Abstract
We previously reported (K. T. Jeang, R. Chun, N. H. Lin, A. Gatignol, C. G. Glabe, and H. Fan, J. Virol. 67:6224–6233, 1993) that human immunodeficiency virus type 1 (HIV-1) Tat and Sp1 form a protein-protein complex. Here, we have characterized the physical interaction and a functional consequence of Tat-Sp1 contact. Using in vitro protein chromatography, we mapped the region in Tat that contacts Sp1 to amino acids 30 to 55. We found that in cell-free reactions, Tat augmented double-stranded DNA-dependent protein kinase (DNA-PK)-mediated Sp1 phosphorylation in a contact-dependent manner. Tat mutants that do not bind Sp1 failed to influence phosphorylation of the latter. In complementary experiments, we also found that Tat forms protein-protein contacts with DNA-PK. We confirmed that in HeLa and Jurkat cells, Tat expression indeed increased the intracellular amount of phosphorylated Sp1 in a manner consistent with the results of cell-free assays. Furthermore, using two phosphatase inhibitors and a kinase inhibitor, we demonstrated a modulation of reporter gene expression as a consequence of changes in Sp1 phosphorylation. Taken together, these findings suggest that activity at the HIV-1 promoter is influenced by phosphorylation of Sp1 which is affected by Tat and DNA-PK.
Tat is a virus-encoded nuclear protein that functions as a transcriptional transactivator of the human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR). The molecular mechanism of Tat action remains incompletely elucidated. Some findings suggest that Tat acts upon the elongating transcript (66, 75, 76, 106), while others demonstrate an effect of Tat on initiation of transcription (6, 51, 52, 70, 80, 92, 101, 104). However, most investigators agree that interaction of host cell factors with Tat is important for regulating expression of the HIV-1 LTR inside cells (reviewed in references 19, 53, 56 and 82).
Tat protein from primary HIV-1 isolates is 101 amino acids in length; some laboratory isolates have a truncated Tat protein of 86 amino acids. Amino acids 1 to 48 compose a highly conserved cysteine-rich tract and core region. These highly conserved regions have been shown by point mutagenesis to be important for activity (82). Amino acids 49 to 58 comprise a basic-charged region necessary for nuclear localization and binding to the HIV leader RNA, TAR (14, 23, 41, 93). It has been hypothesized that binding to TAR tethers Tat to the promoter, allowing it to interact with basal transcription machinery. Many studies using chimeric Tat proteins support this notion. In those assays, Tat function was reconstituted when its activation domain was delivered to the promoter by using heterologous DNA/RNA-binding domains paired with respective cognate binding sites in a TAR-independent manner (4, 63, 94, 99).
A number of cellular proteins have been reported to interact directly with Tat. These proteins include TATA-binding protein (TBP) (65, 104), TAK (43, 44), PKR (8, 79), T3R (21), Tat-binding protein 1 (83, 84), TAP (20, 111, 112), TBP-associated factor TAF55 (11), HT2A (28), Tip60 (62), TFIIH (30, 87), RNA polymerase II (77), and Sp1 (18, 54). A model that incorporates all of these participants is difficult to develop; thus, the mechanistic details of HIV-1 LTR expression remain incompletely understood. One of the cellular factors that interact with Tat is Sp1. Sp1 has been well characterized through genetic and biochemical studies (5, 39, 46, 54, 55, 61, 64, 100, 102). We and others have previously reported on a role for Sp1 in Tat-transactivated expression of the HIV-1 promoter (18, 54). The exact mechanism(s) for how Sp1 could influence Tat action remains to be clarified.
Sp1 is one member of a multigene family (38). It is a 95- to 105-kDa protein that binds DNA through C-terminal zinc finger motifs (59, 60). Sp1 has been shown to interact with TBP (24), TAF110 (34), and RNA polymerase II (107). The activation function of Sp1 has been mapped to its N terminus, which contains glutamine- and serine/threonine-rich domains (16, 17, 60). Jackson et al. have shown that Sp1 is posttranslationally modified by glycosylation and phosphorylation (50). The significance of Sp1 phosphorylation has been extrapolated from observations that dephosphorylated Sp1 when added to in vitro transcription extracts becomes rapidly phosphorylated in a manner that correlates with function (50). It has also been reported that phosphorylated Sp1 binds DNA with reduced affinity, suggesting another route for regulating Sp1 function (2, 73).
Phosphoamino acid analysis reveals that Sp1 is predominantly phosphorylated on serine residues (50). Double-stranded DNA-dependent protein kinase (DNA-PK) (50) has been identified as an Sp1 kinase. DNA-PK is a multiprotein complex comprised of a 350-kDa catalytic subunit, p350, and Ku subunits (p70 and p80), which bind to nucleic acids (36, 58). DNA-PK has also been shown to phosphorylate the carboxy-terminal domain (CTD) of RNA polymerase II (89), and this phosphorylation event is augmented by the proximal presence of transcriptional activator domains (90). These findings suggest a function for DNA-PK in transcription. However, because DNA-PK can phosphorylate many proteins (reviewed in references 1 and 26), its other functional roles are likely to be complex and diverse (27, 67, 72, 103).
Here we show that Tat-Sp1 contact can modulate Sp1 phosphorylation inside cells, and in a DNA-PK-dependent manner, in cell extracts. We found that DNA-PK can bind Tat directly. Furthermore, in agreement with others (105), we show that phosphorylated Sp1 increases the intracellular expression of the HIV LTR. Our findings lead us to propose that Tat and DNA-PK interact to increase the phosphorylation state of Sp1, which results in upregulated expression of the HIV-1 LTR.
MATERIALS AND METHODS
Plasmids.
The 101-amino-acid Tat cDNA from HIV-1 strain SF2 (provided by Ben Berkhout, University of Amsterdam) was used to construct mutants. Mutant cDNAs were prepared by PCR and were expressed in Escherichia coli, using pGEX-2T (Pharmacia, Uppsala, Sweden). His-Tat plasmid was provided by C. Rosen (31). For HIV-1 molecular clones, mutant Tat cDNAs were ligated in frame into nef of pNL4-3 (47, 57). Chloramphenicol acetyltransferase (CAT) reporter constructs and eukaryotic Tat expression vectors have been previously described (5). Gal-Sp1Gln was provided by C. Southgate (101). Point mutant plasmids were constructed by site-directed mutagenesis using a Chameleon kit (Stratagene, La Jolla, Calif.). Sequencing of plasmids were done with Sequenase (Amersham Life Sciences, Cleveland, Ohio).
Preparation of fusion proteins.
E. coli was grown overnight in 50 ml of LB with ampicillin (100 μg/ml). A 500-ml LB-ampicillin flask was inoculated with the overnight culture and was grown for an additional hour at 37°C. Isopropylthiogalactopyranoside was added to a final concentration of 0.1 mM to induce fusion protein expression, and the culture was switched to 30°C for an additional 4 h. Cells were collected by centrifugation in a GSA rotor at 5,800 × g for 10 min at 4°C. Bacterial pellets were lysed either by sonication or by lysozyme digestion. For lysozyme digestion, the pellet was resuspended in 10 ml of buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride (buffer A); 10 mg of lysozyme was added, and the cells were digested for 1 h on ice. Then 10 ml of buffer A supplemented with 20 mM of MgCl2 and 50 U of DNase I (Life Technologies, Gaithersburg, Md.) was added, and the mixture was allowed to incubate for 15 min on ice. For sonication, the bacterial pellet was resuspended in 25 ml of phosphate-buffered saline (PBS) containing 1 mM phenylmethylsulfonyl fluoride and was sonicated (Branson) for 15 pulses (70%) at the maximum microprobe setting. The resulting mixture (either from sonication or from lysozyme digestion) was centrifuged in a GSA rotor at 5,800 × g for 10 min at 4°C. A second centrifugation in a SS-34 rotor at 23,500 × g for 20 min at 4°C clarified the extract of remaining debris.
Fusion protein affinity chromatography.
Truncated variants of HIV (strain SF2) Tat-1 were expressed as glutathione S-transferase (GST) fusion proteins in E. coli DH5α (Life Technologies) or BL21 (Pharmacia). Bacterial lysates were prepared as described above and were incubated with glutathione-Sepharose overnight. The resin was washed extensively with PBS and equilibrated with buffer B (20 mM HEPES-KOH, [pH 7.9], 20 mM KCl, 1 mM MgCl2, 17% glycerol, 2 mM dithiothreitol [DTT]). HeLa cell (Cell Trends, Middletown, Conn.) extracts were prepared as described by Dignam et al. (22), with the following modifications. After the first Dounce homogenization, the mixture was centrifuged once at 1,500 × g. Prior to dialysis with buffer B, ammonium sulfate (0.33 g/ml) was added to precipitate proteins. The pellet was resuspended into 1 packed-cell volume of buffer B and was dialyzed against 100 volumes of buffer B with two changes. Cellular extracts were incubated with the various protein-bound resins overnight at 4°C. The resins were packed into columns, and the columns were washed with buffer B containing 0.1 M KCl. Proteins were eluted in a stepwise fashion with buffer B containing 0.25, 0.5, and 1.0 M KCl. The eluates were desalted and concentrated, using 10,000-molecular-weight-cutoff microconcentration tubes (Amicon, Beverly, Mass.), to a final volume of 100 ml in buffer D.
Western blot analysis.
Column eluates were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to a polyvinylidene difluoride membrane by semidry electroblotting (Millipore, Bedford, Mass.) for 1 h at 400 mA. Rabbit polyclonal antibody against Sp1 (Santa Cruz Biotechnology, Santa Cruz, Calif.) or DNA-PK (Serotec, Washington, D.C.) was used, followed by visualization by chemiluminescence (Tropix, Bedford, Mass.) according to the manufacturer’s protocol.
Confocal microscopy.
HeLa cells were seeded onto glass coverslips and were transfected by using calcium phosphate (3, 37); 48 h later, cells were washed with PBS and fixed for 15 min in 4% paraformaldehyde in PBS, followed by two PBS washes and fixation with methanol for 2 min. Coverslips were incubated overnight in primary antibody. In costaining for Tat and Sp1, anti-Tat rat serum (Anne Gatignol) and anti-Sp1 rabbit serum (Santa Cruz Biotechnology) were used. In costaining for Tat and SC35, anti-Tat rabbit serum (Spring Valley Laboratory) and anti-SC35 mouse serum (Tom Maniatis [29]) were used. Secondary antibodies (Organon Teknika, Durham, N.C.) were Texas red-conjugated goat anti-rat, Texas red-conjugated goat anti-rabbit, fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit, and FITC-conjugated goat anti-mouse.
Virus propagation.
