Abstract
The clinical course of hepatitis C virus (HCV) infections in a chimpanzee cohort was examined to better characterize the outcome of this valuable animal model. Results of a cross-sectional study revealed that a low percentage (39%) of HCV-inoculated chimpanzees were viremic based on reverse transcription (RT-PCR) analysis. A correlation was observed between viremia and the presence of anti-HCV antibodies. The pattern of antibodies was dissimilar among viremic chimpanzees and chimpanzees that cleared the virus. Viremic chimpanzees had a higher prevalence of antibody reactivity to NS3, NS4, and NS5. Since an unexpectedly low percentage of chimpanzees were persistently infected with HCV, a longitudinal analysis of the virological profile of a small panel of HCV-infected chimpanzees was performed to determine the kinetics of viral clearance and loss of antibody. This study also revealed that a low percentage (33%) of HCV-inoculated chimpanzees were persistently viremic. Analysis of serial bleeds from six HCV-infected animals revealed four different clinical profiles. Viral clearance with either gradual or rapid loss of anti-HCV antibody was observed in four animals within 5 months postinoculation. A chronic-carrier profile characterized by persistent HCV RNA and anti-HCV antibody was observed in two animals. One of these chimpanzees was RT-PCR positive, antibody negative for 5 years and thus represented a silent carrier. If extrapolated to the human population, these data would imply that a significant percentage of unrecognized HCV infections may occur and that silent carriers may represent potentially infectious blood donors.
Viral hepatitis represents a major health problem throughout the world. Hepatitis C virus (HCV) infections are particularly serious, since an estimated 70 to 90% of HCV infections become chronic (3, 6, 7, 45, 49, 62, 63). Chronic HCV infection progresses to cirrhosis in at least 20% of infected individuals after 10 to 20 years and is also associated with hepatocellular carcinoma (3, 45). At this point, no vaccine for HCV is available, and antiviral treatments are marginally effective. Interferon is generally used in the treatment of HCV infections. Although interferon treatment is beneficial to some individuals, only 10 to 20% sustain improved biochemical and virological values 6 months posttreatment (45). A better understanding of HCV replication and pathogenesis is essential in combating this disease.
The transmission of HCV is primarily associated with parenteral routes such as blood transfusions and intravenous drug use (5). Mandatory anti-HCV screening of blood donors has significantly decreased the risk of acquiring HCV by transfusion. Sexual transmission is of questionable significance as a route of infection, and if it occurs, the efficiency is very low compared to hepatitis B virus (HBV) or human immunodeficiency virus. Rare instances of perinatal transmission have been documented. However, the route of transmission for many infections is unknown, since over one-third of HCV-infected individuals have no apparent risk factors.
HCV is a member of the Flaviviridae family and possesses a single-stranded RNA genome of positive polarity (13, 26). Other members of the Flaviviridae family include the Pestivirus genus and the Flavivirus genus. The genome organization of HCV is similar to that of the flaviviruses and pestiviruses (13, 42). The 9.4-kb viral RNA has a single large open reading frame which encodes for a polyprotein of approximately 3,010 amino acids.
The viral genome begins with a 5′ noncoding region consisting of about 342 nucleotides. Translation of HCV RNA is presumably cap independent and involves an internal ribosomal entry site located within the 5′ noncoding region (25, 51, 66, 69). Expression of partial and full-length recombinant polyproteins has revealed the organization of the polyprotein (19–21, 26, 38, 54). The structural proteins of HCV are found in the amino-terminal quarter of the polyprotein and are followed by the nonstructural proteins. Individual proteins are cleaved from the polyprotein by host and viral proteases. The structural proteins include the capsid and two envelope glycoproteins, E1 and E2. The nonstructural proteins include NS2, NS3, NS4A, NS4B, NS5A, and NS5B. The NS2 domain and the amino-terminal portion of NS3 form a zinc-dependent metalloproteinase, which cleaves NS2 from the polyprotein. The amino terminus of NS3 encodes a serine proteinase and is involved in cleaving the polyprotein at all sites downstream of NS3, thus releasing the individual proteins. At the carboxy terminus of NS3 are motifs characteristic of nucleoside triphosphatases (NTPases) and helicases which are thought to play a role in viral RNA replication. NS4A is required as a cofactor for NS3 for several cleavages. NS4B is a hydrophobic protein of unknown function, and NS5B contains the GDD motif for the viral RNA polymerase. An untranslated region of approximately 270 nucleotides is present at the 3′ end of the viral genome; it is comprised of a variable region followed by a poly(U)-polypyrimidine stretch of variable length and a highly conserved terminal domain (32, 59). The 3′ end contains secondary structures and is presumably where genomic replication of negative-strand RNA initiates. Partial sequencing of multiple HCV isolates has revealed marked variability, which led to the grouping of various isolates based on genotype (14, 57).
HCV replicates at low levels within hepatocytes. However, the mechanism of HCV replication has not been well established due to several obstacles, such as the lack of a conventional in vitro tissue culture system in which the virus readily replicates. Although HCV replicates at low levels in primary hepatocytes, this in vitro tissue culture system is expensive and difficult to manipulate (35). Low-level replication has also been reported for human lymphocytic cell lines (43, 56). The replication of HCV is probably similar to that of other flaviviruses, which replicate via a negative-strand RNA intermediate. Negative-strand HCV RNA, indicative of active viral replication, has been detected at low levels in the liver of HCV-infected individuals (33). The presence of negative-strand viral RNA in peripheral blood lymphocytes is controversial, since false priming of positive-strand RNA appears to occur under some reverse transcription-PCR (RT-PCR) conditions (22, 35, 41). The low levels at which HCV replicates may be beneficial to maintaining persistent infections.
The hallmark of HCV is its ability to establish persistent infection. Persistent infection is characterized by sustained viremia and may occur in approximately 70 to 90% of HCV-infected humans. These estimations come from numerous studies involving posttransfusion hepatitis, intravenous drug user-(IVDU)-associated hepatitis, and community-acquired hepatitis (3, 4, 6, 7, 45, 49, 62, 63). Viremia is usually associated with anti-HCV antibody. Although the majority of HCV-infected individuals experience persistent infection, the disease may be active or quiescent. Patients with persistent viremia and active disease experience elevated alanine aminotransferase (ALT) levels and ongoing liver damage. In contrast, at least 30% of chronically HCV-infected individuals are asymptomatic and experience quiescent disease (45). Although these individuals resolve hepatitis and have normal ALT levels, viremia persists, and liver disease often develops after many years of asymptomatic infection.
The mechanisms for maintaining viral persistence are unknown. Natural HCV infection does not appear to induce protective immunity, since HCV infection persists despite the presence of virus-specific cytotoxic T cells and circulating antibodies to HCV proteins. HCV variants arise frequently due to the high error rate of the viral RNA-dependent RNA polymerase. Minor variants of the same strain emerge as a result of mutation, and HCV virions circulate as quasispecies, or a heterogeneous population of minor variants along with a predominant species. HCV infection may persist due to the presence of quasispecies or multiple variant genomes that continuously escape neutralization. Several hypervariable regions (HVR) are present within the envelope glycoproteins and may be particularly important in maintaining chronicity (9, 13, 26, 30, 67). The first HVR (HVR-1) within E2 has the most significant divergence. Antibodies elicited against the E2 HVR-1 have been proposed to neutralize the virus, as well as to promote immune selection and genetic drift of the E2 HVR-1 (31, 60, 68). Neutralizing antibodies elicited to a particular HVR of the predominant strain of circulating virus may clear the majority of virus. However, variant viruses exhibiting amino acid changes in the HVR may escape neutralizing antibody and subsequently become the predominant strain of circulating virus. Additional mechanisms for maintaining persistent infection may include immunomodulation by viral proteins, such as the core protein–lymphotoxin-β receptor interaction (40), or the production of defective interfering particles (39).
HCV pathogenesis is difficult to study, since conventional tissue culture systems are not established. Animal models present another challenge in HCV studies. Currently, chimpanzees serve as the only animal model for HCV infection. Advantages of studying HCV infection in chimpanzees include the opportunity to infect chimpanzees with well-characterized HCV inocula and the availability of serial specimens before and after infection. The understanding of HCV infection has been greatly enhanced by the chimpanzee animal model. Many factors involved in HCV infection, such as transmission, genetic drift, clinical outcome of HCV infection, and the role of the immune response, have been examined in chimpanzees (47). As in humans, both viral clearance and persistent viremia in HCV-infected chimpanzees have been observed. The frequencies of persistent infection in chimpanzees and humans appear to differ. However, the actual frequency of persistent infection in chimpanzees is difficult to determine, since most studies have examined only a small panel of animals (2, 8, 16, 18, 23, 48, 53, 64).
This study analyzed (i) the frequency of persistent infection in chimpanzees inoculated with various HCV strains and (ii) the relationship of viremia to anti-HCV antibodies and ALT values. A cross-sectional study was performed on 46 HCV-inoculated chimpanzees, using serum collected an average of 10.6 years postinoculation. An unexpectedly high percentage (61%) of chimpanzees appeared to be convalescent based on RT-PCR negativity. A longitudinal analysis of the virological profile experienced by a panel of six HCV-infected chimpanzees also revealed a high level of convalescence with rapid virus clearance and in some instances rapid loss of antibody. The relevance of these findings to the chimpanzee model and human HCV infections is discussed.
