Abstract
The bovine papillomavirus replication initiator protein E1 is an origin of replication (ori)-binding protein absolutely required for viral DNA replication. In the presence of the viral transcription factor E2, E1 binds to the ori and initiates DNA replication. To understand how the E1 initiator recognizes the ori and how E2 assists in this process, we have expressed and purified a 166-amino-acid fragment which corresponds to the minimal E1 DNA-binding domain (DBD). DNA binding studies using this protein demonstrate that the E1 DBD can bind to the palindromic E1 binding site in several forms but that binding of two monomers, each recognizing one half-site of the E1 palindrome, is the predominant form. This is reminiscent of the binding of the T-antigen DBD to the SV40 ori, and interestingly, the arrangement of E1 binding sites shows striking similarities to the arrangement of T-antigen binding sites in the SV40 ori even though the recognition sequences are unrelated. The E1 DBD is capable of interacting cooperatively with E2; however, the E2 DBD and not the E2 activation domain mediates this interaction. Furthermore, the E2 DBD stimulates binding of two monomers of the E1 DBD to the ori by binding cooperatively with one E1 monomer. Finally, we show that our results concerning the DNA-binding properties of the E1 DBD can be extended to full-length E1.
Papillomaviruses are small DNA viruses which have been studied extensively because they cause disease in humans (49, 58). Infection by human papillomavirus (HPV) produces cutaneous and mucosal squamous epithelial lesions that can eventually become malignant and give rise to, for example, cervical carcinomas. The search for treatment of papillomavirus infections provides one rationale for studying papillomavirus DNA replication. Since the viruses are also known to replicate their DNA only in the S phase of the host cell cycle (11, 37), studies of papillomavirus DNA replication have also proven to be highly informative in the understanding of mechanisms of eukaryotic DNA replication.
Bovine papillomavirus (BPV) has been used as a model system for studying papillomavirus DNA replication, for BPV can be replicated transiently and be stably maintained in cell culture, allowing the study of viral replication in vivo. In addition, an in vitro replication system exists for BPV, making it possible to study both cellular and viral requirements for replication in cell-free environments (4, 33, 36, 41, 44, 56). From in vivo replication studies, two viral proteins, the early viral proteins E1 and E2, have been found to be absolutely essential for DNA replication, (52). These proteins bind to the minimal origin of replication (ori), which contains an A/T-rich region and palindromic binding sites for the E1 and E2 proteins (12, 16, 17, 23, 44, 45, 53, 56).
The 68-kDa E1 protein serves as the viral initiator protein and has ori-specific binding activity, ori distortion activity, and DNA helicase activity (12, 16, 27, 44, 50, 53, 55–57). The E1 protein shows both functional and sequence homology to the simian virus 40 (SV40) T antigen (8, 10, 26, 28, 29, 48). For example, a domain with ATPase and DNA helicase activities with limited sequence homology is present in the C termini of both proteins, while the nuclear localization signal as well as the DNA-binding domain (DBD) are present in the N-terminal half of each protein (10, 21, 22, 28, 39, 48, 50). Multimerization of these initiator proteins results in the assembly of a helicase-active form of the protein, which, for both T antigen and E1, is a hexamer (30, 40, 54).
Despite the analogous roles played by E1 and T antigen in viral DNA replication, significant differences between the two proteins exist. Unlike T antigen, which is the only viral protein necessary for SV40 replication, E1 is not sufficient to initiate BPV replication in vivo; the viral transcription factor E2 is also required (51–53; for a review, see reference 31). The requirement for transcription factors for efficient replication is a common theme in eukaryotic DNA replication (for a review, see reference 9). Some proposed functions for transcription factors in DNA replication are the derepression of chromatin templates by the displacement of nucleosomes, the recruitment of cellular replication factors, and the enhancement of binding by initiator proteins (6, 14, 15, 18). E2 may be involved in all three activities (24, 25, 41), but it is the latter function that seems to best explain the specific and absolute requirement for E2 as an auxiliary factor in papillomavirus DNA replication. Unlike T antigen, E1 is unable to bind to the ori with a high degree of sequence specificity. When challenged with competitor DNA, E1 binds very poorly to the ori (41). However, in the presence of E2, an interaction between E1 and E2 results in the formation of a highly sequence specific complex of E1 and E2 with the ori (E1E2-ori complex) (3, 12, 20, 26, 27, 35, 41, 43, 45, 56).
The formation of the E1E2-ori complex is only one step in the assembly of a final replication-competent complex, for the E1E2-ori complex has no inherent replication-related activity (42). Since the E1E2-ori complex is highly sequence specific, it most likely is involved in ori recognition. In DNA binding studies of the E1E2-ori complex which has been cross-linked by glutaraldehyde, the E1E2-ori complex has been shown to contain only one monomer of E1. Thus, the E1E2-ori complex may be the means of loading additional E1 molecules onto the ori (42). Indeed, recent studies indicate that the E1E2-ori complex serves as a precursor and in fact is a preferred substrate for the formation of an oligomeric E1-ori complex which is capable of origin distortion (38).
The initial binding of E1 to the ori seems to be a critical step in the assembly of a replication-competent complex. To study the DNA binding activity of E1, we have isolated and purified a 166-amino-acid (aa) fragment of E1 that corresponds to the minimal DBD. This fragment is capable of binding to the ori by itself and also binds cooperatively with E2. We show that the E1 DBD binds to the ori as two monomers and that binding of this form of E1 is greatly stimulated by E2. Surprisingly, only the DBD of E2 is responsible for the cooperative interaction with the E1 DBD. By introducing mutations in the E1 binding site, we show that one monomer of E1 binds to one half-site of the palindromic E1 binding site. In addition, the E2 DBD interacts cooperatively with the monomer of E1 bound to the half-site directly adjacent to the E2 binding site. These studies suggest a possible mechanism by which E2 assists in E1 ori-specific binding and provides support for a model for the binding of E1 to the ori DNA. This model shows similarities to binding of T antigen to the SV40 ori.
MATERIALS AND METHODS
Protein expression and purification. (i) E1 DBD.
Escherichia coli BL21(DE3) was transformed with pET expression plasmids (46) containing the E1 (aa 127 to 308, 142 to 308, and 142 to 374). Cultures were grown at room temperature until the optical density at 600 nm reached 0.6 and induced by the addition of 0.4 mM isothiogalactopyranoside (IPTG) for 10 h. Bacterial pellets were resuspended in buffer A (50 mM Tris [pH 7.9], 0.25 M NaCl, 5 mM EDTA, 10 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 10% glycerol), treated with lysozyme (100 μg/ml) for 2 h at 4°C, and lysed in a French press. After the addition of 0.1% Nonidet P-40 (NP-40), the lysate was sonicated and cleared by centrifugation at 20,000 × g (Sorvall SS34 rotor) for 30 min. The supernatant was incubated with glutathione-agarose beads (1 ml/liter of culture) at 4°C for 2 to 8 h. The beads were washed five times with 10 bead volumes of buffer B (50 mM Tris [pH 8.0], 0.2 M NaCl, 5 mM EDTA, 10 mM DTT, 10% glycerol). To remove the glutathione S-transferase (GST) moiety, thrombin (100 U/ml of beads) was added to the beads, and cleavage was allowed to proceed on ice for 10 to 12 h. Thrombin was inactivated by the addition of 1 mM PMSF. The sample was diluted twofold with buffer B, and the supernatant containing the cleaved protein was collected, aliquoted, and frozen in liquid nitrogen.
Purified GST-E1 DBD (aa 142 to 308) used in McKay assays was prepared as described above except that after incubation of the lysate with glutathione-agarose beads, the beads were washed with 20 bead volumes buffer B (1 M NaCl) followed by 20 bead volumes of buffer B (0.2 M NaCl). GST-E1 DBD was then eluted off the beads by the addition of elution buffer (20 mM glutathione, 25 mM Tris [pH 8.0], 0.2 M NaCl, 1 mM EDTA, 5 mM DTT, 10% glycerol; pH adjusted to 8.0 at 4°C). Peak fractions were pooled, aliquoted, and stored at −70°C.
