Abstract
Coreceptor usage by Envs from diverse primary human immunodeficiency virus type 1 isolates was analyzed by a vaccinia virus-based expression and assay system. Usage of recombinant CCR5 and CXCR4 correlated closely with fusogenicity toward macrophages and T-cell lines expressing endogenous coreceptors. Surprisingly, recombinant CCR3 was utilized by most primary and T-cell-line-adapted Envs. Endogenous CXCR4 in macrophages was functional as a coreceptor.
Specific chemokine receptors are now recognized as the elusive coreceptors mediating the membrane fusion events underlying human immunodeficiency virus type 1 (HIV-1) entry into CD4+ target cells (reviewed in reference 5). The first coreceptor, identified by a functional cDNA cloning strategy, was designated fusin to denote its fusion coreceptor activity (24); it was subsequently renamed CXCR4 when it was demonstrated to be a receptor for the CXC chemokine stromal cell-derived factor 1 (7, 40). The molecule was shown to function preferentially for T-cell-line (TCL)-adapted HIV-1 strains (24). This discovery, coupled with an earlier report that the CC chemokines RANTES, MIP-1α, and MIP-1β suppress infection by prototypic macrophage-tropic (M-tropic) HIV-1 isolates (13), led several groups to independently identify a CC chemokine receptor, CCR5, as the major coreceptor for such strains (4, 12, 18, 20, 21). Coreceptor activity has also been observed with other CC chemokine receptors, including CCR3 and CCR2b (12, 20) and CCR8 (4a, 44), as well as with three chemokine receptor-like orphan proteins, one designated STRL33 (33) or BONZO (17) a second designated BOB (17) or GPR15 (23, 30), and a third designated V28 (44). The specificities of different Envs for this array of coreceptors, coupled with the patterns of endogenous coreceptor expression on various CD4+ target cell types, are major determinants mediating the cytotropisms of different HIV-1 strains.
In the present study we examined molecularly cloned Envs from a panel of primary HIV-1 isolates representing diverse phenotypes and genetic subtypes (clades) (26). These isolates were collected at epicenters of the global AIDS pandemic by the World Health Organization and National Institute of Allergy and Infectious Diseases Networks for HIV Isolation and Characterization, with the goal of generating a panel of naturally occurring primary strains to facilitate structure and function studies and vaccine development. Our findings reveal some unexpected activity patterns with recombinant and endogenous coreceptors and highlight the importance of defining the variables that contribute to the contrasting results obtained with different experimental systems.
Identification of functional Envs.
The primary env genes (26) were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (Rockville, Md.), except for clone 92UG037, which was kindly provided by B. Hahn (University of Alabama, Birmingham). Each was cloned into plasmid pCRII, which contains the bacteriophage T7 promoter. For Envs from prototypic strains, we used the following plasmids derived from pSC59, which contains a synthetic early-late vaccinia virus promoter (11a): pCB-41 (8), TCL-adapted LAV Env; pCB-43 (8), M-tropic Ba-L Env; pGA13-89.6 (2a), dual-tropic primary 89.6 Env; and pCB-16 (8), nonfusogenic uncleavable (Unc) Env (8). To prepare effector cells, HeLa cell monolayers were transfected with Env-containing plasmids with DOTAP (Boehringer Mannheim, Indianapolis, Ind.) and then trypsinized and infected in suspension with vP11T7gene1, which contains the bacteriophage T7 RNA polymerase gene linked to a natural late vaccinia virus promoter (1). To assess fusogenic activities, we used a quantitative cell fusion assay as previously described (39), with target cells infected with vCB-21R containing the lacZ gene linked to the T7 promoter (3).
Peripheral blood mononuclear cells (PBMCs) were chosen as targets to identify functional Envs. Of the 31 primary Envs examined, 19 displayed detectable fusion with PBMCs (as well as with the PM1 cell line [data not shown]). Table 1 shows the active Envs, all of which routinely gave values at least threefold over the activity observed with the nonfusogenic Unc Env; their reported non-syncytium-inducing (NSI) versus syncytium-inducing (SI) phenotypes (26) are also indicated. To facilitate comparisons of activities against PBMCs in different experiments, the fusion activity observed with the prototypic LAV Env in each experiment was assigned a value of 100% and the activities of the other Envs were expressed as relative values. The 19 functional primary Envs showed wide variations in relative fusogenic strengths with PBMC targets. Our results are in general agreement with and expand upon a previous study in which the activities of many of these Envs were scored by infectivity assays with pseudotyped virions containing the chloramphenicol acetyltransferase reporter gene (26); we identified five additional fusogenic Envs not analyzed in the previous report (92US711.14, 92US716.6, 93ZR001.3, 92TH022.4, and 93BR019.10).
