Abstract
We have investigated the molecular evidence in favor of the transmission of human immunodeficiency virus (HIV) from an HIV-infected surgeon to one of his patients. After PCR amplification, the env and gag sequences from the viral genome were cloned and sequenced. Phylogenetic analysis revealed that the viral sequences derived from the surgeon and his patient are closely related, which strongly suggests that nosocomial transmission occurred. In addition, these viral sequences belong to group M of HIV type 1 but are divergent from the reference sequences of the known subtypes.
The reports of transmission of human immunodeficiency virus (HIV) from a Florida dentist with AIDS to six of his patients (1, 11) initially raised great concern about the possibility of HIV transmission from health care workers infected with this retrovirus. However, a large retrospective investigation conducted by the Centers for Disease Control and Prevention indicated that this risk of transmission is very small (14). In fact, if we exclude the cases associated with the Florida dental practice, no case of transmission of HIV has been documented among more than 20,000 patients treated by infected surgeons or dentists.
We report here the molecular evidence in favor of the transmission of HIV from an HIV-infected surgeon to one of his patients.
The physician was an orthopedic surgeon working in a public hospital in a western suburb of Paris, France. In 1983, while performing surgery on a multitransfused patient, he reported a percutaneous injury. After the physician experienced several health problems, an HIV serological assay was performed in 1994 and he was found to be HIV seropositive. An epidemiological investigation for which the detailed results will be published elsewhere (8) and the present study were requested by the French Ministry of Health. The epidemiological analysis indicated that the surgeon probably became infected in 1983 and had performed surgical procedures on 3,004 persons since that time. Among these patients, the epidemiological study found only one subject who was HIV seronegative before a prolonged operation performed by the surgeon in 1992 and who is now HIV seropositive. No other risk factors were documented for this patient, a woman born in 1925. Therefore, the nosocomial transmission of HIV from the surgeon to his patient was a possibility worth evaluating by analysis of viral sequences from both persons.
Blood samples were collected at the Hospital Pasteur from both the surgeon and his patient after obtaining their informed consent. The dates of blood collection from these two persons were different to avoid any possibility of mixing up samples. Highly stringent precautions were also in use in our laboratory to prevent the possibility of cross-contamination between the two samples or between them and other viral samples already in our collection. There was strict physical separation between the surgeon’s and his patient’s samples because the experimental procedures performed on the two samples (peripheral blood cell separation, viral isolation, DNA extraction, PCR, and cloning and sequencing of the full-length env genes) were performed more than 3 months apart. In addition, the PCR primers which allowed the amplification of the HIV type 1 (HIV-1) env and gag genes had not previously been used in our laboratory and the use of positive controls in the experiments was excluded to minimize the risk of cross-contamination.
A viral isolate was obtained from the patient and cultivated once with normal donor peripheral blood mononuclear cells (PBMC). A pellet of these cells was kept at −80°C until DNA extraction. The uncultured PBMC from this patient were also kept as a source of DNA for some of the PCR amplifications (Table 1). The surgeon’s blood was collected twice over a 10-month interval, and the DNA was directly purified from a pellet of the isolated, uncultured PBMC. DNA extraction was performed by using a blood DNA extraction kit as recommended by the manufacturer (Qiagen, Chatsworth, Calif.). PCR amplification of the full-length gp160-encoding gene together with part of the nef gene was performed as described by others (4). Part of the gag gene (592 bp) was also amplified by a nested-PCR procedure with outer primers G37 and G40 (16) and inner primers SK39 (12) and SK22 (10). Nested PCR was also used to amplify a 700-bp internal fragment of the HIV-1 env gene (region V3-V5) by using primers, thermal cycling parameters, and limited dilutions of primary PBMC DNA as described by others (2). Following PCR amplification, amplicons were subcloned into pCRII by T/A overhang (Invitrogen, San Diego, Calif.). The nucleotide sequences of the clones were determined with an automated DNA sequencer (model 373A; Applied Biosystems, Inc., Foster City, Calif.), and individual sequence segments were assembled with AssemblyLine software (Kodak Inc., Rochester, N.Y.). The sequences of both strands of DNA were entirely determined at least once. The molecular clones which were obtained from these different samples are described in Table 1.
TABLE 1.
