Abstract
Bactericidal antibiotics can cause metabolic perturbations that contribute to antibiotic-induced lethality. The molecular mechanism underlying these downstream effects remains unknown. Here, we show that ofloxacin, a fluoroquinolone that poisons DNA gyrase, induces a cascade of metabolic changes that are dependent on an active SOS response. We identified the SOS-regulated TisB protein as the unique molecular determinant responsible for cytoplasmic condensation, proton motive force dissipation, loss of pH homeostasis, and H2O2 accumulation in Escherichia coli cells treated with high doses of ofloxacin. However, TisB is not required for high doses of ofloxacin to interfere with the function of DNA gyrase or the resulting rapid inhibition of DNA replication and lethal DNA damage. Overall, the study sheds light on the molecular mechanisms by which ofloxacin affects bacterial cells and highlights the role of the TisB protein in mediating these effects.
The SOS-regulated TisB protein is responsible for cytoplasmic condensation and H2O2 accumulation during ofloxacin treatment.
INTRODUCTION
Antibiotics are essential in human health to fight bacterial infections. As antibiotic resistance determinants are alarmingly spreading, there is an urgent need to expand our current antimicrobial arsenal. To develop effective antimicrobial therapies, a deeper understanding of how bacterial cells respond to drugs at the molecular and physiological levels is mandatory. While the interactions of antibiotics with their primary targets are generally well characterized, the physiological changes triggered by target corruption and how they contribute to growth inhibition or cell death remain less well understood (1–3). Treatment with bactericidal antibiotics (i.e., fluoroquinolones, β-lactams, and aminoglycosides) has been shown to induce common metabolic perturbations and promiscuous production of reactive oxygen species (ROS) (4–9). It has been proposed that ROS contribute to cell death by damaging fatty acids in the inner membrane which leads to cytosol leakage and cytoplasmic condensation (10). Despite the increased descriptive details of the downstream events associated with antibiotic lethality, the molecular mechanism(s) underlying these phenotypes have been elusive.
Fluoroquinolone antibiotics poison DNA gyrase, leading to the generation of double-strand breaks in DNA and the induction of the SOS system. This coordinated cellular SOS response is triggered by a variety of stressors that cause DNA damage and replication blockage (11–14). In Escherichia coli, the SOS regulon is composed of ~50 genes, many of them being involved in DNA repair functions such as homologous recombination, nucleotide excision repair, and translesion DNA synthesis. The SOS response contributes to tolerance to fluoroquinolones by enabling cells to cope with DNA damage during the recovery period following the removal of the antibiotic (15–17). Here, we identified the SOS-regulated protein TisB as the sole molecular determinant responsible for ROS accumulation during treatment with high doses of ofloxacin (OFX). This finding allowed us to show unambiguously that ROS do not contribute to cell death as we could not detect any differences in viability between the wild-type strain and a strain devoid of TisB, reinforcing the idea that poisoning of DNA gyrase is sufficient for cell death. Nevertheless, using a combination of chemical dyes and genetically encoded fluorescent reporters monitoring physiological parameters in single-cell live imaging experiments, we were able to dissect a cascade of downstream events triggered by TisB that occur during the OFX treatment. We observed that treatment with high doses of OFX induces cytoplasmic condensation concomitantly with membrane depolarization and loss of intracellular pH homeostasis. These first physiological changes are followed by an increase in H2O2 at later times during the OFX treatment. Together, our study elucidates the molecular mechanism underlying ROS production upon fluoroquinolone treatment and its potential role in cell death.
RESULTS
A high dose of ofloxacin induces nucleoid compaction and cytoplasmic condensation
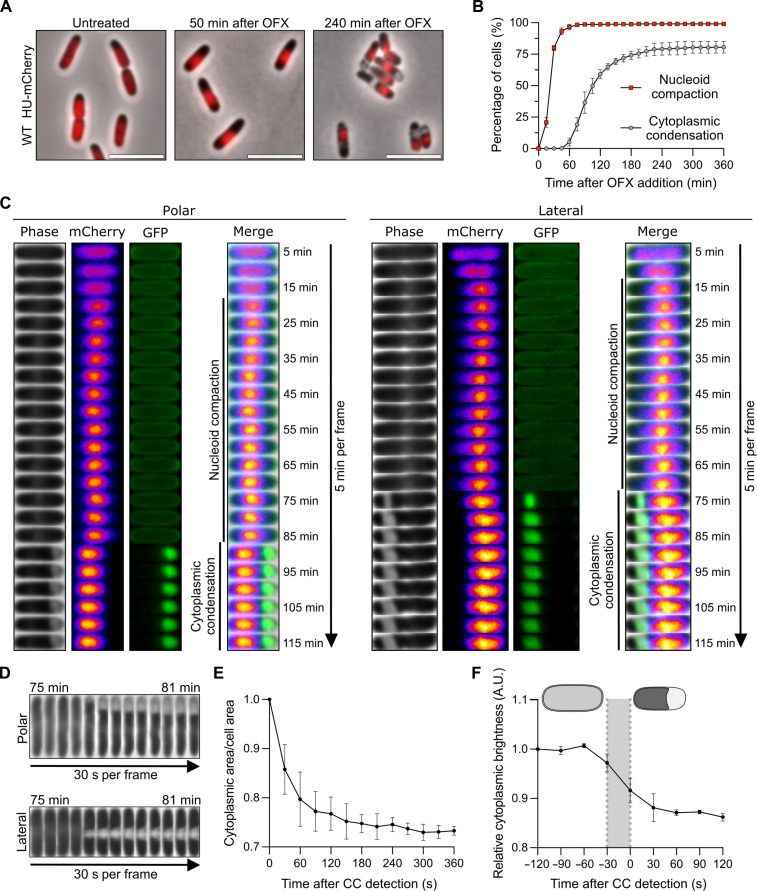
