Abstract
Several studies indicate that paramyxoviruses require a specific cellular factor(s) for transcription of their genomic RNAs. We previously reported that the cellular cytoskeletal protein actin, in its polymeric form, participates in the transcription of human parainfluenza virus type 3 (HPIV3) in vitro. In the present study, we investigated the role of the polymeric form of actin, i.e., the actin microfilaments of the cytoskeletal framework, in the reproduction of HPIV3 in vivo. Pulse-chase labeling analyses indicate that the viral nucleocapsid-associated proteins, NP and P, are present predominantly in the cytoskeletal framework during infection. By in situ hybridization, we found that viral mRNAs and genomic RNA were synthesized from the nucleocapsids that were bound to the cytoskeletal framework. Double immunofluorescent labeling and confocal microscopy of the cytoarchitecture revealed that the viral nucleocapsids are specifically localized on the actin microfilaments. Treatment of cells with the actin-depolymerizing agent, cytochalasin D, resulted in the inhibition of viral RNA synthesis and ribonucleoprotein accumulation. These results strongly suggest that actin microfilaments play an important role in the replication of HPIV3.
Human parainfluenza virus type 3 (HPIV3), a paramyxovirus, is an important pathogen that causes severe respiratory tract illness in children (11). The single-stranded RNA genome of HPIV3, 15,461 nucleotides long, is contained within a helical nucleocapsid (20). Three virus-encoded proteins, the nucleocapsid protein, NP (68 kDa), the phosphoprotein, P (90 kDa), and the RNA polymerase, L (257 kDa), are associated with the nucleocapsid to form a transcribing ribonucleoprotein (RNP) complex (1, 2, 20). NP enwraps the genome RNA, while L and P together constitute the RNA-dependent RNA polymerase complex, similar to that characterized for vesicular stomatitis virus (14), that transcribes the NP-bound genome RNA both in vitro and in vivo. Previous studies indicate that in addition to the RNP-associated viral proteins, cellular actin is necessary for the activation of HPIV3 transcription in vitro (15, 16). Further analyses of this RNP-actin interaction demonstrated that the binding of the polymeric form of actin to the RNP results in an alteration of structure of the RNP from a loosely coiled to a moderately condensed form which appeared to be favorable for transcription (17). Similar involvement of cytoskeletal proteins in transcription is also observed in several other paramyxoviruses, namely, Sendai virus, measles virus, and respiratory syncytial virus, where actin and tubulin have been shown to be involved in the activation of transcription (26, 30, 31). In the case of HPIV3, the specific requirement of the polymeric form of actin in transcription in vitro suggests an interaction between the viral RNP and actin microfilaments in the infected cells. Thus, it appears that paramyxoviruses perhaps use a common strategy for their gene expression by exploiting cellular cytoskeletal components.
Actin is present in nonmuscle cells in two forms, a globular monomeric form that represents the soluble pool of actin and a filamentous form that constitutes the actin microfilaments of the cytoskeletal framework (28). The actin microfilaments representing the polymeric form of actin is present in a dynamic state which is constantly forming and breaking within the cell in response to various external or internal stimuli (28, 35, 37). Many enveloped viruses utilize actin microfilaments during the process of budding and maturation of virus particles from the infected cells (5, 13, 23, 36, 39, 45, 47). Furthermore, in Newcastle disease virus, the RNP is attached to the cytoskeletal framework during RNA synthesis, suggesting involvement of the cytoskeletal component(s) in viral RNA synthesis (24). Thus, it appears that, consistent with the in vitro requirement for cytoskeletal proteins in transcription, paramyxoviruses in general may use the same proteins for their reproduction in vivo.
In this study, we have made an effort to understand the role of the actin microfilaments of the cytoskeletal framework in HPIV3 gene expression in vivo. By biochemical and immunological analyses, we demonstrate that the viral RNPs, following entry, are rapidly associated with the cytoskeletal framework in the infected cells. These RNPs are actively involved in RNA synthesis, as revealed by in situ hybridization. By double immunofluorescent labeling and confocal microscopy, we have demonstrated that the actin microfilaments but not the microtubules are the site of interaction of RNP.
(This work forms part of the dissertation to be submitted by S.G. to the Molecular Virology Program, Case Western Reserve University, Cleveland, Ohio, as partial fulfillment of the requirements for the degree of Doctor of Philosophy.)
MATERIALS AND METHODS
Cells and viruses.
CV-1 (ATCC CCL 185) cells were propagated in monolayers as described previously (15). HPIV3 (HA-1; NIH 47885) was grown in CV-1 cells and purified as described previously (15).
Cell fractionation.
CV-1 cells (4 × 107) were harvested, and the cell pellet was washed with phosphate-buffered saline (PBS). The cell pellet was then gently resuspended in extraction buffer (24) containing 10 mM piperazine-N,N1-bis(2-ethenesulfonic acid) (PIPES; pH 6.8), 100 mM KCl, 2.5 mM MgCl2, 1 mM CaCl2, 0.3 M sucrose, and 1 mM phenylmethylsulfonyl fluoride. Triton X-100 was added to a final concentration of 1%. After gentle mixing, the slurry was left on ice for 3 min and centrifuged at 800 rpm for 3 min. The supernatant was referred to as the soluble (SOL) fraction, and the pellet was referred to as the cytoskeletal (CSK) fraction.
For cells that were grown on coverslips, the extraction buffer (24) was directly added and the coverslips were kept on ice for 3 min. The coverslips were washed with extraction buffer and then with PBS. The material retained on the coverslips were fixed with 1% formaldehyde and used as the CSK fraction.
Protein analysis.
Infected cells were pulse-labeled with 50 μCi of [35S]methionine at 10 μCi/ml for 10 min and chased for 60 min. The labeled proteins were resolved in a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel (29) and detected by fluorography followed by exposure to X-ray film.