Virus was prepared by calcium phosphate transfection of HeLa cells (3, 37). Viral supernatant was harvested 48 h posttransfection and clarified by centrifugation and filtration. Reverse transcriptase (RT) activity was determined for each stock. A total of 5 × 105 C8166-45 cells were infected with equivalent amounts of RT-normalized virus. Virus growth was monitored by collecting medium supernatant every 2 to 3 days for RT measurements. RT assays were performed on 10 μl of supernatant in a 3-h incubation of 50 μl of buffer containing 50 mM Tris-HCl (pH 8.0), 63 mM KCl, 4.2 mM MgCl2, 0.08% Nonidet P-40, 0.85 mM EDTA, 4.2 μg of poly(A) and 0.13 μg of oligo(dT) per ml, 4 mM DTT and 0.5 μCi of [α-32P]dTTP; 10 μl of the reaction mix was spotted onto DEAE filter paper, and activity was determined by scintillation counting.
In vitro kinase assays.
Reaction mixtures contained either HeLa whole-cell protein extracts (∼4 μg) or 250 ng of purified double-stranded DNA-PK (Promega) or immunoprecipitated DNA-PK mixed with 200 ng of purified Sp1 (Promega), 100 ng of double-stranded oligonucleotide containing HIV-1 Sp1 and TATA motif 5′ d(GAT CTG GGC GGG ACT GGG GAG TGG CGA GCC CTC AGA TGC TAC ATA TAA GCA GCT) 3′, 100 ng of control/test proteins, 50 μl of kinase buffer (50 mM Tris-HCl [pH 7.5], 5 mM DTT, 5 mM MnCl2), and 5 μCi of [γ-32P]ATP (43, 44). The reaction was terminated by addition of 100 μl of 2× SDS sample buffer (125 mM Tris-HCl [pH 6.8], 20% glycerol, 2% SDS, 2% β-mercaptoethanol, 0.05% bromophenol blue) with boiling for 3 min. Where wortmannin (Calbiochem, San Diego, Calif.) was added to reaction, it was dissolved in dimethyl sulfoxide (DMSO) and added at the concentrations indicated in the figures. We noted that the specific activity of wortmannin appears to vary greatly from batch to batch and from different vendors, suggesting that different preparations of this reagent contains highly variable amounts of inert carrier material. Hence the drug concentrations described in the experiments should be regarded as mass (active plus inert material) concentrations which do not necessarily reflect true activity units.
Transient expression assays.
HeLa cells were transfected by using calcium phosphate (3, 37). Amounts of DNA used are indicated in the figure legends. In experiments involving okadaic acid, calyculin A or wortmannin (Calbiochem), the agents were dissolved in DMSO and were introduced into the media 3 h before transfection. Drugs were maintained continuously thereafter for the duration of the experiment. CAT assays were performed as described previously (3, 35). Each series of transfections (most were performed at least three times) was normalized based on amount of protein extract, and at least one of the sets of transfections contained a second cotransfected plasmid (i.e., expressing β-globin or lacZ [9, 51]). On repetition, none of the transfections varied in value from each other by more than 20%.
Biosynthetic labeling and immunoprecipitation.
Either 100 μCi of [35S]cysteine and 150 μCi of [35S]methionine or 1 mCi of 32Pi was used in labeling experiments. HeLa, HeLa-Tat, Jurkat, or Jurkat-Tat cells (106 of each) were washed once with PBS; 2 ml of methionine- and cysteine-free or 4 ml of phosphate-free Dulbecco modified Eagle medium or RPMI medium (Specialty Media, Lavallette, Wis.) was used for either HeLa and HeLa-Tat or Jurkat and Jurkat-Tat cells, respectively, in labeling periods of 12 h. The cells were washed with PBS twice and were extracted with 1 ml of radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl [pH 8.0]). A portion of samples was trichloroacetic acid (TCA) precipitated and quantitated by scintillation counting. Immunoprecipitations were performed overnight at 4°C, using 200 ng of anti-Sp1 serum (Santa Cruz Biotechnology) and 15 μl of protein A-agarose (Life Technologies). Beads were washed 10 times with RIPA buffer. SDS sample buffer was added to the protein A agarose pellets, and the mixtures were boiled for 3 min and resolved by SDS-PAGE.
RESULTS
Tat residues 30 to 55 contact Sp1.
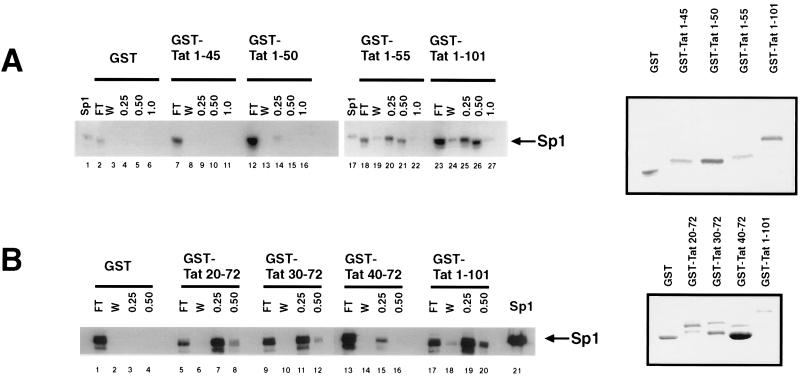
We previously described interaction between HIV-1 Tat and Sp1 in vitro and inside cells (54). To extend further that observation, we used protein affinity chromatography to characterize the region in Tat that contacts Sp1 directly. Various Tat fragments were expressed as GST fusion proteins (Fig. 1, right-hand panels), purified from E. coli, and then bound individually to glutathione-Sepharose. The resulting matrices were challenged with HeLa cellular extracts. Bound proteins were eluted with buffers containing stepwise increasing concentration of KCl. Western blot analysis was used (Fig. 1) to evaluate proteins eluted from the columns. Sp1 was found in the 0.5 M KCl eluates from GST-Tat 1-101 (Tat containing residues 1 to 101) (Fig. 1A, lane 26) and GST-Tat 1-55 (lane 21). By contrast, 0.5 M KCl eluates from GST alone (lane 5), GST-Tat 1-45 (lane 10), or GST-Tat 1-50 (lane 15) did not contain Sp1.
FIG. 1.
Definition of the region within Tat that binds Sp1. (A) Binding of Sp1 to GST, GST-Tat, or GST-Tat C-terminal mutants affixed to a solid matrix was assessed by immunoblotting of column elutions using rabbit polyclonal antibody (left). Protein column chromatography was performed as described in Materials and Methods. FT, flowthrough; W, wash fraction (0.1 M KCl). The 0.25, 0.5, and 1.0 M KCl buffer elutions are indicated; Sp1 indicates the lane in which purified protein (Promega) was loaded as a control. A Coomassie blue-stained gel of the purified GST-Tat fusion proteins used in constructing immobilized column matrices is shown to the right. (B) Western blot identifying Sp1 in elutions from columns constructed using GST, GST-Tat, or GST-Tat N-terminal mutants. A Coomassie blue-stained gel of the GST-Tat fusion proteins is shown to the right.
N-terminal Tat deletion proteins were similarly prepared (Fig. 1B, right), affixed to a solid matrix, and equilibrated with HeLa cell extract. After equilibration, bound proteins were eluted. Western blots of Tat mutant column elutions were compared with elutions from full-length Tat 1-101 (Fig. 1B, left). We found that GST-Tat 20-72, GST-Tat 30-72, and GST-Tat 1-101 (Fig. 1B, lanes 8, 12, 20 respectively) bound Sp1 which was eluted in buffer containing 0.5 M KCl; GST alone failed to bind Sp1 (lanes 3 and 4). GST-Tat 40-72 did bind Sp1; however, this interaction was totally released by 0.25 M KCl (lane 15), suggesting a reduced affinity between Sp1 and Tat 40-72. We constructed two additional GST-Tat fusion proteins (GST Tat 20-58 and GST Tat 30-58). From Western blots of elutions from GST-Tat 20-58 and GST-Tat 30-58 columns, we observed more Sp1 in the 0.25 M KCl than in the 0.5 M KCl fractions (data not shown). These results suggest weakened affinity of these fragments compared to full-length Tat for Sp1. Taken together our data suggest that Tat amino acids 30 to 55 contact Sp1, with sequences outside this region influencing overall binding affinity.
Tat and Sp1 colocalize in the nucleus.
Previously, we demonstrated that Tat and Sp1 can be coimmunoprecipitated from HIV-1-infected cells (54). Others have also shown a Tat-Sp1 association in transcription complexes (18). These results are qualified by the fact that cell lysis and cell content mixing occur during biochemical isolation of protein complexes. More physiological evidence for biological cross talk between Tat and Sp1 in cells would be in the in situ colocalization of both proteins within intact nuclei. To address this, we checked for intracellular co-association by immunoconfocal microscopy.
HeLa cells were transfected with a Tat expression plasmid. The cells were then fixed onto coverslips, stained with antibodies to Tat (rat) and Sp1 (rabbit), and processed with FITC-conjugated second antibodies directed against either rat or rabbit. Figures 2A and D show the patchy distribution pattern of Tat in nuclei. Under these conditions of expression, we do not observe localization of Tat into nucleoli (which we see only upon extreme overexpression). Figure 2E, stained in parallel with Fig. 2D, shows the nuclear presentation of Sp1. As a control, monoclonal antibody staining for an unrelated nuclear antigen, SC35 (29), was performed (Fig. 2B). In Fig. 2F, Tat and Sp1 association was visualized with the colocalized subpopulation highlighted in yellow. A similar computer-assisted colocalization analysis of Tat and SC35 is shown in Fig. 2C. We saw that a significant subset of Tat and Sp1 can be found together in the nucleus (Fig. 2F), at a level substantially greater than that expected for random interactions between two unrelated nuclear factors (e.g., Tat and SC35 [Fig. 2C]). Additional views of cells stained simultaneously with anti-Tat and anti-Sp1 are presented in Fig. 2G, J, K, and L, and computer-assisted colocalizations are shown in Fig. 2H and I.
FIG. 2.
