered by the gut microbiota occurs through different mechanisms, including secretion of antimicrobial products, nutrient competition, support of epithelial barrier integrity and immune activation [19]. In the case of commensal E. coli, the protective mechanisms are thought to rely mainly on bacteriocin synthesis and competition for nutrient resources that can limit the growth of enteric pathogens. As an example, a study from Hudault et al. showed that colonization of germfree mice by an E. coli strain had a protecting effect against Salmonella enterica serovar Typhimurium infection [20]. Competition for the same nutritional substrates between commensal and pathogenic E. coli strains has been also largely studied in the literature. Commensal E. coli consumes monosaccharides such as fucose, mannose, galactose, ribose, arabinose and N-acetyl glucosamine in the mucus layer, which are carbohydrates that are also used by enterohaemorrhagic E. coli (EHEC) O157:H7 during infection. Several in vivo mouse studies demonstrated that commensal E. coli can prevent EHEC infection due to competition for nutrients such as carbohydrates, amino acids and organic acids [21,22,23,24].
2. Proteobacteria/Enterobacteriaceae Expansion in Dysbiosis Associated with Human Disease
2.1. Proteobacteria Bloom: A Marker of Gut Microbiota in Disease
It has been demonstrated that the gut microbiota contributes to health and the well-being of the individual [25,26]. Any disruption in the composition of the gut microbiota, called dysbiosis, has impacts on host health. Dysbiosis can be defined by typical features such as a decrease in microbial diversity and/or richness, an altered ratio of Bacteroidetes/Firmicutes, a decrease in beneficial species (i.e., Bifidobacterium, Faecalibacterium prauznitsii…) and/or an increase in deleterious bacteria such as Proteobacteria. Indeed, Proteobacteria usually represent less than 5% of the healthy gut microbiota, but this phylum can reach 10–90% in the gut of patients suffering from various diseases, especially inflammatory ones [27]. Currently, several studies have shown that the composition of the gut microbiota of patients suffering from different diseases differs comparing to the healthy individual. Intestinal dysbiosis has been observed in various diseases such as obesity, allergic disorders, diabetes, autism, colorectal cancer, Alzheimer’s, Parkinson’s or cardiovascular diseases.
Among the multiple taxonomic variations associated with gut dysbiosis, the most consistent and robust ecological pattern is an expansion of facultative anaerobic bacteria belonging to the phylum Proteobacteria. While information at a lower taxonomic level than the Proteobacteria phylum is not systematically described in the different works, the Enterobacteriaceae family appears to have a prominent place for explaining the expansion of Proteobacteria associated with disease. From these observations, several groups proposed that an enrichment of Proteobacteria/Enterobacteriaceae in the gut is a marker for an unstable microbial community and a potential diagnostic criterion for disease [27,28,29]. Among the works correlating disease state and gut dysbiosis, gut inflammation at low or high levels appears to be associated with expansion of Enterobacteriaceae in various disease contexts such as inflammatory bowel diseases, colorectal cancer or metabolic syndrome [30,31,32,33]. Naturally, a legitimate question concerns the causal relationship between Enterobacteriaceae bloom and disease state, asking what is the cause and what is the consequence. If the answer is not as simple as the question is, numerous works with mechanistic insights have been performed in the last decade to highlight (i) the conditions that could cause proliferation of Enterobacteriaceae in the gut of patients and (ii) the potential consequences of such expansion on health status and disease progression.
2.2. Causes of Enterobacteriaceae Proliferation in Dysbiotic Conditions
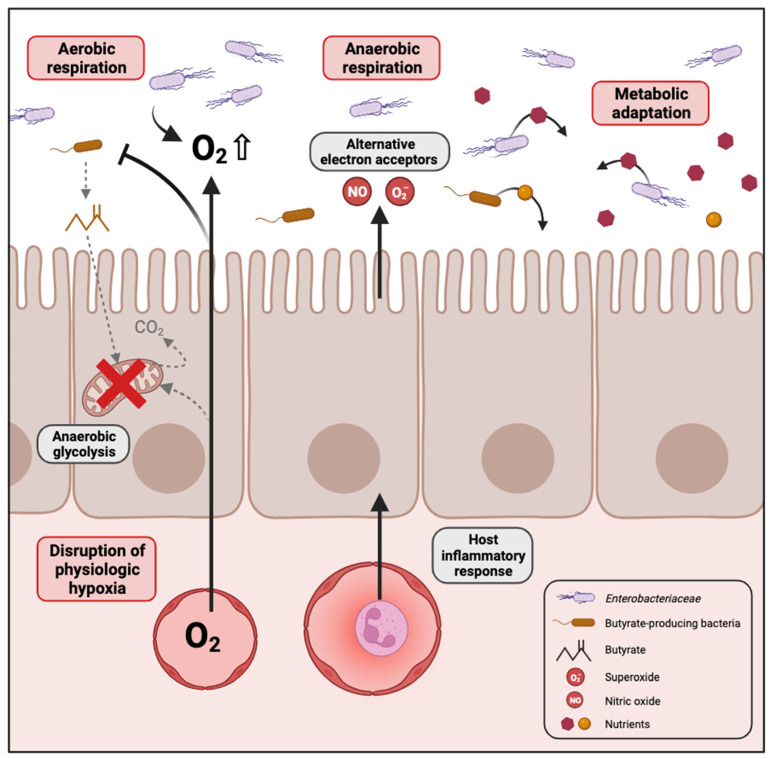
Several mechanisms have been proposed in the literature that could explain the bloom of Enterobacteriaceae in the dysbiotic gut, which are summarized in Figure 1.
Figure 1.
Mechanisms contributing to the expansion of Enterobacteriaceae in the dysbiotic gut. In conditions where high levels of oxygen reach the gut lumen, for example when enterocytes switch from mitochondrial fatty acid β-oxidation to anaerobic glycolysis, the facultative anaerobes of Enterobacteriaceae proliferate through aerobic respiration. In cases of gut inflammation, an increase in the availability of various electron acceptors promotes anaerobic respiration by Enterobacteriaceae. Changes in the gut environment caused by disease-associated dysbiosis can lead to Enterobacteriaceae expansion as a result of their adaptability to metabolize potential arising substrates. Created with Biorender.com.
