ABSTRACT
Background
Inflammatory bowel disease (IBD) is characterized by immune-mediated, chronic inflammation of the intestinal tract. The occurrence of IBD is driven by the complex interactions of multiple factors. The objective of this study was to evaluate the therapeutic effects of IAA in colitis.
Method
C57/BL6 mice were administered 2.5% DSS in drinking water to induce colitis. IAA, Bifidobacterium pseudolongum, and R-equol were administered by oral gavage and fed a regular diet. The Disease Activity Index was used to evaluate disease activity. The degree of colitis was evaluated using histological morphology, RNA, and inflammation marker proteins. CD45+ CD4+ FOXP3+ Treg and CD45+ CD4+ IL17A+ Th17 cells were detected by flow cytometry. Analysis of the gut microbiome in fecal content was performed using 16S rRNA gene sequencing. Gut microbiome metabolites were analyzed using Untargeted Metabolomics.
Result
In our study, we found IAA alleviates DSS-induced colitis in mice by altering the gut microbiome. The abundance of Bifidobacterium pseudolongum significantly increased in the IAA treatment group. Bifidobacterium pseudolongum ATCC25526 alleviates DSS-induced colitis by increasing the ratio of Foxp3+T cells in colon tissue. R-equol alleviates DSS-induced colitis by increasing Foxp3+T cells, which may be the mechanism by which ATCC25526 alleviates DSS-induced colitis in mice.
Conclusion
Our study demonstrates that IAA, an indole derivative, alleviates DSS-induced colitis by promoting the production of Equol from Bifidobacterium pseudolongum, which provides new insights into gut homeostasis regulated by indole metabolites other than the classic AHR pathway.
KEYWORDS: IAA, regulatory T cells, gut microbiome, bifidobacterium pseudolongum, colitis
1. Introduction
Inflammatory bowel disease (IBD) is characterized by immune-mediated chronic inflammation of the intestinal tract, including Ulcerative Colitis (UC) and Crohn’s Disease.1 The number of IBD patient are over 1 million in the USA and 2.5 million in European countries, and the annual cost of the therapy in each patient is beyond 25,000$ in Western world,2 The high annual costs burden the healthcare system. However, the effects of current treatments are limited.3
The occurrence of IBD is driven by the complex interaction of multiple factors, including immunological, microbiome, and environmental factors, as well as genetic risk.1,4 In genetic or chemical induction colitis mouse models, germ-free (GF) mice exhibit milder colitis, indicating the importance of the gut microbiome in IBD progression.4,5 Gut dysbiosis occurs in patients with IBD, especially in those with active disease.4,6 Fecal microbiota transplantation (FMT) from a healthy donor appears to be a new therapeutic option for UC treatment in several clinical trials.7,8 The gut microbiome may maintain homeostasis through metabolites such as short-chain fatty acids (SCFAs) and bile acids.4,9 The functions of these metabolites involve regulation of gut immunity and barrier maintenance.6
Tryptophan is an essential amino acid obtained from diet and can be metabolized into bioactive molecules mainly through three pathways (the kynurenine, serotonin, and indole pathways), among which the indole pathways are catalyzed by the gut microbiome to produce a series of indole metabolites.10 Indole metabolites can bind and activate the aryl hydrocarbon receptor (AhR), which induces the expression of downstream cytokines to maintain gut homeostasis.11 However, the effects of indole metabolites on gut homeostasis are not yet fully understood. Indole-3-acetic acid (IAA) is an indole metabolite. Most studies have focused on the effects of IAA on liver disease.12–14 In our study, we found that IAA can alleviate DSS-induced colitis by promoting the production of Equol from Bifidobacterium pseudolongum, which provides new insights into gut homeostasis regulated by indole metabolites.
2. Results
2.1. IAA alleviates DSS-induced colitis in mice
To evaluate the effect of IAA on colitis, mice were pretreated with IAA for two weeks and then orally treated with 2.5% DSS for seven consecutive days (Figure 1(a)). We found that IAA improved body weight and Disease Activity Index (DAI) scores compared to the DSS group (Figure 1(b–c)). Colon length also significantly increased in the IAA group (Figure 1(d–e)). The pathological scores in colon tissues also suggested a protective effect of IAA against colitis (Figure 1(f–g)).
Figure 1.
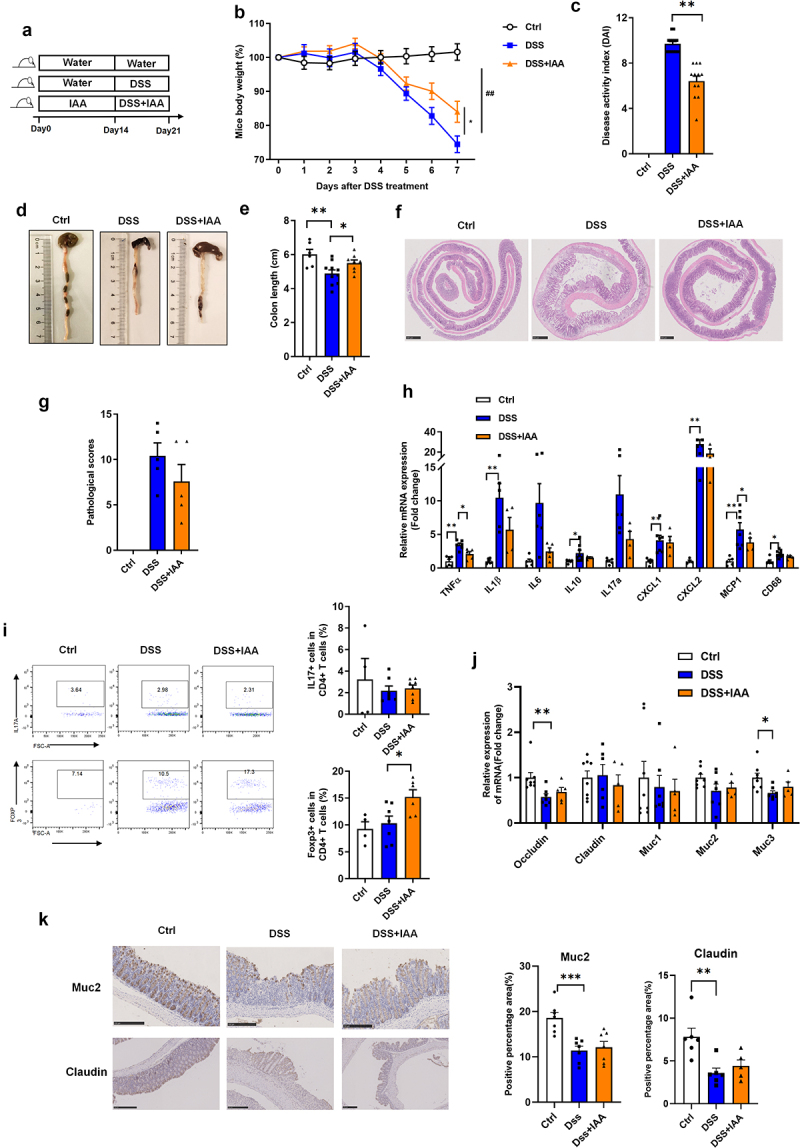
IAA alleviates DSS-induced colitis in mice.