For Western blot analysis, the proteins were resolved in an SDS–10% polyacrylamide gel and transferred to a nylon (GeneScreen) membrane (44). The blot was incubated with the primary antibody, and the complex was detected with horseradish peroxidase-conjugated second antibody and diaminobenzidine as the substrate.
Viral RNA detection in cells.
CV-1 cells were grown on coverslips and were infected with HPIV3 at 1 PFU/cell. The CSK structures were fixed on the coverslips and were hybridized (Boehringer Mannheim) to digoxigenin-labeled sense and antisense NP-mRNA, synthesized in vitro from pGEM-NP plasmid DNA as specified by the manufacturer (Boehringer Mannheim) with T7 and SP6 RNA polymerase, respectively. The hybridization signal was detected with anti-digoxigenin antibody coupled to alkaline phosphatase followed by a color reaction with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate as substrates (Boehringer Mannheim). The slides were visualized under a phase-contrast microscope.
For Northern hybridization, the cells were infected with HPIV3 at 1 PFU/cell and were harvested at 2, 4, 8, 12, 24, and 36 h postinfection. The cells were pelleted by centrifugation at 800 × g and fractionated in extraction buffer containing ribonucleoside vanadyl complex (Sigma Chemical Co.). The RNAs from the CSK and SOL fractions were purified by digestion with proteinase K and then subjected to phenol extraction and ethanol precipitation. The RNA was analyzed in a formaldehyde-agarose gel and transferred to a nylon membrane (GeneScreen). The membrane was hybridized with radiolabeled NP cDNA to detect the virus-specific RNAs. The gel was analyzed with a phosphorimager, and the distribution of RNAs between the SOL and CSK fractions of cells was determined.
The NP cDNA insert was obtained by restriction digestion of an NP cDNA clone in pGEM4Z and eluted from a 1% agarose gel with GeneClean (Bio-Rad). The cDNA was labeled by the random primed labeling reaction (Boehringer Mannheim) with [32P]dCTP and used as a probe in the hybridization reaction.
Plaque assay.
The virus titer was determined by the standard plaque assay technique with CV-1 cell monolayers as previously described (18).
CD treatment.
Cytochalasin D (CD) (Sigma Chemical Co.) was initially resuspended in dimethyl sulfoxide at 5 mg/ml. Subsequent dilutions were made in the culture media.
Isolation of viral RNP complex.
The HPIV3 RNP complex was isolated from infected CV-1 cells by a method described previously (43). CV-1 cells were infected at 5 PFU/cell and harvested 24 h postinfection. The cells were washed with PBS and then with 10 mM phosphate buffer (pH 7.2) containing 0.15 M NaCl and finally disrupted in 10 mM Tris-HCl by sonication. The cell debris were removed by centrifugation at 10,000 × g for 10 min, and the supernatant was further centrifuged at 100,000 × g for 1 h to pellet the RNP. The RNP was suspended in 0.1 ml of buffer containing 10 mM Tris, 1 mM EDTA, and 1 mM dithiothreitol and further purified by centrifugation through a 50% glycerol cushion (0.5 ml) at 150,000 × g in an SW 50.1 centrifuge.
Isolation of total RNA from infected cells.
Total RNA was isolated from HPIV3-infected CV-1 cells by a method described previously (9). The cells were washed with PBS and lysed in solution D (4 M guanidinium thiocyanate, 25 mM sodium citrate [pH 7.0], 0.5% Sarkosyl, 0.1 M 2-mercaptoethanol). RNA was recovered from the lysed cells by phenol-chloroform extraction and ethanol precipitation.
Immunofluorescent staining.
Cells were grown on coverslips and were infected with HPIV3 at 1 PFU/cell. At 12 to 14 h postinfection, the cells were fractionated and stained with rhodamine-conjugated phalloidin in extraction buffer (1:100, vol/vol) in the dark followed by fixation with 3.7% formaldehyde in PBS at room temperature. Microtubules were labeled either directly or after extraction with CSK buffer on coverslips and were treated with monoclonal anti-tubulin antibody (Boehringer Mannheim) followed by fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin G (IgG). The coverslips were mounted on slides with Vectashield (Vector) and visualized in a Leica CISM confocal laser-scanning microscope. For double immunofluorescent staining, the infected cells were fractionated and stained with rhodamine-conjugated phalloidin in extraction buffer (1:100, vol/vol) in the dark followed by fixation with 3.7% formaldehyde in PBS at room temperature. The fixed cells were stained with goat anti-HPIV3 antibody (Biodesign) and anti-goat IgG conjugated with biotin followed by FITC-conjugated avidin. The coverslips were mounted on slides with Vectashield and visualized in a Leica CISM confocal laser-scanning microscope. For simultaneous staining of microtubules and viral antigens, the cells were fractionated with extraction buffer without phalloidin and fixed with 3.7% formaldehyde. The fixed cells were then treated with both anti-tubulin antibody and anti-virus antibody and subsequently with anti-mouse IgG conjugated to FITC for tubulin and anti-goat IgG conjugated to biotin followed by Texas red-conjugated avidin for viral antigens.
RESULTS
Association of HPIV3 RNP with the cytoskeletal framework.