Molecular cloning of HIV-1 DNA sequences
| Subject | Sample
|
No. of molecular clones sequenced (identification codes)b for:
|
|||
|---|---|---|---|---|---|
| Date (mo/day/yr) | Typea | env (full length) | env (partial length) | gag (partial length) | |
| Surgeon | 11/20/1995 | NCP | 3 (FRCNP1–FRCNP3) | 5 (FRCNP7–FRCNP11) | 3 (FRCNPg1 and FRCNPg3) |
| 9/09/1996 | NCP | 3 (FRCNP4–FRCNP6) | 4 (FRCNP12–FRCNP15) | 3 (FRCNPg4–FRCNPg6) | |
| Surgeon’s patient | 2/23/1996 | NCP | 8 (FRMNR4–FRMNR11) | 2 (FRMNRg1–FRMNRg2) | |
| 2/23/1996 | CP | 2 (FRMNR1–FRMNR2) | |||
Source of DNA template for PCR amplification. NCP, uncultured PBMC; CP, cultured PBMC.
Regions of the HIV-1 genome that were sequenced included full-length env followed by the 5′ end of nef (2.8 kbp), partial env (700 bp; V3-V5 region), and partial gag (580 bp; positions 621 to 1205 in HIV-1 strain BRUCG).
Nucleotide sequences were aligned by using CLUSTAL W software (20); some minor manual corrections (gap stripping) were necessary. Phylogenetic analysis was performed with the software package Phylip, edition 3.5 (3). Pairwise evolutionary distances were estimated by using Kimura’s two-parameter method, excluding positions where there was a gap in any sequence. Phylogenetic trees were rooted by using cpzgab as an outgroup and were constructed by the neighbor-joining method. The reliability of the junctions was evaluated by performing 1,000 bootstrap replicates unless otherwise indicated. Other trees were also obtained with the DNAML software (maximum likelihood) from the Phylip package. Trees essentially similar to the ones shown in Fig. 1 were obtained with all the test methods.
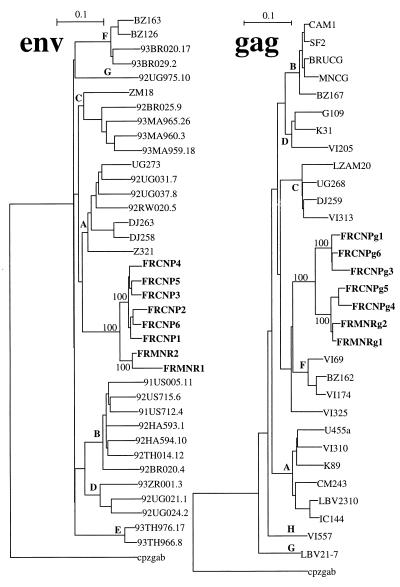
FIG. 1.
Phylogenetic trees for HIV-1 env and gag sequences from the surgeon and his patient and for reference sequences representative of the different HIV-1 subtypes. The trees were obtained by the neighbor-joining method, but other methods produced essentially identical results. The trees are rooted by using cpzgab as the outgroup, and selected bootstrap percentages (from 1,000 replicates) are indicated. The subtypes of HIV-1 sequences are shown at the corresponding node, VI325 being a gag sequence from an unclassified HIV-1 isolate. The scale bars indicate 10% nucleotide distance.
The relationships between the viral sequences from both the surgeon and his patient were examined. The pairwise distances between the sequences within the highly variable V3-V5 region directly amplified from the uncultured PBMC were evaluated (Table 2). There was a significant heterogeneity among the HIV-1 sequences recovered from the surgeon (mean difference: 8.2%), with this heterogeneity increasing for the second time point (mean difference: 9.6%). This high degree of difference was due to the recovery of two sets of sequences from this person which were homologous within each set and heterogeneous when one set was compared to the other one. At least three factors are to be taken into account to explain this high degree of difference, the age (54 years) of the surgeon, the lack of antiretroviral treatment, and the putative duration of infection (12 years). The sequences for the surgeon’s patient were more homogeneous, with a mean of pairwise distances of 5.2%, which is consistent with the hypothesis that this person became infected about 10 years later than her surgeon. Finally, the DNA distances between the HIV-1 sequences from the surgeon and his patient differed by an average of 15.2%.
TABLE 2.
Pairwise distances among proviral HIV-1 env sequencesa
| Subject | Date(s) of samples | Comparison categoryb | No. of comparisons | % Difference
|
|
|---|---|---|---|---|---|
| Mean | Range | ||||
| Surgeon | 1995 | A | 10 | 8.2 | 4.3–11.4 |
| 1996 | A | 6 | 9.6 | 1.2–17.0 | |
| 1995 and 1996 | B | 36 | 10.6 | 1.2–18.9 | |
| Surgeon’s patient | 1996 | A | 28 | 5.2 | 1.2–9.0 |
| Surgeon and surgeon’s patient | 1995 and 1996 | C | 72 | 15.2 | 11.8–18.7 |
Distances between DNA sequences of the env genes from the samples described in Table 1. A matrix of pairwise DNA distances was generated by using CLUSTAL W, excluding positions with gaps and with correction for multiple substitutions. The distances are expressed as percent differences.