(a) Experimental design. Relative mRNA expression of inflammatory factors and mucosal protective factors in colon tissue. (b) Percentage change of body weight during the whole experimental time. (c) Disease activity index evaluated at sacrifice. (d – e). Representative images and length of the colon. (f – g) the image of H&E staining of the colon and pathological score. (h) Relative mRNA expression of inflammatory factors in colon tissue. (i) the proportion of CD45+ CD4+ IL17A+ Th17 cells and CD45+CD4+Foxp3+ Treg cells in colon tissue. (j) Relative mRNA expression of mucosal protective factors in colon tissue. (k) The immunohistochemistry staining of MUC2 protein and Claudin protein in colon tissues.
Data are expressed as mean±SE, n = 5-12,*p<0.05,**p<0.01.
Furthermore, proinflammatory cytokines were also detected to assess the degree of inflammation in colon tissue, and we found that the mRNA levels of proinflammatory cytokines (TNF-α, IL-1β, CXCL1, CXCL2, MCP-1, and CD68) in colon tissue were increased after DSS administration, and the mRNA levels of TNF-α and MCP-1 were significantly reduced after IAA treatment (Figure 1(h)). TH17 and regulated T cells are crucial in the development of IBD,15 we then measured the ratio of TH17 (CD45+ CD4+ IL17A+), and regulated T cells (CD45+ CD4+FOXP3+) in CD4+ T cells in colon tissue after IAA administration. As shown in Figure 1(i), the proportion of Tregs was significantly increased in the IAA treatment group compared with vehicle control mice; however, the proportion of IL17 cells in colon tissue was not affected after IAA administration.
In addition, the mechanical and mucosal barriers are important components of the gut barrier. Next, we assessed the effects of IAA on the mechanical and mucosal barriers; however, the mRNA levels of the tight junction proteins claudin and occludin and the protein level of claudin did not change after IAA treatment. The mRNA levels of mucins Muc1, Muc2, Muc3 and the protein level of Muc2 was also not affected by IAA administration (Figure 1(j–k)).
Furthermore, we tested the safety of IAA. As showed in Figure S1, there was no significant difference of the body weight between control and sole IAA group (Figure S1A). There was also no significant difference in colon length between the two groups (Figure S1(b – c)). The administration of IAA alone did not induce any colon damage (Figure S1(d)).
These results indicate that IAA alleviates DSS-induced colitis in mice, mainly by suppressing the immune inflammatory response, rather than improving the mucosal barrier.
2.2. IAA alleviates DSS-induced colitis in mice through gut microbiome
Alterations in the gut microbiome are associated with IBD development.16 We further explored whether the protective effect of IAA on colitis was mediated by the modulation of the gut microbiome. Bacterial 16sRNA sequencing results of cecal contents showed that IAA administration increased the α-diversity of gut microbiome in DSS-induced colitis mice, especially the Shannon diversity index (Figure 2(a–c)), indicating that IAA may exert its protective effect by altering gut microbiome. The following experiments were designed to test this hypothesis. Mice were pretreated with antibiotic-containing drinking water for consecutive 14 days, and then orally treated with 2.5% DSS for 7 days by gavage with IAA or PBS (Figure 2(d)). We found that the protective effect of IAA on DSS-induced colitis diminished after gut microbiome elimination. As shown in Figure 2(e–h), there were no significant differences in body weight, DAI score, and colon length between the two groups. The protective effect of IAA on colon tissue was also substantially blunted by antibiotics (AB) (Figure 2(i–j)). We also analyzed the bacteria of cecal contents by 16sRNA sequencing, we found after the treatment of antibiotics. The alpha diversity was significantly decreased both in DSS+AB and DSS+AB+IAA group(Figure 2(l–n)). Furthermore, there was also no significant difference in the relative abundance of species between the two groups(Figure 2(k)). These results indicate that IAA exerts a protective effect on the colon by altering the gut microbiome
Figure 2.
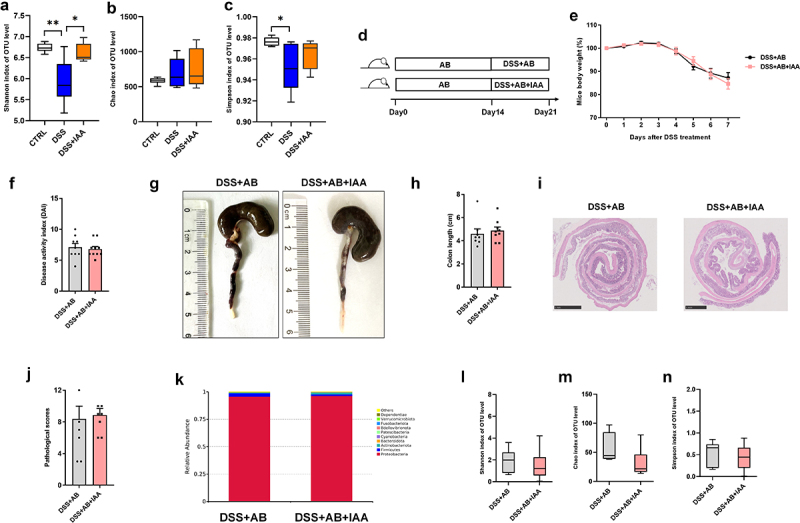
IAA alleviates DSS-induced colitis in mice through gut microbiome.
