Abstract
Intraoperative histology is essential for surgical guidance and decision-making. However, frozen-sectioned hematoxylin and eosin (H&E) staining suffers from degraded accuracy, whereas the gold-standard formalin-fixed and paraffin-embedded (FFPE) H&E is too lengthy for intraoperative use. Stimulated Raman scattering (SRS) microscopy has shown rapid histology of brain tissue with lipid/protein contrast but is challenging to yield images identical to nucleic acid–/protein-based FFPE stains interpretable to pathologists. Here, we report the development of a semi-supervised stimulated Raman CycleGAN model to convert fresh-tissue SRS images to H&E stains using unpaired training data. Within 3 minutes, stimulated Raman virtual histology (SRVH) results that matched perfectly with true H&E could be generated. A blind validation indicated that board-certified neuropathologists are able to differentiate histologic subtypes of human glioma on SRVH but hardly on conventional SRS images. SRVH may provide intraoperative diagnosis superior to frozen H&E in both speed and accuracy, extendable to other types of solid tumors.
Label-free and slide-free virtual histology of brain tissue by stimulated Raman scattering achieves gold-standard H&E quality.
INTRODUCTION
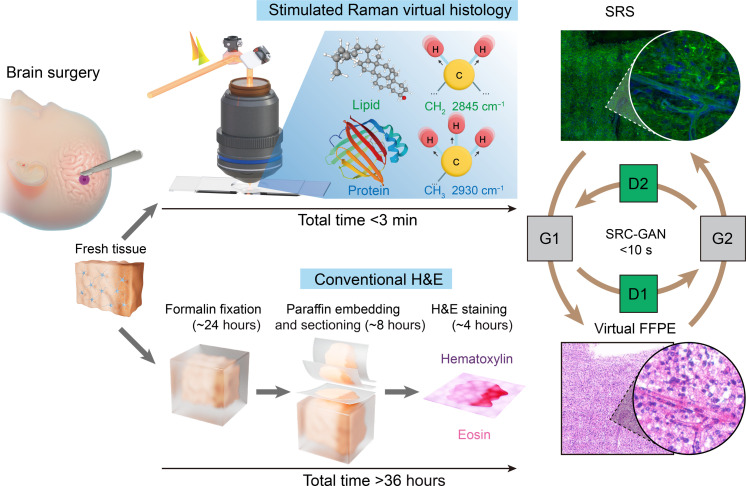
Timely diagnosis of histopathologic subtypes of brain tumors during an operation is critically important to the optimal surgical management (1) but is challenging on the basis of the gross appearance of brain tissues. The gold-standard histologic method requires tissue being formalin-fixed and paraffin-embedded (FFPE), followed by thin sectioning (2 to 10 μm) and staining with hematoxylin and eosin (H&E), which is too slow (>36 hours) to be intraoperatively compatible (Fig. 1). Although current intraoperative H&E gains speed (<1 hour) using frozen section technique, it sacrifices diagnostic accuracy due to the artefacts introduced by freezing and sectioning processes, causing cellular/tissue loss, distortion and folding, as well as staining variances. Therefore, a modality that is stain- and slide-free, while capable of generating FFPE-quality images would be invaluable for rapid intraoperative histology of neurosurgery.
Fig. 1. Deep learning–based SRVH.
Schematic workflows of the conventional FFPE histology with H&E staining (>36 hours), and the rapid (<3 min) label-free SRVH on unprocessed surgical tissue, with the lipid and protein (L&P)–based contrast converted to FFPE-style via SRC-GAN model.
Growing advances in label-free optical imaging techniques have brought new opportunities for biomedical applications including the revolution of histopathology. Multiphoton microscopy provides multimodality images, such as two-photon excited fluorescence, second-harmonic generation, third-harmonic generation, etc. (2). However, these techniques are primarily sensitive to extracellular matrix (collagen fibers, elastin fibers, and lipid droplets), lacking cell nuclear contrast as the key cytologic and histologic information for diagnosis. Whereas the nuclear content could be detected by the strong optical responses to ultraviolet (UV) light excitation, taking advantages of either UV-autofluorescence (3, 4), or UV absorption–based photoacoustic microscopy (UV-PAM) (5). They tend to provide single-channel intensity images with limited chemical specificity, prone to the interference of other UV active fluorophores or pigments (6). Stimulated Raman scattering (SRS) microscopy has enabled histologic imaging of various types of human tissues with high spatial and chemical resolution (7–10), mostly relying on the lipid/protein contrast of cell nuclear, cytoplasm, and extracellular matrix. However, transforming SRS images to pathological interpretable H&E stains remains unsolved because the latter is based on nucleic acid/protein contrast, and the discrepancies of the contrast mechanism prevent the current efforts of using color projection algorithms from yielding FFPE-grade images (11, 12).
The integration of artificial intelligence (AI) with various microscopy techniques has revolutionized the fields of biomedical photonics, enabling the tasks of breaking the limits of image resolution and speed (13, 14), diagnostic classification (15–18), semantic segmentation (19–22), cross-modality transformation (23, 24), and virtual staining (5, 25–27), etc. In particular, virtual histologic stains could be converted from unlabeled UV-autofluorescence (27) and UV-PAM images (5), and virtual FFPE could be transformed from frozen H&E results via deep learning (26). These methods exploit the power of generative adversarial networks (GANs), a type of deep-learning model that is used in the translation between two relevant domains (28–30). Note that conventional GANs are constrained to the pixel-level registered and paired training datasets due to the strongly supervised nature. Therefore, only sectioned tissue slides of unlabeled-stained image pairs are suitable for such algorithms (27). For slide-free images of fresh tissues, weakly supervised CycleGAN architectures are more suited to generate virtual histologic images without the need of well-aligned image pairs (5, 31). However, generic CycleGAN is still insufficient to achieve FFPE-quality images, even for the translation from similar-styled frozen H&E sections (26). It is much more challenging to transform SRS images of unprocessed tissues to FFPE style. First, paraffin-embedding is incompatible with SRS imaging because the Raman background from paraffin severely disrupts the imaged tissue morphologies; hence, obtaining corresponding SRS and FFPE images from the same tissue section is unfeasible. Second, fresh tissue could not provide images of the two modalities from the same image plane. Furthermore, SRS and H&E images differ in multiple aspects such as chemical contrasts, color style, and textures, which cause generic CycleGAN to fail to generate FFPE-quality from SRS images.
In this work, we report a deep-learning strategy tailored for stimulated Raman virtual histology (SRVH) using SRS images of fresh brain tissues and unaligned FFPE sections. We demonstrate that SRVH is capable of rapidly creating FFPE-grade images without labeling or sectioning (~3 min). It not only avoids cryosectioning artefacts but also reveals clinically critical cytologic and histologic features of human brain tumors including nuclear atypia, abnormal vasculature, microvascular proliferation, necrosis, etc. Therefore, SRVH successfully differentiates histologic subtypes such as astrocytoma, oligodendroma, and glioblastoma, as verified by board-certified neuropathologists. Our method holds promise to provide quasi–real-time intraoperative histopathology during brain surgery to improve the surgical care and may be extended to other types of human solid tumors with transfer learnings.