Colocalization of Sp1 and Tat in transfected HeLa cells. (A) Tat localization in HeLa cells transfected with Tat expression plasmid and stained in parallel for Tat (anti-Tat rabbit primary antibody and Texas red-conjugated goat anti-rabbit second antibody) and SC35 (anti-SC35 mouse monoclonal primary antibody and FITC-conjugated goat anti-mouse second antibody). In panel A, only the Tat visualization window is shown. (B) Visualization of SC35 in the cell shown in panel A. (C) Computer colocalization analysis of the signals shown in panels A and B, with Tat staining indicated by red and SC35 staining indicated by green. Areas of colocalization are indicated in yellow. (D) Tat localization in HeLa cells transfected with Tat expression plasmid and stained in parallel for Tat (anti-Tat rat primary antibody and Texas red-conjugated goat anti-rat second antibody) and Sp1 (rabbit anti-Sp1 primary antibody and FITC-conjugated goat anti-rabbit second antibody). Only the Tat window is shown. (E) Visualization of Sp1 in the cell shown in panel D. (F) Colocalization analysis of panels D and E, with Tat staining highlighted by red and Sp1 staining highlighted by green. Areas of colocalization between Tat and Sp1 are indicated in yellow. (G) Additional views of cells transfected with a Tat-expressing plasmid and stained with rat antiserum to Tat and rabbit polyclonal antiserum to Sp1. The confocal image capture window was set for the anti-Tat signal. (H and I) Computer-assisted colocalization of Tat and Sp1 signals shown in black (H) and red (I). (J) Confocal micrograph with fluorescence window adjusted to capture anti-Tat (red), anti-Sp1 (green), and colocalized images of the two proteins (yellow). This field of cells is identical to that in panel G. (K and L) Lower-magnification views of the same field of cells transfected with a Tat-expressing plasmid stained with rat anti-Tat and rabbit anti-Sp1. The fluorescence image capture window was restricted to either anti-Tat (K) or anti-Sp1 (L).
Tat influences phosphorylation of Sp1.
The observed Tat-Sp1 interaction provided an impetus for exploring the possible biological consequence(s) of this complex formation. Others have reported that dephosphorylated Sp1 when added to cell extracts quickly becomes phosphorylated, suggesting phosphorylation as an important step for activation (50). Additionally, many studies have implicated a kinase activity that is associated with Tat (13, 30, 43, 44, 79, 87, 114). Hence, we wondered whether these two observations could be linked (i.e., Tat contacts Sp1 and brings a kinase that phosphorylates Sp1).
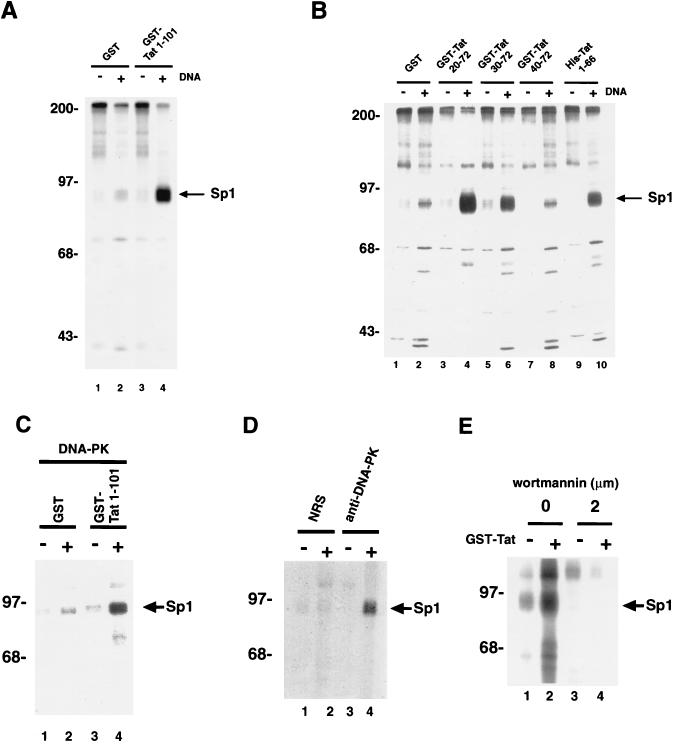
To address this possibility, we performed in vitro kinase assays (Fig. 3), mixing HeLa total cell extract with [γ-32P]ATP and purified Sp1. Where indicated, either control protein (GST) or Tat (GST-Tat) with or without double-stranded DNA oligonucleotides containing an Sp1 motif were added to the mix. We included DNA oligonucleotides because previously it has been reported that Sp1 phosphorylation is enhanced by binding to cognate DNA (37). After incubation, the samples were analyzed by SDS-PAGE. Covalent addition of 32P to Sp1 was visualized by autoradiography and/or Fuji phosphorimaging.
FIG. 3.
Tat affects phosphorylation of Sp1 in vitro. (A) In vitro kinase reaction of Sp1 using HeLa cellular extract. The effect of purified GST-Tat 1-101 on Sp1 phosphorylation is compared to that of purified GST alone. Four micrograms of cell extract was mixed with 100 ng of GST alone or GST-Tat 1-101 and 200 ng of purified Sp1. +’s mark lanes that contain 100 ng of double-stranded oligonucleotides comprised of Sp1 and TATA motifs from HIV-1. After 15 min of incubation on ice, kinase buffer was added, and the samples were incubated for an additional 15 min at room temperature. Reactions were terminated by addition of SDS-PAGE sample buffer and were analyzed by SDS-PAGE (10% gel) followed by autoradiography or Fuji phosphorimaging. The arrow indicates the position of phosphorylated Sp1. Identity of phosphorylated Sp1 was also confirmed separately by direct immunoprecipitation using specific antibody (data not shown). (B) Delineation of the in vitro modulatory effect on Sp1 phosphorylation by four GST-Tat mutants. Note that GST-Tat 20-72, GST-Tat 30-72, and His-Tat 1-66 can all bind Sp1, while GST-Tat 40-72 cannot (Fig. 1B). Phosphorylation of Sp1 was augmented by GST-Tat 20-72, GST-Tat 30-72, and His-Tat 1-66 but not by GST-Tat 40-72. By laser densitometry (Molecular Dynamics), GST-Tat 20-72 showed a 5-fold increase over GST, while GST-Tat 30-72 and His-Tat 1-66 showed increases of 3- and 2.5-fold, respectively. (C) In vitro kinase reactions were performed without (−) or with (+) double-stranded DNA as for panel A except that 250 ng of purified DNA-PK was used in place of HeLa nuclear extract. (D) Verification of the involvement of DNA-PK by direct immunoprecipitation. In vitro kinase reactions were performed as for panel A except that the source of DNA-PK was immunoprecipitation using specific antiserum to DNA-PK (lanes 3 and 4) of HeLa cell extract. As negative controls, immunoprecipitation were performed with normal rabbit serum (NRS; lanes 1 and 2). All four samples contain double-stranded oligonucleotides with Sp1 and TATA motifs from HIV-1. In lanes 1 and 3, GST alone was used in place of GST-Tat 1-101. (E) In vitro kinase reactions were performed as for panel A except that wortmannin (Calbiochem) was added to the incubations in lanes 3 and 4. All samples contain double-stranded oligonucleotides with Sp1 and TATA motifs from HIV-1 and GST alone (−) or GST-Tat 1-101 (+). The arrow points to the migration position of exogenously added purified Sp1. Sizes are indicated in kilodaltons.
When purified Sp1 was added to the reaction containing GST-Tat 1-101, efficient phosphorylation was readily detected (Fig. 3A, lane 4). In comparison, Sp1 in an otherwise identical reaction with control GST protein (lane 2) was approximately fivefold less efficiently (as measured by laser densitometry) phosphorylated. One interpretation of this result is that Sp1 phosphorylation in vitro is augmented by Tat 1-101. Because we have mapped the region in Tat that contacts Sp1 (Fig. 1), we wondered if a correlation could be established between protein-protein binding and phosphorylation. To assess this, we tested four forms of Tat in kinase assays (Fig. 3B). Three (GST-Tat 20-72, GST-Tat 30-72, and His-Tat 1-66) have been shown to bind Sp1, and each of these proteins was found to enhance phosphorylation of Sp1 (Fig. 3B, lanes 4, 6, and 10) relative to the GST control (lane 2). The fourth protein, GST-Tat 40-72, was previously shown to be incapable of Sp1 binding. In a kinase assay, this Tat-protein failed to influence Sp1 phosphorylation (lane 8) above background levels (lane 2).
What might be the kinase responsible for Tat-augmented Sp1 phosphorylation? In other settings, DNA-PK has been found to phosphorylate Sp1 (50). We wondered whether the effect observed with Tat was mediated through DNA-PK. As a first step in answering that question, we reconstituted in vitro reactions containing Sp1, using purified DNA-PK in place of HeLa total-cell extract. We asked if purified DNA-PK alone could reflect the Sp1 kinase activity observed in total HeLa cell extract. To this reconstitution, we added either GST (Fig. 3C, lanes 1 and 2) or GST-Tat 1-101 (lanes 3 and 4). In side-by-side comparisons, the reaction that contained GST-Tat, Sp1, and DNA-PK produced five times more phosphorylated Sp1 than that containing GST, Sp1, and DNA-PK (compare lanes 2 and 4).
Next, we performed immunoprecipitations to verify further DNA-PK as the kinase in Tat-modulated Sp1 phosphorylation. We used specific rabbit polyclonal serum to immunoprecipitate DNA-PK from HeLa extracts (Fig. 3D, lanes 3 and 4). Normal rabbit serum was used in control immunoprecipitations (lanes 1 and 2). The immunoprecipitates were tested in kinase assays with either GST (lanes 1 and 3) or GST-Tat 1-101 (lane 2 and 4) and Sp1 substrate. We found that the DNA-PK captured by specific antiserum effected 20-fold-greater efficiency in phosphorylating Sp1 in the reaction containing Tat compared to the control containing GST alone (compare lanes 2 and 4).
The role of kinases can be studied by using drug inhibitors. Wortmannin has been reported to inhibit potently DNA-PK function (40). To correlate inside cells the role of DNA-PK in Tat-modulated Sp1 phosphorylation, we checked for activity in the presence or absence of wortmannin. HeLa extract was incubated with (Fig. 3E, lanes 3 and 4) or without (lanes 1 and 2) wortmannin in the presence of either GST alone (lanes 1 and 3) or GST-Tat 1-101 (lanes 2 and 4), and phosphate addition to Sp1 was assayed. A dose-dependent reduction of Sp1 phosphorylation in the wortmannin-treated samples was observed (lanes 3 and 4; inhibition of phosphorylation could be observed at apparent drug concentrations as low as 30 nM [data not shown]), which provides supportive (albeit not definitive) evidence for the role of DNA-PK. Thus, taken together, the findings from reconstitution, immunoprecipitation, and drug inhibition assays are all consistent with DNA-PK contributing to Tat-modulated Sp1 phosphorylation.
DNA-PK binds Tat.