TABLE 1.
Fusogenic activities of each Env with cells expressing endogenous coreceptors
| Cladeb | Isolate (World Health Organization)b | MT-2 assay phenotypeb,c | Relative Env-mediated fusion against cells expressing endogenous coreceptors (%)a
|
||
|---|---|---|---|---|---|
| PBMCs | Macrophagesd | Jurkat | |||
| Prototypic | |||||
| B | LAV | SI | 100 | 14 | 100 |
| B | Ba-L | NSI | 72 | 100 | 0 |
| B | 89.6 | SI | 99 | 287 | 274 |
| Primary | |||||
| A | 92UG037.8 | NA | 2 | 17 | 0 |
| B | 91US005.11 | NSI | 74 | 319 | 10 |
| 92US711.14 | NSI | 5 | 3 | 0 | |
| 91US712.4 | SI | 2 | 25 | 0 | |
| 92US715.6 | NSI | 98 | 205 | 2 | |
| 92US716.6 | NSI | 54 | 152 | 2 | |
| 92HT593.1 | NSI | 160 | 104 | 71 | |
| 92TH014.12 | NSI | 94 | 109 | 2 | |
| 92BR020.4 | NSI | 100 | 74 | 1 | |
| C | 93MW965.26 | NSI | 98 | 88 | 1 |
| 92BR025.9 | NSI | 41 | 91 | 1 | |
| D | 92UG021.16 | SI | 74 | 30 | 74 |
| 92UG024.2 | SI | 171 | 104 | 123 | |
| 93ZR001.3 | NA | 107 | 109 | 78 | |
| E | 93TH966.8 | NSI | 19 | 6 | 0 |
| 92TH022.4 | NSI | 95 | 107 | 6 | |
| F | 93BR029.2 | NA | 25 | 51 | 0 |
| G | 92UG975.10 | NSI | 22 | 29 | 2 |
| F/B | 93BR019.10 | NA | 58 | 273 | 6 |
LAV Env fusion with PBMCs and Jurkat cells was arbitrarily set at 100%, as was Ba-L Env fusion with macrophages. The fusion exhibited by each Env against a particular cell type was scored relative to these standards.
From reference 26.
SI, syncytium-inducing; NSI, non-syncytium-inducing; NA, not available.
Env effector cells (103) were mixed with 105 macrophages expressing vaccinia virus-encoded CD4.
CCR5, CXCR4, and CCR3 usage by primary Envs.
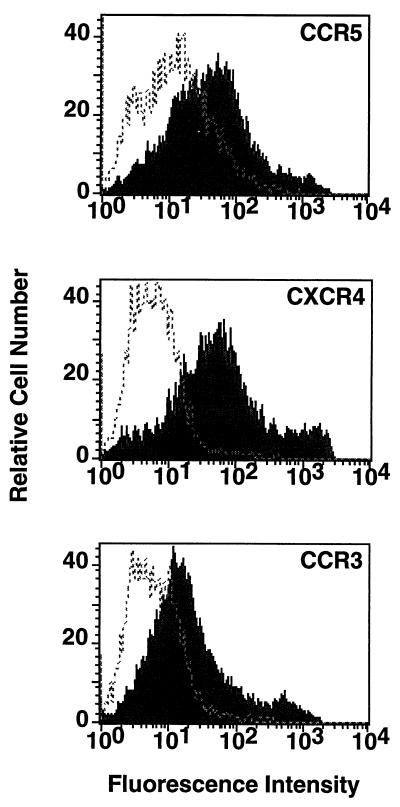
The vaccinia virus system was used to coexpress CD4 and coreceptors on target cells. NIH 3T3 cell monolayers were transfected (with DOTAP) with pSC59-based plasmids containing the chemokine receptor sequences as follows. For CXCR4, pYF1-fusin (24) was used; for CCR5, pGA9-CKR5 (4) was used; and for CCR3, a previously described plasmid (2), herein designated pGA12-CCR3, was used. The cells were then trypsinized and coinfected in suspension with vCB-3 encoding CD4 (9) and vCB-21R. The flow cytometry analysis shown in Fig. 1 indicates that CCR5, CXCR4, and CCR3 all were expressed at the cell surface. The effectiveness of this experimental system for coreceptor expression is critical for the analyses presented below.
