Abstract
Open reading frame IV (ORF-IV) of Borna disease virus (BDV) encodes a protein with a calculated molecular mass of ca. 57 kDa (p57), which increases after N glycosylation to 94 kDa (gp94). The unglycosylated and glycosylated proteins are proteolytically cleaved by the subtilisin-like protease furin. Furin most likely recognizes one of three potential cleavage sites, namely, an arginine at position 249 of the ORF-IV gene product. The furin inhibitor decRVKRcmk decreases the production of infectious BDV significantly, indicating that proteolytic cleavage of the gp94 precursor molecule is necessary for the full biological activity of the BDV glycoprotein.
Borna disease virus (BDV) is the etiological agent of Borna disease (BD), a persistent infection of the central nervous system, and has been recently identified as an enveloped, nonsegmented, negative-strand RNA virus with unique properties of replication (2, 19). On the basis of its genome organization, BDV was classified as the prototype of the new family Bornaviridae within the order Mononegavirales. Studies on the pathogenesis of BD in the Lewis rat have shown that BDV replication does not directly influence vital functions in the infected host; it seems to affect only modulations of high integrative brain functions, like learning (13). Typical signs of BD are caused by a virus-induced T-cell-mediated immune reaction (17, 22).
To date, virus-encoded protein products of five open reading frames (ORFs) have been identified, representing the putative nucleoprotein (encoded by ORF-1), phosphoprotein (encoded by ORF-II), matrix protein (encoded by ORF-III), and glycoprotein (GP, encoded by ORF-IV) of BDV (6, 19, 20) and, recently, the p10 BDV-specific protein (encoded by ORF-x1 [24]). ORF-IV predicts a polypeptide with a mass of approximately 57 kDa (p57). This BDV-specific protein (BDV-GP) has been recently found by vector expression and in persistently BDV-infected cell lines and has a molecular mass of approximately 84 kDa (6), or 94 kDa (20) when glycosylated. The BDV genome encodes at least two glycosylated gene products of 18 (gp18, matrix protein) and 84 or 94 (gp84 or gp94, GPs) kDa.
Apart from their immunogenic significance, GPs from enveloped viruses are of great importance for the uptake of virus particles by the host cell. They are responsible for virus attachment to cellular receptors and are involved in virus penetration by fusion of the virus envelope with cellular membranes. For most viral GPs, fusion and, consequently, virus infectivity depend on proteolytic cleavage of a precursor molecule, which results in the exposure of a hydrophobic fusion-mediating peptide, the fusion peptide. Proteolytic cleavage activation is performed by cellular proteases, and the presence of an appropriate protease determines whether infectious virus is produced in a given cell (12). Most of the cleavable viral GPs processed in a wide range of different host cells possess a cleavage site which contains a multibasic amino acid motif, usually RXK/RR (11). Endoproteases which recognize those substrate motifs belong to the eukaryotic subtilisin family, which includes furin, PC2, PC1/PC3, PC4, PACE4, PC5/PC6, and LPC/PC7. Furin is a ubiquitously occurring, well-characterized class I membrane protein of the exocytotic transport route and accumulates in the trans-Golgi network. A considerable portion of furin is cleaved within the cell and secreted as a truncated, soluble enzyme into the extracellular space (23).
The study presented here was initiated to elucidate the proteolytic processing of the p57/gp94 BDV-GP by furin. Cleavage of the BDV-GP precursor molecule was shown for the in vitro-transcribed and -translated, unglycosylated p57, for glycosylated gp94 expressed by a recombinant vaccinia virus (recVV), and in vivo in BDV-infected rat brain material.
Primary structure of BDV-GP.