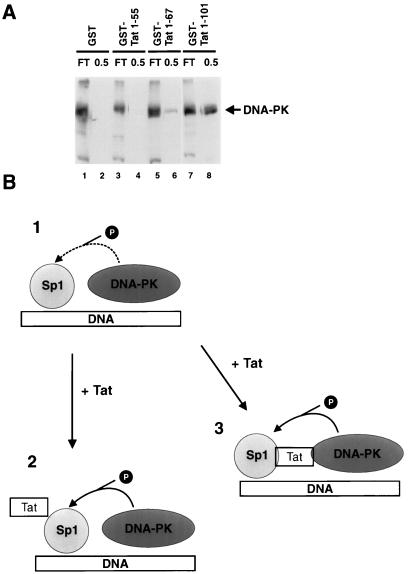
The foregoing results led us to examine whether Tat binds DNA-PK (Fig. 4A). To assess this, HeLa cell extract was bound to and eluted from GST alone, GST-Tat 1-55, GST-Tat 1-67, or GST-Tat 1-101 columns. The column eluates were probed for DNA-PK by Western blotting. From the immunoblots, we found that neither GST-Tat 1-55 (lane 4) nor GST alone (lane 2) showed any affinity for DNA-PK. On the other hand, GST-Tat 1-101 retained DNA-PK (lane 8) slightly better than GST-Tat 1-67 (lane 6). (This pattern was replicated when we chromatographed over the same protein columns purified DNA-PK enzyme purchased from Promega [data not shown]). Previously, we noted that a GST-Tat 1-55 column bound Sp1 effectively (Fig. 1A, lane 21). Juxtaposed with the current results (where GST-Tat 1-55 did not bind DNA-PK), this finding suggests that the contact points for Tat and DNA-PK are different from that for Tat and Sp1. The observation that amino acids in the second coding exon of Tat (i.e., residues 73 to 101) contribute, in part, to the binding of DNA-PK would be consistent with the transcriptional activity of this portion of the Tat protein in the activation of integrated LTR templates (52).
FIG. 4.
DNA-PK binds Tat. (A) HeLa cell extracts were equilibrated with either GST or GST-Tat protein columns. Columns were extensively washed (>20 column volumes) with 0.1 M KCl buffer and then eluted with 0.5 M KCl buffer. Eluates were assayed for DNA-PK by immunoblotting using rabbit polyclonal antibody to DNA-PK (Serotec). Flowthrough (FT) and 0.5 M KCl elutions are indicated. Note that neither GST alone (lane 2) nor GST-Tat 1-55 bound DNA-PK (lane 4); GST-Tat 1-67 (lane 6) bound DNA-PK less strongly than GST-Tat 1-101 (lane 8). (B) Schematic diagram of possible interactions between Sp1, DNA-PK, and Tat. In diagram 1, phosphorylation as indicated by the dotted arrow occurs in the absence of Tat. In diagram 2, Tat interaction with Sp1 induces a conformational change in Sp1, enhancing its ability to become phosphorylated. In diagram 3, Tat bridges Sp1 and DNA-PK, facilitating Sp1 phosphorylation. The ability of Tat to contact directly both Sp1 and DNA-PK is consistent with events in diagram 3.
That Tat could modulate Sp1 phosphorylation and that DNA-PK might be the participating kinase raise several considerations (Fig. 4B). We envision several possible scenarios. In the absence of Tat, Sp1 and DNA-PK could colocalize by bindings to common double-stranded DNAs. In such a manner, the two proteins could be brought into proximity and phosphorylation of Sp1 by DNA-PK could occur (diagram 1). How Tat enhances Sp1 phosphorylation might be explained in two ways. First, direct Tat-Sp1 contact (55) could modify the conformation of Sp1 thereby making it a better kinase substrate (diagram 2). This could occur whether Tat does or does not contact DNA-PK directly. On the other hand, a direct contact of Tat with DNA-PK suggests that Tat might serve to bridge Sp1 and DNA-PK, thus adding strength to the initial complex formed through binding of Sp1 and DNA-PK to double-stranded DNA (diagram 3). The ability of Tat to contact directly both Sp1 and DNA-PK would be consistent with events portrayed in diagram 3.
Phosphatase inhibitors increase Sp1-dependent expression of the HIV-1 LTR.
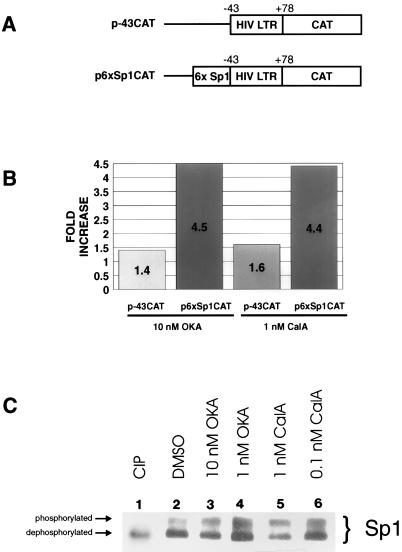
The biological relevance of Sp1 phosphorylation could be indirectly demonstrated with an increased intracellular HIV-1 LTR activity by inhibitors that prevent Sp1 dephosphorylation. To check this, HeLa cells treated with and without phosphatase inhibitors were transfected with a derivative of HIV-1 LTR-CAT (p-43CAT or p6xSp1CAT [Fig. 5A]). p-43CAT contains the minimal HIV-1 TATAA box and TAR sequence fused to a CAT cDNA. p6xSp1CAT is p-43CAT with six Sp1-binding sites placed upstream of TATAA. We treated cells with either okadaic acid or calyculin A, which are potent inhibitors of protein phosphatase 1 (PP1) and protein phosphatase 2A (PP2A). By inhibiting dephosphorylation, the ambient level of phosphorylated Sp1 is expected to be elevated (48). In Western blots, we verified that in mock (DMSO)-treated HeLa cells (Fig. 5C, lane 2), only a minority of steady-state Sp1 (31% as measured by laser densitometry) was phosphorylated. However, when these cells were treated with either 10 nM okadaic acid (lane 3) or 1 nM calyculin A (lane 5), the amount of phosphorylated Sp1 increased by approximately 60%. When lower concentrations of drug (1 nM okadaic acid [lane 4]) or 0.1 nM calyculin A [lane 6]) were used, the percentage of phosphorylated Sp1 (42%) was similar to that from mock-treated cells (lane 2), in agreement with the reported effective drug concentrations needed for okadaic acid (for PP1, 10 to 15 nM; for PP2A, 0.1 nM) and calyculin A (for PP1, 0.5 to 2 nM; for PP2A, 0.1 to 1 nM) (15, 49). Overall, these results are consistent with Sp1 function being regulated through a balance of phosphatases and kinases.
FIG. 5.
Effects of phosphatase inhibitors (okadaic acid and calyculin A) on Sp1-dependent expression. (A) Schematic representations of reporter plasmids. p-43CAT contains the HIV-LTR minimal promoter (nucleotides −43 to +78). p6xSp1CAT contains the HIV-LTR minimal promoter with six consensus Sp1-binding sites positioned upstream of TATAA. (B) Bar graph of CAT assays. HeLa cells were transfected with the indicated plasmids. In all cases, pUC19 was used to normalize for total amounts of DNA. A total of 2.5 μg of DNA was used per transfection. Phosphatase inhibitors were introduced into the media at the indicated concentrations 3 h before transfection and were maintained continuously. CAT activities were visualized by phosphorimaging. Data are averages from triplication (okadaic acid [OKA]) and duplication (calyculin A [CalA]). Amounts of activity were measured by scintillation counting of silica plate slices. (C) Western blot of Sp1 from HeLa cells treated with the indicated phosphatase inhibitors. Samples were separated in an SDS–12.5% (12.4:0.1, acrylamide/bisacrylamide) polyacrylamide gel that enhances migration differences between phosphorylated and nonphosphorylated forms of Sp1. The arrows indicate the relative positions of the two forms of Sp1. Calf intestinal alkaline phosphatase (CIP)-treated extract shows the migration profile of dephosphorylated Sp1 (lane 1). Treatment with phosphatase inhibitors increased the amount of phosphorylated relative to dephosphorylated Sp1.
Next, the relevance of Sp1 phosphorylation on HIV-1 LTR activity was measured by the expression of reporter genes. CAT activities in Fig. 5B illustrate the effect of okadaic acid (performed in triplicate) and calyculin A (performed in duplicate) on the HIV-1 promoter. Both inhibitors marginally (1.4- to 1.6-fold) affected CAT activity from p-43CAT, which is Sp1 independent (Fig. 5B). However, both increased Sp1-dependent expression of p6xSp1CAT 4.5-fold (Fig. 5B). These results are compatible with phosphorylation of Sp1 being one limiting step governing expression of the HIV-1 LTR.
Kinase inhibitors decrease Sp1-dependent expression of the HIV-1 LTR.
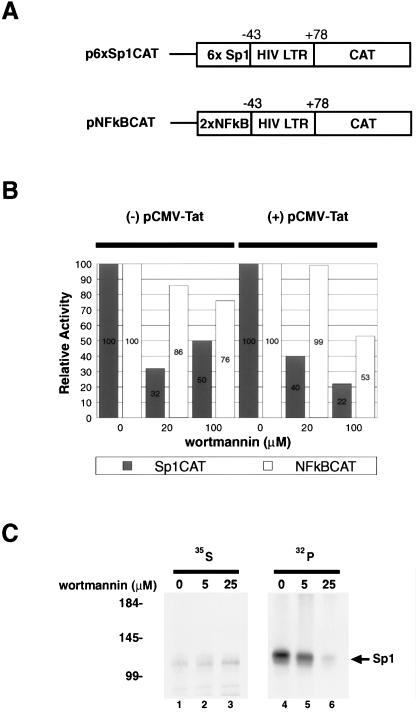
A complementary approach to explore the biological impact of Sp1 phosphorylation on HIV-1 LTR expression is to treat cells with an inhibitor of DNA-PK (e.g., wortmannin [40]). We checked this by transfecting two different reporters (Fig. 6A) into HeLa cells treated with or without wortmannin and then determining CAT activities. Expression of p6xSp1CAT reporter is Sp1 dependent, while expression of pNF-κB-CAT is Sp1 independent. Thus, if our expectations were correct, the former but not the latter would be sensitive to wortmannin. Indeed, we observed differential responses between the two reporters to wortmannin (Fig. 6B). p6xSp1CAT showed threefold-lower basal activity in wortmannin-treated cells than in mock-treated cells. In contrast, expression of pNF-κB-CAT was minimally affected by wortmannin. In the presence of Tat, p6xSp1CAT showed a fivefold reduction with 100 mM wortmannin treatment, compared to a twofold change seen for pNF-κB-CAT (Fig. 6B).
FIG. 6.
Effects of wortmannin, a DNA-PK inhibitor, on Sp1-dependent expression. (A) Schematic representations of reporters. p6xSp1CAT contains the HIV-1 minimal promoter (nucleotides −43 to +78) with six consensus Sp1 binding sites positioned upstream of TATAA. pNF-κBCAT contains the HIV-1 minimal promoter and two NF-κB sites positioned upstream of TATAA. (B) Bar graph (average of two experiments) of CAT assays performed in the presence of different concentrations of wortmannin. Activity measured at 0 mM wortmannin was set as 100%. The reporter plasmids (1 μg) are as in panel A. Assays with (pCMV-Tat; right) and without (-pCMV-Tat; left) the addition of a Tat expression plasmid (100 ng) are shown. (C) Cells were treated with the indicated amounts of wortmannin at the same time as with [35S]methionine-cysteine and 32Pi overnight labeling. HeLa cell extracts were immunoprecipitated with anti-Sp1 serum and resolved by SDS-PAGE (10% gel). Benchmark (Life Technologies) molecular weight marker positions are indicated in kilodaltons on the left. Equivalent amounts of total protein were immunoprecipitated as quantitated by TCA precipitation and scintillation counting.
