Abstract
Differences/Disorders of sex development (DSDs) are conditions in which the development of chromosomal, gonadal, and anatomical sexes is atypical. DSDs are relatively rare, but their incidence is becoming alarmingly common in sub-Saharan Africa (SSA). Their etiologies and mechanisms are poorly understood. Therefore, we have investigated cytogenetic profiles, including telomere dysfunction, in a retrospective cohort of Senegalese DSD patients. Materials and methods: Peripheral blood lymphocytes were sampled from 35 DSD patients (mean age: 3.3 years; range 0–18 years) admitted to two hospital centers in Dakar. Peripheral blood lymphocytes from 150 healthy donors were used as a control. Conventional cytogenetics, telomere, and centromere staining followed by multiplex FISH, as well as FISH with SRY-specific probes, were employed. Results: Cytogenetic analysis identified 19 male and 13 female patients with apparently normal karyotypes, two patients with Turner syndrome, and one patient with Klinefelter syndrome. Additional structural chromosome aberrations were detected in 22% of the patients (8/35). Telomere analysis revealed a reduction in mean telomere lengths of DSD patients compared to those of healthy donors of similar age. This reduction in telomere length was associated with an increased rate of telomere aberrations (telomere loss and the formation of telomere doublets) and the presence of additional chromosomal aberrations. Conclusions: To the best of our knowledge, this study is the first to demonstrate a correlation between telomere dysfunction and DSDs. Further studies may reveal the link between telomere dysfunction and possible mechanisms involved in the disease itself, such as DNA repair deficiency or specific gene mutations. The present study demonstrates the relevance of implementing telomere analysis in prenatal tests as well as in diagnosed genetic DSD disorders.
Keywords: DSDs, chromosomal aberrations, telomere shortening, telomere dysfunctions
1. Introduction
Differences/Disorders of sex development (DSDs) are congenital anomalies characterized by atypical chromosomal, gonadal, and anatomical sex development resulting in ambiguous external and internal genitalia and hormonal dysfunction [1,2]. The term “disorders of sexual development” is currently used to replace terms such as “sexual ambiguity”, “intersex”, “hermaphroditism”, or “pseudo hermaphroditism”. This new terminology was related essentially not only to the potential pejorative of the old [3] but also associated with the new classification during the Pediatric Endocrine Society and European Society for Pediatric Endocrinology (LWEPS-ESPE) conference [1,4]. This classification is based on the chromosomal analysis and clinical features [5,6]. Nevertheless, DSD classification remains very difficult because similar phenotypes can have multiple etiologies [4,7]. Currently, the management of patients is multidisciplinary, involving imaging, genetics, and hormonology [8].
DSDs are rare genetic disorders with incidences varying between 1/4500 and 1/5500 live births worldwide [9,10,11,12]. DSD 46,XX is the most represented variant [13] (1/14,000 to 1/15,000 vs. 1/20,000 for DSD 46,XY) [14,15,16]. However, DSD incidence is becoming alarmingly common in Africa in general (1/3000 in Egypt) and in sub-Saharan Africa (SSA) (1/357 in Ghana), in particular [12,17]. This increased incidence could be related not only to a highly endogenous and inbred population, to the efficacy of prenatal diagnosis in these countries but also to environmental factors. The lack of epidemiological studies and specific structures, as well as cultural barriers, make the treatment and follow-up of these diseases in Africa very difficult.
Unfortunately, major challenges with the diagnosis and management of DSD patients persist in this part of the world. In addition, the etiologies and specific biomarkers related to DSDs remain poorly understood.
DSD patients exhibit a very high risk of gonadal cancers [18], hypogonadism [19,20], lung and breast cancers [21,22], as well as various fertility complications and hormonal insufficiency [23]. Chromosomal instability, a driving force of the progression of malignancy, has been previously described in DSD patients [24]. Previous studies have shown that cells derived from patients with trisomy 13 (Patau syndrome), trisomy 18 (Edwards syndrome), trisomy 21 (Down syndrome), or monosomy X (Turner syndrome) exhibit a significantly higher frequency of sporadically acquired non-specific whole chromosome losses and gains compared to control cases [25,26]. It has also been reported that patients with DSDs have chromosomal aberrations that are often related to the Y chromosome [27].
Telomeres are nucleoprotein complexes located at the ends of eukaryotic chromosomes, and they have a critical role in preserving chromosomal integrity and stability [28,29]. Telomere length is used as a biomarker of biological age [30] and an aging-disease risk factor [31,32,33]. Telomere dysfunction is related to chromosomal instability, either through progressive telomere shortening or telomere aggregation and telomere loss and deletion [34,35]. The loss of telomere functionalities is considered the one major mechanism for the progression of genomic instability [36]. Significant shortening of telomere length and significantly higher frequencies of telomere loss and deletion have been found in peripheral lymphocytes of patients with cancer and genetic diseases compared to healthy donors of the same age [37]. Chromosomal instability is also associated with telomere shortening and loss of telomere functionality that ultimately leads to end-to-end chromosomal fusions, thus contributing to the initiation and progression of cancer [38,39,40].
In this study, cytogenetic analysis has been conducted to evaluate not only structural and numerical chromosomal aberrations but also telomere profiles of DSD patients from SSA. We demonstrate for the first time that telomere instability is a common characteristic of SSA DSD patients. Telomere instability could indeed be playing a role in the formation of additional chromosomal aberrations. To our knowledge, this is the first study to address telomere profiles in a cohort of DSD patients. We hypothesize that chromosomal aberrations in these patients are related to telomere dysfunction.
2. Materials and Methods
2.1. Declaration of Ethics
This study and research protocol were approved by the ethics committee of the Cheikh Anta Diop University of Dakar (Protocol 041512019/CER/UCAD). Patients and their guardians or family members were included in the study only after receiving detailed information about the study and signing an informed consent form. Data were collected and processed in a confidential manner.
2.2. Pediatric Patients
We conducted a retrospective study of 35 pediatric patients with DSD admitted to clinical consultation from November to December 2021 at the Diamniadio Children’s Hospital and the Albert Royer Pediatric Surgery Department of FANN. These are two large hospitals in Dakar that cover the entire Dakar region and surrounding areas. The patients were initially received for clinical examination in the two above-mentioned hospitals and referred to the National Centre of Blood Transfusion (NCBT) in Dakar. Sampling and cytogenetic analyses were performed at the NCBT. All patients had disorders of sexual development ranging from mild hypospadias pubertal delay to overt external genitalia ambiguity.
2.3. Culture of Lymphocytes, Preparation of Metaphases, and Analysis of Karyotypes after G-Banding (GTG-Banding)
Peripheral blood lymphocytes were cultured for 72 h, and standard GTG-Banding was performed according to a previous publication [41]. A total of 50 metaphases from each sample were analyzed at the resolution level of 550 bands. Karyotypes are presented according to ISCN2020 [42].