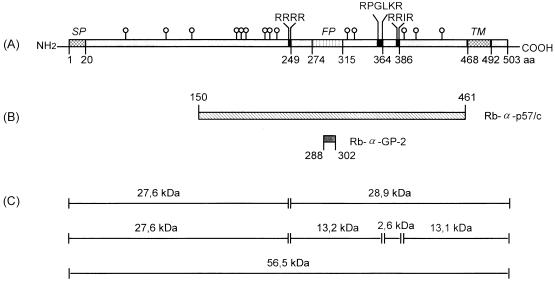
In order to examine the BDV-GP polypeptide, ORF-IV, which codes for BDV-GP, was amplified and then cloned into expression vectors. Total RNA from the wild-type horse BDV isolate H640 was reverse transcribed, and the cDNA was amplified by PCR with the oligonucleotides 5′ACGGGATCCATGCAGCCTTCAATGTCTTT3′ and 5′GTAGAATTCTTATTCCTGCCACCGGCCGAGGCGTC3′. The amplified cDNA was inserted into the BamHI and EcoRI sites of the plasmid pcDNA3 (Invitrogen, NV Leek, The Netherlands) and was used for transformation of Escherichia coli. The GP-specific DNA of pcDNA3-GP was sequenced by the dideoxy chain termination method (18), employing the following oligonucleotides as primers: 5′CTTCCAGATTGACGACTTCT3′, 5′GGTAGCTACAGCGCTGACCG3′, 5′GGACCATACTCTGCAACTCA3′, 5′GGGGTTGTCTGGCAATGC3′, 5′CAATGGGCGGAAGTACTTCC3′, 5′TAATACGACTCACTATAGGG3′ (T7 promoter site), and 5′TAGAAGGCACAGTCGAGG3′. The GP polypeptide, deduced from the full-length ORF-IV gene, consists of 503 amino acids with a molecular mass of approximately 56.5 kDa. Sequence comparison of isolate H640 with BDV strain He/80 (1) revealed 35 differences in the nucleotide sequence, of which only 5 led to amino acid changes, at positions 21 (R to Q), 242 (S to P), 243 (K to R), 245 (R to K), and 311 (F to L). BDV-GP contains two hydrophobic amino acid sequences close to the N and C termini representing the signal sequence (amino acids 1 to 20) and the transmembrane domain (amino acids 468 to 492). Two additional hydrophobic amino acid sequences between positions 274 and 315 and three potential furin recognition motifs at arginine residues 249, 364, and 386 are also present in the GP polypeptide (Fig. 1A).
FIG. 1.
Schematic map of BDV ORF-IV. (A) Diagram of the translation product of ORF-IV, BDV-GP, containing the signal peptide (SP), the transmembrane domain (TM), the putative fusion peptide (FP), potential cleavage sites for the eukaryotic subtilisin-like endoprotease furin (black boxes, amino acid sequences in one-letter code), and N-glycans (○|). (B) BDV-GP-specific fragment and oligopeptide used for the immunization of rabbits. The monospecific antiserum Rb-α-p57/c was obtained by immunization of rabbits with the recombinant GP-specific polypeptide (amino acids 150 to 461), and the monospecific antiserum Rb-α-GP-2 was obtained by immunization with a chemically synthesized oligopeptide with the sequence ASASQFLRGWLNHPD. (C) Fragments, with calculated molecular masses, of nonglycosylated BDV-GP containing the signal peptide predicted by hypothetical furin cleavage sites.
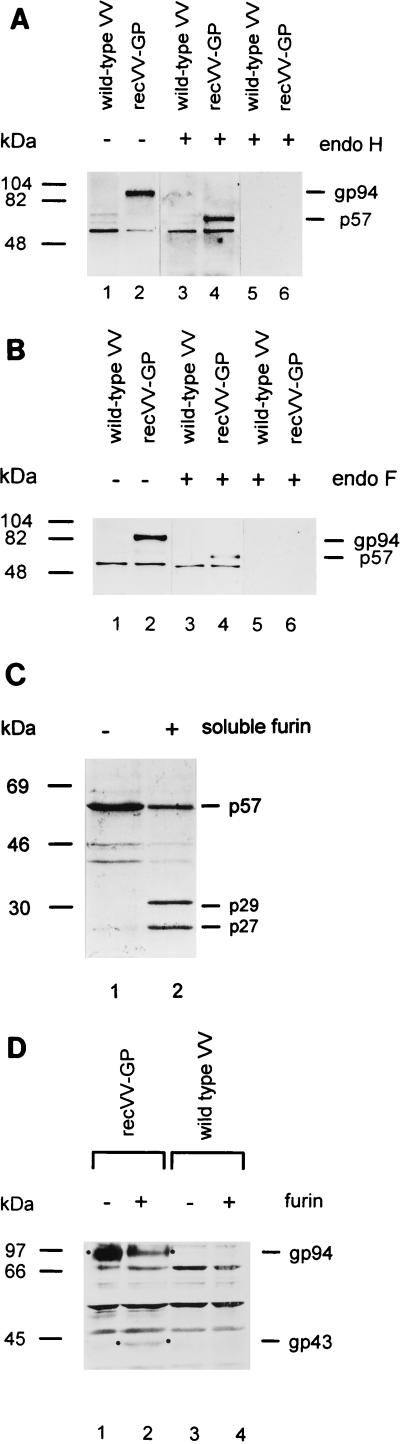
Within the coding sequence, 14 putative attachment sites for N-glycans were found at asparagine residues 63, 109, 139, 192, 196, 202, 221, 230, 235, 321, 328, 388, 404, and 438 (Fig. 1A). To study glycosylation, a eukaryotic expression system was established. A recVV expressing full-length BDV-GP (recVV-GP) was generated by insertion of the GP DNA from vector pcDNA3-GP into the SmaI site of vector pSC11. Transfection and isolation of recombinant viruses were performed as previously described (14). Through glycosylation, the molecular mass of the BDV ORF-IV gene product, p57, increases to about 94 kDa, the mass of the gp94 BDV protein (Fig. 2A and B, lanes 2). Using different deglycosylation enzymes, we could show that the glycans were linked N-glycosidically. The sensitivity of gp94 to endoglycosidase H (endo H) (Fig. 2A, lane 4) and endo F (Fig. 2B, lane 4) suggests that the N-linked carbohydrates consist largely of the high-mannose and/or hybrid oligosaccharide type. The absence of a shift in molecular weight after O-glycosidase digestion (data not shown) suggests that BDV-GP does not contain O-linked carbohydrates. Similar