(a – c) the α-diversity of gut microbiome in fecal content. (d) Experimental design. (e) Percentage change of body weight during the whole experimental time. (f) Disease activity index evaluated at sacrifice. (g – h) Representative images and length of the colon. (i – j) the image of H&E staining of the colon and pathological score. (k) the relative abundance of species in fecal content of DSS+AB and DSS+AB+IAA group. (l – n) the alpha diversity in fecal content of DSS+AB and DSS+AB+IAA group.
Data are expressed as mean±SE, n = 6-10,*p<0.05,**p<0.01.
2.3. Bifidobacterium pseudolongum ATCC25526 alleviates DSS-induced colitis through increasing the Foxp3+T cells
We further investigated how IAA protects against DSS-induced colitis through the gut microbiome. The gut microbiome in cecal contents between DSS and DSS+IAA groups was analyzed using LEfSe Differential Analysis. As shown in Figure 3(a), the abundance of Bifidobacterium pseudolongum (B. pseudolongum) was significantly higher in the IAA group than in the DSS group. To further explore the potential role of B. pseudolongum in colitis, mice were administered two subgroups of B. pseudolongum (ATCC25526 or ATCC25865) by gavage for two weeks, and 2.5% DSS was administered orally for 7 consecutive days (Figure 3(b)). We found that mice gavaged with ATCC25526 showed a significantly higher body weight and lower DAI score than mice gavaged with PBS (Figure 3(c–d)). The colon length of mice gavaged with ATCC25526 was significantly longer than that of mice gavaged with PBS (Figure 3(e–f)). The result of colon tissue pathological H&E staining indicated lower tissue damage, as evaluated by inflammation damage, inflammation infiltration, and crypt damage in the ATCC25526 gavage group (Figure 3(g)). Moreover, we found that the ratio of regulated T cells (CD45+ CD4+FOXP3+) in CD4+ T cells of colon tissue was significantly increased in the ATCC25526 group compared with that in the PBS group, however, the proportion of regulated T cells in the ATCC25865 group did not increase significantly compared with that in the PBS group (Figure 3(h)). Interestingly, the serum and colon tissue level of IL10 in the ATCC25526 group was not significantly higher than that in the PBS group; however, gavage of mice with ATCC25865 significantly increased the serum level of IL10 but not in colon tissue (Figure 3(i)).
Figure 3.
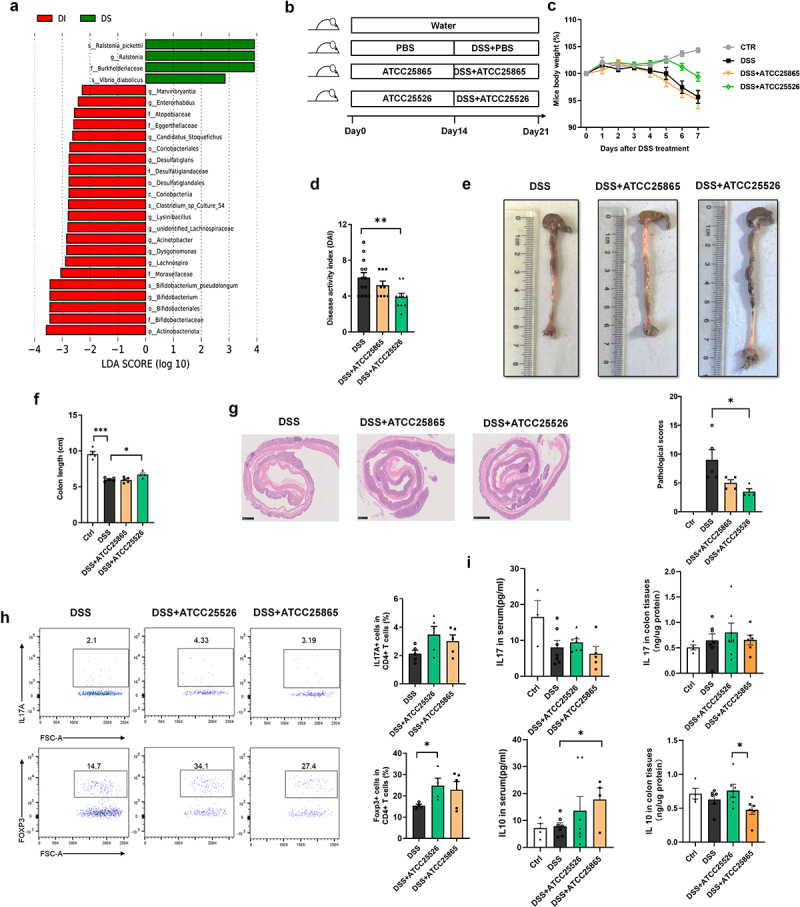
Bifidobacterium pseudolongum ATCC25526 alleviates DSS-induced colitis through increasing the Foxp3+T cells.
(a) The LEfSe differential analysis of gut microbiome between DSS(DS) and DSS+IAA (DI) group.(b) Experimental design.(c) Percentage change of body weight during the whole experimental time.(d) Disease activity index evaluated at sacrifice.(e – f) Representative images and length of the colon.(g) the image of H&E staining of the colon and pathological score.(h) the proportion of CD45+ CD4+ IL17A+ Th17 cells and CD45+CD4+Foxp3+ Treg cells in colon tissue.(i) the level of IL10 and IL17 in serum and colon tissue.
