Abstract
Recently identified human FOXP3lowCD45RA− inflammatory non-suppressive (INS) cells produce proinflammatory cytokines, exhibit reduced suppressiveness, and promote antitumor immunity unlike conventional regulatory T cells (Tregs). In spite of their implication in tumors, the mechanism for generation of FOXP3lowCD45RA− INS cells in vivo is unclear. We showed that the FOXP3lowCD45RA− cells in human tumors demonstrate attenuated expression of CRIF1, a vital mitochondrial regulator. Mice with CRIF1 deficiency in Tregs bore Foxp3lowINS-Tregs with mitochondrial dysfunction and metabolic reprograming. The enhanced glutaminolysis activated α-ketoglutarate–mTORC1 axis, which promoted proinflammatory cytokine expression by inducing EOMES and SATB1 expression. Moreover, chromatin openness of the regulatory regions of the Ifng and Il4 genes was increased, which facilitated EOMES/SATB1 binding. The increased α-ketoglutarate–derived 2-hydroxyglutarate down-regulated Foxp3 expression by methylating the Foxp3 gene regulatory regions. Furthermore, CRIF1 deficiency–induced Foxp3lowINS-Tregs suppressed tumor growth in an IFN-γ–dependent manner. Thus, CRIF1 deficiency–mediated mitochondrial dysfunction results in the induction of Foxp3lowINS-Tregs including FOXP3lowCD45RA− cells that promote antitumor immunity.
CRIF1 deficiency promotes the generation of Foxp3low INS-Tregs via increased glutaminolysis, thereby inhibiting tumor progression.
INTRODUCTION
Regulatory T cells (Tregs) are characterized by the specific expression of the master transcription factor forkhead box protein 3 (Foxp3) and the release of suppressive cytokines (1, 2). Consequently, Tregs play crucial roles in maintaining immune homeostasis and regulating various immune responses. In particular, Tregs can promote tumor progression and growth (3, 4) by suppressing antitumor CD4+ helper T cells and cytotoxic T cells (5, 6).
The Sakaguchi group has classified human Tregs as FOXP3loCD45RA+ naïve Tregs, FOXP3hiCD45RA− effector Tregs, and FOXP3loCD45RA− Tregs (7, 8). While the latter cells were initially thought to largely consist of nonregulatory cells that originate from conventional T cells (7, 8), several studies have shown that they can also originate from Tregs (9–12). Thus, the human FOXP3lowCD45RA− cells in the tumor microenvironment (TME) include both unstable FOXP3low Tregs and activated FOXP3low conventional T cells (non-Tregs) (2, 13). Notably, human FOXP3lowCD45RA− cells are characterized by proinflammatory cytokine production and poor suppressive ability (13, 14), which are similar to murine Foxp3low inflammatory non-suppressive (INS)–Tregs. These Tregs not only express low Foxp3 levels but also secrete proinflammatory cytokines and have reduced suppressive functions (15–20). These cells have also been observed in several settings, including TME, autoimmune conditions, and when the following molecules have been disrupted in mice: caspase recruitment domain-containing membrane-associated guanylate kinase protein 1, von Hippel-Lindau, TRAF3 interacting protein 3 (TRAF3IP3), suppressor of cytokine signaling 1 (SOCS1), and c-Rel (17–21). Significantly, these cells differ from conventional Tregs in that they promote antitumor immunity rather than suppress it (20, 21). Consistently, tumor-infiltrating FOXP3lowCD45RA− T cells are associated with better prognosis in patients with colorectal cancer (CRC) than FOXP3high Tregs (7, 8). Thus, human FOXP3lowCD45RA− T cells, like their similar subset of murine Foxp3low INS-Tregs, promote antitumor immunity. However, the mechanisms that govern the development of these cells in the TME remain to be clarified.
As shown by Treg-specific depletion studies in mice, the suppressive functions of Tregs depend on the metabolic sensor liver kinase B1 (22, 23) and mitochondrial regulatory proteins such as electron transporter complex I and III and mitochondrial transcriptional factor A (24–26). Thus, maintaining mitochondrial fitness is critical for the suppressive function of Foxp3+ Tregs (27, 28). However, whether mitochondria participate in the generation of Foxp3low INS-Tregs including human FOXP3lowCD45RA− cells has not been examined previously.
The present study suggests that mitochondrial function is involved in formation of Foxp3low INS-Treg including FOXP3lowCD45RA− cells. First, we observed that the FOXP3low cells in human TMEs show low expression of CR6-interacting factor 1 (CRIF1), which is a de novo component of the large mitoribosome subunit. It is essential for the translation of oxidative phosphorylation (OXPHOS) polypeptides in mammalian mitochondria and thus plays a key role in mitochondrial biogenesis and metabolism (29, 30). We then found that CRIF1 deficiency in Tregs induced mitochondrial metabolic reprogramming including glutaminolysis, which, in turn, promoted the emergence of Foxp3low Tregs that produced the proinflammatory cytokines interferon-γ (IFN-γ) and interleukin-4 (IL-4). The emergence of IFN-γ–secreting INS-Tregs in the TMEs inhibited tumor growth. Thus, our findings indicate that CRIF1 deficiency–mediated mitochondrial dysfunction results in the induction of Foxp3low INS-Tregs including FOXP3lowCD45RA− cells that promote antitumor immunity.
RESULTS
Human solid tumors are infiltrated with inflammatory FOXP3low T cells that express low levels of the mitochondrial regulator protein CRIF1
A recent study reported that, when human CRCs bear FOXP3low T cells, they have better patient prognosis than when they are infiltrated with the FOXP3high equivalent (8). To test this and to determine whether this is also true for other solid tumors, the FOXP3 expression of the immune cells in the TMEs of 197 non–small cell lung cancer (NSCLC) and 280 CRC samples from patients was determined by immunohistochemistry (fig. S1A). High ratios of FOXP3low cells to total FOXP3+ cells in the TME were associated with high overall survival in both patients with NSCLC and CRC (Fig. 1, A and B, left plots), including when only the large (>5 cm) NSCLCs and pT3 microsatellite-stable CRCs and were examined (Fig. 1, A and B, right plots).
Fig. 1. Human cancers contain inflammatory FOXP3low CD4+ T cells that demonstrate down-regulated mitochondrial biogenesis, including low expression of the mitochondrial regulatory protein CRIF1.
(A and B) Overall survival of NSCLC (left, total cases; right, tumor size ≥ 5 cm) or CRC (left, total cases; right, pT3 microsatellite-stable). (C to K) Analysis of publicly available scRNA-seq data on the TILs in human NSCLCs and CRCs. (C) and (D) Uniform manifold approximation projection (UMAP) visualization of the TILs in the NSCLCs and CRCs. (E) and (F) Volcano plots comparing FOXP3low CD4+ and FOXP3high CD4+ TILs in the NSCLCs and CRCs. (G) and (H) Heatmap showing the gene set variation analysis (GSVA) scores in the TILs from the NSCLCs and CRCs. H, Hallmark; K, Kyoto Encyclopedia of Genes and Genomes; G, Gene Ontology; R, Reactome. TCA, tricarboxylic acid. (I) and (J) Heatmap showing the mitochondrial biogenesis-related gene expressions in the NSCLCs and CRCs. (K) Venn diagram showing down-regulated genes in the heatmap. (L to S) Flow cytometric analysis of the CD4+ T cell subpopulations in human NSCLCs. (L) and (M) Five fractions were defined (n = 27). (N) Frequencies of IFN-γ+ (n = 38), IL-4+ (n = 52), or IL-17A+ (n = 59) cells. (O) and (P) Mitochondrial biogenesis in FOXP3low and FOXP3high CD4+ T cells (n = 18). MFI, mean fluorescence intensity. (Q) and (R) CRIF1 expression in FOXP3low and FOXP3high CD4+ T cells (n = 22). (S) Correlation between CRIF1 and FOXP3 expression in CD4+ T cells (NSCLCs, n = 16; CRCs, n = 18). Dots represent individual cases. The data are pooled from at least three independent experiments and are presented as means ± SEM of biological replicates. Flow cytometry plots are representative of at least two independent experiments. ns, not significant; ***P < 0.001 and ****P < 0.0001. Statistical testing was conducted with the Kaplan-Meier method and the log-rank test (A), Wilcoxon test [(N), (P), and (R)], and Pearson correlation test (S).
The main cellular functions and metabolism of the FOXP3low and FOXP3high CD4+ T cells in human NSCLCs and CRCs were then compared by analyzing publicly available single-cell RNA sequencing (scRNA-seq) data (31, 32). Compared to the FOXP3high CD4+ T cells, the FOXP3low CD4+ T cells exhibited low expression of OXPHOS-related genes (e.g., MT-CO1, MT-ND4, and MT-CYB), low OXPHOS pathway scores, and high expression of proinflammatory cytokine genes (IFNG and IL17A) (Fig. 1, C to H, and fig. S1, B and C). Of all the mitochondrial biogenesis-related differentially expressed genes (DEGs) in the FOXP3low CD4+ T cells (Fig. 1, I and J), GADD45GIP1 was a DEG in both the NSCLCs and CRCs (Fig. 1K). GADD45GIP1 is known as CRIF1.
To see whether these scRNA-seq findings were recapitulated in our NSCLC (n = 18 to 27) and CRC (n = 13 to 27) cohorts, we identified the FOXP3low and FOXP3high CD4+ T cells in the tumor-infiltrating lymphocytes (TILs) by flow cytometry using antibodies against FOXP3 and CD45RA. As described previously, this analysis revealed six TIL fractions that have been denoted I to VI (Fig. 1L and fig. S1D) (7). Fractions IV to VI lack FOXP3 expression, while fractions I, II, and III may, respectively, be the FOXP3lowCD45RA+ naive Tregs, the tumor-promoting FOXP3highCD45RA− effector Tregs, and the FOXP3lowCD45RA− subset described previously (8). On the basis of the literature, the latter could be unstable FOXP3low Tregs and/or activated FOXP3low non-Treg conventional T (Tconv)cells (2, 13). The FOXP3lowCD45RA− subset was the most frequent of the infiltrating FOXP3-expressing CD4+ T cell fractions in both NSCLCs and CRCs; the naive Tregs were infrequent (Fig. 1M and fig. S1E). Consistent with the scRNA-seq data above, the FOXP3low CD4+ T cells from NSCLCs and CRCs produced more proinflammatory cytokines (IFN-γ, IL-4, and IL-17A) than the FOXP3high CD4+ T cells (Fig. 1N and fig. S1F). They also exhibited lower mitochondrial biogenesis, as indicated by low mitochondrially encoded cytochrome c oxidase I (MT-CO1)/succinate dehydrogenase complex flavoprotein subunit A (SDHA) ratio and CRIF1 expression (Fig. 1, O to R, and fig. S1, G to J). Moreover, the FOXP3 expression in the tumor CD4+ T cells was correlated positively with CRIF1 expression (Fig. 1S), meaning that the FOXP3lowCD4+ T cells in the TME of solid lung and colorectal tumors expressed low levels of CRIF1. Thus, CRIF1-mediated mitochondrial functions may participate in the induction of inflammatory FOXP3lowCD45RA− fraction III cells in the TMEs of different human solid tumors.
CRIF1 deficiency induces Foxp3low INS-Tregs that spontaneously generate inflammation
To test whether CRIF1 contributes to the induction of Foxp3low INS-Tregs in vivo, Crif1fl/flFoxp3YFP-Cre [hereafter referred to as CRIF1–conditional knockout (CRIF1-cKO)] mice were generated. Compared to Foxp3YFP-Cre control mice, the CRIF1-cKO mice showed reduced body size and weight, hair loss, and skin erosion (fig. S2, A and B), as well as leukocyte infiltration in various organs, including the ear skin, liver, and lungs (fig. S2C), and died before 9 weeks of age (fig. S2D). Moreover, lymph nodes (LNs) and spleen were enlarged and demonstrated increased total immune cell numbers, while the thymus was smaller and had fewer cells (fig. S2, E and F). The splenic and LN CD4+ T cells from the CRIF1-cKO mice showed increased IFN-γ and IL-4 production (fig. S2G), higher frequencies of memory effector CD4+ T cells, and lower frequencies of naive CD4+ T cells (fig. S2H). Thus, CRIF1 in Tregs plays critical role in maintaining immune cell homeostasis.
With regard to the Tregs, CRIF1-cKO mice showed higher Treg frequencies in the LNs and thymus but not the spleen and higher absolute Treg numbers in the LNs but not the thymus or spleen (Fig. 2, A and B, and fig. S2J). Significantly, the LN and splenic Tregs from CRIF1-cKO mice spontaneously expressed less Foxp3 and exhibited higher percentages of Foxp3low cells than the control Tregs, although this difference was not observed in the thymic Tregs (Fig. 2, A and C, and fig. S2I). Compared to control Tregs from Foxp3YFP-Cre mice, the CRIF1-deficient Tregs did not suppress the proliferation of effector T cells (Fig. 2, D and E). They also demonstrated higher production of IFN-γ and IL-4 upon phorbol 12-myristate 13-acetate (PMA)/ionomycin stimulation (Fig. 2, F and G) or T cell receptor (TCR) stimulation with anti-CD3/CD28 (Fig. 2H). The CRIF1-deficient Tregs also showed spontaneously up-regulated expression of trasncription factors such as T-bet and Gata-3 and representative markers of Tregs including CD25, CD103, and ICOS but down-regulated programmed cell death protein 1 (PD-1) expression (fig. S2, K and L). In CRIF1-cKO mice, minimal Foxp3-YFP expression was detected in the conventional Foxp3− CD4+ and CD8+ T cells. In addition, CRIF1 was not expressed in Tregs, while it exhibited normal expression in conventional T cells (fig. S2, M and N). This indicates that the earlier findings were a result of CRIF1 deletion specifically in Tregs.