A, comparison of sequences from the surgeon or his patient; B, comparison of sequences obtained from the surgeon at different times; C, comparison of sequences from the surgeon with those from his patient.
The phylogenetic relationships among the HIV-1 sequences obtained from these two persons were then determined; representative sequences of different HIV-1 subtypes were used for comparison. The six full-length env sequences from the surgeon and the two from the patient clustered in the same area of the phylogenetic tree and were separate from the reference sequences for the different HIV-1 subtypes (Fig. 1). There was a significant separation of the sequences from these two persons. In contrast, the sequences obtained from the surgeon at an interval of 10 months were mixed and could not be differentiated on the basis of these results. Although both sequences obtained from the surgeon’s patient clustered in the same area, the branch for one of the sequences (FRMNR1) was unusually long. A detailed examination of this sequence revealed that it is characterized by a large number of G→A transitions (13% of the Gs are replaced by As). This hypermutation is particularly pronounced among HIV strains (21), but there is also the possibility that these transitions were generated during the PCR amplification, as reported by others (13). The phylogenetic relationship was also analyzed by a comparison of the gag sequences obtained from both patients (Fig. 1). As with env, the gag sequences clustered and were separate from those of other previously described subtypes. Although the association was not significant, the closest sequences were those belonging to subtype F. However, the high degree of difference between our sequences and those of subtype F (also found in the env tree) do not warrant assigning the former to this subtype.
The analysis of the phylogenetic relationship between the sequences from the surgeon and those from his patient was facilitated because these viral sequences clustered within group M of HIV-1 sequences, but outside of any previously defined subtype. In particular, the sequences do not belong to subtype B, which is the most prevalent in Paris (18). Additional phylogenetic analysis excluded the possibility that these sequences belong to one of the newly reported subtypes (H and I) (7), but the sequences of these subtypes were not included in the analysis shown in Fig. 1 because only partial env sequences are available for them. The possibility that the viral sequences could correspond to recombinant sequences from viruses of different subtypes was evaluated.
Selected full-length env sequences from this study were analyzed by using the Recombinant Identification Program (17) developed at the HIV Sequence Database (Los Alamos, N.Mex.) and available through the Internet (http://hiv-web.lanl.gov). The data obtained with this approach suggested that the HIV sequences under study could be mosaics of sequences from subtypes A and F (data not shown). This possibility was further tested by using the procedure named bootscanning, which allows the resolution of the parentage of HIV genomes (15). After a multiple alignment of test and reference sequences was obtained, the phylogenetic positions of overlapping segments were determined. The significance of the grouping was evaluated by reporting the bootstrap values (Fig. 2). This analysis indicated that only small areas of the sequences significantly clustered with reference sequences A and F. The following is a summary of the results of several bootscanning analyses using different sets of reference sequences. Regions 200 to 400, 675 to 900 and 1000 to 1600 appeared to cluster with subtype A, and regions 1600 to 2000 and 2200 to 2500 appeared to cluster with subtype F. The other regions were found to cluster with various subtypes according to the different sets of reference sequences which were considered (data not shown). The highest bootscan values which were obtained are barely significant, and the trees which were derived from the regions indicated by the bootscanning analysis did not provide consistent results when the method of determining phylogeny (neighbor joining or maximum likelihood) or the set of reference sequences used was varied (data not shown).
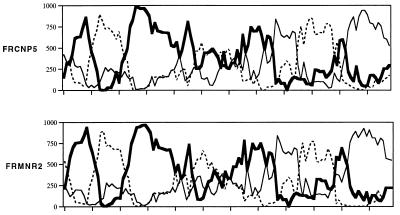
FIG. 2.
Mosaic structure of env genes. The full-length env sequences from the surgeon and his patient were analyzed after alignment with reference subtype HIV-1 sequences. Phylogenetic trees were constructed from sequential 250-bp segments overlapping by 20 bp. The bootstrap values from 1,000 replicates, indicating grouping with subtypes A (thick solid line), F (thin solid line), and C (dashed line), are plotted. The results from two analyses obtained with one sequence from each patient (FRCNP5 and FRMNR2) are shown and are representative of results obtained with the other sequences. Other bootscanning analyses were also performed with other reference and outgroup sequences (data not shown), and the segments of subtype C-related material were not consistently found (data not shown). Recombination breakpoints were identified at the intersection of the lines from the plots.