2.4. Detection of SRY by Fluorescence In Situ Hybridization (FISH)
Fluorescence in situ hybridization (FISH) was performed on interphase cells harvested from freshly collected whole blood and on cultured PHA-stimulated cells. The slide was washed in 2x standard citrate saline (2x SSC) at 37 °C for 30 min. The slides were dehydrated in three alcohol gradients, 70%, 90%, and 100%, for 2 min each and then air dried. In total, 10 μL of the probe was deposited on the slides and then covered with a coverslip. The slides with coverslips were placed on a thermobrite (ThermoFisher, Illkirch, France) for denaturation at 76 °C for 7 min and then hybridized at 37 °C for 24 h. After 24 h, the slides were washed in 1xPBS solution to loosen the coverslips, then immersed in a solution of 0.4x SSC with 0.3% NP-40 for 2 min at 73 °C and in a solution of 2x SSC with 0.1% NP-40 at room temperature for 2 min. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) solution and then rinsed in PBS before mounting the slides with vectashield orp-phenylenediamine (PPD). Two specific probes for the SRY gene were used: the Vysis AneuVysion probe (Vysis LSI SRY/CEP X FISH Probe Kit, Abbott, Des Plaines, IL 60018, USA) and the Cytocell probe (SRY Probe, Cytocell Aquarius, Symex, Bremerhaven, Germany). The Vysis probe consists of two parts: The Xp11.1-q11.1 CEP X (DXZ1) Spectrum Green part specific for the X chromosome and the Yp11.3 LSI SRY Spectrum Orange part specific for the SRY gene. The Cytocell probe is a mixture of three probes: One probe (SRY), Yp11.31, in red (Texas Red), a control probe for the Y chromosome (DYZ1), Yq12 (heterochromatic block) in green (Green) and a control probe for the X centromere (DXZ1), Xp11.1-q11.1 in blue (Aqua). Images were captured using an automated acquisition module Autocapt software (MetaSystems, version 3.9.1) using an automated ZEISS Plan-Apochromat 63×/1.40 oil and CoolCube 4 Digital High-Resolution CCD camera. The analysis was performed on 200 cells for each sample [25].
2.5. Staining of Telomere and Centromere Sequences
Telomere and centromere staining were performed using a Cy-3-labelled PNA probe specific for telomere sequences and a FITC-labelled PNA probe specific for centromere sequences (Cell Environment, Evry, France), as previously described [37].
2.6. Telomeres Length Analysis
Telomere quantification was performed on interphase cells using TeloScore software (Cell Environment, version 1.1.2, Evry, France). Quantitative image acquisition was performed using MetaCyte software (MetaSystem, version 3.9.1, Altlussheim, Germany) and a ZEISS Plan-Apochromat (Zeiss, Oberkochen, Germany) and CoolCube 1 Digital High-Resolution CCD Camera (MetaSystems, Altlussheim, Germany). The exposure and gain settings remained constant between captures. The mean fluorescence intensity (FI) of telomeres was automatically quantified in 10,000 nuclei on each slide. The quantifications were performed on triplicate slides. Telomere length, measured as the mean FI, correlates strongly with telomere length measured by conventional Southern blot analysis using the telomeric restriction fragment (TRF) (R2 = 0.721 and p = 2.128 × 10−8). The mean telomere length is expressed in kb.
2.7. Scoring of Telomere Aberrations
Analysis of metaphase spreads allowed the detection of telomere abnormalities using ChromoScore Software (Cell Environment, version 1.1.2, Evry, France). The images of metaphases were captured using the automated acquisition module Autocapt software (MetaSystems, version 3.9.1) and a ZEISS Plan-Apochromat 63×/1.40 oil (Zeiss, Oberkochen, Germany) and CoolCube 1 Digital High-Resolution CCD Camera (MetaSystems, Altlussheim, Germany) with constant settings for exposure and gain.
Telomere abnormalities scored were (i) sister telomere loss, likely occurring in G2, and defined as a telomere signal-free end at a single chromatid [27], (ii) telomere deletion defined as the loss of two telomere signals on the same chromosome arm (likely resulting from the loss of one telomere in G1/S), an aberration considered to represent double-strand breaks, leading to the activation of DNA damage response. Automatic scoring of these aberrations was performed using ChromoScore software (Cell Environment, version1.1.2, Evry, France). An operator validated and excluded the falsely recorded aberrations.
2.8. Multicolor FISH (M-FISH Technique)
The M-FISH technique employs multicolor probes that make it possible to identify each of the 22 pairs of autosomes as well as the X and Y chromosomes by “painting” them with individual colors. Moreover, fragments of chromosomes translocated into non-homologous chromosomes were also identified using M-FISH.
After telomere quantification and the automatic capture of metaphases with telomere and centromere staining, the slides were washed in 2x SSC for 30 min at 70 °C. After rinsing with 0.1x SSC, the slides were denatured using NaOH and subsequently washed with 0.1x SCC and 2x SSC and sequentially dehydrated in 70%, 95%, and 100% ethanol and air-dried. After denaturation of the M-FISH probe (M-FISH 24XCyte, Metasystems, Altlussheim, Germany) for 5 min at 75 °C, the probe was added to the slides and incubated at 37 °C for two days. The slides were subsequently rinsed with 0.4x SSC for 2 min at 72 °C and then with 2x SSC/0.005% (Tween-20). The slides were counterstained with DAPI and mounted in PPD.
2.9. Statistical Analysis
Data were analyzed using the Wilcoxon-Mann–Whitney rank sum test (comparison of two sub-groups) or the Kruskal–Wallis non-parametric test (comparison of three sub-groups). We tested the null hypothesis that the sub-groups are considered identical populations. A p-value < 0.05 is considered statistically significant to reject the null hypothesis.
3. Results
3.1. Clinical Profile of DSD Patients
This study was performed on 35 retrospective DSD pediatric patients admitted to two hospital centers. Inclusion criteria for patients in this study were age less than 18 years and congenital malformation according to clinical examination. The mean age of these patients at diagnosis was 2.62 years (0–14 years) and 3.33 years (0–18 years) at cytogenetic analysis. Twelve of these DSD patients (34.5%) had non-classified pathologies. Clinical features of all patients are listed in Table 1.
Table 1.
Clinical characteristics of Differences/Disorders of sex development (DSDs) patients before cytogenetic investigations.
| Characteristics | No. of Patients |
|---|---|
| DSDs | 35 |
| Assigned sex | |
| Male | 17 |
| Female | 13 |
| ND | 5 |
| Age (years) | |
| At diagnosis | 2.6 |
| At analysis | 3.5 |
| Type | |
| Clitoral hypertrophy | 8 |
| Micropenis + cryptorchidism | 5 |
| Isolated hypospadias | 4 |
| Isolated cryptorchidism | 2 |
| Isolated micropenis | 1 |
| Hypogonadism + gynecomastia | 1 |
| Short stature + Turner syndrome | 1 |
| Other DSDs with diverse congenital malformations | 12 |
ND: The sex of the patient could not be determined by clinical examination.
3.2. Conventional and Molecular Cytogenetic Investigations
After PHA stimulation of freshly isolated circulating lymphocytes and of cells in culture, conventional and molecular cytogenetics were performed to retrieve chromosomal abnormalities. Table 2 summarizes the results of G-Banding and SRY-specific FISH.
Table 2.
Cytogenetic profiles of DSDs patients.
| Cytogenetic Profile | No. of Patients |
|---|---|
| Nb of analyzed DSDs patients | 35 |
| Conventional cytogenetics | 35 |
| Molecular cytogenetic (SRY) | 35 |
| Karyotype results | |
| 46,XY | 18 |
| 46,XX | 14 |
| 46,XX[12]/45,X[5] | 1 |
| 45,X[28]/46,XY[20] | 1 |
| nuc ish(DXZ1x2,SRYx1)[85/200] | 1 |
| Structural chromosome aberrations | 8 |
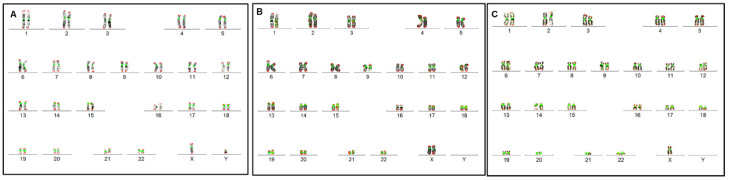
Using conventional cytogenetics, 20 patients had a male karyotype profile (46,XY) without apparent chromosomal abnormalities (Figure 1A). Thirteen patients had a female karyotype (46,XX) without apparent chromosomal abnormalities (Figure 1B). Two patients, YH008 and YH015 (Table 2) (Supplementary Table S1), had mosaic Turner syndrome 46,XX[12]/45,X[5] and 45,X[28]/46,XY[20], respectively (Figure 1C).
