Abstract
Studies of feline leukemia virus (FeLV) have illustrated the importance of the genotype of the infecting virus in determining disease outcome. In FeLV infections, as in other retroviral infections, it is less clear how virus variants that evolve from the transmitted virus affect pathogenesis. We previously reported an analysis of the genotypic changes that occur in the viral envelope gene (env) in cats infected with a prototype transmissible FeLV clone, 61E (J. Rohn, M. Linenberger, E. Hoover, and J. Overbaugh, J. Virol. 68:2458–2467, 1994). In one cat, each variant (81T) had evolved, in addition to scattered amino acid changes, a four-amino-acid insertion with respect to 61E. This insertion was located at the same site in the extracellular envelope glycoprotein where the immunodeficiency-inducing molecular clone 61C possesses a six-amino-acid insertion critical for its pathogenic phenotype, although the sequences of the insertions were distinct. To determine whether acquisition of the four-amino-acid insertion was associated with a change in the replication or cytopathic properties of the virus, we constructed chimeras encoding 81T env genes in a 61E background. One representative chimeric virus, EET(TE)-109, was highly cytopathic despite the fact that it replicated with delayed kinetics in the feline T-cell line 3201 compared to the parental 61E virus. The phenotype of this virus was also novel compared to other FeLVs, including both the parental virus 61E and the immunodeficiency-inducing variant 61C, because infection of T cells was associated with syncytium formation. Moreover, in single-cycle infection assays, the 81T-109 envelope demonstrated receptor usage properties distinct from those of both 61E and 61C envelope. Thus, these studies demonstrate the evolution of a novel T-cell cytopathic and syncytium-inducing FeLV in the host. The 81T virus will be valuable for dissecting the mechanism of T-cell killing by cytopathic variants in the FeLV model.
The diversity of retroviruses may reflect, in part, the ability of these viruses to adapt to different selective pressures applied by the host. For example, virus variants that are selected for transmission may differ from virus variants that are favored for replication within a chronically infected individual. Diversity may also lead to viruses with distinct cytopathic and pathogenic properties; indeed, this paradigm has been supported by data from several animal models of retrovirus infection. For example, infection with one particular cloned feline leukemia virus (FeLV) variant, 61C, leads to fatal immunodeficiency in cats, whereas infection with another variant, FSC, causes aplastic anemia in cats (reviewed in reference 36). The pathogenic properties of these FeLV variants have been mapped to the extracellular envelope glycoprotein (SU), which plays a critical role in receptor recognition and viral host cell range (36). The three previously defined subgroups (A, B, and C) of FeLV most likely define envelope subtypes that use different cellular receptor molecules for viral entry (36).
Subgroup A FeLVs represent the types of FeLV variants that are transmitted in natural infections (36). FeLV-61E is a molecular clone that is a prototype FeLV-A (9); namely, it replicates in a variety of feline cells and tissues, but it is not cytopathic to these host cells and is relatively attenuated in pathogenicity. Thus, infection of cats with FeLV-61E provides a model to study the evolution of pathogenic variants starting with a highly transmissible but generally avirulent cloned virus. We had previously investigated the genetic diversity of FeLV envelope sequences in proviruses from cats inoculated with molecularly cloned 61E (35). From the tumor of one of the cats (Curly 40681), we isolated six highly related clones (81T) that had evolved both scattered mutations as well as a 12-nucleotide (nt) insertion with respect to 61E. The insertion was particularly intriguing because it occurred in the same location of the envelope gene at which there is an 18-nt insertion in the immunodeficiency-inducing 61C variant. This insertion has been identified as a key determinant of the cytopathic and pathogenic properties of the FeLV-FAIDS 61C clone (10, 32).
Both 61E and 61C were isolated simultaneously from a cat that had been infected with FeLV-FAIDS, an isolate that had been obtained from a naturally infected cat that developed a T-cell lymphoma (18, 27). Despite the fact that fatal immunodeficiency is the most common outcome of natural FeLV infections (17), FeLV-FAIDS is the only FeLV isolate examined to date that causes this disease. Therefore, it is unknown whether there are multiple classes of FeLV mutations that might serve as immunodeficiency-inducing determinants, or whether variants bearing essentially the same critical set of mutations are transmitted to, or evolve independently in, each afflicted cat. Indeed, it is also generally unclear how often retroviruses bearing cytopathic determinants arise in vivo and what selective advantage they possess in the host. Although the amino acid sequences encoded within the 81T variant insertions were distinct from that of the 61C SU insertion, we hypothesized that any such disruption in that region of envelope might confer cytopathic properties upon a virus that contained it. In the present study, we sought to test this hypothesis by isolating complete env clones encoding the novel insertion from the tumor DNA of cat 40681 and examining the biological phenotype of viruses bearing the 81T envelope. These studies showed that the 81T virus had evolved from the parental 61E virus into a T-cell-cytopathic and syncytium-inducing virus with altered receptor usage properties. Surprisingly, despite the presence of an insertion at the same position in the envelope protein of the T-cell-cytopathic 81T and 61C variants, the two viruses differed in their replication and interference properties and in their mechanisms of cell killing.
MATERIALS AND METHODS
Detection of 81T env sequences by PCR.
Cat Curly 40681 was inoculated with molecularly cloned virus 61E that was derived from the chronically infected feline fibroblast cell line JOAHE4 and died approximately 14 months after infection with thymic lymphoma, as reported previously (35). Peripheral blood mononuclear cells (PBMC) were isolated by Percoll gradient centrifugation from whole blood of cat 40681 at 9 and 12 months postinfection (p.i.), and total genomic DNA was isolated from these samples by standard methods. To control for extraneous DNA contamination during DNA isolation, genomic DNA from uninfected feline 3201 T cells was isolated in parallel with each sample, and all manipulations were performed in an isolated laboratory as described previously (29). Nested PCR was performed on 100 ng of genomic DNA template (first round) or 0.1 μl of first-round product (second round), using reaction cocktail components and thermal cycling conditions described previously (35). The first-round primer pair, which is specific for exogenous FeLV, consisted of FeLV-pol-5 and FeLV-U3-2B (35). The second-round primer pair, which is specific for 81T env-containing sequences, consisted of FeLV-81T-Ins (5′GGCGCCTGGAGAATCGC), which anneals in the sense orientation to the 12-nt insertion (underlined) and 5 nt of flanking sequences in the 81T-109 env gene (61E nt 7126 to 7130), and FeLV-U3-4 (5′AAACTTCTGCTGTTTCAGCTATA), which anneals in the antisense orientation to nt 8134 to 8156 in the U3 region of 61E. The PCR product was electrophoresed though 1% agarose, stained with ethidium bromide, and photographed under UV light.
Molecular cloning of complete 81T envelope genes.
81T env gene-long terminal repeat (LTR) fragments were amplified by PCR from the thymic tumor genomic DNA of cat Curly 40681. PCR amplification was performed as described (35) but with a different upstream primer that would amplify a larger product that encoded the entire envelope gene. Briefly, FeLV-pol-1 (5′AACCAAGAACCTCGAGCCACGG), which anneals in the sense orientation to the pol gene (61E nt 5807 to 5828), and FeLV-U3-2B (35), which contains an EcoRI tail, were used to amplify a 2.4-kb fragment containing 174 nt of the pol gene, the entire env gene, and 252 nt of the 3′ U3 region of the LTR. This fragment was digested with EcoRI and cloned into M13mp18 cut with SmaI and EcoRI. Nucleotide sequence analysis was performed by the dideoxy chain-terminating method (39).
Chimeric virus construction.
The construction of the EECC chimera, which encodes 5′ LTR and gag-pol genes of 61E and the env gene and 3′ LTR of 61C, has been described previously (27). For construction of 81T envelope chimeras, double-stranded DNA from M13mp18 81T env-LTR clones 81T-102, -106, and -109 was digested with XhoI (corresponding to 61E nt 5818) and RsrII (corresponding to 61E nt 7898) to generate 2.1-kb inserts encoding the entire SU and transmembrane (TM) domains of envelope (corresponding to nt 5818 to 7897 in 61E). A plasmid encoding the 61E genome in pUC18 (p61E) has been described elsewhere (27). p61E was digested with the unique restriction endonucleases XhoI and RsrII, and the desired 10.4-kb fragment (encompassing the plasmid, genomic flanking sequences, and 61E nt 1 to 5817 and 61E nt 7898 to the 3′ end of the provirus) was separated from the internal XhoI-RsrII fragment (nt 5818 to 7897). The two fragments of interest were purified by agarose gel electrophoresis and ligated by standard methods. The resulting constructs were verified by restriction endonuclease digestion and limited nucleotide sequence analysis and were called pEET(TE)-102, -106, and -109, using a modification of the nomenclature described previously (10, 44), where E indicates 61E origin, T indicates 81T origin, and the five letters correspond to 5′ LTR/gag, pol, env SU, env TM, and 3′ LTR, respectively. The numbers 102, 106, and 109 indicate specific 81T envelope subclones.