MATERIALS AND METHODS
Serum samples from chimpanzees.
Serum samples were collected in 1995 from 52 non-A, non-B hepatitis virus (NANBH)- or HCV-inoculated chimpanzees in the Southwest Foundation for Biomedical Research chimpanzee colony and were stored at −80°C. Serum samples used in retrospective studies had been collected twice a year from each animal in the chimpanzee colony and stored at −70°C. The 52 chimpanzees had been inoculated with various strains of NANBH or HCV between 1978 and 1993. Some of the chimpanzees had been inoculated with HBV, but only one HCV-inoculated animal was a chronic carrier of HBV. Additionally, some chimpanzees had been inoculated with human immunodeficiency virus or other viruses. ALT values were determined by standard laboratory procedures at the time the serum samples were obtained.
Extraction of RNA from chimpanzee sera for RT-PCR analysis.
Briefly, 100 μl of serum was mixed with 900 μl of RNAzol B (Biotecx Laboratories) and 100 μl of chloroform. The suspension was clarified by centrifugation at 12,000 × g for 15 min. The aqueous phase was separated, mixed with an equal volume of isopropanol, and centrifuged at 12,000 × g for 15 min to precipitate the RNA. The RNA pellet was washed in 75% ethanol and resuspended in 50 μl of nuclease-free water containing 1 mM dithiothreitol and 800 U of RNasin RNase inhibitor (Promega) per ml.
RT-PCR analysis of sera from HCV-inoculated chimpanzees.
RNA was reverse transcribed and amplified by RT-PCR using the Access RT-PCR system (Promega). Briefly, a 50-μl reaction mix containing nuclease-free water, RT-PCR buffer, 0.2 mM dNTP mix, 1 μM downstream primer, 1 μM upstream primer, 1 mM MgSO4, 5 U of avian myeloblastosis virus reverse transcriptase, 5 U of Thermus flavus DNA polymerase, and 10 μl of the sample RNA was prepared. The downstream primer (5′-TCGCGACCCAACACTACTC-3′) spanned nucleotides 256 to 274, and the upstream primer (5′-GGGGGCGACACTCCACCA-3′) spanned nucleotides 15 to 32. The RT-PCR reaction mixes were incubated at 48°C for 45 min for cDNA synthesis. The reactions were thermal cycled by using the following scheme for amplification: 94°C for 2 min (1 cycle); 94°C for 30 s, 60°C for 1 min, 68°C for 2 min (40 cycles); and 68°C for 7 min (1 cycle).
Some samples negative by one round of RT-PCR were reexamined in a second round of PCR. Briefly, a 100-μl reaction mix containing Vent RT-PCR buffer (New England BioLabs), dNTPs (0.2 mM), downstream primer (1 μg), upstream primer (1 μg), 2 U of Vent DNA polymerase, and DNA (5 μl) from the first-round RT-PCR reaction mix was prepared. The primers used for the second round of RT-PCR were the same as those used for the first round. The reaction cycle included 1.3 min at 94°C, 2 min at 46°C, and 3 min at 72°C. The reaction mixes were held at 72°C for 7 min after 35 cycles to complete the DNA synthesis.
Southern blot hybridization analysis of RT-PCR products.
RT-PCR products were analyzed by electrophoresis on a 1.0% agarose gel in 1× TAE (0.04 M Tris-acetate, 2 mM EDTA) at 80 V for 1.25 h. Gels were stained with ethidium bromide (0.5 μg/ml) for 15 min at room temperature, destained in water for 15 min, and photographed. Depurination was performed by incubating the gels in 0.25 M HCl for 10 min at room temperature, followed by a 30-min incubation in 0.4 M NaOH. DNA was transferred to a Gene Screen Plus hybridization transfer membrane (NEN Research Products), using downward transfer in a TurboBlotter apparatus (Schleicher & Schuell). Membranes were equilibrated in 0.4 N NaOH for 15 min prior to the transfer. Following the transfer, membranes were rinsed in 2× SSPE (0.3 M NaCl, 20 mM NaH2PO4 · H2O, 2 mM EDTA [pH 7.4]) at room temperature. Membranes were placed in a seal bag and incubated with prehybridization mix (50% formamide, 7% sodium dodecyl sulfate [SDS], 0.25 M NaPO4 [pH 7.2], 0.25 M NaCl, 1 mM EDTA) for 4 h at 42°C. The DNA probe was prepared by using a Prime-It II random primer kit (Stratagene) according to the manufacturer’s instructions. Briefly, 25 ng of a gel-purified DNA fragment spanning nucleotides 95 to 274 was heated with oligonucleotide primers in a boiling water bath for 5 min. The DNA fragment was added to buffer containing dATP, dGTP, dTTP, and [α-32P]dCTP (3,000 Ci/mmol; New England Nuclear), and Exo(−) Klenow enzyme (5 U) was used to radioactively label the DNA template. The probe was purified over a G-25 Sephadex Quick Spin column (Boehringer Mannheim Biochemicals). Cerenkov counts were determined in a scintillation counter, and the probe was added to the prehybridization mix at a concentration of 106 cpm/ml. Hybridization was conducted for 16 h at 42°C. Membranes were washed one time at 42°C and two times at 60°C for 10 min in 2× SSC (0.3 M NaCl, 30 mM C6H5O7Na3 · 2H2O [pH 7.0]) containing 0.1% SDS. Membranes were washed three times for 10 min with 0.1× SSC containing 0.1% SDS at 60°C, dried, and exposed to X-ray film for approximately 1 h at −70°C with one intensifying screen.
Anti-HCV testing.
Chimpanzee sera were analyzed for anti-HCV antibody by using a second-generation enzyme-linked immunosorbent assay (ELISA; Ortho Diagnostic Systems, Raritan, N.J.). The ELISA was performed according to the manufacturer’s instructions. The assay kit contains the following recombinant antigens: c100-3 (amino acids [aa] 1569 to 1931, NS3 and NS4), c200 (aa 1192 to 1931, NS3 and NS4), and c22-3 (aa 2 to 120, capsid). Chimpanzee sera were analyzed for anti-HCV antibody to individual HCV proteins by using HCV BLOT 3.0 (Genelabs Diagnostics). The assay was performed according to the manufacturer’s instructions. The assay kit contains the following recombinant antigens: capsid (aa 1 to 150), NS3-1 (aa 1368 to 1492), NS3-2 (aa 1192 to 1367), NS4 (aa 1695 to 1735), and NS5 (aa 2120 to 2623).
RESULTS
Confirmation of HCV infection in a cohort of NANBH- and HCV-inoculated chimpanzees.
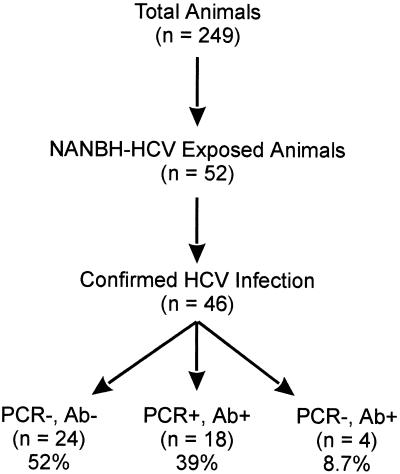
To better understand the characteristics of HCV infections in chimpanzees, a cohort of NANBH- and HCV-exposed animals was examined. The Southwest Foundation for Biomedical Research accommodates 249 chimpanzees, 52 of which had been exposed to various NANBH and HCV inocula during the past 2 to 19 years (Fig. 1). Since some of the animals received uncharacterized inocula, it was first necessary to determine the number of animals with confirmed HCV infections. Initially, serum samples collected in 1995 were examined for the presence of anti-HCV antibody and viral RNA. Anti-HCV antibody and/or HCV RNA were detected in serum samples from 22 chimpanzees. However, no evidence of previous infection was detected in serum samples from the remaining 30 animals. Clinical records from chimpanzees with unconfirmed HCV exposure were examined to determine if the animals were inoculated with a strain of NANBH now known to be HCV. HCV inoculation was confirmed in eight chimpanzees, and the development of hepatitis was confirmed based on increased ALT levels following inoculation. Archived serum samples from the remaining 22 chimpanzees were analyzed for anti-HCV antibody and HCV RNA. HCV infection was confirmed in 6 chimpanzees based on antibody positivity and in 10 chimpanzees based on RT-PCR positivity. Therefore, HCV infection was confirmed in a total of 46 chimpanzees. The remaining six animals were excluded from these studies. These chimpanzees may have been inoculated with a noninfectious inoculum or an inoculum containing a non-A, non-B, non-C hepatitis virus, or the serum samples available for analysis may not have been optimal for the detection of anti-HCV antibody or HCV RNA.
FIG. 1.