FIG. 1.
Surface expression of coreceptors. NIH 3T3 cells were transfected with pSC59-based plasmids containing a synthetic vaccinia virus promoter linked to the indicated coreceptor genes and coinfected with vCB-3 (CD4) and vCB21R-LacZ; control cells were transfected with the empty pSC59 plasmid and infected identically. Following overnight incubation to allow expression of vaccinia virus-encoded proteins, the cells were stained with the corresponding antibodies as follows: for CCR5, rabbit polyclonal antisera against a synthetic peptide representing the CCR5 extracellular N terminus (1:50 dilution) (4); for CXCR4, the 12G5 monoclonal antibody (23 μg/ml) (22), donated by J. Hoxie, University of Pennsylvania; and for CCR3, the 7B11 monoclonal antibody (20 μg/ml) (29), donated by C. Mackay, Leukosite. Detection was achieved with the following secondary antibodies (10 μg/ml; Boehringer Mannheim): for CCR5, fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G; and for CXCR4 and CCR3, fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G. The cells were washed, treated with 1 μg of ethidium bromide homodimer per ml, fixed with 0.1 ml of 4% paraformaldehyde, and analyzed with a FACSCAN flow cytometer (Becton Dickinson, Menlo Park, Calif.). Analyses of forward and side scatter as well as ethidium bromide homodimer fluorescence indicated nearly homogeneous populations of viable cells. Shaded profiles indicate coreceptor-expressing cells; unshaded profiles indicate control cells transfected with the empty pSC59 plasmid. The mean fluorescence intensities with coreceptor-expressing versus control cells were as follows: CCR5, 92 versus 21; CXCR4, 167 versus 4; and CCR3, 57 versus 9. In separate experiments, similar distinctions were seen when coreceptor-expressing cells were stained with either an anticoreceptor monoclonal antibody or an isotype-matched control monoclonal antibody (data not shown).
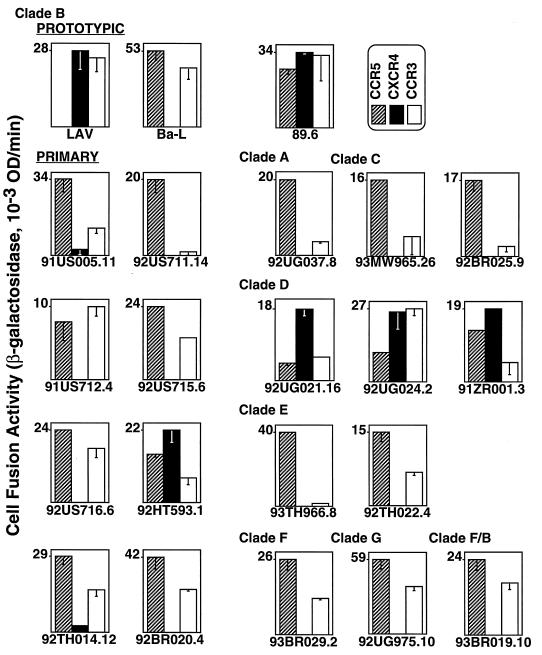
The Envs that displayed fusogenic activity against PBMCs were analyzed for their ability to use specific recombinant chemokine receptors (Fig. 2). Each Env was analyzed for its relative activity with the target cells expressing the indicated recombinant coreceptor. By this type of comparison, the coreceptor preference of each Env could be assessed without complications introduced by the wide variations in the intrinsic fusogenicities of the different Envs discussed above (note the different maximal values on the y axes for each Env shown in Fig. 2). When activities with CCR5 were compared to those with CXCR4, Envs from the prototypic strains showed preferences consistent with previous reports (4, 12, 18, 20, 21, 24). The TCL-adapted LAV Env efficiently used CXCR4 but not CCR5, the M-tropic Ba-L Env used CCR5 but not CXCR4, and the 89.6 Env used each coreceptor with comparable efficiency. The Envs from the primary strains all used CCR5, and most of them used it relatively efficiently. Six primary Envs also showed some activity with CXCR4; four used CXCR4 as well as or better than CCR5 (92HT593.1, 92UG021.16, 92UG024.2, and 93ZR001.3), whereas two used CXCR4 relatively weakly compared to CCR5 (91US005.11 and 92TH014.12). Our results with the vaccinia virus-based cell fusion assay are in agreement with recently published studies with an infectivity assay with pseudotyped virions containing the luciferase reporter gene (6, 17); similar patterns of CCR5 versus CXCR4 preference were found for the 12 primary Envs tested by both assay systems. Although in the present study we did not observe primary Envs with exclusive usage of CXCR4 compared to CCR5, even for isolates previously reported to have this restrictive profile, our finding of Envs with preferential usage of CXCR4 over CCR5 is consistent with other reports (6, 16, 44, 46, 49).