To verify that wortmannin directly affected Sp1 phosphorylation, we labeled HeLa cells overnight with [35S]methionine-cysteine or 32Pi in the presence of drug (Fig. 6C). The next day, protein extracts were immunoprecipitated with anti-Sp1 and analyzed by SDS-PAGE. Less phosphorylated Sp1 was observed in wortmannin-treated (lanes 5 and 6) than in mock-treated (lane 4) cells. The similar intensities of the corresponding 35S-labeled proteins (lanes 1 to 3) indicated that similar amounts of protein were recovered by immunoprecipitation.
Increased Sp1 phosphorylation in Tat-expressing cells.
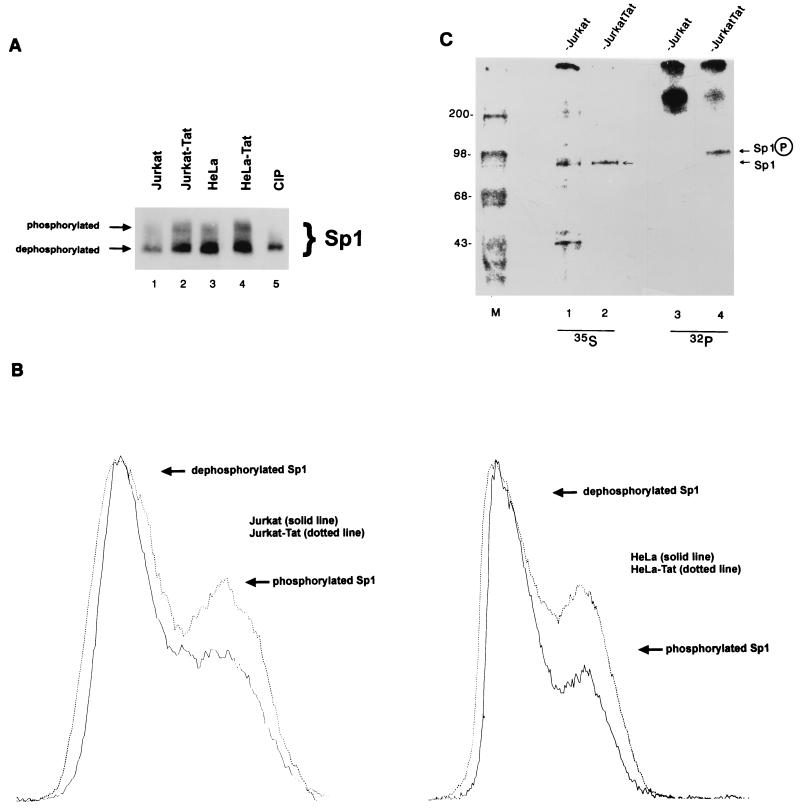
Next, we examined the influence of Tat on intracellular Sp1 phosphorylation. We compared Jurkat and HeLa cells to their Tat-expressing counterparts. Sp1 from Jurkat, Jurkat-Tat, HeLa, or HeLa-Tat cells was analyzed by Western blotting (Fig. 7A). Phosphorylated and dephosphorylated Sp1 moieties were distinguished by a mobility shift in SDS-PAGE (50). Using this approach, we observed distinct changes in the ratio of phosphorylated versus dephosphorylated Sp1 when comparing paired cells that express and do not express Tat (Fig. 7A). This change was quantified by laser densitometry. In Fig. 7B, we show superimposed densitometries from two paired cell lines. In these tracings, the laser-scanned peaks for the dephosphorylated form of Sp1 were equalized. Normalizing for dephosphorylated Sp1 in paired cells, are found the amount of phosphorylated Sp1 to be 24% more in Jurkat-Tat than in Jurkat (Fig. 7B, left) and 60% more in HeLa-Tat than in HeLa (Fig. 7B, right) cells.
FIG. 7.
Increased phosphorylation of Sp1 in Tat-expressing cells. (A) Western analysis of Jurkat, Jurkat-Tat, HeLa, and HeLa-Tat cell extracts, using rabbit anti-Sp1 serum. Blots were developed by using chemiluminescence. Lanes were normalized for duration of exposure. (B) Densitometer (Molecular Dynamics) tracings of Western blot signals comparing the amounts of phosphorylated and dephosphorylated Sp1 in Jurkat (31.6% phosphorylated) and Jurkat-Tat (39.3% phosphorylated; left) and in HeLa (21.5% phosphorylated) and HeLa-Tat (34.4% phosphorylated; right) cells. (C) [35S]methionine-cysteine and 32Pi-labeled HeLa cell proteins were immunoprecipitated with anti-Sp1 serum. Equivalent amounts of total protein were immunoprecipitated as quantitated by TCA precipitation and scintillation counting. Images were visualized with a Fuji phosphorimaging system, and exposures were normalized for total amounts of signal. Greater amounts of phosphorylated Sp1 was observed for Jurkat-Tat than for Jurkat, while amount of 35S labeled-Sp1 was recovered in essentially identical amounts from the two cell lines. M, size markers (positions are indicated in kilodaltons).
The phosphorylation state of Sp1 in Jurkat and Jurkat-Tat cells was also independently verified by radiolabeling with [35S]methionine-cysteine (Fig. 7C, lanes 1 and 2) and 32Pi (lanes 3 and 4) followed by immunoprecipitation with anti-Sp1 serum. In parallel immunoprecipitations, equivalent amounts of 35S-labeled Sp1 were recovered from Jurkat and Jurkat-Tat (Fig. 7C) cells. However, the same immunoprecipitation for phosphorylated species showed that far more 32P-labeled Sp1 was present in Jurkat-Tat than in Jurkat cells. This assay indicates that in a setting where both cells have similar amounts of total Sp1 protein (as evidenced by the 35S-labeled signal), a greater amount of phosphorylated Sp1 species exists in Tat-expressing cells (Fig. 7C).
Point mutation of serine 131 in Sp1 reduces transcriptional activity.
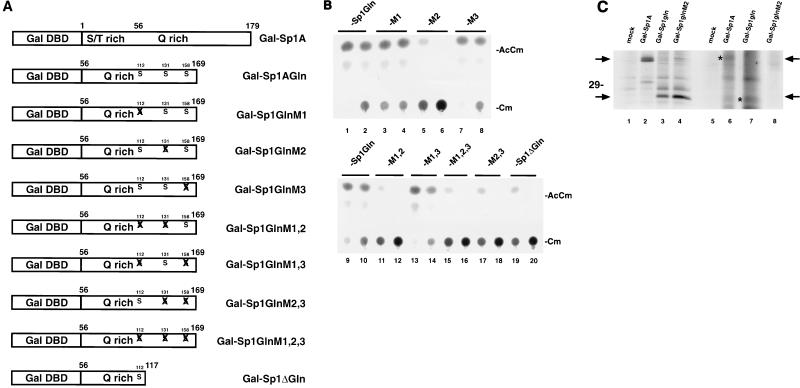
Sp1 is phosphorylated primarily on serines (50). In principle, the functional significance of Sp1 phosphorylation could be explored directly through point mutagenesis of serines. Because full-length Sp1 contains 78 serines, an exhaustive point mutagenesis of this form would be prohibitive. However, since a short N-terminal Sp1 A domain (Sp1A) fused to the Gal DNA-binding domain supports Tat transactivation of a chimeric Gal-HIV-1 LTR reporter (positions 99 to 101), one could examine the role of serine phosphorylation in this context. The minimal Gal-Sp1 fusion (Gal-Sp1Gln) has only three serine residues (Fig. 8). Thus, all of these serines could be mutated singly or in combinations. For this purpose, we generated eight mutant forms of Gal-Sp1Gln (Fig. 8A). Each contained point mutations of serine to alanine; all were confirmed by sequencing (data not shown).
FIG. 8.
A point mutation at serine 131 in Sp1 reduces Tat transactivation. (A) Schematic representations of point mutations. Gal-Sp1A contains the N-terminal A domain of Sp1 (from amino acids 1 to 169), while Gal-Sp1Gln has the N-terminal serine/threonine-rich portion (amino acids 1 to 55) deleted. Relative locations of point mutations are indicated by X. M1, serine 112 to alanine; M2, serine 131 to alanine; M3, serine 158 to alanine. Combinations are indicated as M1,2, M1,3, etc. Gal-Sp1ΔGln is a C-terminal deletion (amino acids 118 to 169 removed) of Gal-Sp1Gln; serines 131 and 158 are absent from this construct. The amino acid position assignments are relative to the Sp1 sequence in GenBank (accession no. J03133). All mutations were verified directly by sequencing (data not shown). DBD, DNA-binding domain. (B) CAT assays of the various Gal-Sp1Gln mutant individually tested in the presence of coexpressed Tat protein. HeLa cells were transfected with Gal-HIV-1 LTR-CAT reporter, pSV-Tat expression plasmid, and one of the Gal-Sp1Gln mutants (-M1, -M2, or -M3). Each set of transfections used either 5 (left) or 1 (right) μg of Gal-Sp1 mutant plasmid. With the lower activity level of Sp1Gln arbitrarily set as 1, the comparable average relative activities from the mutants are as follows: M1, 1.3; M2, 0.05; M3, 1.1; M1,2, 0.02; M1,3, 1.5; M1,2,3, 0.02; M2,3, 0.1; and Sp1ΔGln, 0.1. AcCm, acetylated chloramphenicol; Cm, chloramphenicol. (C) Immunoprecipitation of Gal-Sp1 proteins. HeLa cells transfected with the indicated plasmids were labeled overnight with [35S]methionine-cysteine (lanes 1 to 4) or 32Pi (lanes 5 to 8). The cells were lysed in RIPA buffer and immunoprecipitated with antiserum raised to the Gal DNA-binding domain. The immunoprecipitates were resolved by SDS-PAGE (14% gel) followed by autoradiography. Arrows and asterisks indicate the positions of relevant bands. Note the absence of 32P-labeled immunoprecipitated band in Gal-Sp1GlnM2 (compare lanes 4 and 8).
The Gal-Sp1Gln mutants were transfected individually into cells with pSV-Tat and the Gal-HIV-1 LTR-CAT reporter. Promoter expression was assayed by CAT activities. Various profiles of activity were seen; however, a consistent finding was that reduced expression from Gal-HIV-1 LTR-CAT was found in all Gal-Sp1Gln constructs that had a point mutation in serine 131, (e.g., M2, M1,2, M1,2,3, and M2,3 [Fig. 8B, lanes 6, 12, 16, and 18]). These results, although not directly addressing phosphorylation, point to the selective importance of serine 131 over serine 112 or serine 158 for Sp1 and Tat function.