Treatment of E. coli cells with nalidixic acid and ciprofloxacin (quinolone and fluoroquinolone, respectively) has been reported to induce cellular phenotypes such as nucleoid compaction and cytoplasmic condensation, the latter being characterized by the appearance of clear regions inside the cell (18, 19). To confirm that OFX induces similar cellular alterations, we used E. coli cells encoding a translational HU-mCherry fusion as a proxy for nucleoid organization and dynamics (20). In growing cells, the HU-mCherry fluorescence occupies a large part of the cell volume (Fig. 1A). Upon addition of OFX [5 μg/ml; corresponding to 83-fold the minimal inhibitory concentration (MIC)], the fluorescence coalesces to form a bright mass indicative of nucleoid compaction (Fig. 1A). Nucleoid compaction occurs rapidly after OFX addition in nearly every cell, with ~97% of the population showing nucleoid compaction after 60 min of treatment (Fig. 1B and movie S1). At later times of treatment, cytoplasmic condensation was observed as described by others (Fig. 1, A and B) (10, 18, 19). The number of cells showing cytoplasmic condensation gradually increases with time and reaches a maximum after 180 min in the presence of OFX with ~75% of the population showing the phenotype (Fig. 1B). Using a green fluorescent protein (GFP) construct specifically labeling the periplasm (fig. S1A) (21), we confirmed that the bright regions observed during cytoplasmic condensation are periplasmic in nature (Fig. 1C and movie S1). These periplasmic expansions were detected at two preferential locations: either at one of the cell poles (quoted as “polar’) or close to the mid-cell position (quoted as “lateral’) (Fig. 1C), with the polar localization being prevalent (71.2 ± 8.3% of the cytoplasmic condensed cells after 330 min of OFX exposure) (fig. S1B). During time-lapse image analysis, conversion of polar periplasmic expansions to lateral location and vice versa or total reversion was occasionally observed. However, these events are infrequent (less than 7.9 ± 3.8%) as the ratio of polar to lateral periplasmic expansions varied very little over time in the presence of OFX (fig. S1B). Cell-length distribution and demographic analyses revealed that polar and lateral periplasmic expansions present similar distributions, indicating that their location is unlikely to be dependent on the cell cycle (fig. S1, C and D). Time-lapse microscopy performed at higher temporal resolution (one frame every 30 s) showed that cytoplasmic condensation occurs rapidly (within 1 min) (Fig. 1D and movie S2). This phenotype coincides with a reduction of the ratio between the cytoplasm and cell area (~30%) (Fig. 1E), a reduction of the cytosol brightness (Fig. 1F), and displacement of the nucleoid most likely due to the reduction of the volume of the cytoplasm (Fig. 1C and fig. S1D). Together, these data show that OFX treatment leads to a condensation of the cytoplasmic content as described by Collins and coworkers for similar doses of ciprofloxacin (10). It is likely that the molecular mechanisms underlying both phenotypes are different since nucleoid compaction and cytoplasmic condensation appear at different frequencies and timing during the OFX treatment (Fig. 1B). While nucleoid compaction has been shown to be dependent on the activation of the SOS response, more specifically on the RecA and SMC-like RecN proteins (22–24), the molecular mechanism responsible for cytoplasmic condensation was previously unknown.
Fig. 1. Ofloxacin induces DNA compaction and cytoplasmic condensation.
Results are representative of three biological replicates. The data points are means ± SD. (A) Representative images of wild-type (WT) cells encoding a hupA-mCherry fusion before, 45, and 240 min after the addition of OFX. Cells were spotted on pads containing OFX. Image acquisition started 5 min after spotting with additional imaging every 5 min. The HU-mCherry fusion is highlighted in red. Scale bars, 5 μm. (B) Quantification of DNA compaction and cytoplasmic condensation (CC) during the OFX treatment. Cells from (A) were grown in the microfluidic device for 60 min followed by perfusion with a medium containing OFX for 360 min. Image acquisition was performed every 15 min. A total of at least 709 cells were analyzed. (C) Kymographs of representative cells forming polar (left) or lateral (right) periplasmic expansions during OFX treatment. WT cells encoding a hupA-mCherry and an isopropyl-β-D-thiogalactopyranoside (IPTG)–inducible SSdsbA-sfgfp fusion were grown in the presence of IPTG and spotted on pads containing OFX and IPTG. Image acquisition started 15 min after spotting with additional imaging every 5 min. Phase contrast, mCherry (fire), green fluorescent protein (GFP; green), and merge of channels are shown. (D to F) WT cells were treated with OFX for 60 min and spotted on pads containing OFX. Image acquisition started 5 min after spotting with additional imaging every 30 s. (D) Kymographs of representative cells forming polar (top) and lateral (bottom) periplasmic expansions. (E) Cytoplasmic area/cell area ratio of individual cells after CC detection. A total of 100 cells were analyzed. (F) Relative cytoplasmic brightness of individual cells before and after CC detection. The gray area represents the timing of CC formation. A.U., arbitrary unit. A total of 50 cells were analyzed.