RESULTS
Dual-color SRS imaging of fresh brain tissue
To enable high-quality label-free histologic imaging of fresh unprocessed brain tissue, rapid two-color SRS microscopy techniques have been developed, such as dual-phase lock-in detection and deep learning–based single-shot femtosecond-SRS methods (24). Here, we mainly applied dual-phase technique to collect brain tissue images, although single-shot femtosecond-SRS could provide equivalent results (fig. S1). Two-color SRS microscopy captures raw images at two Raman frequencies (2845 cm−1 for CH2 and 2930 cm−1 for CH3 vibrations), allowing the extraction of lipid (green colored) and protein (blue colored) as the contrast basis for conventional SRS histology of tissues (Fig. 1), including but not limited to the brain (15, 16, 22, 24). However, generating FFPE-quality virtual H&E images from SRS is difficult for conventional CycleGAN algorithm, with mapping errors due to weakly supervised limitations. Despite previous trials of using linear color projection algorithms to mimic the lookup tables (LUTs) of H&E (11, 12, 18), substantial deviations from true H&E staining remain unsolved because of the distinct differences of contrast mechanism between SRS and H&E.
Creating SRC-GAN model to generate virtual H&E from SRS images
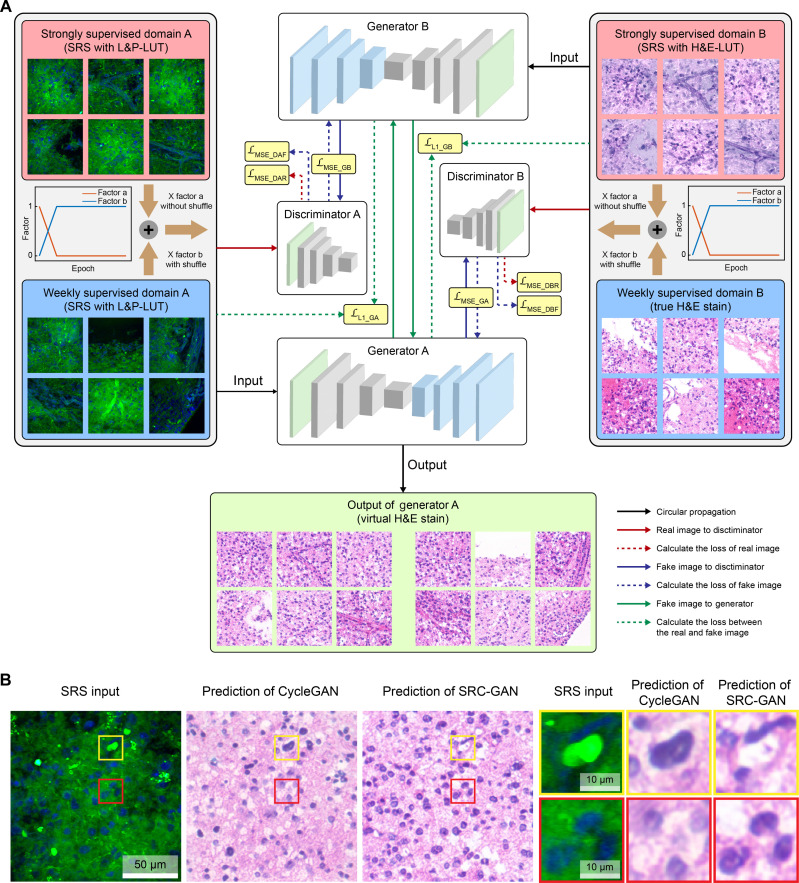
We constructed an improved semi-supervised learning model, named stimulated Raman CycleGAN (SRC-GAN), to convert lipid/protein-based SRS images to H&E stains (Fig. 2A). The core architecture of SRC-GAN consists of two sets of generators and discriminators (fig. S2). The generator is responsible for generating the target domain image, with enhanced performance by integrating with ResNet-based blocks (32) and a padding layer. The discriminator uses a fully convolutional architecture known as Patch-GAN (30), which enables more accurate image evaluation by mapping from a 512–by–512 input to a 62–by–62 array, rather than a single scalar output representing real or virtual image (fig. S2). Such pixel-level judgment capability allows the discriminator to assess image authenticity in finer subareas. Unlike conventional CycleGANs, SRC-GAN uses a label mask instead of a single scalar label to train the generator. The label mask is used to calculate the mean squared error loss (LMSE) along with the decision mask of the discriminator output. In addition, a self-regulated loss function (LL1) is applied to promote the generalizability of the generator by supervising the input and the loop generation output, which enhance the clarity of the generator output.
Fig. 2. Model architecture for stimulated Raman virtual staining.
(A) Detailed diagram of the semi-supervised SRC-GAN model consisting of two generators and two discriminators. The training data gradually evolve from paired SRS images with L&P- and H&E-LUT to fully unpaired images of SRS and true H&E stains. (B) The accurate conversion of cell nucleus and lipid bodies by SRC-GAN, in comparison with the mapping errors by conventional CycleGAN.
One key challenge of generic CycleGAN is that the weakly supervised learning approach tends to yield severe mapping errors for the conversion of SRS to H&E because of the large differences of their color styles and contrast mechanisms. For instance, lipid droplets in SRS images are constantly mis-mapped to cell nuclei in the output virtual H&E images (Fig. 2B), whereas the cell nuclei may be mis-mapped to common voids—usually represent lipid bodies in H&E due to the deparaffination process. Therefore, we introduce strongly supervised substructures using paired training set of SRS images with two color styles (fig. S3A), one with the common lipid/protein LUT (L&P-LUT) and the other with H&E-LUT as developed previously (11, 17, 18, 24). Although H&E-LUT still differs from true H&E staining in many details, it provides a color style closer to H&E that leads the model to the correct mapping direction. We further implement a gradually weakening semi-supervised algorithm, where the dynamic training set evolves with the growing number of epochs (Fig. 2A). The model was initially trained using paired L&P- and H&E-LUT images. As the training progressed, the training dataset gradually transitioned to partially unpaired images and finally to the completely unpaired dataset of L&P-SRS and true H&E images. This progressive transition enables the model to converge toward the desired optimization goal during the training stages.
The loss function curves of the generator and discriminator obtained from the training process show two intersections (circles in fig. S3B), demonstrating the adversarial process of the two supervised modes during the model transformation iterations. The output images at different iterations showed a transition from H&E-LUT to virtual H&E images (fig. S3C). For example, the blood vessels gradually transformed from the initial dark purple to the final pinkish H&E-staining color, and the cell nuclei converged to the effect of hematoxylin stains. The well-agreements between the output images and H&E stains indicate that SRC-GAN model could nonlinearly project SRS images to virtual histologic stains with high accuracy.