Current HCV status of the chimpanzees at the Southwest Foundation for Biomedical Research. During the past 2 to 19 years, 52 of the 249 chimpanzees were inoculated with various NANBH inocula. HCV infection was confirmed in 46 animals. Serum samples collected in 1995 were analyzed for HCV RNA by RT-PCR analysis and for anti-HCV antibody by ELISA.
Evaluation of viral persistence and immune response to HCV proteins in a cohort of HCV-inoculated chimpanzees.
The relationship of anti-HCV status and ALT levels to viremia was examined in the 46 HCV-infected chimpanzees. Current status with regard to the circulating virus was determined by RT-PCR analysis of serum samples collected in 1995. The average duration of time since inoculation was 10.6 years. Serum samples from 18 (39%) of the chimpanzees were RT-PCR positive, while viral RNA was not detected in 28 (61%) of the chimpanzees (Fig. 1). The RT-PCR primers were from a highly conserved area of the 5′ noncoding region known to detect all HCV genotypes. In addition, no correlation was observed between the inoculating strain and viral persistence, since a number of both RT-PCR-positive and RT-PCR-negative animals had received the well-characterized Hutchinson strain of HCV 1a genotype. The initial RT-PCR analysis involved a single round of RT-PCR followed by Southern hybridization. Although this method detects an estimated 10 molecules of HCV, HCV may exist below the level of detection. Additional analysis of the chimpanzees that were antibody positive but RT-PCR negative (Ab+PCR−) was conducted by using a second round of PCR followed by Southern hybridization. All samples tested by a second round of PCR were negative. Convalescence was supported by the observation that most RT-PCR-negative animals were also anti-HCV antibody negative by ELISA (86%; 24/28). Thus, the RT-PCR-negative animals had most likely cleared the viral infection. The average duration of time since infection was similar for RT-PCR-positive (9.6 years) and RT-PCR-negative (11.6 years) animals. The high percentage of possible convalescence was unexpected, since only 20% of HCV-inoculated humans are expected to convalesce (3, 4, 6, 7, 45, 49, 62, 63).
The same serum samples were analyzed for anti-HCV antibody by using a second-generation ELISA and a third-generation recombinant immunoblot assay (RIBA). Anti-HCV antibody was detected by ELISA in 22 (48%) of the HCV-inoculated chimpanzees. Immunoblot analysis was performed on chimpanzee sera that were positive by ELISA, and 55, 91, 64, 73, and 73% were reactive with capsid, NS3-1, NS3-2, NS4, and NS5, respectively (Table 1). The low percentage of chimpanzees reactive to the HCV capsid protein has been described previously (8, 23, 34).
TABLE 1.
HCV-inoculated chimpanzee colony
| Group | Chimp no. | ALTa | RIBA analysisb
|
||||
|---|---|---|---|---|---|---|---|
| Capsid | NS3-1 | NS3-2 | NS4 | NS5 | |||
| Ab+PCR+ (n = 18) | −43 | N | − | 1+ | 1+ | 2+ | 3+ |
| x62 | 59 | 2+ | 3+ | 1+ | 2+ | 3+ | |
| x81 | 62 | − | 3+ | 1+ | 2+ | 3+ | |
| x83 | 51c | − | 2+ | 1+ | 2+ | − | |
| x99 | N | 2+ | 4+ | 1+ | 2+ | 3+ | |
| x119 | N | 3+ | − | 1+ | − | 3+ | |
| x123 | N | − | 4+ | 1+ | 3+ | 4+ | |
| x130 | N | 3+ | 4+ | 1+ | 2+ | 2+ | |
| x134 | 55c | 1+ | − | − | − | 3+ | |
| x174 | 72 | 2+ | 4+ | 2+ | 2+ | 3+ | |
| x183 | 73 | 1+ | 2+ | − | − | − | |
| x196 | 49c | − | 4+ | 1+ | 2+ | 3+ | |
| x204 | N | − | 3+ | − | 4+ | 4+ | |
| x216 | N | 1+ | 4+ | 1+ | 1+ | 1+ | |
| x258 | N | 1+ | 2+ | − | 2+ | − | |
| x304 | 45c | 2+ | 2+ | 1+ | 2+ | 3+ | |
| x341 | N | − | 3+ | 1+ | 2+ | 3+ | |
| x342 | N | − | 4+ | 2+ | 2+ | 4+ | |
| Percentage | 44 | 56 | 89 | 78 | 83 | 83 | |
| Ab+PCR− (n = 4) | x30 | N | − | 1+ | − | − | − |
| x65 | N | − | 3+ | − | 1+ | − | |
| x108 | N | 1+ | 2+ | − | − | − | |
| x189 | 61 | 2+ | 2+ | − | − | 2+ | |
| Percentage | 25 | 50 | 100 | 0 | 25 | 25 | |
| Ab−PCR− (n = 24)d | |||||||
Based on a minimum of eight samples over 4 years. The mean ALT value is indicated if it was over the normal cutoff. N, normal ALT value.
Serum samples were analyzed by RIBA for reactivity to capsid, NS3-1, NS3-2, NS4, and NS5; the intensity of the reaction is designated negative (−) to 4+ as indicated by the manufacturer. Chimpanzee x30 was indeterminate by RIBA.
The mean was not above the normal cutoff value of 55, but at least two samples were elevated.
These animals were RT-PCR negative and were negative for anti-HCV by ELISA, and so RIBA analysis was not performed. No ALT elevations were observed in this group.
RT-PCR positivity correlated with anti-HCV antibody positivity, since 100% (18/18) of the RT-PCR-positive chimpanzees were also anti-HCV antibody positive. Additionally, antibody positivity correlated with RT-PCR positivity, as 82% (18/22) of the antibody-positive chimpanzees were also RT-PCR positive (Fig. 1; Table 1). However, 18% (4/22) of the antibody-positive samples were HCV RNA negative. Ab+PCR− serum samples have been observed by other investigators (17, 58, 65). Chimpanzees with an Ab+PCR− status may have low levels of RNA that are undetectable by RT-PCR or a sustained anti-HCV antibody response in the absence of HCV RNA.
The percentages of chimpanzees with antibody reactivity to capsid and NS3-1 were similar in both Ab+PCR+ and Ab+PCR− groups (Table 1). However, these groups differed in the percentages of chimpanzees with antibody reactivity to NS3-2, NS4, and NS5. In the Ab+PCR+ group of chimpanzees, 78% (14/18) were anti-NS3-2 antibody positive, while an anti-NS3-2 antibody response was not observed in Ab+PCR− chimpanzees (0/4). Anti-NS4 and anti-NS5 antibody responses were observed in 83% (15/18) and 25% (1/4) of the Ab+PCR+ and Ab+PCR− chimpanzees, respectively. Therefore, antibody against NS3-2, NS4, and NS5 were observed more frequently in chimpanzees with viremia.
The relationship between antibody and RT-PCR positivity to ALT levels was examined as well. Elevated ALT levels in HCV-infected humans are usually defined as 2 to 2.5 times the upper limit of normal. Since ALT levels are rarely this high in HCV-infected chimpanzees after the acute-phase episode, the ALT level was considered elevated if the mean ALT value was above the upper limit of normal, or if two or more elevated ALT values were observed, but the mean ALT was within the normal range. The mean ALT values were calculated from at least two ALT values per year for the past 4 years. In the Ab+PCR+ group of chimpanzees, 44% (8/18) had elevated ALT values; one chimpanzee in the Ab+PCR− group had elevated ALT values; and all 24 Ab−PCR− chimpanzees had normal ALT values (Table 1).
Evaluation of viral persistence in six HCV-inoculated chimpanzees.
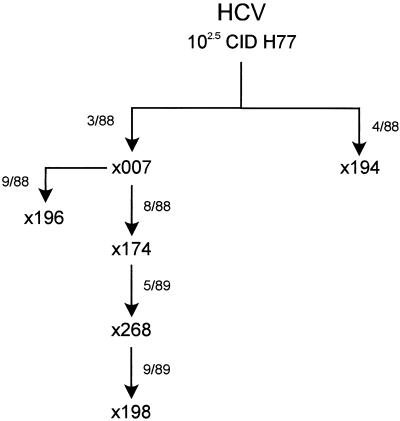
Since an unexpectedly high percentage of chimpanzees appeared to be convalescent in the cross-sectional study, a longitudinal study of the full clinical spectrum experienced by six HCV-infected chimpanzees was performed. These chimpanzees were not selected to represent different disease outcomes but were selected based on exposure to a common inoculum. The Hutchinson strain of HCV was serially inoculated into six chimpanzees between 1988 and 1989 (Fig. 2). Serial bleeds prior to infection and for 5 to 8 years after infection were examined for the presence of HCV RNA. All six chimpanzees were successfully infected, since HCV RNA was initially detected in the earliest tested serum sample (approximately 1 to 2 months postinoculation). In two chimpanzees (33%), HCV RNA was persistently detected in serial serum samples, including the most recent samples collected at 6 and 7 years postinoculation. In contrast, HCV RNA remained undetectable by RT-PCR analysis after 3 to 5 months postinoculation in serum samples collected from the other four chimpanzees (67%). Therefore, the percentages of chimpanzees with persistently detectable HCV RNA after HCV infection were similar in the cross-sectional (39%) and longitudinal (33%) studies.