Cell culture.
The feline T-lymphoma cell line 3201 was maintained as described previously, with the addition of 0.25 μg of amphotericin B per ml to the media (44). AH927 feline fibroblasts cells, both uninfected and infected, were maintained in complete minimal essential medium as described previously for AH927 cells (44), with the addition of 0.25 μg of amphotericin B per ml to the medium. To select for drug-resistant AH927 cells, G418 (Geneticin; Gibco BRL) was added at a concentration of 0.65 mg/ml. For all G418 selections, the active drug concentration ranged from 589 to 743 μg/mg. All cell lines derived from D17 dog osteosarcoma cells were maintained in complete minimal essential medium supplemented with 0.8 mg of G418, 155 U of hygromycin B, and 0.25 μg of amphotericin B per ml. PA317 murine cells expressing LAPSN (LAPSN/PA317 [20]) were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, 0.25 μg amphotericin B per ml, 2 mM l-glutamine, and 1 mg of G418 per ml.
Infection studies.
To generate virus stocks, proviral clones were transfected by electroporation into the feline T-cell line 3201. Viral replication and spread were assessed by an enzyme-linked immunosorbent assay (ELISA) for FeLV p27gag (Virachek/FeLV; Synbiotics, San Diego, Calif.) in the culture supernatant. Cell-free supernatants were harvested from infected cells 2 days after p27gag production had reached saturating levels, and the 50% tissue culture infectious dose (TCID50) was determined as described previously (44). Briefly, viral supernatants were serially diluted and were used to infect 3201 cells in quadruplicate; the TCID50 was considered the concentration at which at least 50% percent of the cultures were ELISA positive for p27gag after 4 weeks of culture. Virus supernatants were used to infect 3201 cells in triplicate at a multiplicity of infection (MOI) of 0.005 or 0.001. For analysis of SU cell surface expression, a lower dose of the viruses, the amount of which was normalized between viruses for reverse transcriptase (RT) activity, were used for one infection.
Virus-induced cytopathic effects (CPE) were assessed by counting viable cells, which were identified by trypan blue dye exclusion, every 2 to 4 days and correcting for passage dilution to obtain total viable cell number. At each passage, viable cells were diluted to the same density, 5 × 105 cells/ml in 5 ml. Cells undergoing CPE were not passaged; rather, at each time point, these cells were centrifuged at 800 × g for 5 min and resuspended in 5 ml of fresh medium. In such cases, the concentration of certain cultures frequently fell below 5 × 105 cells/ml. Syncytium induction at each time point was quantitated by counting the number of large, multinucleated syncytia (>5 fused cells) visible in one defined field of the microscope prior to any physical disturbance of the cultures. This number was divided by the density (all viable cells per milliliter) of the culture at that time point, and the resulting quotient was termed the syncytium index.
RT assays.
For analysis of RT activity, cell-free viral supernatants were harvested and stored at −70°C. Ten-microliter aliquots of supernatant, and various dilutions thereof, were assayed essentially as described previously (14); briefly, we measured the ability of lysed virion preparations to catalyze the incorporation of radiolabeled nucleotide, using a polyribonucleotide template to which an oligodeoxyribonucleotide primer was annealed. Intensity of radioactivity was quantitated by PhosphorImager analysis, and the assigned PhosphorImager units (PIU) were adjusted by subtracting background radiation on the filter. Dilutions producing PIU in the linear range were adjusted to reflect PIU/microliter of supernatant for all samples. To take into account large differences in cell density during windows of cytopathicity, RT activity was also normalized by dividing adjusted PIU/microliter by viable cells/milliliter at the time of harvest.
Flow cytometry analysis.
To analyze SU cell surface expression over the course of infection in T cells, flow cytometry was performed at daily intervals, from days 2 to 16 post p.i. infection. To stain cells for fluorescence-activated cell sorting (FACS) analysis, 2 × 105 3201 T cells were washed three times in WB (Hanks balanced salt solution plus 2% fetal calf serum and 0.08% sodium azide). Cells were resuspended in 50 μl of WB containing a 1:50 dilution of monoclonal antibody C11D8 (15), specific for Env-SU, and incubated for 25 min at 37°C. Cells were then washed three times in WB, resuspended in 50 μl of WB plus a 1:20 dilution of goat anti-mouse fluorescein isothiocyanate-conjugated immunoglobulin G2b (Southern Biotechnologies), and incubated on ice in the dark for 15 min. Cells were washed three times with WB, resuspended in 150 μl of WB, and then fixed with 150 μl of 2% paraformaldehyde in phosphate-buffered saline. Flow cytometry was performed on a Becton Dickinson FACS Star Plus.
Southern analysis.
Infected cells were harvested, pelleted by centrifugation, and stored at −70°C. Total genomic DNA was isolated and analyzed by Southern blot with the exogenous FeLV LTR-specific probe exU3 as described previously (28, 35). Briefly, in this analysis, digestion of either unintegrated or integrated proviral DNA with the restriction endonuclease KpnI yields a 3.4-kb internal fragment that hybridizes to the exU3 probe in the case of 61E and EET(TE)-109 and a 2.1-kb fragment in the case of EECC. Additionally, linear or circular unintegrated viral DNA (UVD) of all three viruses will produce an approximately 0.4 or 0.5-kb 5′-terminal LTR fragment, respectively.
Pseudotyped virus construction.
A packaging-deficient EET(TE)-109 proviral genome was constructed by using a strategy described previously to generate a 61E packaging-deficient virus (5). Briefly, this construct, called pEET(TE)-109ΔΨ, is identical to pEET(TE)-109 except that it lacks a region of 107 bp upstream of the gag gene that includes a sequence (Ψ) required for specific encapsidation of the FeLV genome into virions (5). pEET(TE)-109ΔΨ was verified by restriction endonuclease digestion.
The pLAPSN vector has been described previously (23); it is a murine leukemia virus (MuLV)-based vector that contains two marker genes, alkaline phosphatase (AP) and neomycin phosphotransferase (neo), the latter of which confers resistance to the drug G418. pLAPSN also contains an MuLVΨ sequence that provides a signal for encapsidation into MuLV (23) and FeLV (5) virions. D17-LAPSN is a D17 cell line stably expressing the pLAPSN vector; it was made by infecting D17 cells with PG13-derived virus (LAPSN pseudotyped by gibbon ape leukemia virus [22]) as described previously (5). Cell clones were isolated, and those expressing high amounts of RNA that hybridized to a neo probe were identified by Northern analysis. EET(TE)-109ΔΨ and a vector encoding hygromycin resistance (pCMVhph [2]) were cotransfected into a D17-LAPSN cell line expressing high levels of vector RNA, and stable cell lines (called D17-EET(TE)-109ΔΨ/LAPSN) expressing both LAPSN and EET(TE)-109ΔΨ were selected and isolated as described previously (5). We identified cell clones that produced high amounts of virus [called EET(TE)-109ΔΨ/LAPSN virus] by screening for efficient transfer of the pLAPSN vector to naive 3201 cells. For this screen, 3201 cells were infected for 2 days with cell-free supernatant from candidate D17-EET(TE)-109ΔΨ/LAPSN cell lines, dotted onto glass slides, and allowed to air dry, and AP staining was performed essentially as described previously (13). D17 cell lines expressing LAPSN plus 61E-ΔΨ or EECC-ΔΨ have been described previously (5, 24); viruses derived from these cell lines are called 61E-ΔΨ/LAPSN or EECC-ΔΨ/LAPSN, respectively.
The infectious titer of cell-free supernatants from cells expressing viral/LAPSN pseudotypes was determined in both AH927 and 3201 cells, using resistance to G418 as a marker. For the adherent AH927 cells, 2 × 105 cells were plated in 6-cm-diameter dishes and infected the next day in the presence of Polybrene (4 μg/ml). Cultures were split 1:10 into medium containing G418 (0.65 mg/ml) the next day, maintained for 10 to 12 days, then stained with 2% methylene blue in 50% ethanol. The number of CFU per ml was determined by scoring total drug-resistant colonies. For 3201 suspension cells, limiting dilutions of virus were used to infect 3201 cells in the presence of Polybrene (4 μg/ml). Replicate cultures were split into medium containing G418 (3 mg/ml), maintained with periodic replacement of media without cell passage for approximately 1 month, and scored for the ability to repopulate the culture after escaping from drug-mediated killing. G418-resistant TCID50 was calculated from the dilution of virus that allowed 50% of the cultures to repopulate with drug-resistant cells.