Cytoplasmic condensation by ofloxacin is mediated by the SOS-regulated TisB protein
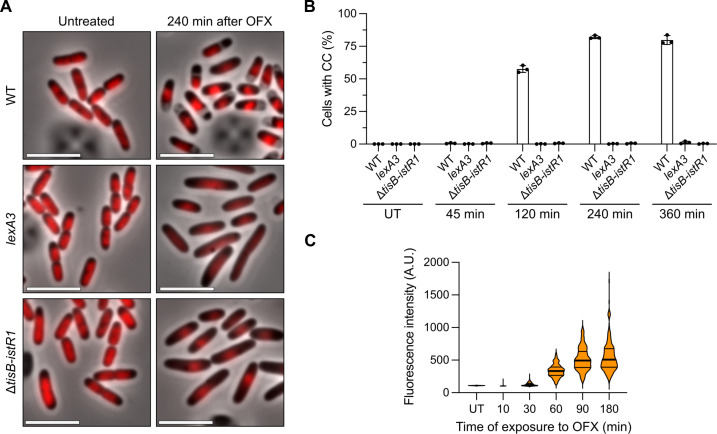
To characterize the molecular mechanism underlying cytoplasmic condensation induced by OFX, we first assessed the phenotype of the LexA3 mutant strain, which is unable to mount the SOS response (25, 26). We confirmed that nucleoid compaction is perturbed in the LexA3 mutant (22) and showed that cytoplasmic condensation did not occur in the mutant strain (Fig. 2A). Since SOS induction is needed for cytoplasmic condensation, we reasoned that the SOS-controlled tisB-istR1 toxin-antitoxin (TA) system might be involved as the TisB toxin was previously shown to interact with and corrupt the E. coli inner membrane (27–29). This type I TA system comprises the istR1 RNA antitoxin (inhibitor of SOS-induced toxicity by RNA) and the TisB protein (toxicity-induced by SOS) (30, 31). The expression of the two components is differentially regulated with the tisB gene being under the control of the SOS response and istR1 being constitutively expressed (30, 31). Additional posttranscriptional regulation prevents toxin translation in steady-state conditions (32–34). Upon SOS induction, the inhibitory effect of istR1 is bypassed and the TisB protein is produced. In the tisB-istR1 deletion mutant, OFX-mediated cytoplasmic condensation was abolished, while nucleoid compaction remained unchanged (Fig. 2, A and B, and fig. S2A), confirming that the molecular mechanisms underlying the two cellular phenotypes are different, although both being controlled by the SOS response. Complementation experiments were performed with a single-copy number plasmid carrying either the entire tisB-istR1 system or the tisB gene alone under the control of its native promotor (fig. S2, A and B). Both constructs restored the wild-type phenotype, showing that the TisB toxin is required for cytoplasmic condensation after OFX exposure. We further confirmed that tisB is expressed under our conditions using a transcriptional fusion coupling the expression of GFP to the tisB promoter and comprising the tisB mRNA region recognized by the istR1 antitoxin to keep the regulation of tisB expression as close as possible to that of the native context (35). Fluorescence gradually increased with time to reach a maximum after 90 min in the presence of OFX and remained stable over time (Fig. 2C), paralleling the gradual slowdown of detection of cells showing cytoplasmic condensation (Fig. 1B).
Fig. 2. The SOS-induced TisB toxin is responsible for cytoplasmic condensation during ofloxacin treatment.
Results are representative of three biological replicates. (A and B) WT, lexA3, and ΔtisB-istR1 cells encoding a hupA-mCherry fusion were grown in the microfluidic device for 60 min followed by perfusion with a medium containing OFX for 360 min. Image acquisition was performed every 15 min. (A) Representative images of the time-lapse experiment before and 240 min after the addition of OFX. Scale bars, 5 μm. (B) Percentage of cells in which CC was detected after addition of OFX. Between 297 and 1121 cells were analyzed for each sample. The data points are means ± SD. (C) Induction of transcriptional tisB reporter during OFX treatment. WT cells encoding a ptisB-gfp fusion were treated with OFX. At indicated times, cells were spotted on pads and imaged. Between 193 and 975 cells were analyzed for each sample. The data points are median (solid line) ± quartiles (dashed line).