2.2.1. Disruption of Physiologic Hypoxia
The gastrointestinal tract in humans is hypoxic, characterized by an oxygen gradient extending from the colonic mucosal surface to the gut lumen. This oxygen variation influences the structure of the intestinal microbial community, favouring the establishment of predominantly obligate anaerobes during homeostasis [34,35]. Furthermore, the microbiome plays a key role in sustaining the hypoxic environment within the intestine, which is essential for nutrient absorption, intestinal barrier function and the activation of innate and adaptive immune responses in mucosal cells. Emerging evidence supports that oxygen diffusion within the gut lumen can be a driver of gut dysbiosis [36]. Indeed, an increase in the oxygen level in the gut would destabilize the oxygen-sensitive strict anaerobes and promote the expansion of facultative anaerobes [37]. The majority of Proteobacteria are facultative anaerobes, meaning they have the ability to thrive in the presence of oxygen. This confers a notable competitive edge over beneficial obligate anaerobes in environments where oxygen is present. For instance, antibiotic treatment or inflammation are conditions known to promote Enterobacteriaceae expansion. Consistent with this idea, metagenomic analysis in the murine model of colitis identified oxygen respiration as a dominant signature associated with commensal E. coli expansion in the gut [38,39].
Butyrate, a short-chain fatty acid, is generated as a metabolite by obligate anaerobes during the breakdown of dietary fiber in the colon. Interestingly, it has been shown that elevated Proteobacteria levels and diminished populations of butyrate-producing bacteria serve as a microbial signature for dysbiosis in various chronic diseases such as inflammatory bowel disease [40], irritable bowel syndrome [41], colorectal cancer [31], diabetes [42] and obesity [43]. For instance, streptomycin treatment leads to a depletion of bacteria belonging to the class Clostridia which are important producers of butyrate. In normal conditions, colonocytes are hypoxic because they consume oxygen through mitochondrial β-oxidation of microbiota-derived butyrate to CO2, which represents their main pathway for producing energy [44]. Depletion of butyrate-producing bacteria by antibiotic treatment or other dysbiotic conditions would reduce luminal butyrate levels, resulting in a metabolic reorientation of intestinal epithelial cells towards anaerobic glycolysis, a process that does not consume oxygen [45]. The consequent increase in the amount of oxygen emanating from the colonic surface drives an expansion of facultative anaerobes such as Enterobacteriaceae in the gut [46]. Colonocyte metabolism serves as a pivotal regulator for gut microbiota, orchestrating a transition between balanced, homeostatic communities and imbalanced, dysbiotic states.
2.2.2. Inflammation Generates Electron Acceptors for Anaerobic Respiration
Intestinal inflammation also contributes to an increased availability of various electron acceptors that promote anaerobic respiration by Enterobacteriaceae [47]. The host inflammatory response produces reactive nitrogen species and reactive oxygen species that, upon diffusion into the gut lumen, will react with organic sulphides, tertiary amines or other molecules to form S-oxides, N-oxides and nitrate. Enterobacteriaceae can use these molecules as terminal electron acceptors for anaerobic respiration by expressing DMSO, TMAO and nitrate reductases, respectively [48,49]. For instance, E. coli encodes two DMSO reductases (dmsABC, ynfFGH), three TMAO reductases (torCAD, torYZ, yedYZ) and three nitrate reductases (narGHJI, narZYWV, napFDAGHBC). Such a rich environment for alternative electron acceptors therefore confers a growth advantage for Enterobacteriaceae in inflammatory conditions. Indeed, anaerobic respiration is considered a process more efficient for energy production than fermentation made by other members of the gut microbial community.
2.2.3. Metabolic Adaptation to Dysbiotic Conditions
Dysbiosis often causes significant changes in the gut environment and may influence the diversity and concentrations of nutrients available in the lumen that the different members of the gut microbiota can utilize with variable efficiency. Enterobacteriaceae and particularly E. coli present a dynamic genome structure leading to a high level of genome plasticity. Moreover, Enterobacteriaceae have been shown to be the major source of variable genes in the gut microbiome between healthy individuals, whereas they represent only a minor population, indicating that this family brings variability in gut microbial gene function [50]. Consistent with these observations, changes in the gut environment caused by disease-associated dysbiosis can lead to Enterobacteriaceae expansion as a result of a better adaptability to metabolize potential arising substrates. Different studies have already explored microbial metabolic adaptation, and some nutrient sources available to Enterobacteriaceae during intestinal dysbiosis are summarized in Table 1. Although some of this research has been conducted with enteric pathogens, it is reasonable to believe that most traits can be transposed to commensal Enterobacteriaceae.
Table 1.
Studies reporting nutrient sources available for Enterobacteriaceae during intestinal dysbiosis.
| Nutrient | Bacterial Species | References |
|---|---|---|
| Ethanolamine |
S. Typhimurium 1 C. rodentium 2 |
[51] [52] |
| Lactate | S. Typhimurium | [53] |
| Glucarate/galactarate |
S. Typhimurium Commensal E. coli |
[54] |
| 1,2-propanediol | S. Typhimurium | [55] |
| Succinate | S. Typhimurium | [56] |
| L-Serine | Adherent-invasive E. coli C. rodentium |
[57] [58] |
| Glycerol | Commensal E. coli | [59] |
| Carnitine and Acylcarnitines | E. coli | [60] |
1 Salmonella enterica serovar Typhimurium. 2 Citrobacter rodentium.
Glycerol in cystic fibrosis patients
Cystic fibrosis is a genetic disease affecting the normal functions of multiple organs including the lungs, pancreas and gastrointestinal tract with a pronounced inability of patients to digest and absorb proteins and lipids [61]. A gut dysbiosis notably characterized by a bloom of Enterobacteriaceae is associated with cystic fibrosis [62]. Indeed, E. coli that normally represents less than 1% of the gut microbiota can reach more than 90% in the fecal microbiota of young children with cystic fibrosis, compared with age-matched healthy controls [63]. In 2018, Matamouros et al. isolated E. coli strains from fecal samples of young children suffering from this disease and showed an increased growth rate in the presence of glycerol as a sole carbon source [59]. Glycerol is a major component of fatty acids of intestinal fat, present in high concentrations in the gut of patients as a result of fat malabsorption. In addition, cystic fibrosis and control E. coli isolates have differential gene expression when grown in the presence of glycerol, likely resulting in a better growth rate of the patient strains. The authors suggest that E. coli expansion in the gut of cystic fibrosis children can be explained through adaptation and/or selection of E. coli strains possessing a high ability to metabolize glycerol [59].