Superinfection interference assays.
Cell lines chronically infected with EET(TE)-109 or EECC were created by infecting AH927 cells with viral supernatant from transfected 3201 T cells at a high MOI (≥1) in the presence of Polybrene. These cells were maintained until they were judged to be chronically infected as assessed by ELISA detection of p27gag protein in the culture supernatant. The derivation of chronically 61E infected AH927 cells, JOAHE4, was described previously; this cell line was also the source of virus that infected the cat from which 81T was isolated (35). Flow cytometric analysis of chronically infected target cells was performed by methods similar to those described above for 3201 T cells, using monoclonal antibody C11D8 for the primary stain and fluorescein-conjugated anti-murine immunoglobulin G antibody as the secondary stain. Superinfection interference assays in chronically infected AH927 cell lines were performed essentially identically to the manner of determining the infectious G418-resistant titer of LAPSN-containing pseudotyped virus supernatants (described above). All infections were carried out with approximately 5,000 CFU of virus on approximately 4 × 105 cells.
Nucleotide sequence accession numbers.
The sequences of the env genes of 81T-102, 81T-106, and 81T-109 are available from GenBank under accession no. U70377, U70378, and U58951, respectively.
RESULTS
PCR cloning of complete env genes from cat 40681.
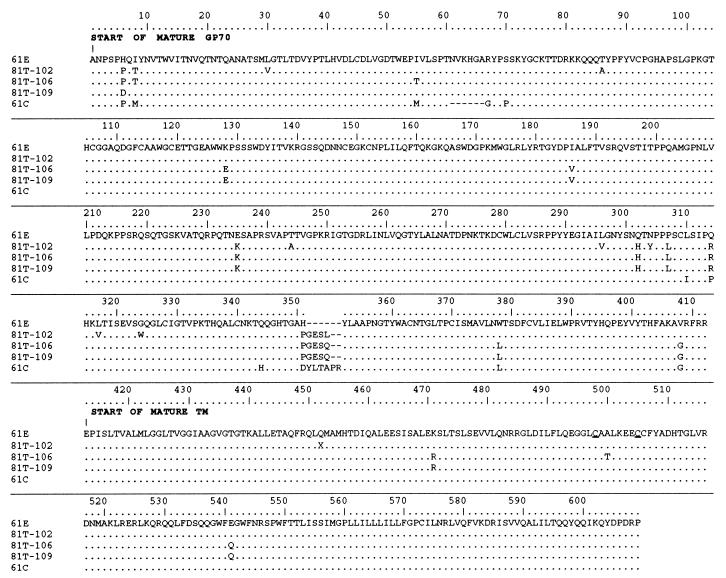
Although we previously obtained clones representing FeLV env and U3 sequences from cat 40681 tumor (35), these clones lacked most of the sequences encoding the N-terminal envelope leader signal peptide. Therefore, to obtain a representative clone with a complete 81T env gene, we amplified a larger 3′ genomic fragment from cat 40681 tumor DNA. We cloned and determined the partial nucleotide sequence of eight clones; of these, one resembled the parental 61E, and the other seven contained insertions similar to the previously isolated 81T clones (GESL or GESQ). Taken together with previous data (35), the results indicated that 13 of 14 clones isolated from cat 40681 tumor contained the 12-nt insertion. In Fig. 1, The predicted amino acid sequences of three representative clones whose nucleotide sequences were determined throughout the entire SU and TM regions of envelope (81T-102, -106, and -109) are aligned and compared to the amino acid sequence of the inoculated parental virus 61E. The 81T variants are highly related to 61E but have several mutations in common with one another. Comparing the amino acid sequences of 81T-102, -106, and -109 with those of the six previously published 81T variant envelopes (35), we determined that nine of nine envelope proteins shared a mutation at position 6 and a cluster of changes at positions 302, 307, and 314. Adjacent to the four-amino-acid insertion, there was also an amino acid change in each clone (position 351) compared to 61E. Additionally, some 81T clones encoded amino acid changes that were found in the same sites in 61C (positions 6, 378, and 408), perhaps reflecting some convergent evolution.
FIG. 1.
The deduced amino acid sequence of the envelope proteins of 81T-102, -106, and -109 variants compared to that of 61E. The sequence of 61C is also aligned for comparison. The sequences begin at the first amino acid of the mature SU and include the complete SU and TM regions. Dots indicate identity, letters indicate amino acid differences, and X denotes a premature stop codon. The conserved cysteines that flank the putative cysteine loop in TM (6) are underlined.
Origin of the 81T virus.
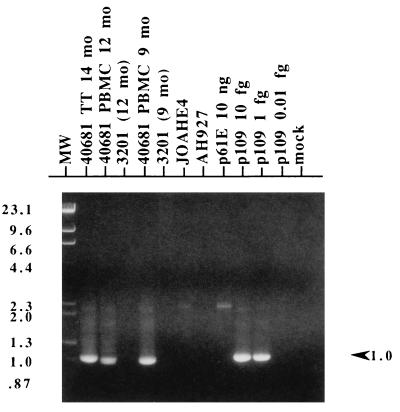
To determine whether FeLV proviral genomes bearing the novel 12-nt insertion evolved directly from 61E during the course of infection, we performed a nested PCR strategy that used a primer specific for the 81T insertion in the second round of amplification. We examined both the 61E-infected AH927 cell line (JOAHE4), which was used to generate the inoculum for cat 40681, as well as PBMC derived from this cat at 9 and 12 months p.i. As shown in Fig. 2, 81T-specific amplification products were present in all samples derived from cat 40681, including blood drawn at the earliest available time point (9 months p.i.). In contrast, specific product was not seen in JOAHE4 DNA from approximately 5 × 104 cells. These results were confirmed with Southern blot analysis of PCR products with the exU3 probe (data not shown). The PCR strategy was sufficiently sensitive to reproducibly detect approximately 1 to 10 copies (0.01 to 0.1 fg) of pEET(TE)-109 plasmid DNA. These data suggest that variants bearing an 81T envelope were not present in the inoculum but instead evolved from a parental 61E virus at some point prior to 9 months p.i.
FIG. 2.
PCR analysis of cells from cat 40681. Nested PCR with a first-round primer pair specific for exogenous FeLV (FeLV-pol-5/FeLV-U3-2B) and a second-round primer pair specific for 81T variants (FeLV-81T-Ins/FeLV-U3-4) was performed on 100 ng of genomic DNA template (approximately 5 × 104 cells) isolated from the following cat 40681 samples: thymic tumor (TT) harvested at necropsy (14 months [mo] p.i.) and PBMC from 12 and 9 months p.i. Additionally, 100 ng of genomic DNA from JOAHE4, the cell line used to generate the inoculum for cat 40681, was tested. Control templates included genomic DNA from the following sources: 3201 feline T cells that had been isolated in parallel with cat 40681 PBMC samples at 12 and 9 months p.i. (3201 [12 mo] and 3201 [9 mo], respectively), and uninfected AH927 feline fibroblasts. Plasmid control templates included 10 ng of p61E and a dilution series (10, 1, and 0.1 fg) of pEET(TE)-109 (p109). A reaction mixture containing water instead of DNA was included (mock). Second-round PCR product was electrophoresed through 1% agarose, stained with ethidium bromide, and photographed under UV light. Multiple amplifications using different primer pairs yielded similar results. Mobility of the molecular weight standards (MW), which are a mixture of phage lambda DNA cut with HindIII and phage φX174 DNA cut with HincII, are indicated with their corresponding sizes in kilobases on the left figure. On the right, the arrowhead indicates the position and size of the expected 1-kb amplimer.
Replication and cytopathicity of EET(TE)-109 in feline T cells.