results were previously obtained by Schneider et al. (20) and by Gonzalez-Dunia et al. (6) using a recombinant protein expressed by Semliki Forest and vaccinia virus vectors, respectively. For these experiments, CV-1 cells were infected with recVV-GP and wild-type VV at a multiplicity of infection of 10. At 18 h postinfection, CV-1 cells infected with recVV-GP (Fig. 2A and B, lanes 2, 4, and 6) or wild-type VV (Fig. 2A and B, lanes 1, 3, and 5) were solubilized in Laemmli buffer, and proteins were precipitated with 10% trichloroacetic acid, treated with endo H or endo F (15), and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
FIG. 2.
Analysis of unglycosylated and glycosylated BDV-GP. (A and B) Deglycosylation of BDV-GP by endo H (A) and endo F (B). CV-1 cells were infected with recVV-GP and wild-type VV. BDV-GP was detected in immunoblots with the antiserum Rb-α-p57/c (lanes 1 to 4). As a control, the respective preimmune serum (lanes 5 and 6) was employed. The prestained molecular mass markers (Bio-Rad, Munich, Germany) were phosphorylase b (104 kDa), bovine albumin (82 kDa), and ovalbumin (48 kDa). (C) Cleavage of in vitro-translated unglycosylated BDV-GP. Full-length mRNA of ORF-IV of BDV was transcribed and translated in a rabbit reticulocyte lysate in the presence of [35S]methionine. The translation product was incubated with soluble furin (lane 2) and without furin (lane 1). Radioactively labeled proteins were electrophoretically separated on SDS-PAGE (10% polyacrylamide) gels and visualized by autora- diography. The molecular mass markers (Amersham Buchler, Braunschweig, Germany) were bovine albumin (69 kDa), ovalbumin (46 kDa), and carbonic anhydrase (30 kDa). (D) Coexpression of VV-expressed glycosylated BDV-GP with furin. CV-1 cells were infected with recVV-GP alone (lane 1), recVV-GP and VV expressing furin (lane 2), wild-type VV alone (lane 3), and wild-type VV and VV expressing furin (lane 4) at a multiplicity of infection of 10. The cells were treated with sample buffer and subjected to SDS-PAGE. The proteins were blotted onto Immobilon-P membranes (Millipore, Bedford, Mass.) and incubated with Rb-α-GP-2 antiserum. BDV-specific bands were detected by chemiluminescence (ECL detection kit; Amersham). The molecular mass markers (Bio-Rad) were rabbit phosphorylase (97 kDa), bovine albumin (66 kDa), and ovalbumin (45 kDa).
Cleavage of unglycosylated BDV-GP by furin.
Full-length unglycosylated BDV-GP with the expected size of ca. 57 kDa including the signal peptide was obtained by in vitro transcription and translation of ORF-IV in reticulocyte lysates (Fig. 2C, lane 1). ORF-IV of BDV was transcribed from plasmid pcDNA3-GP and translated in a rabbit reticulocyte lysate in the presence of 100 μCi of [35S]methionine. The unglycosylated GP polypeptide (50 μl) was treated with soluble furin obtained from the supernatants of CV-1 cells (1 μl of a 100-fold-concentrated solution) which were infected with a recVV expressing bovine furin according to the method of Hallenberger et al. (9). The translation product was incubated with soluble furin in the presence of 10 mM CaCl2 for 30 min at 37°C (Fig. 2C, lane 2) or incubated under the same conditions without furin (Fig. 2C, lane 1). As shown in Fig. 2C, p57 was cleaved into two fragments with molecular masses of 27 kDa (p27) and 29 kDa (p29) (Fig. 2C, lane 2). This indicates that the cleavage site is most likely located at arginine 249 (Fig. 1C). Since only these two cleavage products were found, it can be assumed that the other two potential furin recognition sites were not accessible to furin (Fig. 1A and C). Cleavage at arginine 364 or arginine 386 would result in fragments with considerably different molecular weights (Fig. 1C).