FIG. 2.
Serial passage of the Hutchinson strain of HCV in chimpanzees. Chimpanzees x007 and x194 were inoculated with 102.5 CID50 of the Hutchinson strain (H77) of HCV in March and April 1988 (3/88 and 4/88), respectively. Acute-phase plasma from x007 was inoculated into x174 in August 1988. Concentrated tissue culture medium from hepatocytes obtained from x007 liver was used to inoculate x196 in September 1988 (27, 28). Acute-phase plasma from x174 was concentrated and inoculated into x268 in May 1989. Chimpanzee x198 was inoculated with acute-phase plasma from x268 in September 1989.
Evaluation of ALT levels and anti-HCV status in chimpanzees with persistent viremia.
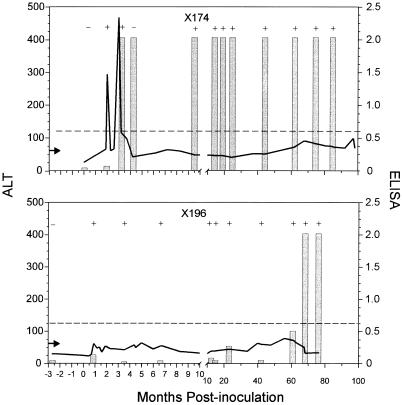
Analyses of serial bleeds for ALT levels, HCV RNA, and anti-HCV antibody status from the two chimpanzees with persistent viremia (x174 and x196) were performed to better understand the clinical course of the HCV infection in these animals. Although persistent viremia was observed in both chimpanzees, major differences in ALT levels and anti-HCV status were observed. One chimpanzee had significant elevations of ALT values and an early and long-lived anti-HCV antibody response, while the other animal had primarily normal ALT levels and an undetectable anti-HCV antibody response for 5 years.
Chimpanzee x174 was inoculated with acute phase plasma from x007 (Fig. 2). A biphasic pattern of peak ALT elevation was observed between 2 and 3 months postinoculation and was followed by periods of slightly elevated ALT levels intermittent with periods of normal ALT levels (Fig. 3). Consistently elevated ALT values were observed in sera examined after 62 months postinoculation. The first serum sample tested, excluding the prebleed, was HCV RNA positive based on RT-PCR analysis. All subsequent serum samples examined were RT-PCR positive with the exception of one sample collected at 4.2 months postinoculation. Anti-HCV antibody was first detected in serum collected from x174 at 3 months postinoculation, and all subsequent serum samples collected throughout the 7-year analysis were strongly anti-HCV antibody positive by ELISA (Fig. 3). Immunoblot analysis was performed on serial serum samples from x174 to determine the relationship of an antibody response elicited against capsid, NS3-1, NS3-2, NS4, and NS5 to viral RNA persistence (Table 2). An antibody response elicited against NS3-1 and NS4 was observed in serum collected at 2.6 months postinoculation. An additional antibody response elicited against NS3-2 was detected at 3.1 months postinoculation. Antibody specific for all five proteins was detected in serum samples collected at 75 and 85 months postinoculation.
FIG. 3.
ALT levels and anti-HCV status in HCV-inoculated chimpanzees with persistent viremia. The presence (+) or absence (−) of HCV RNA, detected by RT-PCR, is indicated. The arrow indicates the ALT upper normal limit (55 U/liter). The solid line indicates the ALT values. The bars represent the anti-HCV ELISA (Ortho HCV 2.0) OD values. The dashed line indicates the ELISA cutoff OD value.
TABLE 2.
Antibody responses in chronic and convalescent chimpanzees
| Group | Chimp no. | Mo PIa | HCV RNAb | RIBA analysisc
|
||||
|---|---|---|---|---|---|---|---|---|
| Capsid | NS3-1 | NS3-2 | NS4 | NS5 | ||||
| With persistent HCV infection | x174 | 0 | − | − | − | − | − | − |
| 2.6 | + | − | 1+ | − | 2+ | − | ||
| 3.1 | + | − | 2+ | 2+ | 2+ | − | ||
| 75 | + | 2+ | 4+ | 2+ | 2+ | 3+ | ||
| 85 | + | 2+ | 4+ | 2+ | 2+ | 3+ | ||
| x196 | −2.7 | − | − | − | − | − | − | |
| 62 | + | − | − | − | − | 1+ | ||
| 68 | + | − | 4+ | 1+ | 2+ | 3+ | ||
| 77 | + | − | 4+ | 1+ | 2+ | 3+ | ||
| With viral clearance | x194 | −0.2 | − | − | − | − | 1+ | − |
| 2 | + | − | 1+ | − | 2+ | − | ||
| 4.8 | − | − | 2+ | − | 2+ | − | ||
| 66d | − | − | 2+ | − | 2+ | − | ||
| x198 | −0.2 | − | − | − | − | − | − | |
| 2.9 | + | − | 3+ | − | 2+ | 2+ | ||
| 64d | − | − | 2+ | − | 1+ | − | ||
| x268 | −0.2 | − | − | − | − | − | − | |
| 2.6 | + | 1+ | − | − | − | − | ||
| 3.3 | − | 1+ | 2+ | − | 1+ | − | ||
| 7.3d | − | − | 2+ | − | − | − | ||
| 61d | − | − | 1+ | − | − | − | ||
| x007 | −2.8 | − | − | − | − | − | − | |
| 1.8 | + | − | − | − | 2+ | − | ||
| 2.2 | − | − | 1+ | − | 2+ | − | ||
| 6.9d | − | − | 1+ | − | − | − | ||
| 67d | − | − | − | − | − | − | ||
Negative values indicate prebleeds taken before inoculation.
Detected by RT-PCR as described in Materials and Methods.
Serum samples were analyzed by RIBA; the intensity of the reaction is designated negative (−) to 4+ as indicated by the manufacturer.
Sample taken at a time when the animal was no longer ELISA positive.
Chimpanzee x196 was inoculated with concentrated tissue culture medium from hepatocytes obtained from x007 by liver wedge surgery (27, 28). A significant ALT elevation was not observed during the acute phase of infection for x196, and ALT levels were primarily below the upper limit of normal, although slightly elevated ALT levels were observed intermittently throughout the analysis (Fig. 3). The first serum sample tested, excluding the prebleed, was HCV RNA positive based on RT-PCR analysis. HCV RNA was consistently detected in all subsequent serum samples analyzed throughout 77 months postinoculation. An anti-HCV antibody response recently emerged in x196 but was undetectable for the first 5 years postinoculation. Since x196 was persistently HCV RNA positive, x196 was considered to be a long-term antibody-negative silent carrier. An anti-HCV antibody response which was negative based on the ELISA cutoff value was first detected at 62 months (5 years) postinoculation (Fig. 3). Immunoblot analysis of this serum sample revealed that the first detectable anti-HCV response was elicited against NS5 (Table 2). A stronger anti-HCV antibody response, which was considered positive based on the ELISA cutoff value, was observed in serum samples collected at 68 and 77 months postinoculation. Immunoblot analysis of these serum samples revealed that antibody responses were elicited against all antigens except capsid.
Evaluation of ALT levels and anti-HCV status in chimpanzees with viral clearance.
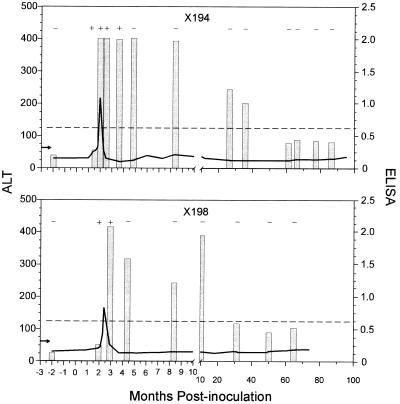
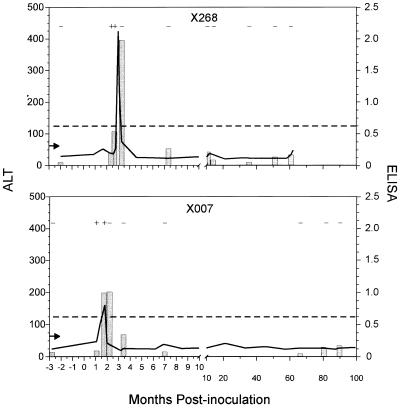
The four chimpanzees that cleared the virus were examined by using the same approach as used for the persistently infected animals to determine if viral clearance was associated with an antibody response elicited to any particular HCV protein(s). Similarities were observed in the patterns of viral clearance and ALT levels. However, two very different antibody profiles were observed. A typical convalescent antibody profile was observed in two animals (x194 and x198), with a gradual reduction in anti-HCV antibody throughout 7 years of analysis (Fig. 4). In contrast, a rapid loss of antibody occurred in the other two convalescent animals (x007 and x268) and represented an atypical convalescent antibody profile (Fig. 5).