To correlate a mutation at serine 131 with a reduction in phosphorylation (and not a trivial reduction in protein stability), HeLa cells were transfected with either Gal-Sp1Gln or mutant Gal-Sp1GlnM2. Transfected cells were labeled overnight with either [35S]methionine-cysteine or 32Pi. Cell extracts were immunoprecipitated with antiserum to the Gal DNA-binding domain. In immunoprecipitations of [35S]methionine-cysteine samples, a radiolabeled band (Fig. 8C, lanes 3 and 4) consistent with the size for Gal-Sp1Gln was seen. This band was not present in either mock-transfected (lane 1) or Gal-Sp1A-transfected (lane 2) samples. The fact that the intensities of the two samples were approximately equivalent suggests that comparable steady-state amounts of Gal-Sp1GlnM2 and Gal-Sp1Gln proteins are present in cells. Thus, the reduced activity from Gal-Sp1GlnM2 is unlikely to be from an absence or instability of protein. Next, when the comparable part of the gel for immunoprecipitated 32P-labeled samples was examined, a band found in the Gal-Sp1Gln (lane 7) sample was not seen in the Gal-Sp1GlnM2 (lane 8) sample. This observation is compatible with the phosphorylation site in Gal-Sp1 Gln being serine 131.
The role of serine 131 as a phosphoacceptor of DNA-PK-mediated kinase activity was assayed further in vitro. We expressed and purified GST-fused versions of the Sp1Gln, Sp1GlnM2, and Sp1ΔGln proteins (Fig. 9A). Each was independently assayed in kinase reactions using purified DNA-PK enzyme (Fig. 9B) without (left) or with (right) the additional of 100 ng of double-stranded DNA oligonucleotides. Consistent with the findings presented above, in a DNA-dependent manner, Sp1Gln was strongly phosphorylated whereas Sp1GlnM2 and Sp1ΔGln were not (Fig. 9B, lanes 1 to 3). It should be noted that Sp1GlnM2 differs from Sp1Gln only in the mutation at serine 131.
FIG. 9.
Mutation at serine 131 affects DNA-PK-mediated phosphorylation of Sp1. Sp1Gln, Sp1GlnM2, and Sp1ΔGln proteins were expressed and purified as GST fusion proteins. (A) Purified proteins stained by Coomassie brilliant blue. Arrow points to the migration position of Sp1Gln and Sp1GlnM2. Sp1ΔGln (lane 3) migrates with an apparent size that is approximately 5 kDa smaller than either Sp1Gln (lane 1) or Sp1GlnM2 (lane 2). (B) DNA-PK (purified enzyme purchased from Promega)-mediated transfer of 32P from [γ-32P]ATP to Sp1Gln (lane 1), Sp1GlnM2 (lane 2), or Sp1ΔGln (lane 3) in the absence (left) or presence (right) of double-stranded oligonucleotides. Arrow points to phosphorylated Sp1Gln protein seen only in the reaction supplemented with DNA (lane 1, right).
Expression of inactive Tat mutants that bind Sp1 interferes with virus growth.
In principle, if Tat-Sp1 interaction is biologically important for HIV-1, then overexpression of inactive Tat mutants that retain the ability to contact Sp1 should dominantly interfere with viral propagation. Considering the relative hierarchy of Tat-Sp1 binding in vitro (Tat 30-72 > Tat 20-58 > Tat 30-58 [Fig. 1]), we wished to examine the biological relevance of this contact during virus replication in cells. cDNAs for different portions of Tat were individually positioned into the nef reading frame of a replication-competent HIV-1 molecular clone (47, 57) (Fig. 10A). Four Tat chimeric viruses (Tat 20-58, Tat 30-58, Tat 30-72, and Tat 30-72i; Fig. 10A) were generated. Three viruses express truncated Tat peptides; the fourth (Tat 30-72i) has a cDNA for Tat 30-72 placed into nef in a reversed orientation, serving as a negative control.
FIG. 10.
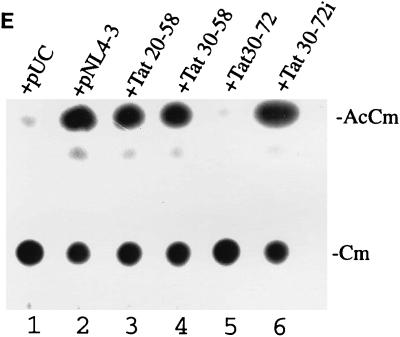
Correlation between mutant Tat proteins that bind Sp1 and interference with HIV-1 growth in C8166-45 T-cells. (A) Schematic representation of virus constructions. Mutant Tat (m-Tat) cDNAs were positioned in frame into nef of pNL4-3. Note that these chimeric proviruses produce both wild-type (wt-Tat) and mutant Tat proteins. (B) RT growth curves for viruses propagated in C8166-45. Data are representative of points from experiment 2 (Fig. 9C). (C) Summary from three separate experiments of the times of peak RT produced by the different viruses. (D) Interpretive model of the competition between mutant and wild-type Tat proteins for binding to Sp1. (E) Transdominant inhibitory activity of Tat fragments expressed from proviral constructions. In this assay, 0.1 μg of pLTR-CAT was transfected into HeLa cells with 2 μg of pUC19 alone (lane 1) or with 0.1 μg of pSV-Tat plus 2 μg of either pNL4-3 provirus (lane 2), Tat 20-58 provirus (lane 3), Tat 30-58 provirus (lane 4), Tat 30-72 provirus (lane 5), or Tat 30-72i provirus (lane 6). CAT activities were assayed 48 h after transfection. Under these transfection conditions, activity from 0.1 μg of pLTR-CAT is saturated by cotransfection with 0.1 μg of pSV-Tat since when 0.1 μg of pLTR-CAT and 0.1 μg of pSV-Tat were transfected together (data not shown), CAT activity was equivalent to that shown in lane 2. AcCm, acetylated chloramphenicol; Cm, chloramphenicol.
Virus stocks for each chimeric genome were generated by plasmid transfections into HeLa cells. Equal amounts of infectious virus, normalized by RT, were used to infect C8166-45 cells. Virus replication was monitored by assaying for supernatant RT production, and the infection profiles from five viruses (including wild-type pNL4-3) are graphed in Fig. 10B. Figure 10C summarizes the results from three independent experiments. The control Tat 30-72i virus grew slightly more slowly than wild-type NL4-3. This small change presumably occurred as a result of insertion into nef enlarging the viral genome. Interestingly, Tat 30-58 virus, expressing the 30-58 peptide, grew very similarly to Tat 30-72i. On the other hand, Tat 20-58 showed a 4- to 6-day and Tat 30-72 showed a 6- to 15-day growth delay.
These growth results for Tat mutant viruses (Tat 30-72 > Tat 20-58) can be interpreted in two ways. We think it less likely that the differences resulted from binding competition for TAR RNA, since all peptides contain the same TAR RNA-binding motif and should bind TAR with similar affinities (9, 10). We favor reduced viral growth as being explained by competitive binding for Sp1 by inactive Tat fragments inside cells (schematically modeled in Fig. 10D). Currently, we do not exclude that additional effects could emerge from interference by Tat mutants on Tat–DNA-PK or Sp1–Tat–DNA-PK interactions.
A separate series of transfections was performed to verify that the Tat mutant fragments expressed from the different proviruses indeed exerted a trans-inhibitory effect. In this approach, an LTR-CAT reporter was transfected into HeLa cells in the presence of pUC19 alone (Fig. 10E, lane 1) or pSV-Tat plus a proviral plasmid (pNL4-3) [lane 2], pTat 20-58 [lane 3], pTat 30-58, [lane 4], pTat 30-72 [lane 5], pTat 30-72i [lane 6]). Results of CAT assays performed 48 h later are consistent with a strong trans-inhibitory effect from the mutant Tat fragment expressed from the pTat 30-72 provirus, suggesting that the reduced replication capacity of this virus (Fig. 10C) is due more to a trans-rather than to a cis-inhibitory effect that occurred as a consequence of insertion into nef.
DISCUSSION
Phosphorylation is one mechanism that variously regulates gene expression (45, 48). Many examples illustrate the importance of phosphorylation in gene activity. One example is the NF-κB proteins. Phosphorylation of IκB, the inhibitory subunit for NF-κB, in the cytoplasm (33, 74, 97) leads to its degradation (25, 42), resulting in nuclear translocation and activity of NF-κB. Another example is Jun, where phosphorylation influences DNA binding (7, 86). A third example is described by CREB, whose activity is stimulated by phosphorylation (78, 108). It has been proposed that phosphorylation enhances the ability of CREB to interact with other factors (12, 68, 69, 78, 110).
In the case of Sp1, ample evidence suggests that DNA-bound Sp1 interacts with coactivators to activate transcription (91). Many factors, including TBP (24), TAF110 (34), nuclear protein p74 (81), RelA (p65) (88, 98), YY1 (71, 95), TAF55 (11), and RNA polymerase II CTD (107), interact with Sp1. Exactly how these interactions are regulated and how they might relate to transcription have not been clearly defined. Potentially, phosphorylation plays a role in Sp1 activity since there is strong circumstantial evidence that phosphorylated Sp1 represents the active moiety in transcription (2, 50, 73, 105).
We and others have previously demonstrated that in the HIV-1 system, Sp1 and Tat can form a protein-protein complex (18, 54). Indeed, various studies have shown that Sp1 contributes to basal (5, 55, 96, 102, 113) and Tat-activated (5, 39, 44, 55, 61, 63, 100, 102) expression of the viral LTR. What are the physical and biological consequences of Tat-Sp1 interaction? Here, we show that Tat-Sp1 interaction affects the phosphorylation state of Sp1 and that inactive Tat mutants that interrupt Tat-Sp1 interaction affect HIV-1 replication in cell cultures.