TisB-dependent cytoplasmic condensation is specific to SOS inducers
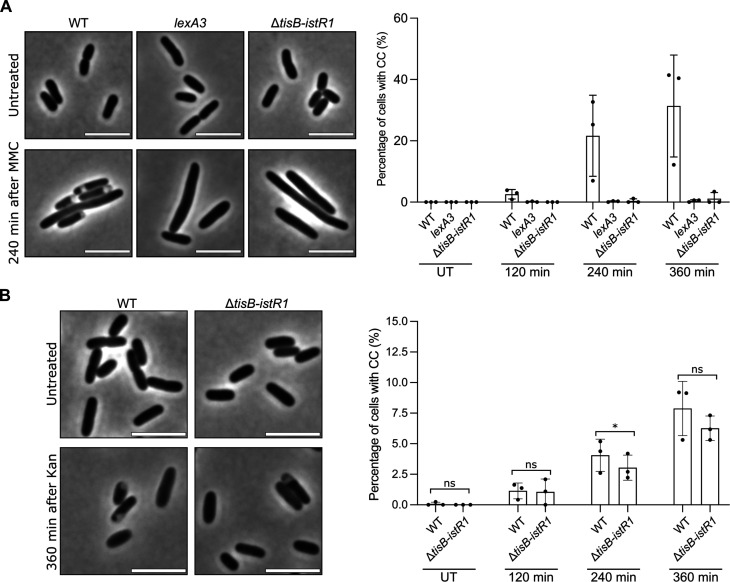
A variety of treatments and conditions [nalidixic acid, ciprofloxacin, kanamycin (Kan), osmotic shock, and nutrient downshift, among others] have been shown to induce phenotypes resembling cytoplasmic condensation that we observed after OFX (10, 18, 19, 36, 37). Since the expression of tisB is controlled by the SOS response, we first assessed whether mitomycin C (MMC), a compound inducing SOS by other means than fluoroquinolones (38), leads to cytoplasmic condensation. MMC (20 μg/ml, 10-fold the MIC) induces cytoplasmic condensation in a TisB-dependent manner (Fig. 3A). It was recently shown that the aminoglycoside Kan also induces the same phenotype (10). We found that treating wild-type cells with Kan also induced cytoplasm condensation, although at a lower frequency than OFX does (less than 10% of the cells after 360 min of treatment) (Fig. 3B). A similar phenotype was observed in the ΔtisB-istR1 cells (Fig. 3B), indicating that cytoplasmic condensation induced by OFX and Kan does not rely on the same molecular mechanism. We also tested whether TisB is involved in plasmolysis following an osmotic shock. We observed that TisB was involved neither in sucrose-induced plasmolysis nor in its reversion as both the wild-type and the ΔtisB-istR1 mutant showed the same phenotype (fig. S3A). Recently, Shi et al. (37) showed that nutrient starvation induces cytoplasm shrinkage which translates into periplasmic expansions. Transferring wild-type or ΔtisB-istR1 cells growing in LB medium in poor M9 medium resulted in periplasmic expansions in both strains; however, expansions appeared less pronounced than what was observed for OFX or sucrose. Shifting back the cells to LB reversed the phenotype (fig. S3B). Thus, TisB-induced cytoplasmic condensation appears to be specific to OFX and MMC, two DNA-damaging agents that induce the SOS response.
Fig. 3. TisB-dependent cytoplasmic condensation is specific to SOS inducers.
At indicated times, cells were spotted on pads and imaged. Results are representative of three biological replicates. The data points are means ± SD. (A) Left: Representative images of WT, lexA3, and ΔtisB-istR1 cells before (untreated, UT) and 240 min after the addition of MMC. Scale bars, 5 μm. Right: Percentage of cells in which CC was detected after the addition of MMC. Between 190 and 716 cells were analyzed for each sample. (B) Left: Representative images of time-course analysis of WT and ΔtisB-istR1 cells before (UT) and 240 min after the addition of kanamycin (Kan). Scale bars, 5 μm. Right: Percentage of cells in which CC was detected after the addition of Kan. Between 239 and 2233 cells were analyzed for each sample. Fisher’s exact test was used to determine significance (ns, not significant P > 0.05, *P < 0.05).