FIG. 2.
Fusogenic activities of each Env with recombinant coreceptors. Target NIH 3T3 cells coexpressing coreceptors and CD4 (and containing the lacZ gene linked to the T7 promoter) as well as control cells lacking coreceptors were prepared as described in the legend for Fig. 1. Effector HeLa cells were transfected with plasmids encoding the indicated Envs and then infected with vP11T7gene1 (T7 polymerase). After overnight incubation to allow recombinant protein expression, cells were mixed and fusion was scored after 3 h. The low background values for each Env obtained with the control target cells expressing CD4 but no coreceptors were subtracted to give the data shown. Error bars indicate the sample standard deviations of the mean values obtained from duplicate samples. OD, optical density.
In contrast with these concordant results for CCR5 and CXCR4 usage, our findings with CCR3 show major differences from studies by other groups. We observed efficient CCR3 usage by most primary Envs (exceptions are 92US711.14 and 93TH966.8) as well as by the TCL-adapted LAV Env (Fig. 2) (and the IIIB Env but not the RF Env [2]). These positive results differ markedly from published findings on assays of HIV infectivity or cell fusion. While CCR3 has been reported to function for some M-tropic isolates and the dual-tropic 89.6 isolate (12, 16, 18, 20, 28), activity was not found for any of the TCL-adapted strains examined, including IIIB and NL4-3 (which contains the LAV Env), and was observed only infrequently for primary NSI or SI isolates (6, 12, 16, 18–21, 28, 49). We believe that the major basis for these discrepancies lies in the varied effectiveness of coreceptor expression by different experimental methods. Particular difficulties have been encountered in attempts to express CCR3 (but not CCR5 or CXCR4) from nuclear promoters (12, 37a, 41). In the published experiments showing infrequent CCR3 usage with nuclear expression systems, surface levels of this coreceptor either were not measured (6, 16, 18–21, 49) or were found at only barely detectable levels (12). With the vaccinia virus system, which drives transient gene expression from promoters in the cytoplasm, we have clearly demonstrated CCR3 surface expression (Fig. 1). We also observed that when the vaccinia virus inhibitor cytosine arabinoside (AraC) was used to greatly reduce surface CCR3 expression, LAV Env-mediated fusion was still detected, albeit at a reduced level; thus, the coreceptor activity of CCR3 is revealed by the vaccinia virus system, even under suboptimal expression conditions. Our interpretation that the restrictive use of recombinant CCR3 previously reported by many groups is due to inadequate expression of this coreceptor is supported by a recent study describing experimental variables that influence CCR3 expression from a nuclear promoter. Under conditions in which surface CCR3 was elevated to significant levels, cell fusion was revealed with many Envs, including that from the TCL-adapted IIIB (but not RF) strain (44).
Correlation between coreceptor usage profiles and fusion specificities for natural target cells: evidence for functional CXCR4 on macrophages.