To examine the biological properties of the 81T envelope, we prepared chimeric proviruses containing the entire 81T-102, -106, and -109 env genes in a background of the 61E provirus [pEET(TE)-102, -106, and -109, respectively]. These chimeras, along with plasmids encoding the noncytopathic parental virus 61E and the cytopathic FeLV-FAIDS chimera EECC, which encodes the 61C envelope (27), were transfected into the feline T-cell line 3201. The 3201 T-cell line was chosen for this analysis because the ability of a given FeLV to induce CPE in these cells has been strongly correlated with the capacity of that virus to induce fatal immunodeficiency disease in vivo (10, 28, 32). We did not detect viral p27gag production by either EET(TE)-102 and EET(TE)-106 viral clones after more than a month of culturing, suggesting that they were replication defective in this cell type. Consistent with this observation, there is a mutation in 81T-102 resulting in a premature termination codon in the extracellular portion of TM (Fig. 1). In the case of EET(TE)-106, a probable cause for its inability to replicate is not as apparent, but this clone does contain a potentially disruptive mutation in the putative cysteine loop in TM (Fig. 1), a region previously determined to be important for envelope protein processing (6, 44). In contrast, EET(TE)-109 virus, which contains an insertion identical to the one in 81T-106, was replication competent. This virus was chosen as a representative infectious 81T virus for all further experiments.
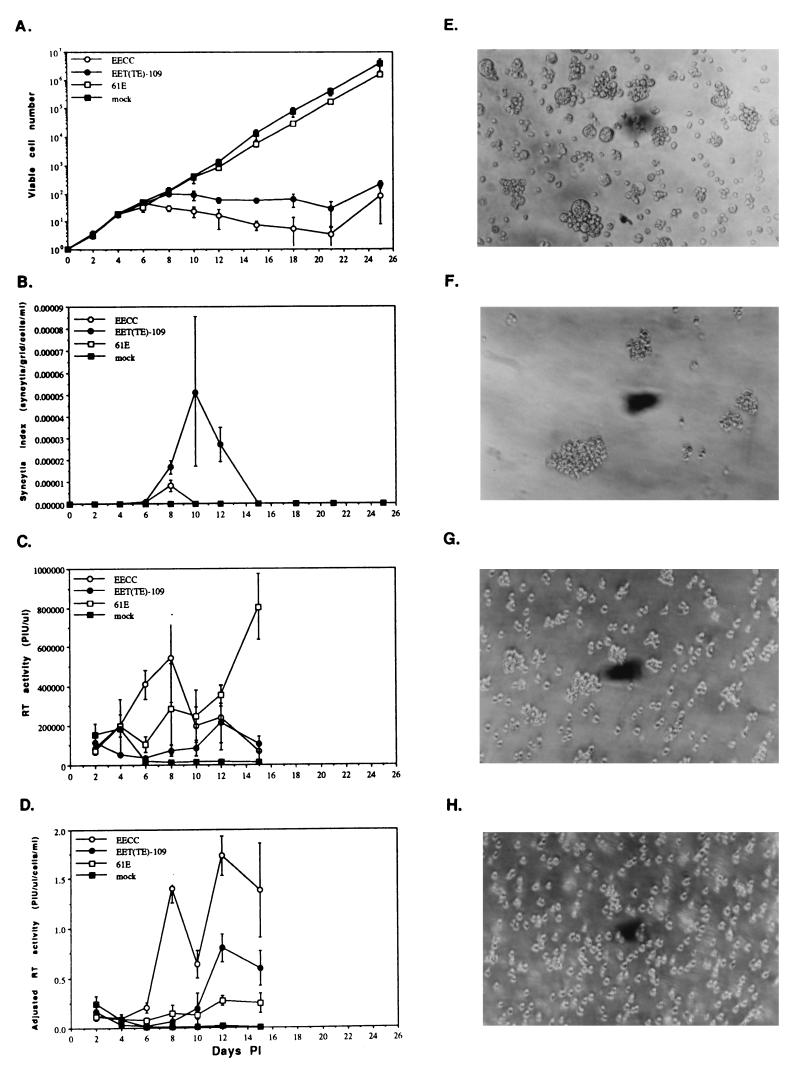
Virus supernatants were used to infect naive 3201 T cells, and the viability of cells in infected cultures was monitored over approximately 4 weeks. As shown in Fig. 3A, EET(TE)-109 virus, like EECC, caused considerable cell killing, whereas the parental virus 61E, as established previously (10), had no significant effect on viability. Thus, the envelope of 81T-109 was sufficient to confer cytopathicity onto an otherwise noncytopathic virus. Loss of cell viability in EET(TE)-109-infected cultures was somewhat delayed (2 days) with respect to EECC-infected cells. Interestingly, the manifestation of 81T-109-induced CPE was distinct from that of EECC or from that of any other FeLV molecular clone yet described. As shown in Fig. 3E, EET(TE)-109 induced widespread syncytium formation in infected cultures, whereas EECC (Fig. 3F) induced characteristic aggregation (reference 19 and our unpublished observations) and only extremely limited, smaller-syncytium formation. Cultures of 61E-infected (Fig. 3G) or mock-infected (Fig. 3H) 3201 cells manifested neither of these types of CPE. We quantitated the number of large syncytia at each time point as a function of cell density, and these results are shown in Fig. 3B. Comparison of Fig. 3A and B reveals that syncytium induction in EET(TE)-109-infected cells occurred concomitant with the loss of cell viability.
FIG. 3.
CPE and viral replication kinetics after infection of 3201 feline T cells with molecularly cloned viruses. Infection was performed in triplicate with an MOI of 0.001. All replicate values were averaged, and error bars indicate the extent of sample standard deviation. The ordinates are as follows: (A) viable cell number, as assessed by the exclusion of the vital dye trypan blue (log scale); (B) syncytium index, which is the quotient of the number of large syncytia visible in one defined field of the microscope divided by the cell density at that time point (linear scale); (C) total RT activity in the culture supernatants, expressed in terms of arbitrary adjusted PIU/microliter (linear scale); and (D) PIU/microliter, divided by the cell density to account for large differences in number of virus-producing cells in cytopathic and noncytopathic infections (linear scale). (E to H) Phase-contrast photomicrographs (magnification, ×200) showing morphological appearance of 3201 T cells infected with various FeLVs at day 8 p.i. Cultures were infected with EET(TE)-109 (E) EECC (F), or 61E (G) or were mock infected (H). The results in panels A and B and E to H are representative of six, and those in panels C and D are representative of two, similar, independent infections.
To assess the efficiency of virus replication, we examined RT activity of the culture supernatants over time. Figure 3C depicts total RT activity as a function of days p.i. Figure 3D shows the same data normalized for cell density at the time of harvest, which takes into account the fact that during CPE, there were frequently very few viable cells to secrete virus particles into the supernatant. These data show that cultures infected with EECC virus reached maximal RT activity rapidly during the course of infection, with a peak level of RT activity achieved by approximately day 8. In contrast, cultures infected with EET(TE)-109 demonstrated significantly slower replication kinetics, with peak RT activity on approximately day 12. In addition to the delay of peak RT activity, the total amount of RT activity exhibited by EET(TE)-109-infected cells at its peak was significantly lower than that shown by EECC-infected cells during their time of maximum production. The EET(TE)-109 virus replicated with kinetics more similar to that of 61E virus; in fact, the maximal level of RT activity in 61E-infected cells on day 15 was higher than that exhibited in cells infected with EET(TE)-109 (Fig. 3C), although the RT equivalents per cell were slightly lower in 61E-infected cells at these later times p.i. (Fig. 3D). These results suggest that the EET(TE)-109 virus though cytopathic, appeared to replicate and/or spread in T cells more slowly than EECC, similarly to 61E.
Kinetics of virus spread measured by FACS analysis of envelope-SU expression.