TisB-mediated cytoplasmic condensation is associated with membrane depolarization and loss of pH homeostasis
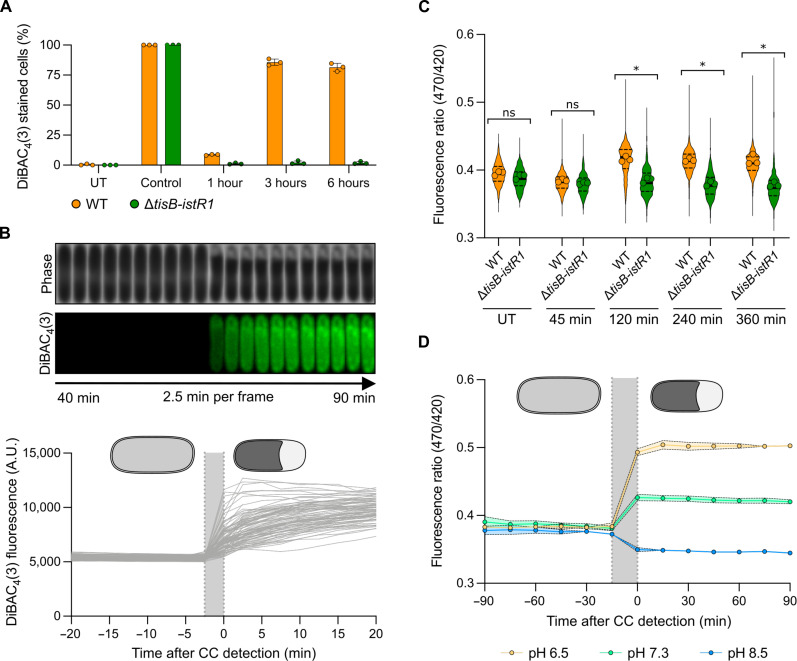
The 29–amino acid hydrophobic peptide TisB has been reported to target the inner membrane and to induce membrane depolarization in cells when overexpressed from heterologous promoters or when treated at high doses of ciprofloxacin (27, 30, 39). We confirmed that, under the conditions tested here, wild-type cells treated with OFX are stained by the ΔΨ-sensitive dye bis-(1,3-dibutylbarbituric acid) trimethine oxonol [DiBAC4(3)] (40), while ΔtisB-istR1 mutant cells are not (Fig. 4A and fig. S4A), confirming that tisB expression leads to membrane depolarization. Moreover, time-lapse microscopy of wild-type cells treated with OFX showed DiBAC4(3) staining, concomitantly with the detection of cytoplasmic condensation (Fig. 4B). Of note, the vast majority of wild-type cells that are stained with DiBAC4(3) are cells with detectable cytoplasmic condensation. However, ~10% of DiBAC4(3)-stained cells do not show detectable periplasmic expansions (fig. S4B), indicating that either membrane depolarization can occur independently of cytoplasmic condensation or that this phenotype sometimes fails to be detected for technical reasons (e.g., resolution of our phase-contrast microscopy setup).
Fig. 4. TisB-dependent cytoplasmic condensation is concomitant with membrane depolarization and loss of ΔpH.
Results are representative of three biological replicates. The gray area represents the timing of CC formation. (A) Percentage of DiBAC4(3)-stained cells during OFX treatment. WT and ΔtisB-istR1 cells were treated with OFX. At indicated times, samples were incubated with DiBAC4(3) and spotted on pads. Heat shock was used as a positive control. At least 376 cells were analyzed for each sample. Data points are means ± SD. (B) Representative kymograph of DiBAC4(3)-stained cells (top) and DiBAC4(3) fluorescence of individual cells before and after CC detection (bottom). WT cells were spotted on pads containing DiBAC4(3) and OFX. Image acquisition started 7.5 min after spotting with additional imaging every 2.5 min. A total of 100 cells were analyzed. Data points are means ± SD. (C) pHluorin2 fluorescence ratio during OFX treatment. WT and ΔtisB-istR1 cells containing the pHluorin2 reporter were grown in the microfluidic device for 60 min followed by perfusion with a medium containing OFX for 360 min. Image acquisition was performed every 15 min. At least 178 cells were analyzed for each sample. Data points are the mean of individual replicate (symbol), median (solid line) ± quartiles (dashed line). Multiple comparison Kruskal-Wallis with false discovery rate (FDR) approach was used to determine significance based on replicates (FDR-adjusted ns P > 0.05 and *P < 0.05). (D) pHluorin2 fluorescence ratio during OFX treatment at different pH. WT cells containing the pHluorin2 reporter were grown in a medium adjusted to different pH, introduced into a microfluidic device, and perfused with the respective pH-adjusted medium for 60 min followed by perfusion with the respective pH-adjusted medium containing OFX for 360 min. Image acquisition was performed every 15 min. At least 20 cells were analyzed for each sample. Data points are means ± SD.