We next tested the ability of Envs from the primary isolates to mediate fusion with natural human target cell types known to be differentially capable of supporting infection by different classes of HIV-1 isolates. Effector cells expressing the indicated vaccinia virus-encoded Envs (and T7 RNA polymerase) were mixed with either primary macrophages or the Jurkat TCL (containing the lacZ gene). Macrophages were prepared by countercurrent centrifugation, elutriation of PBMCs, and differentiation of the monocyte fraction for 20 days in bacteriological plates in the absence of exogenous cytokines according to a previously described method (32); we have previously verified the macrophage phenotypic markers and homogeneity of these cells (10). In Table 1, the data for each cell type were normalized: a value of 100% was assigned to the β-galactosidase activity obtained with Ba-L Env in macrophages or LAV Env in Jurkat cells. Nearly all of the primary Envs displayed some capacity to mediate fusion with macrophages; only 2 of 19 gave <10% of the activity observed with the prototypic M-tropic Ba-L Env (92US711.14 and 93TH966.8; note that these Envs were inherently weak as judged by their low fusogenic activities against PBMC targets). By contrast, a minority of the primary Envs showed significant activity with Jurkat cells; only 4 of 19 gave >10% of the activity observed with the prototypic TCL-adapted LAV Env (92HT593.1, 92UG021.6, 92UG024.2, and 93ZR001.3). The four primary Envs that mediated fusion with Jurkat cells and efficiently used CXCR4 (but not the other Envs) contained basic amino acid residues at positions 11 and 25 or 27 of the V3 loop (26); such basic residues in V3 have previously been associated with TCL tropism (see citations in reference 5).
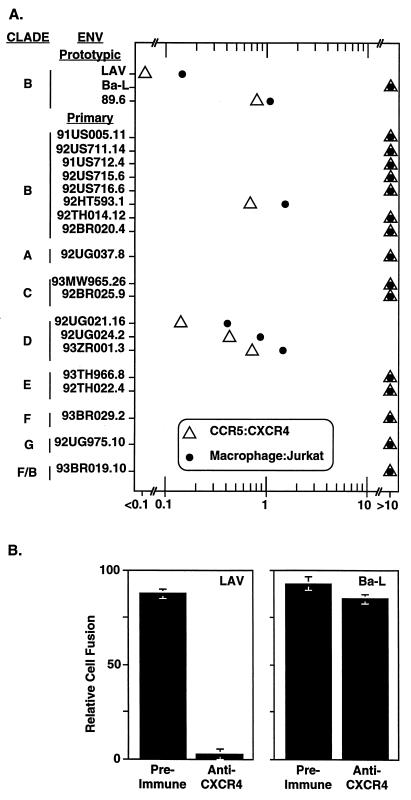
For each Env, the fusion activities shown in Fig. 2 were used to compute the CCR5-to-CXCR4 activity ratios; the relative fusion activities in Table 1 were used to derive the macrophage-to-Jurkat-cell activity ratios. The data shown in Fig. 3A reveal a close relationship between the coreceptor usage of each primary Env and its ability to mediate fusion with natural target cells. Most (15 of the 19) primary Envs strongly preferred CCR5 to CXCR4 and gave high relative activities with macrophages compared to Jurkat cells. The four primary Envs that gave comparable fusion values with CCR5 and CXCR4 also gave comparable relative fusion activities with macrophages and Jurkat cells. Only the prototypic LAV Env, derived from a strongly TCL-adapted strain, functioned well with CXCR4 but completely failed to use CCR5; this Env gave the lowest macrophage-to-Jurkat-cell activity score, although significant fusion with macrophages was observed (see below).
FIG. 3.
(A) Correlation between coreceptor usage profiles and fusion specificities for natural target cells. For each Env, the ratio of the fusion activity with CCR5 versus CXCR4 was calculated from the data shown in Fig. 2 (open triangles); the ratio of the relative fusion activity obtained with macrophage versus Jurkat cell targets was calculated from the data in Table 1 (closed circles). All ratio values below 0.1 or above 10 were grouped together. (B) Functional CXCR4 coreceptor on macrophages. Macrophages coinfected with vTF7-3 (T7 RNA polymerase) and vCB-3 (CD4) were preincubated for 45 min at 37°C without antibody or with 1 mg of preimmune or immune immunoglobulin per ml purified from a rabbit immunized with a peptide representing the extracellular N terminus of CXCR4 (24). Effector HeLa cells were coinfected with vCB21RLacZ and either vCB-41 (LAV Env), vCB-43 (Ba-L Env), or vCB-16 (Unc Env). The Ba-L Env infection was performed with AraC to reduce the fusion activity so that it was comparable to that of the LAV Env; similar results were obtained when the Ba-L Env was expressed without AraC (i.e., no inhibition; data not shown). For the LAV and Ba-L Envs, the minimal values obtained with Unc Env were subtracted and the results are expressed as the percentage of activity obtained in the absence of antibody (set at 100%). Error bars indicate the sample standard deviations of the mean values obtained from duplicate samples.