Fig. 2. CRIF1-deficient Tregs exhibit the distinctive characteristics of INS-Tregs.
Comparison of Crif1fl/flFoxp3YFP-Cre and control Foxp3YFP-Cre mice. (A to H) Foxp3-YFP+ Treg phenotypes. (A) and (B) Treg frequencies in the splenic and LN CD4+ T cell population [left two plots in (B)] and absolute Treg numbers [right two plots in (B)]. (C) Foxp3-YFP expression intensity in splenic and LN Foxp3-YFP+ Tregs. (D) and (E) Ability of Tregs to suppress effector T cell proliferation. (F) and (G) Frequencies of splenic Tregs that spontaneously produce IFN-γ, IL-4, and IL-17A. (H) Cytokine production of Tregs that underwent TCR stimulation for 2 days, as shown by enzyme-linked immunosorbent assay of the supernatants. (I to N) Adoptive transfer of Foxp3-YFP+ Tregs into Rag1-KO mice that did or did not lack Ifng and IL4 expression. (I) Schematic depiction of the experiment. (J) Foxp3-YFP expression intensity in the total CD4+ T cells in the spleen and LNs. (K) Frequencies of Tregs in the spleen and LNs that produced IFN-γ, IL-4, or IL-17A. (L) Gross appearance of the spleen and LNs. (M) Absolute cell numbers in the spleen and LNs. (N) Representative histological images of the ear skin, lung, and liver. Scale bars, 200 μm (for the lungs) and 100 μm (for the ear skin and liver). Dots represent individual mice (n = 4 to 13 per group). The data are pooled from at least two independent experiments and are presented as means ± SEM of biological replicates. Gross and histological images and flow cytometry plots are representative of at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Statistical testing was conducted with an unpaired two-tailed t test [(B), (C), (E), (G), and (H)], and one-way analysis of variance (ANOVA) [(J), (K), and (M)]. i.v., intravenous.
To determine whether the Foxp3lowCRIF1-deficient Tregs that spontaneously expressed IFN-γ and IL-4 originated from Foxp3 expressing Tregs, we first used the ROSA26tdTomato fate–mapping mouse system. In these mice, all cells originating from parent Tregs express tdTomato, and they can be divided into Tregs that maintain Foxp3 expression (Foxp3-YFP+tdTomato+ cells) and those that have lost Foxp3 expression (Foxp3-YFP−tdTomato+ cells, referred to as ex-Tregs). The effector T cells are Foxp3-YFP−tdTomato− cells. Compared to control Foxp3YFP-Cre mice, the CRIF1-cKO mice had similarly low frequencies of Tregs but much higher frequencies of ex-Tregs; this was observed in both the spleen and LNs (fig. S3, A and B). Moreover, both the Tregs and ex-Tregs were much more likely to spontaneously express IFN-γ or IL-4 when they came from CRIF1-cKO mice (fig. S3, C and D).
Next, we created mixed bone marrow chimera mice that bore wild-type (WT) CD45.1+ Tregs and CD45.2+ CRIF1-deficient Tregs. In these mixed bone marrow chimera mice, CD4+ T cells were equally derived from bone marrow cells of both WT and knockout (KO) mice (fig. S4, A and B). The CD45.2+ CRIF1-deficient Tregs exhibited significantly lower expression of Foxp3 than the WT Tregs in both spleen and LNs (fig. S4, C and D) but were more prone to express IFN-γ and IL-4 (fig. S4, E and F). In contrast, no alterations were observed in thymic Tregs and Foxp3−CD4+ conventional T cells in the thymus, spleen, and LNs (fig. S4, C to F). Thus, CRIF1 deficiency leads to the development of Foxp3low INS-Tregs in the periphery without affecting the initial development of Tregs.
To confirm the proinflammatory activity of CRIF1-deficient Tregs, Foxp3-YFP+ Tregs were sorted from CRIF1-cKO and Foxp3YFP-Cre mice and adoptively transferred into Rag1-KO mice (Fig. 2I). Compared to the control Tregs from Foxp3YFP-Cre mice, the Tregs from CRIF1-cKO mice showed down-regulated Foxp3 expression in the spleen and LNs (Fig. 2J and fig. S4G). Moreover, they demonstrated up-regulated proinflammatory cytokine production (Fig. 2K). This inflammation associated with markedly increased spleen and LN size in the Rag1-KO mice (Fig. 2L). To test whether the excessive Treg production of IFN-γ and IL-4 was responsible, the CRIF1-cKO mice were crossed with Ifng KO and/or Il4 KO mice, and the Tregs from these triple-KO mice were adoptively transferred into Rag1-KO mice. Compared to CRIF1-cKO Treg transfer, triple-KO Treg transfer yielded significantly less spleen/LN augmentation and inflammation in the skin, lung, and liver, whereas the transfer of double-KO Tregs from Ifng KO CRIF1-cKO and Il4 KO CRIF1-cKO mice did not alter the inflammation (Fig. 2, I to N, and fig. S4H). Thus, CRIF1 deficiency induces Foxp3low INS-Tregs that promote spontaneous inflammation in an IFN-γ– and IL-4–dependent manner in vivo.
CRIF1-deficient Foxp3low INS-Tregs exhibit metabolic reprogramming characterized by increased glutaminolysis
Analysis of mitochondria in Tregs from both control Foxp3YFP-Cre and CRIF1-cKO mice revealed a notable decrease in various parameters due to CRIF1 deficiency. This reduction manifested in diminished mitochondrial biogenesis (Fig. 3, A and B), reduced mitochondrial mass (Fig. 3, C and D), impaired mitochondrial translation (Fig. 3, E and F), lower levels of electron-transport complexes (Fig. 3, G and H), and decreased adenosine triphosphate (ATP) production (Fig. 3I). Together, these findings indicate the presence of mitochondrial dysfunction in CRIF1-deficient Tregs. Their morphology was also aberrant (Fig. 3J), and they demonstrated increased mitochondrial reactive oxygen species and membrane potential (Fig. 3, C and D). Seahorse analysis indicated that the CRIF1-deficient Tregs had a low oxygen consumption rate (OCR) but a high extracellular acidification rate (ECAR) (Fig. 3K). Consistent with this, the CRIF1-deficient Tregs demonstrated increased 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl) amino]-D-glucose (2-NBDG) uptake and Glut1 expression after TCR stimulation (Fig. 3, L and M). Thus, CRIF1 deficiency in Tregs impaired the mitochondria, namely, it reduced biogenesis and translation and reprogrammed Treg metabolism toward glycolysis.
Fig. 3. CRIF1-deficient Tregs exhibit mitochondrial dysfunction. Splenic Foxp3-YFP+ Tregs from Crif1fl/flFoxp3YFP-Cre and control Foxp3YFP-Cre mice were assessed.
(A and B) Mitochondrial biogenesis was measured by determining the MT-CO1/SDHA ratio by flow cytometry. (C and D) Mitochondrial reactive oxygen species, mass, and membrane potential were measured by flow cytometry with MitoSOX, MitoTracker Red, and MitoTracker Orange, respectively. (E and F) Immunofluorescence staining to determine mitochondrial translation. The incorporation of l-homopropargylglycine (HPG) was calculated as a ratio of HPG intensity to the TOM20 stained mitochondrial area. Scale bars, 5 μm; a.u., arbitrary unit. (G) Western blot to measure the mitochondrial electron transport complex levels. (H) Flow cytometric expression of the mitochondrial complex I protein MT-ND1 and complex V protein ATP5A1. (I) Adenosine triphosphate (ATP) levels measured by mass spectrometry (MS). (J) Representative images of electron microscopy showing the morphology of the mitochondria. (K) Seahorse analysis of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone. (L and M) Flow cytometric 2-NBDG uptake and GLUT1 expression after TCR stimulation. Dots represent individual mice (n = 3 to 6 per group). Data are pooled from at least two independent experiments and are presented as means ± SEM of biological replicates. Flow cytometry plots, electron microscopy images, immunofluorescence images, and Western blot images are representative of at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Statistical testing was conducted with an unpaired two-tailed t test.
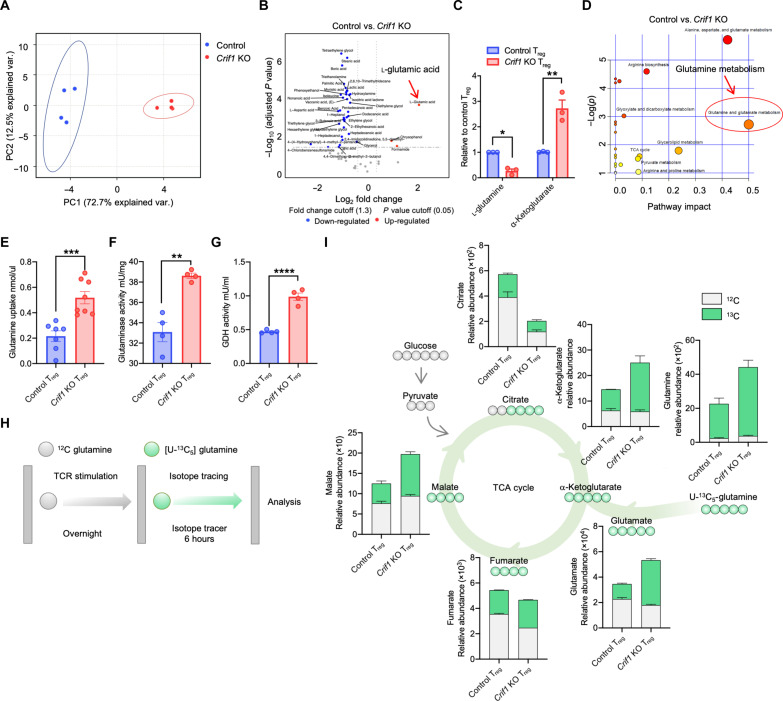
To investigate how CRIF1 regulates the metabolic fitness in Tregs, the metabolites in the Tregs from CRIF1-cKO and control Foxp3YFP-Cre mice were measured. Principal components analysis revealed substantial differences between the two Treg types (Fig. 4A). Untargeted and targeted analyses showed, respectively, that CRIF1-deficient Tregs had higher l-glutamic acid and α-ketoglutarate (α-KG) levels (Fig. 4, B and C, and fig. S5A). Moreover, pathway analysis showed that glutamine metabolism was higher in CRIF1-deficient Tregs (Fig. 4D). The CRIF1-deficient Tregs exhibited enhanced glutamine uptake and glutaminase and glutamine dehydrogenase enzyme activity (Fig. 4, E to G) despite the fact that the two cell types had similar expression levels of glutamine transporters and glutaminolysis-related enzymes (fig. S5, B and C). This was consistent with stable isotope-tracing analysis, which showed that CRIF1-deficient Tregs exhibited increased conversion of uniformed labeled [U-13C5] l-glutamine to α-KG (Fig. 4, H and I). Thus, the glutaminolysis pathway is enhanced in CRIF1-deficient Foxp3low INS-Tregs.
Fig. 4. CRIF1-deficient Tregs demonstrate up-regulation of the glutaminolysis pathway. FACS-sorted Foxp3-YFP+ Tregs from Foxp3YFP-Cre and Crif1fl/flFoxp3YFP-Cre mice were assessed.
(A to D) Metabolite analysis. (A) Principal components analysis. (B) Untargeted metabolite analysis. (C) Targeted metabolite analysis. Two metabolites related to the glutaminolysis pathway were analyzed. (D) Pathway analysis. Red circle and arrow indicate glutamine metabolism. (E to G) Glutaminolysis pathway–related enzyme activity and glutamine uptake. (E) Glutamine uptake. (F) Glutaminase activity. (G) Glutamate dehydrogenase (GDH) activity. (H and I) 13C-stable isotope tracer studies. (H) Schematic depiction of the isotope tracing of metabolites. (I) [U-13C5] glutamine isotope tracing. Dots represent technical replicates (C) or individual mice (E to G) (n = 4 to 8 per group). The data are pooled from at least two independent experiments and are presented as means ± SEM of biological replicates [(C) and (E) to (G)]. Alternatively, they represent at least two independent experiments (I). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Statistical testing was conducted with an unpaired two-tailed t test.