FIG. 4.
ALT levels and anti-HCV status in HCV-inoculated chimpanzees with a gradual reduction in anti-HCV antibody. For details, see the legend to Fig. 3.
FIG. 5.
ALT levels and anti-HCV status in HCV-inoculated chimpanzees with a rapid loss of anti-HCV antibody. For details, see the legend to Fig. 3.
Chimpanzee x194 was inoculated with 102.5 50% chimpanzee infective doses (CID50) of the Hutchinson strain of HCV (Fig. 2). A sharp elevation in ALT was observed around 2 months postinoculation, which returned to normal within 2 weeks (Fig. 4). Excluding the prebleed, the first four serum samples tested were HCV RNA positive based on RT-PCR analysis. However, serum collected from x194 was HCV RNA negative at 4.8 months postinoculation and at all subsequent time points examined throughout 7 years of analysis. A gradual reduction in anti-HCV antibody was observed in x194. Anti-HCV antibody was first detected by ELISA in a serum sample collected at 2 months postinoculation. Although x194 was no longer viremic at 4.8 months postinoculation, a serum sample collected at 36 months postinoculation was still anti-HCV antibody positive by ELISA. Samples examined beyond 61 months were ELISA negative by the cutoff; however, anti-HCV antibody appeared to be sustained at low levels, since even the last serum sample examined from x194, over 7 years postinoculation, still had an elevated optical density (OD) reading by ELISA. Immunoblot analysis was performed on serial serum samples from x194 to determine if a relationship existed between antibody response to individual antigens and clearance of viremia. The serum sample collected from x194 prior to HCV inoculation contained antibody slightly reactive with NS4. Antibody responses elicited against NS3-1 and NS4 were observed in serum samples collected from x194 at 2, 4.8, and 66 months postinoculation (Table 2). The similar RIBA patterns at 4.8 and 66 months fail to explain the loss of ELISA reactivity in the later sample and exemplify the differences in these two assays.
Chimpanzee x198 was inoculated with acute-phase plasma from x268 (Fig. 2). A sharp elevation in the ALT level was observed in x198 at about 2 months postinoculation (Fig. 4). The ALT level returned to normal within 2 weeks and was consistently below the upper limit of normal throughout the analysis. The first two postinoculation serum samples tested were HCV RNA positive. A serum sample collected from x198 at 4.3 months postinoculation and all samples examined at later time points were HCV RNA negative based on RT-PCR analysis. As in x194, anti-HCV antibody in x198 gradually declined following the loss of RT-PCR positivity. Anti-HCV antibody was first detected by ELISA in a serum sample collected at 2.9 months postinoculation. Although x198 was no longer viremic, a serum sample collected at 10 months postinoculation was anti-HCV antibody positive by ELISA. Serum samples collected at 32 to 64 months postinoculation were just below the cutoff value for the ELISA, suggesting that anti-HCV antibody was long lived. Immunoblot analysis revealed that an antibody response elicited to NS3-1, NS4, and NS5 was detected in serum collected 2.9 months postinoculation (Table 2). Anti-NS5 antibody was presumably lost in the absence of viremia and was not detected in serum collected at 64 months postinoculation.
Chimpanzee x268 was inoculated with concentrated acute-phase plasma from x174 (Fig. 2). The ALT level peaked at 3 months postinoculation and returned to normal after about 1.5 months (Fig. 5). The first two postinoculation serum samples tested were HCV RNA positive based on RT-PCR analysis. Chimpanzee x268 apparently cleared the virus at 3.3 months postinoculation, because HCV RNA was not detected in any serum examined from x268 between 3.3 and 61 months postinoculation. The first two serum samples collected from x268 at 2.6 and 3.3 months postinoculation were anti-HCV antibody positive by ELISA. However, the anti-HCV antibody declined rapidly. Serum samples collected from x268 at 7.3 and 10 months postinoculation were antibody negative by ELISA and had OD readings well below the cutoff value. By RIBA, chimpanzee x268 initially elicited an antibody response against capsid at 2.6 months postinoculation (Table 2). Antibodies specific for capsid, NS3-1, and NS4 were detected at 3.3 months postinoculation. Serum collected from x268 at 7.3 and 61 months postinoculation contained antibody specific for NS3-1, but antibodies specific for capsid and NS4 were no longer present.
Chimpanzee x007, like x194, was inoculated with 102.5 CID50 of the Hutchinson strain of HCV (Fig. 2). The ALT level peaked at about 2 months postinoculation and returned to normal within 2 weeks (Fig. 5). The first two postinoculation serum samples tested were HCV RNA positive. However, serum collected from x007 was HCV RNA negative at 2.2 months postinoculation and at all subsequent time points examined. Serum samples collected from x007 at 1.8 and 2.2 months postinoculation were anti-HCV antibody positive by ELISA; however, the ELISA reactivity fell rapidly. Serum samples collected from x007 at 3.4 and 6.9 months postinoculation had OD readings of 0.365 and 0.083, respectively, and were anti-HCV antibody negative by ELISA. All serum samples subsequently collected were anti-HCV antibody negative by ELISA. Immunoblot analysis revealed that x007 elicited an antibody response against NS4 at 1.8 months postinoculation and against NS3-1 and NS4 at 2.2 months postinoculation (Table 2). Serum collected from x007 at 6.9 months postinoculation contained antibody specific for NS3-1, and anti-HCV antibodies were not detected in serum collected at 67 months postinoculation.
Relationship of antibody response to capsid, NS3-1, NS3-2, NS4, and NS5 to viremia.
Immunoblot analysis of the latest bleeds from the two chimpanzees with persistent viremia indicated that x174 and x196 elicited an antibody response to NS3-1, NS3-2, NS4, and NS5 (Table 2). An anticapsid antibody response was observed in x174 but not in x196. These chimpanzees remained RT-PCR positive regardless of the humoral immune response elicited to the HCV recombinant proteins, indicating that persistence was not due to the lack of a humoral immune response to these antigens.
Immunoblot analysis of serum samples from the four convalescent chimpanzees revealed that none of the chimpanzees elicited an anti-NS3-2 antibody response. An anti-NS4 antibody response was observed in all six chimpanzees but only transiently in the two convalescent animals with rapid loss of antibody. Only one convalescent chimpanzee, x198, elicited an anti-NS5 antibody response. The antibody response to NS5 in x198 was transient and was detectable during the viremic phase at 3 months postinoculation but not at 64 months postinoculation. The longitudinal analysis of a small number of animals suggests that most animals destined to clear the virus fail to respond to NS3-2 and NS5. The lack of responses to NS3-2 and NS5 in the convalescent chimpanzees may be due to insufficient time to elicit an immune response prior to viral clearance. Examination of the RIBA reactivities in the cross-sectional analysis of the 22 antibody-positive animals also suggested that chronically infected animals have reactivity to NS3-2 and NS5 (78 and 83%), while most ELISA-positive convalescent animals do not (0 and 25%). In the cross-sectional study, the probability of detecting antibodies to NS3-2 and NS5 in chronically infected chimpanzees was greater than that of ELISA-positive convalescent animals (P = 0.0096 and P = 0.046 respectively; Fisher’s exact test). ELISA-negative convalescent animals were not examined by RIBA because ELISA-negative animals are unlikely to be positive in the assay. Therefore, an anti-NS3-2 or an anti-NS5 antibody response may be a marker for chronicity. However, in the cross-sectional study, it could not be determined whether convalescent animals failed to respond to these antigens or lost the response following viral clearance.
DISCUSSION
Currently, the chimpanzee serves as the only animal model for HCV infection. Although the chimpanzee animal model has been valuable in examining many factors in HCV infection, the characteristics of HCV infection in chimpanzees compared to humans is not well understood (47). To better understand the chimpanzee animal model for HCV infection, a cross-sectional analysis of a cohort of 46 HCV-infected chimpanzees and a longitudinal analysis of 6 chimpanzees were performed.
An unexpectedly high percentage of chimpanzees appeared to convalesce from HCV infection in both the cross-sectional (61%) and longitudinal (67%) studies. The high percentage of convalescence observed in chimpanzees did not appear to be correlated with the inocula, the duration of time since inoculation, or the presence of anti-HCV antibody. Viral persistence or clearance did not appear to be correlated with a particular strain of HCV, since both RT-PCR-positive and RT-PCR-negative chimpanzees received the well-characterized Hutchinson strain of HCV as an inoculum. Since the Hutchinson strain of HCV was serially passaged in the chimpanzees examined in the longitudinal study, some argument could be made for the attenuation of the virus by animal passage. However, x007 and x194, both convalescent animals, received the unpassaged Hutchinson inocula. The average durations of time since infection were also similar for both RT-PCR-positive and RT-PCR-negative chimpanzees. As in human studies, a good correlation between anti-HCV antibody and HCV RNA was observed in chimpanzees. However, the high percentage of convalescence in chimpanzees could not be associated with a humoral immune response elicited to a specific HCV antigen in the RIBA.