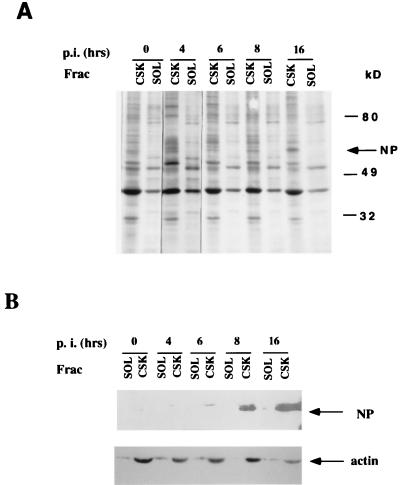
In a previous study, we demonstrated that the cytoskeletal protein actin specifically binds to HPIV3 RNP to activate viral transcription in vitro (17). Ultrastructural analysis of the RNP revealed that the binding of the polymeric form of actin mediates a conformational change of the RNP from a loosely coiled to a moderately condensed form (17) that appeared to be involved in the transcription activation process. This led us to postulate that the CSK framework containing the polymeric form of actin as a major component might play a direct role in HPIV3 gene expression. We therefore investigated whether viral RNP associates with the CSK framework and, more specifically, with actin microfilaments during infection. CV-1 cells, which are highly permissive for HPIV3 growth in culture medium, were selected for this study. The cells were infected with HPIV3 and were pulse-labeled at various times postinfection. The cells were lysed in extraction buffer containing 1% Triton X-100 and separated into CSK and SOL fractions. Polypeptides from each fraction were analyzed in SDS-polyacrylamide gels and visualized by autoradiography. As shown in Fig. 1A, the major RNP-associated protein, NP, was clearly associated with the CSK fraction as early as 6 h postinfection and remained bound to the CSK at least up to 16 h postinfection. Western blot analysis with anti-RNP and anti-actin antibodies confirmed the presence of NP and actin in the CSK fraction (Fig. 1B). Similar analysis with anti-HPIV3 antibody demonstrated the presence of P in the same CSK fraction (data not shown). These results indicate that the viral RNP associates with the CSK framework during the early period, i.e., the RNA-synthetic phase of the virus life cycle.
FIG. 1.
Protein distribution between the SOL fraction and CSK framework. (A) CV-1 cells were infected with HPIV3 and, at different times postinfection (p.i.) (as indicated), pulse-labeled with [35S]methionine for 10 min and chased for 60 min. The labeled cells were fractionated into CSK and SOL fractions with an extraction buffer as described by Hamaguchi et al. (24). The fractions were analyzed by SDS-polyacrylamide gel electrophoresis followed by fluorography and autoradiography. The migration positions of molecular size markers in kilodaltons are shown on the right. (B) Western blot analysis of the proteins in soluble and cytoskeletal fractions. The distribution of HPIV3 NP and the cytoskeletal component actin between the SOL and CSK fractions was determined by the same procedure as described above, except the cells were not labeled and the NP and actin were analyzed by Western blotting with anti-RNP that detects primarily NP and anti-actin antibodies, respectively.
Localization of viral RNAs on the CSK framework.
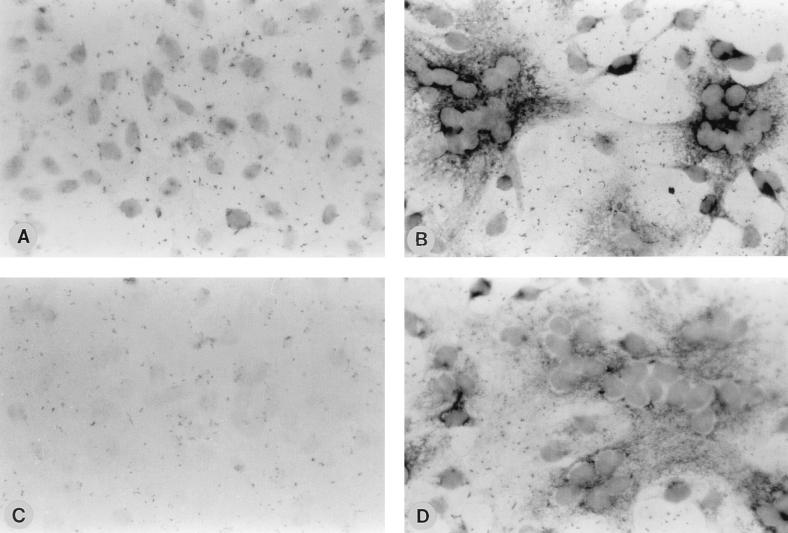
Association of viral RNP with the CSK framework during the early period of infection raised the question whether viral RNA synthesis also occurs on the CSK structure. To investigate this possibility, we performed in situ hybridization of CV-1 cells that were grown on coverslips and were infected with HPIV3. At 12 h postinfection, the cells were extracted with CSK buffer, with which the CSK structure was isolated on the coverslip and was then fixed. The CSK structure on the coverslip was hybridized with digoxigenin-labeled NP-mRNA sense and antisense riboprobes, and the signal was detected by the color reaction as described in Materials and Methods. As shown in Fig. 2, the viral mRNAs (Fig. 2B) and genomic RNA (Fig. 2D) were clearly detected on the CSK framework of the infected cells whereas no such signals were seen in mock-infected cells (Fig. 2A and C). The presence of viral RNAs on the CSK structure suggests that the RNA synthesis occurs on the CSK framework and that actin microfilaments may be directly involved in the RNA synthetic process. To confirm this observation biochemically, we examined the distribution of viral RNA between the CSK and SOL fractions in cells harvested at different times postinfection. The total RNA was isolated from each fraction and analyzed by Northern blotting with 32P-labeled NP cDNA as the probe. As shown in Table 1, the viral NP mRNA was present on the CSK framework during the early period of infection and after 16 h postinfection the RNAs were released into the cytosol. These results strongly suggest that initial synthesis of viral RNA occurs in the CSK framework by CSK-associated RNP and that subsequently the RNP is released into the cytosolic fraction.
FIG. 2.
Detection of HPIV3 RNAs in the CSK framework. CV-1 cells, grown on coverslips, were infected with HPIV3 at 1 PFU/cell. The cells were treated with CSK buffer and hybridized with genome sense or antisense NP RNA labeled with digoxigenin. The coverslips were treated with anti-digoxigenin antibody coupled to alkaline phosphatase (Boehringer Mannheim), and the signal was detected with nitroblue tetrazolium and X-phosphate as the substrate. The coverslips were then mounted on slides and visualized under a phase-contrast microscope. Similarly treated mock-infected CV-1 cells served as the control. Mock-infected CV-1 cells (A) and HPIV3-infected cells (B) hybridized with antisense NP RNA (T7 transcript of NP cDNA clone in pGEM4Z) and mock infected CV-1 cells (C) and HPIV3-infected cells (D) hybridized with NP mRNA (SP6 transcript of NP clone in pGEM4Z) are shown.