Data are expressed as mean±SE, n = 3-13,*p<0.05,**p<0.01,***p<0.001
These results indicate that Bifidobacterium pseudolongum ATCC25526 alleviates DSS-induced colitis by increasing the ratio of Foxp3+T cells in the colon tissue.
2.4. R-equol alleviates DSS-induced colitis through increasing the Foxp3+-T cells
Gut homeostasis can be altered by the gut microbiome through its metabolites, which are key mediators between the gut microbiome and the host.10 To explore whether ATCC25526 ameliorated DSS-induced colitis through metabolites, the cecal contents from the DSS+PBS and DSS+ATCC25526 groups were collected for untargeted metabolomic analysis. As shown in Figure 4(a), the metabolite R-equol was significantly increased in the DSS+ATCC25526 group. Equol is a small molecular compound produced by the metabolism of soybean isoflavones by the gut microbiome in the small intestine and colon, with estrogen-like and antioxidant effects.17,18 Bifidobacterium breve and Bifidobacterium longum are involved in the metabolism of equol from daidzein in humans.19 Therefore, we speculated that ATCC25526 alleviates DSS-induced colitis by producing equol from soybean isoflavones in food.
Figure 4.
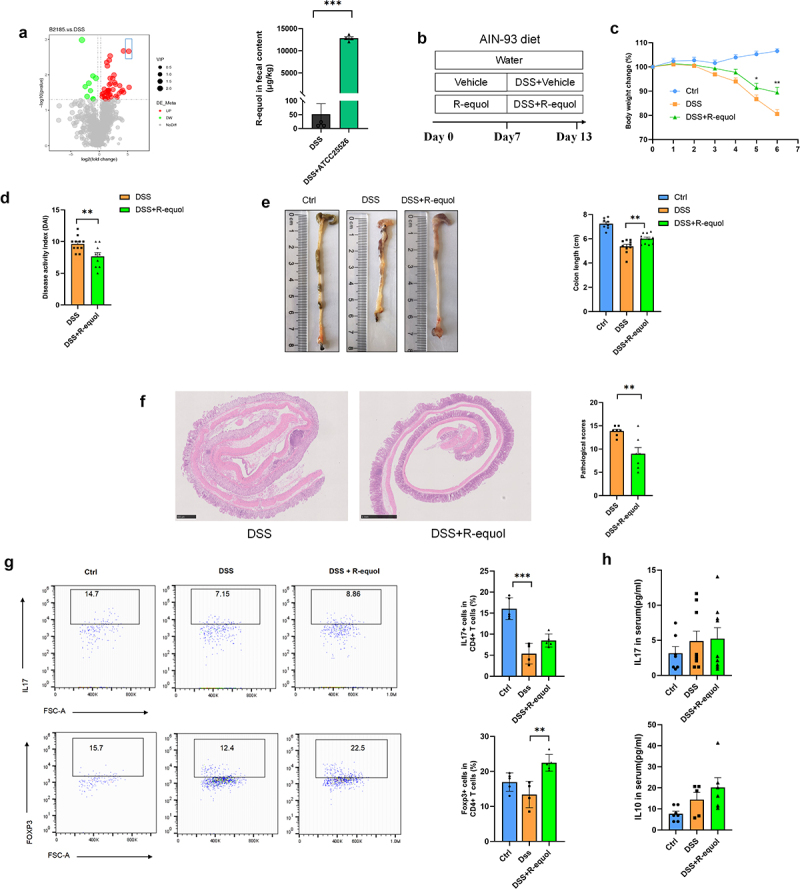
R-equol alleviates DSS-induced colitis through increasing the Foxp3+T cells.
(a) the volcano plot of differential metabolites and the level of R-equol in fecal contents between DSS+ATCC25526 and vehicle group. (b) Experimental design. (c) Percentage change of body weight during the whole experimental time. (d) Disease activity index evaluated at sacrifice. (e) Representative images and length of the colon. (f) the image of H&E staining of the colon and pathological score. (g) the proportion of CD45+ CD4+ IL17A+ Th17 cells and CD45+CD4+Foxp3+ Treg cells in colon tissue. (h) the level of IL10 and IL17 in serum.
Data are expressed as mean±SE, n = 4-11, **p<0.01,***p<0.001
The soy-free diet AIN93 was used to exclude the interference of soy in the regular diet. As shown in Figure 4(b), mice were pretreated with R-equol by oral gavage for 1 week and then orally treated with 2.5% DSS for six consecutive days. Mice in the DSS+R-equol group showed a significantly higher body weight and lower DAI score than those in the DSS group (Figure 4C-D). The colon length of mice gavaged with R-equol was significantly longer than that of mice in the DSS group (Figure 4(e)). Pathological H&E staining of colon tissue indicated lower tissue damage, as evaluated by inflammation damage, inflammation infiltration, and crypt damage in the DSS+R-equol group than in the DSS group (Figure 4(f)). Furthermore, the ratio of regulated T cells (CD45+ CD4+FOXP3+) in CD4+ T cells of colon tissue was significantly higher in the DSS + R-equol group than in the vehicle group (Figure 4(g)). However, the serum level of IL10 in the DSS+R-equol group was not significantly higher than that in the vehicle group (Figure 4(h)). These results suggest that R-equol alleviates DSS-induced colitis by increasing Foxp3+T cells, which may be the mechanism by which ATCC25526 alleviates DSS-induced colitis in mice.