Although we cannot exclude the possibility that our sequences correspond to recombinants between F and A subtypes, which is a combination that was not previously reported (9), the reference sequences obtained from the database are too distant to allow the precise analysis of their parentage. The other possibility, which is that our sequences belong to an as yet undefined HIV-1 subtype, is more likely. It is also of interest to note that the bootscanning profiles obtained with the sequences from the surgeon and his patient were highly similar (Fig. 2), which strengthened our conclusions concerning their relationship.
Our results indicate that the virus sequences from the surgeon and his patient are highly related. As did those of the epidemiological investigation, our results strongly support the conclusion that there was a transmission of HIV from the surgeon to his patient during a surgical procedure. At the time of the surgical procedures, the plasmatic viral load of the surgeon was probably high and his blood was therefore highly infectious. Indeed, he was having opportunistic infections suggestive of AIDS and, not knowing his serological status, was not under any antiretroviral treatment. It is not possible for us to discuss the geographical origin of these viruses because the source of the surgeon’s infection is unknown. Indeed, it is thought that the surgeon got infected in 1983 as a consequence of a needle stick injury while operating on a multitransfused patient who later died and whose HIV serological status was unknown.
In this study, evidence of nosocomial HIV transmission was found in only one case among several thousands of patients operated on by the infected surgeon (8). This result confirms the extensive reports in the literature (5, 6, 14, 19) indicating that there is a very low risk of HIV transmission from HIV-infected surgeons to their patients.
Nucleotide sequence accession numbers.
GenBank accession numbers for the full-length env sequences (six from the surgeon and two from his patient) obtained in this study are as follows: FRCNP1, U85912; FRCNP2, U85913; FRCNP3, U85914; FRCNP4, U85915; FRCNP5, U85916; FRCNP6, U85917; FRMNR1, U85918; FRMNR2, U85919. GenBank accession numbers for the partial gag sequences are as follows: FRCNPg1, AF037316; FRCNPg3, AF037317; FRCNPg4, AF037318; FRCNPg5, AF037319; FRCNPg6, AF037320; FRMNRg1, AF037321; FRMNRg2, AF037322.
Acknowledgments
We thank Simon Wain-Hobson and Lisa Chakrabarti for helpful discussions concerning phylogenetic analysis and Mika Salminen, Thomas Leitner, and Brian T. Foley for discussion about the recombinant nature of the analyzed sequences. We are also thankful to our colleagues who conducted the epidemiological study and who provided their results: F. Lot, J. C. Desenclos, and J. Drucker from the Réseau National de Santé Publique; J. C. Séguier, S. Fégueux, P. Simon and P. Van Amerogen from the Hospital in Saint Germain en Laye; P. Astagneau, G. Brücker, and M. Aggoune from the Centre de Coordination de Lutte contre les Infections Nosocomiales de Paris-Nord; and M. Ruch and A. Bernoux from the Direction Départementale des Affaires Sanitaires et Sociales des Yvelines.