Cleavage of glycosylated BDV-GP by furin.
The monospecific antisera Rb-α-p57/c and Rb-α-GP-2, used to detect BDV-GP, were obtained by immunization of rabbits with a recombinant GP-specific polypeptide (for Rb-α-p57/c) and a chemically synthesized oligopeptide (for Rb-α-GP-2). As indicated in Fig. 1B, the GP-specific polypeptide represents amino acids 150 to 461 (p57/c, expressed as a fusion protein with glutathione S-transferase [GST]) and the oligopeptide comprises amino acids 288 to 302 (peptide GP-2 within the C-terminal part of BDV-GP). The cDNA representing the BDV-GP fragment (amino acids 150 to 461) was cloned into the BamHI and EcoRI sites of pGEX-2T (Pharmacia, Freiburg, Germany) and the construct pGEX-2T-GP150-461, expressed in E. coli as a GST fusion protein. The unglycosylated polypeptide GP150-461 was purified with a GST purification module (Pharmacia). The peptide GP-2, which contains the amino acid sequence ASASQFLRGWLNHPD, was chemically synthesized and coupled to keyhole limpet hemocyanin (15), and the Rb-α-GP-2 serum was generated by standard procedures.
recVV-GP was used in order to study furin accessibility of glycosylated BDV-GP (Fig. 2D). CV-1 cells were infected with recVV-GP or wild-type VV. Eighteen hours after VV infection, the activity of the endogenous protease furin was strongly suppressed in CV-1 cells and furin-mediated cleavage was not observed anymore (21). When these cells were infected for 18 h with recVV-GP alone, the GP-specific rabbit Rb-α-GP-2 antiserum (Fig. 1B) detected a virus-specific protein with an apparent molecular mass of 94 kDa, the gp94 BDV protein (Fig. 2D, lane 1). Proteolytic processing of viral GPs by furin has been shown previously after coinfection of CV-1 cells with recVVs expressing various viral GPs and furin (11). In this approach, the Rb-α-GP-2 serum detected, in addition to gp94, a virus-specific product with a molecular mass of ca. 43 kDa (gp43) in immunoblots (Fig. 2D, lane 2). On the basis of the specificity of the Rb-α-GP-2 serum, the small polypeptide gp43 most likely represents the membrane-anchored, C-terminal part of the furin-cleaved precursor gp94. Similarly, when Rb-α-p57/c serum (Fig. 1B) and a pooled polyclonal BDV-specific rat serum, BDV-Se, were used in immunoblot analyses, two bands, corresponding to gp94 and gp43, were again found (data not shown). The failure to detect the second cleavage product of gp94, in contrast to the results obtained with unglycosylated p57, might be due to the release of the N-terminal part into the supernatant or to comigration of the N- and C-terminal fragments of gp94 on polyacrylamide gels. Only one cleavage site, most likely arginine 249, was found to be accessible by furin, regardless of the presence or absence of carbohydrate side chains. The calculated molecular masses of the unglycosylated (Fig. 1C) and glycosylated cleavage products and the recognition of gp43 by the Rb-α-GP-2 antiserum (Fig. 1B) support this assumption.
Detection of gp94 and gp43 in BDV-infected rat brain material.