Validating the performance of SRC-GAN on brain tissues
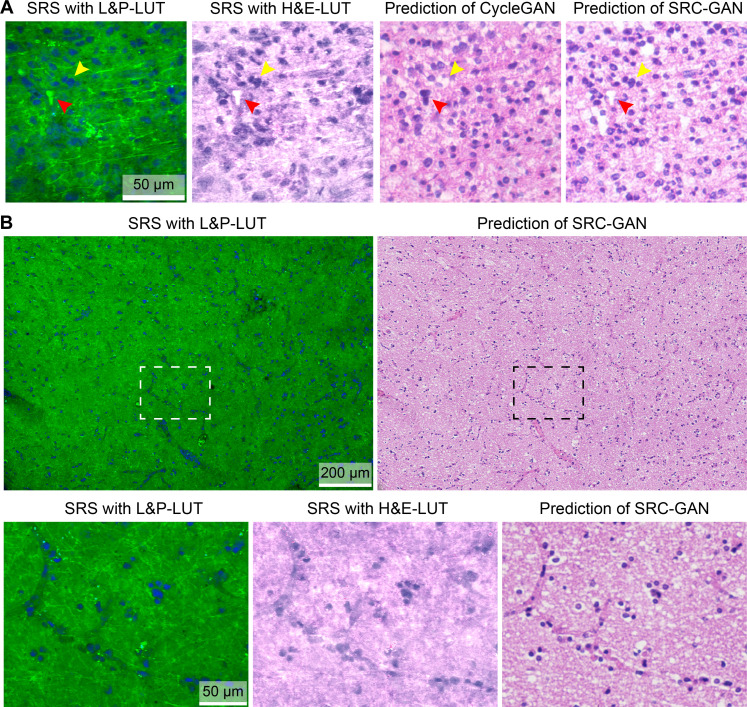
We then evaluate the accuracy and reliability of SRC-GAN by comparing the prediction outputs with generic CycleGAN, along with the images of L&P-LUT and H&E-LUT (Fig. 3A). On fresh brain tissues, SRC-GAN correctly produces important histoarchitectures (cell nuclei, blood vessels, etc.) in terms of both morphology and color, without the mapping errors of CycleGAN, and generates much more superior virtual staining effects comparing with the H&E-LUT images (Fig. 3B). However, fresh tissue results could not provide ground truths to assess the accuracy of model predictions.
Fig. 3. Generating virtual stains from SRS images of brain tissues.
(A) The output image of SRC-GAN compared with those of the generic CycleGAN and SRS images of L&P- and H&E-LUT, showing conversions of lipid bodies (red arrow) and cells (yellow arrow). (B) A typical fresh brain tissue imaged with SRS and transformed to virtual stain; detailed comparisons of cells and microvasculatures are shown in the magnified images.
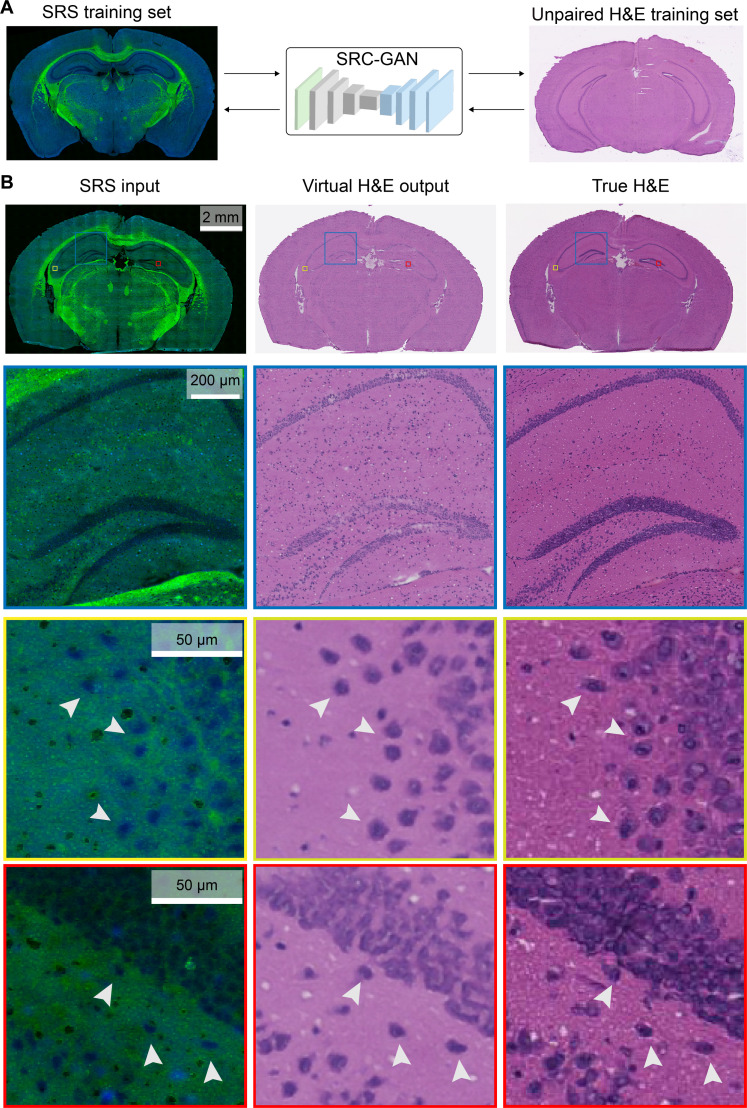
To enable ground-truth validation, we performed SRS imaging on thin frozen sections of mouse brain and subsequently stained the same tissue sections with H&E to serve as the ground truths. Note that the training process uses separate datasets of unpaired SRS and H&E images of different frozen tissue sections (Fig. 4A). The input SRS image of a whole mouse brain coronal section is transformed to virtual H&E via SRC-GAN and compared with the true H&E stain of the same section (Fig. 4B). Near-perfect match between the prediction and ground truth could be seen, not only the overall agreement of the whole brain section (first row of Fig. 4B) but also the magnified view of brain structures such as the hippocampus region (second row of Fig. 4B), and prediction accuracy at the single-cell level is demonstrated (third row of Fig. 4B). The slight color differences between the predicted and true H&E are from the different thicknesses of the SRS-imaged section (10 μm) and the training H&E section (6 μm).
Fig. 4. Ground-truth validation of virtual H&E using thin frozen tissue sections.
(A) The use of unpaired sections of SRS and H&E images as the training dataset for SRC-GAN model. (B) The output virtual stains are compared with the true H&E of the same tissue section imaged with SRS. Typical mouse brain histoarchitectures are clearly revealed by SRS and converted to virtual H&E with cell-to-cell accuracy (arrowheads).
Moreover, we extracted the feature layer of the generator during the translation process and flattened it into a vector with reduced dimension (to n = 3) using t-distributed stochastic neighbor embedding (t-SNE). Three distinct subgroups were clustered in the reduced space to represent images with low, medium, and high nuclear densities (fig. S4), indicating that the model perception of mapping in heterogeneous patterns is sensitive enough to differentiate them. We further calculate the nuclear density and protein/lipid ratio of the three subtypes as depicted in the histograms (fig. S4) and show a clear trend that higher nuclear density corresponds to higher protein/lipid ratio, and vice versa. It implies that the multichemical information obtained from SRS plays an important role in the accurate transformation to virtual H&E.