TABLE 1.
Distribution of HPIV3-specific RNAs between the CSK and SOL fractions from infected CV-1 cells
p.i., postinfection.
The amounts of HPIV3-specific RNAs in the CSK and SOL fractions were determined by Northern hybridization with 32P-labeled NP cDNA as the probe and quantitated by phosphorimager analysis as described in Materials and Methods. The data represent the mean of three sets of experiment with a variability of <10%.
Effect of CD on the production of HPIV3 virions.
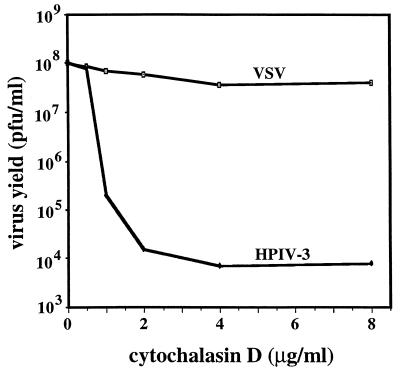
To determine the role of the CSK framework in HPIV3 reproduction, we disrupted the CSK framework with CD, a potent actin-depolymerizing agent which has been shown to disrupt the microfilaments in a dose-dependent manner (7, 33, 38, 40, 42). We preferred CD over CB in these studies because the latter is also known to inhibit hexose transport, causing disturbances in sugar metabolism (27). Confluent monolayers of CV-1 cells were infected with HPIV3 in the presence or absence of different concentrations of CD. At 24 h postinfection, the progeny virions were collected from the medium and quantitated by a standard plaque assay. As shown in Fig. 3, the virus yield was reduced in the presence of CD in a dose-dependent manner with increasing concentrations up to 8 μg/ml. At 4 μg/ml, a reduction by about 4 log units was observed as compared to the control HPIV3 production in the absence of CD. These data indicate that CD has a drastic effect on HPIV3 production and thus confirms the direct involvement of actin microfilaments in this process. When vesicular stomatitis virus was grown under similar conditions, the virus titer was not affected to any significant extent even at a CD concentration of 8 μg/ml (Fig. 3). These results are not unexpected because neither the actin microfilaments nor the CSK has been implicated in VSV reproduction (22).
FIG. 3.
Inhibition of HPIV3 replication by CD. CV-1 cells were infected with HPIV3 at 5 PFU/cell in the presence of different concentrations of CD, as indicated. At 24 h post infection, the progeny virus released into the medium was collected and measured by plaque assay. CV-1 cells were infected with vesicular stomatitis virus (VSV) at 5 PFU/cell in the presence of CD. At 12 h postinfection, the progeny virus released into the medium was measured.
Effect of CD on the intracellular accumulation of viral RNP and mRNAs.
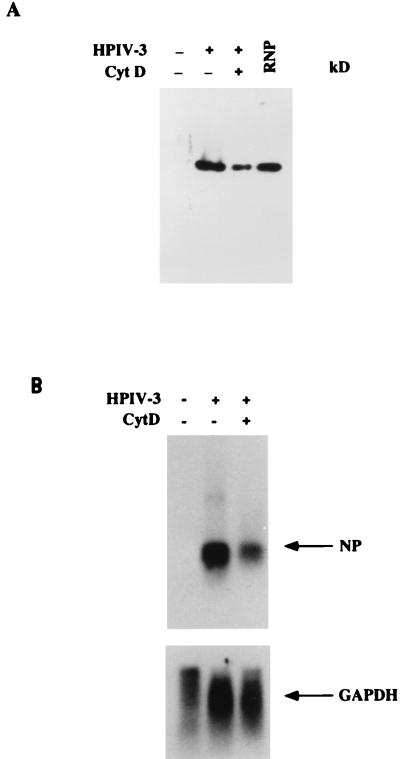
We next examined whether the observed inhibition of progeny virion release in the presence of CD was due to the inhibition of intracellular RNP and mRNA syntheses. Viral RNPs were isolated from cells grown in the absence or presence of CD (8 μg/ml) and were resolved in SDS-polyacrylamide gels. The NP protein in the RNP was detected by Western blotting with anti-RNP antibody. As shown in Fig. 4A), upon treatment with CD, the intracellular RNP production decreased by 60% compared to the control. Similarly, when RNP-associated proteins were analyzed in the total-cell extract, comparable inhibition was observed, indicating that the inhibition of RNP production was at the level of protein synthesis rather than at the level of assembly (data not shown). To examine whether the level of viral mRNAs in CD-treated cells also decreased, total RNA was isolated from HPIV3-infected cells, grown in the presence or absence of CD, and analyzed by Northern blotting with a 32P-labeled NP cDNA probe. As shown in Fig. 4B, the presence of CD in the culture medium resulted in an inhibition of viral mRNA synthesis by about 70% compared to the control. These results suggest that HPIV3 RNA synthesis is significantly affected by CD and, consistent with previous findings (17), that the polymeric form of actin plays a critical role in the RNA synthesis. However, it remains unknown whether the CSK framework also participates in viral mRNA translation, although such involvement has not been reported previously.
FIG. 4.
Inhibition of intracellular RNP synthesis by CD. (A) CV-1 cells were infected with HPIV3 in the presence or absence of 8 mg of CD per ml. Intracellular RNP was isolated by the method described by Toneguzzo and Ghosh (43). The level of NP was determined by Western blot analysis with anti-RNP antibody. RNP; purified viral RNP was used as control. (B) Inhibition of mRNA synthesis by CD. CV-1 cells were infected with HPIV3 in the presence or absence of 8 mg of CD per ml. Total RNA was isolated from the cells, and the levels of NP mRNA were determined by Northern blot analysis with 32P-labeled NP cDNA. The same blot was hybridized with 32P-labeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA and used as an internal control.