The electrical potential ΔΨ established across the inner membrane, reported by DiBAC4(3), is one of two components of the proton motive force (PMF), the other being ΔpH reflecting the difference between the cytoplasmic and external pH (41). We therefore asked whether TisB-dependent membrane depolarization would affect pH homeostasis. To monitor the intracellular pH at the single-cell level, we used the pHluorin2 reporter (42). The emission of this constitutively expressed GFP-based probe is pH-dependent when excited at 470 nm and pH-independent when excited at 420 nm, and can hence be used as a ratiometric tool to measure intracellular pH (for more details and calibration curves of pHluorin2, see the Supplementary Materials) (42). In untreated MOPS-glucose condition, the fluorescence ratio (470/420 nm) of the wild-type and ΔtisB-istR1 strains was similar (0.395 ± 0.026 and 0.390 ± 0.037 for wild-type and ΔtisB-istR1, respectively) (Fig. 4C). After 2 hours of OFX treatment, the fluorescence ratio increased in the wild-type cells and remained stable for the remaining time of treatment, indicating a drop of intracellular pH (0.417 ± 0.0376 and 0.384 ± 0.033 for wild-type and ΔtisB-istR1, respectively). ΔtisB-istR1 cells exhibited a stable fluorescence ratio during the treatment highlighting that these cells are capable of maintaining their ΔpH (0.412 ± 0.034 and 0.379 ± 0.042 for wild-type and ΔtisB-istR1, respectively) (Fig. 4C and fig. S4, C and D). As observed for cytoplasmic condensation, the timing of intracellular pH decrease in the wild-type strain varies from cell to cell (fig. S4D) and is concomitant with the detection of cytoplasmic condensation (Fig. 4D, green symbols). To determine whether the modest cytoplasm acidification observed in OFX-treated wild-type cells is an active mechanism or rather an equilibration of protons from the slightly more acidic external MOPS to the cytosol (measured pH of MOPS 0.4% glucose is 7.3, intracellular pH is between 7.4 and 7.8) (41), wild-type cells were grown and OFX-treated in MOPS medium adjusted to pH 6.5 and 8.5, respectively. The fluorescence ratio of the ΔtisB-istR1 cells remained constant throughout the OFX treatment at both pH 6.5 and 8.5, similar to what we observed for the experiment performed in MOPS glucose at pH 7.3 (Fig. 4C and fig. S4, E and F). This is in stark contrast to the wild-type cells, in which a decrease or an increase of intracellular pH is observed 4 hours after OFX addition in medium adjusted to pH 6.5 or 8.5, respectively (fig. S4, E and F). This suggests that ΔpH is not maintained in OFX-treated wild-type cells. Furthermore, the intracellular pH changes of wild-type cells treated in acidic or alkaline conditions could be linked to the detection of cytoplasmic condensation (Fig. 4D). Together, these data indicate that TisB expression impedes the electrical potential (ΔΨ) as well as the proton gradient (ΔpH) established across the inner membrane, both effects contributing to PMF dissipation upon OFX treatment.
A pkatG::gfp transcriptional reporter is not induced by OFX due to inhibition of transcription
Several indications that pointed toward TisB being responsible for ROS accumulation have been obtained earlier by the Berghoff group using the fluorogenic dye 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) which is oxidized by various ROS to generate the fluorescent dichlorofluorescein (DCF) (43). We first thought to probe ROS accumulation using a classical transcriptional fusion between the promoter of the katG gene and the GFP-encoding gene (pkatG-gfpmut2) (44). The katG gene of E. coli is part of the OxyR regulon and encodes a bifunctional enzyme displaying both catalase and peroxidase activity (45, 46). pkatG-gfpmut2 expression was activated by exogenous addition of H2O2 in a dose-dependent manner (from 10 μM H2O2) showing that the pkatG-gfpmut2 reporter monitors oxidative stress (fig. S5A). OFX treatment for 3 hours did not lead to the activation of the pkatG-gfpmut2 reporter in the wild-type strain (fig. S5B). As it was reported that fluoroquinolones inhibit transcription (47), we reasoned that the lack of pkatG-gfpmut2 induction in OFX-treated cells might be due to the inability to sustain active gene expression. We therefore monitored the ability of the wild-type strain to incorporate radioactive [3H]-uridine and [35S]-methionine during OFX treatment. Both [3H]-uridine and [35S]-methionine incorporation were inhibited by OFX addition. RNA synthesis was rapidly shut down by OFX, with 95% inhibition observed after 30 min of treatment, in both the wild-type and ΔtisB-istR1 strains (fig. S5C). Protein synthesis was less affected, although translation efficiency decreased to ~50% after 30 min in the presence of OFX and as much as 75% after 5 hours of treatment (fig. S5D). The ΔtisB-istR1 mutant exhibited transcription and translation shutdown similar to those seen in the wild-type strain, although protein synthesis was reproducibly less affected between 90 and 180 min of OFX treatment as compared to the wild-type strain. Whether this reflects some TisB-dependent regulation remains to be tested. Together, these data indicate that the transcriptional pkatG-gfpmut2 reporter might not be suitable for ROS detection in our conditions and confirm that OFX inhibits gene expression.