The cross-sectional study revealed four Ab+PCR− chimpanzees. Second-round RT-PCR was performed after the initial RT-PCR to confirm that these animals were negative by RT-PCR. However, levels of HCV RNA below the sensitivity of the assay may exist, and these animals may actually still harbor the virus at low levels. Elevated ALT levels were observed in one Ab+PCR− animal (x189), suggesting that this animal may still be viremic. Long-lasting anti-HCV antibody responses appeared to be rare in the chimpanzees, since most RT-PCR-negative animals were also anti-HCV antibody negative. However, a long-lasting anti-HCV antibody response was observed by RIBA in the absence of viremia in x194 and x198 in the longitudinal analysis for approximately 5 years after viral clearance, and the Ab+PCR− chimpanzees in the cross-sectional study may likewise have a lasting anti-HCV antibody response in the absence of viremia.
The pattern of antibodies was dissimilar among Ab+PCR+ and Ab+PCR− chimpanzees. A higher percentage of persistently infected, Ab+PCR+ chimpanzees than of Ab+PCR− chimpanzees elicited an antibody response to NS3-2, NS4, and NS5. This finding must be interpreted with caution due to the low numbers of Ab+PCR− animals. However, persistently infected chimpanzees may respond more frequently to NS3-2, NS4, and NS5 due to constant or higher levels of antigenic stimulation. If NS3-2, NS4, and NS5 are poor immunogens, Ab+PCR− chimpanzees may have cleared the virus prior to eliciting an immune response against these antigens. Alternatively, short-lived anti-NS3-2, anti-NS4, and anti-NS5 antibody responses may have been elicited in these animals. The longitudinal analysis revealed that anti-NS3-2 antibody responses were never detected in the chimpanzees that cleared the virus, suggesting that a response was never elicited. An anti-NS4 antibody response was observed in all six chimpanzees but only transiently in the two convalescent animals with rapid loss of antibody, suggesting that both short- and long-lived antibody responses to NS4 can be elicited. An anti-NS5 antibody response was initially observed in one chimpanzee that cleared the virus but was not detected in a later serum sample, suggesting that the anti-NS5 antibody response was not long lived. Similarly, Farci and coworkers observed that an anti-NS5 antibody response persisted in animals with chronic infection but reappeared and then disappeared in rechallenged chimpanzees with transient viremia (16).
Based on data from eight previous chimpanzee studies which used RT-PCR analysis to determine if HCV-inoculated chimpanzees experienced self-limited or persistent infection, the average frequency of persistent infection observed in chimpanzees is estimated at 58% (2, 8, 16, 18, 23, 48, 53, 64). However, the frequency of persistent infection in chimpanzees is difficult to determine based on multiple early studies, since different RT-PCR methods were used and since only a small panel (average = 8, range = 4 to 19) of chimpanzees was examined. Although more studies are necessary to evaluate the rate of persistent infection in HCV-inoculated chimpanzees, the rate of persistent infection in the human population appears to be much higher and has been estimated at 70 to 90% (3, 4, 6, 7, 45, 49, 62, 63). The surprisingly low percentage of chimpanzees with persistent HCV infections compared to previous studies involving human populations may be explained if chimpanzees experience a different clinical course than the human population. Alternatively, the full clinical spectrum of HCV-infected humans may not be observed in studies that select individuals based on virological or disease status.
An estimated 20% of HCV-infected humans are expected to convalesce, in contrast to approximately 60% of HCV-infected chimpanzees in this study. If the percentage of chimpanzees that convalesce can be extrapolated to the human population, the frequency of humans that clear the virus may be several times higher than estimated. Since most HCV infections are asymptomatic, and many individuals have normal ALT levels throughout the infection (3, 15, 44, 55), humans that clear the virus and become antibody negative would not be detected in clinical studies or as HCV-exposed blood donors. The selection criteria used for NANBH studies, the potential for rapid loss of antibody in convalescent humans, and the potential presence of silent HCV carriers in the human population may cumulatively lead to the underestimation of HCV infection in the human population. In fact, individuals with rapid viral clearance and antibody loss, as seen in 33% of longitudinally examined chimpanzees, would be virtually invisible in clinical studies without very closely spaced serial bleeds from the time of exposure.
The frequency of persistent HCV infection has been estimated at approximately 80% based on selected populations of individuals diagnosed with posttransfusion NANBH (1, 7, 17, 37, 49). The diagnosis of NANBH was primarily based on elevated ALT levels (24). Generally, individuals were suspected to have hepatitis if there were at least two consecutive serum samples within a 2-week interval in which one ALT level was above the normal range and the second was at least 2 to 2.5 times the upper limit of normal. The elevation in ALT was considered relevant if it occurred from days 11 through 180 after the transfusion, and NANBH was diagnosed if hepatitis could not be attributed to hepatitis A virus infection, HBV infection, or other causative agents. In contrast, for studies performed on chimpanzees, the frequency of persistent infection is based on all inoculated animals. The criteria for selection of humans in the posttransfusion hepatitis studies may have excluded a significant percentage of HCV-infected patients who did not experience ALT levels above 2 to 2.5 times the upper limit of normal. If individuals without significant ALT elevations are more apt to clear their infections, the frequency of HCV infection and viral clearance may be higher than expected. In our previous analysis of 50 NANBH-inoculated chimpanzees, 48% of the chimpanzees would have been excluded based on ALT levels that did not meet the criteria for selection in human posttransfusion studies (34). Additionally, this study and others have documented acute HCV infection in the absence of ALT elevations in the acute phase of disease (17, 47, 49). The frequency of persistent HCV infection has also been estimated based on cohorts of individuals with community-acquired hepatitis and IVDUs. However, the selection of individuals participating in the community-acquired hepatitis study was based on elevated ALT levels and the diagnosis of NANBH (6). In contrast, Thomas and coworkers performed a prospective study of HCV infection in a cohort of IVDUs in which participants were not selected based on the diagnosis of NANBH but were eligible if they had a history of injecting illicit drugs within the previous 10 years (61, 62). A high rate of RT-PCR positivity was also observed in the IVDU cohort and may be explained by either a high rate of persistent infection or by convalescence followed by continuous reexposure and reinfection with HCV. Studies in chimpanzees have demonstrated that immunity to HCV appears to be insufficient to protect against reinfection (16, 46–48). Therefore, it is unclear if the lower percentage of chimpanzees with persistent HCV infection compared to the human population is due to differences in the clinical courses or if the full clinical spectrum of HCV-infected humans is not observed in studies that select individuals based on disease status or high-risk activities.
The frequency of HCV infection may also be underestimated in the human population if similarities in antibody profiles exist between humans and chimpanzees that clear the virus. Both rapid and gradual loss of anti-HCV antibody titer was observed in the chimpanzees that cleared the virus. If rapid loss of antibody occurs in the human population, as in the chimpanzees, the duration of time that HCV infection could be detected would be greatly reduced, and the frequency of HCV infection and viral clearance may be underestimated. Resolved HCV infections in seronegative humans may be more common than is generally suspected, since the loss of anti-HCV antibody has been observed in HCV-infected humans (36, 49), and since HCV-specific cytotoxic T-lymphocyte responses are occasionally detected in the normal control population (11, 50). The frequency of HCV infection may also be underestimated if a significant percentage of silent carriers exist within the human population. Chimpanzee x196 was anti-HCV antibody negative and HCV RNA positive for 5 years. If extrapolated to the human population, this finding suggests that a significant percentage of human chronic carriers may go unrecognized. Silent carriers have been documented in HCV-infected humans (10, 29, 52). However, it is difficult to determine the actual percentage of silent carriers in the human population without performing longitudinal RT-PCR studies on thousands of individuals or at least numerous high-risk individuals (IVDUs). Due to the very rapid loss of RT-PCR reactivity seen in four of six animals, longitudinal studies in humans would need to examine very closely spaced serial bleeds.
The characteristics of HCV infection in the chimpanzee animal model is important to assess, since this animal will most likely be used to examine potential therapies and vaccines. The results of such experiments must be carefully interpreted due to the high percentage of naturally occurring viral clearance (60%). Regardless of whether HCV-infected chimpanzees are truly representative of HCV infection in humans, the chimpanzee animal model will be valuable in understanding the mechanism of viral clearance. Early vaccine trials for HCV in chimpanzees attributed clearance of viremia to attenuation of the infection due to partial protection by the vaccine (12). Such interpretations are surely complicated by the findings presented in this study.
ACKNOWLEDGMENTS
This work was supported by grant AI40035 from the National Institutes of Health.
We thank Mark Sharp for many helpful discussions of the data and for help with the statistical analysis.