(ii) E2 DBD.
E. coli BL21(DE3) was transformed with the pET11C E2 DBD (aa 323 to 410) expression plasmid (2). Liquid cultures were inoculated and grown at 18°C. When an optical density of 0.5 at 600 nm was reached, the cultures were induced with 0.4 mM IPTG and grown for an additional 12 h at 18°C. Bacterial pellets were resuspended in lysis buffer (25 mM Tris [pH 7.5], 0.1 M NaCl, 0.5 mM EDTA, 10 mM DTT, 1 mM PMSF) and treated with lysozyme (100 μg/ml). After the addition of 0.1% NP-40, the lysate was sonicated, cleared by centrifugation, and directly applied to a 1-ml S-Sepharose column. After a wash with 10 column volumes of buffer A (20 mM morpholinepropanesulfonic acid [MES; pH 6.2], 0.2 M NaCl, 10 mM DTT, 1 mM PMSF), the bound protein was eluted with buffer A containing 1 M NaCl. The eluate was diluted threefold with buffer B (50 mM MES [pH 6.5], 0.1 M NaCl, 1 mM DTT, 1 mM PMSF) and loaded onto a 1-ml Mono-S column. The protein was eluted with a 10-ml gradient of between 0.1 and 1 M NaCl in buffer B. Peak fractions were pooled with the addition of 10% glycerol, aliquoted, and frozen in liquid nitrogen.
(iii) Full-length E1 and E2.
The expression and purification of E1 and E2 have been described previously (43).
Plasmid constructs.
The different E1 fragments were cloned in pET11C-GST (1). The E1 constructs were generated by PCR amplification of the appropriate segment by using a 5′ primer containing an XbaI restriction site and a 3′ primer containing a BamHI restriction site. The PCR fragments were digested with XbaI and BamHI and ligated into pET11C-GST digested with XbaI and BamHI.
All ori constructs have been described previously (2, 43, 51–53).
Probes.
For gel mobility shift assays, McKay assays, and diethyl pyrocarbonate (DEPC) interference analysis, probes containing the BPV minimal ori with the low-affinity E2 binding site replaced with the BS9 high-affinity E2 binding site (51) or with a high-affinity E2 binding site from the human papillomavirus type 11 (HPV-11) ori (BS12H) (2) were generated by PCR amplification of ori constructs cloned into pUC19, using the universal primer USP or RSP, 5′ radiolabeled with [γ-32P]ATP by T4 polynucleotide kinase.
Gel mobility shift assays.
Probe containing the BPV minimal ori (5,000 cpm/sample) was mixed with the E1 and/or E2 protein together with 20 ng of nonspecific competitor DNA (pUC119) in 10 μl of binding buffer (20 mM potassium phosphate [pH 7.4], 0.1 M NaCl, 1 mM EDTA, 0.1% NP-40, 3 mM DTT, 0.7 mg of bovine serum albumin/ml, 10% glycerol). After incubation for 30 min at room temperature, the samples were immediately loaded on 5, 6, or 8% 40:1 (acrylamide/bisacrylamide) polyacrylamide gels and subjected to polyacrylamide gel electrophoresis (PAGE) in 0.5× Tris-borate-EDTA. The gels were then dried and subjected to autoradiography.
McKay assay.
The assay is a modified form of the immunoprecipitation assay developed by McKay (32). Binding reactions as described above for gel mobility shift assays were performed. For these binding reactions, the E1 proteins contained an N-terminal GST fusion. After incubation for 30 min at room temperature, 2.5 μl of glutathione-agarose beads in 50 μl binding buffer was added. After 20 min of end-over-end mixing, the beads were washed three times with 200 μl of binding buffer. Then 100 μl of stop buffer was added (1% sodium dodecyl sulfate [SDS], 50 mM EDTA, 0.1 M NaCl, 25 μg of tRNA/ml) together with 5 μg of mussel glycogen carrier. After phenol-chloroform extraction, the samples were ethanol precipitated. The DNA pellets were resuspended in formamide loading buffer and loaded onto a 6% denaturing polyacrylamide gel.
DEPC interference.
Probes (2 × 106 cpm in 10 μl of TE buffer [10 mM Tris, pH 7.5, and 0.1 mM EDTA]) containing the wild-type BPV minimal ori or the BPV minimal ori with a mutation in the E1 binding site (7942 probe) were modified as described by Sturm et al. (47). Gel slices containing free and bound probe from binding reactions were excised and electroeluted. The eluted samples were extracted with phenol-chloroform and ethanol precipitated. The DNA was cleaved at modified bases with piperidine, which was subsequently removed by multiple rounds of butanol extraction. Scission products were analyzed on an 8% sequencing gel.
RESULTS
The minimal DBD of E1 is contained between residues 142 and 308.
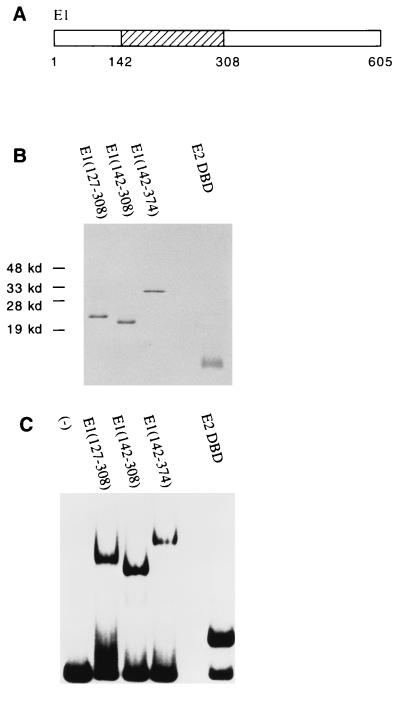
To study how E1 binds and recognizes the ori and how its binding activity is affected by E2, we wanted to define a minimal region of E1 that was competent for DNA binding. Previous studies had shown that residues 121 to 311 were sufficient for ori-specific binding (21). We identified by deletion mapping a fragment containing residues 142 to 308 which is capable of binding alone to the ori (Fig. 1A). A 20-aa deletion either C terminal to residue 142 or N terminal to residue 308 abolished DNA binding completely (13). To perform a careful analysis of the binding properties of the E1 DBD in the presence and absence of E2 or the E2 DBD, we expressed in E. coli and purified to apparent homogeneity different fragments containing the E1 DBD as well as the E2 DBD (Fig. 1B). The purified proteins were then tested for DNA-binding activity in a gel mobility shift assay using a probe containing the BPV minimal ori with the low-affinity E2 binding site replaced with a high-affinity E2 binding site (BS12H). Previously, the detection of E1-ori and E1E2-ori complexes by gel mobility shift assays was possible only by the addition of a cross-linking agent, glutaraldehyde, before resolving the complexes on agarose gels (27, 41, 43). With the E1 DBD fragments, however, we can observe complex formation by PAGE. The use of cross-linker was also no longer necessary, allowing us to study the binding properties of the E1 DBD in the presence or absence of E2 directly without selection for protein-DNA complexes that can be efficiently cross-linked. Figure 1C shows that the E1 fragments E1(127–308), E1(142–308), and E1(142–374) bound DNA with similar efficiencies.
FIG. 1.
DNA-binding activities of the E1 and E2 DBDs. (A) The E1 DBD maps to residues 142 to 308. (B) Analysis by SDS-PAGE (15% gel) and Coomassie staining of 1 μg of protein after the final step of purification. E1(127–308), E1(142–308), and E1(142–374) were expressed as N-terminal GST fusions in E. coli. After affinity purification using glutathione-agarose beads, the E1 fragments were treated with thrombin to remove the GST. The E2 DBD(323–410) was also expressed in E. coli but without any GST fusion. Extracts were then loaded directly onto an S-Sepharose column. Peak fractions were pooled and further purified on a Mono-S column. (C) DNA-binding activity of the purified E1 or E2 DBD protein. Gel mobility shift assays were performed with a probe containing the BPV minimal ori with a high-affinity E2 binding site (BS12H). Ten-microliter binding reaction mixtures containing 12 ng of E1(127–308), 2 ng of E1(142–308), 5 ng of E1(142–374), or 36 pg of E2 DBD(325–410) were incubated for 30 min at room temperature and analyzed on a 5% polyacrylamide gel.