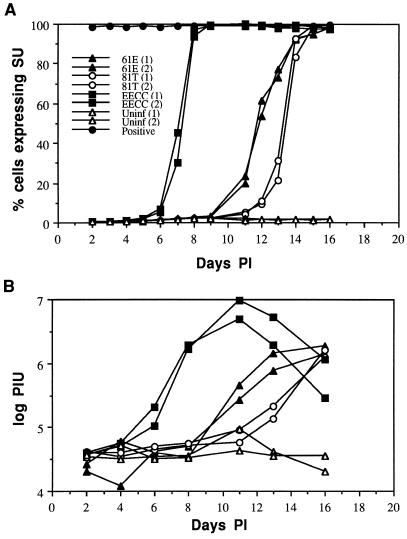
While analysis of virus expression by RT assay provides a picture of virus replication in the total cell culture, it does not show the pattern of virus spread and infection of new target cells throughout the culture. Moreover, interpretations of RT data are somewhat complicated when the infection is cytopathic and total cell numbers are decreasing in some infections and not in others. Thus, to compare the rates of virus replication and spread during infection with 61E, EECC, and EET(TE)-109 variants, we determined the percentage of infected 3201 T cells as a function of time by analyzing cells for envelope-SU expression by flow cytometry at daily intervals p.i. The results of this analysis are shown in Fig. 4A for duplicate infections starting with equivalent levels of each virus, as determined by RT assay. EECC virus replicated with rapid kinetics and spread to essentially 100% of cells within 7 to 8 days p.i. In contrast, infection and spread to new cells were significantly delayed for both EET(TE)-109 and 61E, and the cultures did not become chronically infected until about 14 to 16 days p.i. Interestingly, despite the fact that EET(TE)-109 is a T-cell-cytopathic virus, it spread with somewhat delayed kinetics compared to the noncytopathic 61E parental virus (Fig. 4A). Figure 4B shows the corresponding RT levels in these cultures over time. In general, the peak of RT activity was detected within 2 days after the culture became completely infected, as measured by SU cell surface expression. The relative orders in which the viruses achieved complete infection of the culture were similar in experiments using different virus stocks at an MOI of 0.001 (the same virus stocks and dose used in the experiments in Fig. 3). In this experiment (data not shown), EECC spread most rapidly, and 61E spread more rapidly than EET(TE)-109 in 3201 T cells. At this dose, complete infection of the culture (90 to 100% of cells expressing SU) was achieved in 6 to 10 days, depending on the virus; this was just prior to maximal RT expression for each virus (e.g., Fig. 3C), which again corroborates the data shown in Fig. 4.
FIG. 4.
Analysis of virus spread by flow cytometric analysis of cells expressing envelope SU protein. 3201 T cells were infected with 61E, EECC, or EET(TE)-109 in duplicate. Equivalent doses of virus, as measured by RT assay, were used for all infections. (A) Cell surface expression of SU was monitored by using antibody C11D8. The percentage of cells expressing SU above background is plotted against days p.i. (B) Total RT activity in the culture supernatants, expressed in terms of arbitrary adjusted PIU/microliter (log scale), versus days p.i. The symbols corresponding to the virus used for each infection are designated in panel A and apply to the data in both panels. 81T refers to the EET(TE)-109 virus encoding the 81T envelope protein. The numbers 1 and 2 in parentheses were used to discriminate data from duplicate infections with the same virus. “Positive” in panel A represents data from a chronically 61E infected 3201 T-cell line that was used as a positive control at each time point. Data from duplicate uninfected cultures [Uninf (1) and (2)] are also shown.
Analysis of viral DNA copy number during the course of infection.
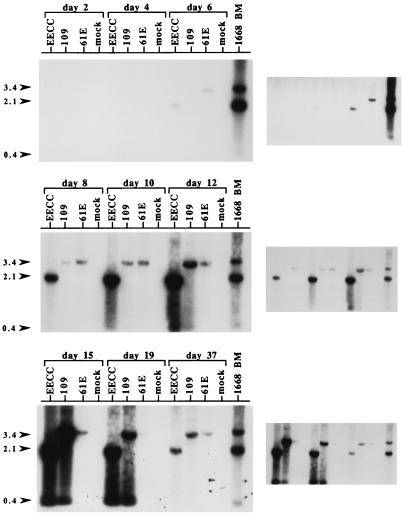
To obtain an alternate assessment of the replication properties of the EET(TE)-109 virus, we harvested genomic DNA over the course of EET(TE)-109 infection and analyzed proviral and UVD copy number by Southern blotting. As shown in Fig. 5, the EECC proviral load accumulated the most rapidly, whereas the EET(TE)-109 proviral load accumulated more slowly but reached levels approximately equal to that of EECC-infected cultures by 15 days p.i. In contrast, the proviral load of 61E-infected cultures also accumulated slowly but stabilized at a level much lower than did the cytopathic viruses, which is consistent with the replication data presented in Fig. 3D. EECC- and EET(TE)-109-infected cultures that survived CPE and eventually repopulated the cultures (day 37) contained a proviral copy number much lower than the levels associated with maximum cytopathicity, which in this particular experiment occurred on days 10 to 19 p.i. (data not shown). However, the proviral load of EET(TE)-109 and EECC was higher than that of 61E at day 37. Interestingly, as has been shown previously for EECC (10), cytopathicity in EET(TE)-109-infected 3201 cells was associated with UVD accumulation (days 12 to 19). The relative amount of UVD in cytopathic EET(TE)-109-infected cells was comparable to that of EECC by day 15.
FIG. 5.
Southern blot analysis of proviral load and UVD during infection. 3201 T cells were infected with EECC, EET(TE)-109 (109), or 61E at an MOI of 0.005 or were mock infected, and genomic DNA was harvested at days 2, 4, 6, 8, 10, 12, 15, 19, and 37 p.i. In this experiment, which is similar to the one depicted in Fig. 3, cytopathicity was first apparent at approximately day 10. Twelve micrograms of DNA was digested with KpnI and analyzed by Southern blotting with probe exU3. Also included as a positive control was 4 μg of genomic DNA from the bone marrow (BM) of cat 1668, which died with immunosuppression and T-cell lymphoma after experimental infection with a mixture of 61E and 61B. This DNA sample was previously shown to harbor high levels of UVD in a similar analysis (28). The positions of the 3.4-kb fragments [corresponding to the internal KpnI fragment of 61E and EET(TE)-109] and 2.1-kb fragments (corresponding to the internal KpnI fragment of EECC) are indicated by arrowheads on the left. Also indicated with an arrowhead (labeled 0.4) is the position of the UVD fragments. The panels at the right indicate a longer exposure (days 2 to 6 p.i.) and shorter exposures (days 8 to 37 p.i.) of each Southern blot shown on the left.
Receptor usage properties of the 81T-109 envelope.
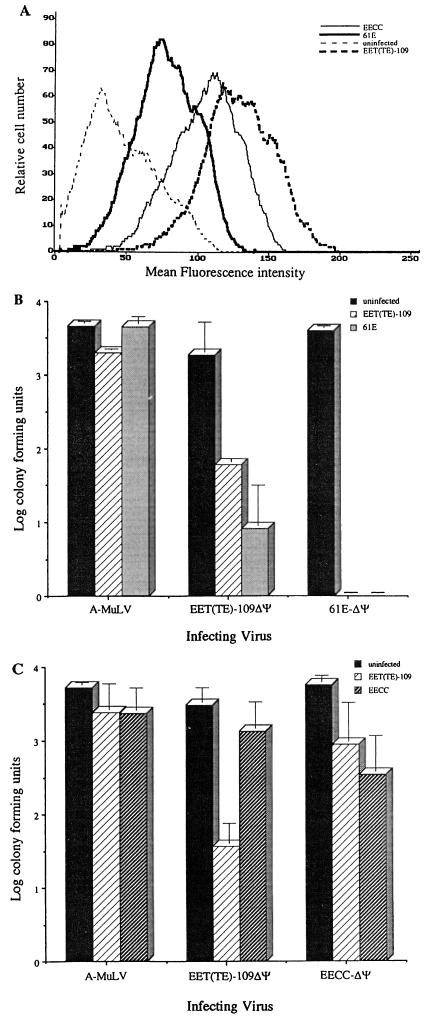
To examine the interference properties of the 81T-109 envelope in a single-cycle infection assay, we used a panel of viruses pseudotyped with various envelope proteins and assayed their ability to enter cells by monitoring expression of a selectable marker (neo) encoded by the packaged RNA. The targets for these infections were chronically infected feline fibroblasts. To confirm that the target cells lines were expressing envelope, we performed flow cytometric analysis of the chronically infected cell targets with the FeLV SU-specific antibody C11D8. In this analysis (Fig. 6A), 61E, EECC, and EET(TE)-109-infected AH927 cells each appeared as a fairly homogeneous population of cells shifted to the right compared to uninfected AH927 cells, which exhibited a low level of autofluorescence. Such a lack of bimodal distribution indicates that the majority of the cells in the population expressed SU to some degree. EET(TE)-109-infected cells exhibited a higher degree of SU-specific staining than did 61E- or EECC-infected cells.
FIG. 6.