REFERENCES
- 1.Aach R D, Stevens C E, Hollinger F B, Mosley J W, Peterson D A, Taylor P E, Johnson R G, Barbosa L H, Nemo G J. Hepatitis C virus infection in post-transfusion hepatitis. N Engl J Med. 1991;325:1325–1329. doi: 10.1056/NEJM199111073251901. [DOI] [PubMed] [Google Scholar]
- 2.Abe K, Inchauspe G, Shikata T, Prince A M. Three different patterns of hepatitis C virus infection in chimpanzees. Hepatology. 1992;15:690–695. doi: 10.1002/hep.1840150423. [DOI] [PubMed] [Google Scholar]
- 3.Alter H J. To C or not to C: these are the questions. J Am Soc Hematol. 1995;85:1681–1695. [PubMed] [Google Scholar]
- 4.Alter H J, Purcell R H, Shih J W, Melpolder J C, Houghton M, Choo Q L, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494–1500. doi: 10.1056/NEJM198911303212202. [DOI] [PubMed] [Google Scholar]
- 5.Alter M J. Epidemiology of hepatitis C in the west. Semin Liver Dis. 1995;15:5–14. doi: 10.1055/s-2007-1007259. [DOI] [PubMed] [Google Scholar]
- 6.Alter M J, Margolis H S, Krawczynski K, Judson F N, Mares A, Alexander W J, Hu P Y, Miller J K, Gerber M A, Sampliner R E, Meeks E L, Beach M J. The natural history of community-acquired hepatitis C in the United States. N Engl J Med. 1992;327:1899–1905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 7.Barrera J M, Bruguera M, Ercilla M G, Gil C, Celis R, Gil M P, Del Valle Onorato M, Rodés J, Ordinas A. Persistent hepatitis C viremia after acute self-limiting posttransfusion hepatitis C. Hepatology. 1995;21:639–644. [PubMed] [Google Scholar]
- 8.Beach M J, Meeks E L, Mimms L T, Vallari D, DuCharme L, Spelbring J, Taskar S, Schleicher J B, Krawczynski K, Bradley D W. Temporal relationships of hepatitis C virus RNA and antibody responses following experimental infection of chimpanzees. J Med Virol. 1992;36:226–237. doi: 10.1002/jmv.1890360314. [DOI] [PubMed] [Google Scholar]
- 9.Bukh J, Purcell R H, Miller R H. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci USA. 1993;90:8234–8238. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukh J, Wantzin P, Krogsgaard K, Knudsen F, Purcell R H, Miller R H Copenhagen Dialysis HCV Study Group. High prevalence of hepatitis C virus (HCV) RNA in dialysis patients: failure of commercially available antibody tests to identify a significant number of patients with HCV infection. J Infect Dis. 1993;168:1343–1348. doi: 10.1093/infdis/168.6.1343. [DOI] [PubMed] [Google Scholar]
- 11.Cerny A, McHutchison J G, Pasquinelli C, Brown M E, Brothers M A, Grabscheid B, Fowler P, Houghton M, Chisari F V. Cytotoxic T lymphocyte response to hepatitis C virus-derived peptides containing the HLA A2.1 binding motif. J Clin Invest. 1995;95:521–530. doi: 10.1172/JCI117694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choo Q-L, Kuo G, Ralston R, Weiner A, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C, Kansopon J, McFarland J, Tabrizi A, Ching K, Moss B, Cummins L B, Houghton M, Muchmore E. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci USA. 1994;91:1294–1298. doi: 10.1073/pnas.91.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choo Q-L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dusheiko G, Schmilovitz-Weiss H, Brown D, McOmish F, Yap P-L, Sherlock S, McIntyre N, Simmonds P. Hepatitis C virus genotypes: an investigation of type-specific differences in geographic origin and disease. Hepatology. 1994;19:13–18. [PubMed] [Google Scholar]
- 15.Esteban J I, Lopez-Talavera J C, Genesca J, Madoz P, Viladomiu L, Muniz E, Martin-Vega C, Rosell M, Allende H, Vidal X, Gonzalez A, Hernandez J M, Esteban R, Guardia J. High rate of infectivity and liver disease in blood donors with antibodies to hepatitis C virus. Ann Intern Med. 1991;115:443–449. doi: 10.7326/0003-4819-115-6-443. [DOI] [PubMed] [Google Scholar]
- 16.Farci P, Alter H J, Govindarajan S, Wong D C, Engle R, Lesniewski R R, Mushahwar I K, Desai S M, Miller R H, Ogata N, Purcell R H. Lack of protective immunity against reinfection with hepatitis C virus. Science. 1992;258:135–140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 17.Farci P, Alter H J, Wong D, Miller R H, Shih J W, Jett B, Purcell R H. A long-term study of hepatitis C virus replication in non-A, non-B hepatitis. N Engl J Med. 1991;325:98–104. doi: 10.1056/NEJM199107113250205. [DOI] [PubMed] [Google Scholar]
- 18.Farci P, London W T, Wong D C, Dawson G J, Vallari D S, Engle R, Purcell R H. The natural history of infection with hepatitis C virus (HCV) in chimpanzees: comparison of serologic responses measured with first- and second-generation assays and relationship to HCV viremia. J Infect Dis. 1992;165:1006–1011. doi: 10.1093/infdis/165.6.1006. [DOI] [PubMed] [Google Scholar]
- 19.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunji T, Kato N, Hijikata M, Hayashi K, Saitoh S, Shimotohno K. Specific detection of positive and negative stranded hepatitis C viral RNA using chemical RNA modification. Arch Virol. 1994;134:293–302. doi: 10.1007/BF01310568. [DOI] [PubMed] [Google Scholar]
- 23.Hilfenhaus J, Krupka U, Nowak T, Cummins L B, Fuchs K, Roggendorf M. Follow-up of hepatitis C virus infection in chimpanzees: determination of viraemia and specific humoral immune response. J Gen Virol. 1992;73:1015–1019. doi: 10.1099/0022-1317-73-4-1015. [DOI] [PubMed] [Google Scholar]
- 24.Hollinger F B, Mosley J W, Szmuness W, Aach R D, Melnick J L, Afifi A, Stevens C E, Kahn R A. Non-A, non-B hepatitis following blood transfusion: risk factors associated with donor characteristics. In: Szmuness W, Alter H J, Maynard J E, editors. Viral hepatitis 1981 international symposium. Philadelphia, Pa: The Franklin Institute Press; 1989. pp. 361–376. [Google Scholar]
- 25.Honda M, Ping L H, Rijnbrand R C A, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 26.Houghton M, Weiner A, Han J, Kuo G, Choo Q-L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of disease. Hepatology. 1991;14:381–388. [PubMed] [Google Scholar]
- 27.Jacob J R, Burk K H, Eichberg J W, Dreesman G R, Lanford R E. Expression of infectious viral particles by primary chimpanzee hepatocytes isolated during the acute phase of non-A, non-B hepatitis. J Infect Dis. 1990;161:1121–1127. doi: 10.1093/infdis/161.6.1121. [DOI] [PubMed] [Google Scholar]
- 28.Jacob J R, Sureau C, Burk K H, Eichberg J W, Dressman G R, Lanford R E. In vitro replication of non-A, non-B hepatitis virus. In: Hollinger F B, Lemon S M, Margolis H S, editors. Viral hepatitis and liver disease. Baltimore, Md: Williams & Wilkins; 1991. pp. 387–392. [Google Scholar]
- 29.Kao J H, Lai M Y, Hwang Y T, Yang P M, Chen P J, Sheu J C, Wang T H, Hsu H C, Chen D S. Chronic hepatitis C without anti-hepatitis C antibodies by second-generation assay—a clinicopathologic study and demonstration of the usefulness of a third-generation assay. Digest Dis Sci. 1996;41:161–165. doi: 10.1007/BF02208599. [DOI] [PubMed] [Google Scholar]
- 30.Kato N, Ootsuyama Y, Tanaka T, Nakagawa M, Nakazawa T, Muraiso K, Ohkoshi S, Hijikata M, Shimotohno K. Marked sequence diversity in the putative envelope proteins of hepatitis C viruses. Virus Res. 1992;22:107–123. doi: 10.1016/0168-1702(92)90038-b. [DOI] [PubMed] [Google Scholar]
- 31.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanford R E, Chavez D, Chisari F, Sureau C. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J Virol. 1995;69:8079–8083. doi: 10.1128/jvi.69.12.8079-8083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanford R E, Notvall L, Barbosa L H, Eichberg J W. Evaluation of a chimpanzee colony for antibodies to hepatitis C virus. J Med Virol. 1991;34:148–153. doi: 10.1002/jmv.1890340303. [DOI] [PubMed] [Google Scholar]
- 35.Lanford R E, Sureau C, Jacob J R, White R, Fuerst T R. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology. 1994;202:606–614. doi: 10.1006/viro.1994.1381. [DOI] [PubMed] [Google Scholar]
- 36.Lefrère J J, Guiramand S, Lefrère F, Mariotti M, Aumont P, Lerable J, Petit J C, Girot R, Morand-Joubert L. Full or partial seroreversion in patients infected by hepatitis C virus. J Infect Dis. 1997;175:316–322. doi: 10.1093/infdis/175.2.316. [DOI] [PubMed] [Google Scholar]