In this work, we make three salient points. First, we observed a good correlation between inactive Tat mutants that compete with wild-type Tat for Sp1 contact and ones that repress HIV-1 replication in cultured T cells (Fig. 1 and 10). A simple corollary of such finding, which does not exclude others, is that wild-type Tat-Sp1 contact is physiologically important for productive HIV-1 infection. Relevant to this idea is the in situ observation that within intact primate cell nuclei, a population of Tat and Sp1 proteins colocalizes. Second, we observed a contribution of phosphorylated Sp1 for Tat transactivation activity. Although performed on a subfragment of transcriptionally active Sp1, the experiments in Fig. 8 and 9 indicate that within a defined context, serine 131 phosphorylation is important for supporting Tat-activated transcription from the HIV-1 promoter. There are 78 serines in full-length Sp1. Presumably, phosphorylation at other residues can differentially affect other functional activities. Third, we propose a regulatory loop in which Tat serves to influence the phosphorylation state of Sp1. In vitro kinase assays demonstrated that the physical presence of Tat augmented significantly phosphorylation of purified Sp1 in a manner mediated through the enzyme DNA-PK (Fig. 3). Relevant to this result, we found that Tat makes protein-protein contact with DNA-PK (Fig. 4). Further compatible evidence from cell cultures includes the observations that the subpopulation of phosphorylated Sp1 (as opposed to total Sp1) is significantly greater in Tat-expressing than in Tat-nonexpressing (Fig. 7) cells and that phosphatase (Fig. 5) and kinase inhibitors (Fig. 6) when used in cultured cells provide results consistent with phosphorylated Sp1 as being important for HIV-1 LTR expression. Overall, a model that integrates these various findings suggests the coalescence of a multifactor complex that includes Tat, Sp1, and DNA-PK at the HIV-1 promoter (Fig. 4).
Sp1 serves basal and activated functions at the HIV-1 promoter (5, 102, 107). In the absence of Tat, the HIV-1 LTR has a clear dependence on Sp1 for basal expression. At a later stage of virus replication, Sp1 cooperates synergistically with Tat to enhance further transcription from the LTR. Thus, it is conceivable that basal Sp1 activity and Sp1-Tat activity reflect two distinct processes (107). Consistent with this idea is the observation that Sp1-dependent (basal) transcription does not require an RNA polymerase II with intact CTD (13, 32) whereas Tat-activated transcription does require a wild-type form of RNA polymerase II (13, 85, 109). Thus, accordingly, Sp1 phosphorylation as influenced by Tat might be required for the latter but not the former interaction. The mechanistic puzzle of how Tat-Sp1 interaction increases transcription might be explained by the concept that Tat, by virtue of increasing Sp1 phosphorylation, alters interactions between Sp1 and components of the basal transcription machinery, leading to activated gene expression.
ACKNOWLEDGMENTS
We thank Dong-Yan Jin and Hua Xiao for discussions and critical reading of the manuscript. We are grateful to Michelle Van for assistance in preparing the manuscript.
This work was supported in part by funds from the AIDS Targeted Antiviral Program of the Office of the Director, NIH.
REFERENCES
- 1.Anderson C W, Lees-Miller S P. The nuclear serine/threonine protein kinase DNA-PK. Crit Rev Eukaryotic Gene Expression. 1992;2:283–314. [PubMed] [Google Scholar]
- 2.Armstrong S A, Barry D A, Leggett R W, Mueller C R. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J Biol Chem. 1997;272:13489–13495. doi: 10.1074/jbc.272.21.13489. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 4.Berkhout B, Gatignol A, Rabson A B, Jeang K T. TAR-independent activation of the HIV-1 LTR: evidence that tat requires specific regions of the promoter. Cell. 1990;62:757–767. doi: 10.1016/0092-8674(90)90120-4. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout B, Jeang K T. Functional roles for the TATA promoter and enhancers in basal and Tat-induced expression of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1992;66:139–149. doi: 10.1128/jvi.66.1.139-149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohan C A, Kashanchi F, Ensoli B, Buonaguro L, Boris-Lawrie K A, Brady J N. Analysis of Tat transactivation of human immunodeficiency virus transcription in vitro. Gene Expr. 1992;2:391–407. [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle W J, Smeal T, Defize L H, Angel P, Woodgett J R, Karin M, Hunter T. Activation of protein kinase C decreases phosphorylation of c-Jun at sites that negatively regulate its DNA-binding activity. Cell. 1991;64:573–584. doi: 10.1016/0092-8674(91)90241-p. [DOI] [PubMed] [Google Scholar]
- 8.Brand S R, Kobayashi R, Mathews M B. The Tat protein of human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase, PKR. J Biol Chem. 1997;272:8388–8395. doi: 10.1074/jbc.272.13.8388. [DOI] [PubMed] [Google Scholar]
- 9.Chang Y-N, Jeang K-T. The basic RNA-binding domain of HIV-2 Tat determines preferential trans-activation of a TAR2 containing LTR. Nucleic Acids Res. 1992;20:5465–5472. doi: 10.1093/nar/20.20.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y.-N., and K.-T. Jeang. 1993. Unpublished observations.
- 11.Chiang C-M, Roeder R G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 12.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 13.Chun R F, Jeang K T. Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T-cell lymphotropic virus I and HIV-1. J Biol Chem. 1996;271:27888–27894. doi: 10.1074/jbc.271.44.27888. [DOI] [PubMed] [Google Scholar]
- 14.Churcher M J, Lamont C, Hamy F, Dingwall C, Green S M, Lowe A D, Butler J G, Gait M J, Karn J. High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J Mol Biol. 1993;230:90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- 15.Cicirelli M F. Inhibitors of protein serine/threonine phosphatases. Focus. 1992;14:16–20. [Google Scholar]
- 16.Courey A J, Holtzman D A, Jackson S P, Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell. 1989;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 17.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 18.Cujec T P, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin B M. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullen B R. Does HIV-1 Tat induce a change in viral initiation rights? Cell. 1993;73:417–420. doi: 10.1016/0092-8674(93)90126-b. [DOI] [PubMed] [Google Scholar]
- 20.Desai K, Loewenstein P M, Green M. Isolation of a cellular protein that binds to the human immunodeficiency virus Tat protein and can potentiate transactivation of the viral promoter. Proc Natl Acad Sci USA. 1991;88:8875–8879. doi: 10.1073/pnas.88.20.8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desai-Yajnik V, Hadzic E, Modlinger P, Malhotra S, Gechlik G, Samuels H H. Interactions of thyroid hormone receptor with the human immunodeficiency virus type 1 (HIV-1) long terminal repeat and the HIV-1 Tat transactivator. J Virol. 1995;69:5103–5112. doi: 10.1128/jvi.69.8.5103-5112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A, Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emili A, Greenblatt J, Ingles C J. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol Cell Biol. 1994;14:1582–1593. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finco T S, Beg A A, Baldwin A S., Jr Inducible phosphorylation of I kappa B alpha is not sufficient for its dissociation from NF-kappa B and is inhibited by protease inhibitors. Proc Natl Acad Sci USA. 1994;91:11884–11888. doi: 10.1073/pnas.91.25.11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finnie N, Gottlieb T, Hartley K, Jackson S P. Transcription factor phosphorylation by the DNA-dependent protein kinase. Biochem Soc Trans. 1993;21:930–935. doi: 10.1042/bst0210930. [DOI] [PubMed] [Google Scholar]
- 27.Finnie N J, Gottlieb T M, Blunt T, Jeggo P A, Jackson S P. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc Natl Acad Sci USA. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fridell R A, Harding L S, Bogerd H P, Cullen B R. Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- 29.Fu X D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Martinez L F, Mavankal G, Neveu J M, Lane W S, Ivanov D, Gaynor R B. Purification of a Tat-associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 1997;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gentz R, Chen C H, Rosen C A. Bioassay for trans-activation using purified human immunodeficiency virus tat-encoded protein: trans-activation requires mRNA synthesis. Proc Natl Acad Sci USA. 1989;86:821–824. doi: 10.1073/pnas.86.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerber H P, Hagmann M, Seipel K, Georgiev O, West M A, Litingtung Y, Schaffner W, Corden J L. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh S, Baltimore D. Activation in vitro of NF-kappa B by phosphorylation of its inhibitor I kappa B. Nature. 1990;344:678–682. doi: 10.1038/344678a0. [DOI] [PubMed] [Google Scholar]
- 34.Gill G, Pascal E, Tseng Z H, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gottlieb T M, Jackson S P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 37.Graham F L, Eb A J. Transformation of rat cells by DNA of human adenovirus 5. Virology. 1973;54:536–539. doi: 10.1016/0042-6822(73)90163-3. [DOI] [PubMed] [Google Scholar]
- 38.Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrich D, Garcia J, Wu F, Mitsuyasu R, Gonazalez J, Gaynor R. Role of SP1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1989;63:2585–2591. doi: 10.1128/jvi.63.6.2585-2591.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartley K O, Gell D, Smith G C, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 41.Hauber J, Malim M H, Cullen B R. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J Virol. 1989;63:1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henkel T, Machleidt T, Alkalay I, Kronke M, Ben-Neriah Y, Baeuerle P A. Rapid proteolysis of I kappa B-alpha is necessary for activation of transcription factor NF-kappa B. Nature. 1993;365:182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- 43.Herrmann C H, Rice A P. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- 44.Herrmann C H, Rice A P. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill C S, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specifity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 46.Huang L M, Jeang K T. Increased spacing between Sp1 and TATAA renders human immunodeficiency virus type 1 replication defective: implication for Tat function. J Virol. 1993;67:6937–6944. doi: 10.1128/jvi.67.12.6937-6944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang L M, Joshi A, Willey R, Orenstein J, Jeang K T. Human immunodeficiency viruses regulated by alternative trans-activators: genetic evidence for a novel non-transcriptional function of Tat in virion infectivity. EMBO J. 1994;13:2886–2896. doi: 10.1002/j.1460-2075.1994.tb06583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 49.Ishihara H, Martin B L, Brautigan D L, Karaki H, Ozaki H, Kato Y, Fusetani N, Watabe S, Hashimoto K, Uemura D, Hartshorne D J. Calyculin A and okadaic acid: inhibitors of protein phosphatase activity. Biochem Biophys Res Commun. 1989;159:871–877. doi: 10.1016/0006-291x(89)92189-x. [DOI] [PubMed] [Google Scholar]
- 50.Jackson S P, MacDonald J J, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- 51.Jeang K T, Berkhout B. Kinetics of HIV-1 long terminal repeat trans-activation. Use of intragenic ribozyme to assess rate-limiting steps. J Biol Chem. 1992;267:17891–17899. [PubMed] [Google Scholar]
- 52.Jeang K T, Berkhout B, Dropulic B. Effects of integration and replication on transcription of the HIV-1 long terminal repeat. J Biol Chem. 1993;268:24940–24949. [PubMed] [Google Scholar]