Virtual FFPE reveals key diagnostic features of fresh brain tissues
The above demonstrations on frozen sections are mainly for the purpose of ground-truth validation, since paraffin embedded sections could not be used for SRS histology. However, the final goal of our work is to achieve FFPE-quality virtual H&E using SRS images of fresh tissue. This could be realized by training unpaired SRS images of fresh tissues and FFPE of different ones. Hence, the SRC-GAN model and training process are essentially the same as that used in frozen sections. Only the training data of frozen-section SRS are replaced by fresh-tissue SRS, and frozen-H&E is replaced by FFPE-H&E, which is expected to share identical model performance, but the output results now converge to FFPE style. The whole-slide images (WSIs) of virtual-FFPE showed high-quality conversion results at different scales (fig. S5 and movie S1).
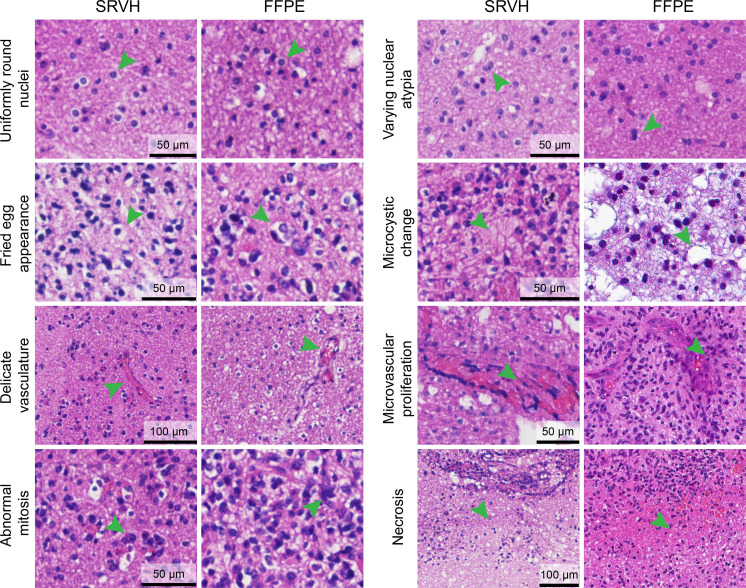
Although our method of generating virtual FFPE lacks the ground truth for the aforementioned reasons, its quality could be assessed by comparing with true FFPE from the same patient cases. Almost all the clinically vital histologic features of glial tumors are revealed in the SRVH images that appear undifferentiable from true FFPE (Fig. 5). These include the cytologic features of uniformly round nuclei, fried-egg appearance, nuclear atypia, and microcystic change. Other histologic features could also be clearly visualized, such as abnormal mitosis, delicate vasculatures, microvascular proliferation, and necrosis. Aside from tumors, other brain features such as cerebellum could also be demonstrated as virtual stains (fig. S6).
Fig. 5. FFPE-quality virtual histology provides key diagnostic features of brain tumor.
SRVH reveals critical cytologic and histologic features for the diagnosis of brain tumor subtypes, including various cellular and nuclear atypia, microcytic change, abnormal vasculature, microvascular proliferation, and necrosis, etc.
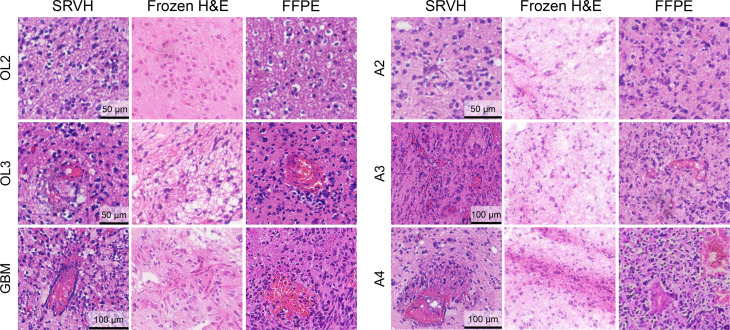
On the basis of these histopathological details, SRVH enabled extended diagnostic power of brain tumors to differentiate important subtypes of glioma (Fig. 6). The features of uniformly round nuclei and fried-egg shapes allow the identification of oligodendroma, the features of nuclear atypia and microcystic change of cytoplasm for the diagnosis of astrocytoma, and the appearance of necrosis and microvascular proliferation for the recognition of glioblastoma. Even finer histologic grades within oligodendroma (grades 2 and 3) and astrocytoma (grades 2 and 4) could be identified by the high-resolution histologic images generated via SRVH. For direct comparison, FFPE and frozen H&E results of the corresponding patients are presented along with SRVH images of different subtypes of brain tumors (Fig. 6). As can be seen, SRVH resembles the results of FFPE, while frozen H&E displays degraded histologic features, such as tissue loss, cell rupture, and blurred tissue morphology, due to the formation of ice crystals and sectioning artefacts. We also simulated intraoperative histology of resection margins on unprocessed surgical tissues, showing the transition of histologic features from the intratumor site to peritumor and further to extratumor (fig. S7).
Fig. 6. SRVH differentiates brain tumor subtypes.
FFPE-quality virtual H&E identifies tumor subtypes and allows the further diagnosis of various tumor grades, such as grades 2 and 3 of oligodendroma, grades 2 and 4 of astrocytoma, and glioblastoma. In contrast, frozen H&E shows degraded histology with freezing and sectioning artefacts.
Blind evaluation of diagnostic efficacy for subtypes of glioma
With the capability of revealing key histologic features of glioma, we next evaluated the diagnostic efficacy of SRVH in the slide-free and stain-free workflow. Fresh surgical tissues (typical 4 mm–by–4 mm size) were loaded between two coverslips with an ~300-μm-thick spacer and directly imaged with SRS to generate SRVH images (within 3 min) as virtually stained WSIs. Neighboring tissues of the same patient cases were sent for FFPE and frozen section–based H&E staining. Blind validations by three board-certified neuropathologists were conducted on mixed and randomly ordered WSIs of SRVH, FFPE, and frozen H&E from 43 patient cases of central nervous system tumors. Without any pre-knowledge or training of the virtual staining technique, the pathologists were asked to rate the WSIs into four categories: astrocytoma, oligodentroma, high-grade glioma, and other tumors. Diagnostic results from the hospital served as the ground truths for all the tests. The results are summarized in Table 1, indicating that the pathologists were able to differentiate glioma subtypes on SRVH, with diagnostic accuracies approaching that of FFPE and notably higher than that of frozen H&E. Comparing with the gold-standard FFPE, current SRVH technique still has limited resolution on certain subcellular features, such as detailed morphologies of nuclear and cytoplasm. Nevertheless, SRVH provides sufficient cytologic and histologic information with higher speed and accuracy than frozen H&E and may become a superior tool for intraoperative diagnosis.