The E1 DBD binds cooperatively with E2.
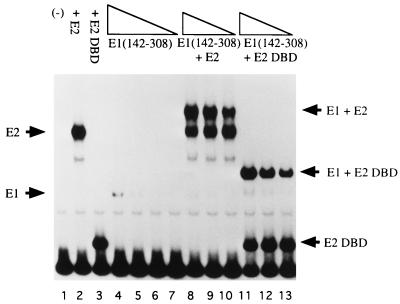
We have shown previously that two separate protein-protein interactions are responsible for the formation of the E1E2-ori complex (2). These two interactions consist of one interaction between E1 and the activation domain of E2 and a separate interaction between E1 and the E2 DBD. To determine whether the E1 DBD could interact with E2 and form a cooperative complex on DNA, we used gel mobility shift assays to detect the formation of E1 DBD-E2-ori complexes. We used a probe containing the BPV minimal ori with the low-affinity E2 binding site replaced by a high-affinity E2 binding site (BS12H) (Fig. 2). The use of a high-affinity E2 binding site allows us to detect an E2 complex by PAGE at low concentrations of protein. The formation of E1 DBD-E2-ori complexes can also be achieved with the wild-type low-affinity E2 binding site at higher concentrations of E2.
FIG. 2.
The E1 DBD binds cooperatively with E2. In 10-μl binding reactions, 0.8, 0.4, 0.2, and 0.1 ng of E1(142–308) were incubated alone (lanes 4 to 7) or in the presence of either 0.1 ng of full-length E2 (lanes 8 to 10) or 18 pg of the E2 DBD (lanes 11 to 13). The probe used in the binding reactions contained the BPV minimal ori with a high-affinity E2 binding site (BS12H). After 30 min of incubation at room temperature, the reaction mixtures were loaded directly onto a 5% polyacrylamide gel.
E1(142–308) forms a complex at high protein concentration (lane 4). Binding of the E1 DBD to the ori was strongly stimulated by the addition of full-length E2 (compare lanes 5 to 7 with lanes 8 to 10). The binding of E1 and E2 to the ori is cooperative since binding of E1 in the presence of E2 can be observed at levels where E1 alone shows no detectable binding. To determine if the DBD of E2 was sufficient to stimulate binding by the E1 DBD, binding reactions containing the E1 DBD and the E2 DBD were also performed. The concentration of the E2 DBD was chosen such that the proportion of probe bound was similar to that bound by full-length E2 (compare lane 2 with lane 3). The DBD of E2 was sufficient to interact cooperatively with the E1 DBD, stimulating the binding of the E1 DBD to levels similar to that in the presence of full-length E2 (compare lanes 11 to 13 to lanes 8 to 10). These results suggested that the interaction between the E1 DBD and E2 may be restricted to the DBD of E2.
The DBD of E1 fails to interact with the E2 activation domain but interacts with the DBD of E2.
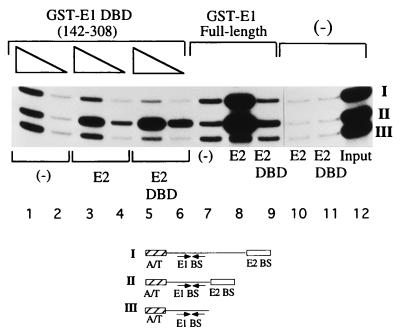
To determine which domain of E2 interacts with the E1 DBD, we performed a McKay assay which allowed us to specifically distinguish between the interaction of E1 with either the activation or DNA-binding domain of E2. The assay is based on previous observations (2). The interaction between full-length E1 and the E2 DBD occurs only when the E2 DBD is bound proximal to the E1 binding site. Consequently, this interaction cannot be detected when the E2 binding site is located distal to the E1 binding site. However, the interaction between the E2 activation domain and E1 can still occur from a distal site. The two interactions involving the E2 activation and DNA-binding domains and E1 can, therefore, be distinguished in a McKay assay through the differential stimulation of E1 binding by E2 to three different probes: one probe containing an E2 binding site distal to the E1 binding site (probe I), one probe containing an E2 binding site proximal to the E1 binding site (probe II), and a third probe containing only the E1 binding site (negative control) (probe III). After incubation with GST-E1 in the presence or absence of E2, probes bound by GST-E1 were recovered by using glutathione-agarose beads and subsequently analyzed by PAGE.
As shown in Fig. 3, lanes 7 to 9, full-length E2 stimulated binding of GST-E1 (full length) to probes I and II, containing the E2 binding site in the distal and proximal positions. Recovery of these two probes compared to probe III (no E2 binding site) was stimulated 10- and 20-fold, respectively. The E2 DBD stimulated binding of GST-E1 (full length) only to probe II, which contains the E2 binding site in the proximal position (lane 9); the degree of stimulation by the E2 DBD was substantially lower (2.5-fold). Thus, the presence of the activation domain in E2 affects E1 binding in two ways: (i) the stimulation of binding to the proximal site probe is much greater with full-length E2 than with the E2 DBD, and (ii) stimulation of binding can be observed from a distal E2 binding site. These results are consistent with our previous observations (2).
FIG. 3.
The E1 DBD interacts with the E2 DBD but not with the E2 activation domain. Three different probes containing a high-affinity E2 binding site distal to the E1 binding site (I), a high-affinity E2 binding site proximal to the E1 binding site (II), and only an E1 binding site (III) were mixed with either 0.5 (lane 1) or 0.25 (lane 2) ng of GST-E1(142–308) in 10-μl binding reactions; 2.0 ng of E2 (lanes 3 and 4) or 0.80 ng of the E2 DBD (lanes 5 and 6) was incubated with GST-E1(142–308) in the binding reaction. Probes bound by GST-E1 DBD were recovered by using glutathione-agarose beads. The recovered probes were analyzed on a 6% urea gel. Control reactions containing GST-E1 (full length) (6 ng) alone (lane 7) or together with full-length E2 (lane 8) or the E2 DBD (lane 9) were performed simultaneously.
When the McKay assay was performed with GST-E1(142–308) in the absence of E2, all three probes were recovered equally well (lanes 1 and 2). In the presence of full-length E2, recovery of the probe containing the proximal E2 binding site was stimulated threefold, but recovery of the probe containing the distal E2 binding site was not stimulated (compare II and III, lanes 3 and 4). This result differs significantly from what we observe with full-length E1, which could stimulate binding of E1 to the probe containing the distal E2 binding site (lane 8). However, this result is virtually identical to that observed with full-length E1 and the E2 DBD, indicating that the contribution from the E2 activation domain was negligible (lane 9). To test this directly, we incubated GST-E1(142–308) with the DBD of E2. As with full-length E2, stimulation of binding was observed only for probe II. Furthermore, the stimulation of binding of GST-E1(142–308) by full-length E2 was similar to that by the E2 DBD, which is in striking contrast to the results observed with full-length E1 (compare the ratio of II to III in lanes 3, 5, and 8). These results suggest that GST-E1(142–308) interacts exclusively with the DBD of the E2 protein and is unable to interact with the E2 activation domain.
The E1 DBD binds as two monomers to the ori in the presence of E2.
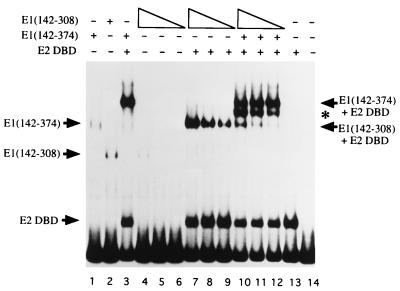
The E1 DBD can form multiple complexes with the ori in a concentration-dependent manner (5). These complexes are detectable in gel mobility shift assays. In the absence of E2, E1 forms one predominant complex. In the presence of E2, a different predominant complex forms. We wanted to determine the stoichiometry of E1 binding in these predominant complexes. Since we know that truncated E1 DBD fragments exist as monomers in solution based on light-scattering and coimmunoprecipitation experiments (data not shown), we decided to perform the following mixing experiment. We generated the predominant E2-containing complexes formed with two E1 DBD fragments of different sizes E1(142–308) and E1(142–374) and analyzed them by gel mobility shift assays. The rationale for this mixing experiment is as follows. If E1 binds as a monomer, two discrete complexes with different mobilities each corresponding to binding of one E1 fragment would be observed. If E1 binds as two monomers, in addition to the parent complexes, a novel complex with intermediate mobility corresponding to binding of a heterodimer is expected to form. Higher-order complexes would be expected to give rise to a greater number of intermediates.