Intestinal mucosa-derived substrates
In the case of diseases marked by an important gut inflammation, alteration of the gut environment can generate a unique set of carbon sources available to Enterobacteriaceae to outgrow other members of the gut microbiota. For instance, inflammation causes the shedding of epithelial cells in the lumen and alterations of the mucus layer. Components derived from the epithelial cell membrane such as phosphatidylcholine and phosphatidylethanolamine increase in this context. Ethanolamine (EA) is derived from phosphatidylethanolamine and can be used as a source of carbon and/or nitrogen by a variety of species in the Firmicutes, Actinobacteria and Proteobacteria phyla, as well as by pathogenic species such as Salmonella and Pseudomonas [64]. Utilization of EA involves breaking ethanolamine into ammonia and acetaldehyde thanks to a specific ethanolamine ammonia lyase [39,64]. The ammonia resulting from this reaction can be used as a cellular supply of reduced nitrogen, and the acetaldehyde is converted to acetyl-CoA that can be used in various bacterial metabolic cycles such as the tricarboxylic acid cycle, the glyoxylate cycle or lipid biosynthesis [64].
The mucus layer of the gut epithelium is composed of mucin proteins that are glycosylated with five major sugars that can be potential nutrients for bacteria: N-acetyl-D-galactosamine, N-acetyl-D-glucosamine, N-acetylneuraminic acid (sialic acid), L-fucose and D-galactose. In cases of gut inflammation, there is an increase in the production and secretion of mucins as a mechanism of maintaining the integrity of the mucus layer. In a study using a mouse model of induced gut inflammation, it has been reported that E. coli outgrowth in the inflamed gut consequently leads to an increase in the commensal species expressing sialidases. Such increased activity leads to the release of high amounts of free sialic acid which is efficiently consumed by E. coli [65]. In another study, an increase in glycosidase-encoding genes in the microbiome of inflamed gut samples of patients was found to be consistent with an enhanced mucin-degrading activity. Mucin-degrading bacteria such as Mucispirillum schaedleri, Akkermansia muciniphila and Bacteroides acidifaciens expand and mediate the release of less complex sugars (lactose, melibiose, raffinose and galactinol) from mucins [39]. These saccharides and metabolites accumulate and may lead to a decrease in commensal bacteria from the Bacteroidia and Clostridia classes in the inflamed gut, but also confer a growth advantage to Enterobacteriaceae as well as to pathogens such as S. Typhimurium and Clostridium difficile [39,66]. Overall, the composition and abundance of mucosal populations, also known as mucosal biofilm, seems to correlate with health status. Whereas healthy mucosal biofilm is of low microbial density (105–106 cells per mL) and is mostly composed of Firmicutes, Bacteroidetes and some oxygen-tolerant microbes such as lactic acid bacteria and Proteobacteria members, clinical observations from patients with chronic intestinal diseases (IBS, IBD, colorectal cancer) have revealed dense polymicrobial biofilms in the mucus with an increased proportion of Enterobacteriaceae. These modified mucosal biofilms have been proposed to be an early warning signal of developing disease and could be considered as the tipping point between a healthy and a diseased state of the gut mucosa [67].
Diet-derived substrates
It is now well admitted that diet impacts the composition of gut microbiota. For example, a Western-style High-Fat Diet (HFD) is known to induce Enterobacteriaceae proliferation in the gut. The enrichment of Enterobacteriaceae, largely represented by Escherichia coli, has been correlated with impaired glucose homeostasis [68]. In mice receiving an HFD treatment, but not a standard diet, the presence of E. coli significantly increased body weight and adiposity and induced impaired glucose tolerance. This work demonstrates the role of commensal E. coli in glucose homeostasis and energy metabolism in response to an HFD, indicating contributions of commensal bacteria to the pathogenesis of obesity and type 2 diabetes. An HFD also promotes cardiovascular diseases, in part because of high levels of choline which is converted to trimethylamine (TMA) by gut microbiota. Using a mouse model of diet-induced obesity, Yoo et al. show that chronic exposure to an HFD increases Escherichia coli choline catabolism by altering the intestinal epithelial physiology [69]. An HFD impaired the bioenergetics of mitochondria in the colonic epithelium to increase the luminal bioavailability of oxygen and nitrate, thereby intensifying respiration-dependent choline catabolism of E. coli. In turn, E. coli choline catabolism increased levels of circulating trimethylamine N-oxide, which is a potentially harmful metabolite generated by gut microbiota.
Another example concerns patients suffering from Crohn’s disease (CD), an inflammatory disease of the gut mucosa, associated with a dysbiosis characterized by a high prevalence of a particular type of strain called Adherent and Invasive Escherichia coli (AIEC) in the inflamed gut mucosa. These strains possess the ability to adhere to and invade intestinal epithelial cells and to replicate within macrophages [70]. A recent work by Kitamoto et al. shows that the amino acid availability is modified in the inflamed gut and that AIEC reprogram their metabolic pathways to use L-serine in order to gain a fitness advantage over resident microbiota [57]. L-serine is acquired from the diet and can be used as a source of energy after conversion into pyruvate, which is a substrate necessary for glucogenesis and the tricarboxylic acid cycle [71]. While this metabolic pathway has a minor role on AIEC fitness in the healthy gut, this amino acid is a key resource for AIEC expansion during CD intestinal dysbiosis. The described study also successfully demonstrated that restriction of dietary amino acid intake, particularly L-serine, prevents the expansion of Enterobacteriaceae in the inflamed gut. Understanding the specific nutrients that Enterobacteriaceae use during dysbiosis is essential for developing innovative strategies to restore a balanced gut microbiota.