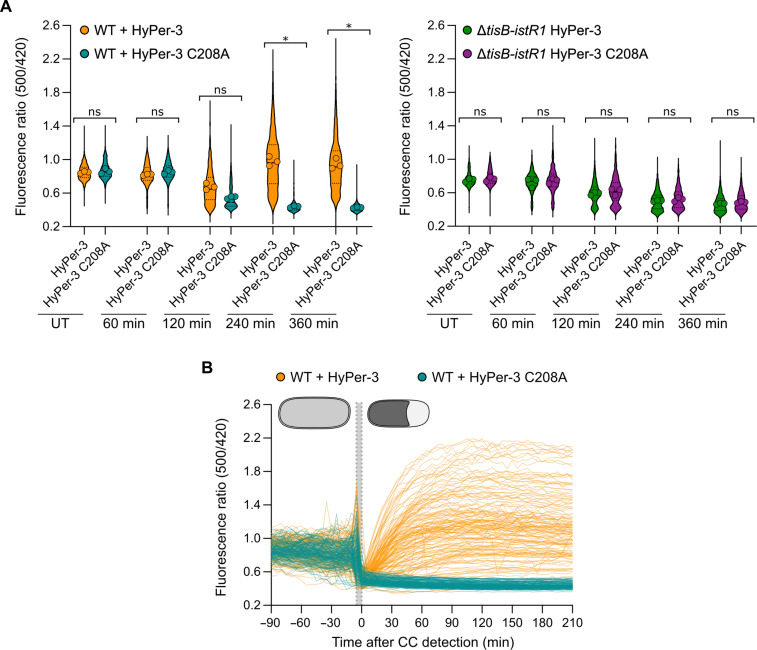
TisB-mediated cytoplasmic condensation is associated with ROS accumulation
To circumvent the effect of OFX on gene expression, we implemented the HyPer-3 biosensor, which consists of a circularly permutated YFP integrated into the H2O2-sensing domain of the E. coli transcription factor OxyR (48). HyPer-3 is constitutively expressed and reports H2O2 within seconds (48). As a control, an H2O2-insensitive version of HyPer-3 was constructed by mutationally replacing cysteine-208 with alanine to prevent intramolecular disulfide-bond formation (HyPer-3 C208A) (for more details and calibration curves of HyPer-3 and HyPer-3 C208A, see the Supplementary Materials). The mutant was also convenient as it was pH-sensitive and allowed us to support the pHluorin2 data. Before OFX addition, the fluorescence ratio (500/420 nm) of the HyPer-3 and HyPer-3 C208A reporters was comparable in the wild-type and the ΔtisB-istR1 strains (0.850 ± 0.012 and 0.863 ± 0.029 for HyPer-3 and HyPer-3 C208A in wild-type, respectively and 0.746 ± 0.249 and 0.751 ± 0.022 for HyPer-3 and HyPer-3 C208A in ΔtisB-istR1, respectively) (Fig. 5A and movie S3). After 120 min in the presence of OFX, the fluorescence ratio of the HyPer-3 reporter in the wild-type cells continuously increased, reflecting an increase of intracellular H2O2. On the other hand, wild-type cells expressing the HyPer-3 C208A manifested a drop in the fluorescence ratio 120 min after the addition of OFX, confirming the drop of intracellular pH observed with the pHluorin2 reporter (Fig. 5A). The fluorescence ratio of the HyPer-3 and HyPer-3 C208A reporters in the ΔtisB-istR1 strain remained stable for the first 120 min after addition of OFX and decreased for the remaining duration of the treatment (Fig. 5A). On the basis of the pHluorin2 data (Fig. 4C), it is unlikely that this reflects a drop of intracellular pH. Whether this observation illustrates some specific physiological changes remains to be investigated. From this experiment, we conclude that TisB is required for H2O2 accumulation and intracellular pH changes observed in the wild-type cells during the OFX treatment. When monitoring the fluorescence ratio of HyPer-3 and HyPer-3 C208A during an OFX treatment in individual wild-type cells, the fluorescence ratio of both reporters dropped simultaneously with the detection of cytoplasmic condensation, again highlighting the link between cytoplasmic condensation and loss of pH homeostasis (Figs. 4D and 5B and fig. S6A). While the fluorescence ratio of the HyPer-3 C208A did not change after the appearance of cytoplasmic condensation, the fluorescence ratio of the HyPer-3 reporter continuously increased up to 90 min after the formation of cytoplasmic condensation, indicating that H2O2 is steadily generated after cytoplasmic condensation (Fig. 5C and fig. S6A). We confirmed these results using the ROS-sensitive H2DCFDA dye. Only cytoplasmic condensed wild-type cells became DCF-stained (fig. S6, B and C), while most of the ΔtisB-istR1 cells were not stained by DCF after exposure to OFX (1.9 ± 1.13% of DCF-stained ΔtisB-istR1 cells after 360 min of OFX) (fig. S5C). Together, these results suggest that H2O2 accumulation is dependent on TisB-mediated cytoplasmic condensation and that it occurs later during OFX treatment (10). ROS have previously been shown to be a common mechanism of bactericidal antibiotics (4–9). When monitoring the fluorescence ratio of HyPer-3 reporter in a wild-type strain during a Kan treatment, we detected H2O2 accumulation 120 min after the onset of the treatment (fig. S6, D and E). Kan-induced H2O2 was however independent of TisB as the ΔtisB-istR1 mutant exhibits similar ROS accumulation as to what we observed in the wild-type strain (fig. S6, D and E), highlighting that the underlying molecular mechanisms of ROS formation are fundamentally different between an OFX and Kan treatment.
Fig. 5. TisB-dependent cytoplasmic condensation leads to delayed H2O2 accumulation.