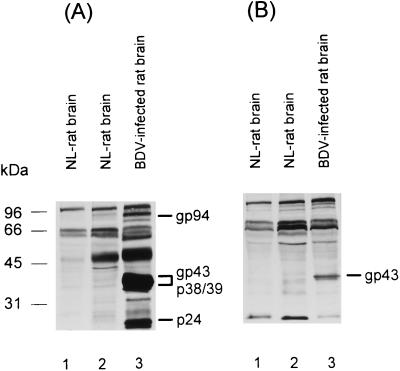
Both gp94 and gp43 were also produced in BDV-infected rat brain. This was shown when BDV-infected brain homogenates from rats euthanatized 68 days after intracerebral infection with the He/80 strain of BDV were subjected to SDS-PAGE and analyzed in immunoblots employing BDV-Se and Rb-α-p57/c sera as well as respective rat and rabbit control sera (Fig. 3). BDV-infected and uninfected rat brains were homogenized in Tris buffer containing Triton X-100 and sodium deoxycholate according to the method of Haas et al. (7). BDV-Se reacted with at least four virus-specific proteins, including gp94 and gp43. The proteins p38/39 and p24, usually found in infected brain material, correspond to ORF-I and ORF-II gene products, respectively (Fig. 3A, lane 3). Rb-α-p57/C serum preferentially recognized gp43, which sometimes became visible as a double band (Fig. 3B, lane 3). Since this serum is potentially capable of recognizing both fragments of gp94, the double band might represent the N- and C-terminal fragments of BDV-GP, which might run very close to each other in SDS-PAGE. As expected, none of these proteins was found in uninfected rat brain homogenates (Fig. 3, lanes 1 and 2) or in BDV-infected brain when control sera were used (data not shown).
FIG. 3.
Cleavage of BDV-GP in rat brain. Brain homogenates from BDV-infected (lanes 3) and uninfected (NL [normal]) (lanes 1 and 2) Lewis rats were analyzed. The proteins were separated by SDS-PAGE and immunoblotted, using a reconvalescent-phase BDV-specific rat serum (A) and the Rb-α-p57/c antiserum (B). The molecular mass markers (Bio-Rad) include those used for Fig. 2D and carbonicanhydrase (31 kDa).
For immunocytochemical detection of BDV-specific proteins, brain material from BDV-infected (91 days postinfection) and uninfected rats was formalin fixed and paraffin embedded. The primary antisera were diluted 1:500 in Tris-buffered saline (TBS). Before use, the Rb-α-p57/c was absorbed to uninfected rat brain material. The secondary antibodies were biotinylated horse anti-mouse antibody (Camon, Wiesbaden, Germany), diluted 1:110 in TBS, and biotinylated goat anti-rabbit antibody (Camon), also diluted 1:110 in TBS. An avidin-biotin-peroxidase complex was added and 3,3-diaminobenzidine-tetrahydrochloride was used as a substrate.
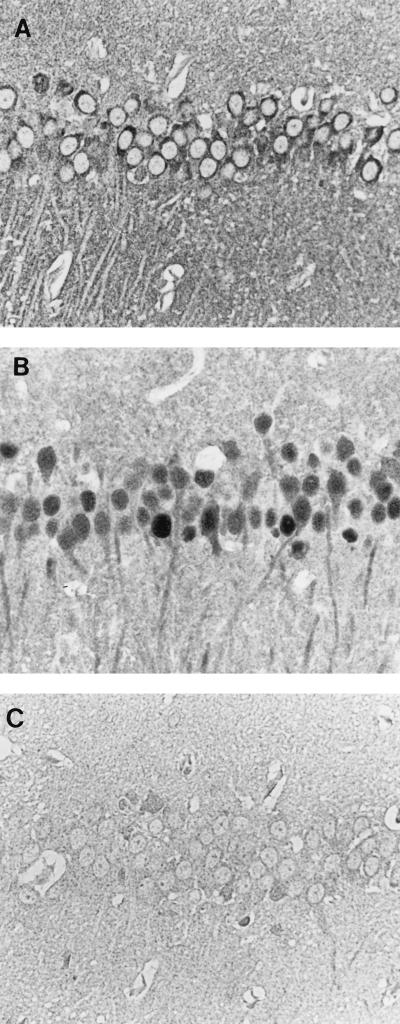
Interestingly, when brain sections from BDV-infected rats were stained with the Rb-α-p57/c serum (Fig. 4A) or the mouse monoclonal antibody Bo18 (Fig. 4B), directed against the p38/p39 BDV protein (7), significantly fewer GP-positive neuronal cells than p38/p39-positive cells were detected. Only in the hippocampus was GP expression comparable to that of the p38/p39 BDV protein. The controls, incubation of preimmune serum with BDV-infected rat brain (Fig. 4C) and of Rb-α-p57/c with uninfected rat brain (data not shown), produced negative results. GP was found only in the cytoplasm (Fig. 4A), whereas p38/p39 was found in the nuclei and cytoplasms of almost all neurons (Fig. 4B). The restricted expression of BDV-GP in infected brain may explain the small quantities of this GP found in brain homogenates and may indicate that only a few neuronal cells are capable of productive BDV replication. The late appearance of neutralizing antibodies, occasionally found after BDV infection (13), could also be explained by this finding.
FIG. 4.