2.3. Consequences of Enterobacteriaceae Proliferation in Disease Progression
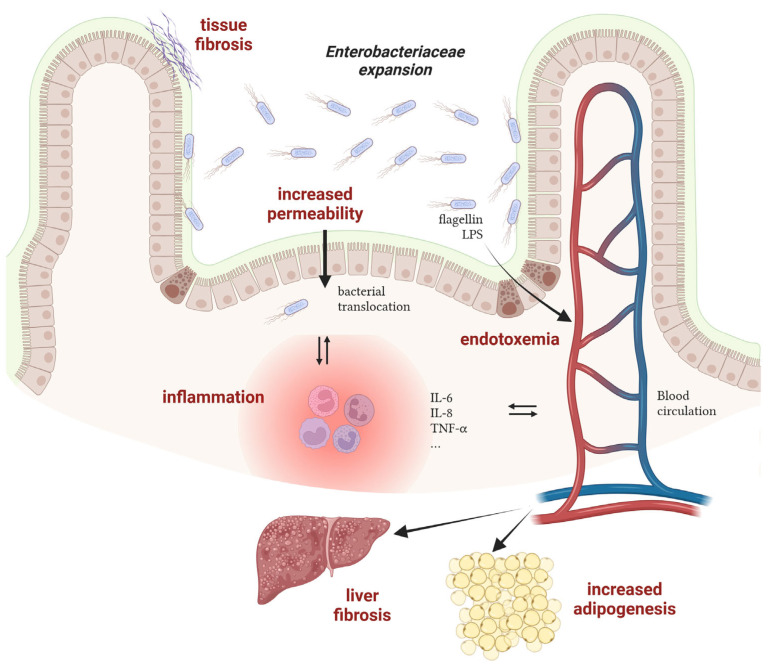
Although the conditions promoting the bloom of Enterobacteriaceae have been well investigated these last years, the consequences of such an imbalance on the host are often unknown or underestimated. Nevertheless, works with mechanistic insights into the processes connecting gut dysbiosis and disease are emerging and will allow researchers to unravel whether unbalanced microbial communities participate in disease progression. Hereafter are elements suggesting the harmful role of a bloom of Enterobacteriaceae on host health in the case of Crohn’s disease and obesity-associated diseases (Figure 2).
Figure 2.
Mechanisms of action of Enterobacteriaceae in the progression of inflammatory bowel diseases and obesity-associated diseases. LPS: lipopolysaccharides. Created with Biorender.com.
Crohn’s disease
The relationship between AIEC and intestinal inflammation in CD patients has been extensively investigated in the last 20 years and it is now well admitted that these bacteria participate in the induction and/or maintenance of disease symptoms. Experimental data from CD patients or from AIEC-infected rodents have revealed an increased permeability of the intestinal epithelium in the presence of AIEC [72]. Such dysfunction of the intestinal barrier may cause microbial translocation and trigger an immune response, leading to inflammation. Moreover, adhesion to and invasion of epithelial cells by AIEC also contribute to inflammation, as revealed by increased expression levels of multiple pro-inflammatory effectors such as IL-6, IL-8 and TNF-α [73]. Replication of AIEC within macrophages also leads to TNF-α stimulation [74]. Concomitantly with inflammation, chronic colonization of the intestinal mucosa by AIEC contributes to tissue fibrosis via collagen deposition in the extracellular matrix, potentially leading to bowel obstruction in CD patients [75,76]. Besides AIEC and their contribution to disease, other pathobionts with increased prevalence in CD patients have been identified but their role remains elusive to date [77].
Obesity-associated diseases
Overweight and obesity, which have tripled worldwide over the last 40 years, are major risk factors for several chronic diseases, including cardiovascular diseases, diabetes, musculoskeletal disorders, nonalcoholic fatty liver disease (NAFLD) and cancers. In most cases, a major gut microbiota alteration is observed in obese patients with a potential impact on the progression of obesity-associated diseases [78]. For instance, a causal link between a strain of Enterobacter cloacae (Enterobacteriaceae family) and obesity has been deciphered in a work published by the group of L. Zhao [33]. During a clinical study, they found that the gut microbiota of a morbidly obese volunteer was 35% composed of bacteria of the genus Enterobacter. After a specific diet program, the weight loss of the patient was concomitant with the disappearance of Enterobacter in the gut microbial community. Moreover, colonization of germ-free mice with the Enterobacter cloacae strain isolated from the obese human gut leads to the development of obesity in animals fed on a high-fat diet. A second study also demonstrated that E. cloacae administration to mice increases subcutaneous fat mass and promotes liver fibrosis, a sign of hepatic damages [79]. In a similar way, supplementation of high-fat diet treated mice with a strain of E. coli induces adipogenesis, reduces glucose tolerance and promotes a low-grade systemic inflammation, suggesting a role of commensal bacteria in the pathogenesis of obesity and type 2 diabetes (T2D) [68]. At the molecular level, flagellin, the monomer of the bacterial locomotor appendage, has been shown to induce a pro-inflammatory response in pancreatic beta cells in obese individuals with T2D. Because Enterobacteriaceae are more abundant in the gut of individuals with T2D, this flagellin-dependent induction is massive and leads to beta cell dysfunction, a key abnormality leading to hyperglycemia and T2D [80]. In the case of NAFLD, the lipopolysaccharide (LPS) and its endotoxic activity from E. cloacae, Klebsiella pneumoniae or E. coli have been found to participate in liver dysfunction in mice mono-associated with these Enterobacteriaceae strains [81]. Altogether, these works reveal that specific bacterial species belonging to the Enterobacteriaceae family may causatively contribute to the development of disease in human when their population levels are deregulated in dysbiotic conditions.
3. Conclusions
Taxonomic analyses of the gut microbiota from healthy individuals or patients suffering from various intestinal or extra-intestinal diseases reveal that gut dysbiosis in patients are often characterized by an expansion of Proteobacteria, particularly Enterobacteriaceae, making this trait a potential marker to detect the onset of disease. Recent studies also decipher some conditions or parameters associated with dysbiosis that could explain Enterobacteriaceae proliferation in the gut and how such an imbalance could affect host health and disease progression. A better understanding of the mechanisms governing disease state and gut dysbiosis, especially Enterobacteriaceae bloom, would improve the development of new approaches for the manipulation of gut bacterial communities in order to sustain or restore homeostasis and improve host health.