Our results also show a good, though not absolute, correlation between coreceptor usage and the infection phenotypes for the 15 primary strains previously characterized as NSI or SI in the MT-2 assay (Table 1), similar to findings of others (6, 16, 44, 46, 49). Of the Envs from the 12 isolates designated NSI, 11 showed a strong preference for CCR5 over CXCR4; the exception was 92HT593.1, which used both coreceptors comparably. Of the Envs from the three strains characterized as SI, two used both CXCR4 and CCR5; the exception was 91US712.4, which was previously designated SI but was specific for CCR5. It should be noted that the NSI versus SI characterization of these isolates was performed with uncloned virus populations; thus, it is possible that the few examples in which the phenotypes did not correlate with the CCR5/CXCR4 usage profiles were due to cloning of Envs from minor variants within these populations.
The results presented above can be considered in terms of the patterns of endogenous coreceptor expression in the human target cell types. Jurkat cells and many other continuous human TCLs express CXCR4 (24, 34, 35, 38) but negligible levels of CCR5 (4, 45); this expression pattern is consistent with the ability of many human TCLs to support TCL-adapted and dual-tropic isolates but not M-tropic strains. Interpreting findings with primary macrophages is more complex. CCR5 has been detected in these cells by both mRNA and protein analyses (4, 35, 37, 43, 48), and various lines of evidence suggest that CCR5 expression in macrophages accounts for their susceptibility to M-tropic HIV-1 isolates (4, 15, 42, 46, 48). However, the resistance of macrophages to infection by TCL-adapted strains is puzzling, since CXCR4 mRNA and protein have been detected in these cells (2a, 35, 37), although the levels are low compared to TCLs and can vary with culture conditions (35). Suggestions have been offered to explain anomalies of infection resistance of certain cell types despite coreceptor expression, including the possibility that virus entry mechanisms are fundamentally distinct in different CXCR4+ target cells (37) or that cell type-dependent variations in processing or presentation of a chemokine receptor influence its functionality as an HIV coreceptor (19, 46). These hypotheses fail to explain the inability of TCL-adapted strains to productively infect macrophages, since Envs from such isolates can mediate cell fusion with these cells (albeit less efficiently than Envs from M-tropic isolates [Table 1] [8]). To test whether this fusion is mediated by CXCR4, we examined the effects of a polyclonal anti-CXCR4 antibody on fusion by a TCL-adapted Env with macrophage targets. The results, shown in Fig. 3B, indicate that LAV Env-mediated fusion was strongly inhibited by the anti-CXCR4 antibody; by contrast, fusion mediated by the M-tropic Ba-L Env was unaffected. These results clearly demonstrate that the CXCR4 endogenously expressed on macrophages can function as an HIV-1 coreceptor.
Multiple factors probably contribute to the contrasting activities of TCL-adapted Envs with macrophages in the cell fusion assay versus the minimal activity of the corresponding HIV strains in productive infection assays. First, the resistance of macrophages to infection by TCL-adapted strains is not absolute; low-level infection has been reported by several groups (14, 25, 27, 35, 46, 47). Second, it has been demonstrated recently that the CXCR4 level in macrophages declines dramatically during 5 days in culture (35); this coreceptor may therefore be more available in the short-term cell fusion assay (3 h) than in the longer-term infectivity assays (several days). Third, the surface densities of coreceptors and/or CD4 on different target cell types may critically influence permissiveness (31, 44, 48), and threshold effects might have different consequences for cell fusion versus virus infection assay systems; however, we have shown that vaccinia virus-mediated augmentation of the low endogenous CD4 level on macrophages enhances overall cell fusion but does not influence the relative activities of TCL-adapted versus M-tropic Envs (8). Finally, while numerous studies have suggested that restriction at the level of the membrane fusion reactions involved in virus entry is a major factor underlying the inability of these cells to support productive replication by TCL-adapted HIV-1 strains, blocks at postentry steps of the viral replication cycle may also contribute to the limited productive replication of TCL-adapted strains in macrophages (see citations in reference 5). This notion has been suggested for both HIV-1 and the related simian immunodeficiency virus; curiously, Env (36) and coreceptors (11) have been reported to contribute to infection efficiency and tropism by unidentified postentry mechanisms.