Detection of the GP and nucleoprotein of BDV in BDV-infected rat brain. (A and B) Immunocytochemical detection of BDV-GP with Rb-α-p57/c antiserum (A) and of BDV nucleoprotein (p38/p39) with the monoclonal antibody Bo18 (B) in pyramidal cells of the hippocampus. BDV-GP is localized only in the cytoplasm and neuropil, whereas the nucleoprotein is found in the nuclei, cytoplasms and neuronal processes of infected cells and in the neuropil. (C) BDV-infected rat brain incubated with preimmune rabbit serum. The sections were counterstained with Papanicolaou stain. Magnification, ×350.
Inhibition of BDV infectivity by the protease inhibitor decRVKRcmk.
To substantiate our findings that furin proteolytically activates the precursor gp94, we examined whether peptidylchloromethylketones can inhibit the production of infectious BDV. We used the decanoylated arginyl-valyl-lysyl-arginyl chloromethylketone (decRVKRcmk), which penetrates into cells and therefore enhances the efficacy of the inhibitor (5). As demonstrated previously, the inhibitor prevents cleavage, GP-mediated membrane fusion, and, therefore, the production of infectious virus by mimicking the sequence motif at the cleavage site. This was demonstrated for hemagglutinin of pathogenic avian influenza viruses (21) and gp160 of human immunodeficiency virus (8). Under the conditions described in the legend to Fig. 5, production of infectious virus was reduced approximately 90-fold in the presence of decRVKRcmk (Fig. 5A). The incomplete suppression is most likely due to the instability of the inhibitor in cell cultures, as has been found in other virus systems (5).
FIG. 5.
Effect of the furin inhibitor decRVKRcmk on BDV infectivity. (A) Rabbit brain embryo cells were infected at 105 50% tissue culture infective doses (TCID50) per ml with a Vero cell-adapted BDV strain in the presence or absence of 30 μM decRVKRcmk as described previously (5). After 5 days of incubation at 37°C, infectivity of BDV in respective cell lysates was determined after sonication by serial dilutions of the samples on rabbit brain embryo cells. After 11 days of incubation, evaluation was carried out by indirect immunofluorescence with polyclonal reconvalescent-phase rat serum (10). The data presented are means and standard deviations of three independent experiments. (B) Furin treatment of decRVKRcmk-incubated BDV isolates restores infectivity. The experiment was performed as described for panel A, except that cell lysates from decRVKRcmk-treated and untreated BDV-infected cells (200 μl each) were incubated with 10 μl of soluble furin for 30 min at 37°C in the presence of 10 mM CaCl2 before both samples were titrated for TCID50 determination.
Furthermore, when BDV isolated from decRVKRcmk-treated cells was incubated with soluble furin, infectivity increased and BDV titers rose to near control levels (Fig. 5B). This indicates that the number of virus particles present in the lysate of decRVKRcmk-treated cells seems to be similar to that of virus particles present in untreated BDV-infected cell lysates. Similarly, when BDV isolated from decRVKRcmk-treated cells was incubated with trypsin (10 μg per ml), its infectivity increased about 60-fold (data not shown). This indicates that another arginine-specific protease can substitute for furin in vitro.
Conclusions.
This study shows that the subtilisin-like protease furin is able to proteolytically process unglycosylated and glycosylated precursor molecules of BDV-GP, most likely using arginine 249 as its cleavage site. Two cleavage products, p27 and p29, were found when unglycosylated p57 protein was analyzed, and only one product, gp43, was found when glycosylated gp94 was analyzed. Most probably, cleavage of gp94 by furin results in two proteins with similar molecular masses, about 43 to 50 kDa. The additional two furin recognition sites at arginine 364 and arginine 386 are unlikely to be cleaved because no corresponding fragments were found. In addition, the cleavage site arginine 386 may be not accessible to furin for several reasons: the tyrosine following the cleavage motif is an unusual amino acid to find at the P+1 position, and the next amino acid is asparagine with an N-linked sugar; both amino acids may block furin accessibility. Furthermore, the furin recognition motifs RXXXKR and RXXR (16), found at arginines 364 and 386, respectively, are rare in furin-mediated cleavage of viral GPs (11).
Gonzalez-Dunia et al. (6) reported that expression of BDV ORF-IV results in two virus-specific polypeptides with molecular masses of about 84 kDa (gp84) and 43 kDa (gp43), where gp84 corresponds to the full-length glycosylated BDV-GP and the polypeptide gp43 corresponds to BDV-GP’s C-terminal portion. They suggested that gp43 is generated either by cleavage of gp84 by cellular proteases or by internal initiation of GP-specific mRNA. These findings were extended in our study, and furin-mediated cleavage was clearly demonstrated.