Author Contributions
All of the authors contributed to proofreading and validation of the proposed review. M.I.M.d.G., A.B.-D. and G.J. wrote the review. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript.
Funding Statement
This work was supported by funding from the National Research Institute for Agriculture, Food and Environment (INRAE). Maria Ines Moreira de Gouveia was a PhD Research Fellow funded by the French Ministry of Education and Research.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Sakkas H., Bozidis P., Touzios C., Kolios D., Athanasiou G., Athanasopoulou E., Gerou I., Gartzonika C. Nutritional Status and the Influence of the Vegan Diet on the Gut Microbiota and Human Health. Medicina. 2020;56:88. doi: 10.3390/medicina56020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinninella E., Raoul P., Cintoni M., Franceschi F., Miggiano G.A.D., Gasbarrini A., Mele M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms. 2019;7:14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huttenhower C., Gevers D., Knight R., Abubucker S., Badger J.H., Chinwalla A.T., Creasy H.H., Earl A.M., Fitzgerald M.G., Fulton R.S., et al. Structure, Function and Diversity of the Healthy Human Microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinson J.N.V., Pinkham N.V., Peters G.W., Cho H., Heng J., Rauch M., Broadaway S.C., Walk S.T. Rethinking Gut Microbiome Residency and the Enterobacteriaceae in Healthy Human Adults. ISME J. 2019;13:2306–2318. doi: 10.1038/s41396-019-0435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tenaillon O., Skurnik D., Picard B., Denamur E. The Population Genetics of Commensal Escherichia coli. Nat. Rev. Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 6.Alm E.W., Walk S.T., Gordon D.M. Population Genetics of Bacteria. ASM Press; Washington, DC, USA: 2014. [(accessed on 20 February 2024)]. The Niche of Escherichia coli. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1128/9781555817114.ch6. [Google Scholar]
- 7.Marin J., Clermont O., Royer G., Mercier-Darty M., Decousser J.W., Tenaillon O., Denamur E., Blanquart F. The Population Genomics of Increased Virulence and Antibiotic Resistance in Human Commensal Escherichia coli over 30 Years in France. Appl. Environ. Microbiol. 2022;88:e0066422. doi: 10.1128/aem.00664-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon D.M., O’Brien C.L., Pavli P. Escherichia coli Diversity in the Lower Intestinal Tract of Humans. Environ. Microbiol. Rep. 2015;7:642–648. doi: 10.1111/1758-2229.12300. [DOI] [PubMed] [Google Scholar]
- 9.Calderón D., Cárdenas P.A., Prado-Vivar B., Graham J.P., Trueba G. A Longitudinal Study of Dominant E. coli Lineages and Antimicrobial Resistance in the Gut of Children Living in an Upper Middle-Income Country. J. Glob. Antimicrob. Resist. 2022;29:136–140. doi: 10.1016/j.jgar.2022.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Priya S., Blekhman R. Population Dynamics of the Human Gut Microbiome: Change Is the Only Constant. Genome Biol. 2019;20:150. doi: 10.1186/s13059-019-1775-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinson J.N.V., Walk S.T. Escherichia coli Residency in the Gut of Healthy Human Adults. EcoSal Plus. 2020;9:10–1128. doi: 10.1128/ecosalplus.esp-0003-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomas J., Reygner J., Mayeur C., Ducroc R., Bouet S., Bridonneau C., Cavin J.B., Thomas M., Langella P., Cherbuy C. Early Colonizing Escherichia coli Elicits Remodeling of Rat Colonic Epithelium Shifting toward a New Homeostatic State. ISME J. 2015;9:46–58. doi: 10.1038/ismej.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller N.T., Bakacs E., Combellick J., Grigoryan Z., Dominguez-Bello M.G. The Infant Microbiome Development: Mom Matters. Trends Mol. Med. 2015;21:109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang M., Cao Y., Guo Z.F., Chen M., Chen X., Guo Z. Menaquinone Biosynthesis in Escherichia coli: Identification of 2-Succinyl-5-Enolpyruvyl-6-Hydroxy-3-Cyclohexene-1-Carboxylate as a Novel Intermediate and Re-Evaluation of MenD Activity. Biochemistry. 2007;46:10979–10989. doi: 10.1021/bi700810x. [DOI] [PubMed] [Google Scholar]
- 15.Karl J.P., Meydani M., Barnett J.B., Vanegas S.M., Barger K., Fu X., Goldin B., Kane A., Rasmussen H., Vangay P., et al. Fecal Concentrations of Bacterially Derived Vitamin K Forms Are Associated with Gut Microbiota Composition but Not Plasma or Fecal Cytokine Concentrations in Healthy Adults. Am. J. Clin. Nutr. 2017;106:1052–1061. doi: 10.3945/ajcn.117.155424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang H., Kang J., Zhang D. Microbial Production of Vitamin B12: A Review and Future Perspectives. Microb. Cell Fact. 2017;16:15. doi: 10.1186/s12934-017-0631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Degnan P.H., Taga M.E., Goodman A.L. Vitamin B12 as a Modulator of Gut Microbial Ecology. Cell Metab. 2014;20:769–778. doi: 10.1016/j.cmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnúsdóttir S., Ravcheev D., De Crécy-Lagard V., Thiele I. Systematic Genome Assessment of B-Vitamin Biosynthesis Suggests Cooperation among Gut Microbes. Front. Genet. 2015;6:148. doi: 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ducarmon Q.R., Zwittink R.D., Hornung B.V.H., van Schaik W., Young V.B., Kuijper E.J. Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol. Mol. Biol. Rev. 2019;83:e00007-19. doi: 10.1128/MMBR.00007-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudault S., Guignot J., Servin A.L. Escherichia coli Strains Colonising the Gastrointestinal Tract Protect Germfree Mice against Salmonella Typhimurium Infection. Gut. 2001;49:47–55. doi: 10.1136/gut.49.