The results of the mixing experiment are shown in Fig. 4. In the absence of the E2 DBD, the two different E1 DBD proteins form complexes which migrate with different mobilities (compare lanes 1 and 2). The addition of the E2 DBD stimulates binding by both E1 fragments and in each case gives rise to a characteristic new complex (lanes 3 and 7 to 9, respectively). When the two E1 DBD proteins are mixed, both of the original complexes are observed. In addition, an intermediate complex migrating between the two original complexes is observed (lanes 10 to 12), suggesting that the E1 DBD does not bind as a monomer to the E1 binding site in the presence of the E2 DBD. The fact that only one intermediate species can be detected indicates that most likely two E1 DBD molecules are bound to the ori. When this mixing experiment was performed in the absence of the E2 DBD, a similar single intermediate between the predominant form of the E1(142–374)-ori and E1(142–308)-ori complexes was seen (data not shown), indicating that also in the predominant complex observed in the absence of E2, two monomers of the E1 DBD are present.
FIG. 4.
The E1 DBD binds as two monomers to the minimal ori. To determine if the E1 DBD binds as two monomers on the E1 palindrome, three quantities of E1(142–308), 0.4, 0.2, and 0.1 ng, were incubated together with 0.5 ng of E1(142–374), 18 pg of the E2 DBD, and probe containing the BPV minimal ori with a high-affinity E2 binding site in 10-μl binding reactions (lanes 10 to 12). The asterisk marks the mobility of an intermediate complex formed when E1(142–374) and E1(142–308) were mixed in the presence of the E2 DBD (lanes 3 and 7 to 9, respectively). The levels of binding by E1(142–308) in the absence of the E2 DBD and E1(142–374) are shown in lanes 4 to 6. Lanes 1 and 2 are markers for the binding of E1(142–374) and E1(142–308), respectively.
Formation of the E1 DBD-ori and E1 DBD-E2 DBD-ori complexes on mutant oris.
The results of the mixing experiment suggested that the E1 DBD binds as two monomers to the ori. A likely possibility is that each monomer recognizes and binds to one half-site of the palindromic E1 binding site. Because the binding of two monomers is predominant, it is likely that the two E1 monomers are stabilized on the DNA by protein-protein interactions. Also, binding of two monomers of the E1 DBD is stimulated by the E2 DBD.
If one E1 molecule binds to each half-site of the E1 palindrome, then a mutation in one half-site should impair binding of two monomers of the E1 DBD but might still allow binding of one E1 monomer to the wild-type half-site. Furthermore, because of the linear arrangement of the two E1 binding sites and the E2 binding site, it is possible that mutations in the two half-sites will have different effects on the complexes formed in the presence of E2. A mutation in the half-site distal to the E2 binding site might allow an E1 monomer to bind cooperatively with the E2 DBD on the half-site proximal to the E2 binding site, while a mutation in the proximal half-site might not. If an interaction between the two monomers of E1 is important for binding of two monomers to the ori, then increasing the spacing between the two half-sites would prevent the binding of two monomers of E1 even though the two wild-type half-sites are still present. However, a complex containing a monomer of the E1 DBD and the E2 DBD may still form.
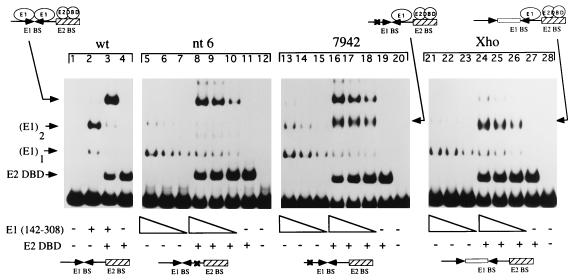
To test this model, we performed gel mobility shift assays (Fig. 5) using a wild-type ori and three previously characterized mutant oris (43): (i) an ori with a point mutation in the half-site proximal to the E2 binding site (nt 6), (ii) an ori with a point mutation in the half-site distal to the E2 binding site (7942), or (iii) an ori with an 8-bp XhoI linker insertion between the two half-sites (Xho). On the wild-type probe, the E1 DBD forms a predominant complex containing two monomers and a less abundant complex which, based on conclusions drawn from the experiments in Fig. 4, most likely contains one monomer (lane 2). The mutant oris behave differently. All of the mutant probes show reduced binding as expected. Four times more E1 were used in the first lane of each set compared to lane 2 containing the wild-type probe. The predominant band for all mutant probes is a band that comigrates with the monomer band in lane 2 (lanes 5 to 7, 13 to 15, and 21 to 23). This complex most likely contains a monomer of the E1 DBD bound to one wild-type half-site. The levels of monomeric E1 binding are similar for all three type of mutant probes, suggesting that E1 has similar affinities for the two half-sites. A fainter band comigrating with the predominant complex in lane 2 probably corresponds to the binding of two monomers, indicating that the two point mutants reduce but do not abolish binding of two E1 monomers (lanes 5 to 7 and 13 to 15), consistent with the low sequence specificity of E1. The XhoI linker insertion, however, inhibits the formation of this complex (lanes 21 to 23).
FIG. 5.
Gel shift analysis of complexes formed on mutant ori. Assays to determine the ability to form complexes in the presence of the E1 DBD alone or in the presence of both the E1 and E2 DBDs were performed with a probe containing the wild-type (wt) minimal ori (lanes 1 to 4), a probe containing a single-point mutation in the E1 half-site proximal to the E2 binding site (nt 6; lanes 5 to 12), a probe containing a single-point mutation in the E1 half-site distal to the E2 binding site (7942; lanes 13 to 20), and probe in which the two half-sites are separated by an 8 bp XhoI linker (Xho; lanes 21 to 28). All four probes contained a high-affinity E2 binding site (BS9). Lane 2 contains 1 ng of E1(142–308); lane 3 contains 0.5 ng of E1(142–308) and 18 pg of the E2 DBD. For the mutant probes, three twofold dilutions of E1(142–308), corresponding to 5, 2.5, and 1.3 ng, were used both in the presence and in the absence of the E2 DBD; 18 pg of the E2 DBD was used in all E2-containing reactions. Binding reactions were in a final volume of 10 μl and incubated at room temperature for 30 min. The reactions were subsequently loaded on a 6% native polyacrylamide gel.
At high concentrations of the E1 DBD in the presence of the E2 DBD, all three mutant probes show a characteristic behavior. On the mutant probe containing a mutation in the half-site proximal to the E2 binding site (nt 6), a complex corresponding to the two monomers of E1 bound together with the E2 DBD is seen (compare lanes 8 to 10 with lane 3). This complex is also observed on the 7942 probe (lanes 16 to 18), which suggests that the interaction between E1 and E2 can compensate for a mutation in the E1 binding site as has been observed previously (41). It is likely that protein-protein interactions between two E1 monomers further stabilize binding. A very faint band corresponding to two monomers of E1, in the absence and presence of E2, is also observed on the Xho probe, which supports this conclusion (lanes 21 to 28). Interestingly, on the 7942 and Xho probes, but not on the nt 6 probe, a new complex is apparent. This complex is intermediate in mobility between the E1 monomer and the complex containing E2 together with two monomers of E1 (lanes 16 to 18 and 24 to 26). A common feature of the two probes on which this complex forms is that both the Xho and 7942 probes have a wild-type half-site adjacent to the E2 binding site. Therefore, this novel complex most likely corresponds to a monomer of the E1 DBD bound to the proximal half-site together with the E2 DBD. That this complex is absent on the nt 6 probe indicates that the E2 DBD can interact only with an E1 monomer bound to the proximal half-site (lanes 8 to 10). Binding of the E1 monomer is clearly stimulated by the presence of the E2 DBD, indicating that the interaction between the E1 monomer and the E2 DBD is cooperative (compare lanes 13 to 15 with lanes 16 to 18).