Superinfection interference patterns of the 81T-109 versus 61E and 61C envelope proteins. (A) Flow cytometric analysis of chronically 61E, EECC, and EET(TE)-109 infected AH927 cells compared with uninfected AH927 cells, using C11D8, a murine monoclonal antibody against FeLV SU. The ordinate indicates relative cell number; the abscissa indicates mean fluorescence intensity. (B) Superinfection interference of pseudotyped viruses in the 61E- and EET(TE)-109-infected feline fibroblasts analyzed in panel A. (C) Superinfection interference of pseudotyped viruses in the EECC- and EET(TE)-109-infected feline fibroblasts analyzed in panel A. In panels B and C, the height of the bars indicates the log of the number of CFU scored after infection at an MOI of approximately 0.0125. Pseudotyped viruses used for challenge bear either the amphotropic MuLV (A-MuLV), 81T-109 [EET(TE)-109ΔΨ], 61E (61E-ΔΨ), or 61C (EECC-ΔΨ) envelope and are labeled under each set of three bars. The legend to the right indicates the target cell lines for these infection studies. These data are an average of three independent experiments; the error bars indicate the sample standard deviation between the experiments. Similar results were obtained from multiple infection experiments using different stocks and target cells at different passage numbers, with the exception of one experiment in which we detected a low level of 61E-ΔΨ infection in EET(TE)-109-infected AH927 cells. However, 61E infection of these cells was not detected in additional experiments using up to 100-fold more virus.
We compared the interference properties of EET(TE)-109ΔΨ and 61E-ΔΨ to determine whether the 81T variant had altered receptor specificity compared to the parental virus from which it had evolved. An average of data from multiple superinfection interference assays using these target cells is presented in Fig. 6B. The control A-MuLV/LAPSN virus infected 61E- and EET(TE)-109-infected and uninfected AH927 cells with approximately equal efficiency, indicating no cross-interference among these viral envelope proteins. However, both 61E- and EET(TE)-109-infected cells were able to completely block infection by 61E-ΔΨ/LAPSN virus at this MOI (0.0125; 5 × 103 CFU), indicating that the 61E and 81T-109 envelope proteins can establish superinfection interference against viruses with a 61E envelope. This pattern of superinfection interference was observed using as much as 5 × 105 CFU per infection (MOI = 1.25); even at this dose, EET(TE)-109-infected cells were completely resistant to 61E viral challenge. Cells infected with 61E could be infected at very low levels by 61E virus at an MOI of greater than 1; 30 to 100 foci were detected when 5 × 105 CFU of 61E-ΔΨ/LAPSN virus particles was used for challenge. This very low level of infection with homologous virus challenge may reflect the lower levels of envelope expression in the 61E-infected target cells compared to the EET(TE)-109-infected target cells (Fig. 6A), which were completely resistant to 61E infection at all MOIs tested.
Interestingly, in the reciprocal experiment using a virus with the 81T envelope for challenge (Fig. 6B), 61E-infected cells demonstrated only partial interference when challenged by EET(TE)-109ΔΨ/LAPSN virus, with infection approximately 2.5 logs lower than the level of infection seen in AH927 cells. In repeated parallel experiments, 61E-infected cells were able to completely inhibit infection by viruses bearing the 61E envelope at this MOI (>3.5-log reduction [Fig. 6B]). Additionally, cells chronically infected with EET(TE)-109 were unable to completely block homologous challenge (approximately 1.5-log reduction), even though they expressed very high levels of envelope on the cell surface, as judged by FACS analysis (Fig. 6A). These results suggest that the 81T-109 envelope protein may recognize an additional, lower-affinity receptor on fibroblasts that is not recognized by the 61E envelope protein.
Our previous studies showed that 61E and 61C viruses do not exhibit any reciprocal interference, suggesting that they may use different receptor molecules (24). Here, we examined the interference between 81T and 61C envelope in AH927 cells infected with these two cytopathic FeLVs (Fig. 6C). As reported previously (24), AH927 cells chronically infected with EECC were infectable by EECC-ΔΨ/LAPSN virus, although there was an approximately 16-fold reduction in infection compared to uninfected cells (Fig. 6C). There was no significant interference (≤2-fold) against EET(TE)-109 infection in EECC-infected cells. Cells infected with EET(TE)-109 were partially resistant to homologous virus challenge (approximately 2 logs), but there was very modest interference (6.4-fold) to EECC infection in these cells. Thus, the 81T envelope is distinct from the 61C envelope in its ability to establish significant interference to reinfection, although both envelope proteins were impaired in the ability to establish complete superinfection interference to infection by homologous virus in this assay.
DISCUSSION
These data comprise the first report of a T-cell-cytopathic FeLV having evolved directly from a noncytopathic FeLV during a single in vivo passage. Our previous understanding of FeLV immunodeficiency-inducing determinants was gleaned from studies of proviruses, like 61C, that were derived from a single isolate, FeLV-FAIDS. Because immunodeficiency is such a common outcome of natural FeLV infection, we were concerned that previous assumptions about the pathogenic mechanisms of FeLV-induced immunodeficiency could be biased toward this small subset of FeLV variants. In this context, one might consider, for example, how our understanding of human immunodeficiency virus (HIV)-induced cytopathology and pathogenesis might be affected if all studies of HIV type 1 were focused on a single viral isolate. In the present study, we characterized the evolution and cytopathic properties a variant, 81T, that was isolated from the tumor DNA of a cat infected with 61E. Our data suggest that while the 81T and 61C envelope proteins contain an insertion in the same site, and also share some phenotypic characteristics, they differ in many important aspects.
A virus encoding the 81T envelope protein in the context of the parental 61E proviral genome was highly cytopathic in feline 3201 T cells. Intriguingly, EET(TE)-109 virus induced syncytium formation in 3201 cells, a phenotype that has not been reported for other molecularly cloned FeLV variants. Virally induced membrane fusion is thought to occur due to interactions between viral envelope glycoprotein and the cellular receptor molecule (45). While syncytium formation is common in lentiviruses that induce immunodeficiency and has been correlated with pathogenicity (3, 7, 12, 42, 43), syncytium induction by mammalian type C retroviruses is relatively rare (1, 30, 31, 37, 46, 47). Because the commencement of EET(TE)-109-induced cell killing and syncytium formation were temporally associated, syncytium formation may play an important role in the cytopathic effects that occur during EET(TE)-109 infection.
Although the EET(TE)-109 virus was only modestly delayed in inducing the onset of CPE in 3201 T cells compared to EECC, the replication kinetics of EET(TE)-109 were significantly attenuated with respect to EECC in the same cell type. The replication kinetics of EET(TE)-109 were more similar to those of the noncytopathic 61E virus. In fact, a detailed analysis of virus spread to new target cells over the course of infection suggests that the 61E virus replicates with more rapid kinetics than the cytopathic EET(TE)-109 variant in 3201 T cells. A major difference that we observed between 61E and EET(TE)-109 infection was that when all of the target cells became infected with 61E virus, the cells were resistant to reinfection; specifically, in 61E-infected T cells, the proviral copy number did not increase once 100% of the cells became infected. In contrast, the proviral load continued to increase in cultures infected with EET(TE)-109 virus, and the proviral copy number in EET(TE)-109 chronically infected T cells was approximately 20- to 50-fold higher than in chronically 61E infected cells at 2 to 3 weeks p.i. EET(TE)-109 infection in T-cells was also characterized by high levels of UVD accumulation, which is a hallmark of superinfection. The interference studies also show that cells chronically infected with EET(TE)-109 cannot establish complete superinfection interference against challenge by pseudotyped viruses bearing the 81T-109 envelope protein.
As first postulated by Temin, superinfection is thought to play a central role in the cytopathicity of retroviruses, although the mechanism of cell killing remains unknown (40). There is also good evidence that the cytopathicity of FeLV variants bearing the 61C envelope is caused by superinfection (8, 10, 19, 25, 26, 33). Interestingly, CPE were evident several days before superinfection by EET(TE)-109 virus was apparent. It seems plausible, therefore, that syncytium-associated cell killing by 81T-109 virus may be responsible for the initial CPE observed in EET(TE)-109 infection and that CPE associated with high levels of replication and reinfection may enhance cell killing at later times in infection. Thus, EET(TE)-109 virus may kill cells by two distinct mechanisms: single-cell killing and killing by cell fusion. This is in contrast to EECC, which appears to cause CPE in T cells by an as yet undefined mechanism of single-cell killing associated with rapid replication kinetics and/or superinfection.