2.5. Bifidobacterium pseudolongum ATCC25526 alleviates DSS-induced colitis through increasing the Foxp3+-T cells by R-equol
To further explore whether B.pseudolongum ATCC25526 alleviates DSS-induced colitis through R-equol, mice were pretreated with B.pseudolongum ATCC25526 or PBS for 2 weeks by oral gavage with an AIN 93 diet or regular diet, and 2.5%DSS was administered orally for 6 days (Figure 5(a)). The difference in the use of diets, including AIN 93 and regular diet, was used to evaluate whether B.pseudolongum ATCC25526 still relieved experimental colitis without soybean isoflavones. As shown in Figure 5(b), there was no difference in body weight between the DSS + PBS and DSS+ ATCC25526 groups. The DAI score and colon length were also not improved in the DSS+ATCC25526 group fed the AIN93 diet compared with the DSS+PBS group (Figure 5(c–e)). However, the pathological scores of colon tissue were significantly lower in the DSS+ATCC25526 group than in the DSS+PBS group in the AIN93 diet (Figure 5(f)). The serum level of IL17 and IL10 were not notably increased in the DSS+ATCC25526 group compared with those in the DSS+ATCC25526 group (Figure 5(g)). The ratio of regulated T cells (CD45+ CD4+FOXP3+) in CD4+ T cells of colon tissue was not increased in the DSS+ ATCC25526 group compared with that in the DSS+PBS group (Figure 5(h)). These results suggest that the AIN93 diet diminished the protective effect of ATCC25526 in DSS-induced colitis.
Figure 5.
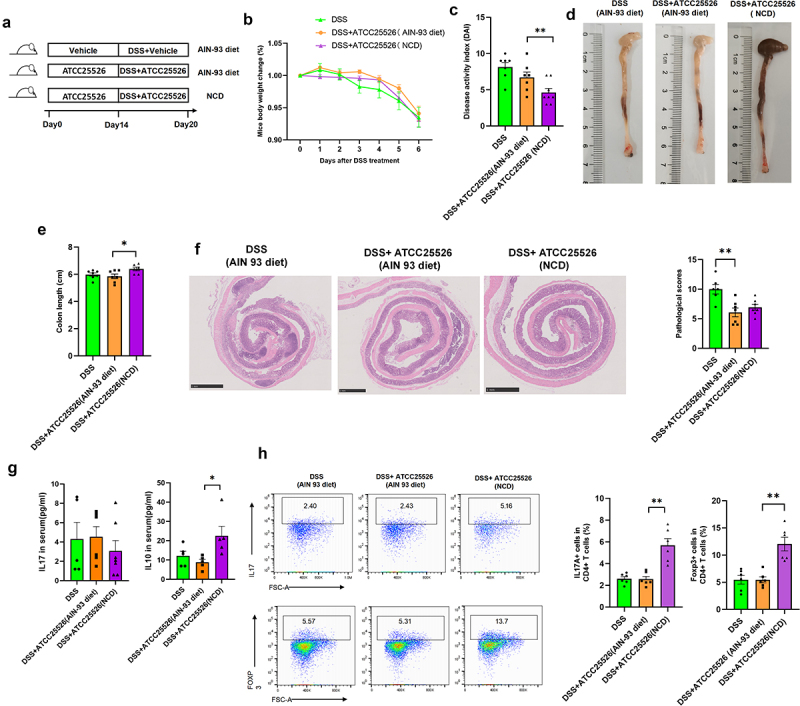
Bifidobacterium pseudolongum ATCC25526 alleviates DSS-induced colitis through increasing the Foxp3+T cells by R-equol.
(a) Experimental design (NCD normal chow diet). (b) Percentage change of body weight during the whole experimental time. (c) Disease activity index evaluated at sacrifice. (d – e) Representative images and length of the colon. (f) the image of H&E staining of the colon and pathological score. (g) the level of IL10 and IL17 in serum. (h) the proportion of CD45+ CD4+ IL17A+ Th17 cells and CD45+CD4+Foxp3+ Treg cells in colon tissue.
Data are expressed as mean±SE, n = 5-8,*p<0.05,**p<0.01.
In the ATCC25526 treatment groups, compared with the AIN93 diet, mice fed a regular diet showed significantly lower DAI scores and longer colon lengths (Figure 5(c–e)). However, there was no difference in the pathological scores of colon tissue between the regular diet and AIN93 diet groups (Figure 5(f)). The serum level of IL10 in the regular diet group was significantly higher than compared in the AIN93 diet (Figure 5(g)). Moreover, the ratio of regulated T cells (CD45+CD4+FOXP3+) in CD4+ T cells of the colon tissue was also higher in the regular diet group than in the AIN93 diet group (Figure 5(h)).
The above results indicate that when soybeans were removed from the diet, the protective effect of B.pseudolongum on the gut was weakened, and they failed to increase the number of Tregs. After soybeans were added to the diet, the elevated effect of B. pseudolongum on Tregs was restored.
3. Discussion
Our study demonstrated the potential protective effects of IAA on DSS-induced colitis in mice. In this study, we focused on the effect of IAA on intestinal immunity, especially in Foxp3+-T regulatory cells. Tregs are key mediators in the regulation of immune tolerance in the intestinal tissue and can maintain immune homeostasis through multiple pathways. Transfer treatment of Tregs in mice can alleviate colitis, indicating the potential protective effect of Tregs in IBD.20 In our study, the proportion of Tregs in the intestinal tissue was significantly increased after IAA administration in a DSS-induced colitis mouse model, suggesting that IAA improves colitis by affecting Tregs to regulate intestinal immunity. IL-10 secreting Tregs are a subgroup of Treg cells that mainly rely on the secretion of the anti-inflammatory cytokine IL-10 to regulate intestinal immunity.21 However, in our study, the trend of serum IL-10 was not consistent with that of Tregs, indicating that the increased Tregs in our study may not be IL-10 secreting Tregs.