REFERENCES
- 1.Ciesielski C A, Marianos D W, Schochetman G, Witte J J, Jaffe H W. The 1990 Florida dental investigation. Ann Intern Med. 1994;121:886–890. doi: 10.7326/0003-4819-121-11-199412010-00011. [DOI] [PubMed] [Google Scholar]
- 2.Delwart E L, Shpaer E G, Louwagie J, McCutcham F E, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationship determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 3.Fesenstein J. PHYLIP (phylogenic inference package), 3.5c ed. Seattle, Wash: Department of Genetics, University of Washington; 1992. [Google Scholar]
- 4.Gao F, Morrison S G, Robertson D L, Thornton C L, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp P M, Shaw G M, Hahn B H the WHO and NIAID Networks for HIV Isolation and Characterization. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes E C, Zhang L Q, Simmonds P, Smith Rogers A, Leigh Brown A J. Molecular investigation of human immunodeficiency virus (HIV) infection in a patient of an HIV-infected surgeon. J Infect Dis. 1993;167:1411–1414. doi: 10.1093/infdis/167.6.1411. [DOI] [PubMed] [Google Scholar]
- 6.Jaffe H W, McCurdy J M, Kalish M L, Liberti T, Metellus G, Bowman B H, Richards S B, Neasman A R, Witte J J. Lack of HIV transmission in the practice of a dentist with AIDS. Ann Intern Med. 1994;121:855–859. doi: 10.7326/0003-4819-121-11-199412010-00005. [DOI] [PubMed] [Google Scholar]
- 7.Kostrikis L G, Bagdades E, Cao Y, Zhang L, Dimitriou D, Ho D D. Genetic analysis of human immunodeficiency virus type 1 strains from patients in Cyprus: identification of a new subtype designated subtype 1. J Virol. 1995;69:6122–6130. doi: 10.1128/jvi.69.10.6122-6130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lot, F., J.-C. Seguier, S. Fegueux, P. Astagneau, P. Simon, M. Aggoune, P. Van Amerogen, M. Ruch, G. Brucker, and J.-C. Desenclos. HIV transmission from an orthopedic surgeon to a patient in France. Submitted for publication. [DOI] [PubMed]
- 9.McCutchan F E, Salminen M O, Carr J K, Burke D S. HIV-1 genetic diversity. AIDS. 1996;10:S13–S20. [PubMed] [Google Scholar]
- 10.McCutchan F E, Ungar B L P, Hegerich P, Roberts C R, Fowler A K, Hira S K, Perine P L, Burke D S. Genetic analysis of HIV-1 isolates from Zambia and an expanded phylogenetic tree for HIV-1. J Acquired Immune Defic Syndr. 1992;5:441–449. [PubMed] [Google Scholar]
- 11.Myers G. Molecular investigation of HIV transmission. Ann Intern Med. 1991;121:889–890. doi: 10.7326/0003-4819-121-11-199412010-00012. [DOI] [PubMed] [Google Scholar]
- 12.Ou C Y, Kwok S, Mitchell S W, Mack D H, Sninsky J J, Krebs J W, Feorino P, Warfield D, Schochetman G. DNA amplification for direct detection of HIV-1 in DNA of peripheral blood mononuclear cells. Science. 1988;239:295–297. doi: 10.1126/science.3336784. [DOI] [PubMed] [Google Scholar]
- 13.Overbaugh J, Jackson S M, Papenhausen M D, Rudensey L M. Lentiviral genomes with G-to-A hypermutation may result from Taq polymerase errors during polymerase chain reaction. AIDS Res Hum Retroviruses. 1996;12:1605–1613. doi: 10.1089/aid.1996.12.1605. [DOI] [PubMed] [Google Scholar]
- 14.Robert L M, Chamberland M E, Cleveland J L, Marcus R, Gooch B F, Srivastava P U, Culver D H, Jaffe H W, Marianos D W, Panlilio A L, Bell D M. Investigations of patients of health care workers infected with HIV. Ann Intern Med. 1995;122:653–657. doi: 10.7326/0003-4819-122-9-199505010-00002. [DOI] [PubMed] [Google Scholar]
- 15.Salminen M O, Carr J K, Burke D S, McCutchan F E. Identification of breakpoints in intergenotypic recombinants of HIV-1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 16.Salminen M O, Carr J K, Robertson D L, Hegerich P, Gotte D, Koch C, Sanders-Buell E, Gao F, Sharp P M, Hahn B H, Burke D S, McCutchan F E. Evolution and probable transmission of intersubtype recombinant human immunodeficiency virus type 1 in a Zambian couple. J Virol. 1997;71:2647–2655. doi: 10.1128/jvi.71.4.2647-2655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siepel A C, Halpern A L, Macken C, Korber B T M. A computer program designed to rapidly screen for HIV-1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses. 1995;11:1413–1425. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 18.Simon F, Loussert-Ajaka I, Damond F, Saragosti S, Barin F, Brun-Vézinet F. HIV type 1 diversity in northern Paris, France. AIDS Res Hum Retroviruses. 1996;12:1427–1433. doi: 10.1089/aid.1996.12.1427. [DOI] [PubMed] [Google Scholar]
- 19.Smith-Rogers A, Froggatt III J W, Townsend T, Gordon T, Leigh-Brown A J, Holmes E C, Zhang L Q, Moses H., III Investigation of potential HIV transmission to the patients of an HIV-infected surgeon. JAMA. 1993;269:1795–1801. [PubMed] [Google Scholar]
- 20.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vartanian J P, Meyerhans A, Sala M, Wain-Hobson S. G→A hypermutation of the HIV-1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc Natl Acad Sci USA. 1994;91:3092–3096. doi: 10.1073/pnas.91.8.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]