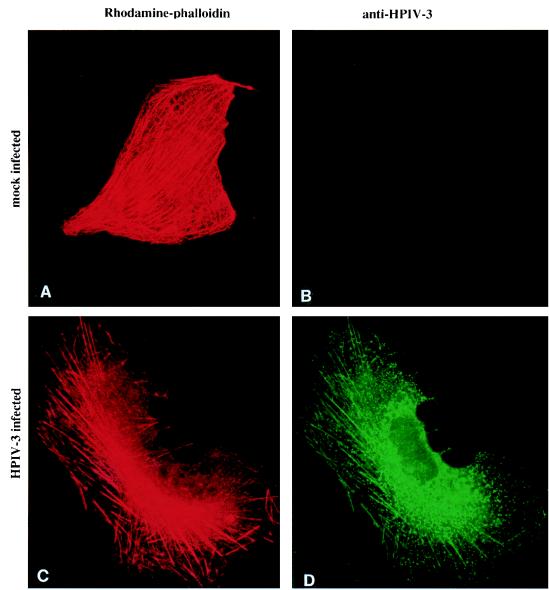
Colocalization of viral RNP with actin microfilaments.
To directly investigate the involvement of actin microfilaments in the interaction with viral RNP, we used double immunofluorescent labeling and confocal microscopy. Cells were treated with CSK buffer to isolate the CSK framework. In the isolated CSK framework, the actin microfilaments were labeled with rhodamine-conjugated phalloidin (Fig. 5A and C) and viral antigens were similarly labeled with anti-HPIV3 antibody followed by anti-goat IgG conjugated to FITC (Fig. 5B and D). As is evident from Fig. 5, the distribution of viral RNP closely resembled that of the actin microfilaments (compare Fig. 5C and D), suggesting an association between actin microfilaments and the viral RNP. When we performed the immunostaining with anti-RNP, a similar distribution of viral antigens was observed, which confirmed the presence of RNP (data not shown).
FIG. 5.
Association of HPIV3 RNP with the CSK framework in vivo. CV-1 cells grown on coverslips were infected with HPIV3 at 1 PFU/cell, and at 12 h postinfection the cells were treated with CSK buffer containing rhodamine-conjugated phalloidin. The cells were washed with the same buffer followed by PBS and fixed. The fixed CSK structures were treated with anti-HPIV3 antibody that detects the RNP-associated proteins NP and P. The coverslips were then treated with biotin-conjugated anti-goat antibody followed by fluorescein-conjugated avidin. Similar staining of mock-infected cells served as control. Mock-infected (A) or HPIV3-infected (C) CV-1 cells treated with CSK buffer containing rhodamine-phalloidin and mock-infected (B) or HPIV3-infected (D) CV-1 cells treated with anti-HPIV3 antibody followed by biotin-conjugated anti-goat immunoglobulin plus fluorescein-conjugated avidin are shown.
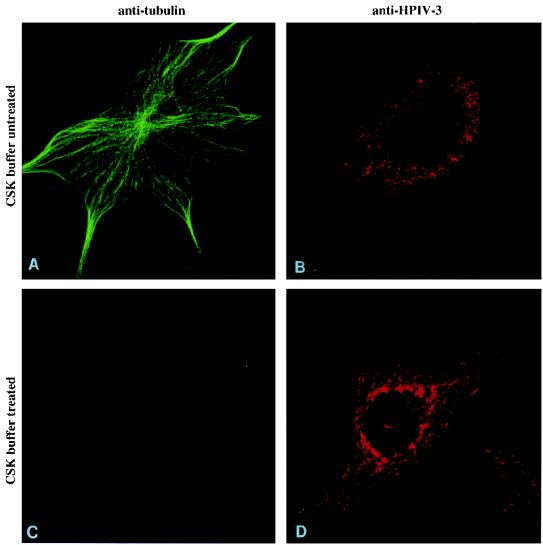
To determine the specificity of this interaction, we examined the association of viral RNP with microtubules containing tubulin, which are also a part of the cytoskeletal framework. When similar double immunofluorescent labeling and confocal microscopy with anti-tubulin antibody were used, no similarity in the distribution of microtubules (Fig. 6A) and viral RNP (Fig. 6B) was observed. Moreover, when the cells were extracted with CSK buffer, the microtubules disintegrated and were removed (Fig. 6C), as described in earlier studies (4), whereas the viral antigens remained on the cytoskeletal framework (Fig. 6D). These findings clearly indicate that the viral proteins are not associated with tubulin-containing microtubules in the infected cells but, rather, associate with the actin microfilaments. The slightly diffused staining of viral proteins (Fig. 6) was possibly due to their colocalization with actin microfilaments that are partially depolymerized in the absence of phalloidin in the staining buffer.
FIG. 6.
Specificity of the interaction between the RNP and actin microfilaments. To determine the specificity of the RNP-actin interaction, the association of RNP with another cytoskeletal protein, tubulin, was examined. CV-1 cells, grown on coverslips, were infected with HPIV3 at 1 PFU/cell and at 12 h postinfection either incubated in PBS (A and B) or treated with CSK buffer (C and D) for 3 min on ice. The cells were washed with PBS and fixed. The coverslips were treated with anti-tubulin or anti-HPIV3 antibody. They were then treated with fluorescein-conjugated anti-mouse IgG for anti-tubulin (A and C) or biotin-conjugated anti-goat antibody followed by Texas red-conjugated avidin for HPIV3 antigens (B and D).