- 37.Lelie P N, Cuypers H T M, Reesink H W, van der Poel C L, Winkel I, Bakker E, van Exel-Oehlers P J, Vallari D, Allain J P, Mimms L. Patterns of serological markers in transfusion-transmitted hepatitis C virus infection using second-generation HCV assays. J Med Virol. 1992;37:203–209. doi: 10.1002/jmv.1890370310. [DOI] [PubMed] [Google Scholar]
- 38.Lohmann, V., J. O. Koch, and R. Bartenschlager. 1996. Processing pathways of the hepatitis C virus proteins. J. Hepatol. 24(Suppl. 2):11–19. [PubMed]
- 39.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto M, Hsieh T-Y, Zhu N, VanArsdale T, Hwang S B, Jeng K-S, Gorbalenya A E, Lo S-Y, Ou J-H, Ware C F, Lai M M. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxic-B receptor. J Virol. 1997;71:1301–1309. doi: 10.1128/jvi.71.2.1301-1309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGuinness P H, Bishop G A, McCaughan G W, Trowbridge R, Gowans E J. False detection of negative-strand hepatitis C virus RNA. Lancet. 1994;343:551–552. [PubMed] [Google Scholar]
- 42.Miller R H, Purcell R H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci USA. 1990;87:2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizutani T, Kato N, Saito S, Ikeda M, Sugiyama K, Shimotohno K. Characterization of hepatitis C virus replication in cloned cells obtained from a human T-cell leukemia virus type 1-infected cell line, MT-2. J Virol. 1996;70:7219–7223. doi: 10.1128/jvi.70.10.7219-7223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morales T G, Sampliner R E, Bhattacharyya A, Alter M J. Liver histology in anti-HCV-positive persons with normal or minimally elevated aminotransferases. J Clin Gastroenterol. 1995;21:301–305. doi: 10.1097/00004836-199512000-00011. [DOI] [PubMed] [Google Scholar]
- 45.National Institutes of Health. National Institutes of Health Consensus development conference panel statement: management of hepatitis C. Hepatology. 1997;26:2S–10S. doi: 10.1002/hep.510260701. [DOI] [PubMed] [Google Scholar]
- 46.Okamoto H, Mishiro S, Tokita H, Tsuda F, Miyakawa Y, Mayumi M. Superinfection of chimpanzees carrying hepatitis C virus of genotype II/1b with that of genotype III/2a or I/1a. Hepatology. 1994;20:1131–1136. [PubMed] [Google Scholar]
- 47.Prince A M, Brotman B. The biology of hepatitis C virus infection. Curr Stud Hematol Blood Transf. 1994;61:195–207. doi: 10.1159/000423276. [DOI] [PubMed] [Google Scholar]
- 48.Prince A M, Brotman B, IIuima T, Pascual D, Jaffery M, Inchauspe G. Immunity in hepatitis C infection. J Infect Dis. 1992;165:438–443. doi: 10.1093/infdis/165.3.438. [DOI] [PubMed] [Google Scholar]
- 49.Prince A M, Brotman B, Inchauspe G, Pascual D, Nasoff M, Hosein B, Wang C Y. Patterns of prevalence of hepatitis C virus infection in posttransfusion non-A, non-B hepatitis. J Infect Dis. 1993;167:1296–1301. doi: 10.1093/infdis/167.6.1296. [DOI] [PubMed] [Google Scholar]
- 50.Rehermann B, Chang K M, McHutchison J, Kokka F, Houghton M, Rice C M, Chisari F V. Differential cytotoxic T-lymphocyte responsiveness to the hepatitis B and C viruses in chronically infected patients. J Virol. 1996;70:7092–7102. doi: 10.1128/jvi.70.10.7092-7102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sallie R, King R, Silva E, Tibbs C, Johnson P, Williams R. Community prevalence of hepatitis C viraemia: a polymerase chain reaction study. J Med Virol. 1994;43:111–114. doi: 10.1002/jmv.1890430202. [DOI] [PubMed] [Google Scholar]
- 53.Schlauder G G, Leverenz G J, Amann C W, Lesniewski R R, Peterson D A. Detection of the hepatitis C virus genome in acute and chronic experimental infection in chimpanzees. J Clin Microbiol. 1991;29:2175–2179. doi: 10.1128/jcm.29.10.2175-2179.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selby M J, Berger K, Kuo G, Glazer E, Eckart M, Lee C, Chien D, Kuo C, Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993;74:1103–1113. doi: 10.1099/0022-1317-74-6-1103. [DOI] [PubMed] [Google Scholar]
- 55.Shakil A O, Conry-Cantilena C, Alter H J, Hayashi P, Kleiner D E, Tedeschi V, Krawczynski K, Conjeevaram H S, Sallie R, Di Bisceglie A M. Volunteer blood donors with antibody to hepatitis C virus: clinical, biochemical, virologic, and histologic features. Ann Intern Med. 1995;123:330–337. doi: 10.7326/0003-4819-123-5-199509010-00002. [DOI] [PubMed] [Google Scholar]
- 56.Shimizu Y K, Iwamoto A, Hijikata M, Purcell R H, Yoshikura H. Evidence for in vitro replication of hepatitis C virus genome in a human T-cell line. Proc Natl Acad Sci USA. 1992;89:5477–5481. doi: 10.1073/pnas.89.12.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simmonds P, Alberti A, Alter H J, Bonino F, Bradley D W, Brechot C, Brouwer J T, Chan S-W, Chayama K, Chen D-S, Choo Q-L, Colombo M, Cuypers H T M, Date T, Dusheiko G M, Esteban J I, Fay O, Hadziyannis S J, Han J, Hatzakis A, Holmes E C, Hotta H, Houghton M, Irvine B. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology. 1994;19:1321–1324. [PubMed] [Google Scholar]
- 58.Suzuki E, Kaneko S, Udono T, Tanoue T, Hayashi Y, Yoshihara N, Murakami S, Hattori N, Hayashi M, Sasaoka S, Mitani T, Kurono M, Sawai K, Kobayashi K. Absence of nonpercutaneous transmission of hepatitis C virus in a colony of chimpanzees. J Med Virol. 1993;39:286–291. doi: 10.1002/jmv.1890390406. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka T, Kato N, Cho M J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taniguchi S, Okamoto H, Sakamoto M, Kojima M, Tsuda F, Tanaka T, Munekata E, Muchmore E E, Peterson D A, Mishiro S. A structurally flexible and antigenically variable N-terminal domain of the hepatitis C virus E2/NS1 protein: implication for an escape from antibody. Virology. 1993;195:297–301. doi: 10.1006/viro.1993.1378. [DOI] [PubMed] [Google Scholar]
- 61.Thomas D L, Shih J W, Alter H J, Vlahov D, Cohn S, Hoover D R, Cheung L, Nelson K E. Effect of human immunodeficiency virus on hepatitis C virus infection among injecting drug users. J Infect Dis. 1996;174:690–695. doi: 10.1093/infdis/174.4.690. [DOI] [PubMed] [Google Scholar]
- 62.Thomas D L, Vlahov D, Solomon L, Cohn S, Taylor E, Garfein R, Nelson K E. Correlates of hepatitis C virus infections among injection drug users. Medicine (Baltimore) 1995;74:212–220. doi: 10.1097/00005792-199507000-00005. [DOI] [PubMed] [Google Scholar]
- 63.van der Poel C L, Cuypers H T M, Reesink H W, Weiner A J, Quan S, di Nello R, van Boven J J P, Winkel I, Mulder-Folkerts D, Exel-Oehlers P J, Schaasberg W, Leentvaar-Kuypers A, Polito A, Houghton M, Lelie P N. Confirmation of hepatitis C virus infection by new four-antigen recombinant immunoblot assay. Lancet. 1991;337:317–319. doi: 10.1016/0140-6736(91)90942-i. [DOI] [PubMed] [Google Scholar]
- 64.Van Doorn L-J, Quint W, Tsiquaye K, Voermans J, Paelinck D, Kos T, Maertens G, Schellekens H, Murray K. Longitudinal analysis of hepatitis C virus infection and genetic drift of the hypervariable region. J Infect Dis. 1994;169:1226–1235. doi: 10.1093/infdis/169.6.1226. [DOI] [PubMed] [Google Scholar]
- 65.Vrielink H, Reésink H W, Zaaijer H L, Scholten E, Kremer L C M, Cuypers H T M, Lelie P N, Van Oers M H J, van der Poel C L. Look-back of anti-HCV ELISA-positive, HCV-RNA PCR-negative donors and recipients of their blood products. Vox Sang. 1997;72:67–70. doi: 10.1046/j.1423-0410.1997.7220067.x. [DOI] [PubMed] [Google Scholar]
- 66.Wang C, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q L, Houghton M, et al. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 68.Weiner A J, Geysen M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, Brunetto M, Barr P J, Miyamura T, McHutchinson J, Houghton M. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: Potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoo B J, Spaete R R, Geballe A P, Selby M, Houghton M, Han J H. 5′ end-dependent translation initiation of hepatitis C viral RNA and the presence of putative positive and negative translational control elements with the 5′ untranslated region. Virology. 1992;191:889–899. doi: 10.1016/0042-6822(92)90264-p. [DOI] [PubMed] [Google Scholar]