Table 1. Blind evaluations of SRVH, frozen H&E, and FFPE staining on subtypes of glioma.
| Imaging modality | Histologic subtype | Pathologist 1 | Pathologist 2 | Pathologist 3 | Accuracy (%) | |||
|---|---|---|---|---|---|---|---|---|
| Correct | Incorrect | Correct | Incorrect | Correct | Incorrect | |||
| SRVH | ||||||||
| Astrocytoma | 6 | 0 | 6 | 0 | 5 | 1 | 94.4 | |
| Oligodendroma | 5 | 0 | 5 | 0 | 4 | 1 | 93.3 | |
| High-grade glioma | 23 | 3 | 21 | 5 | 20 | 6 | 82.1 | |
| Other tumors | 4 | 2 | 5 | 1 | 5 | 1 | 77.8 | |
| Total | 38 | 5 | 37 | 6 | 34 | 9 | 84.5 | |
| Accuracy (%) | 88.4 | 86.0 | 79.1 | |||||
| FFPE | ||||||||
| Astrocytoma | 7 | 0 | 6 | 1 | 7 | 0 | 95.2 | |
| Oligodendroma | 7 | 0 | 7 | 0 | 6 | 1 | 95.2 | |
| High-grade glioma | 20 | 2 | 19 | 3 | 18 | 4 | 86.4 | |
| Other tumors | 6 | 1 | 5 | 2 | 7 | 0 | 85.7 | |
| Total | 40 | 3 | 37 | 6 | 38 | 5 | 89.1 | |
| Accuracy (%) | 93.0 | 86.0 | 88.4 | |||||
| Frozen H&E | ||||||||
| Astrocytoma | 4 | 3 | 3 | 4 | 4 | 3 | 52.4 | |
| Oligodendroma | 5 | 2 | 4 | 3 | 4 | 3 | 61.9 | |
| High-grade glioma | 15 | 7 | 14 | 8 | 12 | 10 | 62.1 | |
| Other tumors | 3 | 1 | 3 | 1 | 2 | 2 | 66.7 | |
| Total | 27 | 13 | 24 | 16 | 22 | 18 | 60.8 | |
| Accuracy (%) | 67.5 | 60.0 | 55.0 | |||||
DISCUSSION
Rapid intraoperative histopathology plays a crucial role during surgery, and extensive efforts have been made to develop AI-based instant histopathology toward this goal. In the current study, we have developed a modified CycleGAN model tailored for SRS images to achieve label-free and slide-free virtual FFPE histology without the need of complex tissue processing. The advantage of SRS in fast chemical imaging is fully exploited to generate high-quality H&E results interpretable to professional pathologists. Compared with imaging modalities that generate virtual stains from single-channel grayscale images (5, 17, 18, 24, 27), SRS provides dual-chemical information (lipid and protein) for the accurate transformation to dual-stain (H&E) results, avoiding the mapping errors using single-channel SRS images (fig. S8). The success of our model is mainly attributed to the design of strongly supervised structure that bridges L&P and H&E color styles, which takes advantage of the linear projection algorithms (11, 12) and generates FFPE-quality histology to reveal detailed H&E features directly accessible to human diagnosis. Although conventional SRS images could be integrated with convolutional neural network (CNN) for AI-assisted classification (21), the images themselves are noninterpretable to ordinary pathologists, not to mention glioma subtyping, which relies on subtle histologic differences (fig. S9).
Aside from the demonstrated capabilities, our method could be further improved in several aspects. The current model has reached decent accuracy at the cellular level as verified on identical sections, but the detailed subcellular information is difficult to retain, such as chromatin density and distribution. Higher spatial resolution, improved training data, and more advanced deep-learning model may be required to push the virtual staining toward subcellular accuracy. More histologic features (calcification, vascular hyperplasia, etc.) of brain tissues may be identified using enlarged training dataset and optimized model architecture. Several technical advances are needed to increase the histologic imaging speed. The current acquisition time for a 512 × 512 pixels image frame (~400 μm by 400 μm) is about 1 s (Materials and Methods), and the current SRC-GAN conversion time is ~24 frames/s. Hence, a typical 4 mm–by–4 mm–sized tissue is estimated to cost <3 min to generate virtual FFPE image. Faster laser scanning device (such as a resonant galvo mirror), better stitching strategy (33), and upgraded computational hardware will further improve the speed to cope with larger-area specimens.
Volumetric histology provides three-dimensional (3D) tissue structures exceeding the information acquired by conventional histology of a certain section. 3D tissue imaging could be realized by stitching a stack of 2D images taken on either physical sections (34) or optical sections (35). SRS is usually limited to shallow imaging depth of ~100 μm in turbid tissues. To extend the optical imaging depth, various tissue-clearing techniques have been developed, most of which involve the removal of lipid contents (36). Here, we adapted a clearing method for SRS imaging without the extraction of lipids (37) by the use of high-concentration urea and CHAPS (see Materials and Methods). A quick proof-of-principle demonstration showed SRVH for a brain tissue block of 363 by μm 363 μm by 500 μm (fig. S10), revealing continuous variation of brain histopathology at various depths and the 3D relationship of the connecting histoarchitectures (movie S2). Future efforts will be emphasized on developing fast tissue clearing methods (38, 39) and rapid volumetric imaging techniques for practical applications.
The proposed method may be extended to other imaging modalities and histopathology techniques, including immunohistochemistry, spatial-omics, etc. Predictions of immunohistochemistry and mutations from H&E-staining results have been reported (40), and similar approach of conversion from SRS images to virtual immunostains may also be achievable with careful experimental design. Multimodal imaging can provide information of more histologic features (collagen fibers, elastin fiber, NADH, etc.) (2, 41), which may enable the model to learn more diverse histoarchitectures for highly heterogeneous tissue types such as breast tumor (15). Last, SRVH could be applied to other types of human solid tumors such as gastric cancer and lung cancer as shown in our preliminary results (fig. S11) and may be generalizable with transfer learning algorithms.
In summary, we have developed an SRVH method to produce FFPE-quality images on unprocessed fresh brain tissues within a few minutes. With the SRS-tailored deep-learning model, virtual FFPE images could reveal detailed histologic features of brain tumors that allow accurate diagnosis of tumor subtypes. Our method provides potentials for rapid intraoperative histology, as well as 3D histopathology extendable to other types of solid tumors.
MATERIALS AND METHODS
Sample preparation
The study was approved by the Institutional Ethics Committee of Huashan Hospital with written informed consent (approval no. KY2022-762). All human tissue samples were collected from patients who underwent brain tumor resection. The harvested fresh samples were transferred to the imaging laboratory under 4°C within 2 hours, sealed between two coverslips with a spacer to generate uniform thickness (~300 μm) slices for direct SRS imaging. Total 47 patients were recruited, including 7 cases of astrocytoma, 7 cases of oligodendroma, 26 cases of high-grade glioma, and 7 others. Among these, 15 cases generated SRVH images for training the SRC-GAN network and 28 separated cases for testing; all the 43 cases were used in the blind evaluation for pathologists. Most of the cases imaged with SRS have corresponding FFPE and frozen H&E images, with a few lacking frozen sections (Table 1). For frozen sections of mouse brain, whole brains were snap-frozen in liquid nitrogen and then immersed in optimal cutting temperature compound and stored at −80°C. Frozen tissues were sectioned to 10-μm-thick sections for SRS imaging and subsequent H&E staining of the same sections for ground-truth validation of the model. Unpaired sections of 6-μm thickness were H&E stained for training purpose.