As determined by amino acid sequencing, the N-terminal fragment generated by cleavage contains 16 cysteine residues and nine putative glycosylation sites, which predicts an estimated molecular mass of about 50 kDa. The C-terminal fragment contains two cysteine residues in the ectodomain and one cysteine in the membrane anchor. Of five consensus sequences for N-glycans, only four are likely to be occupied. The proline following asparagine at position 404 might prevent glycosylation, as was demonstrated for influenza virus hemagglutinin (3). The calculated molecular mass of the C-terminal fragment of gp94 is about 43 kDa.
When BDV isolated from decRVKRcmk-treated cells was incubated with soluble furin or trypsin, virus infectivity increased to levels similar to those in nontreated BDV-infected cells. This indicates that proteolytic cleavage of BDV-GP is a prerequisite for virus infectivity. Although not directly shown, it can be assumed that cleavage activation coincides with the activation of virus-triggered membrane fusion. Through the cleavage event, a bipartite hydrophobic fusion peptide (amino acids 274 to 294 and 303 to 315) separated by an octapeptide of a predicted turn structure may be exposed in the C-terminal fragment. This putative fusion peptide starts 25 amino acids in the C-terminal direction from the putative cleavage site (arginine 249) and most likely represents a loop held together by a disulfide bond between cysteine 272 and cysteine 317. A similar fusion domain has been described for the transmembrane proteins of the Ebola/Marburg and avian sarcoma viruses (4). It can therefore be assumed that membrane fusion is mediated by a fusion peptide of BDV-GP which is exposed after furin cleavage. Assuming that an intramolecular disulfide bond between cysteine 272 and cysteine 317 of the membrane-anchored C-terminal fragment exists, no additional cysteine will be available for intermolecular linkage. We suggest, therefore, that the two GP fragments are not linked to each other by disulfide bridges. Which of the gp94 fragments is responsible for binding to a cellular receptor remains to be elucidated.
The data presented here strongly indicate that BDV-GP needs to be proteolytically cleaved in order to express all of its biological properties. The possibility that, besides furin, other proteases might be able to activate gp94 cannot be excluded. Other subtilisin-like proteases, particularly PC4, PACE4, PC5/PC6, and LPC/PC7, all of which are normally present in the constitutive pathway, and others not belonging to the subtilisin family, such as trypsin-like enzymes, could be candidates for such proteases. Since BDV is a highly neurotropic virus, subtilisin-like proteases PC1/PC3 and PC2, constituents of the regulatory pathway of neuroendocrine cells, should also be considered. Coexpression of BDV-GP with the respective proprotein convertases is necessary for gaining further insight into the activation of BDV in the central nervous systems of infected animals.
Acknowledgments
This study was supported by Sonderforschungsbereiche (SFB) 272 (J.A.R. and I.P.), SFB 535 (J.A.R.), and SFB 286 (W.G.) and by grant Ga 282/2-2 (W.G.) from the Deutsche Forschungsgemeinschaft.
We acknowledge the excellent technical assistance of S. Berghöfer, H. Eckhardt, K. Haberzettl, and M. Sordel. We thank K. Frese, S. Herzog, and W. Schäfer for their help. We are very grateful to R. Rott and H.-D. Klenk for their interest, critical discussions, and reading of the manuscript.