Figure 1.
G-banded karyotypes supplemented with centromere and telomere-specific FISH. (A) Normal male karyotype (46,XY) from a DSD patient with ambiguous genitalia but no apparent chromosomal abnormalities. (B) Normal female karyotype (46,XX) from DSDs patients with ambiguous genitalia without visible chromosomal abnormalities. (C) Karyotype of a DSD patient with ambiguous genitalia and a monosomy X (45,X, Turner syndrome).
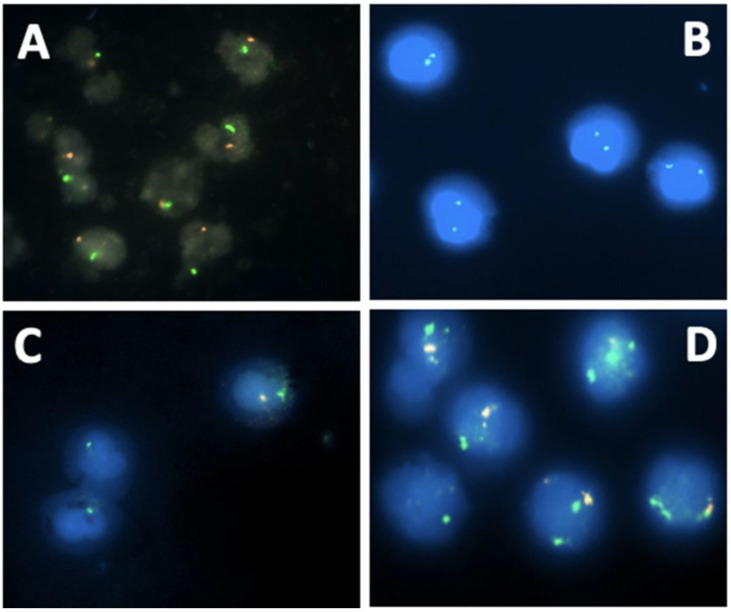
Molecular analysis using co-hybridization with FISH probes specific for SRY and X centromere sequences, respectively, was used to validate the conventional cytogenetic data (Figure 2). A male profile with corresponding signals was found in 19 patients (Figure 2A). The female profile with corresponding signals was found in 13 patients (Figure 2B). We have also confirmed the Turner syndrome profile for two patients (YH008 and YH015) detected by conventional cytogenetics (Figure 2C). In addition, Klinefelter syndrome was also found in patient YH009 (Figure 2D). The latter anomaly had not been identified using conventional cytogenetics.
Figure 2.
DSDs profiles using molecular cytogenetics with specific probes for SRY (red signal) and X centromere (green signal) (A) Normal FISH signals of a male DSD patient (one green signal and one red signal); (B) Normal FISH signals of female DSD patient (two green signals); (C) Mosaic male Turner syndrome. Two cell populations: one with a red signal (appears whitish) and a green signal, and the other with only a green signal due to the loss of the Y chromosome; (D) Mosaic Klinefelter syndrome. Two cell populations: one with one green signal and one red signal (normal cells) and the second cell population with two green signals and one red signal.
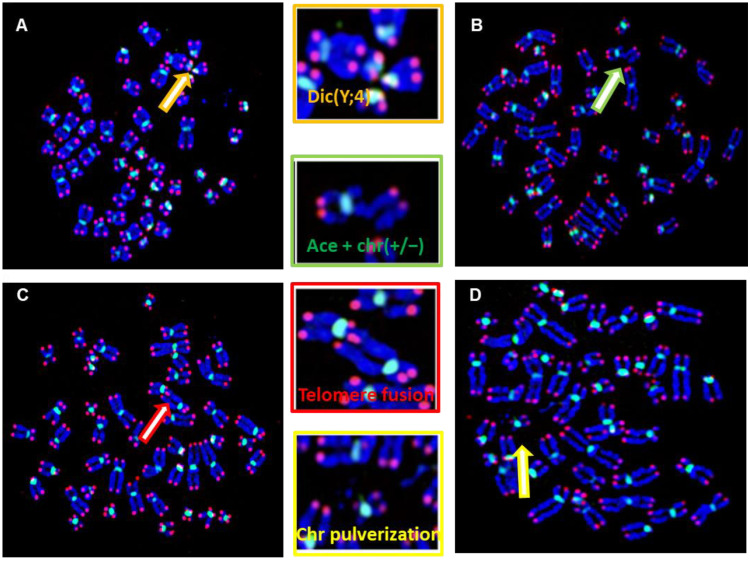
Using telomere and centromere-specific probes, several additional structural chromosome aberrations were identified in metaphases from eight of the patients (22%), such as a dicentric chromosome (Figure 3A) and acentric chromosomes with chromosome deletions (2 patients) (Figure 3B). Telomere fusions (3 patients) (Figure 3C) were observed in addition to chromosome fragmentations (2 patients) (Figure 3D), as well as chromosomal breaks (3 patients).
Figure 3.
Structural chromosome aberrations in DSD patients were identified by telomere (red signal) and centromere (green signal) staining. (A) Metaphase showing a dicentric chromosome, dic(Y;4) between chr 7 with interstitial telomeres, demonstrating that telomere fusion is the origin of the formation of aberration. (B) Double Strand breaks (DSB) resulting in the formation of an acentric chromosome and chromosome deletion (ace(+/−) with chr(+/−)). These aberrations have been observed in 4 patients (YH014, YH020, YH032, YH033). (C) Telomere fusion between two different chromosomes (D) DNA pulverization observed in two patients (YH016 and YH017). Metaphase of a DSD with DNA fragmentation observed in patients YH004 and YH033. (C) Chromatid break observed in several DSDs involving different chromosomes. (D) Metaphase showing a fusion of two chromosomes from the chromatids of each chromosome with interstitial telomeres, observed in DSDs (YH016, YH017).