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang D.E., Smalley D.J., Tucker D.L., Leatham M.P., Norris W.E., Stevenson S.J., Anderson A.B., Grissom J.E., Laux D.C., Cohen P.S., et al. Carbon Nutrition of Escherichia coli in the Mouse Intestine. Proc. Natl. Acad. Sci. USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabich A.J., Jones S.A., Chowdhury F.Z., Cernosek A., Anderson A., Smalley D., McHargue J.W., Hightower G.A., Smith J.T., Autieri S.M., et al. Comparison of Carbon Nutrition for Pathogenic and Commensal Escherichia coli Strains in the Mouse Intestine. Infect. Immun. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leatham M.P., Banerjee S., Autieri S.M., Mercado-Lubo R., Conway T., Cohen P.S. Precolonized Human Commensal Escherichia coli Strains Serve as a Barrier to E. coli O157:H7 Growth in the Streptomycin-Treated Mouse Intestine. Infect. Immun. 2009;77:2876–2886. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maltby R., Leatham-Jensen M.P., Gibson T., Cohen P.S., Conway T. Nutritional Basis for Colonization Resistance by Human Commensal Escherichia coli Strains HS and Nissle 1917 against E. coli O157:H7 in the Mouse Intestine. PLoS ONE. 2013;8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Hara A.M., Shanahan F. The Gut Flora as a Forgotten Organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin N.R., Whon T.W., Bae J.W. Proteobacteria: Microbial Signature of Dysbiosis in Gut Microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 28.Rizzatti G., Lopetuso L.R., Gibiino G., Binda C., Gasbarrini A. Proteobacteria: A Common Factor in Human Diseases. BioMed. Res. Int. 2017;2017:9351507. doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter S.E., Bäumler A.J. Why Related Bacterial Species Bloom Simultaneously in the Gut: Principles Underlying the “like Will to like” Concept. Cell. Microbiol. 2014;16:179–184. doi: 10.1111/cmi.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seksik P., Rigottier-Gois L., Gramet G., Sutren M., Pochart P., Marteau P., Jian R., Doré J. Alterations of the Dominant Faecal Bacterial Groups in Patients with Crohn’s Disease of the Colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang T., Cai G., Qiu Y., Fei N., Zhang M., Pang X., Jia W., Cai S., Zhao L. Structural Segregation of Gut Microbiota between Colorectal Cancer Patients and Healthy Volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krogius-Kurikka L., Lyra A., Malinen E., Aarnikunnas J., Tuimala J., Paulin L., Mäkivuokko H., Kajander K., Palva A. Microbial Community Analysis Reveals High Level Phylogenetic Alterations in the Overall Gastrointestinal Microbiota of Diarrhoea-Predominant Irritable Bowel Syndrome Sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fei N., Zhao L. An Opportunistic Pathogen Isolated from the Gut of an Obese Human Causes Obesity in Germfree Mice. ISME J. 2013;7:880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Espey M.G. Role of Oxygen Gradients in Shaping Redox Relationships between the Human Intestine and Its Microbiota. Free Radic. Biol. Med. 2013;55:130–140. doi: 10.1016/j.freeradbiomed.2012.10.554. [DOI] [PubMed] [Google Scholar]
- 35.Albenberg L., Esipova T.V., Judge C.P., Bittinger K., Chen J., Laughlin A., Grunberg S., Baldassano R.N., Lewis J.D., Li H., et al. Correlation between Intraluminal Oxygen Gradient and Radial Partitioning of Intestinal Microbiota. Gastroenterology. 2014;147:1055–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henson M.A., Phalak P. Microbiota Dysbiosis in Inflammatory Bowel Diseases: In Silico Investigation of the Oxygen Hypothesis. BMC Syst. Biol. 2017;11:145. doi: 10.1186/s12918-017-0522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rigottier-Gois L. Dysbiosis in Inflammatory Bowel Diseases: The Oxygen Hypothesis. ISME J. 2013;7:1256–1261. doi: 10.1038/ismej.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes E.R., Winter M.G., Duerkop B.A., Spiga L., Furtado de Carvalho T., Zhu W., Gillis C.C., Büttner L., Smoot M.P., Behrendt C.L., et al. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe. 2017;21:208–219. doi: 10.1016/j.chom.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stecher B. The Roles of Inflammation, Nutrient Availability and the Commensal Microbiota in Enteric Pathogen Infection. Microbiol. Spectr. 2015;3:297–320. doi: 10.1128/microbiolspec.MBP-0008-2014. [DOI] [PubMed] [Google Scholar]
- 40.Morgan X.C., Tickle T.L., Sokol H., Gevers D., Devaney K.L., Ward D.V., Reyes J.A., Shah S.A., LeLeiko N., Snapper S.B., et al. Dysfunction of the Intestinal Microbiome in Inflammatory Bowel Disease and Treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carroll I.M., Ringel-Kulka T., Siddle J.P., Ringel Y. Alterations in Composition and Diversity of the Intestinal Microbiota in Patients with Diarrhea-Predominant Irritable Bowel Syndrome. Neurogastroenterol. Motil. 2012;24:521-e248. doi: 10.1111/j.1365-2982.2012.01891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsen N., Vogensen F.K., Van Den Berg F.W.J., Nielsen D.S., Andreasen A.S., Pedersen B.K., Al-Soud W.A., Sørensen S.J., Hansen L.H., Jakobsen M. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu L., Baker S.S., Gill C., Liu W., Alkhouri R., Baker R.D., Gill S.R. Characterization of Gut Microbiomes in Nonalcoholic Steatohepatitis (NASH) Patients: A Connection between Endogenous Alcohol and NASH. Hepatology. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 44.Litvak Y., Byndloss M.X., Tsolis R.M., Bäumler A.J. Dysbiotic Proteobacteria Expansion: A Microbial Signature of Epithelial Dysfunction. Curr. Opin. Microbiol. 2017;39:1–6. doi: 10.1016/j.mib.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 45.Litvak Y., Byndloss M.X., Bäumler A.J. Colonocyte Metabolism Shapes the Gut Microbiota. Science. 2018;362:eaat9076. doi: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andoh A., Nishida A. Alteration of the Gut Microbiome in Inflammatory Bowel Disease. Digestion. 2023;104:16–23. doi: 10.1159/000525925. [DOI] [PubMed] [Google Scholar]