The increased proinflammatory cytokine production by CRIF1-deficient Foxp3low INS-Tregs reflects α-KG–mTORC1 axis–mediated up-regulation of EOMES and SATB1 expression
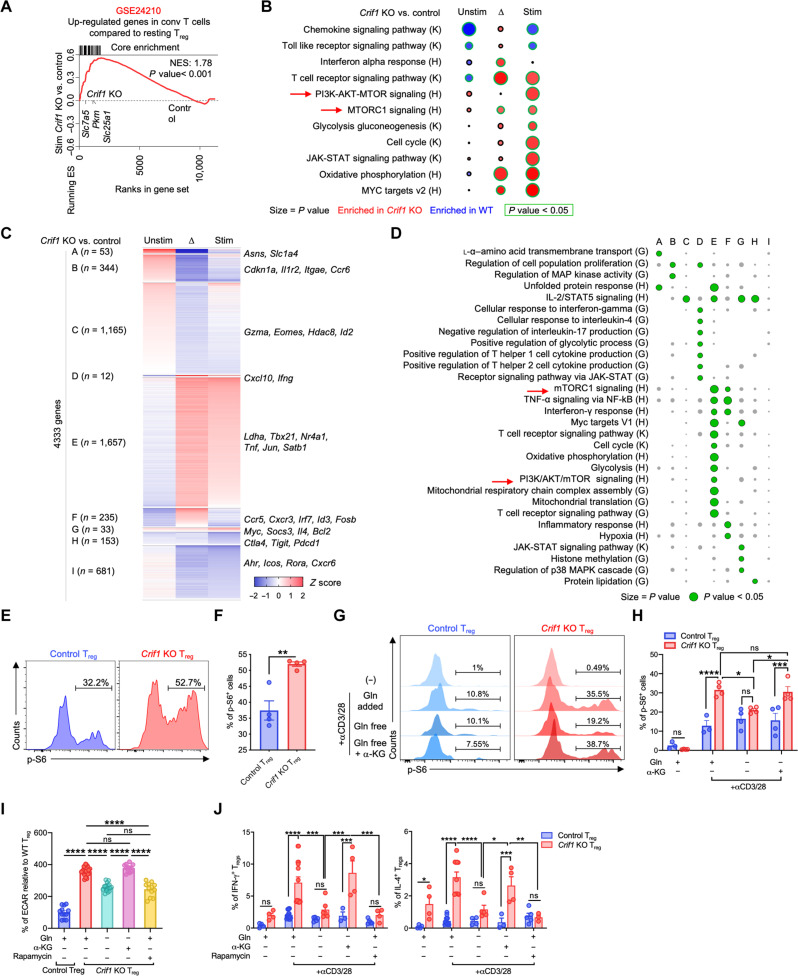
To investigate the link between the metabolic reprograming and proinflammatory functions of CRIF1-deficient Tregs, the Tregs from CRIF1-cKO and control Foxp3YFP-Cre mice underwent TCR stimulation or were left unstimulated, and the four cell types were subjected to transcriptome analysis (fig. S6, A to G). Gene set enrichment analysis (GSEA) revealed an enrichment of gene sets displaying up-regulated expression in conventional CD4+ T cells within CRIF1-deficient Tregs compared to control Tregs under TCR stimulation. This implies that the global gene expression patterns of CRIF1-deficient Tregs were more closely resemble those of conventional CD4+ T cells, particularly in contrast to control Tregs (Fig. 5A) (33). Bulk RNA-seq and self-organizing map (SOM) group analysis showed that the mammalian target of rapamycin (mTOR) pathway, which plays critical role in the metabolic reprograming in Tregs, (34) was significantly up-regulated in TCR-stimulated CRIF1-deficient Tregs relative to TCR-stimulated control Tregs (Fig. 5, B to D). Flow cytometric analysis then showed that TCR-stimulated CRIF1-deficient Tregs had significantly higher phosphorylated S6 (p-S6) ribosomal protein levels than stimulated control Tregs (Fig. 5, E and F). p-S6 is a marker of mTOR pathway activity (34). Moreover, incubating the CRIF1-deficient Tregs in glutamine-free medium reduced elevated p-S6 levels, ECAR, and IFN-γ and IL-4 production, and these effects were reversed by adding α-KG. However, these treatments had minimal effects on the same functions in control Tregs (Fig. 5, G to J). Treating the TCR-stimulated CRIF1-deficient Tregs with the mTOR inhibitor rapamycin also decreased ECAR as well as IFN-γ and IL-4 production, whereas it had little effect on control Tregs (Fig. 5, I and J). Several studies suggest that the phosphoinositide 3-kinase (PI3K)–AKT-forkhead box O (FOXO) axis regulates the IFN-γ production in Tregs (35, 36). However, the levels of p-AKT were comparable in CRIF1-deficient and WT Tregs (fig. S6, H and I). Thus, the glutaminolysis-mTOR axis regulates the proinflammatory cytokine production of CRIF1-deficient Tregs.
Fig. 5. The α-KG–mTORC1 axis regulates the proinflammatory cytokine production in CRIF1-deficient Tregs.
Foxp3-YFP+ Tregs from Foxp3YFP-Cre and Crif1fl/flFoxp3YFP-Cre mice were assessed. (A to D) Transcriptome analysis of TCR-stimulated and unstimulated Tregs. (A) and (B) Enrichment plot for showing overall character of CRIF1-deficient Tregs (A) and bubble plot (B) for gene set enrichment analysis (GSEA) of bulk RNA-seq data. ES, enrichment score; NES, normalized enrichment score. In (B), the bubbles encircled with green had P values of <0.05. Unstim, unstimulated; stim, overnight TCR stimulation; ∆, stim/unstim value. K, Kyoto Encyclopedia of Genes and Genomes KEGG; H, Hallmark. (C) and (D) Heatmap of self-organizing map (SOM)–clustered RNA-seq data (C) and bubble plot of the GSEA of each cluster (D). G, Gene Ontology biological process. TNF-α, tumor necrosis factor–α; JAK-STAT, Janus kinase–signal transducer and activator of transcription; MAP, mitogen-activated protein; MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor κB. (E and F) Percentages of phosphorylated S6+ (p-S6+) cells after TCR stimulation. (G to J) Effect of culture in glutamine-free RPMI 1640, adding 2 mM glutamine (Gln), 1 mM α-KG, or treatment with 100 nM rapamycin on TCR-stimulated functions. (G) and (H) Percentages of cells with p-S6 expression. (I) ECAR levels after d-(+)-glucose treatment. (J) The percentages of Tregs with cytokine production. The data are pooled from at least two independent experiments and are presented as means ± SEM of biological replicates. Flow cytometry plots are representative of at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Statistical testing was conducted with an unpaired two-tailed t test (F), two-way ANOVA [(H) and (J)], and one-way ANOVA (I).
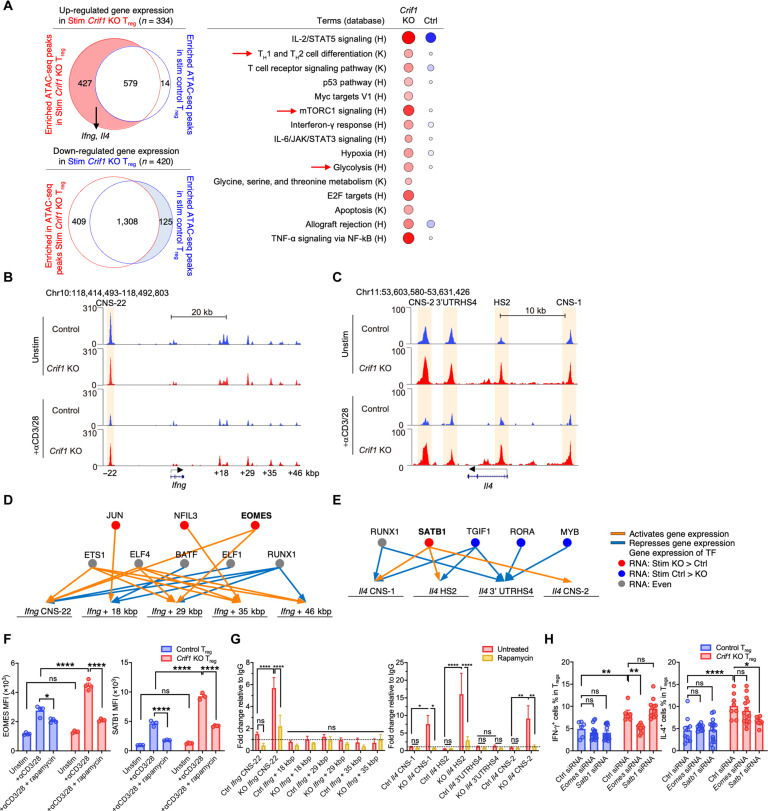
We next investigated the impact of CRIF1 deficiency on chromatin openness in Tregs. Thus, TCR-stimulated and unstimulated Tregs from CRIF1-cKO and control Foxp3YFP-Cre mice were subjected to assays for transposase accessible chromatin sequencing (ATAC-seq). The CRIF1-deficient Tregs displayed more differential accessible ATAC peaks compared to control Tregs regardless of whether they underwent TCR stimulation (fig. S6, J to M). Thus, CRIF1 deficiency increased Treg chromatin openness in general and in the regulatory regions of genes. Integration of the ATAC peaks corresponding to the genes that were up-regulated in TCR-stimulated CRIF1-deficient and control Tregs showed that, of the 1020 ATAC peaks identified, 41.9% were enriched in only the TCR-stimulated CRIF1-deficient Tregs, and these were associated with the mammalian target of rapamycin complex 1 (mTORC1) pathway, glycolysis, and proinflammatory cytokines such as Ifng and Il4 (Fig. 6A). Consistent with these findings, the ATAC peaks that were enriched in the stimulated CRIF1-deficient Tregs relative to stimulated control Tregs were associated more strongly with consensus-binding motifs for the inflammation-related transcription factors FOS, JUN, and activator protein-1 (AP-1) (fig. S6N) (37).
Fig. 6. The up-regulated IFN-γ and IL-4 gene transcription in CRIF1-deficient Tregs is associated with chromatin openness and transcription-factor expression.
Foxp3-YFP+ Tregs from Foxp3YFP-Cre and Crif1fl/flFoxp3YFP-Cre mice were assessed. (A to E) Integration of RNA-seq and ATAC-seq data of TCR-stimulated and unstimulated Tregs. (A) Left: Venn diagram showing the number of ATAC peaks that corresponded to genes that were up-regulated (top) or down-regulated (bottom) in the TCR-stimulated CRIF1-deficient (red) and control (blue) Tregs. (Right) Gene Ontology analysis. The size and color density of circles represent the number of genes and P values, respectively. (B) and (C) ATAC-seq peaks in and around the Ifng (B) and (C) Il4 genes in TCR-stimulated and unstimulated Tregs. (D) and (E) Modeling of the binding of transcription factors to the Ifng (D) and Il4 (E) genes. (F to H) Relationship between EOMES/SATB1 and IFN-γ/IL-4 expression. (F) Flow cytometry of EOMES and SATB1 expression in TCR-stimulated and unstimulated Tregs. Rapamycin was added to some TCR-stimulated Tregs. Dashed line indicates immunoglobulin G (IgG) control. (G) ChIP-qPCR analysis of the binding of EOMES and SATB1 to the Ifng and Il4 regulatory regions in TCR-stimulated Tregs. Rapamycin was added to some cells. (H) Flow cytometry analysis of the cytokine production of TCR-stimulated Tregs after Eomes or Satb1 were knocked down. The data are presented as means ± SEM of biological replicates and are representative of at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Statistical testing was conducted with two-way ANOVA [(F) to (H)].
Examination of the ATAC-seq data relating to the Ifng and Il4 genes revealed that the CRIF1-deficient Tregs showed more chromatin openness than the control Tregs in conserved noncoding DNA sequence 22 (CNS22), which is the region of the Ifng gene that potentially regulates IFN-γ transcription (38); however, this was only observed after TCR stimulation (Fig. 6B). Relative to the control Tregs, the CRIF1-deficient Tregs also demonstrated increased chromatin openness in the regions that regulate Il4 (39); this was noted in both the absence and presence of TCR stimulation (Fig. 6C). Thus, the differential chromatin openness mediated by CRIF1 may contribute to the IFN-γ and IL-4 production of Tregs.
Notably, deleting the key transcription factors for IFN-γ and IL-4 (T-bet and Gata-3, respectively) did not affect the production of these cytokines in CRIF1-deficient Tregs (fig. S6O). On the basis of previous chromatin immunoprecipitation (ChIP) sequencing data (40–42), we speculated that the transcription factors eomesodermin (EOMES) and special AT-rich sequence-binding protein 1 (SATB1) could be responsible for the IFN-γ and IL-4 production in CRIF1-deficient Tregs, respectively. Combined analysis using RNA-seq and ATAC-seq data, we noted that EOMES and SATB1, respectively, associated with the activated Ifng and Il4 expression in the CRIF1-deficient Tregs (Fig. 6, D and E). Consistent with this, CRIF1-deficient Tregs expressed more EOMES and SATB1 than control Tregs in both the absence and presence of TCR stimulation (Fig. 6F and fig. S7A). Moreover, ChIP–quantitative polymerase chain reaction (qPCR) showed that, after TCR stimulation, EOMES and SATB1 bound more often to the regulatory regions of Ifng and Il4 in CRIF1-deficient Tregs than in control Tregs (Fig. 6G). Small interfering RNA (siRNA) knockdown of Eomes and Satb1, achieving 50 to 60% efficiency, led to a decrease in the PMA/ionomycin-stimulated production of IFN-γ and IL-4 by CRIF1-deficient Tregs (Fig. 6H and fig. S7B). Notably, rapamycin treatment reduced the EOMES and SATB1 expression and their binding to Ifng and Il4 regulatory regions in CRIF1-deficient Tregs (Fig. 6, F and G). Thus, the α-KG–mTORC1 axis promotes the proinflammatory cytokine production in CRIF1-deficient Foxp3low INS-Tregs by up-regulating EOMES and SATB1 expression, thereby increasing their binding to the regulatory regions of the cyokine genes.
Foxp3 down-regulation in CRIF1-deficient Foxp3low INS-Tregs associates with high intracellular 2-HG and Foxp3 regulatory region methylation
The lower Foxp3 expression of the TCR-stimulated CRIF1-deficient Tregs was not due to changes in the glutamine–α-KG axis (fig. S8A) or chromatin openness: The control Tregs demonstrated similar chromatin openness in the Foxp3 regulatory region regardless of TCR stimulation (fig. S8B). However, analysis of the methylation status showed that TCR-stimulated CRIF1-deficient Tregs exhibited increased methylation in the Foxp3 promoter and CNS1 (fig. S8C). Thus, increased methylation may contribute to the reduced expression of Foxp3 in CRIF1-deficient Tregs.