3.3. Quantification of Telomere Length of DSD Patients
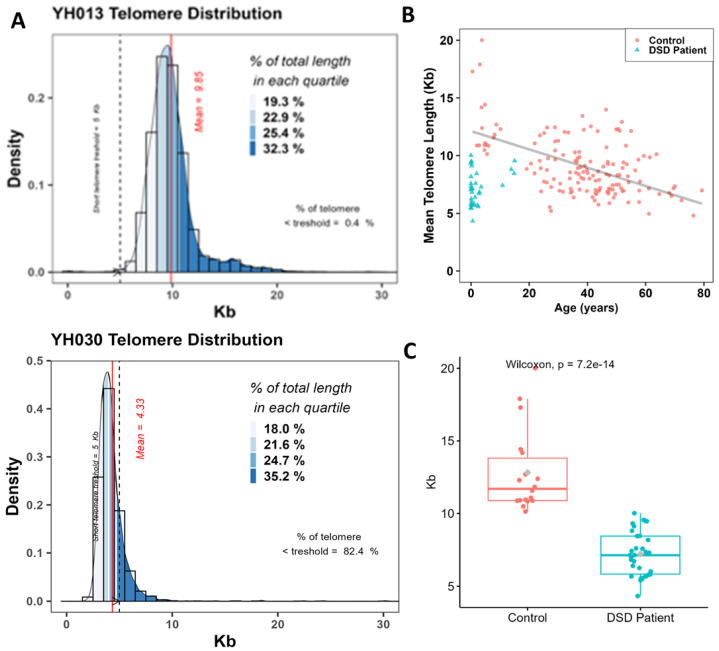
To understand the origin and mechanisms underlying the formation of these additional aberrations, we assessed the telomere lengths of circulating lymphocytes of the DSD patients using an automated approach based on cytogenetic preparations and FISH (Aging kit Cell Environment). This approach permits not only the assessment of mean telomere length but also the intercellular variation and proportion of cells with extreme telomere shortening (<5 kb) in vast numbers of interphase nuclei (Figure 4A). A large cohort of 150 healthy donors (0.5–79 years of age) served as controls in this study. Telomere length in healthy donors was age-dependent and characterized by high inter-individual variation (R2 = 0.316 and p = 2.48 × 10−10) (Figure 4B). The spontaneous decrease in telomere length in healthy donors was 79 bp per year. Interestingly, there was a significant difference (p < 10−6) between the mean telomere length of DSD patients and that of healthy donors of similar age, being 6.99 kb (4.33–9.85 kb) for DSD patients and 10.5 kb (5.2–13.9 kb) for healthy donors, respectively (Figure 4C).
Figure 4.
Quantification of telomere length in circulating lymphocytes of DSDs patients. (A) Distribution of the telomere lengths in circulating lymphocytes of patients YH013 and YH030 that exhibit the most extreme telomere lengths in our DSDs cohort. The mean telomere length is presented by the red line and the frequency of cells with extreme telomere shortening (<5 kb) is presented as a dashed blue line. The different quartiles of fluorescence signal intensities in telomeres are also shown. (B) Telomere length (kb) as a function of age in lymphocytes from healthy donors (150 donors, mean age 36 years, range 0.5–79 years) (red circles) and in DSDs patients (blue triangle). (C) A significant difference between the means of telomere length of DSD patients and those from healthy donors with similar ages.
3.4. Telomere Dysfunction of DSDs Patients
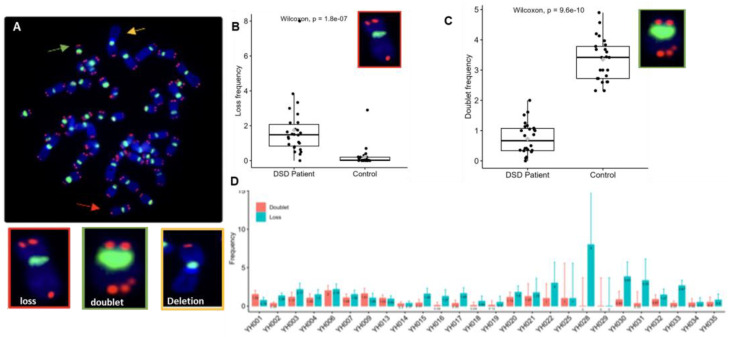
Telomere dysfunction relates to any telomere structural aberration that effectively abolishes the presence of a functional telomere, resulting in chromosome end-to-end fusion, dicentric chromosome formation, and ongoing chromosomal instability. In metaphases of DSDs patients, telomere loss (Figure 5A) was significantly more extensive than in those of healthy donors of similar age 1.42 (range 0–8) per cell vs. 0.52 (range 0–3.23) per cell, (p < 10−7), respectively (Figure 5B). In contrast, telomere doublet formation was significantly lower in DSD patients (0.8, range 0–1.7 per cell) than in healthy donors (5.51, range 1.29–12.17 per cell) (p < 10−10), respectively (Figure 5C). In addition, high inter-individual variation in the frequencies of telomere losses and doublets was recorded in DSD patients (Figure 5D).
Figure 5.
Telomere aberrations are identified by assessing telomeres on individual chromosomes in metaphases of circulating lymphocytes. (A) Representative metaphase after telomere (red) and centromere (green) staining showing different types of telomere aberrations: telomere loss (red arrow), telomere doublet (green arrow), and telomere deletion (yellow arrow). (B,C) Frequencies of telomere losses and telomere doublet formations per cell in DSDs patients and in the control panel with similar age. A significant increase in the frequency of telomere loss was observed in DSDs patients in comparison to the control group (p < 1.8 × 10−7). A significant decrease in telomere doublet formation in DSDs patients was observed compared to that in the control (p < 9.6 × 10−10). (D) The frequency of telomere loss and telomere doublets in each patient shows high inter-individual variation. One hundred metaphases were analyzed per patient.
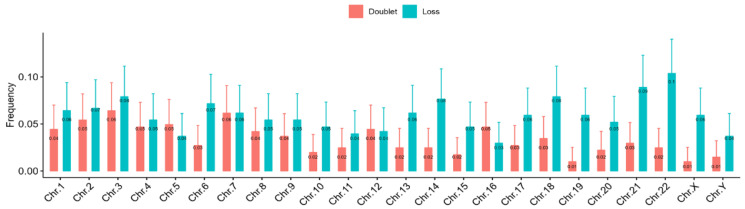
A closer inspection of the data on telomere aberrations in individual chromosomes revealed that many chromosomes, especially chromosomes 21 and 22, were more frequently affected than others (Figure 6).
Figure 6.
The pooled frequencies of telomere aberrations (loss and doublet) per chromosome of all the DSDs patients reveal that the most abundant aberrations are in chromosomes 3, 14, 21, and 22.
Furthermore, we quantitated telomere length and the frequency of telomere losses and telomere doublet formations in DSDs patients with or without additional chromosome aberrations to assess a putative correlation between telomere dysfunction and the formation of chromosomal aberrations. Telomere loss in patient cells with additional chromosome aberrations (1.60 telomere loss/cell) was not significantly higher than in those with normal karyotypes (1.32 telomere loss/cell). Of note, dicentric chromosomes, a driving force of chromosomal instability, were identified in two patients with structural chromosome aberrations. Interstitial telomeres were observed in dicentric chromosomes, demonstrating the role of telomere dysfunction in their formations.
In addition, we observed a high frequency of micronuclei in these DSDs, which is relevant given that micronuclei can originate from those with additional chromosomal aberrations and chromosomal pulverizations. The chromosomal and telomere profiles of each patient are listed in Table 3, in addition to clinical features.
Table 3.