SRS microscopy and data acquisition
In our SRS imaging setup, a femtosecond laser (OPO, Insight DS+, Newport) with a tunable pump beam (680 to 1300 nm, ~150 fs) and a fixed Stokes beam (1040 nm, ~200 fs) was used as the light source to generate the SRS signal. For use in the “spectral focusing” mode, both the pump and Stokes beams were linearly chirped to picoseconds through SF57 glass rods to supply the resolvable spectral condition. The Stokes beam, then, was modulated by an electro-optical modulator at 20 MHz and overlapped temporally and spatially with the pump beam through a dichroic mirror (DMSP1000, Thorlabs). The collinearly overlapped beams were tightly focused onto the sample using a water immersion objective lens (UPLSAPO 25XWMP2, NA 1.0 water, Olympus) after entering into a laser scanning microscope (FV1200, Olympus). The transmitted stimulated Raman loss signal was detected by a Si photodiode and demodulated by a lock-in amplifier (HF2LI, Zurich Instruments) to generate an SRS image. The target Raman wave number was adjusted by changing the time delay between the pump and the Stokes beams. For histopathological imaging, two-time delays corresponding to 2845 and 2930 cm−1 were set to acquire raw images that could be further decomposed to the distributions of lipid and protein. All images were of 512 × 512 pixels with pixel dwell time of 2 μs. The spatial resolution is ~350 nm. Laser powers at the sample were kept as pump ~40 mW and Stokes ~40 mW.
WSI acquisition and image processing
For SRS imaging, two-color WSIs were generated to include lipid and protein contrast, which was performed as follows: (i) A motorized 2D translational stage was used for mosaic imaging by acquiring a series of images tiles, which were stitched into large flattened images at 2845 and 2930 cm−1, and further generated lipid and protein channels. (ii) Two types of LUTs were used to color-code the dual-channel SRS images. In the L&P-LUT, the lipid channel was mapped to green, and the protein channel was mapped to blue. In the H&E-LUT, the lipid channel was mapped to pink, and the protein channel was mapped to dark purple. (iii) Each pair of the two-channel images was merged into a single WSI.
For imaging H&E-stained tissue slides, we used a bright-field microscope with a charge-coupled device camera and a motorized sample stage to stitch the mosaic images into a large WSI.
Network architecture
In this study, we used a modified CycleGAN architecture (fig. S2) with a pair of generators and discriminators to learn the transformation from a label-free SRS input image to the corresponding H&E stain. Unlike the typical CycleGAN architecture that consists of a full convolutional layer generator and conventional CNN discriminator without edge processing, we improved the network of conversion histopathology images with several key modifications.
For the generator part, kernels were used with the size of 3 × 3. In particular, we added the padding layer to the first and penultimate layers to solve the problem of poor image edge mapping quality. In addition, a residual architecture layer was added before deconvolution to enable the model with both linear and nonlinear mapping capabilities.
For the discriminator part, we used a fully convolutional architecture instead of a combination of convolution and fully connected architecture with 4 × 4–sized kernels. The decision label and output dimensions were kept the same, allowing input images of different sizes.
Semi-supervised algorithm design
We designed a semi-supervised algorithm based on CycleGAN, which converts L&P-LUT images to FFPE virtual stains using H&E-LUT images as the intermediate transition, where L&P-LUT represents the conventional color-coding of SRS with lipid (green) and protein (blue) information, and H&E-LUT represents the color projection to H&E-like style but still differs from true H&E. Python language (https://python.org) and the deep learning framework of Pytorch (https://pytorch.org) were used to build the algorithm.
We split the dataset into two domains called domain X and domain Y after slicing the large image into 450 × 450–pixel tiles, and each domain was composed of strongly and weakly supervised parts, respectively. The strong supervision part consists of paired L&P- and H&E-LUT images, while the weak supervision part consists of unpaired L&P-LUT and true H&E images.
During model training process, the proportion of two parts in the training set was adjusted by two factors, represented as a factor a and factor b, respectively
where epoch is the iteration of model training. At the beginning of model training, the strongly supervised part occupies the main part of the training set. As the epoch increases, the weakly supervised part gradually takes over the training dominant.
Hyperparameters of model and training details
During the training of the model, the loss values of many loss functions need to be calculated to calculate gradients and update model parameter weights. For the generator, the image generated by the generator should be judged to be real by the discriminator; meanwhile, regular supervision was introduced to increase the generalization ability of the model, which made the output images look more like real histopathological images. The loss function is, hence, calculated as
where X and Y represent domain A and domain B, GA and GB represent generator A and generator B, and DA and DB represent discriminator A and discriminator B. The model weight parameters of the generator, then, could be optimized after the gradient descent direction is calculated by LG. As for the discriminator, it should attempt to recognize whether the patch is generated by the generator or a real image. To promote the judgment of the discriminator, the loss function of that is
where the label-mask instead of single-label is used. The model weight parameters of the discriminator, then, could be optimized after the gradient descent direction is calculated by and , respectively.
Generation of the whole-slide virtual staining image
Since our SRC-GAN model is based on patch level, the generation of the whole-slide virtual histologic image from the converted small tiles was performed as follows: (i) The full SRS image of L&P-LUT was sliced by a sliding window with 450 × 450 pixels starting from the top left corner. (ii) Then, the patch obtained by sliding the window was fed to the SRC-GAN model to generate a virtual H&E patch. (iii) The sliding window was moved with a step size of 150 pixels, and step (ii) was repeated to obtain a series of patches. (iv) The patches were filled into the mosaic matrix according to their numbers, and the duplicate areas were normalized to synthesize an entire virtual staining image.
Ground-truth validation of SRC-GAN
Since paraffin-embedded tissue sections are improper for SRS histologic imaging, we chose to conduct ground-truth validation on thin frozen sections of mouse brain because of the rich and well-known histoarchitectures. SRS and frozen H&E images from different brain sections (unpaired data) were used for model training, whereas those from the same sections (paired data) were used for validation. To get paired images of the identical tissue sections, they were first noninvasively imaged with SRS to generate input data for the model and subsequently stained with H&E to serve as the ground truth. Therefore, virtual H&E converted from SRS images could be compared with true H&E from the same brain tissues.