Cellular levels of the metabolites 2-hydroxyglutarate (2-HG), S-adenosylmethionine (SAM), and S-adenosylhomocysteine (SAH) can shape the degree of DNA methylation (43, 44). Compared to control Tregs, CRIF1-deficient Tregs had higher levels of 2-HG and lower levels of SAM and SAH (fig. S8D). Upon treatment with 2-HG, control Tregs demonstrated heightened methylation in the Foxp3 promoter and CNS1 comparable to CRIF1-deficient Tregs. However, 2-HG did not alter cytokine production by control Tregs or affect methylation in CRIF1-deficient Tregs, and CNS2 methylation was increased only in conventional CD4+ T cells (fig. S8, E and F). These findings suggest that the elevated levels of 2-HG down-regulate Foxp3 expression in CRIF1-deficient Foxp3low INS-Tregs by modulating methylation in the Foxp3 promoter and CNS1.
Furthermore, to explore the potential correlation between decreased Foxp3 expression and heightened proinflammatory cytokine production in CRIF1-deficient Tregs, we used a green fluorescent protein (GFP)–tagged retrovirus (RV) transduction system to enhance Foxp3 expression (fig. S8, G and H). While the IFN-γ production of control Tregs decreased following Foxp3 overexpression, CRIF1-deficient Tregs exhibited no impact on proinflammatory cytokine production in the Foxp3 overexpression system, and there was no alteration in EOMES and SATB1 expression (fig. S8, I and J). These data suggest that the diminished Foxp3 expression and the increased proinflammatory cytokine production are independently regulated in CRIF1-deficient Tregs.
CRIF1 deficiency–induced Foxp3low INS-Tregs promote antitumor immunity in an IFN-γ–dependent manner
To investigate whether CRIF1 deficiency induces Foxp3low INS-Tregs in TME, WT C57BL/6 mice were inoculated with mouse TC-1 (a lung cancer cell line) or MC-38 (a colorectal adenocarcinoma cell line) tumor cells, and the tumors were subjected 17 days later to digestion and flow cytometry. The murine TMEs bore Foxp3low Tregs that demonstrated low mitochondrial biogenesis and CRIF1 expression and higher proinflammatory cytokine production relative to the infiltrating Foxp3high Tregs (Fig. 7, A to E, and fig. S9, A to E). Thus, Foxp3low INS-Tregs infiltrated both tumor types.
Fig. 7. CRIF1-deficient Foxp3low INS-Tregs promote IFN-γ–dependent antitumor immunity.
(A to E) Solid tumors were induced in WT mice with TC-1 tumor cells and the infiltrating CD4+ T cells were subjected to flow cytometry. (A) Foxp3 expression of CD4+ T cells. (B) and (C) Mitochondrial biogenesis. (D) and (E) CRIF1 expression. (F to P) Solid tumors were induced in Foxp3EGFP-cre-ERT2 and Crif1fl/flFoxp3EGFP-cre-ERT2 mice with TC-1 cells and the mice were treated intraperitoneally with tamoxifen. (F) Treatment schedule. (G) and (H) Foxp3 expression intensity of the tumor-infiltrating Tregs. (I) and (J) Cytokine production of the tumor-infiltrating Tregs. (K) and (L) Foxp3 expression intensity (K) and cytokine production (L) of splenic and LN Tregs. (M) Tumor volume. (N) Tumor weight at day 17. (O) Gross tumor images at day 17. (P) Flow cytometric frequencies of immune cells in the TC-1 tumor model. Myeloid cells (left) and lymphoid cells (right). Myeloid and lymphoid cells are gated from live CD45+ cells and lymphoid cells, respectively. (Q) Cytokine production of tumor-infiltrating CD4+ and CD8+ T cells. (R to V) Solid tumors were induced in Foxp3EGFP-cre-ERT2 and Crif1fl/flFoxp3EGFP-cre-ERT2 mice with TC-1 cells and the mice were treated intraperitoneally with tamoxifen with or without blocking anti–IFN-γ, anti–IL-4, or isotype-control antibodies. (R) Tumor volume. (S) Tumor weight at day 17. (T) Gross tumor images at day 17. (U) and (V) Cytokine production of tumor-infiltrating Tregs (T) and CD4+ and CD8+ T cells (U). Dots represent individual mice (n = 5 to 16 per group). The data are pooled from at least two independent experiments and are presented as means ± SEM of biological replicates. Flow cytometry plots are representative of at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Statistical testing was conducted with Wilcoxon test [(C) and (E)], an unpaired two-tailed t test [(H) to (Q)], and one-way ANOVA [(R), (S), (U), and (V)]. PMN, polymorphonuclear.
Next, Crif1fl/flFoxp3EGFPcre-ERT2 and Foxp3EGFPcre-ERT2 mice were inoculated with TC-1 or MC-38, and, starting on day 3, the mice were injected intraperitoneally with tamoxifen (Fig. 7F). The loss of CRIF1 expression decreased the Foxp3 expression of the tumor-infiltrating Tregs (Fig. 7, G and H, and fig. S9, G and H) and increased the frequencies of IFN-γ– and IL-4–producing Tregs (Fig. 7, I and J, and fig. S9, I and J). In the non-tumor lymphoid organs of Crif1fl/flFoxp3EGFPcre-ERT2 mice, the abrogation of Crif1 expression did not alter Foxp3 expression intensity or cytokine production (Fig. 7, K and L, and fig. S9, K and L). Thus, systemic CRIF1 deficiency in Tregs primarily induced Foxp3low INS-Tregs in the TME. Down-regulating Crif1 expression significantly reduced tumor volume and weight (Fig. 7, M to O, and fig. S9, M to O), which showed that CRIF1 deficiency–induced Foxp3low INS-Tregs inhibited tumor growth. CRIF1 deficiency also reduced the frequencies of tumor-associated macrophages (TAMs) and dendritic cells (DCs) in the TC-1 tumors while slightly reducing the tumor Treg frequencies (Fig. 7P and fig. S9O). Notably, CRIF1 deficiency increased the IFN-γ and IL-4 production of CD4+ and CD8+ T cells (Fig. 7Q and fig. S9P), which could reflect the lower suppressive activities of the infiltrating Tregs. Moreover, when the Crif1fl/flFoxp3EGFPcre-ERT2 mice were treated with IFN-γ–blocking antibody as well as tamoxifen, the volume and weight of the tumors were restored (Fig. 7, R to V, and fig. S9, Q to U). By contrast, IL-4–blocking antibody did not have this effect (Fig. 7U and fig. S9T). Treatment with the IFN-γ antibody also reduced the frequencies of IFN-γ–producing CD4+ and CD8+ T cells (Fig. 7V and fig. S9U). Thus, CRIF1 deficiency–induced Foxp3low INS-Tregs exert an antitumor effect in an IFN-γ–dependent manner.
DISCUSSION
Our study showed that human FOXP3lowCD45RA− cells reduced expression of CRIF1 in TME and CRIF1 deficiency in murine Foxp3high Tregs induced metabolic reprograming that caused them to turn into Foxp3low INS-Tregs in mice. As shown by our study and others, these cells produce proinflammatory cytokines, which causes them to lose ability to suppress effector T cells (24–26). Notably, recent studies showed that, while the absence of mitochondrial regulatory proteins causes Tregs to lose suppressive ability regardless of Foxp3 expression, they did not demonstrate up-regulated production of proinflammatory cytokines (27, 28). Because this is the distinguishing feature of INS-Tregs, it was therefore thought that mitochondria play only limited roles in the generation of Foxp3low INS-Tregs including human FOXP3lowCD45RA− cells. This notion is disputed by our study, which shows a causal link between mitochondrial function and the formation of Foxp3low INS-Tregs in vivo.
Our study also showed that CRIF1-deficient Foxp3low INS-Tregs produced not only IFN-γ but also IL-4 and that their transfer into Rag1-KO mice induced systemic organ inflammation. Although T-bet and Gata3 are the key transcription factors for IFN-γ and IL-4, respectively, their in vivo deletion did not abrogate the IFN-γ/IL-4 production by the CRIF1-deficient Foxp3low INS-Tregs. Rather, we found that the transcription factors for these cytokines were EOMES and SATB1, respectively: They were up-regulated in CRIF1-deficient Foxp3low INS-Tregs and demonstrated increased binding to the regulatory regions of Ifng and Il4, respectively. This is significant because EOMES and SATB1 are better known for key roles in the development and function of other immune cells, especially CD8+ T and CD4+ T cells (42, 45). Moreover, although recent studies suggest that EOMES and SATB1 are expressed in IL-10–producing T regulatory type 1 cells and Tregs, respectively (46, 47), our study shows clearly that they are responsible for production of IFN-γ and IL-4 in Foxp3low INS-Tregs. These findings may be important for advancing our understanding of Foxp3low INS-Treg biology.
Notably, we observed that ~50% of the murine CRIF1-deficient Tregs produced either IFN-γ or IL-4. Similarly, the FOXP3lowCD45RA− cells in our human cancer cohorts secreted IFN-γ, IL-4, or IL-17A. These observations suggest that Foxp3low INS-Tregs exhibit functional heterogeneity in both humans and mice in terms of cytokine production patterns. Such functional heterogeneity may be attributable to EOMES and SATB1 expression patterns in the mouse Treg population. Another, not mutually exclusive, explanation is that Treg functional heterogeneity is driven by TCR specificity, which is known to regulate the transcriptomic phenotypes and functions of Tregs (48, 49).
We found that enhancement of glutaminolysis activated α-KG, which promoted the inflammatory cytokine production of CRIF1-deficient Tregs due to the activation of the mTORC1: This glutaminolysis-mediated activation of α-KG–mTORC1 axis elevated the expression of EOMES and SATB1. Significantly, Zeng et al. (34) have demonstrated that disrupting mTORC1 through Treg-specific deletion of Raptor leads to notable loss of suppressive function of Tregs, accompanied by disturbances in cholesterol and lipid metabolism and the onset of inflammatory disorder. However, Raptor-deficient Tregs did not exhibit proinflammatory cytokine production or low Foxp3 expression. Thus, it is most likely that Raptor deficiency–induced disruption and CRIF1 deficiency–mediated activation of mTORC1 might differentially influence functional phenotypes of Tregs through distinct mechanistic pathways, and fine-tuning of mTORC1 signaling may play critical role in the homeostasis and suppressive function of Tregs. Moreover, Matias et al. (50) showed that exogenous α-KG treatment during Treg differentiation results in the development of IFN-γ–producing Foxp3low Tregs by increasing OXPHOS and the generation of lipids. In contrast, CRIF1-deficient Tregs displayed a decrease in OCR, and α-KG treatment enhanced IFN-γ production by activating the mTORC1 axis. Therefore, it is likely that exogenous and endogenous α-KG might contribute to IFN-γ production in Tregs via distinct mechanisms, depending on the status of Treg differentiation and metabolic conditions. In addition, α-KG was implicated in the reduced expression of Foxp3 in CRIF1-deficient Tregs: 2-HG, derived from α-KG, methylated the promoter and CNS1 regions of the Foxp3 gene in CRIF1-defcient Tregs. Consistent with our findings, it has been reported that the promoter and CNS1 regions play a crucial role in the regulating Foxp3 expression in Tregs (51–53), although methylation of the CNS2 region is known to be critical for Treg instability (54). The fact that α-KG regulated the proinflammatory cytokine production and Foxp3 expression of CRIF1-deficient Foxp3low INS-Tregs via independent mechanisms is supported by studies showing that, while Nrp1, Foxo1, or Eos-deficient Tregs exhibit IFN-γ production and loss of suppressive activity, their Foxp3 expression is maintained (35, 55, 56).
Several studies suggest that the PI3K-AKT-FOXO axis regulates the IFN-γ production in Tregs (35, 36). However, in our study, the CRIF1-deficient and control Tregs had similar levels of p-Akt. Thus, this axis is not involved in the proinflammatory cytokine production of Foxp3low INS-Treg.
It was notable that both the human FOXP3lowCD45RA− cells and the murine Foxp3low CD4+ T cells in the TMEs of lung and CRCs exhibited reduced mitochondrial biogenesis, lower CRIF1 expression, and higher proinflammatory cytokine production. This suggested that CRIF1 may contribute to the induction of Foxp3low INS-Tregs within TME specifically. Our experiments with tamoxifen-inducible CRIF1-KO mice showed that the loss of CRIF1 expression generated Foxp3low INS-Tregs in only the TME: These cells were rare in the lymphoid tissues. Moreover, we found that the emergence of CRIF1-deficient Foxp3low INS-Tregs in the inducible mouse system inhibited tumor growth in an IFN-γ–dependent manner. This association between CRIF1 deficiency in FOXP3lowCD45RA− cells and antitumor activity was also observed for human cancers: CRIF1 expression in human TME CD4+ T cells correlated positively with FOXP3 expression, and higher numbers of Foxp3low cells in the human cancer cells were associated with better prognosis. These findings together suggest that factors in the TME may specifically induce the local formation of CRIF1-deficient IFN-γ–expressing Foxp3low INS-Tregs. It is not clear which TME factors drive the Treg changes in our model but several studies have reported various pathways by which TMEs induce antitumor IFN-γ+ Tregs in both mice and humans (8, 55, 57, 58). In particular, FOXP3low cells that relate to the intestinal bacterium Fusobacterium nucleatum have been shown to contribute to the antitumor immunity in human CRCs (8). However, we noted that human NSCLCs, which are influenced less strongly by microbiota than CRCs, also bear proinflammatory FOXP3low Tregs that display reduced mitochondrial biogenesis and CRIF1 expression. Further studies are needed to identify the TME factor(s) that induce CRIF1 deficiency in Tregs. Nonetheless, these findings together suggest that CRIF1-mediated induction of proinflammatory Foxp3low INS-Tregs in the TME can induce effective antitumor responses in both mice and humans. This supports the notion that targeting Foxp3low INS-Tregs may be a promising strategy for cancer immunotherapy.