The gut microbiome plays an important role in shaping the Treg cell pool. The number of Tregs in GF mice was lower than that in specific pathogen-free (SPF) mice. The mechanism mainly involves the TCR repertoire of Tregs in the gut microbiome.22,23 In addition, the gut microbiome can also modulate Treg homeostasis through metabolites such as bile acids24 and short-chain fatty acids (SCFAs).25 In our study, the use of antibiotics abolished the protective effect of IAA against colitis, indicating an important role of IAA in the gut microbiome for intestinal protection. Furthermore, we found that the abundance of B. pseudolongum in feces after IAA administration was significantly increased. B. pseudolongum is a species of Bifidobacterium that is considered an anti-inflammatory bacterium in osteomyelitis.26 Moreover, B. pseudolongum has the potential to alleviate intestinal barrier function by modulating the PPARγ/STAT3 pathway in DSS-induced colitis.27 However, the mechanisms by which B. pseudolongum alleviates colitis are not fully understood.
The gut microbiome may maintain homeostasis through its metabolites. In our study, we found that R-equol was significantly increased in feces after B. pseudolongum treatment. When soybeans were removed from the diet, the protective effect of B.pseudolongum on the gut was weakened, and the number of Tregs failed to increase. Equol is a metabolite produced by the metabolism of soybean isoflavones by the gut microbiome, which has estrogen-like effects.17 The structure of equol is similar to that of 17-β-estradiol, which can bind to two estrogen ligand-bound nuclear receptors, ERα and β,28 and the membrane-bound G protein-coupled receptor 30(GPR30).29 Research has shown that estrogen can promote the differentiation of Tregs, as decreased levels of Tregs are associated with spontaneous abortion.30 Moreover, ERα can bind to the promoter of Foxp3, which is a vital transcription factor for Treg differentiation, and upregulates its expression.31 In the gut tissue, ERβ is the most abundant isoform in both sexes.32,33 Mice with ERβ deficiency are more susceptible to inducible colitis.34 Activation of ERβ ameliorates DSS-induced colitis and upregulates the proportion of Treg cells among CD4+T cells.35 Therefore, we speculate that the protective effect of equol may act as an ER agonist to improve intestinal resistance to injury by increasing the proportion of Tregs among CD4+ T cells.
In conclusion, our study demonstrated that IAA, an indole derivative, alleviates DSS-induced colitis by promoting the production of Equol from Bifidobacterium pseudolongum, which provides a new insight into gut homeostasis regulated by indole metabolites other than the classic AHR pathway.
4. Material and methods
4.1. Animals and treatment
Healthy, 6–8-week-old male C57BL/6J mice were purchased from Vital River Laboratory Animal Technology (Beijing, China). Mice were maintained on a 12/12 h light-dark cycle with free access to water and diet, and were reared for 1 week before the experiments. The mouse colitis model was induced by adding 2.5% DSS (molecular weight 36,000–50,000 kDa, MP Biomedicals, Santa Ana, United States) to drinking water. Animal experiments were conducted in accordance with the Institutional Animal Care and Use Committee of Capital Medical University and performed in accordance with the ARRIVE guidelines.36
DAI was used to evaluate disease activity, including percentage of weight loss, fecal sparsity, and fecal blood.37 The mice were deeply anesthetized with pentobarbital by intraperitoneal injection (60 mg/kg). Serum samples, cecal contents, and colon tissues were collected.
4.2. Preparation of bifidobacterium pseudolongum
Bifidobacterium pseudolongum was purchased from the China General Microbiological Culture Collection Center (CGMCC). One subgroup number was 1.2203 and the other subgroup number was 1.2285. The bacteria were cultured in Bifidobacterium Medium (Hope Bio-Technology Co., Ltd., Qingdao, China) under an atmosphere of 10% H2, 10% CO2, and 80% N2 for 24 h. The cultures were centrifuged at 1200 × g for 5 min at 4°C, washed twice with sterile anaerobic PBS, and resuspended at a final concentration of 1 × 109 CFU/200 μL under strict anaerobic conditions.
4.3. Histological and Immunohistochemical Analysis
Colon tissues were removed after the mice were deeply anesthetized. They were rinsed thoroughly with phosphate-buffered saline for sectioning and colon tissue pieces were fixed in 4% paraformaldehyde. The degree of colitis was evaluated by hematoxylin and eosin (H&E) staining. The degree of tissue damage was evaluated using a pathological scoring system (0–5 score): loss of the epithelial surface, destruction of crypts, and infiltration of immunocytes.
The staining of MUC2 (anti-MUC2, GB11344, Servicebio, Wuhan, China) in colon tissue for immunohistochemical analysis was performed following a standard protocol. MUC2-positive areas were quantified calculating 10 fields per mouse.
4.4. Real-time quantitative polymerase chain reaction (RT-PCR)
Total RNA was extracted from colon tissues using TRIzol reagent (GenStar, Beijing, China). Reverse transcription of each sample was performed using the HiScript II 1st Strand cDNA Synthesis Kit (R211; Vazyme, Nanjing, China). RT-PCR was performed using the Taq Pro Universal SYBR qPCR Master Mix (P211, Vazyme, Nanjing, China). The primer sequences are listed in Supplementary Material Table S2.
4.5. Flow cytometry
Fresh colon tissue was dissociated into a single-cell suspension for the detection of the T cell proportion by flow cytometry. Briefly, the tissue was cut into pieces and incubated with RPMI 1640 medium (BBI, Shanghai, China), which contained 5 mmol/L DTT (Beyotime Biotechnology, Shanghai, China), 1 mmol/L EDTA (TGREGA, Beijing, China), and 5% HyClone™ fetal bovine serum (Cytiva, United States) for 40 min at 37°C. After washing with medium twice, the colon tissue was incubated with 1 mg/mL collagenase I and 0.1 mg/mL DNase I in RPMI 1640 medium for 30 min, and the cell suspension was then filtered through a 70 μm nylon mech into a 50 ml centrifuge tube. After centrifugation at 500 g for 10 min, the supernatant was aspirated. The cell pellet was a single cell that we needed. Details of the straining method are described in an article by Han 2022.38 After sample preparation, a Flow Cytometer (BD Biosciences) was used for sample analysis. The FlowJo_V10 software was used for data analysis. Tregs were identified as CD45+ CD4+ FOXP3+ cells. Th17 cells were identified as CD45+ CD4+ IL17A+.