Tissue clearing
The fresh brain tissues were manually sliced to a thickness of ~1 mm and then immersed in the clearing solution at room temperature, which was prepared by diluting CHAPS (V900480, Sigma-Aldrich) in 8 M urea (after reconstitution with 16 ml of high-purity water, U4883, Sigma-Aldrich) to final concentration of 1% w/v. After 5 days, the samples were transferred to a homemade chamber (perforated glass slides) and mounted in the clearing solution for SRS microscopy. By stepping through the z axis of the sample with a step size of 4 μm, we could obtain an image stack that carried depth information.
Signal compensation
Because SRS signal decays with increasing depth into the tissue, we compensated the decayed signal for 3D volumetric imaging. An exponential-decay fitting function was used to normalize signals in the restrained depth, and the original SRS signal was divided by the decay function to yield corrected signal for 3D reconstruction.
Statistical analysis
The t-SNE dimensionality reduction, histogram of nuclear density, and protein/lipid ratio were calculated to evaluate the performance of the model. The analyses were run on Matlab R2023a for MacOS (https://mathworks.com/products/matlab.html).
Acknowledgments
We thank W. Min and M. Wei from Columbia University for helping with tissue clearing protocol.
Funding: We acknowledge the financial supports from the National Key R&D Program of China (2021YFF0502900); National Natural Science Foundation of China (61975033); Municipal Natural Science Foundation of Shanghai (23dz2260100); Shanghai Municipal Science and Technology Project (22Y11907500); Shanghai Municipal Science and Technology Major Project (2018SHZDZX01); and Beijing Xisike Clinical Oncology Research Foundation (Y-zai2021/qn-0204).
Author contributions: M.J., Z.L., J.A., and Y.M. designed the study; L.C. prepared the tissue samples; Z.L., Y.C., and J.A. performed stimulated Raman scattering microscopy experiments; Z.L. performed deep learning algorithms and training; H.C., J.X., and X.L. performed histologic diagnosis; M.J., L.C., and Z.L. wrote the manuscript with contributions from all the authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Codes and data for training the deep-learning model are available under open access at https://zenodo.org/doi/10.5281/zenodo.10622491 and available from the corresponding author upon request.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Legends for moives S1 and S2
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Eisenhardt L., Cushing H., Diagnosis of intracranial tumors by supravital technique. Am. J. Pathol. 6, 541–552.7 (1930). [PMC free article] [PubMed] [Google Scholar]
- 2.Zipfel W. R., Williams R. M., Christie R., Nikitin A. Y., Hyman B. T., Webb W. W., Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc. Natl. Acad. Sci. U.S.A. 100, 7075–7080 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fereidouni F., Harmany Z. T., Tian M., Todd A., Kintner J. A., McPherson J. D., Borowsky A. D., Bishop J., Lechpammer M., Demos S. G., Levenson R., Microscopy with ultraviolet surface excitation for rapid slide-free histology. Nat. Biomed. Eng. 1, 957–966 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy P., Claude J. B., Tiwari S., Barulin A., Wenger J., Ultraviolet nanophotonics enables autofluorescence correlation spectroscopy on label-free proteins with a single tryptophan. Nano Lett. 23, 497–504 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Cao R., Nelson S. D., Davis S., Liang Y., Luo Y., Zhang Y., Crawford B., Wang L. V., Label-free intraoperative histology of bone tissue via deep-learning-assisted ultraviolet photoacoustic microscopy. Nat. Biomed. Eng. 7, 124–134 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C., Zhang Y. S., Yao D. K., Xia Y., Wang L. V., Label-free photoacoustic microscopy of cytochromes. J. Biomed. Opt. 18, 20504 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freudiger C. W., Min W., Saar B. G., Lu S., Holtom G. R., He C. W., Tsai J. C., Kang J. X., Xie X. S., Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 322, 1857–1861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji M., Lewis S., Camelo-Piragua S., Ramkissoon S. H., Snuderl M., Venneti S., Fisher-Hubbard A., Garrard M., Fu D., Wang A. C., Heth J. A., Maher C. O., Sanai N., Johnson T. D., Freudiger C. W., Sagher O., Xie X. S., Orringer D. A., Detection of human brain tumor infiltration with quantitative stimulated Raman scattering microscopy. Sci. Transl. Med. 7, 309ra163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji M., Orringer D. A., Freudiger C. W., Ramkissoon S., Liu X., Lau D., Golby A. J., Norton I., Hayashi M., Agar N. Y., Young G. S., Spino C., Santagata S., Camelo-Piragua S., Ligon K. L., Sagher O., Xie X. S., Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci. Transl. Med. 5, 201ra119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji M., Arbel M., Zhang L., Freudiger C. W., Hou S. S., Lin D., Yang X., Bacskai B. J., Xie X. S., Label-free imaging of amyloid plaques in Alzheimer's disease with stimulated Raman scattering microscopy. Sci. Adv. 4, eaat7715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin K. S., Francis A. T., Hill A. H., Laohajaratsang M., Cimino P. J., Latimer C. S., Gonzalez-Cuyar L. F., Sekhar L. N., Juric-Sekhar G., Fu D., Intraoperative assessment of skull base tumors using stimulated Raman scattering microscopy. Sci. Rep. 9, 20392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarri B., Poizat F., Heuke S., Wojak J., Franchi F., Caillol F., Giovannini M., Rigneault H., Stimulated Raman histology: One to one comparison with standard hematoxylin and eosin staining. Biomed. Opt. Express 10, 5378–5384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang H., Li Y., Fung A. A., Bagheri P., Hoang K., Skowronska-Krawczyk D., Chen X., Wu J. Y., Bintu B., Shi L., Super-resolution SRS microscopy with A-PoD. Nat. Methods 20, 448–458 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu S., Shi W., Luo T., He Y., Zhou L., Yang J., Yang Z., Liu J., Liu X., Guo Z., Yang C., Liu C., Huang Z. L., Ries J., Zhang M., Xi P., Jin D., Li Y., Field-dependent deep learning enables high-throughput whole-cell 3D super-resolution imaging. Nat. Methods 20, 459–468 (2023). [DOI] [PubMed] [Google Scholar]