In conclusion, our results indicate that CRIF1-mediated mitochondrial and metabolic homeostasis plays a critical regulatory role in the induction of Foxp3low INS-Tregs including FOXP3lowCD45RA− cells in TME that promote antitumor immunity (fig. S10).
MATERIALS AND METHODS
Patients and sample collection
Immunohistochemistry analysis was conducted on tumor tissues from 197 patients with NSCLC [from Seoul National University Hospital (SNUH)] and 280 patients with CRC (from Asan Medical Center, Seoul, Korea) (tables S1 and S2). These formalin-fixed paraffin-embedded (FFPE) tumor tissues were used to construct tissue microarrays. The SNUH Institutional Review Boards waived the need for informed consent for participation because the immunohistochemistry analysis was a retrospective study using archived material that did not pose any additional risk to the patient. The tumor, node, and metastasis stage of each case was determined using on the seventh American Joint Committee on Cancer (AJCC) for lung cancer and the eighth AJCC for colon cancer (59, 60). The clinicopathological and survival data of the patients were reviewed by an oncologist (T.M.K.) and two pathologists (S.K. and J.K.). Flow cytometric analyses were conducted on tumor samples from 91 patients with NSCLC and 50 patients with CRC who underwent surgical resection at SNUH (tables S3 and S4 and data S1). The fresh NSCLC samples were acquired within 3 hours after surgery. The CRC samples were frozen tumor samples that were provided by the SNUH Cancer Tissue Bank. Patients who had a history of previous cancer, neoadjuvant chemotherapy, or immunotherapy were excluded. All patients provided a written informed consent according to Institutional Review Board–approved protocols. Flow cytometry and immunohistochemistry analyses were conducted in accordance with the Declaration of Helsinki and World Medical Association recommendations and were approved by the Institutional Review Board of SNUH (no. H-1408-007-598).
Mice
C57BL/6 (B6) mice (7 to 8 weeks old) were purchased from Koatech (Pyeongtaek, Korea). Crif1fl/fl mice were a generous gift from M. Shong (Chungnam University, Korea) and were crossed with Foxp3YFP-Cre (Jackson strain no. 016959) or Foxp3EGFP-cre-ERT2 (Jackson strain no. 016961) mice. Other purchased mice were B6 CD45.1 congenic (Jackson strain no. 002014), T-betfl/fl (Jackson strain no. 022741), Ifng KO (Jackson strain no. 002287), Il4 KO (Jackson strain no. 002253), Rag1 KO (Jackson strain no. 002216), and ROSA26tdtomato mice (Jackson strain no. 007909) mice. All of these Jackson strain mice were purchased from Jackson Laboratory. Gata3fl/fl mice were a gift from Professor G. Ryol Lee (Sogang University, Korea). All strains were maintained on a B6 background and crossed as necessary to generate the desired genotypes. Age- and gender-matched male and female mice between 4 and 8 weeks of age were used in the study. Littermates were randomly assigned to experimental groups. All mice were bred and maintained under specific pathogen–free conditions at the Clinical Research Institute, SNUH. All in vivo experiments were approved by the Institutional Animal Care and Use Committee of the Clinical Research Institute, SNUH, and were conducted in accordance with the relevant guidelines and regulations.
Primary cultures and cell lines
Primary murine CD4+Foxp3-YFP+ Tregs from the spleens and LNs were fluorescence-activated cell sorting (FACS)–sorted with BD FACSAria III and cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, 2 mM glutamine, 10 mM Hepes, and 55 μM β-mercaptoethanol with and without TCR stimulation. TCR stimulation involved incubating the cells in plates coated with anti-CD3 antibody (1 μg/ml) and soluble anti-CD28 antibody (1 μg/ml) with IL-2 (5 ng/ml; PeproTech) for 1 to 2 days. For the glutamine-free medium experiment, glutamine-free RPMI 1640 was used with the same additives described above. To conduct the Treg suppression assay, conventional Foxp3-YFP−CD4+ T cells and Tregs were FACS-sorted from the spleen and LNs. Conventional Foxp3-YFP−CD4+ T cells were stained by CellTrace Violet and cocultured with TCR stimulation at T:Treg ratios of 1:0, 8:1, 4:1, 2:1, and 1:1 for 3 days. The proliferation of the conventional Foxp3-YFP−CD4+ T cells was assessed by flow cytometry. The murine colon adenocarcinoma cell line MC-38 and the immortalized primary murine lung epithelial cell line TC-1 were cultured in Dulbecco’s modified Eagle medium (DMEM) and RPMI 1640 in the presence of 10% FBS and 1% penicillin-streptomycin, respectively.
Histology and immunohistochemistry
Histology was conducted on FFPE tissue blocks that had been sectioned and stained with hematoxylin and eosin. Microscopic evaluations were performed at least three times for each individual experiment. Immunohistochemistry was performed with rabbit anti-CD8 monoclonal (Thermo Fisher Scientific) and rabbit anti-FOXP3 monoclonal (Abcam) antibodies and the Benchmark XT autostainer (Ventana Medical Systems). The numbers of CD8+, FOXP3low, and FOXP3high TILs per square millimeter were automatically counted via modified nuclear immunohistochemistry algorithms using Aperio ImageScope v12.4.3 (Aperio Technologies).
scRNA-seq data analysis
Processed scRNA-seq data of patients with lung cancer or CRC were downloaded from the Gene Expression Omnibus database by using the accession codes GSE131907 and GSE132465 (31, 32). Subsequent data analyses were performed in R 4.0.5 with Seurat R package version 4.0.2 (61).
With regard to the lung cancer data, the cells were first filtered using the criteria outlined in the source publication of the original sequencing data. A subset of tumor-infiltrating T lymphocytes (n = 18,587) was generated on the basis of the metadata provided with the source publication. After normalization with the “LogNormalize” method, 5000 variable features were selected with “vst” method. The data were then centered, and principal components analysis was performed. Cells were clustered using the top 20 dimensions and the “resolution = 1.3” parameter. The resulting clusters were visualized with uniform manifold approximation projection. Average gene expression counts were calculated using the “AverageExpression” function, and the cell type of each cluster was annotated by referencing the marker genes and cell-type lists used in the source publication. The DEGs in the FOXP3high and FOXP3low CD4+ T cell clusters were determined by using the “FindMarkers” function. Differential expression was considered significant when the fold change was >1.5 and the adjusted P value (q value) was <0.05.
The overall analysis of the CRC sequencing data was similar to that described for the lung cancer data. Thus, the tumor-infiltrating T cell subset (n = 23,115) was first generated from high-quality filtered cells using the annotation information provided in the source publication. The gene expression counts were normalized, and the top 2000 variable features were selected. The cells were clustered using the top 20 dimensions and the “resolution = 1.0” parameter.
To evaluate the relative enrichment of gene sets across cell subsets, gene set variation analysis (GSVA) was performed using the Bioconductor R package GSVA version 1.38.2 as previously described (62). Gene sets representing cell functions and metabolic pathway activities were curated from previous publications and the Molecular Signatures’ Database v7.5.1 (63, 64).
Flow cytometry
To prepare the samples, the fresh human lung cancer tissues, which weighed more than 0.1 g and were obtained within 3 hours after surgery, were immediately placed in ice-cold RPMI 1640 medium. The human CRC samples, which had been stored in liquid nitrogen, were rapidly thawed and washed in phosphate-buffered saline (PBS). Mouse tumors were obtained as described further below. The human tumor samples were then mechanically chopped with a blade, incubated at 37°C in RPMI 1640 supplemented with collagenase IV (1 mg/ml; Sigma-Aldrich) in a shaking incubator for 20 min, passed through a 70-μm cell strainer, and washed with PBS. Murine tumors were chopped with a blade and dissociated with collagenase (1 mg/ml; Sigma-Aldrich) and deoxyribonuclease 1 (100 μg/ml; Sigma-Aldrich) in RPMI 1640 medium for 25 to 30 min in a shaking incubator at 37°C. The dissociated samples were then passed through a 100-μm strainer. Murine thymus, spleen, and LNs were mashed through a 40-μm cell strainer, subjected to red blood cell lysis, and washed with PBS. All single-cell suspensions from the human and murine tissues were then passed through a 35-μm nylon mesh filter and stained with the Zombie Aqua Fixable Viability Kit (BioLegend) and surface or intracellular antibodies. If necessary, the cells were fixed and permeabilized with eBioscience Foxp3/Transcription Factor Fixation/Permeabilization solution (Invitrogen). For intracellular cytokine staining, 1 × 106 to 2 × 106 cells were stimulated in RPMI 1640 medium containing PMA (200 ng/ml), ionomycin (1 μg/ml), and GolgiStop (1 μl/ml) for between 3 hours 30 min and 4 hours.
Live cells are gated from Zombie Aqua negative cells, and the mouse tumor samples were gated for the following myeloid subpopulations in live CD45+ cells: eosinophil (CD11b+Siglec-F+), TAMs (CD11b+F4/80+), M1 TAMs (CD11b+F4/80+MHC-II+CD206−), M2 TAMs (CD11b+F4/80+MHC-II−CD206+), DCs (MHC-II+CD11c+), polymorphonuclear myeloid-derived suppressor cells (MDSCs) (CD11b+Ly-6G+), and monocytic MDSCs (CD11b+Ly-6C+). The samples were also gated for the following lymphoid subpopulations in live CD45+ lymphocytes: natural killer (NK) cells (NK1.1+TCR-Β−), CD4+ T cells (CD4+TCR-Β+), CD8+ T cells (CD8+TCR-Β+), and Tregs (Foxp3-GFP+CD4+TCR-Β+).
Electron microscopy
Electron microscopy was performed according to the previously established protocol (65). The FACS-sorted Treg pellet was fixed with 2.5% glutaraldehyde and then treated with 2% OsO4 in 0.1 M phosphate or cacodylate buffer for 1.5 hours at room temperature. Thin sections generated using an ultramicrotome (RMC MTXL) were collected on a copper grid. Appropriate areas for thin sections were cut at 65 nm and subsequently stained with saturated 6% uranyl acetate and 4% lead citrate at 80 kV before examination using a transmission electron microscope (JEM-1400).
Mitochondrial translation immunofluorescence
Tregs were cultured overnight with TCR stimulation, seeded onto glass coverslips, and cultured with methionine-free DMEM. Cytosolic translation was halted with cycloheximide (25 μg/ml) for 20 min, followed by adding 500 μM l-homopropargylglycine for 30 min. The cells were then incubated in buffer A (10 mM NaCl, 10 mM Hepes, 300 mM sucrose, 5 mM KCl, and 0.015% digitonin) for 2 min on ice, followed by 15-s incubation with buffer A without digitonin. The coverslips were fixed in 4% paraformaldehyde containing 4% sucrose for 30 min at room temperature and then incubated with blocking solution [5% bovine serum albumin (BSA) and 0.1% Triton X-100] for 20 min. After a brief wash with 3% BSA in 0.1 M pH 7.3 PBS, the cells were clicked for 20 min using the Click-iT Cell Reaction Buffer Kit and labeled with 3 μM Alexa Fluor 647 azide. After another quick wash with 3% BSA in PBS, the cells were incubated sequentially with primary translocase of outer mitochondrial membrane 20 (TOM20) antibody and secondary goat anti-mouse immunoglobulin G (IgG) Alexa Fluor 405 antibodies (both diluted in blocking solution at a 1:1000 ratio) for 45 min. Last, the samples were mounted on the glass slides using mounting medium. Images were acquired using a spinning disk confocal microscope (Nikon) with an oil-immersion objective lens (Plan Apo 60× numerical aperture 1.40); a Neo sCMOS camera (Andor Technology); and 405-, 561-, or 647-nm lasers at room temperature.
Western blot
Cells were homogenized using a TissueLyser II (Qiagen) in lysis buffer containing 50 mM tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA (pH 8.0), and 0.1% Triton X-100. The supernatants were obtained by centrifugation at 16,000g for 15 min, and the protein concentrations were measured using a protein assay dye. Protein (5 μg) from each sample was loaded onto 10% polyacrylamide gels and subjected to electrophoresis. The separated proteins were transferred onto a 0.45-μm nitrocellulose membrane at 400 mA for 2 hours. The membrane was blocked with 5% skimmed milk for 1 hour and then incubated with primary antibodies overnight at 4°C. After incubation with a horseradish peroxidase (HRP)–conjugated secondary antibody for 2 hours at room temperature, the membrane was visualized using the WesternBright ECL Spray (Advansta). The following antibodies were used for detection: CRIF1 antibody (Santa Cruz Biotechnology), Total OXPHOS Rodent WB Antibody Cocktail (Abcam), glyceraldehyde-3-phosphate dehydrogenase antibody (Cell Signaling Technology), HRP-linked anti-mouse IgG antibody (Cell Signaling Technology), and HRP-conjugated goat anti-rabbit IgG (H+L) (Bio-Rad).