Given these variables, we acknowledge the previous caution (19) that reactivity of a particular Env with a recombinant coreceptor does not necessarily imply that the corresponding virus will productively infect any CD4+ cell type expressing that coreceptor, even if the fusion and entry step can occur. Resolution of the discrepancies obtained in alternate expression and assay systems must await detailed analyses of the multiple variables that differentially influence the readouts. These issues assume increasing importance as studies are extended to quantitate the coreceptor usage patterns of diverse Envs with natural CD4+ target cells and are critical for applying knowledge of coreceptors to the broader problems of HIV transmission and pathogenesis and to the development of novel vaccine and therapeutic strategies.
Acknowledgments
This study was funded in part by the NIH Intramural AIDS Targeted Antiviral Program. H.A.B. is an Advanced Scholar in the HHMI-NIH Research Scholars Program. G.A. is partially supported by the Dr. Nathan Davis Award from the American Medical Association Education and Research Foundation to E. A. Berger.
We thank B. Hahn for donation of the 92UG037 Env clone, J. Hoxie for the 12G5 monoclonal antibody, and C. Mackay for the 7B11 monoclonal antibody.
REFERENCES
- 1.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Berger E A, Murphy P M, Pease J E. Determinants of HIV-1 coreceptor function on CC chemokine receptor 3: importance of both extracellular and transmembrane/cytoplasmic regions. J Biol Chem. 1997;272:20420–20426. doi: 10.1074/jbc.272.33.20420. [DOI] [PubMed] [Google Scholar]
- 2a.Alkhatib, G., and E. A. Berger. Unpublished data.
- 3.Alkhatib G, Broder C C, Berger E A. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J Virol. 1996;70:5487–5494. doi: 10.1128/jvi.70.8.5487-5494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 4a.Bazan, H., G. Alkhatib, H. Tiffany, T. Bonner, P. Murphy, and E. Berger. Submitted for publication.
- 5.Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11(Suppl. A):S3–S16. [PubMed]
- 6.Bjorndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyö E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 8.Broder C C, Berger E A. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broder C C, Dimitrov D S, Blumenthal R, Berger E A. The block to HIV-1 envelope glycoprotein-mediated membrane fusion in animal cells expressing human CD4 can be overcome by a human cell component(s) Virology. 1993;193:483–491. doi: 10.1006/viro.1993.1151. [DOI] [PubMed] [Google Scholar]
- 10.Broder C C, Kennedy P E, Michaels F, Berger E A. Expression of foreign genes in cultured human primary macrophages using recombinant vaccinia virus vectors. Gene. 1994;142:167–174. doi: 10.1016/0378-1119(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 11.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Chakrabarti S, Sisler J R, Moss B. Compact, synthetic vaccinia virus early/late promoter for protein expression. BioTechniques. 1997;23:1094–1097. doi: 10.2144/97236st07. [DOI] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L J, Mackay C R, Larosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Cocchi F, DeVico A L, Garzinodemo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 14.Collman R, Hassan N F, Walker R, Godfrey B, Cutilli J, Hastings J C, Friedman H, Douglas S D, Nathanson N. Infection of monocyte-derived macrophages with human immunodeficiency virus type 1 (HIV-1): monocyte-tropic and lymphocyte-tropic strains of HIV-1 show distinctive patterns of replication in a panel of cell types. J Exp Med. 1989;170:1149–1163. doi: 10.1084/jem.170.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connor R I, Paxton W A, Sheridan K E, Koup R A. Macrophages and CD4+ T lymphocytes from two multiply exposed, uninfected individuals resist infection with primary non-syncytium-inducing isolates of human immunodeficiency virus type 1. J Virol. 1996;70:8758–8764. doi: 10.1128/jvi.70.12.8758-8764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng H, Unutmaz D, Kewalramani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 18.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Dimarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 19.Dittmar M T, McKnight A, Simmons G, Clapham P R, Weiss R A, Simmonds P. HIV-1 tropism and co-receptor use. Nature. 1997;385:495–496. doi: 10.1038/385495a0. [DOI] [PubMed] [Google Scholar]