A monomer of the E1 DBD binds to one half-site of the palindromic E1 binding site.
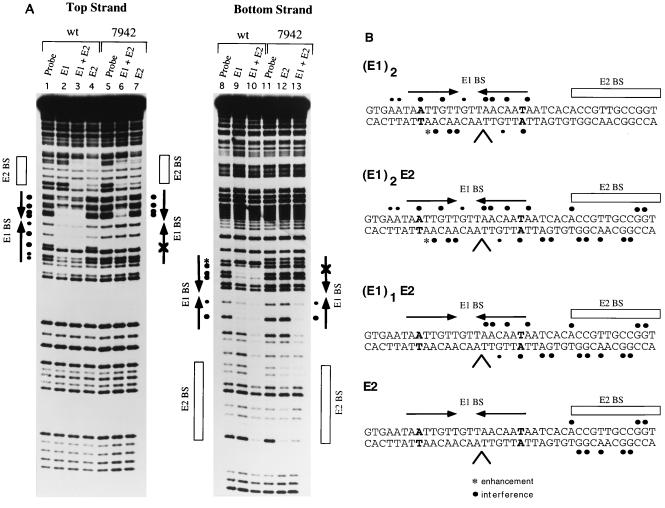
To verify the conclusions from the gel shift assay in Fig. 5, we performed DEPC interference analysis for several of these complexes. We have previously shown for the E1E2-ori complex that base positions that give rise to interference when modified by DEPC occur over both half-sites of the E1 palindrome (43). The wild-type and 7942 probes were treated with DEPC, which modifies A and G residues, and then used in a gel mobility shift assay. Protein-DNA complexes corresponding to the two E1 monomers and the two E1 monomers complexed with the E2 DBD were generated on the wild-type probe. The monomer form of E1 complexed with the E2 DBD was generated on the 7942 probe. In addition, complexes containing only E2 were generated for both probes. The complexes were extracted from the gel, cleaved with piperidine, and analyzed on a sequencing gel. The results for both strands of the DNA are shown in Fig. 6A. On the top strand, the complex containing two monomers of the E1 DBD shows strong interference over both half-sites of the palindrome (lane 2). For this form of E1 complexed with the E2 DBD, there are additional interferences over the E2 binding site which result from the binding of E2 (compare lane 3 to lane 4, which shows the interference produced by E2 alone). However, the interferences over the E1 binding site are identical to those produced by E1 alone. On the mutant 7942 ori, the complex containing a monomer of the E1 DBD and the E2 DBD shows interferences over the E1 binding site, but only on the half-site proximal to the E2 binding site (lane 6), suggesting that one monomer of the E1 DBD recognizes and binds to the wild-type half-site. The interferences over the E1 binding site are not due to the E2 DBD since the interference pattern shown by the E2 DBD alone is exclusively located over the E2 binding site (lane 7).
FIG. 6.
(A) One monomer of the E1 DBD binds to one half-site of the E1 palindrome. DEPC interference analysis was performed on both strands of the probe containing the wild-type minimal ori (wt) and probe containing the ori with a single-point mutation in nucleotide position 7942, which is distal to the E2 binding site. The wt and 7942 probes were treated with DEPC, which modifies A and G residues, and then used in binding reactions. Complexes corresponding to the binding of two E1 monomers (lanes 2 and 9) and two E1 monomers together with the E2 DBD (lanes 3 and 10) were generated on the wild-type probe, as well as the monomer of E1 complexed with the E2 DBD on the 7942 probe (lanes 6 and 13). Complexes containing only the E2 DBD were also generated for both probes (lanes 4, 7, and 12). Complexes were separated on a 6% polyacrylamide gel, the recovered probes were cleaved with piperidine, and the products were analyzed on an 8% sequencing gel. (B) Summary of the DEPC interference analysis. Interferences are shown for the complex containing two E1 monomers, the E1 monomer and E2 DBD, and the E2 DBD alone. The position of the mutation present in either the 7942 probe or nt 6 probe is shown in bold. The caret indicates the position of the XhoI linker insertion.
On the bottom strand, the binding of two monomers of E1 shows virtually the same interference pattern over both half-sites in the presence or absence of the E2 DBD domain (lane 9). The combined binding of both the E1 DBD and the E2 DBD to the wild-type probe, however, produces interferences at two positions not seen by either the E1 DBD alone or the E2 DBD alone, suggesting that either the E1 or E2 DBD or both may have additional DNA contacts when the two proteins bind simultaneously. On the mutant probe, the complex containing only a monomer of the E1 DBD shows interferences only on the wild-type half-site of the palindrome; no interferences are observed on the mutated half-site (lane 13). The E2 DBD again shows interferences only over the E2 binding site (lane 12).
Figure 6B summarizes the results from the interference analysis. Interestingly, the interference pattern produced by the E1 DBD is virtually identical to that produced by full-length E1 when complexed with E2 (43). This observation strongly suggests that the E1 DBD binds in the same way as that of full-length E1 in the E1E2-ori complex. The interference produced by the complex generated on the 7942 probe shows interferences on only one half-site of the E1 palindrome, consistent with the observation that this complex contains one monomer of E1 (Fig. 5).
Full-length E1 does not bind to the ori as a monomer.
Previous results using various assays have indicated that in the cross-linked E1E2-ori complex, E1 was present as a monomer. The sum of the gel shift and interference analyses performed here clearly indicate that the DBD of E1 forms a stable complex containing two molecules of E1 in the presence of E2. The interference pattern for this complex is virtually identical to that observed with the full-length E1E2-ori complex, suggesting that full-length E1 may in fact bind as two monomers to the E1 palindrome. To resolve this apparent discrepancy, we decided to perform a similar mixing experiment as described above for the E1 DBD, but using full-length E1 in combination with truncated E1(142–308). The results of this experiment are shown in Fig. 7. Full-length E1 alone under these conditions does not give rise to a detectable complex (lanes 1 to 4). In the presence of full-length E2, two prominent complexes are formed. The faster-migrating complex corresponds to E2 alone (compare lanes 5 to 8 with lane 21). The slower-migrating complex most likely corresponds to the previously characterized E1E2-ori complex. Truncated E1(142–308) alone gives rise to a single complex (lanes 9 and 10). In the presence of full-length E2, a complex migrating slightly more slowly than the E2 complex is formed (lanes 11 and 12).
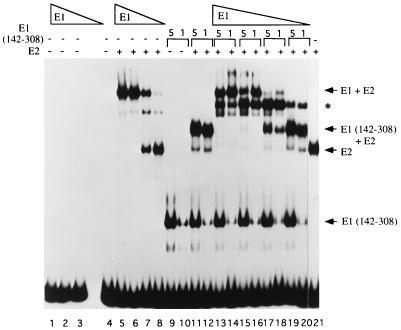
FIG. 7.
Full-length E1 also binds as two monomers to the minimal ori. A mixing experiment similar to that shown in Fig. 4 was performed with full-length E1. Four reactions containing 5, 1.3, 0.6, and 0.3 ng of full-length E1 were incubated alone (lanes 1 to 4), together with 0.1 ng of full-length E2 (lanes 5 to 8), or with two different quantities of truncated E1(142–308) (2 and 0.4 ng) in the presence of 0.1 ng of full-length E2 (lanes 13 to 20) in 10-μl binding reactions containing probe with the BS12H high-affinity E2 binding site. The asterisk indicates the position of the intermediate complex which formed when E1 and truncated E1(142–308) were mixed together in the presence of E2. Lanes 9 and 10 contain truncated E1(142–308) alone at the two concentrations used in the mixing experiment; lanes 11 and 12 contain the two concentrations of E1(142–308) in the presence of the E2 DBD. After incubation for 30 min at room temperature, the reactions were analyzed by SDS-PAGE (6% gel).