ECAR and OCR
The ECAR and OCR of Tregs were measured using a XF-24 and XF-96 analyzer (Seahorse Biosciences) in accordance with the manufacturer’s instructions. Thus, FACS-sorted Tregs were stimulated with TCR overnight and plated in Cell-Tak–coated XF-24 or XF-96 plates (Corning). We used 1.0 × 106 to 1.5 × 106 cells per well for the XF-24 assays and 3 × 105 to 5 × 105 cells per well for the XF-96 assays. OCR was measured with final concentrations of 1 μM oligomycin, 1 μM carbonyl cyanide p-trifluoromethoxyphenylhydrazone, and 2 μM antimycin A, while ECAR was measured with final concentrations of 10 mM glucose, 2 μM oligomycin, and 100 mM 2-deoxy-d-glucose.
Enzyme-linked immunosorbent assay
Cytokine levels in culture supernatants were evaluated by following the manufacturer’s instructions. The following reagents were used for enzyme-linked immunosorbent assay: BD Pharmingen Purified Rat Anti-Mouse IFN, Biotin Anti-Mouse IFN-γ, BD Pharmingen Purified Rat Anti-Mouse IL-4, BD Pharmingen Biotin Rat Anti-Mouse IL-4, and Streptavidin-HRP (all from BD Biosciences).
Bone marrow chimeric mice
To generate bone marrow chimeras, bone marrow cells (2 × 106 to 3 × 106) isolated from the femurs and tibias of WT CD45.1 congenic and CD45.2 Crif1fl/flFoxp3-Cre mice were cotransferred intravenously into Rag1-KO mice with 1:1 ratio that had been irradiated at a total dose of 600 rad in two doses (300 rad) administered 3 hours apart. The reconstituted Rag1 KO mice were maintained on antibiotic water (neomycin sulfate, 0.5 mg/ml) for 2 weeks and euthanized 8 weeks after transfer.
Treg-adoptive transfer into Rag1 KO mice
To adoptively transfer Tregs, 0.8 × 106 to 1.0 × 106 FACS-sorted WT or CRIF1-deficient Tregs were transferred intravenously into Rag1-KO mice. The mice were euthanized 5 weeks after transfer.
Metabolomics and 13C-stable isotope tracer studies
Metabolites were extracted from Tregs as previously described (66). Briefly, cells were subjected to extraction in methanol/water (v/v, 4:1) with the internal standard DL-norvaline, vortexed, and sonicated in an ice bath (Diagenode, Bioruptor) at high intensity with a 30-s on and 30-s off cycle for 10 min. The extracts were then centrifuged at 21,000g for 10 min at 4°C, and the supernatants were dried with a vacuum concentrator (N-BIOTEK). Two derivatization steps were then performed: (i) 30 μl of MOX solution [10 mg of methoxyamine (Sigma-Aldrich) in 1 ml of pyridine (Sigma-Aldrich)] was added, followed by vortexing for 10 s and incubation at 37°C for 30 min and (ii) 70 μl of N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA) (Sigma-Aldrich) was added, followed by incubation at 60°C for 1 hour. Gas chromatography–mass spectrometry (GC-MS) or GC–tandem MS (MS/MS) analysis was performed using the Agilent 7890/7000 GC Triple Quadrupole mass spectrometer system. Gas chromatography was conducted using the Agilent J&W HP-5 ms UI 15 m–by–0.25 mm–by–0.25 μm (P/N 19091S-431UI) capillary column. Helium at a constant column flow rate of 1.5 ml min−1 served as the carrier gas. The initial oven temperature was 60°C, which rose to 320°C at a rate of 10°C min−1. The injector temperature was set to 280°C, and the samples were injected in the splitless mode with the auto sampler Agilent 7683A Injector. For untargeted analysis, the mass spectrometer was operated in the electron ionization mode at 70 eV and full-scan mode. Agilent MassHunter Data Acquisition Software (version B.04.00, Agilent Technologies) was used for data acquisition, and the metabolites were quantified by using Unknowns Analysis from Agilent MassHunter Quantitative Analysis (version 10.0, Agilent Technologies). MetaboAnalyst (version 5.0) was used for pathway analysis. For targeted analysis, the mass spectrometer was operated in multiple reaction monitoring mode, and the MassHunter Workstation Software for Quantitative Analysis (version 10.0, Agilent Technologies) was used for quantitative analysis. Peaks were assigned, quantified, and normalized to protein content and internal standard (ISTD).
For the measurement of ATP, 2-HG, SAM, and SAH, ultra high performance liquid chromatography (UHPLC) separation was implemented using a Thermo Scientific UltiMate 3000 RS pump equipped with Acquity UPLC BEH C18, 1.7 μm, 2.1 mm by 100 mm, operating at a flow rate of 300 μl/min and a column temperature at 35°C. The LC solvents used were as follows: solvent A, consisting of 10 mM ammonium formate in 50% ACN and 0.1% formic acid (A), and solvent B, consisting of 2 mM ammonium formate in ACN/isopropyl alcohol (IPA)/H2O 10:88:2 and 0.02% formic acid (B). The separation was achieved using linear gradient starting from 35 and increasing to 60% B for 4.0 min. Subsequently, the gradient was increase to 85% at 12 min, followed by an increase to 100% at 21 min. The system was held at 100% B for 3 min and then decrease to 35% at 24.1 min. Last, the system was equilibrated for additional 3.9 min. MS analysis was performed using a Thermo Scientific Q Exactive Orbitrap Plus mass spectrometer in electrospray ionization positive and negative modes. A full scan was conducted in the range of mass/charge ratio of 250 to 1200 with a resolution 70,000. The automatic gain control (AGC) target was set at 1 × 106 ions, and the maximum ion injection time (IT) was 100 ms. Data-dependent MS/MS was carried out using a “Top4” data-dependent mode, with the following parameters: resolution of 17,500; AGC of 1 × 105 ions; maximum IT of 50 ms; 1.0-u isolation window; normalized collision energies of 20, 30, 40; and a dynamic exclusion time of 6 s. The ionization parameters for the source were set as follows: spray voltage at 3.0 kV, capillary temperature at 370°C, and S-lens level at 45.
For 13C-stable isotope-tracing analysis, Tregs were stimulated overnight with anti-CD3/CD28 in RPMI 1640 medium as previously described. The medium was then changed to glutamine-free RPMI 1640 medium containing 4 mM [U-13C5] glutamine (Cambridge Isotope Laboratories, CLM-1822) for 6 hours, after which the metabolites were extracted and analyzed using GC-MS/MS. The peaks were assigned and corrected for the natural abundance distribution of each isotopologue.
Glutamine uptake and enzyme activity assay
FACS-sorted Tregs were cultured overnight with TCR stimulation in 2 mM glutamine-containing RPMI 1640. Glutamine uptake was determined by measuring the glutamine in the supernatants according to the manufacturer’s protocol and subtracting this value from 2 mM. The glutaminase and glutamate dehydrogenase activity in the Tregs was also determined according to the manufacturer’s protocol.
Quantitative real-time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) and transcribed into cDNA using M-MLV transcriptase (Promega). The primer list for real-time PCR was listed in table S5.
Transcriptomic analysis
Transcriptomics sample preparation
RNA was extracted from the FACS-sorted Treg pellet by using the QIAzol Lysis Reagent (Qiagen) according to the manufacturer’s instructions. Total RNA integrity was assessed using an Agilent Technologies 2100 Bioanalyzer and assigned an RNA integrity number value. Total RNA-seq libraries were prepared by using the Illumina Truseq Stranded Total RNA Sample prep kit with Ribo-zero human/mouse/rat according to the manufacturer’s protocol (Illumina). To deplete ribosomal RNA, 400 ng of total RNA was treated with biotinylated probes that selectively bind ribosomal RNA (rRNA) species, followed by purification. The rRNA-depleted total RNA was then fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were converted into first-strand cDNA using reverse transcriptase and random primers and then synthesized into second-strand cDNA using DNA polymerase I and ribonuclease H. The resulting cDNA fragments were adenylated and ligated with an adaptor. The final cDNA library was purified and enriched with PCR. The amplified libraries were validated using TapeStation (Agilent Technologies) and then indexed, tagged, and polled in equimolar amounts. RNA-seq was performed with an Illumina NovaSeq 6000 system using the protocols provided by the manufacturer (Illumina) for 2 × 100 sequencing. Before sequencing, the quality of the pooled libraries was confirmed by qPCR using the KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems).
To align the sequence reads to the mm10 mouse genome, the HISAT2 software was used with default settings. The “featureCount” function from the Rsubread R package was then used to quantify fragment overlap with exons. The General Transfer Format file from GENCODE vM25 was used for the mouse genome. To identify expressed genes, we excluded those with a count per million value less than 1 in at least one replicate by using the edgeR R package. Next, a generalized linear model was used to assess the significance of differences between the two groups for each gene; this was conducted by estimating the dispersion.
Transcriptome data analysis
TCR-stimulated and unstimulated CRIF1-deficient and control Tregs were compared in terms of gene expression and functionality via three steps. First, the genes whose expression was altered by TCR stimulation relative to no stimulation were identified in both CRIF1-deficient Tregs (denoted KO stim/unstim or ΔKO) and control Tregs (denoted WT stim/unstim or ΔWT) based on a fold-change value of group averages of >1.5 and a P value of <0.05. Second, the genes whose expression was altered in TCR-stimulated CRIF1-deficient Tregs compared to TCR-stimulated control Tregs (denoted stim KO/WT) were identified, again by looking for genes with >1.5-fold change difference in group averages. This was repeated for unstimulated CRIF1-deficient and unstimulated control Tregs (denoted unstim KO/WT). Third, we integrated transcriptomic alterations to identify a gene set responding to TCR stimulation in both CRIF1-deficient and WT Tregs by GSEA. To achieve this, we selected genes showing differential expression in DEGs or differential changes and separated them into segments such as “before TCR stimulation,” “during TCR stimulation,” and “after TCR stimulation.” These segments were then clustered using the SOM with the Kohonen R package.
The unstim KO/WT, stim KO/WT, and ΔKO/WT gene sets were subjected to GSEA version 4.1.0 (64) and EnrichR overrepresentation analysis of DEGs to identify enriched biological functions based on the weighted expression patterns. Gene sets from Hallmark, Kyoto Encyclopedia of Genes and Genomes pathway, and Gene Ontology were used. Multidimensional scaling (MDS) was used to condense the overall transcriptional patterns of expressed genes into a two-dimensional representation.
Assays for transposase accessible chromatin sequencing
ATAC-seq library preparation
ATAC-seq was performed according to a previously described protocol (67). Briefly, 50,000 live FACS-sorted Tregs were washed with cold 1× PBS at 4°C and centrifuged at 500g for 5 min at 4°C. The resulting pellets were resuspended in 50 μl of cold lysis buffer [10 mM tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40, 0.1% Tween 20, and 0.01% digitonin] and incubated on ice for 3 min. The cells were then washed with 1 ml of wash buffer [10 mM tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, and 0.1% Tween 20], centrifuged at 500g for 10 min to isolate the nuclei, which were then resuspended in transposase-reaction mix (25 μl of 2× TD buffer, 2.5 μl of Tn5 transposase (Illumina), and 22.5 μl of nuclease-free water) and incubated at 37°C with gentle agitation for 30 min. The DNA of the tagmentation reaction was purified using a Qiagen PCR Purification Kit (Qiagen), followed by PCR amplification using the indexing primers from the Nextera XT DNA Library Preparation Kit (Illumina). The optimal number of cycles for each sample was determined via qPCR to prevent saturation of the indexed DNA library. The resulting library was quantified by using a Bioanalyzer (Agilent Technologies), and each library was confirmed by qPCR using the KAPA library quantification kit (Roche Applied Science). The indexed library pool was sequenced using Illumina NovaSeq 6000 for 150 base pairs (bp), and the resulting reads were analyzed using CASAVA (v. 1.8.2) base-calling software (Illumina). The quality of pair-ended read sequences was assessed by using the FASTQC software.
Read alignment to the mm10 mouse genome was conducted by using Bowtie2 software with the “-X 2000” option, which resulted in a Binary Alignment Map (BAM) file. The BAM file was converted to the Browser Extensible Data (BED) format by using the “bamToBed” function in BEDtools software. The alignment positions of the fragments were corrected by shifting +4 bp for the positive strands and −5 bp for the negative strands to compensate for 9 bp; this was conducted with the “makeTagDirectory” function in the Homer-Suite, which created an intermediate folder. Genome browser tracks were generated for each sample by using the “MakeUCSCfile” function in the Homer Suite with the “--bigwig” option.
ATAC-seq data processing
Analysis began with merging the aligned reads into a single file. The “findPeaks” function in the Homer-Suite was then used to compare the enrichment of ATAC peaks with fixed coordinates. Further processing was conducted as previously described (68). Specifically, the ATAC-peak area was calculated by summing the enrichment scores within individual base pairs of the peak. The ATAC peaks of samples were then normalized by using the average heights of 286 control ATAC peaks. The normalized ATAC-peak areas were compared in terms of fold change of averages and P values determined with Mann-Whitney test. ATAC peaks were considered to differ significantly when fold change was >1.5 and P value was <0.05. To further analyze the genomic features of the differential ATAC peaks in Tregs, we used the “annotatePeaks.pl” function in Homer-Suite and updated the target gene of the ATAC peak. The target gene of the ATAC peak was updated from the nearest gene to the corresponding gene locus. Those ATAC peaks were then classified into nine categories: (i) promoter transcription start site (TSS) (from −2,000 bp upstream of the TSS to the TSS), (ii) distal promoter (from −10,000 to −2000 bp upstream of the TSS), (iii) 5′ untranslated regions (5′UTRs), (iv) first introns (from the first position of the first intron to the last position of the first intron), (v) other intron regions except the first intron, (vi) exon; (vii) 3′UTR, (viii) transcription terminal site, and (ix) the intergenic regions located between the gene loci. To predict the potential difference in gene expression regulation enriched in CRIF1-deficient and WT Tregs at a global scale, the transcriptional factors of differential ATAC peaks were identified by using the “findmotifs.pl” function in Homer-Suite. In addition, the matched motifs in the given regulatory regions of given genes were detected by using the “annotatedPeaks.pl” function. Their potential impact on gene expression was estimated on the basis of the relevant literature.