- 20.Doranz B J, Rucker J, Yi Y J, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 21.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y X, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4(+) cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 22.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by Fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 23.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 25.Fouchier R A M, Brouwer M, Kootstra N A, Huisman H G, Schuitemaker H. HIV-1 macrophage tropism is determined at multiple levels of the viral replication cycle. J Clin Invest. 1994;94:1806–1814. doi: 10.1172/JCI117529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao F, Morrison S G, Robertson D L, Thornton C L, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp P M, Shaw G M, Hahn B H the WHO and NIAID Networks for HIV Isolation and Characterization. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gartner S, Markovits P, Markovitz D M, Kaplan M H, Gallo R C, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 28.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 29.Heath H, Qin S, Rao P, Wu L, Larosa G, Kassam N, Ponath P D, Mackay C R. Chemokine receptor usage by human eosinophils. The importance of CCR3 demonstrated using an antagonistic monoclonal antibody. J Clin Invest. 1997;99:178–184. doi: 10.1172/JCI119145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heiber M, Marchese A, Nguyen T, Heng H H, George S R, O’Dowd B F. A novel human gene encoding a G-protein-coupled receptor (GPR15) is located on chromosome 3. Genomics. 1997;32:462–465. doi: 10.1006/geno.1996.0143. [DOI] [PubMed] [Google Scholar]
- 31.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lazdins J K, Woods-Cook K, Walker M, Alteri E. The lipophilic muramyl peptide MTP-PE is a potent inhibitor of HIV replication in macrophages. AIDS Res Hum Retroviruses. 1990;6:1157–1161. doi: 10.1089/aid.1990.6.1157. [DOI] [PubMed] [Google Scholar]
- 33.Liao F, Alkhatib G, Peden K W C, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loetscher M, Geiser T, O’Reilly T, Zwahlen R, Baggiolini M, Moser B. Cloning of a human seven-transmembrane domain receptor, LESTR, that is highly expressed in leukocytes. J Biol Chem. 1994;269:232–237. [PubMed] [Google Scholar]
- 35.McKnight A, Wilkinson D, Simmons G, Talbot S, Picard L, Ahuja M, Marsh M, Hoxie J A, Clapham P R. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J Virol. 1997;71:1692–1696. doi: 10.1128/jvi.71.2.1692-1696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori K, Ringler D J, Desrosiers R C. Restricted replication of simian immunodeficiency virus strain 239 in macrophages is determined by env but is not due to restricted entry. J Virol. 1993;67:2807–2814. doi: 10.1128/jvi.67.5.2807-2814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriuchi H, Moriuchi M, Combadiere C, Murphy P M, Fauci A S. CD8+ T-cell-derived soluble factor(s), but not β-chemokines RANTES, MIP-1α, and MIP-1β, suppress HIV-1 replication in monocyte/macrophages. Proc Natl Acad Sci USA. 1996;93:15341–15345. doi: 10.1073/pnas.93.26.15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Murphy, P. M. Personal communication.
- 38.Nomura H, Nielsen B W, Matsushima K. Molecular cloning of cDNAs encoding a LD78 receptor and putative leukocyte chemotactic peptide receptors. Int Immunol. 1993;5:1239–1249. doi: 10.1093/intimm/5.10.1239. [DOI] [PubMed] [Google Scholar]
- 39.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 41.Ponath P D, Qin S, Post T W, Wang J, Wu L, Gerard N P, Newman W, Gerard C, Mackay C R. Molecular cloning and characterization of a human eotaxin receptor expressed selectively on eosinophils. J Exp Med. 1996;183:2437–2448. doi: 10.1084/jem.183.6.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J D, Guo H-H, Du J-D, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R G. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the delta CCR5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raport C J, Gosling J, Schweickart V L, Gray P W, Charo I F. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1β, and MIP-1α. J Biol Chem. 1996;271:17161–17166. doi: 10.1074/jbc.271.29.17161. [DOI] [PubMed] [Google Scholar]
- 44.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;35:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 46.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valentin A, Albert J, Fenyö E M, Asjö B. Dual tropism for macrophages and lymphocytes is a common feature of primary human immunodeficiency virus type 1 and 2 isolates. J Virol. 1994;68:6684–6689. doi: 10.1128/jvi.68.10.6684-6689.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L Q, Huang Y X, He T, Cao Y Z, Ho D D. HIV-1 subtype and second-receptor use. Nature. 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]