To perform the mixing experiment, decreasing concentrations of full-length E1 were mixed with two different concentrations of truncated E1(142–308) (lanes 13 to 20). At the higher concentrations of full-length E1, the most prominent band corresponds to full-length E1 complexed with E2 (lanes 13 to 16). In addition, another prominent complex migrates intermediate to the position of the full-length E1E2-ori complex and the E1(142–308)-ori complex (marked with an asterisk). Surprisingly, the complex containing truncated E1(142–308) and E2 is absent (lanes 13 to 16). This complex only appears at the lower concentrations of E1 when little full-length E1E2-ori complex formation occurs (lanes 17 to 20). Most likely, this is caused by a preferential interaction between full-length E1 and E2. Truncated E1(142–308) interacts less strongly with E2 due to the failure to interact with the activation domain of E2 (Fig. 3). Nonetheless, the appearance of a mixed complex with intermediate mobility to the two parent complexes (lanes 17 and 18) clearly demonstrates that E1 is not present as a monomer in the full-length E1E2-ori complex. Interestingly, there is a relatively high level of mixed complex formation, which indicates that a single molecule of full-length E1 is sufficient for efficient interaction with the E2 activation domain. The band migrating just below the mixed complex most likely corresponds to the complex containing one monomer of full-length E1 and E2.
DISCUSSION
We have previously demonstrated that full-length E1 can bind cooperatively with E2 to form the E1E2-ori complex. Our present study of the E1 DBD is consistent with those studies and helps further clarify how E1 may recognize and bind to the ori. Using E1 DBD fragments, we can now analyze ori complexes without cross-linking. In this way, we have shown that the E1 DBD(142–308) can bind to the ori in various forms, including as one and two monomers. The predominant form appears to be a form where a monomer binds to each half-site of the palindrome. This is also the preferred form of E1 in the E1E2-ori complex. From DEPC interference analysis, the interference pattern produced by the E1 DBD is virtually identical to that of full-length E1 when complexed with E2. The binding properties of the E1 DBD, therefore, are likely to reflect those of the full-length protein. Indeed, the mixing experiment using full-length E1 indicates that full-length E1 binds as two monomers in the E1E2-ori complex, at least when the complex is analyzed without cross-linking. Mutational analysis of the E1 binding site has shown that as part of the E1E2-ori complex, E1 requires the sequence element ATNGTTNNNAACNAT (43). It was previously believed that only one E1 molecule bound to this sequence element (43); however, in light of the results of the experiments with the E1 DBD presented here, this sequence element likely corresponds to two binding sites for E1. From DEPC interference analysis of the monomer of the E1 DBD complexed with E2 on the 7942 mutant probe as well as from previous mutational data of the ori (43), one E1 binding site most likely consists of the sequence element AACAAT. This sequence is also present in the other half-site, and the DEPC interference pattern for binding of two monomers of the E1 DBD suggests that it probably constitutes a second binding site for another monomer of E1. The placement of these two sequence elements relative to each other is important, since insertion of the XhoI linker disrupts binding of two E1 monomers, suggesting that protein-protein interactions between two DNA-bound E1 monomers stabilize DNA binding. These findings are consistent with the studies by Mendoza et al., who demonstrated that the two half-sites of the palindromic E1 binding site are separable and can function independently, but that binding is cooperative when the two half-sites are not separated (34). By comparing sequences between the BPV-1 ori and the upstream regulatory region, to which E1 has been shown to be capable of binding (34, 55), Mendoza et al. proposed that the sequence APyAAPy was a recognition sequence for E1 (34). This element is contained in the sequence we have shown to be required for the binding of one monomer of E1.
E1 is also capable of forming a larger complex on the origin, the E1-ori complex (27, 41, 42, 44, 45). In hydroxy-radical footprinting studies, strong protections seen with the E1-ori complex duplicate those seen with the E1E2-ori complex, but the protections are shifted by three nucleotides relative to the E1E2-ori footprint (42). This has been interpreted to mean that additional binding sites are used for binding of additional E1 molecules in the E1-ori complex compared to the E1E2-ori complex. Mutagenesis of the E1 binding site also indicated that the formation of the E1-ori complex may require additional binding sites within the E1 palindrome since mutations at certain positions within the palindrome affected the formation of the E1-ori complex but not the E1E2-ori complex (43). Taken together, both the hydroxy-radical footprinting and mutagenesis data indicated that an additional sequence element which is recognized by E1 is present in the E1 palindrome. This sequence element, GTTGTTNNNAATAAT, is shifted by three nucleotides relative to that used by E1 in the E1E2-ori complex (43). It contains two hexanucleotide sequences which are similar to the two used by the binding of two monomers of E1 but differ only by two transition changes. Studies by Holt and Wilson, who mutated individual bases in the E1 palindrome, found that most transitions have small effects on E1 binding (17).
Thus, the palindromic E1 binding site may contain four binding sites for E1. The result of this study suggest that E1 binds as two monomers to the sequence element ATNGTTNNNAACNAT in the E1E2-ori complex. By analogy, the second sequence element, GTTGTTNNNAATAAT, may be used for the binding of two additional molecules of E1 to form the E1-ori complex. This would mean that the E1-ori complex in fact contains four molecules of E1. However, in a previous study of the cross-linked E1E2-ori and E1-ori complexes, we estimated by molecular weight determination that the cross-linked E1-ori complex consisted of three molecules of E1 whereas the cross-linked E1E2-ori complex contained only one monomer of E1 (42). These discrepancies with the data from the present study could be due to the use of a cross-linker which may fail to cross-link the entire complex. Thus, the cross-linked complexes observed with the full-length E1 protein on agarose gels may not directly correlate with the complexes observed with the E1 DBD on PAGE in the absence of cross-linker. The direct relationship between the cross-linked complexes observed on agarose gels and the complexes detected by PAGE is, therefore, unclear.
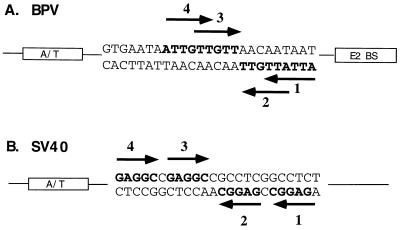
Based on the results of this study of the E1 DBD, we believe that the E1 palindrome contains at least four binding sites for E1. The arrangement of these putative binding sites is shown in Fig. 8A. We have shown through interference analysis that the E1 DBD binds as two monomers to two separate half-sites, designated 2 and 4. Each half-site contains the sequence AACNAT. We propose that there are additional binding sites for E1, sites 1 and 3, which are nearly identical in sequence. This model for binding of E1 shows striking similarities to that which has been proposed for SV40 T antigen. The SV40 ori contains four T-antigen binding sites, each containing the pentanucleotide recognition sequence GAGGC, arranged in the manner shown in Fig. 8B. The T-antigen DBD like that of E1, exists as a monomer in solution, but it binds as two monomers preferentially to two separate half-sites, designated sites 1 and 3, which are oriented in a head-to-head direction (19). If these two sites are mutated, the T-antigen DBD then binds to sites 2 and 4. Joo et al. (19) have proposed that T antigen binds initially as a dimer to sites 1 and 3 but that the binding of the first two monomers of T antigen nucleates and stabilizes the binding of additional molecules of T antigen. In the same way, two E1 molecules bind preferentially to sites 2 and 4, and once bound, protein-protein interactions may mediate the binding of additional E1 monomers to sites 1 and 3. Even though sites 1 and 3 overlap sites 2 and 4, the binding of two E1 molecules to overlapping E1 binding sites is still structurally possible if an E1 molecule can recognize and bind to separate sequences on different strands of the DNA. It is striking that the arrangement of the E1 and T-antigen binding sites are similar even though the actual recognition sequences are different and the DBDs of the two proteins show little or no sequence homology. This suggests that BPV and SV40 share a common mechanism for generating a multimeric complex on the ori from monomeric initiator proteins.
FIG. 8.