MDS was used to generate a two-dimensional representation of the overall ATAC-peak areas. To visualize genome browser tracks, the normalized bigWig files were deposited in CyVerse, and the UCSC genome browser was used.
Chromatin immunoprecipitation–quantitative polymerase chain reaction
ChIP-qPCR was performed by using the Pierce Agarose ChIP kit according to the manufacturer’s instructions. Briefly, Tregs that were TCR-stimulated for 1 day were fixed with 1% formaldehyde and treated with 1× glycine. The cells were washed with cold PBS, lysed, and treated with micrococcal nuclease. The reaction was stopped with the MNase Stop Solution. The nuclear extract was obtained by using Nuclear Extraction Buffer and centrifuging the mixture at 9000g for 5 min. To generate immunoprecipitates, the diluted nuclear extract was incubated overnight at 4°C on a rocking platform with either normal rabbit IgG, 2 μg of anti-EOMES antibody, or 2 μg of anti-SATB1 antibody. Each sample was washed and eluted by incubation at 65°C for 40 min in a heat block. The eluted samples were used for real-time qPCR, and 10% of the total input samples were used for normalization of each sample. The primers for the regulatory regions of Ifng and Il4 genes were designed with Primer BLAST (table S6).
siRNA knockdown
Tregs that underwent TCR-stimulation for 2 days were transfected with siRNA by using the jetPRIME reagent according to the manufacturer’s protocol. Thus, 20 nM control siRNA, mixed mouse Eomes siRNA, and mouse Satb1 siRNAs were incubated with the jetPRIME reagent for 15 min at room temperature and then added to the Tregs. The cells were harvested after 2 days of incubation. For intracellular cytokine staining, the Tregs were restimulated with PMA/ionomycin and GolgiStop.
Foxp3 gene DNA methylation
Foxp3 gene DNA methylation was assessed by using the Active motif MeDIP kit according to the manufacturer’s instructions. Briefly, DNA from overnight TCR-stimulated Tregs was sonicated for 8 min with 20-s on/off cycles at 30% power. Immunoprecipitation was performed by mixing 300 to 500 ng of sonicated DNA with mouse anti–5-methylcytosine monoclonal antibody and magnetic beads in a shaking incubator overnight at 4°C. The DNA bound to magnetic beads was isolated, and qPCR was performed. To assess the effect of (2S)-Tetrahydro-5-oxo-2-furancarboxylic acid (TFMB-(S)–2-HG) treatment, WT Tregs were treated with 2 mM 2-HG and overnight TCR stimulation. The primers used for the experiments were reported previously (table S7) (69). Data analysis was conducted in accordance with the manufacturer’s recommended protocol. This procedure entailed evaluating precipitated DNA through a standard curve and calculating the percentage of the initial input.
Retroviral transduction
The murine Foxp3 coding sequence was incorporated into a RV expression vector known as pMIG. To generate the RV, Plat-E cells were transfected with either the Empty-pMIG or Foxp3-pMIG plasmid. Subsequent to transfection, culture supernatants were collected at 24 and 48 hours, filtered, and stored at 4°C until required. Isolated Tregs were resuspended in R10 medium, comprising RPMI 1640 containing 10% FBS, 2 mM GlutaMAX (Gibco), and penicillin and streptomycin (100 U/ml; Biowest). This medium was supplemented with recombinant mouse IL-2 (10 ng/ml; PeproTech), 50 μM β-mercaptoethanol (BME; Gibco), and nonessential amino acids (NEAA; Gibco). Cells (1 × 106 to 1.5 × 106) were seeded in 24-well plates and stimulated with plate-bound anti-CD3 (1 μg/ml; 17A2; BioXcell) and anti-CD28 antibodies (37.51; BioXcell). Transduction of cells occurred 24 and 36 hours after stimulation by centrifugation for 90 min at 2000 rpm at 37°C with RV supernatant. This supernatant was supplemented with 50 μM BME and polybrene (10 μg/ml; Santa Cruz Biotechnology). On the day following the second transduction, Tregs were transferred into 12-well plates in D10 medium [DMEM containing 10% FBS, 2 mM GlutaMAX (Gibco), and penicillin and streptomycin (100 U/ml; Biowest)]. This medium contained 50 μM BME, recombinant mouse IL-2 (10 ng/ml; PeproTech), and NEAA. The cells were then incubated for 1 day before undergoing analysis for the expression of transcription factors and cytokines via flow cytometry.
Mouse tumor model
Tumors were generated by subcutaneously injecting the flank of 8- to 12-week-old mice with 5 × 105 cells of MC-38 or TC-1 cells. The mice were euthanized on day 17, and the single-cell suspensions generated from the tumors were subjected to surface and intracellular staining for FACS analysis. To induce CRIF1 depletion in Tregs during tumor growth, the conditional Crif1fl/flFoxp3EGFPcre-ERT2 and Foxp3EGFPcre-ERT2 KO mice were injected intraperitoneally on days 3, 4, 7, 8, and 9 after tumor inoculation with 100 μl of tamoxifen (20 mg/ml) that had been dissolved in corn oil with 5% ethanol. To block specific cytokines during tumor growth, the mice were injected intraperitoneally on days 4, 8, 11, and 14 after tumor inoculation with Armenian hamster IgG (0.2 mg per mouse; Bio X cell), anti–IFN-γ (Bio X cell), or anti–IL-4 antibody (Bio X cell).
Statistical analyses
All statistical analyses were performed using Prism software v8. Group comparisons were conducted using unpaired two-tailed t tests for two group comparisons and either one-way analysis of variance (ANOVA) or two-way ANOVA for comparisons involving multiple groups. In terms of the human and mouse tumor data, groups were compared in terms of mitochondrial biogenesis and CRIF1 expression by paired Wilcoxon matched-pairs signed-rank test. The correlation between CRIF1 and FOXP3 expression in human tumor samples was evaluated by using Pearson correlation test. P values of <0.05 were considered significant. Survival analysis was performed using the Kaplan-Meier method and the log-rank test. In all analyses, two-sided P values of <0.05 were considered to be significant. Statistical analyses of immunohistochemistry data were performed by using R statistical software version 4.1.1.
Acknowledgments
We would like to thank the Hyehwa Forum members and Professor G. Ryol Lee (Sogang University, Korea) for helpful discussions and providing Gata3fl/fl mice, respectively. We also thank the members in the Department of Thoracic Surgery in SNUH for efforts to provide human lung cancer samples.
Funding: This research was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government Ministry of Science and ICT (MSIT) (grant nos. RS-2023-00217571 and NRF-2020R1A2C2008312).
Author contributions: In vitro and in vivo experiments: S.L., G.K., H.J.Y., Y.-J.K., J.T., and E.C. Providing CRIF1 flox mice: M.S. Analysis of the publicly available data and performed the experiments on the human tumor samples: S.G.S. Histological and clinical data analyses: S.K., J.K., and Y.K.J. review and analysis of the clinical data: T.M.K. Providing the tissue microarrays of the CRCs: Ji Hoon Kim. (AMC). Analyzing and interpreting the data: Y.S.C., S.C., H.M.S., K.C.J., H.Y.K., and Ji Hyung Kim. (Korea University). Writing: S.L., S.G.S., and D.H.C. Design and supervision: D.H.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The dataset analyzed in this study is available in the NCBI Gene Expression Omnibus (GSE233903).
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 to S7
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Josefowicz S. Z., Lu L. F., Rudensky A. Y., Regulatory T cells: Mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakaguchi S., Mikami N., Wing J. B., Tanaka A., Ichiyama K., Ohkura N., Regulatory T cells and human disease. Annu. Rev. Immunol. 38, 541–566 (2020). [DOI] [PubMed] [Google Scholar]
- 3.O’Callaghan D. S., Rexhepaj E., Gately K., Coate L., Delaney D., O’Donnell D. M., Kay E., O’Connell F., Gallagher W. M., O’Byrne K. J., Tumour islet Foxp3+ T-cell infiltration predicts poor outcome in nonsmall cell lung cancer. Eur. Respir. J. 46, 1762–1772 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Saleh R., Elkord E., FoxP3+ T regulatory cells in cancer: Prognostic biomarkers and therapeutic targets. Cancer Lett. 490, 174–185 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Guo J., Zhou X., Regulatory T cells turn pathogenic. Cell. Mol. Immunol. 12, 525–532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li C., Jiang P., Wei S., Xu X., Wang J., Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 19, 116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyara M., Yoshioka Y., Kitoh A., Shima T., Wing K., Niwa A., Parizot C., Taflin C., Heike T., Valeyre D., Mathian A., Nakahata T., Yamaguchi T., Nomura T., Ono M., Amoura Z., Gorochov G., Sakaguchi S., Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 30, 899–911 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Saito T., Nishikawa H., Wada H., Nagano Y., Sugiyama D., Atarashi K., Maeda Y., Hamaguchi M., Ohkura N., Sato E., Nagase H., Nishimura J., Yamamoto H., Takiguchi S., Tanoue T., Suda W., Morita H., Hattori M., Honda K., Mori M., Doki Y., Sakaguchi S., Two FOXP3+CD4+ T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat. Med. 22, 679–684 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Wing J. B., Kitagawa Y., Locci M., Hume H., Tay C., Morita T., Kidani Y., Matsuda K., Inoue T., Kurosaki T., Crotty S., Coban C., Ohkura N., Sakaguchi S., A distinct subpopulation of CD25(−) T-follicular regulatory cells localizes in the germinal centers. Proc. Natl. Acad. Sci. U.S.A. 114, E6400–E6409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonseca V. R., Agua-Doce A., Maceiras A. R., Pierson W., Ribeiro F., Romão V. C., Pires A. R., da Silva S. L., Fonseca J. E., Sousa A. E., Linterman M. A., Graca L., Human blood T(fr) cells are indicators of ongoing humoral activity not fully licensed with suppressive function. Sci. Immunol. 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira R. C., Simons H. Z., Thompson W. S., Rainbow D. B., Yang X., Cutler A. J., Oliveira J., Castro Dopico X., Smyth D. J., Savinykh N., Mashar M., Vyse T. J., Dunger D. B., Baxendale H., Chandra A., Wallace C., Todd J. A., Wicker L. S., Pekalski M. L., Cells with Treg-specific FOXP3 demethylation but low CD25 are prevalent in autoimmunity. J. Autoimmun. 84, 75–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuadrado E., van den Biggelaar M., de Kivit S., Chen Y. Y., Slot M., Doubal I., Meijer A., van Lier R. A. W., Borst J., Amsen D., Proteomic analyses of human regulatory T cells reveal adaptations in signaling pathways that protect cellular identity. Immunity 48, 1046–1059.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Wing J. B., Tanaka A., Sakaguchi S., Human FOXP3+ regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity 50, 302–316 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Togashi Y., Shitara K., Nishikawa H., Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat. Rev. Clin. Oncol. 16, 356–371 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Dominguez-Villar M., Hafler D. A., Regulatory T cells in autoimmune disease. Nat. Immunol. 19, 665–673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandiyan P., Zhu J., Origin and functions of pro-inflammatory cytokine producing Foxp3+ regulatory T cells. Cytokine 76, 13–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J. H., Elly C., Park Y., Liu Y. C., E3 ubiquitin ligase VHL regulates hypoxia-inducible factor-1α to maintain regulatory T cell stability and suppressive capacity. Immunity 42, 1062–1074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X., Teng X. L., Wang F., Zheng Y., Qu G., Zhou Y., Hu Z., Wu Z., Chang Y., Chen L., Li H. B., Su B., Lu L., Liu Z., Sun S. C., Zou Q., Metabolic control of regulatory T cell stability and function by TRAF3IP3 at the lysosome. J. Exp. Med. 215, 2463–2476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi R., Nishimoto S., Muto G., Sekiya T., Tamiya T., Kimura A., Morita R., Asakawa M., Chinen T., Yoshimura A., SOCS1 is essential for regulatory T cell functions by preventing loss of Foxp3 expression as well as IFN-γ and IL-17A production. J. Exp. Med. 208, 2055–2067 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grinberg-Bleyer Y., Oh H., Desrichard A., Bhatt D. M., Caron R., Chan T. A., Schmid R. M., Klein U., Hayden M. S., Ghosh S., NF-κB c-Rel Is crucial for the regulatory T cell immune checkpoint in cancer. Cell 170, 1096–1108.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Pilato M., Kim E. Y., Cadilha B. L., Prüßmann J. N., Nasrallah M. N., Seruggia D., Usmani S. M., Misale S., Zappulli V., Carrizosa E., Mani V., Ligorio M., Warner R. D., Medoff B. D., Marangoni F., Villani A.-C., Mempel T. R., Targeting the CBM complex causes Treg cells to prime tumours for immune checkpoint therapy. Nature 570, 112–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang K., Blanco D. B., Neale G., Vogel P., Avila J., Clish C. B., Wu C., Shrestha S., Rankin S., Long L., KC A., Chi H., Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature 548, 602–606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He N., Fan W., Henriquez B., Yu R. T., Atkins A. R., Liddle C., Zheng Y., Downes M., Evans R. M., Metabolic control of regulatory T cell (Treg) survival and function by Lkb1. Proc. Natl. Acad. Sci. U.S.A. 114, 12542–12547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angelin A., Gil-de-Gómez L., Dahiya S., Jiao J., Guo L., Levine M. H., Wang Z., Quinn W. J. III, Kopinski P. K., Wang L., Akimova T., Liu Y., Bhatti T. R., Han R., Laskin B. L., Baur J. A., Blair I. A., Wallace D. C., Hancock W. W., Beier U. H., Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 25, 1282–1293.