We hypothesize that the predicted four-amino-acid insertion (GESQ) in the C-terminal portion of the 81T-109 SU could function as one cytopathic determinant because the principal cytopathic and pathogenic determinant of 61C includes a predicted six-amino-acid insertion (YLTAPR) in the same site (10, 27, 32). Similarly, other cytopathic FeLV-FAIDS variants (61B and 82K) also contain related but distinct predicted six-amino-acid insertions (YLAAPR and YRAAPR, respectively) in this site that likely serve as similar pathogenic determinants (27, 28). Taken together, such observations suggest that the primary sequence of the insertion may not be as important as the potential disruption an insertion might have on the envelope protein structure. It is possible that the specific sequences of the insertion contribute to the unique ability of 81T envelope to cause cell-cell fusion in T cells. Alternatively, there may be additional determinants for syncytium formation by 81T. Because the sequences of the 81T-109, 61C, and 61E env genes differ by very few nucleotides, it should be possible to map the determinant(s) for the T-cell killing and syncytium formation phenotypes and to determine whether they can be decoupled.
Studies of the interference properties of 61E and 81T viruses suggest that the SU of the 81T virus may recognize the 61E receptor as well as a second receptor protein. The fact that AH927 cells chronically infected with 81T-109 were able to completely block infection by pseudotyped viruses bearing the 61E envelope is strongly suggestive that these two viruses can utilize the same receptor molecule to enter fibroblast cells. However, EET(TE)-109 can enter 61E-infected cells that are completely resistant to an equivalent dose of homologous virus challenge, although there is only partial resistance to EET(TE)-109 infection. This result is consistent with a model whereby viruses with an 81T-109 envelope can gain entry by an alternative mechanism, such as binding to a lower-affinity secondary receptor that is not recognized by 61E. These data are less consistent with a model based on differences in envelope-receptor affinity for a single receptor because the complete resistance to 61E infection in cells expressing 61E SU strongly implies that the 61E receptor is no longer available for attachment on the surface of these cells. Moreover, if 81T SU had a higher affinity than 61E for a common receptor, we would expect cells infected with EET(TE)-109 to be resistant to infection mediated by both 81T and 61E envelope, and they are not. Rather, the fact that EET(TE)-109-infected cells can completely block 61E infection but not infection with homologous virus is consistent with a model of two receptors. Based on these data, we hypothesize that there is one common receptor for 61E and 81T SU, as well as a second receptor recognized for attachment and entry of virus encoding the 81T SU that is not blocked by 81T SU in a manner that promotes infection interference.
The inability of 81T to establish complete homologous virus interference is reminiscent of the properties of the immunodeficiency-inducing 61C variant. Viruses encoding the 61C envelope have a limited ability to establish superinfection interference against homologous virus challenge, and it has been suggested that the lack of interference against reinfection may contribute to the cytopathic properties of the 61C variant (10, 19, 24, 33). Because there is incomplete superinfection interference even against homologous challenge with 81T and 61C virus, our experiments designed to address the interference properties of 61C and 81T SU were inconclusive. Several studies have shown that 61E and 61C viruses have nonreciprocal interference patterns (19, 24, 33). Additional studies from our laboratory using a similar single-cycle infection assay to monitor viral entry suggest that the 61C and the 61E viruses recognize different receptors on AH927 cells (24). Taken together, our data are consistent with a model in which 81T-109 is a dualtropic virus that can recognize two different receptor proteins, whereas 61E and 61C are able to recognize only one or the other of these receptors. However, both kinetic analysis of viral replication and interference studies suggest that the 81T virus may not efficiently recognize the 61C receptor. In support of this model of dual-receptor recognition by FeLV, we have recently shown that while all subgroup B FeLV variants can recognize the Pit1 receptor, some variants of FeLV subgroup B can also enter cells by using a second receptor, Pit2 (4). These FeLV-B variants recognize the Pit2 receptor with reduced efficiency compared to the Pit1 receptor (4), which also provides a paradigm for the model proposed for the 81T variant. Thus, studies of FeLV-B viruses provide direct evidence for dual-receptor recognition by FeLV; by analogy, our model of dual-receptor recognition by 81T SU may be most clearly addressed when the receptor molecules are identified.
Both the 81T clones and the FeLV-FAIDS isolate, which was the original biological source of 61C, were derived from cat thymic lymphoma tissue. It is intriguing that both animals developed a lymphoproliferative disease, because 61C causes a lymphoid depletion disorder in infected cats, and our observation of EET(TE)-109-induced T-cell CPE strongly suggests that the 81T-109 virus would cause some T-cell-related immunosuppressive disorders as well. In this regard, it should be noted that cat 40681, from which 81T-109 was cloned, had other FeLV-associated changes, including proviral rearrangement and enhanced expression of flvi-2 (20, 21) and a population of recombinant FeLVs that had transduced a potential oncogene, feline Notch2 (34). Because immune surveillance is likely required to eliminate cells that have become transformed or malignant, an immunosuppressed state could facilitate tumor formation in cats, like 40681, that harbor FeLV-associated genetic rearrangements and potentially oncogenic viral variants. Indeed, two common complications in human HIV-infected individuals are Kaposi’s sarcoma and B-cell lymphoma (reviewed in reference 16). Similarly, some monkeys infected with simian immunodeficiency virus (SIV) develop B-cell lymphoma (reviewed in reference 11).
The experiments described here were designed, in part, to examine the properties of viral variants that are selected during the course of pathogenic retroviral infections. At present, our understanding of the aspects of viral fitness that drive evolution and selection in retroviral infection is limited. Our experiments suggest that T-cell cytopathic viruses bearing the 81T-109 envelope evolved in the cat from a noncytopathic virus, 61E. We speculate that functional changes in the 81T-109 envelope could have conferred a selective advantage in vivo to viruses that contained it. Viruses that could circumvent 61E interference, even at reduced efficiency, would be expected to be strongly selected in a cat persistently infected with 61E virus, because these variants could spread throughout the 61E-saturated target cell population.
These results complement and extend our knowledge of the somewhat paradoxical phenomenon whereby the cytopathicity of SIV and HIV tends to increase during the course of infection (3, 7, 12, 38, 41–43). Our system offers the advantage of studying this phenomenon using a relatively simple, molecularly defined retrovirus. Furthermore, studies using 61E as the inoculum are particularly valuable because this virus exemplifies the type of FeLV that is ubiquitous among cats and that is most frequently transmitted (9). Therefore, observations on the pathogenicity of variants that evolve from 61E may be more relevant than studies using acutely cytopathic laboratory-derived isolates that might otherwise be unlikely to be transmitted in nature.
ACKNOWLEDGMENTS
We thank Mike Linenberger for collaborative studies with cat Curly 40681, Maxine Linial for providing pCMVhph, A. Dusty Miller for providing LAPSN/PA317 cells and the PG13 virus, Christopher Grant (Custom Monoclonals) for supplying antibody C11D8, Dana DeVange for experimental assistance, and Cara Burns and Jason Kimata for critical reviews of the manuscript.
This research was supported by Public Health Service grant CA 51080. J.O. is a Scholar of the Leukemia Society of America. J.L.R. was supported in part by a Helen Riaboff Whitely Graduate Fellowship Award.
Footnotes
This work is dedicated to the memory of our colleague and friend, Samuel Rudolph Gwynn.