Comparison of the arrangements of T-antigen binding sites in the SV40 ori with that of the proposed E1 binding sites in the BPV minimal ori. (A) The BPV minimal ori may contain four E1 binding sites. Binding sites 2 and 4, arranged in a head-to-head orientation, are used for the binding of two E1 monomers in the E1E2-ori complex. Binding sites 2 and 4, which are also oriented head to head, overlap binding sites 1 and 3 by three nucleotides. (B) The SV40 ori consists of four T-antigen binding sites of which T antigen binds primarily to sites 1 and 3, which are also oriented head to head.
One major difference between E1 and T antigen is the requirement for E2 for in vivo replication. It is clear that a critical function of E2 as an auxiliary factor in DNA replication is to interact with E1 to form the E1E2-ori complex. This complex, in turn, serves as a precursor for a higher-order form of E1 with ori melting activity (38). We have previously shown that the interaction between the E1 and E2 proteins has two components. Both the DBD and the activation domain of E2 are independently capable of interacting with E1. Our present study demonstrates that the E1 DBD interacts with the E2 DBD as strongly as full-length E1, indicating that the sequences required for this interaction are completely contained within the E1 DBD. The failure of the E1 DBD to interact with the E2 activation domain suggests that a region outside the E1 DBD domain is required for this interaction. The observation that two different regions in E2 can interact with separate regions of E1 could explain some apparently contradictory results concerning the sequences in E1 required for interaction with E2. Thorner et al. found that the N-terminal portion of E1 contained between residues 1 to 423 was sufficient to interact with E2 and exhibit origin-specific binding (50). In another study, Leng et al. narrowed this region to aa 121 to 311 (21). In light of our results, it is likely that the interactions observed in these two studies involved the E2 DBD only. Sarafi and McBride, however, found a predominant interaction that required a region of E1 C terminal to the DBD (39). This region, in accordance with our results, may be involved in the interaction with the activation domain of E2.
The existence of two separate interacting domains in both the E1 and E2 proteins could very likely have functional significance. It has previously been shown that the activation domain of E2 is sufficient for DNA replication (2). The requirement for the E2 DBD is conditional; the interaction between E1 and the E2 DBD is necessary only in the context of the BPV ori in which the E2 binding site is proximal to the E1 binding site. Our studies confirm that the interaction between the E1 DBD and the E2 DBD is sufficient to stimulate binding of E1 to the ori; however, the significance of the interaction is still obscure. A possible role for the interaction between the E1 and E2 DBDs is to provide an additional interaction between E1 and E2 in order to compensate for the very low affinity of the proximal E2 binding site. Another possibility is that the E1-E2 interaction is a two-step process and that the interaction between the E2 DBD and E1 is required before a productive interaction between the E2 activation domain and E1 can occur. This possibility is supported by a previous observation that replacement of the BPV E2 DBD with the highly homologous HPV-11 E2 DBD disrupted both interactions and abolished replication (2).
Nonetheless, the interaction between E1 and E2 ultimately results in the formation of a highly specific E1E2-ori complex on the origin. Our studies suggest that the E2 DBD stabilizes binding of an E1 monomer bound to the half-site proximal to the E2 binding site. Thus, it is possible that the first step in ori recognition is the interaction between E2 and a monomer of E1 either in solution or on the DNA. This possibility is further supported by the results of the mixing experiment shown in Fig. 8 with full-length E1, E1(142–308), and full-length E2. In this mixing experiment, a significant amount of mixed complex was observed, probably due to a direct interaction between the full-length E1 monomer and both the activation domain and DBD of E2. In addition, since we observe very little complex containing a monomer of either full-length E1 or truncated E1(142–308) together with E2, the interaction between a monomer of E1 and E2 is probably immediately followed by the cooperative binding of a second E1 monomer. This would explain the predominance of E1 binding as two monomers. Both E1 and E1(142–308) exhibit these properties, strongly indicating that sequences within E1(142–308) are sufficient for cooperative binding of two E1 monomers.
Our studies suggest that the formation of the E1E2-ori complex is a complicated process entailing interactions between the DBDs of both E1 and E2, between the C terminus of E1 and the E2 activation domain, and between two E1 molecules. Presumably, the formation of higher-order E1 complexes would involve additional interactions between E1 molecules. Further studies of these interactions will be required to understand how complexes competent for initiation of DNA replication are generated.
ACKNOWLEDGMENTS
We thank B. Henry and C. Sanders for critical reading of the manuscript.
This work was supported by National Institutes of Health grant CA 13106 to A.S.
REFERENCES
- 1.Aurora R, Herr W. Segments of the POU domain influence one another’s DNA-binding specificity. Mol Cell Biol. 1992;12:455–467. doi: 10.1128/mcb.12.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg M, Stenlund A. Functional interactions between papillomavirus E1 and E2 proteins. J Virol. 1997;71:3853–3863. doi: 10.1128/jvi.71.5.3853-3863.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blitz I L, Laimins L. The 68-kilodalton E1 protein of bovine papillomavirus is a DNA-binding phosphoprotein which associates with the E2 transcriptional activator in vitro. J Virol. 1991;65:649–656. doi: 10.1128/jvi.65.2.649-656.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonne-Andrea C, Santucci S, Clertant P. Bovine papillomavirus E1 protein can, by itself, efficiently drive multiple rounds of DNA synthesis in vitro. J Virol. 1995;69:3201–3205. doi: 10.1128/jvi.69.5.3201-3205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, G., and A. Stenlund. Unpublished results.
- 6.Cheng I, Workman J I, Kingston R E, Kelly T J. Regulation of DNA replication in vitro by the transcriptional activation domain of GAL4-VP16. Proc Natl Acad Sci USA. 1992;89:589–593. doi: 10.1073/pnas.89.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang C M, Ustav M, Stenlund A, Ho T F, Broker T R, Chow L T. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc Natl Acad Sci USA. 1992;89:5799–5803. doi: 10.1073/pnas.89.13.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clertant P, Seif I. A common function for polyoma virus large-T and papillomavirus E1 proteins? Nature. 1984;311:276–279. doi: 10.1038/311276a0. [DOI] [PubMed] [Google Scholar]
- 9.DePamphilis M L. How transcription factors regulate origins of DNA replication in eukaryotic cells. Trends Cell Biol. 1993;3:161–167. doi: 10.1016/0962-8924(93)90137-p. [DOI] [PubMed] [Google Scholar]
- 10.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert D M, Cohen S N. Bovine papillomavirus plasmids replicate randomly in mouse fibroblasts throughout S phase of the cell cycle. Cell. 1987;50:59–68. doi: 10.1016/0092-8674(87)90662-3. [DOI] [PubMed] [Google Scholar]
- 12.Gillette T G, Lusky M, Borowiec J A. Induction of structural changes in the bovine papillomavirus type 1 origin of replication by the viral E1 and E2 proteins. Proc Natl Acad Sci USA. 1994;91:8846–8850. doi: 10.1073/pnas.91.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillitzer, E., and A. Stenlund. Unpublished results.