- 15.Yang Y., Liu Z., Huang J., Sun X., Ao J., Zheng B., Chen W., Shao Z., Hu H., Yang Y., Ji M., Histological diagnosis of unprocessed breast core-needle biopsy via stimulated Raman scattering microscopy and multi-instance learning. Theranostics 13, 1342–1354 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L., Wu Y., Zheng B., Su L., Chen Y., Ma S., Hu Q., Zou X., Yao L., Yang Y., Chen L., Mao Y., Chen Y., Ji M., Rapid histology of laryngeal squamous cell carcinoma with deep-learning based stimulated Raman scattering microscopy. Theranostics 9, 2541–2554 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollon T. C., Lewis S., Pandian B., Niknafs Y. S., Garrard M. R., Garton H., Maher C. O., McFadden K., Snuderl M., Lieberman A. P., Muraszko K., Camelo-Piragua S., Orringer D. A., Rapid intraoperative diagnosis of pediatric brain tumors using stimulated Raman histology. Cancer Res. 78, 278–289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orringer D. A., Pandian B., Niknafs Y. S., Hollon T. C., Boyle J., Lewis S., Garrard M., Hervey-Jumper S. L., Garton H. J. L., Maher C. O., Heth J. A., Sagher O., Wilkinson D. A., Snuderl M., Venneti S., Ramkissoon S. H., McFadden K. A., Fisher-Hubbard A., Lieberman A. P., Johnson T. D., Xie X. S., Trautman J. K., Freudiger C. W., Camelo-Piragua S., Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat. Biomed. Eng. 1, 0027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C., Lu M. Y., Williamson D. F. K., Chen T. Y., Schaumberg A. J., Mahmood F., Fast and scalable search of whole-slide images via self-supervised deep learning. Nat. Biomed. Eng. 6, 1420–1434 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollon T., Jiang C., Chowdury A., Nasir-Moin M., Kondepudi A., Aabedi A., Adapa A., Al-Holou W., Heth J., Sagher O., Lowenstein P., Castro M., Wadiura L. I., Widhalm G., Neuschmelting V., Reinecke D., von Spreckelsen N., Berger M. S., Hervey-Jumper S. L., Golfinos J. G., Snuderl M., Camelo-Piragua S., Freudiger C., Lee H., Orringer D. A., Artificial-intelligence-based molecular classification of diffuse gliomas using rapid, label-free optical imaging. Nat. Med. 29, 828–832 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollon T. C., Pandian B., Adapa A. R., Urias E., Save A. V., Khalsa S. S. S., Eichberg D. G., D'Amico R. S., Farooq Z. U., Lewis S., Petridis P. D., Marie T., Shah A. H., Garton H. J. L., Maher C. O., Heth J. A., McKean E. L., Sullivan S. E., Hervey-Jumper S. L., Patil P. G., Thompson B. G., Sagher O., McKhann G. M. II, Komotar R. J., Ivan M. E., Snuderl M., Otten M. L., Johnson T. D., Sisti M. B., Bruce J. N., Muraszko K. M., Trautman J., Freudiger C. W., Canoll P., Lee H., Camelo-Piragua S., Orringer D. A., Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat. Med. 26, 52–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ao J., Shao X., Liu Z., Liu Q., Xia J., Shi Y., Qi L., Pan J., Ji M., Stimulated Raman scattering microscopy enables Gleason scoring of prostate core needle biopsy by a convolutional neural network. Cancer Res. 83, 641–651 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manifold B., Men S., Hu R., Fu D., A versatile deep learning architecture for classification and label-free prediction of hyperspectral images. Nat. Mach. Intell. 3, 306–315 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z., Su W., Ao J., Wang M., Jiang Q., He J., Gao H., Lei S., Nie J., Yan X., Guo X., Zhou P., Hu H., Ji M., Instant diagnosis of gastroscopic biopsy via deep-learned single-shot femtosecond stimulated Raman histology. Nat. Commun. 13, 4050 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai B., Yang X., Li Y., Zhang Y., Pillar N., Ozcan A., Deep learning-enabled virtual histological staining of biological samples. Light Sci. Appl. 12, 57 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozyoruk K. B., Can S., Darbaz B., Basak K., Demir D., Gokceler G. I., Serin G., Hacisalihoglu U. P., Kurtulus E., Lu M. Y., Chen T. Y., Williamson D. F. K., Yilmaz F., Mahmood F., Turan M., A deep-learning model for transforming the style of tissue images from cryosectioned to formalin-fixed and paraffin-embedded. Nat. Biomed. Eng. 6, 1407–1419 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Rivenson Y., Wang H., Wei Z., de Haan K., Zhang Y., Wu Y., Gunaydin H., Zuckerman J. E., Chong T., Sisk A. E., Westbrook L. M., Wallace W. D., Ozcan A., Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat. Biomed. Eng. 3, 466–477 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Creswell A., White T., Dumoulin V., Arulkumaran K., Sengupta B., Bharath A. A., Generative adversarial networks: An overview. IEEE Signal Process. Mag. 35, 53–65 (2018). [Google Scholar]
- 29.I. J. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville, Y. Bengio, in Generative adversarial nets. Proceedings of the 27th International Conference on Neural Information Processing Systems (NIPS) (ACM, 2014), volume 2, pp. 2672–2680. [Google Scholar]
- 30.P. Isola, J.-Y. Zhu, T. Zhou, A. A. Efros, in Image-to-image translation with conditional adversarial networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2017), pp. 5967–5976. [Google Scholar]
- 31.J.-Y. Zhu, T. Park, P. Isola, A. A. Efros, in Unpaired image-to-image translation using cycle-consistent adversarial networks. 2017 IEEE International Conference on Computer Vision (ICCV) (IEEE, 2017), pp. 2242–2251. [Google Scholar]
- 32.K. He, X. Zhang, S. Ren, J. Sun, in Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2016), pp. 770–778. [Google Scholar]
- 33.Zhang B., Sun M., Yang Y., Chen L., Zou X., Yang T., Hua Y., Ji M., Rapid, large-scale stimulated Raman histology with strip mosaicing and dual-phase detection. Biomed. Opt. Express 9, 2604–2613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li A., Gong H., Zhang B., Wang Q., Yan C., Wu J., Liu Q., Zeng S., Luo Q., Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science 330, 1404–1408 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y., Zhang M., Zhang W., Zhou Y., Chen L., Liu Q., Wang P., Chen R., Duan X., Chen F., Deng H., Wei Y., Fei P., Zhang Y. H., Isotropic super-resolution light-sheet microscopy of dynamic intracellular structures at subsecond timescales. Nat. Methods 19, 359–369 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Chung K., Wallace J., Kim S. Y., Kalyanasundaram S., Andalman A. S., Davidson T. J., Mirzabekov J. J., Zalocusky K. A., Mattis J., Denisin A. K., Pak S., Bernstein H., Ramakrishnan C., Grosenick L., Gradinaru V., Deisseroth K., Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei M., Shi L., Shen Y., Zhao Z., Guzman A., Kaufman L. J., Wei L., Min W., Volumetric chemical imaging by clearing-enhanced stimulated Raman scattering microscopy. Proc. Natl. Acad. Sci. U.S.A. 116, 6608–6617 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu X., Huang L., Zheng Y., Song Y., Xu Q., Wang J., Si K., Duan S., Gong W., Ultrafast optical clearing method for three-dimensional imaging with cellular resolution. Proc. Natl. Acad. Sci. U.S.A. 116, 11480–11489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J., Lin P., Tan Y., Cheng J. X., Volumetric stimulated Raman scattering imaging of cleared tissues towards three-dimensional chemical histopathology. Biomed. Opt. Express 10, 4329–4339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coudray N., Ocampo P. S., Sakellaropoulos T., Narula N., Snuderl M., Fenyo D., Moreira A. L., Razavian N., Tsirigos A., Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X., Nadiarynkh O., Plotnikov S., Campagnola P. J., Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat. Protoc. 7, 654–669 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Legends for moives S1 and S2
Movies S1 and S2