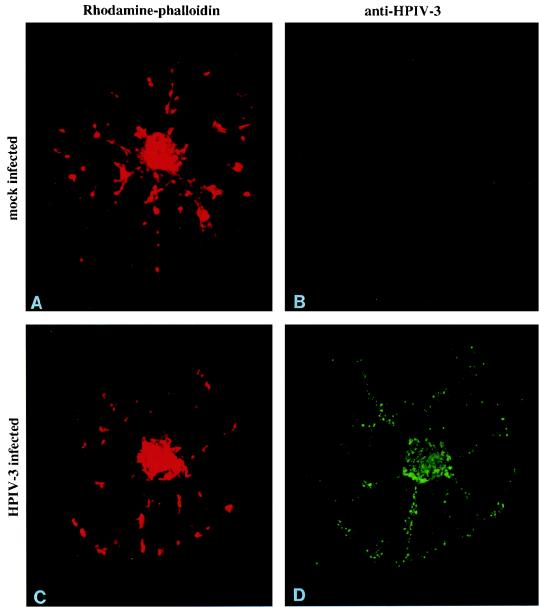
We further examined the role of actin microfilaments by using CD. As shown in Fig. 7, treatment of cells with CD severely disrupted the distribution pattern of actin microfilaments (compare Fig. 5A and 7A), forming distinct patches. Interestingly, the viral antigens also colocalized with the remnants of actin filaments, indicating that the viral RNP remains associated with actin in the disrupted microfilaments (Fig. 7C and D). Taken together, the above findings indicate that the association of viral RNP with the actin microfilaments is strong and specific.
FIG. 7.
Effect of CD on the distribution of RNP in vivo. CV-1 cells, grown on coverslips, were infected with HPIV3 at 1 PFU/cell in the presence of 8 μg of CD per ml. At 12 h postinfection, the cells were extracted with CSK buffer containing rhodamine-conjugated phalloidin. The CSK structures remaining on the coverslips were washed with the CSK buffer followed by PBS. The CSK structures were fixed and treated with anti-HPIV3 antibody followed by biotin-conjugated anti-goat Ig and fluorescein-conjugated avidin. Similarly treated HPIV3-infected CV-1 cells served as controls. Mock-infected (A) or HPIV3-infected (C) CV-1 cells treated with 8 μg of CD per ml were treated with CSK buffer containing rhodamine-conjugated phalloidin. Mock-infected (B) or HPIV3-infected (D) CV-1 cells were treated with anti-HPIV3 antibody followed by biotin-conjugated anti-goat Ig plus fluorescein conjugated avidin.
DISCUSSION
In the present study, we have demonstrated that the cytoskeletal framework plays an important role in the replication of HPIV3 in vivo. The major finding of our study is that the viral RNP associates with the cytoskeletal framework at an early step during infection, i.e., the primary transcription phase (Fig. 1) and that actin microfilaments but not microtubules of the cytoskeletal framework are the sites of interaction (Fig. 5 through 7). The actin microfilaments were found to be the site of viral RNA synthesis, as revealed by in situ hybridization (Fig. 2) and further confirmed by the findings that CD, which specifically disrupts actin microfilaments (40), inhibited viral RNA synthesis and RNP accumulation in cells (Fig. 4). Furthermore, double immunofluorescent labeling and confocal microscopy revealed colocalization of RNP with the actin microfilaments. Disruption of microfilaments with CD resulted in the distribution of viral antigens together with the remnants of microfilaments (Fig. 7). These results clearly indicate that the polymeric form of actin is required for HPIV3 multiplication in cells, which concurs with our previous observation that actin is necessary for in vitro transcription by purified viral RNP (16).
The requirement for specific proteins of the CSK framework in the transcription of paramyxoviruses in vitro have been well documented (16, 21, 25, 26, 30–32). For example, HPIV3 and respiratory syncytial virus both require cellular actin for transcription of genomic RNA in vitro (16, 26). In contrast, Sendai virus and measles virus transcription was activated by tubulin (30, 31), while canine distemper virus required hsp70, which also associates with the CSK framework (32). These findings strongly suggest that the CSK framework plays a role in viral gene expression in vivo. The involvement of the CSK framework in the virus life cycle has also been suggested, although indirectly, by several other studies (10). For example, RNA and DNA viruses including Newcastle disease virus (24), measles virus (5, 39), vesicular stomatitis virus (8), rubella virus (6), vaccinia virus (12), and Autographa californica nuclear polyhedrosis virus (46) have been shown to interact with the CSK during their life cycle (46). Although this interaction is believed to be involved in various activities such as viral genome replication, assembly and release of progeny virions, and protein synthesis, its critical role in the virus life cycle has not yet been established. The HPIV3 system appears to be unique in that the actin microfilaments seem to be directly involved in the viral RNA synthesis in vivo (Fig. 2 and 4). Actin microfilaments are one of the major components of the CSK framework and are involved in such normal cellular processes as adhesion, motility, division, and intracellular transport of organelles (28, 35, 37, 38, 40). Our findings that CD completely inhibited HPIV3 progeny virus release whereas intracellular RNP accumulation was inhibited by about 40% suggest that actin microfilaments are perhaps also involved in the maturation and budding of HPIV3. Thus, it would be interesting to elucidate the various steps of morphogenesis of the HPIV3 virion with reference to its interaction with the CSK framework. Conversely, it is of interest to determine whether other paramyxoviruses, such as Newcastle disease virus, utilize the CSK components, namely, actin, tubulin, or vimentin for RNA synthesis.