- 14.Guo Z S, DePamphilis M L. Specific transcription factors stimulate simian virus 40 and polyomavirus origins of DNA replication. Mol Cell Biol. 1992;12:2514–2524. doi: 10.1128/mcb.12.6.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Z, Brinton B T, Greenblatt J, Hassel J A, Ingles C L. The transactivator proteins VP16 and Gal 4 bind replication factor A. Cell. 1993;73:1223–1232. doi: 10.1016/0092-8674(93)90650-f. [DOI] [PubMed] [Google Scholar]
- 16.Holt S E, Schuller G, Wilson V G. DNA binding specificity of the bovine papillomavirus E1 protein is determined by the sequences contained within an 18-base-pair inverted repeat element at the origin of replication. J Virol. 1993;68:1094–1102. doi: 10.1128/jvi.68.2.1094-1102.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt S E, Wilson V G. Mutational analysis of the 18-base-pair inverted repeat element at the bovine papillomavirus origin of replication: identification of critical sequences for E1 binding and in vivo replication. J Virol. 1995;69:6525–6532. doi: 10.1128/jvi.69.10.6525-6532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito K, Asano M, Hughes P, Kohzaki H, Masutani C, Hanaoka F, Kerppola T, Curran T, Murakami Y, Ito Y. c-Jun stimulates origin-dependent DNA unwinding by polyomavirus large T antigen. EMBO J. 1996;15:5636–5646. [PMC free article] [PubMed] [Google Scholar]
- 19.Joo W S, Luo X, Denis D, Kim H Y, Rainey G J, Jones C, Sreekumar K R, Bullock P A. Purification of the simian virus 40 (SV40) T-antigen DNA-binding domain and characterization of its interactions with the SV40 origin. J Virol. 1997;71:3972–3985. doi: 10.1128/jvi.71.5.3972-3985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Moal M A, Yaniv M, Thierry F. The bovine papillomavirus type 1 (BPV1) replication protein E1 modulates transcriptional activation by interacting with BPV1 E2. J Virol. 1994;68:1085–1093. doi: 10.1128/jvi.68.2.1085-1093.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leng X, Ludes-Meyers J H, Wilson V G. Isolation of an amino-terminal region of bovine papillomavirus type 1 E1 protein that retains origin binding and E2 interaction capacity. J Virol. 1997;71:848–852. doi: 10.1128/jvi.71.1.848-852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lentz M R, Pak D, Mohr I, Botchan M R. The E1 replication protein of bovine papillomavirus type 1 contains an extended nuclear localization signal that includes a p34cdc2 phosphorylation site. J Virol. 1993;67:1414–1423. doi: 10.1128/jvi.67.3.1414-1423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li R, Knight J, Bream G, Stenlund A, Botchan M. Specific recognition nucleotides and their DNA context determine the affinity of E2 protein for 17 binding sites in the BPV genome. Genes Dev. 1989;3:510–526. doi: 10.1101/gad.3.4.510. [DOI] [PubMed] [Google Scholar]
- 24.Li R, Botchan M. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 25.Li R, Botchan M R. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc Natl Acad Sci USA. 1994;91:7051–7055. doi: 10.1073/pnas.91.15.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lusky M, Fontane E. Formation of the complex of bovine papillomavirus E1 and E2 proteins is modulated by E2 phosphorylation and depends upon sequences within the carboxyl terminus of E1. Proc Natl Acad Sci USA. 1991;88:6363–6367. doi: 10.1073/pnas.88.14.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lusky M, Hurwitz J, Seo Y S. The bovine papillomavirus E2 protein modulates the assembly of but is not stably maintained in a replication-competent multimeric E1-replication origin complex. Proc Natl Acad Sci USA. 1994;91:8895–8899. doi: 10.1073/pnas.91.19.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacPherson P, Thorner L, Parker M, Botchan M. The bovine papillomavirus E1 protein has ATPase activity essential to viral DNA replication and efficient transformation in cells. Virology. 1994;204:403–408. doi: 10.1006/viro.1994.1544. [DOI] [PubMed] [Google Scholar]
- 29.Mansky K C, Batiza A, Lambert P F. Bovine papillomavirus type 1 E1 and simian virus 40 large T antigen share regions of sequence similarity required for multiple functions. J Virol. 1997;71:7600–7608. doi: 10.1128/jvi.71.10.7600-7608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of soluble hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 31.McBride A A, Romanczuk H, Howley P M. The papillomavirus E2 regulatory proteins. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- 32.McKay R D G. An immunoassay for the interaction between an SV40 T antigen related protein and DNA. J Mol Biol. 1981;266:18411–18414. [Google Scholar]
- 33.Melendy T, Sedman J, Stenlund A. Cellular factors required for papillomavirus DNA replication. J Virol. 1995;69:7857–7867. doi: 10.1128/jvi.69.12.7857-7867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendoza R, Gandhi L, Botchan M R. E1 recognition sequences in the bovine papillomavirus type 1 origin of DNA replication: interaction between half-sites of the inverted repeats. J Virol. 1995;69:3789–3798. doi: 10.1128/jvi.69.6.3789-3798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohr I J, Clark R, Sun S, Androphy E J, MacPherson P, Botchan M. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 36.Muller F, Seo Y S, Hurwitz J. Replication of bovine papillomavirus type 1 origin-containing DNA in crude extracts and with purified factors. J Biol Chem. 1994;269:17086–17094. [PubMed] [Google Scholar]
- 37.Ravnan J B, Gilbert D M, Ten Hagen K G, Cohen S N. Random-choice replication of extrachromosomal bovine papillomavirus (BPV) molecules in heterogeneous, clonally derived BPV-infected lines. J Virol. 1992;66:6946–6952. doi: 10.1128/jvi.66.12.6946-6952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders, C., and A. Stenlund. Submitted for publication.
- 39.Sarafi T R, McBride A A. Domains of the BPV-1 E1 replication protein required for origin-specific DNA-binding and interaction with the E2 transactivator. Virology. 1995;211:385–396. doi: 10.1006/viro.1995.1421. [DOI] [PubMed] [Google Scholar]
- 40.Sedman, J., and A. Stenlund. Unpublished results.
- 41.Sedman J, Stenlund A. Cooperative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sedman J, Stenlund A. The initiator protein E1 binds to the bovine papillomavirus origin of replication as a trimeric ring-like structure. EMBO J. 1996;15:5085–5092. [PMC free article] [PubMed] [Google Scholar]
- 43.Sedman T, Sedman J, Stenlund A. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J Virol. 1997;71:2887–2896. doi: 10.1128/jvi.71.4.2887-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seo Y S, Muller F, Lusky M, Hurwitz J. Bovine papillomavirus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc Natl Acad Sci USA. 1993;90:702–706. doi: 10.1073/pnas.90.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seo Y S, Muller F, Lusky M, Gibbs E, Kim H Y, Phillips B, Hurwitz J. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc Natl Acad Sci USA. 1993;90:2865–2869. doi: 10.1073/pnas.90.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Studier F, Moffatt B. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 47.Sturm R, Baumruker T, Franza B R, Jr, Herr W. A 100-kD HeLa cell octamer binding protein (OBP100) interacts differently with two separate octamer-related sequences within the SV40 enhancer. Genes Dev. 1987;1:1147–1160. doi: 10.1101/gad.1.10.1147. [DOI] [PubMed] [Google Scholar]
- 48.Sun S, Thorner L, Lentz M, MacPherson P, Botchan M. Identification of a 68-kilodalton nuclear ATP-binding phosphoprotein encoded by bovine papillomavirus type 1. J Virol. 1990;64:5093–5105. doi: 10.1128/jvi.64.10.5093-5105.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Syrjanen L, Gissman L, Koss L G, editors. Papillomaviruses and human disease. New York, N.Y: Springer-Verlag; 1987. [Google Scholar]
- 50.Thorner L K, Lim D L, Botchan M R. DNA-binding domain of bovine papillomavirus type 1 E1 helicase: structural and functional aspects. J Virol. 1993;67:6000–6014. doi: 10.1128/jvi.67.10.6000-6014.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ustav E, Ustav M, Szymanski P, Stenlund A. The bovine papillomavirus origin of replication requires a binding site for the E2 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:898–902. doi: 10.1073/pnas.90.3.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ustav M, Ustav E, Szymanski P, Stenlund A. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 1991;10:4321–4329. doi: 10.1002/j.1460-2075.1991.tb05010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson V G, Ludes-Meyers M J. A bovine papillomavirus E1-related protein binds specifically to bovine papillomavirus DNA. J Virol. 1991;65:5314–5322. doi: 10.1128/jvi.65.10.5314-5322.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang L, Li R, Mohr I, Clark R, Botchan M R. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991;353:628–633. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 57.Yang L, Mohr I, Fouts E, Lim D A, Nohaile M, Botchan M. The E1 protein of the papillomavirus BPV-1 is an ATP dependent DNA helicase. Proc Natl Acad Sci USA. 1993;90:5086–5090. doi: 10.1073/pnas.90.11.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.zur Hausen H. Viruses in human cancers. Science. 1991;254:1167–1173. doi: 10.1126/science.1659743. [DOI] [PubMed] [Google Scholar]