Clinical and cytogenetic characteristics of the used DSDs cohort.
| ID | Assigned SEXE | Age Month | Reason for Consultation |
FISH (SRY) | Karyotypes | Telomere Length (kb) | Telomere Loss/Cell | Telomere Doublet/Cell | Additional Chromosome Aberrations |
|---|---|---|---|---|---|---|---|---|---|
| YH001 | M | 0 | Micropenis and cryptorchidism | XY (SRY+) | 46,XY | 9.31 | 0.78 | 1.52 | |
| YH002 | M | 178 | Micropenis and cryptorchidism) | XY (SRY+) | 46,XY | 8.5 | 1.35 | 0.35 | |
| YH003 | F | 3 | Prader Type I | XX (SRY−) | 46,XX | 9.15 | 2.17 | 1.17 | |
| YH004 | M | 0 | ND | XY (SRY+) | 46,XY | 7.66 | 1.5 | 1.05 | DNA fragmentation Dicentric with interstitial telomeres |
| YH005 | M | 4 | Clitoral hypertrophy | XX (SRY−) | 46,XX | 6.32 | NA | NA | |
| YH006 | M | 33 | ND | XY (SRY+) | 46,XY | 7.57 | 2.19 | 2 | |
| YH007 | M | 19 | Hypospadias | XY (SRY+) | 46,XY | 8.09 | 1.52 | 1.08 | |
| YH008 | M | 120 | Micropenis and cryptorchidism | 28% XY/57% X (SRY+) | 45,X[28]/46,XY[20] | 6.09 | NA | NA | |
| YH009 | M | 7 | Hypospadias | ish(DXZ1x2,SRYx1)[85/200] (SRY+) | 46,XY[115]/47,XXY[85] | 8.21 | 1.06 | 1.71 | |
| YH010 | F | 42 | Clitoral hypertrophy | XX (SRY−) | 46,XX | 7.29 | 0 | 0 | |
| YH011 | M | 1 | ND | XY (SRY+) | 46,XY | 5.42 | 0 | 0 | |
| YH012 | M | 166 | Hypogonadism and gynecomastia | XY (SRY+) | 46,XY | 8.12 | 0 | 0 | |
| YH013 | F | 3 | Clitoral hypertrophy | XY (SRY+) | 46,XY | 9.85 | 0.94 | 1.03 | |
| YH014 | F | 0 | Clitoral hypertrophy | XX (SRY−) | 46,XX | 9.47 | 0.37 | 0.3 | Chromosomal breakage 46,XX,del(15)(q10q26) |
| YH015 | F | 180 | Statural retardation and Turner syndrome | 71% XX/29% X (SRY−) | 46,XX[12]/45,X[5] | 9.55 | 1.59 | 0.42 | |
| YH016 | ND | ND | XX (SRY−) | 46,XX | 5.56 | 1.28 | 0.09 | chromatid fusion of ch 8 and 10 | |
| YH017 | M | 13 | ND | XY (SRY+) | 46,XY | 5.83 | 1.65 | 0.35 | chromatid fusion of ch 15 and 4 chromatid fusion of ch 15 and 9 |
| YH018 | ND | 0 | ND | XY (SRY+) | 46,XY | 6.71 | 0.6363636364 | 0.09090909091 | |
| YH019 | ND | 0 | ND | XX (SRY−) | 46,XX | 8.18 | 0.5 | 0.125 | |
| YH020 | M | 130 | Hypospadias | XY (SRY+) | 46,XY | 7.42 | 1.8 | 1.13 | Chromosome 2 breakage |
| YH021 | ND | 0 | ND | XY (SRY+) | 46,XY | 5.5 | 1.75 | 1.25 | |
| YH022 | M | 27 | Micropenis and cryptorchidism | XY (SRY+) | 46,XY | 5.7 | 3 | 1 | |
| YH023 | F | 23 | Clitoral hypertrophy | XX (SRY−) | 46,XX | 6.34 | NA | NA | |
| YH024 | M | 11 | Micropenis and cryptorchidism | XY (SRY+) | 46,XY | 5.67 | NA | NA | |
| YH025 | F | 17 | external Genitalia Anomaly | XX (SRY−) | 46,XX | 6.39 | 1 | 1 | |
| YH026 | F | 0 | Clitoral hypertrophy | XX (SRY−) | 46,XX | 7.02 | NA | NA | |
| YH027 | F | 0 | XX (SRY−) | 46,XX | 5.8 | NA | NA | ||
| YH028 | ND | 0 | Polymalformation | XY (SRY+) | 46,XY | 6.42 | 8 | 0 | |
| YH029 | M | 1 | ND | XY (SRY+) | 46,XY | 6.25 | NA | NA | |
| YH030 | M | 7 | Ovotestis | XX (SRY−) | 46,XX | 4.33 | 3.83 | 0.8 | Acentric chromosome |
| YH031 | F | 30 | Prader Type II | XY (SRY+) | 46,XY | 5.57 | 3.333 | 0.33 | |
| YH032 | M | 1 | hypospadias | XY (SRY+) | 46,XY | 7.14 | 1.47 | 0.87 | Chromosome breakage |
| YH033 | F | 35 | ND | XX (SRY−) | 46,XX | 6.84 | 2.67 | 0.374 | DNA Fragmentation |
| YH034 | F | 3 | ND | XX (SRY−) | 46,XX | 6.01 | 0.5 | 0.42 | |
| YH035 | F | 2 | Micro penis | XY (SRY+) | 46,XY | 5.68 | 0.8 | 0.5 |
4. Discussion
Rapid and precise diagnosis of DSDs patients is considered a major public health and societal challenge, particularly in light of the increasing prevalence of DSDs in, e.g., SSA countries in general and in Senegal in particular. Our study is a first step in the design of novel biomarkers for the identification and diagnosis of DSDs patients in order to optimize patient care. Furthermore, it will contribute to our understanding of mechanisms involved in the development of DSDs. Currently, the analysis of karyotypes is the primary and most accurate approach to diagnoseDSDs in clinical settings. In the present study, we have extended the classical cytogenetic analyses by including, for the first time, the analysis of telomeres to assess the telomere and chromosomal profiles in a Senegalese cohort of 35 DSDs patients. Our study has highlighted the implication of telomere dysfunction in the development of DSDs, thus adding a novel biomarker for the identification and diagnosis of DSD patients.
The high prevalence of DSDs in SSA countries is ascribed to two major causes: (i) consanguinity and endogamy in these populations [2], and (ii) the excessive use of insecticides such as Dichlorodiphenyltrichloroethane (DDT) in agriculture or farming in order to eradicate tropical endemic diseases [43]. Consanguinity and endogamy are well-known causes of many genetic disorders, including DSDs [44]. DDT is recognized as an endocrine disruptor causing a reduction in sperm quality and increased risk of congenital diseases [45]. Accumulating evidence suggests that prenatal exposure to DDT is considered a risk factor in the incidence of DSDs. The presence of chromosomal aberrations in DSDs patients could be associated with genotoxic stress.
Conventional cytogenetic analysis of our cohort identified two patients with mosaic Turner syndrome (45,X/46,XY, and 45,X/46,XX). Turner syndrome is a chromosomal DSD caused by the monosomy of the X chromosome [46]. The mosaic subtypes of Turner syndrome are relatively rare [47]. We confirmed our findings by employing SRY-specific probes. By this approach, we also identified a mosaic Klinefelter syndrome (KS) (46,XY/47,XXY), which had not been detected by conventional cytogenetic, possibly because only 20 metaphases are being analyzed for conventional karyotyping, but 200 nuclei with the automated FISH approach. KS is the most frequently observed chromosomal DSD, with an estimated frequency of 1/500 to 1/1000 [48]. The mosaic subtype accounts for some 10–20% of KS cases [49].
Using conventional and molecular cytogenetics, structural chromosomal aberrations were identified in only three of the DSD patients (3/35; 8.5%). This rate is less than that previously described in other registers and in other countries (varying between 13% and 15%), thus underscoring a significant contribution of the etiology to the occurrence of DSD. Our findings highlight the limitations of using conventional karyotyping in clinical genetics and call for new genetic tests to identify additional biomarkers characteristic for these diseases [50].