Actin microfilaments have recently been shown to be involved in the spread of vaccinia virus between cells (12). Immunofluorescence and video microscopy revealed that virus particles move through the tip of actin-rich projections that extends from an infected cell into an uninfected cell. A similar role of the CSK in the cell-to-cell transmission of human immunodeficiency virus has been demonstrated (34). It is therefore possible that HPIV3 not only utilizes actin for RNA synthesis but also recruits and exploits actin to facilitate the spread of virus from cell to cell. Our data indicate that HPIV3 RNP-associated proteins maintain their association with the CSK framework, beginning with their syntheses and at least up to 16 h postinfection (Fig. 1). Thus, it seems that the viral proteins, once attached to the CSK framework following infection, do not leave this cytoarchitecture until budding. This intimate relationship between the viral proteins and actin microfilaments suggests that the microfilaments may also play a role in the translation of HPIV3 mRNAs. Furthermore, it is apparent that the virus contains a protein(s) that interacts directly or indirectly with actin. It would therefore be interesting to determine whether actin binds to NP, the RNA polymerase complex proteins L and P, or the RNP complex. Alternatively, other cellular proteins such as glyceraldehyde-3-phosphate dehydrogenase and La protein, which have recently been shown to interact with HPIV3 cis-acting RNA elements and to be packaged within the matured virions, may be involved in recruiting actin (19). Regarding the mode of action of actin microfilaments, it is tempting to speculate that these microfilaments form the sites of interaction between the viral RNA polymerase and the putative host factor(s). Many cellular proteins, including some transcription factors, interact with actin (3, 41), which may be exploited by the virus for its transcription/replication without having the proteins packaged within the virions. Given that actin alters the HPIV3 RNP structure from a loosely coiled to a moderately condensed form (17), it may facilitate the interaction between RNA polymerase and the putative host factors even when they bind to cis-acting elements at some distance apart on the genome template. Further studies on these interactions will shed light on this important area of host-virus interaction.
ACKNOWLEDGMENTS
We thank Laura Tripepi for secretarial assistance.
This work was supported by U.S. Public Health Service grant AI32027 (to A.K.B.)
REFERENCES
- 1.Banerjee A K. Transcription and replication of rhabdoviruses. Microbiol Rev. 1987;51:66–87. doi: 10.1128/mr.51.1.66-87.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee A K, Barik S, De B P. Gene expression of non-segmented RNA viruses. Pharmacol Ther. 1991;51:47–70. doi: 10.1016/0163-7258(91)90041-j. [DOI] [PubMed] [Google Scholar]
- 3.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 4.Bell P B, Jr, Lindorth M, Fredriksson B A. Preparation of cytoskeletons of cells in culture for high resolution scanning and scanning transmission electron microscopy. Scanning Microsc. 1988;2:1647–1661. [PubMed] [Google Scholar]
- 5.Bohn W, Rutter G, Hohenberg H, Mannweiler K, Nobis P. Involvement of actin microfilaments in the budding of measles virus: studies on cytoskeletons of infected cells. Virology. 1986;149:91–106. doi: 10.1016/0042-6822(86)90090-5. [DOI] [PubMed] [Google Scholar]
- 6.Bowden D S, Pederson J S, Toh B H, Westaway E G. Distribution by immunofluorescence of viral products and actin containing cytoskeletal filaments in rubella virus infected cells. Arch Virol. 1987;92:211–219. doi: 10.1007/BF01317478. [DOI] [PubMed] [Google Scholar]
- 7.Brown S S, Spudich J A. Mechanism of action of cytochalasin: evidence that it binds to actin filament ends. J Cell Biol. 1981;88:487–491. doi: 10.1083/jcb.88.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cervera M, Dreyfuss G, Penman S. Messenger RNA is translated when associated with the cytoskeletal framework in normal and VSV-infected HeLa cells. Cell. 1981;23:113–120. doi: 10.1016/0092-8674(81)90276-2. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Ciampor F. The role of cytoskeleton and nuclear matrix in virus replication. Acta Virol. 1988;32:168–189. [PubMed] [Google Scholar]
- 11.Collins P L, Chanock R M, Mackintosh K. The paramyxovirus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. New York, N.Y: Raven Press; 1996. pp. 1205–1241. [Google Scholar]
- 12.Cudmore S, Cossart P, Griffiths G, Way M. Actin based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 13.Damsky C H, Sheffield J B, Tuszynski G P, Warren L. Is there a role for actin in virus budding? J Cell Biol. 1977;75:593. doi: 10.1083/jcb.75.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De B P, Banerjee A K. Requirements and functions of vesicular stomatitis virus L and NS proteins in transcription process in vitro. Biochem Biophys Res Commun. 1985;126:40–49. doi: 10.1016/0006-291x(85)90568-6. [DOI] [PubMed] [Google Scholar]
- 15.De B P, Galinski M S, Banerjee A K. Characterization of an in vitro system for the synthesis of mRNA from human parainfluenza virus type 3. J Virol. 1990;64:1135–1142. doi: 10.1128/jvi.64.3.1135-1142.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De B P, Lesoon A, Banerjee A K. Human parainfluenza virus type 3 transcription in vitro: role of cellular actin in mRNA synthesis. J Virol. 1991;65:3268–3275. doi: 10.1128/jvi.65.6.3268-3275.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De B P, Burdsall A L, Banerjee A K. Role of cellular actin in human parainfluenza virus type 3 genome transcription. J Biol Chem. 1993;268:5703–5710. [PubMed] [Google Scholar]
- 18.De B P, Gupta S, Gupta S, Banerjee A K. Cellular protein kinase C ζ regulates HPIV-3 replication. Proc Natl Acad Sci USA. 1995;92:5204–5208. doi: 10.1073/pnas.92.11.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De B P, Gupta S, Zhao H, Drazba J A, Banerjee A K. Specific interaction in vitro and in vivo of glyceraldehyde-3-phosphate dehydrogenase and LA protein with cis acting RNAs of human parainfluenza virus-3. J Biol Chem. 1996;271:24728–24735. doi: 10.1074/jbc.271.40.24728. [DOI] [PubMed] [Google Scholar]