- 47.Winter S.E., Thiennimitr P., Winter M.G., Butler B.P., Huseby D.L., Crawford R.W., Russell J.M., Bevins C.L., Adams L.G., Tsolis R.M., et al. Gut Inflammation Provides a Respiratory Electron Acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winter S.E., Winter M.G., Xavier M.N., Thiennimitr P., Poon V., Keestra A.M., Laughlin R.C., Gomez G., Wu J., Lawhon S.D., et al. Host-Derived Nitrate Boosts Growth of E. coli in the Inflamed Gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winter S.E., Lopez C.A., Bäumler A.J. The Dynamics of Gut-Associated Microbial Communities during Inflammation. EMBO Rep. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradley P.H., Pollard K.S. Proteobacteria Explain Significant Functional Variability in the Human Gut Microbiome. Microbiome. 2017;5:36. doi: 10.1186/s40168-017-0244-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiennimitr P., Winter S.E., Winter M.G., Xavier M.N., Tolstikov V., Huseby D.L., Sterzenbach T., Tsolis R.M., Roth J.R., Bäumler A.J. Intestinal Inflammation Allows Salmonella to Use Ethanolamine to Compete with the Microbiota. Proc. Natl. Acad. Sci. USA. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rowley C.A., Sauder A.B., Kendall M.M. The Ethanolamine-Sensing Transcription Factor EutR Promotes Virulence and Transmission during Citrobacter Rodentium Intestinal Infection. Infect. Immun. 2020;88 doi: 10.1128/IAI.00137-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gillis C.C., Hughes E.R., Spiga L., Winter M.G., Zhu W., Furtado de Carvalho T., Chanin R.B., Behrendt C.L., Hooper L.V., Santos R.L., et al. Dysbiosis-Associated Change in Host Metabolism Generates Lactate to Support Salmonella Growth. Cell Host Microbe. 2018;23:54–64.e6. doi: 10.1016/j.chom.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faber F., Tran L., Byndloss M.X., Lopez C.A., Velazquez E.M., Kerrinnes T., Nuccio S.P., Wangdi T., Fiehn O., Tsolis R.M., et al. Host-Mediated Sugar Oxidation Promotes Post-Antibiotic Pathogen Expansion. Nature. 2016;534:697–699. doi: 10.1038/nature18597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Faber F., Thiennimitr P., Spiga L., Byndloss M.X., Litvak Y., Lawhon S., Andrews-Polymenis H.L., Winter S.E., Bäumler A.J. Respiration of Microbiota-Derived 1,2-Propanediol Drives Salmonella Expansion during Colitis. PLoS Pathog. 2017;13:e1006129. doi: 10.1371/journal.ppat.1006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spiga L., Winter M.G., Furtado de Carvalho T., Zhu W., Hughes E.R., Gillis C.C., Behrendt C.L., Kim J., Chessa D., Andrews-Polymenis H.L., et al. An Oxidative Central Metabolism Enables Salmonella to Utilize Microbiota-Derived Succinate. Cell Host Microbe. 2017;22:291–301.e6. doi: 10.1016/j.chom.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitamoto S., Alteri C.J., Rodrigues M., Nagao-Kitamoto H., Sugihara K., Himpsl S.D., Bazzi M., Miyoshi M., Nishioka T., Hayashi A., et al. Dietary L-Serine Confers a Competitive Fitness Advantage to Enterobacteriaceae in the Inflamed Gut. Nat. Microbiol. 2020;5:116–125. doi: 10.1038/s41564-019-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu W., Miyata N., Winter M.G., Arenales A., Hughes E.R., Spiga L., Kim J., Sifuentes-Dominguez L., Starokadomskyy P., Gopal P., et al. Editing of the Gut Microbiota Reduces Carcinogenesis in Mouse Models of Colitis-Associated Colorectal Cancer. J. Exp. Med. 2019;216:2378–2393. doi: 10.1084/jem.20181939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matamouros S., Hayden H.S., Hager K.R., Brittnacher M.J., Lachance K., Weiss E.J., Pope C.E., Imhaus A.F., McNally C.P., Borenstein E., et al. Adaptation of Commensal Proliferating Escherichia coli to the Intestinal Tract of Young Children with Cystic Fibrosis. Proc. Natl. Acad. Sci. USA. 2018;115:1605–1610. doi: 10.1073/pnas.1714373115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lemons J.M.S., Conrad M., Tanes C., Chen J., Friedman E.S., Roggiani M., Curry D., Chau L., Hecht A.L., Harling L., et al. Enterobacteriaceae Growth Promotion by Intestinal Acylcarnitines, a Biomarker of Dysbiosis in Inflammatory Bowel Disease. Cell. Mol. Gastroenterol. Hepatol. 2024;17:131–148. doi: 10.1016/j.jcmgh.2023.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wouthuyzen-Bakker M., Bodewes F.A.J.A., Verkade H.J. Persistent Fat Malabsorption in Cystic Fibrosis; Lessons from Patients and Mice. J. Cyst. Fibros. 2011;10:150–158. doi: 10.1016/j.jcf.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 62.Caley L.R., White H., de Goffau M.C., Floto R.A., Parkhill J., Marsland B., Peckham D.G. Cystic Fibrosis-Related Gut Dysbiosis: A Systematic Review. Dig. Dis. Sci. 2023;68:1797–1814. doi: 10.1007/s10620-022-07812-1. [DOI] [PubMed] [Google Scholar]