For a reliable and precise analysis of additional chromosomal aberrations in DSD patients, we employed telomere and centromere staining. Using that approach, we identified additional non-clonal chromosomal aberrations in 22% of the DSD patients, including dicentric chromosomes, DNA pulverization, acentric chromosomes, terminal deletions, and chromatid breaks. Dicentric chromosomes are considered the best biomarker for irradiation-induced DNA damage as well as for chromosomal instability [37]. In our cohort, we detected a dic(Y;4) with interstitial telomere sequences, indicating that the formation of this dicentric chromosome is related to telomere dysfunction. Several DSDs studies have previously reported the implication of the Y chromosome in the formation of a dicentric [51,52,53,54,55,56]. In addition, chromosome pulverization was observed in two patients in our cohort as the consequence of telomere dysfunction and probably of micronuclei formation and chromothripsis mechanisms [57]. We demonstrate that our protocol for telomere and centromere staining offers improved detection of all chromosomal aberrations, clonal and non-clonal ones. Employment of this technique in SSA cytogenetic laboratories will constitute a major step forward in the management of DSDs patients.
Analysis of telomere lengths and telomere aberrations revealed shortening of and accumulation of aberrations of telomeres in the DSDs patients. A link between reduced fertility and telomere dysfunction has been reported previously [38,58]. Reduced telomere length has been described in patients with chromosomal DSDs, including KS associated with acute lymphoblastic leukemia (ALL). It was suggested that telomere dysfunction in these KS patients may contribute to the pathogenesis of ALL [24]. However, no studies have addressed a direct link between decreased telomere length and the prevalence of DSDs.
Here, we demonstrate for the first time a significant age-independent reduction in mean telomere length in our cohort of DSDs patients compared to that observed in healthy donors of similar age. Our data reveal an accelerated aging process in DSD patients, thus opening new horizons in our understanding of these disorders in terms of possible mechanisms.
In addition to telomere length, we studied telomere aberrations involving losses and doublet formations. The frequency of telomere aberrations in the DSDs cohort was also independent of age. However, we recorded a more frequent loss of telomeres than the occurrence of doublets in contrast to the healthy controls. The analysis of telomere aberrations in each chromosome revealed a higher rate of these aberrations in chromosomes 21 and 22 compared to other chromosomes. These findings call for further investigation.
In this paper, we have conducted a comprehensive and in-depth cytogenetic study, including telomere analysis of patients with DSD from West Africa, in particular from Senegal. Although this study suffers from limitations in terms of the complexity of DSDs, the difficulty in precise diagnosis, and the role of a specific environment, it nevertheless allowed us to gain more insight into African DSDs that may present distinct genetic features related to the environment and the management methods in these regions. However, analysis of a large prospective cohort with access to complete clinical and biological parameters is required to validate the present results and, thus, the application of these techniques for future genetic diagnosis of DSDs. The techniques employed for cytogenetic diagnosis in the present cohort are accessible, and their application is feasible in Senegal. Hence, the development of these techniques will be an indispensable tool for the management of DSDs, which still constitute a major challenge in hospitals today.
5. Conclusions
The results of this study have allowed the establishment of a complete cytogenetic diagnosis, including telomere analysis, and also defined specific features of DSDs that have not been previously reported. Indeed, our data demonstrate for the first time that telomere shortening and telomere aberrations represent one of the most common cytogenetic features of DSDs. The results show a major involvement of small acrocentric chromosomes, especially chromosomes 21 and 22, in telomere aberrations, allowing us to better understand the mechanisms resulting in DSDs. The sequential analysis of telomere length and aberrations for DSDs patients may contribute to our knowledge and better understanding of molecular mechanisms of the accelerated aging process and the implication of DNA repair mechanisms and specific mutations in these diseases.
Acknowledgments
The authors warmly thank the patients and their parents for consenting to participate in this study and make it feasible.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biomedicines12030565/s1, Table S1: Clinical and cytogenetic characteristics of the DSD patient cohort.
Author Contributions
Conceptualization: R.M. and M.G.; methodology: H.Y., R.M. and M.S.; software: B.C. and A.D.; validation: H.Y. and N.R.D.; formal analysis: A.D., B.C. and E.J.; investigation: H.Y. and E.J.; resources: A.P., L.H., A.S. and N.A.N., data curation: B.C. and H.Y.; writing: H.Y., M.G. and R.M.; writing—original draft preparation: H.Y.; writing—review and editing: S.J. and P.V.; visualization: P.C. and R.M.; supervision: M.G.; project administration: M.S.; funding acquisition: M.G. and R.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the ethics committee of the Cheikh Anta Diop University of Dakar (Protocol 041512019/CER/UCAD).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
Authors Haifaou Younoussa, Ahmed Said, Philippe Voisin and Radhia M’kacher were employed by the company Cell Environment. Authors Andreas Plesch and Leonhard Heidingsfelder were employed by the company MetaSystems. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The companies Cell Environent and MetaSystems had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Funding Statement
This research was funded by the L’OREAL Foundation for Women in Science 2020 for the scholarship of Haifaou Younoussa. This work was supported by a French State grant managed by the Agence Nationale de la Recherche (ANR) under the third program of investments for the future (PIA), integrated into France 2030, with reference ANR-21-RHUS-0013 (RHU REVEAL).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Lee P.A., Houk C.P., Ahmed S.F., Hughes I.A. International Consensus Conference on Intersex organized by the Lawson Wilkins Pediatric Endocrine Society and the European Society for Paediatric Endocrinology Consensus statement on management of intersex disorders. International Consensus Conference on Intersex. Pediatrics. 2006;118:e488–e500. doi: 10.1542/peds.2006-0738. [DOI] [PubMed] [Google Scholar]
- 2.Bashamboo A., McElreavey K. Consanguinity and disorders of sex development. Hum. Hered. 2014;77:108–117. doi: 10.1159/000360763. [DOI] [PubMed] [Google Scholar]