- 20.Galinski M S, Wechsler S L. The molecular biology of the Paramyxovirus genus. In: Kingsbury D W, editor. Paramyxoviruses. New York, N.Y: Plenum Publishing Corp.; 1991. pp. 41–82. [Google Scholar]
- 21.Garcia-Barreno B, Jorcano J L, Aukenbauer T, Lopez-Galindez C, Melero J A. Participation of cytoskeletal intermediate filaments in the infectious cycle of human respiratory syncytial virus (RSV) Virus Res. 1988;9:307–322. doi: 10.1016/0168-1702(88)90090-1. [DOI] [PubMed] [Google Scholar]
- 22.Genty N, Bussereau F. Is cytoskeleton involved in vesicular stomatitis virus reproduction? J Virol. 1980;34:777–781. doi: 10.1128/jvi.34.3.777-781.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffin J A, Compans R W. Effect of cytochalasin B on maturation of enveloped viruses. J Exp Med. 1979;150:379–391. doi: 10.1084/jem.150.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamaguchi M, Nishikawa K, Toyoda T, Yoshida T, Hanaichi T, Nagai Y. Transcriptive complex of Newcastle disease virus: structural and functional assembly associated with the cytoskeletal framework. Virology. 1985;147:295–308. doi: 10.1016/0042-6822(85)90132-1. [DOI] [PubMed] [Google Scholar]
- 25.Hill V M, Summers D F. A minor microtubule-associated protein is responsible for the stimulation of vesicular stomatitis virus transcription in vitro. J Virol. 1990;71:289–298. doi: 10.1099/0022-1317-71-2-289. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y T, Romito R R, De B P, Banerjee A K. Characterization of the in vitro system for the synthesis of mRNA from human respiratory syncitial virus. Virology. 1993;193:862–867. doi: 10.1006/viro.1993.1195. [DOI] [PubMed] [Google Scholar]
- 27.Kletzien R F, Perdue J F. The inhibition of sugar transport in chick embryo fibroblast by cytochalasin B. Evidence for a membrane-specific effect. J Biol Chem. 1973;248:711–719. [PubMed] [Google Scholar]
- 28.Korn E D. Actin polymerization and its regulation by proteins in non-muscle cells. Physiol Rev. 1982;62:672–737. doi: 10.1152/physrev.1982.62.2.672. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Moyer S A, Baker S C, Lessard J L. Tubulin: a factor necessary for the synthesis of both Sendai virus and vesicular stomatitis virus RNAs. Proc Natl Acad Sci USA. 1986;83:5405–5409. doi: 10.1073/pnas.83.15.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moyer S A, Baker S C, Horikami S M. Host cell proteins required for measles virus reproduction. J Gen Virol. 1990;71:775–783. doi: 10.1099/0022-1317-71-4-775. [DOI] [PubMed] [Google Scholar]
- 32.Oglesbee M J, Liu Z, Kenney H, Brooks C L. The highly inducible member of the 70kDa family of heat shock proteins increases canine distemper virus polymerase activity. J Gen Virol. 1996;77:2125–2135. doi: 10.1099/0022-1317-77-9-2125. [DOI] [PubMed] [Google Scholar]
- 33.Ohmori H, Toyama S, Toyama S. Direct proof that the primary site of action of cytochalasin on cell motility process is actin. J Cell Biol. 1992;116:933–941. doi: 10.1083/jcb.116.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearce-Pratt R, Malamud D, Phillips D M. Role of the cytoskeleton in cell-to-cell transmission of human immunodeficiency virus. J Virol. 1994;68:2898–2905. doi: 10.1128/jvi.68.5.2898-2905.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard T D, Cooper J A. Actin and actin binding proteins. A critical evaluation of mechanisms and functions. Annu Rev Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki H, Nakamura M, Ohiro T, Matsuda Y, Yuda Y, Nonomura Y. Myosin-actin interaction plays an important role in HIV-1 release from host cells. Proc Natl Acad Sci USA. 1995;92:2026–2030. doi: 10.1073/pnas.92.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schliwa M, Van Jerklom J. Structural interaction of cytoskeletal components. J Cell Biol. 1981;90:222–235. doi: 10.1083/jcb.90.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spooner B S, Yamada K M, Wessels N K. Microfilaments and cell locomotion. J Cell Biol. 1971;49:595. doi: 10.1083/jcb.49.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stallcup K C, Raine C S, Fields B N. Cytochalasin B inhibits the maturation of measles virus. Virology. 1983;124:59–74. doi: 10.1016/0042-6822(83)90290-8. [DOI] [PubMed] [Google Scholar]
- 40.Sundell C L, Singer R H. Requirement of microfilament in sorting of actin messenger RNA. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka Y, Watanabe T, Chiba N, Niki M, Kuroiwa Y, Nishihira T, Satomi S, Ito Y, Satake M. The protooncogene product, PEBP2beta/CBFbeta, is mainly located in the cytoplasm and has an affinity with the cytoskeletal structures. Oncogene. 1997;15:677–683. doi: 10.1038/sj.onc.1201235. [DOI] [PubMed] [Google Scholar]
- 42.Tanenbaum J. Approaches to the molecular biology of cytochalasin action. In: Tannenbaum S W, editor. Cytochalasins: biochemical and biological aspects. New York, N.Y: Elsevier/North-Holland; 1978. pp. 521–559. [PubMed] [Google Scholar]
- 43.Toneguzzo F, Ghosh H P. Characterization and translation of methylated and unmethylated vesicular stomatitis virus mRNA synthesized in vitro by ribonucleoprotein particles from vesicular stomatitis virus-infected cells. J Virol. 1976;17:477–491. doi: 10.1128/jvi.17.2.477-491.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyrrel D L J, Erhn A. Transmembrane communication in cells chronically infected with measles virus. J Cell Biol. 1979;81:396. doi: 10.1083/jcb.81.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volkman L E, Goldsmith P A, Hess R T. Evidence for microfilament involvement in budded Autographa californica nuclear polyhedrosis virus production. Virology. 1987;156:32–39. doi: 10.1016/0042-6822(87)90433-8. [DOI] [PubMed] [Google Scholar]
- 47.Wang E, Wolf B A, Lamb R A, Choppin P W, Goldberg A R. The presence of actin in enveloped viruses. In: Goldman R, Pollard T, Rozenbaum J, editors. Cell motility. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1976. pp. 589–599. [Google Scholar]