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinberg S. E., Singer B. D., Steinert E. M., Martinez C. A., Mehta M. M., Martínez-Reyes I., Gao P., Helmin K. A., Abdala-Valencia H., Sena L. A., Schumacker P. T., Turka L. A., Chandel N. S., Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature 565, 495–499 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Z., Ye J., Dean J. W., Bostick J. W., Weinberg S. E., Xiong L., Oliff K. N., Chen Z. E., Avram D., Chandel N. S., Zhou L., Requirement of mitochondrial transcription factor a in tissue-resident regulatory T Cell maintenance and function. Cell Rep. 28, 159–171.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howie D., Cobbold S. P., Adams E., Bokum A. T., Necula A. S., Zhang W., Huang H., Roberts D. J., Thomas B., Hester S. S., Vaux D. J., Betz A. G., Waldmann H., Foxp3 drives oxidative phosphorylation and protection from lipotoxicity. JCI Insight 2, e89160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almeida L., Dhillon-LaBrooy A., Carriche G., Berod L., Sparwasser T., CD4+ T-cell differentiation and function: Unifying glycolysis, fatty acid oxidation, polyamines NAD mitochondria. J. Allergy Clin. Immunol. 148, 16–32 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Kim S. J., Kwon M. C., Ryu M. J., Chung H. K., Tadi S., Kim Y. K., Man Kim J., Lee S. H., Park J. H., Kweon G. R., Ryu S. W., Jo Y. S., Lee C. H., Hatakeyama H., Goto Y. I., Yim Y. H., Chung J., Kong Y. Y., Shong M., CRIF1 is essential for the synthesis and insertion of oxidative phosphorylation polypeptides in the mammalian mitochondrial membrane. Cell Metab. 16, 274–283 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Koc E. C., Cimen H., Kumcuoglu B., Abu N., Akpinar G., Haque M. E., Spremulli L. L., Koc H., Identification and characterization of CHCHD1, AURKAIP1, and CRIF1 as new members of the mammalian mitochondrial ribosome. Front. Physiol. 4, 183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim N., Kim H. K., Lee K., Hong Y., Cho J. H., Choi J. W., Lee J. I., Suh Y. L., Ku B. M., Eum H. H., Choi S., Choi Y. L., Joung J. G., Park W. Y., Jung H. A., Sun J. M., Lee S. H., Ahn J. S., Park K., Ahn M. J., Lee H. O., Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 11, 2285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee H. O., Hong Y., Etlioglu H. E., Cho Y. B., Pomella V., van den Bosch B., Vanhecke J., Verbandt S., Hong H., Min J. W., Kim N., Eum H. H., Qian J., Boeckx B., Lambrechts D., Tsantoulis P., de Hertogh G., Chung W., Lee T., An M., Shin H. T., Joung J. G., Jung M. H., Ko G., Wirapati P., Kim S. H., Kim H. C., Yun S. H., Tan I. B. H., Ranjan B., Lee W. Y., Kim T. Y., Choi J. K., Kim Y. J., Prabhakar S., Tejpar S., Park W. Y., Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 52, 594–603 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Collison L. W., Chaturvedi V., Henderson A. L., Giacomin P. R., Guy C., Bankoti J., Finkelstein D., Forbes K., Workman C. J., Brown S. A., Rehg J. E., Jones M. L., Ni H. T., Artis D., Turk M. J., Vignali D. A. A., IL-35-mediated induction of a potent regulatory T cell population. Nat. Immunol. 11, 1093–1101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng H., Yang K., Cloer C., Neale G., Vogel P., Chi H., mTORC1 couples immune signals and metabolic programming to establish Treg-cell function. Nature 499, 485–490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouyang W., Liao W., Luo C. T., Yin N., Huse M., Kim M. V., Peng M., Chan P., Ma Q., Mo Y., Meijer D., Zhao K., Rudensky A. Y., Atwal G., Zhang M. Q., Li M. O., Novel Foxo1-dependent transcriptional programs control Treg cell function. Nature 491, 554–559 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitz A., de Marcken M., Gautron A. S., Mitrovic M., Hafler D. A., Dominguez-Villar M., AKT isoforms modulate Th1-like Treg generation and function in human autoimmune disease. EMBO Rep. 17, 1169–1183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang A., al-Kuhlani M., Johnston S. C., Ojcius D. M., Chou J., Dean D., Transcription factor complex AP-1 mediates inflammation initiated by Chlamydia pneumoniae infection. Cell. Microbiol. 15, 779–794 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aune T. M., Collins P. L., Collier S. P., Henderson M. A., Chang S., Epigenetic activation and silencing of the gene that encodes IFN-gamma. Front. Immunol. 4, 112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubo M., The role of IL-4 derived from follicular helper T (TFH) cells and type 2 helper T (TH2) cells. Int. Immunol. 33, 717–722 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Zhang J., le Gras S., Pouxvielh K., Faure F., Fallone L., Kern N., Moreews M., Mathieu A. L., Schneider R., Marliac Q., Jung M., Berton A., Hayek S., Vidalain P. O., Marçais A., Dodard G., Dejean A., Brossay L., Ghavi-Helm Y., Walzer T., Sequential actions of EOMES and T-BET promote stepwise maturation of natural killer cells. Nat. Commun. 12, 5446 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Istaces N., Splittgerber M., Lima Silva V., Nguyen M., Thomas S., le A., Achouri Y., Calonne E., Defrance M., Fuks F., Goriely S., Azouz A., EOMES interacts with RUNX3 and BRG1 to promote innate memory cell formation through epigenetic reprogramming. Nat. Commun. 10, 3306 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai S., Lee C. C., Kohwi-Shigematsu T., SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 38, 1278–1288 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Du X., Hu H., The roles of 2-hydroxyglutarate. Front. Cell Dev. Biol. 9, 651317 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sibani S., Melnyk S., Pogribny I. P., Wang W., Hiou-Tim F., Deng L., Trasler J., James S. J., Rozen R., Studies of methionine cycle intermediates (SAM, SAH), DNA methylation and the impact of folate deficiency on tumor numbers in Min mice. Carcinogenesis 23, 61–65 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Kakugawa K., Kojo S., Tanaka H., Seo W., Endo T. A., Kitagawa Y., Muroi S., Tenno M., Yasmin N., Kohwi Y., Sakaguchi S., Kowhi-Shigematsu T., Taniuchi I., Essential roles of SATB1 in specifying T lymphocyte subsets. Cell Rep. 19, 1176–1188 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Roessner P. M., Llaó Cid L., Lupar E., Roider T., Bordas M., Schifflers C., Arseni L., Gaupel A. C., Kilpert F., Krötschel M., Arnold S. J., Sellner L., Colomer D., Stilgenbauer S., Dietrich S., Lichter P., Izcue A., Seiffert M., EOMES and IL-10 regulate antitumor activity of T regulatory type 1 CD4+ T cells in chronic lymphocytic leukemia. Leukemia 35, 2311–2324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beyer M., Thabet Y., Müller R. U., Sadlon T., Classen S., Lahl K., Basu S., Zhou X., Bailey-Bucktrout S. L., Krebs W., Schönfeld E. A., Böttcher J., Golovina T., Mayer C. T., Hofmann A., Sommer D., Debey-Pascher S., Endl E., Limmer A., Hippen K. L., Blazar B. R., Balderas R., Quast T., Waha A., Mayer G., Famulok M., Knolle P. A., Wickenhauser C., Kolanus W., Schermer B., Bluestone J. A., Barry S. C., Sparwasser T., Riley J. L., Schultze J. L., Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat. Immunol. 12, 898–907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shevyrev D., Tereshchenko V., Treg heterogeneity, function, and homeostasis. Front. Immunol. 10, 3100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zemmour D., Zilionis R., Kiner E., Klein A. M., Mathis D., Benoist C., Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol. 19, 291–301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matias M. I., Yong C. S., Foroushani A., Goldsmith C., Mongellaz C., Sezgin E., Levental K. R., Talebi A., Perrault J., Rivière A., Dehairs J., Delos O., Bertand-Michel J., Portais J. C., Wong M., Marie J. C., Kelekar A., Kinet S., Zimmermann V. S., Levental I., Yvan-Charvet L., Swinnen J. V., Muljo S. A., Hernandez-Vargas H., Tardito S., Taylor N., Dardalhon V., Regulatory T cell differentiation is controlled by αKG-induced alterations in mitochondrial metabolism and lipid homeostasis. Cell Rep. 37, 109911 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colamatteo A., Carbone F., Bruzzaniti S., Galgani M., Fusco C., Maniscalco G. T., Rella F. D., de Candia P., De Rosa V., Molecular mechanisms controlling Foxp3 expression in health and autoimmunity: From epigenetic to post-translational regulation. Front. Immunol. 10, 3136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng Y., Josefowicz S., Chaudhry A., Peng X. P., Forbush K., Rudensky A. Y., Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y., Lu Y., Wang S., Han Z., Zhu F., Ni Y., Liang R., Zhang Y., Leng Q., Wei G., Shi G., Zhu R., Li D., Wang H., Zheng S. G., Xu H., Tsun A., Li B., USP21 prevents the generation of T-helper-1-like Treg cells. Nat. Commun. 7, 13559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Someya K., Nakatsukasa H., Ito M., Kondo T., Tateda K. I., Akanuma T., Koya I., Sanosaka T., Kohyama J., Tsukada Y. I., Takamura-Enya T., Yoshimura A., Improvement of Foxp3 stability through CNS2 demethylation by TET enzyme induction and activation. Int. Immunol. 29, 365–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Overacre-Delgoffe A. E., Chikina M., Dadey R. E., Yano H., Brunazzi E. A., Shayan G., Horne W., Moskovitz J. M., Kolls J. K., Sander C., Shuai Y., Normolle D. P., Kirkwood J. M., Ferris R. L., Delgoffe G. M., Bruno T. C., Workman C. J., Vignali D. A. A., Interferon-γ Drives treg fragility to promote anti-tumor immunity. Cell 169, 1130–1141.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan F., Yu H., Dang E. V., Barbi J., Pan X., Grosso J. F., Jinasena D., Sharma S. M., McCadden E. M., Getnet D., Drake C. G., Liu J. O., Ostrowski M. C., Pardoll D. M., Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science 325, 1142–1146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Overacre-Delgoffe A. E., Vignali D. A. A., Treg fragility: A prerequisite for effective antitumor immunity? Cancer Immunol. Res. 6, 882–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakagawa H., Sido J. M., Reyes E. E., Kiers V., Cantor H., Kim H. J., Instability of helios-deficient tregs is associated with conversion to a T-effector phenotype and enhanced antitumor immunity. Proc. Natl. Acad. Sci. U.S.A. 113, 6248–6253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edge S. B., Compton C. C., The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 17, 1471–1474 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Amin M. B., Greene F. L., Edge S. B., Compton C. C., Gershenwald J. E., Brookland R. K., Meyer L., Gress D. M., Byrd D. R., Winchester D. P., The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 67, 93–99 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. III, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hanzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo Y., Xu C., Wang B., Niu Q., Su X., Bai Y., Zhu S., Zhao C., Sun Y., Wang J., Liu M., Sun X., Song G., Cui H., Chen X., Huang H., Wang H., Han M., Jiang E., Shi L., Feng X., Single-cell transcriptomic analysis reveals disparate effector differentiation pathways in human Treg compartment. Nat. Commun. 12, 3913 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ko J. S., Koh J. M., So J. S., Jeon Y. K., Kim H. Y., Chung D. H., Palmitate inhibits arthritis by inducing t-bet and gata-3 mRNA degradation in iNKT cells via IRE1α-dependent decay. Sci. Rep. 7, 14940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faubert B., Boily G., Izreig S., Griss T., Samborska B., Dong Z., Dupuy F., Chambers C., Fuerth B. J., Viollet B., Mamer O. A., Avizonis D., DeBerardinis R. J., Siegel P. M., Jones R. G., AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 17, 113–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., Orloff L. A., Risca V. I., Kundaje A., Khavari P. A., Montine T. J., Greenleaf W. J., Chang H. Y., An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shin H. M., Kim G., Kim S., Sim J. H., Choi J., Kim M., Kwon M., Ye S. K., Lee D. S., Cho S. W., Kim S. T., Lee J., Kim H. R., Chromatin accessibility of circulating CD8+ T cells predicts treatment response to PD-1 blockade in patients with gastric cancer. Nat. Commun. 12, 975 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang R., Qu C., Zhou Y., Konkel J. E., Shi S., Liu Y., Chen C., Liu S., Liu D., Chen Y., Zandi E., Chen W., Zhou Y., Shi S., Hydrogen sulfide promotes Tet1- and Tet2-mediated Foxp3 demethylation to drive regulatory t cell differentiation and maintain immune homeostasis. Immunity 43, 251–263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 to S7
Legend for data S1
Data S1