- 3.Bouwman H., Kylin H., Sereda B., Bornman R. High levels of DDT in breast milk: Intake, risk, lactation duration, and involvement of gender. Environ. Pollut. 2012;170:63–70. doi: 10.1016/j.envpol.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Houk C.P., Lee P.A. Consensus Statement on Terminology and Management: Disorders of Sex Development. Sex Dev. 2008;2:172–180. doi: 10.1159/000152032. [DOI] [PubMed] [Google Scholar]
- 5.Hughes I.A. Disorders of sex development: A new definition and classification. Best Pract. Res. Clin. Endocrinol. Metab. 2008;22:119–134. doi: 10.1016/j.beem.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Aaronson I.A., Aaronson A.J. How should we classify intersex disorders? J. Pediatr. Urol. 2010;6:443–446. doi: 10.1016/j.jpurol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Amolo P., Laigong P., Omar A., Drop S. Etiology and Clinical Presentation of Disorders of Sex Development in Kenyan Children and Adolescents. Int. J. Endocrinol. 2019;2019:2985347. doi: 10.1155/2019/2985347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerrero-Fernández J., Azcona San Julián C., Barreiro Conde J., Bermúdez De La Vega J.A., Carcavilla Urquí A., Castaño González L.A., Martos Tello J.M., Rodríguez Estévez A., Yeste Fernández D., Martínez Martínez L., et al. Guía de actuación en las anomalías de la diferenciación sexual (ADS)/desarrollo sexual diferente (DSD) An. Pediatría. 2018;89:315.e1–315.e19. doi: 10.1016/j.anpedi.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 9.Sax L. How common is lntersex? A response to Anne Fausto-Sterling. J. Sex Res. 2002;39:174–178. doi: 10.1080/00224490209552139. [DOI] [PubMed] [Google Scholar]
- 10.Thyen U., Lanz K., Holterhus P.-M., Hiort O. Epidemiology and initial management of ambiguous genitalia at birth in Germany. Horm. Res. 2006;66:195–203. doi: 10.1159/000094782. [DOI] [PubMed] [Google Scholar]
- 11.Ganie Y., Aldous C., Balakrishna Y., Wiersma R. Disorders of sex development in children in KwaZulu-Natal Durban South Africa: 20-year experience in a tertiary centre. J. Pediatr. Endocrinol. Metab. JPEM. 2017;30:11–18. doi: 10.1515/jpem-2016-0152. [DOI] [PubMed] [Google Scholar]
- 12.Ameyaw E., Asafo-Agyei S.B., Hughes I.A., Zacharin M., Chanoine J.-P. Incidence of disorders of sexual development in neonates in Ghana: Prospective study. Arch. Dis. Child. 2019;104:636–638. doi: 10.1136/archdischild-2019-316986. [DOI] [PubMed] [Google Scholar]
- 13.Alkhzouz C., Bucerzan S., Miclaus M., Mirea A.-M., Miclea D. 46,XX DSD: Developmental, Clinical and Genetic Aspects. Diagnostics. 2021;11:1379. doi: 10.3390/diagnostics11081379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pang S.Y., Wallace M.A., Hofman L., Thuline H.C., Dorche C., Lyon I.C., Dobbins R.H., Kling S., Fujieda K., Suwa S. Worldwide experience in newborn screening for classical congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Pediatrics. 1988;81:866–874. doi: 10.1016/S0022-5347(17)42164-1. [DOI] [PubMed] [Google Scholar]
- 15.Kim K.S., Kim J. Disorders of sex development. Korean J. Urol. 2012;53:1–8. doi: 10.4111/kju.2012.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wisniewski A.B., Batista R.L., Costa E.M.F., Finlayson C., Sircili M.H.P., Dénes F.T., Domenice S., Mendonca B.B. Management of 46,XY Differences/Disorders of Sex Development (DSD) Throughout Life. Endocr. Rev. 2019;40:1547–1572. doi: 10.1210/er.2019-00049. [DOI] [PubMed] [Google Scholar]
- 17.Mazen I., Hiort O., Bassiouny R., El Gammal M. Differential Diagnosis of Disorders of Sex Development in Egypt. Horm. Res. 2008;70:118–123. doi: 10.1159/000137657. [DOI] [PubMed] [Google Scholar]
- 18.Pyle L.C., Nathanson K.L. A Practical Guide for Evaluating Gonadal Germ Cell Tumor Predisposition in Differences of Sex Development. Am. J. Med. Genet. C Semin. Med. Genet. 2017;175:304–314. doi: 10.1002/ajmg.c.31562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayers K.L., Bouty A., Robevska G., van den Bergen J.A., Juniarto A.Z., Listyasari N.A., Sinclair A.H., Faradz S.M.H. Variants in congenital hypogonadotrophic hypogonadism genes identified in an Indonesian cohort of 46,XY under-virilised boys. Hum. Genomics. 2017;11:1. doi: 10.1186/s40246-017-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viswanathan V., Eugster E.A. Etiology and Treatment of Hypogonadism in Adolescents. Pediatr. Clin. N. Am. 2011;58:1181–1200. doi: 10.1016/j.pcl.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganmore I., Smooha G., Izraeli S. Constitutional aneuploidy and cancer predisposition. Hum. Mol. Genet. 2009;18:R84–R93. doi: 10.1093/hmg/ddp084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji J., Zöller B., Sundquist J., Sundquist K. Risk of solid tumors and hematological malignancy in persons with Turner and Klinefelter syndromes: A national cohort study. Int. J. Cancer. 2016;139:754–758. doi: 10.1002/ijc.30126. [DOI] [PubMed] [Google Scholar]
- 23.Gomes N.L., Chetty T., Jorgensen A., Mitchell R.T. Disorders of Sex Development—Novel Regulators, Impacts on Fertility, and Options for Fertility Preservation. Int. J. Mol. Sci. 2020;21:2282. doi: 10.3390/ijms21072282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjeldsen E. Congenital Aneuploidy in Klinefelter Syndrome with B-Cell Acute Lymphoblastic Leukemia Might Be Associated with Chromosomal Instability and Reduced Telomere Length. Cancers. 2022;14:2316. doi: 10.3390/cancers14092316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reish O., Brosh N., Gobazov R., Rosenblat M., Libman V., Mashevich M. Sporadic aneuploidy in PHA-stimulated lymphocytes of Turner’s syndrome patients. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2006;14:527–534. doi: 10.1007/s10577-006-1050-9. [DOI] [PubMed] [Google Scholar]
- 26.Reish O., Regev M., Kanesky A., Girafi S., Mashevich M. Sporadic Aneuploidy in PHA-Stimulated Lymphocytes of Trisomies 21, 18, and 13. Cytogenet. Genome Res. 2011;133:184–189. doi: 10.1159/000323504. [DOI] [PubMed] [Google Scholar]
- 27.Zhong Q., Layman L.C. Genetic considerations in the patient with Turner syndrome--45,X with or without mosaicism. Fertil. Steril. 2012;98:775–779. doi: 10.1016/j.fertnstert.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callén E., Surrallés J. Telomere dysfunction in genome instability syndromes. Mutat. Res. Mutat. Res. 2004;567:85–104. doi: 10.1016/j.mrrev.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Gadji M., Mathur S., Bélanger B., Jangamreddy J.R., Lamoureux J., Tsanaclis A.M.C., Fortin D., Drouin R., Mai S. Three-Dimensional Nuclear Telomere Profiling as a Biomarker for Recurrence in Oligodendrogliomas: A Pilot Study. Int. J. Mol. Sci. 2020;21:8539. doi: 10.3390/ijms21228539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferlin A., Rampazzo E., Rocca M.S., Keppel S., Frigo A.C., De Rossi A., Foresta C. In young men sperm telomere length is related to sperm number and parental age. Hum. Reprod. 2013;28:3370–3376. doi: 10.1093/humrep/det392. [DOI] [PubMed] [Google Scholar]