4.6 Analysis of Inflammatory cytokines
The IL10 and IL17A level in mouse serum were detected using ELISA kits (IL10:HEA056Mu, IL17A:HEA063Mu, Cloud-Clone, Wuhan, China) according to the manufacturer’s instructions.
4.7. 16S rRNA gene sequencing and analysis
The analysis of the gut microbiome was performed as in our previous study.38 After DNA samples were extracted from cecal contents, the V3–V4 hypervariable regions of 16S rRNA genes were amplified, and PCR was conducted using Phusion High-Fidelity PCR Master Mix (New England Biolabs, USA). Sequencing libraries were generated using the Illumina TruSeq DNA PCR-Free Library Preparation Kit (Illumina, USA), following the manufacturer’s recommendations, and index codes were added. Library quality was assessed on a Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina Nova Seq platform and 250 bp paired-end reads were generated. Sequences with > 97% similarity were assigned to the same OTUs.
4.8. Untargeted metabolomics
Tissues (100 mg) were individually ground with liquid nitrogen, and the homogenate was resuspended in prechilled 80% methanol by vortexing. The samples were incubated on ice for 5 min and then centrifuged at 15,000 g at 4°C for 20 min. Some of the supernatant was diluted to a final concentration of 53% methanol using LC-MS grade water. The samples were subsequently transferred to fresh Eppendorf tubes and centrifuged at 15,000 g at 4°C for 20 min. Finally, the supernatant was injected into the LC-MS/MS system analysis.39
UHPLC-MS/MS analyses were performed using a Vanquish UHPLC system (ThermoFisher, Germany) coupled with an Orbitrap Q ExactiveTMHF-X mass spectrometer (Thermo Fisher,Germany) at Novogene Co., Ltd. (Beijing, China). Statistical analyses were performed using statistical software R (R version R-3.4.3), Python (Python 2.7.6 version), and CentOS (CentOS release 6.6). These metabolites were annotated using the KEGG, HMDB, and LIPID Map databases.
5. Statistical analysis
All results are shown as mean ±SEM (Prism 8; GraphPad Software). The data were analyzed by unpaired t-test using SPSS software (SPSS 19.0, Chicago, USA). At least three independent experiments were performed for each group. p < 0.05 was considered statistically significant. Metabolites with VIP > 1 and p-value <0.05, and fold change ≥ 2 or FC ≤ 0.5 were considered differential metabolites. Volcano plots were used to filter metabolites of interest based on log2(Fold Change) and -log10(p-value) of metabolites using ggplot2 in R language.
Supplementary Material
Acknowledgments
We appreciate Xiang Zhou for advice on histopathology.
Funding Statement
This work was supported by National Natural Science Foundation of China [Grant no. 82300733]; National Natural Science Foundation of China [Grant no. 82330017]; National Natural Science Foundation of China [Grant no. 81930015] and Golden Seed Research Fund [Grant no. CYJZ202222].
Disclosure statement
No potential conflict of interest was reported by the author (s).
Ethics approval and consent to participate
Animal experiments were conducted in accordance with the Institutional Animal Care and Use Committee of Capital Medical University, and were performed in accordance with the ARRIVE guidelines.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2024.2329147.
References
- 1.Agrawal M, Allin KH, Petralia F, Colombel JF, Jess T.. Multiomics to elucidate inflammatory bowel disease risk factors and pathways. Nat Rev Gastro Hepat. 2022;19(6):399–13. doi: 10.1038/s41575-022-00593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastro Hepat. 2015;12(12):720–7. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 3.Mao R, Chen M. Precision medicine in IBD: genes, drugs, bugs and omics. Nat Rev Gastro Hepat. 2022;19(2):81–2. doi: 10.1038/s41575-021-00555-w. [DOI] [PubMed] [Google Scholar]
- 4.Lee M, Chang EB. Inflammatory bowel diseases (IBD) and the microbiome-searching the crime scene for clues. Gastroenterology. 2021;160(2):524–537. doi: 10.1053/j.gastro.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126(6):1620–33. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 6.Shan Y, Lee M, Chang EB. The gut microbiome and inflammatory bowel diseases. Annu Rev Med. 2022;73(1):455–68. doi: 10.1146/annurev-med-042320-021020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RWL, Connor S, Ng W, Paramsothy R. et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389(10075):1218–28. doi: 10.1016/S0140-6736(17)30182-4. [DOI] [PubMed] [Google Scholar]
- 8.Paramsothy S, Nielsen S, Kamm MA, Deshpande NP, Faith JJ, Clemente JC, Paramsothy R, Walsh AJ, van den Bogaerde J, Samuel D. et al. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology. 2019;156(5):1440–54.e2. doi: 10.1053/j.gastro.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Russo E, Giudici F, Fiorindi C, Ficari F, Scaringi S, Amedei A. Immunomodulating activity and therapeutic effects of short chain fatty acids and tryptophan post-biotics in Inflammatory bowel disease. Front Immunol. 2019;10:2754. doi: 10.3389/fimmu.2019.02754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastro Hepat. 2020;17(4):223–37. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 11.Sun M, Ma N, He T, Johnston LJ, Ma X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit Rev Food Sci Nutr. 2020;60(10):1760–8. doi: 10.1080/10408398.2019.1598334. [DOI] [PubMed] [Google Scholar]