- 53.Jeang, K. T., Y. Chang, B. Berkhout, M. L. Hammarskjold, and D. Rekosh. 1991. Regulation of HIV expression: mechanisms of action of Tat and Rev. AIDS 5(Suppl 2):S3–14. [PubMed]
- 54.Jeang K T, Chun R, Lin N H, Gatignol A, Glabe C G, Fan H. In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J Virol. 1993;67:6224–6233. doi: 10.1128/jvi.67.10.6224-6233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones K A, Kadonaga J T, Luciw P A, Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science. 1986;232:755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- 56.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 57.Joshi A, Jeang K-T. Reduction in growth temperature minimizes instability of large plasmids containing HIV-1 proviral genomes. BioTechniques. 1993;14:883–884. [PubMed] [Google Scholar]
- 58.Kaczmarski W, Khan S A. Lupus autoantigen Ku protein binds HIV-1 TAR RNA in vitro. Biochem Biophys Res Commun. 1993;196:935–942. doi: 10.1006/bbrc.1993.2339. [DOI] [PubMed] [Google Scholar]
- 59.Kadonaga J T, Carner K R, Masiarz F R, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 60.Kadonaga J T, Courey A J, Ladika J, Tjian R. Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science. 1988;242:1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- 61.Kamine J, Chinnadurai G. Synergistic activation of the human immunodeficiency virus type 1 promoter by the viral Tat protein and cellular transcription factor Sp1. J Virol. 1992;66:3932–3936. doi: 10.1128/jvi.66.6.3932-3936.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 63.Kamine J, Subramanian T, Chinnadurai G. Sp1-dependent activation of a synthetic promoter by human immunodeficiency virus type 1 Tat protein. Proc Natl Acad Sci USA. 1991;88:8510–8514. doi: 10.1073/pnas.88.19.8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamine J, Subramanian T, Chinnadurai G. Activation of a heterologous promoter by human immunodeficiency virus type 1 Tat requires Sp1 and is distinct from the mode of activation by acidic transcriptional activators. J Virol. 1993;67:6828–6834. doi: 10.1128/jvi.67.11.6828-6834.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kashanchi F, Piras G, Radonovich M F, Duvall J F, Fattaey A, Chiang C M, Roeder R G, Brady J N. Direct interaction of human TFIID with the HIV-1 transactivator tat. Nature. 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 66.Kato H, Sumimoto H, Pognonec P, Chen C H, Rosen C A, Roeder R G. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 1992;6:655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- 67.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 68.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 69.Kwok R P S, Laurance M E, Lundblad J R, Goldman P S, Shih H-M, Connor L M, Marriot S J, Goodman R H. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 70.Laspia M F, Rice A P, Mathews M B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- 71.Lee J S, Galvin K M, Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc Natl Acad Sci USA. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M, Day R S, Barron G M, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 73.Leggett R W, Armstrong S A, Barry D, Mueller C R. Sp1 is phosphorylated and its DNA binding activity down-regulated upon terminal differentiation of the liver. J Biol Chem. 1995;270:25879–25884. doi: 10.1074/jbc.270.43.25879. [DOI] [PubMed] [Google Scholar]
- 74.Link E, Kerr L D, Schreck R, Zabel U, Verma I, Baeuerle P A. Purified I kappa B-beta is inactivated upon dephosphorylation. J Biol Chem. 1992;267:239–246. [PubMed] [Google Scholar]
- 75.Lu X, Welsh T M, Peterlin B M. The human immunodeficiency virus type 1 long terminal repeat specifies two different transcription complexes, only one of which is regulated by Tat. J Virol. 1993;67:1752–1760. doi: 10.1128/jvi.67.4.1752-1760.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marciniak R A, Sharp P A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mavankal G, Ignatius Ou S H, Oliver H, Sigman D, Gaynor R B. Human immunodeficiency virus type 1 and 2 Tat proteins specifically interact with RNA polymerase II. Proc Natl Acad Sci USA. 1996;93:2089–2094. doi: 10.1073/pnas.93.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mayall T P, Sheridan P L, Montminy M R, Jones K A. Distinct roles for P-CREB and LEF-1 in TCR-alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 79.McMillan N A J, Chun R F, Sidervoski D P, Galabru J, Toone W M, Samuel C E, Mak T W, Hovanessian A G, Jeang K-T, Williams B R G. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology. 1995;213:413–424. doi: 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 80.Muesing M A, Smith D H, Capon D J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 81.Murata Y, Kim H G, Rogers K T, Udvadia A J, Horowitz J M. Negative regulation of Sp1 trans-activation is correlated with the binding of cellular proteins to the amino terminus of the Sp1 trans-activation domain. J Biol Chem. 1994;269:20674–20681. [PubMed] [Google Scholar]
- 82.Myers G, Wain-Hobson S, Henderson L E, Korber B, Jeang K-T, Pavlakis G N. Human Retroviruses and AIDS 1994. Los Alamos, N.Mex: Los Alamos National Laboratory; 1994. [Google Scholar]
- 83.Nelbock P, Dillon P J, Perkins A, Rosen C A. A cDNA for a protein that interacts with the human immunodeficiency virus Tat transactivator. Science. 1990;248:1650–1653. doi: 10.1126/science.2194290. [DOI] [PubMed] [Google Scholar]
- 84.Ohana B, Moore P A, Ruben S M, Southgate C D, Green M R, Rosen C A. The type 1 human immunodeficiency virus Tat binding protein is a transcriptional activator belonging to an additional family of evolutionarily conserved genes. Proc Natl Acad Sci USA. 1993;90:138–142. doi: 10.1073/pnas.90.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Okamoto H, Sheline C T, Corden J L, Jones K A, Peterlin B M. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA. 1996;93:11575–11579. doi: 10.1073/pnas.93.21.11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Papavassiliou A G, Bohmann K, Bohmann D. Determining the effect of inducible protein phosphorylation on the DNA-binding activity of transcription factors. Anal Biochem. 1992;203:302–309. doi: 10.1016/0003-2697(92)90317-z. [DOI] [PubMed] [Google Scholar]
- 87.Parada C A, Roeder R G. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 88.Perkins N D, Agranoff A B, Pascal E, Nabel G J. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol Cell Biol. 1994;14:6570–6583. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peterson S R, Dvir A, Anderson C W, Dynan W S. DNA binding provides a signal for phosphorylation of the RNA polymerase II heptapeptide repeats. Genes Dev. 1992;6:426–438. doi: 10.1101/gad.6.3.426. [DOI] [PubMed] [Google Scholar]
- 90.Peterson S R, Jesch S A, Chamberlin T N, Dvir A, Rabindran S K, Wu C, Dynan W S. Stimulation of the DNA-dependent protein kinase by RNA polymerase II transcriptional activator proteins. J Biol Chem. 1995;270:1449–1454. doi: 10.1074/jbc.270.3.1449. [DOI] [PubMed] [Google Scholar]
- 91.Pugh B F, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 92.Rice A P, Mathews M B. Transcriptional but not translational regulation of HIV-1 by the tat gene product. Nature. 1988;332:551–553. doi: 10.1038/332551a0. [DOI] [PubMed] [Google Scholar]
- 93.Ruben S, Perkins A, Purcell R, Joung K, Sia R, Burghoff R, Haseltine W A, Rosen C A. Structural and functional characterization of human immunodeficiency virus Tat protein. J Virol. 1989;63:1–8. doi: 10.1128/jvi.63.1.1-8.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Selby M J, Peterlin B M. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell. 1990;62:769–776. doi: 10.1016/0092-8674(90)90121-t. [DOI] [PubMed] [Google Scholar]
- 95.Seto E, Lewis B, Shenk T. Interaction between transcription factors Sp1 and YY1. Nature. 1993;365:462–464. doi: 10.1038/365462a0. [DOI] [PubMed] [Google Scholar]
- 96.Sheridan P L, Sheline C T, Cannon K, Voz M L, Pazin M J, Kadonaga J T, Jones K A. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 97.Shirakawa F, Mizel S B. In vitro activation and nuclear translocation of NF-κB catalyzed by cyclic AMP-dependent protein kinase and protein kinase C. Mol Cell Biol. 1989;9:2424–2430. doi: 10.1128/mcb.9.6.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sif S, Gilmore T D. Interaction of the v-Rel oncoprotein with cellular transcription factor Sp1. J Virol. 1994;68:7131–7138. doi: 10.1128/jvi.68.11.7131-7138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Southgate C, Zapp M L, Green M R. Activation of transcription by HIV-1 Tat protein tethered to nascent RNA through another protein. Nature. 1990;345:640–642. doi: 10.1038/345640a0. [DOI] [PubMed] [Google Scholar]
- 100.Southgate C D, Green M R. The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes Dev. 1991;5:2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- 101.Southgate C D, Green M R. Delineating minimal protein domains and promoter elements for transcriptional activation by lentivirus Tat proteins. J Virol. 1995;69:2605–2610. doi: 10.1128/jvi.69.4.2605-2610.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sune C, Garcia-Blanco M A. Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1995;69:6572–6576. doi: 10.1128/jvi.69.10.6572-6576.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Taccioli G E, Gottlieb T M, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann A R, Alt F W, Jackson S P, Jeggo P A. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 104.Veschambre P, Simard P, Jalinot P. Evidence for functional interaction between the HIV-1 Tat transactivator and the TATA box binding protein in vivo. J Mol Biol. 1995;250:169–180. doi: 10.1006/jmbi.1995.0368. [DOI] [PubMed] [Google Scholar]
- 105.Vlach J, Garcia A, Jacque J M, Rodriguez M S, Michelson S, Virelizier J L. Induction of Sp1 phosphorylation and NF-kappa B-independent HIV promoter domain activity in T lymphocytes stimulated by okadaic acid. Virology. 1995;208:753–761. doi: 10.1006/viro.1995.1207. [DOI] [PubMed] [Google Scholar]
- 106.Wu-Baer F, Sigman D, Gaynor R B. Specific binding of RNA polymerase II to the human immunodeficiency virus trans-activating region RNA is regulated by cellular cofactors and Tat. Proc Natl Acad Sci USA. 1995;92:7153–7157. doi: 10.1073/pnas.92.16.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xiao H, Lis J T, Jeang K T. Promoter activity of Tat at steps subsequent to TATA-binding protein recruitment. Mol Cell Biol. 1997;17:6898–6905. doi: 10.1128/mcb.17.12.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamamoto K K, Gonzalez G A, Biggs III W H, Montminy M R. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988;334:494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 109.Yang X, Herrmann C H, Rice A P. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yin M J, Gaynor R B. HTLV-1 21 bp repeat sequences facilitate stable association between Tax and CREB to increase CREB binding affinity. J Mol Biol. 1996;264:20–31. doi: 10.1006/jmbi.1996.0620. [DOI] [PubMed] [Google Scholar]
- 111.Yu L, Loewenstein P M, Zhang Z, Green M. In vitro interaction of the human immunodeficiency virus type 1 Tat transactivator and the general transcription factor TFIIB with the cellular protein TAP. J Virol. 1995;69:3017–3023. doi: 10.1128/jvi.69.5.3017-3023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu L, Zhang Z, Loewenstein P M, Desai K, Tang Q, Mao D, Symington J S, Green M. Molecular cloning and characterization of a cellular protein that interacts with the human immunodeficiency virus type 1 Tat transactivator and encodes a strong transcriptional activation domain. J Virol. 1995;69:3007–3016. doi: 10.1128/jvi.69.5.3007-3016.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou Q, Sharp P A. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou Q, Sharp P A. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]