- 31.The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.M’kacher R., Bennaceur-Griscelli A., Girinsky T., Koscielny S., Delhommeau F., Dossou J., Violot D., Leclercq E., Courtier M.H., Béron-Gaillard N., et al. Telomere Shortening and Associated Chromosomal Instability in Peripheral Blood Lymphocytes of Patients With Hodgkin’s Lymphoma Prior to Any Treatment Are Predictive of Second Cancers. Int. J. Radiat. Oncol. 2007;68:465–471. doi: 10.1016/j.ijrobp.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 33.Girinsky T., M’Kacher R., Lessard N., Koscielny S., Elfassy E., Raoux F., Carde P., Santos M.D., Margainaud J.-P., Sabatier L., et al. Prospective Coronary Heart Disease Screening in Asymptomatic Hodgkin Lymphoma Patients Using Coronary Computed Tomography Angiography: Results and Risk Factor Analysis. Int. J. Radiat. Oncol. 2014;89:59–66. doi: 10.1016/j.ijrobp.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 34.Maciejowski J., de Lange T. Telomeres in cancer: Tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017;18:175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dewhurst S.M., Yao X., Rosiene J., Tian H., Behr J., Bosco N., Takai K.K., de Lange T., Imieliński M. Structural variant evolution after telomere crisis. Nat. Commun. 2021;12:2093. doi: 10.1038/s41467-021-21933-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muraki K., Nyhan K., Han L., Murnane J.P. Mechanisms of telomere loss and their consequences for chromosome instability. Front. Oncol. 2012;2:135. doi: 10.3389/fonc.2012.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M’kacher R., Colicchio B., Borie C., Junker S., Marquet V., Heidingsfelder L., Soehnlen K., Najar W., Hempel W.M., Oudrhiri N., et al. Telomere and Centromere Staining Followed by M-FISH Improves Diagnosis of Chromosomal Instability and Its Clinical Utility. Genes. 2020;11:475. doi: 10.3390/genes11050475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M’kacher R., Colicchio B., Marquet V., Borie C., Najar W., Hempel W.M., Heidingsfelder L., Oudrhiri N., Jawhari M.A., Wilhelm-Murer N., et al. Telomere aberrations, including telomere loss, doublets, and extreme shortening, are increased in patients with infertility. Fertil. Steril. 2021;115:164–173. doi: 10.1016/j.fertnstert.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Murnane J.P. Telomere dysfunction and chromosome instability. Mutat. Res. Mol. Mech. Mutagen. 2012;730:28–36. doi: 10.1016/j.mrfmmm.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nassour J., Radford R., Correia A., Fusté J.M., Schoell B., Jauch A., Shaw R.J., Karlseder J. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature. 2019;565:659–663. doi: 10.1038/s41586-019-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M’Kacher R., Colicchio B., Junker S., El Maalouf E., Heidingsfelder L., Plesch A., Dieterlen A., Jeandidier E., Carde P., Voisin P. High Resolution and Automatable Cytogenetic Biodosimetry Using In Situ Telomere and Centromere Hybridization for the Accurate Detection of DNA Damage: An Overview. Int. J. Mol. Sci. 2023;24:5699. doi: 10.3390/ijms24065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGowan-Jordan J., Hastings R.J., Moore S. Iscn 2020: An International System for Human Cytogenomic Nomenclature 2020. Cytogenet. Genome Res. 2020;160:7–8. doi: 10.1159/000516655. [DOI] [PubMed] [Google Scholar]
- 43.Riana Bornman M.S., Bouwman H. Environmental pollutants and diseases of sexual development in humans and wildlife in South Africa: Harbingers of impact on overall health? Reprod. Domest. Anim. Zuchthyg. 2012;47((Suppl. S4)):327–332. doi: 10.1111/j.1439-0531.2012.02094.x. [DOI] [PubMed] [Google Scholar]
- 44.Barriga H.H.A., Velásquez F.C., Carrillo C.B., Tacuri A.P., Hurtado M.B., Loarte T.V., Rondón L.O., Linares M.O., Abuhadba E.A.R. Variantes en el número de copias y consanguinidad parental en neonatos de altura con anomalías congénitas en Perú. Rev. Fac. Cienc. Médicas. 2022;79:132. doi: 10.31053/1853.0605.v79.n2.34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaspari L., Sampaio D.R., Paris F., Audran F., Orsini M., Neto J.B., Sultan C. High prevalence of micropenis in 2710 male newborns from an intensive-use pesticide area of Northeastern Brazil. Int. J. Androl. 2012;35:253–264. doi: 10.1111/j.1365-2605.2011.01241.x. [DOI] [PubMed] [Google Scholar]
- 46.Syndrome de Turner Protocole National de Diagnostic et de Soins. [(accessed on 3 January 2023)]. Available online: https://www.has-sante.fr/upload/docs/application/pdf/2021-11/pnds_turner_29_10.pdf.
- 47.Ennazk L., Mghari G.E., Ansari N.E. Le DSD mosaïque (45,X0, 46,XY), un sous-type peu connu du syndrome de Turner. Ann. Endocrinol. 2016;77:469. doi: 10.1016/j.ando.2016.07.649. [DOI] [Google Scholar]
- 48.Bojesen A., Juul S., Gravholt C.H. Prenatal and postnatal prevalence of Klinefelter syndrome: A national registry study. J. Clin. Endocrinol. Metab. 2003;88:622–626. doi: 10.1210/jc.2002-021491. [DOI] [PubMed] [Google Scholar]
- 49.Bonomi M., Rochira V., Pasquali D., Balercia G., Jannini E.A., Ferlin A., Balercia G., Bonomi M., Calogero A., Corona G., et al. Klinefelter syndrome (KS): Genetics, clinical phenotype and hypogonadism. J. Endocrinol. Investig. 2017;40:123–134. doi: 10.1007/s40618-016-0541-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Achermann J.C., Domenice S., Bachega T.A.S.S., Nishi M.Y., Mendonca B.B. Disorders of sex development: Effect of molecular diagnostics. Nat. Rev. Endocrinol. 2015;11:478–488. doi: 10.1038/nrendo.2015.69. [DOI] [PubMed] [Google Scholar]
- 51.DesGroseilliers M., Beaulieu Bergeron M., Brochu P., Lemyre E., Lemieux N. Phenotypic variability in isodicentric Y patients: Study of nine cases. Clin. Genet. 2006;70:145–150. doi: 10.1111/j.1399-0004.2006.00654.x. [DOI] [PubMed] [Google Scholar]
- 52.Abdelmoula N.B., Amouri A. [Dicentric Y chromosome] Ann. Biol. Clin. 2005;63:363–375. [PubMed] [Google Scholar]
- 53.Roubin M., de Grouchy J., Chauveau P., Rappaport R., Pellerin D. [Dicentric Y chromosome in a male pseudohermaphrodite 45,X/46,X, dic (Y)/47, XYY] Ann. Genet. 1977;20:185–189. [PubMed] [Google Scholar]
- 54.Alexander D.S., Soudek D., Laraya P. Unstable dicentric iso(Yq) chromosome in a pseudohermaphrodite. Am. J. Med. Genet. 1978;1:265–269. doi: 10.1002/ajmg.1320010302. [DOI] [PubMed] [Google Scholar]
- 55.Reshmi S.C., Miller J.L., Deplewski D., Close C., Henderson L.J., Littlejohn E., Schwartz S., Waggoner D.J. Evidence of a mechanism for isodicentric chromosome Y formation in a 45,X/46,X,idic(Y)(p11.31)/46,X,del(Y)(p11.31) mosaic karyotype. Eur. J. Med. Genet. 2011;54:161–164. doi: 10.1016/j.ejmg.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 56.Ponzio G., DeMarchi M., Carbonara A., Godano A., Massara F. Dicentric Y chromosome in a patient with gonadal dysgenesis and seminoma. Hum. Genet. 1981;58:282–284. doi: 10.1007/BF00294924. [DOI] [PubMed] [Google Scholar]
- 57.Ernst A., Jones D.T.W., Maass K.K., Rode A., Deeg K.I., Jebaraj B.M.C., Korshunov A., Hovestadt V., Tainsky M.A., Pajtler K.W., et al. Telomere dysfunction and chromothripsis. Int. J. Cancer. 2016;138:2905–2914. doi: 10.1002/ijc.30033. [DOI] [PubMed] [Google Scholar]
- 58.Vasilopoulos E., Fragkiadaki P., Kalliora C., Fragou D., Docea A.O., Vakonaki E., Tsoukalas D., Calina D., Buga A.M., Georgiadis G., et al. The association of female and male infertility with telomere length (Review) Int. J. Mol. Med. 2019;44:375–389. doi: 10.3892/ijmm.2019.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.