- 63.Hoffman L.R., Pope C.E., Hayden H.S., Heltshe S., Levy R., McNamara S., Jacobs M.A., Rohmer L., Radey M., Ramsey B.W., et al. Escherichia coli Dysbiosis Correlates with Gastrointestinal Dysfunction in Children with Cystic Fibrosis. Clin. Infect. Dis. 2014;58:396–399. doi: 10.1093/cid/cit715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garsin D.A. Ethanolamine Utilization in Bacterial Pathogens: Roles and Regulation. Nat. Rev. Microbiol. 2010;8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang Y.L., Chassard C., Hausmann M., Von Itzstein M., Hennet T. Sialic Acid Catabolism Drives Intestinal Inflammation and Microbial Dysbiosis in Mice. Nat. Commun. 2015;6:8141. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ng K.M., Ferreyra J.A., Higginbottom S.K., Lynch J.B., Kashyap P.C., Gopinath S., Naidu N., Choudhury B., Weimer B.C., Monack D.M., et al. Microbiota-Liberated Host Sugars Facilitate Post-Antibiotic Expansion of Enteric Pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tytgat H.L.P., Nobrega F.L., van der Oost J., de Vos W.M. Bowel Biofilms: Tipping Points between a Healthy and Compromised Gut? Trends Microbiol. 2019;27:17–25. doi: 10.1016/j.tim.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 68.Ju T., Bourrie B.C.T., Forgie A.J., Pepin D.M., Tollenaar S., Sergi C.M., Willing B.P. The Gut Commensal Escherichia coli Aggravates High-Fat-Diet-Induced Obesity and Insulin Resistance in Mice. Appl. Environ. Microbiol. 2023;89:e01628-22. doi: 10.1128/aem.01628-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoo W., Zieba J.K., Foegeding N.J., Torres T.P., Shelton C.D., Shealy N.G., Byndloss A.J., Cevallos S.A., Gertz E., Tiffany C.R., et al. High-Fat Diet-Induced Colonocyte Dysfunction Escalates Microbiota-Derived Trimethylamine N-Oxide. Science. 2021;373:813–818. doi: 10.1126/science.aba3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chervy M., Barnich N., Denizot J. Adherent-Invasive E. coli: Update on the Lifestyle of a Troublemaker in Crohn’s Disease. Int. J. Mol. Sci. 2020;21:3734. doi: 10.3390/ijms21103734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pizer L.I., Potochny M.L. Nutritional and Regulatory Aspects of Serine Metabolism in Escherichia. J. Bacteriol. 1964;88:611–619. doi: 10.1128/jb.88.3.611-619.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Denizot J., Sivignon A., Barreau F., Darcha C., Chan H.F.C., Stanners C.P., Hofman P., Darfeuille-Michaud A., Barnich N. Adherent-Invasive Escherichia coli Induce Claudin-2 Expression and Barrier Defect in CEABAC10 Mice and Crohn’s Disease Patients. Inflamm. Bowel Dis. 2012;18:294–304. doi: 10.1002/ibd.21787. [DOI] [PubMed] [Google Scholar]
- 73.Guo Z., Cai X., Guo X., Xu Y., Gong J., Li Y., Zhu W. Let-7b Ameliorates Crohn’s Disease-Associated Adherent-Invasive E. coli Induced Intestinal Inflammation via Modulating Toll-Like Receptor 4 Expression in Intestinal Epithelial Cells. Biochem. Pharmacol. 2018;156:196–203. doi: 10.1016/j.bcp.2018.08.029. [DOI] [PubMed] [Google Scholar]
- 74.Glasser A.L., Boudeau J., Barnich N., Perruchot M.H., Colombel J.F., Darfeuille-Michaud A. Adherent Invasive Escherichia coli Strains from Patients with Crohn’s Disease Survive and Replicate within Macrophages without Inducing Host Cell Death. Infect. Immun. 2001;69:5529–5537. doi: 10.1128/IAI.69.9.5529-5537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Small C.L.N., Reid-Yu S.A., McPhee J.B., Coombes B.K. Persistent Infection with Crohn’s Disease-Associated Adherent-Invasive Escherichia coli Leads to Chronic Inflammation and Intestinal Fibrosis. Nat. Commun. 2013;4:1957. doi: 10.1038/ncomms2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ellermann M., Gharaibeh R.Z., Fulbright L., Dogan B., Moore L.N., Broberg C.A., Lopez L.R., Rothemich A.M., Herzog J.W., Rogala A., et al. Yersiniabactin-Producing Adherent/Invasive Escherichia coli Promotes Inflammation-Associated Fibrosis in Gnotobiotic Il10−/− Mice. Infect. Immun. 2019;87 doi: 10.1128/IAI.00587-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gilliland A., Chan J.J., De Wolfe T.J., Yang H., Vallance B.A. Pathobionts in Inflammatory Bowel Disease: Origins, Underlying Mechanisms, and Implications for Clinical Care. Gastroenterology. 2023;166:44–58. doi: 10.1053/j.gastro.2023.09.019. [DOI] [PubMed] [Google Scholar]
- 78.Hamjane N., Mechita M.B., Nourouti N.G., Barakat A. Gut Microbiota Dysbiosis–Associated Obesity and Its Involvement in Cardiovascular Diseases and Type 2 Diabetes. A Systematic Review. Microvasc. Res. 2024;151:104601. doi: 10.1016/j.mvr.2023.104601. [DOI] [PubMed] [Google Scholar]
- 79.Keskitalo A., Munukka E., Toivonen R., Hollmén M., Kainulainen H., Huovinen P., Jalkanen S., Pekkala S. Enterobacter Cloacae Administration Induces Hepatic Damage and Subcutaneous Fat Accumulation in High-Fat Diet Fed Mice. PLoS ONE. 2018;13:e0198262. doi: 10.1371/journal.pone.0198262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scheithauer T.P.M., Herrema H., Yu H., Bakker G.J., Winkelmeijer M., Soukhatcheva G., Dai D., Ma C., Havik S.R., Balvers M., et al. Gut-Derived Bacterial Flagellin Induces Beta-Cell Inflammation and Dysfunction. Gut Microbes. 2022;14:2111951. doi: 10.1080/19490976.2022.2111951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fei N., Bruneau A., Zhang X., Wang R., Wang J., Rabot S., Gérard P., Zhao L. Endotoxin Producers Overgrowing in Human Gut Microbiota as the Causative Agents for Nonalcoholic Fatty Liver Disease. MBio. 2020;11 doi: 10.1128/mBio.03263-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.