- 12.Hendrikx T, Duan Y, Wang Y, Oh JH, Alexander LM, Huang W, Stärkel P, Ho SB, Gao B, Fiehn O. et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 2019;68(8):1504–15. doi: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji Y, Gao Y, Chen H, Yin Y, Zhang W. Indole-3-acetic acid alleviates nonalcoholic fatty liver disease in mice via attenuation of hepatic lipogenesis, and oxidative and inflammatory stress. Nutrients. 2019;11(9):2062. doi: 10.3390/nu11092062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Sun S, Liang L, Lou C, He Q, Ran M, Zhang L, Zhang J, Yan C, Yuan H. et al. Role of the aryl hydrocarbon receptor and gut microbiota-derived metabolites indole-3-acetic acid in sulforaphane alleviates hepatic steatosis in mice. Front Nutr. 2021;8:756565. doi: 10.3389/fnut.2021.756565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geremia A, Biancheri P, Allan P, Corazza GR, Di Sabatino A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev. 2014;13(1):3–10. doi: 10.1016/j.autrev.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ. et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4(2):293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayo B, Vázquez L, AB F. Equol: a bacterial metabolite from the daidzein isoflavone and its presumed beneficial health effects. Nutrients. 2019;11(9):2231. doi: 10.3390/nu11092231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Setchell KD, Clerici C. Equol: history, chemistry, and formation. J Nutr. 2010;140(7):1355s–1362s. doi: 10.3945/jn.109.119776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mustafa SE, Mustafa S, Abas F, Manap M, Ismail A, Amid M, Elzen S. Optimization of culture conditions of soymilk for equol production by Bifidobacterium breve 15700 and Bifidobacterium longum BB536. Food Chem. 2019;278:767–772. doi: 10.1016/j.foodchem.2018.11.107. [DOI] [PubMed] [Google Scholar]
- 20.Clough JN, Omer OS, Tasker S, Lord GM, Irving PM. Regulatory T-cell therapy in Crohn’s disease: challenges and advances. Gut. 2020;69(5):942–52. doi: 10.1136/gutjnl-2019-319850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28(4):546–58. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh C-S. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobse J, Li J, Rings E, Samsom JN, Goettel JA. Intestinal regulatory T cells as specialized tissue-restricted immune cells in intestinal immune homeostasis and disease. Front Immunol. 2021;12:716499. doi: 10.3389/fimmu.2021.716499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, Geva-Zatorsky N, Jupp R, Mathis D, Benoist C. et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature. 2020;577(7790):410–415. doi: 10.1038/s41586-019-1865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shim JA, Ryu JH, Jo Y, Hong C. The role of gut microbiota in T cell immunity and immune mediated disorders. Int J Biol Sci. 2023;19(4):1178–91. doi: 10.7150/ijbs.79430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bui TI, Gill AL, Mooney RA, Gill SR, Auchtung JM. Modulation of gut microbiota metabolism in obesity-related type 2 diabetes reduces osteomyelitis severity. Microbiol Spectr. 2022;10(2):e0017022. doi: 10.1128/spectrum.00170-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo W, Mao B, Cui S, Tang X, Zhang Q, Zhao J, Zhang H. Protective effects of a novel probiotic bifidobacterium pseudolongum on the intestinal barrier of colitis mice via modulating the Pparγ/STAT3 pathway and intestinal microbiota. Foods. 2022;11(11):1551. doi: 10.3390/foods11111551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paterni I, Granchi C, Katzenellenbogen JA, Minutolo F. Estrogen receptors alpha (ERα) and beta (ERβ): subtype-selective ligands and clinical potential. Steroids. 2014;90:13–29. doi: 10.1016/j.steroids.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carmeci C, Thompson DA, Ring HZ, Francke U, Weigel RJ. Identification of a gene (GPR30) with homology to the G-protein-coupled receptor superfamily associated with estrogen receptor expression in breast cancer. Genomics. 1997;45(3):607–17. doi: 10.1006/geno.1997.4972. [DOI] [PubMed] [Google Scholar]
- 30.Wang WJ, Liu FJ, Xin L, Hao CF, Bao HC, Qu QL, Liu X-M. Adoptive transfer of pregnancy-induced CD4+CD25+ regulatory T cells reverses the increase in abortion rate caused by interleukin 17 in the CBA/J×BALB/c mouse model. Hum Reprod. 2014;29(5):946–952. doi: 10.1093/humrep/deu014. [DOI] [PubMed] [Google Scholar]
- 31.Adurthi S, Kumar MM, Vinodkumar HS, Mukherjee G, Krishnamurthy H, Acharya KK, Bafna UD, Uma DK, Abhishekh B, Krishna S. et al. Oestrogen Receptor-α binds the FOXP3 promoter and modulates regulatory T-cell function in human cervical cancer. Sci Rep. 2017;7(1):17289. doi: 10.1038/s41598-017-17102-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science. 2005;307(5715):1625–30. doi: 10.1126/science.1106943. [DOI] [PubMed] [Google Scholar]
- 33.Warner M, Huang B, Gustafsson J-A. Estrogen Receptor β as a Pharmaceutical Target. Trends Pharmacol Sci. 2017;38(1):92–9. doi: 10.1016/j.tips.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Saleiro D, Murillo G, Benya RV, Bissonnette M, Hart J, Mehta RG. Estrogen receptor-β protects against colitis-associated neoplasia in mice. Int J Cancer. 2012;131(11):2553–61. doi: 10.1002/ijc.27578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo D, Liu X, Zeng C, Cheng L, Song G, Hou X, Zhu L, Zou K. Estrogen receptor β activation ameliorates DSS-induced chronic colitis by inhibiting inflammation and promoting treg differentiation. Int Immunopharmacol. 2019;77:105971. doi: 10.1016/j.intimp.2019.105971. [DOI] [PubMed] [Google Scholar]
- 36.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8(6):e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang YJ, Kim WK, Han DH, Lee K, Ko G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes. 2019;10(6):696–711. doi: 10.1080/19490976.2019.1589281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han X, Li M, Sun L, Liu X, Yin Y, Hao J, Zhang W. p-Hydroxybenzoic Acid Ameliorates Colitis by Improving the Mucosal Barrier in a Gut Microbiota-Dependent Manner. Nutrients. 2022;14(24):5383. doi: 10.3390/nu14245383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Want EJ, Masson P, Michopoulos F, Wilson ID, Theodoridis G, Plumb RS, Shockcor J, Loftus N, Holmes E, Nicholson JK. et al. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat Protoc. 2013;8(1):17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