REFERENCES
- 1.Cubitt B, Oldstone C, de la Torre J C. Sequence and genome organization of Borna disease virus. J Virol. 1994;68:1382–1396. doi: 10.1128/jvi.68.3.1382-1396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de la Torre J C. Molecular biology of Borna disease virus: prototype of a new group of animal viruses. J Virol. 1994;68:7669–7675. doi: 10.1128/jvi.68.12.7669-7675.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feldmann H, Kretzschmar E, Klingeborn B, Rott R, Klenk H-D, Garten W. The structure of serotype H10 hemagglutinin of influenza A virus: comparison of an apathogenic avian and a mammalian strain pathogenic for mink. Virology. 1988;165:428–437. doi: 10.1016/0042-6822(88)90586-7. [DOI] [PubMed] [Google Scholar]
- 4.Gallaher W R. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell. 1996;85:477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- 5.Garten W, Stieneke A, Shaw E, Wikstrom P, Klenk H-D. Inhibition of proteolytic activation of influenza virus hemagglutinin by specific peptidyl chloroalkyl ketones. Virology. 1989;172:25–31. doi: 10.1016/0042-6822(89)90103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Dunia D, Cubitt B, Grässer F A, de la Torre J C. Characterization of Borna disease virus p56 protein, a surface glycoprotein involved in virus entry. J Virol. 1997;71:3208–3218. doi: 10.1128/jvi.71.4.3208-3218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas B, Becht H, Rott R. Purification and properties of an intranuclear virus-specific antigen from tissue infected with Borna disease virus. J Gen Virol. 1986;67:235–241. doi: 10.1099/0022-1317-67-2-235. [DOI] [PubMed] [Google Scholar]
- 8.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk H-D, Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 9.Hallenberger S, Moulard M, Sordel M, Klenk H-D, Garten W. The role of eukaryotic subtilisin-like endoproteases for the activation of human immunodeficiency virus glycoproteins in natural host cells. J Virol. 1997;71:1036–1045. doi: 10.1128/jvi.71.2.1036-1045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herzog S, Rott R. Replication of Borna disease virus in cell cultures. Med Microbiol Immunol. 1980;168:153–158. doi: 10.1007/BF02122849. [DOI] [PubMed] [Google Scholar]
- 11.Klenk H-D, Garten W. Activation cleavage of viral spike proteins. In: Wimmer E, editor. Cellular receptors for animal viruses, monograph 28. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 241–280. [Google Scholar]
- 12.Klenk H-D, Rott R. The molecular biology of influenza virus pathogenicity. Adv Virus Res. 1988;34:247–281. doi: 10.1016/S0065-3527(08)60520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig, H., K. Furuya, L. Bode, N. Klein, R. Dürrwald, and D. S. Lee. 1993. Biology and neurobiology of Borna disease viruses (BDV), defined by antibodies, neutralizability and their pathogenic potential. Arch. Virol. 7(Suppl.):111–133. [DOI] [PubMed]
- 14.Mackett M, Smith G L, Moss B. General method for production and selection of infectious vaccinia virus recombinants expressing foreign genes. J Virol. 1984;49:857–864. doi: 10.1128/jvi.49.3.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munk K, Pritzer E, Kretzschmar E, Gutte B, Garten W, Klenk H-D. Carbohydrate masking of an antigenic epitope of influenza virus hemagglutinin independent of oligosaccharide size. Glycobiology. 1992;2:233–240. doi: 10.1093/glycob/2.3.233. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama K, Watanabe T, Nakagawa T, Kim W S, Nagahama M, Hosaka M, Hatsuzawa K, Kondoh-Hashiba K, Murakami K. Consensus sequence for precursor processing at mono-arginyl sites. Evidence for the involvement of a Kex2-like endoprotease in precursor cleavages at both dibasic and mono-arginyl sites. J Biol Chem. 1992;267:16335–16340. [PubMed] [Google Scholar]
- 17.Richt J A, VandeWoude S, Zink M C, Clements J E, Herzog S, Stitz L, Rott R, Narayan O. Infection with Borna disease virus: molecular and immunobiological characterization of the agent. Clin Infect Dis. 1992;14:1240–1250. doi: 10.1093/clinids/14.6.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneemann A, Schneider P A, Lamb R A, Lipkin W I. The remarkable coding strategy of Borna disease virus: a new member of the nonsegmented negative strand RNA viruses. Virology. 1995;210:1–8. doi: 10.1006/viro.1995.1311. [DOI] [PubMed] [Google Scholar]
- 20.Schneider P A, Hatalski C G, Lewis A J, Lipkin W I. Biochemical and functional analysis of the Borna disease virus G protein. J Virol. 1997;71:331–336. doi: 10.1128/jvi.71.1.331-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stieneke-Gröber A, Vey M, Angliker H, Shaw E, Thomas G, Roberts C, Klenk H-D, Garten W. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11:2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stitz L, Dietzschold B, Carbone K M. Immunopathogenesis of Borna disease. Curr Top Microbiol Immunol. 1995;190:75–92. doi: 10.1007/978-3-642-78618-1_5. [DOI] [PubMed] [Google Scholar]
- 23.Vey M, Schäfer W, Berghöfer S, Klenk H-D, Garten W. Maturation of the trans-Golgi network protease furin: compartmentalization of zymogen activation, substrate cleavage, and C-terminal truncation. J Cell Biol. 1994;127:1829–1842. doi: 10.1083/jcb.127.6.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wehner T, Ruppert A, Herden C, Frese K, Becht H, Richt J A. Detection of a novel Borna disease virus encoded 10 kilodalton protein in infected cells and tissues. J Gen Virol. 1997;78:2459–2466. doi: 10.1099/0022-1317-78-10-2459. [DOI] [PubMed] [Google Scholar]