REFERENCES
- 1.Anderson K B, Skov H. Retrovirus-induced cell fusion is enhanced by protease treatment. J Gen Virol. 1989;70:1921–1927. doi: 10.1099/0022-1317-70-7-1921. [DOI] [PubMed] [Google Scholar]
- 2.Aronoff R, Linial M. Specificity of retroviral RNA packaging. J Virol. 1991;65:71–80. doi: 10.1128/jvi.65.1.71-80.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asjo B, Morfeldt-Manson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyo E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;2:660–662. [PubMed] [Google Scholar]
- 4.Boomer S, Eiden M, Burns C C, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor interactions. J Virol. 1997;71:8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns C, Moser M, Banks J, Alderete J, Overbaugh J. Identification and deletion of sequences required for feline leukemia virus RNA packaging and construction of a high-titer feline leukemia virus packaging cell line. Virology. 1996;222:14–20. doi: 10.1006/viro.1996.0393. [DOI] [PubMed] [Google Scholar]
- 6.Burns C C, Poss M L, Thomas E, Overbaugh J. Mutations within a putative cysteine loop of the transmembrane protein of an attenuated immunodeficiency-inducing feline leukemia virus variant inhibit envelope protein processing. J Virol. 1995;69:2126–2132. doi: 10.1128/jvi.69.4.2126-2132.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 8.De Noronha C M, Reinhart T A, Mullins J I. Generation and role of defective proviruses in cytopathic feline leukemia virus (FeLV-FAIDS) infections. J Virol. 1996;70:359–367. doi: 10.1128/jvi.70.1.359-367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donahue P R, Hoover E A, Beltz G A, Riedel N, Hirsch V M, Overbaugh J, Mullins J I. Strong sequence conservation among horizontally transmissible, minimally pathogenic feline leukemia viruses. J Virol. 1988;62:722–731. doi: 10.1128/jvi.62.3.722-731.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donahue P R, Quackenbush S L, Gallo M V, deNoronha C M C, Overbaugh J, Hoover E A, Mullins J I. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol. 1991;65:4461–4469. doi: 10.1128/jvi.65.8.4461-4469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feichtinger H, Kaaya E, Putkonen P, Li S L, Ekman M. Malignant lymphoma associated with human AIDS and with SIV-induced immunodeficiency in macaques. AIDS Res Hum Retroviruses. 1992;8:339–348. doi: 10.1089/aid.1992.8.339. [DOI] [PubMed] [Google Scholar]
- 12.Fenyö E M, Morfeldt-Manson L, Chiodi F, Lind B, von Gegerfelt A, Albert J, Olausson E, Asjö B. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol. 1988;62:4414–4419. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields-Berry S C, Halliday A L, Cepko C L. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc Natl Acad Sci USA. 1992;89:693–697. doi: 10.1073/pnas.89.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goff S, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant C K, Ernisse B J, Jarrett O, Jones F R. Feline leukemia virus envelope gp70 of subgroups B and C defined by monoclonal antibodies with cytotoxic and neutralizing functions. J Immunol. 1983;131:3042–3048. [PubMed] [Google Scholar]
- 16.Gurley R J. The clinical spectrum of HIV infection. In: Gallo R C, Wong-Staal F, editors. Retrovirus biology and human disease. San Diego, Calif: Academic Press; 1990. pp. 359–381. [Google Scholar]
- 17.Hardy W D. Feline oncoretroviruses. In: Levy J A, editor. The Retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 109–180. [Google Scholar]
- 18.Hoover E A, Mullins J I, Quackenbush S L, Gasper P W. Experimental transmission and pathogenesis of immunodeficiency syndrome in cats. Blood. 1987;70:1880–1892. [PubMed] [Google Scholar]
- 19.Kristal B S, Reinhart T A, Hoover E A, Mullins J I. Interference with superinfection and with cell killing and determination of host range and growth kinetics mediated by feline leukemia virus surface glycoproteins. J Virol. 1993;67:4142–4153. doi: 10.1128/jvi.67.7.4142-4153.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy L S, Lobelle-Rich P A, Overbaugh J. flvi-2, a target of retroviral insertional mutagenesis in feline thymic lymphosarcomas, encodes bmi-1. Oncogene. 1993;8:1833–1838. [PubMed] [Google Scholar]
- 21.Levy L S, Lobelle-Rich P A, Overbaugh J, Abkowitz J L, Fulton R, Roy-Burman P. Coincident involvement of flvi-2, c-myc, and novel env genes in natural and experimental lymphosarcomas induced by feline leukemia virus. Virology. 1993;196:892–895. doi: 10.1006/viro.1993.1553. [DOI] [PubMed] [Google Scholar]
- 22.Miller A D, Garcia J V, Lynch S N von, C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moser, M., C. Burns, S. Boomer, and J. Overbaugh. The host range and interference properties of two closely related feline leukemia virus variants suggest that they use distinct receptors. Virology, in press. [DOI] [PubMed]
- 25.Mullins J I, Chen C S, Hoover E A. Disease-specific and tissue-specific production of unintegrated feline leukaemia virus variant DNA in feline AIDS. Nature. 1986;319:333–336. doi: 10.1038/319333a0. [DOI] [PubMed] [Google Scholar]
- 26.Mullins J I, Hoover E A, Quackenbush S L, Donahue P R. Disease progression and viral genome variants in experimental feline leukemia virus-induced immunodeficiency syndrome. J Acquired Immune Defic Syndr. 1991;4:547–557. [PubMed] [Google Scholar]
- 27.Overbaugh J, Donahue P R, Quackenbush S L, Hoover E A, Mullins J I. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science. 1988;239:906–910. doi: 10.1126/science.2893454. [DOI] [PubMed] [Google Scholar]
- 28.Overbaugh J, Hoover E A, Mullins J I, Burns D P W, Rudensey L, Quackenbush S L, Stallard V, Donahue P R. Structure and pathogenicity of individual variants within an immunodeficiency disease-inducing isolate of FeLV. Virology. 1992;188:558–569. doi: 10.1016/0042-6822(92)90510-v. [DOI] [PubMed] [Google Scholar]
- 29.Overbaugh J, Rudensey L M. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J Virol. 1992;66:5937–5948. doi: 10.1128/jvi.66.10.5937-5948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park B H, Lavi E, Blank K J, Gaulton G N. Intracerebral hemorrhages and syncytium formation induced by endothelial cell infection with a murine leukemia virus. J Virol. 1993;67:6015–6024. doi: 10.1128/jvi.67.10.6015-6024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinter A, Chen T-E, Lowey A, Cortez N G, Silagi S. Ecotropic murine leukemia virus-induced fusion of murine cells. J Virol. 1986;57:1048–1054. doi: 10.1128/jvi.57.3.1048-1054.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quackenbush S L, Donahue P R, Dean G A, Myles M H, Ackley C D, Cooper M D, Mullins J I, Hoover E A. Lymphocyte subset alterations and viral determinants of immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol. 1990;64:5465–5474. doi: 10.1128/jvi.64.11.5465-5474.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinhart T A, Ghosh A K, Hoover E A, Mullins J I. Distinct superinfection interference properties yet similar receptor utilization by cytopathic and noncytopathic feline leukemia viruses. J Virol. 1993;67:5153–5162. doi: 10.1128/jvi.67.9.5153-5162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohn J, Lauring A, Linenberger M, Overbaugh J. Transduction of Notch2 in feline leukemia virus-induced thymic lymphoma. J Virol. 1996;70:8071–8080. doi: 10.1128/jvi.70.11.8071-8080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohn J L, Linenberger M L, Hoover E A, Overbaugh J. Evolution of feline leukemia virus variant genomes with insertions, deletions, and defective envelope genes in infected cats with tumors. J Virol. 1994;68:2458–2467. doi: 10.1128/jvi.68.4.2458-2467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohn J L, Overbaugh J. Pathogenic feline retroviruses: feline leukemia virus and feline immunodeficiency virus. In: Chen I S Y, Ahmed R, editors. persistent viral infections, in press. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 37.Rowe W P, Pugh W E, Hartley J W. Plaque assay techniques for murine leukemia viruses. Virology. 1970;42:1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- 38.Rudensey L M, Kimata J T, Benveniste R E, Overbaugh J. Progression to AIDS in macaques is associated with changes in the replication, tropism, and cytopathic properties of the simian immunodeficiency virus variant population. Virology. 1995;207:528–542. doi: 10.1006/viro.1995.1113. [DOI] [PubMed] [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Temin H M. Mechanisms of cell killing/cytopathic effects by nonhuman retroviruses. Rev Infect Dis. 1988;10:399–405. doi: 10.1093/clinids/10.2.399. [DOI] [PubMed] [Google Scholar]
- 41.Tersmette M, de Goede R E Y, Al B J M, Winkel I N, Gruters R A, Cuypers H T, Huisman H G, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolate: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tersmette M, Gruters R A, de Wolf F, de Goede R E Y, Lange J M A, Schellekens P T A, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tersmette M, Lange J M, de Goede R E, de Wolf F, Eeftink-Schattenkerk J K, Schellekens P T, Coutinho R A, Huisman J G, Goudsmit J, Miedema F. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;1:983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 44.Thomas E, Overbaugh J. Delayed cytopathicity of a feline leukemia virus variant is due to four mutations in the transmembrane protein gene. J Virol. 1993;67:5724–5732. doi: 10.1128/jvi.67.10.5724-5732.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White J. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 46.Wilson C A, Marsh J W, Eiden M V. The requirements for viral entry differ from those for virally induced syncytium formation in NIH 3T3/DTras cells exposed to Moloney murine leukemia virus. J Virol. 1992;66:7262–7269. doi: 10.1128/jvi.66.12.7262-7269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong P K Y, Yuen P H, Kaufman S J. Induction of syncytia by Moloney murine leukemia virus in myoblasts defective in differentiation. J Virol. 1977;21:319–327. doi: 10.1128/jvi.21.1.319-327.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]