Cells were grown in the microfluidic device for 60 min followed by perfusion with a medium containing OFX for 360 min. Image acquisition was performed every 5 min. Results are representative of three biological replicates. (A) Fluorescence ratio (500/420 nm) of individual WT (left) and ΔtisB-istR1 cells (right) containing the HyPer-3 or HyPer-3 C208A reporter. Between 206 and 1449 cells were analyzed for each sample. Data points are the mean of individual replicate (symbol), median (solid line) ± quartiles (dashed line). Multiple comparison Kruskal-Wallis was used to determine significance based on replicates (FDR-adjusted ns P > 0.05 and *P < 0.05). (B) Fluorescence ratio (500/420 nm) of individual WT cells containing the HyPer-3 (orange, 91 cells) or HyPer-3 C208A (blue, 59 cells) reporter normalized by the time of CC detection. The gray area represents the timing of CC formation.
Ofloxacin-mediated TisB downstream effects are associated with neither cell envelope damage nor lethality
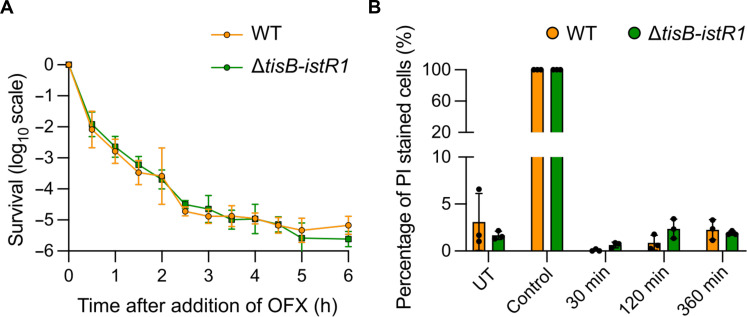
It was previously proposed that cytoplasmic condensation and ROS are markers of cell death induced by bactericidal antibiotics such as aminoglycosides and fluoroquinolones (2, 10, 49). If this were the case under our conditions, then we would expect to observe a higher survival rate for the ΔtisB-istR1 mutant compared to the wild-type strain. Both strains exhibited similar growth (fig. S7A) and OFX MIC (0.06 μg/ml). The time-kill curve obtained for the ΔtisB-istR1 mutant treated with OFX for 6 hours showed a classical biphasic shape which superimposes with that of the wild-type strain (Fig. 6A). This indicates that the killing of neither susceptible cells nor persister cells is affected by tisB expression, consistent with our previous observation (16). Longer treatment time did not reveal any difference in survival between the wild-type and the ΔtisB-istR1 strains (fig. S7B). We also monitored propidium iodide (PI) staining in the wild-type and ΔtisB-istR1 strains. We observed that less than 5% of the cells were PI-stained in both strains during the OFX treatment (Fig. 6B), which is comparable to previous data obtained with the wild-type strain treated with ciprofloxacin at 10-fold the MIC (50). Together, these data show that TisB-mediated phenotypes do not obviously contribute to OFX-mediated cell death and are consistent with the conclusion that DNA gyrase corruption is sufficient to lead to lethality. DNA gyrase cycling on DNA and poisoned by (fluoro)quinolones constitute roadblocks that physically prevent the progression of replication and transcription forks (47, 51–55). We therefore tested the kinetics of DNA replication inhibition caused by OFX in both strains. As we observed for [3H]-uridine incorporation, when cells were treated with OFX, the incorporation of [3H]-thymidine was inhibited at the same rate and to a comparable extent in the wild-type and ΔtisB-istR1 strains, further pointing to replication blockage by OFX-stalled DNA gyrase complexes as the major contributor of ofloxacin lethality (fig. S7C).
Fig. 6. Cytoplasmic condensation is not implicated in OFX-mediated lethality.
Cells were grown to an OD600nm ~ 0.3 and treated with OFX. Results are representative of three biological replicates. Data points are means ± SD. (A) Time-kill curve of WT and ΔtisB-istR1 strains. Culture samples were diluted and plated on LB agar plates. (B) Quantification of propidium iodide (PI)–stained WT and ΔtisB-istR1 cells. At indicated times, culture samples were incubated with PI for 15 min, spotted on agarose pads, and imaged. Isopropanol (70%) was used as positive control. Syto9 was used as a counterstain. Between 338 and 5830 cells were analyzed for each sample.
DISCUSSION
Bactericidal antibiotics targeting different cellular processes have been proposed to trigger common metabolic perturbations as a consequence of their interactions with their primary targets (6, 10). Despite years of debate and experiments, several questions remain regarding the nature of these downstream events and the underlying molecular determinant(s), as well as their precise contribution to antibiotic lethality. Also conceptually, one would predict that antibiotics from different classes (i.e., targeting unrelated cellular processes) would generate at least partially differentiated and specific downstream effects. In this work, we set up a workflow allowing us to determine the sequential appearance of cellular phenotypes triggered by antibiotics at the single-cell level using a combination of chemical dyes and genetically encoded reporters. We observed multiple downstream effects that arise during treatment with high doses of OFX and identified the SOS-regulated TisB protein as the unique molecular determinant underlying these phenotypes. While a Kan treatment results in similar phenotypes as observed during the OFX treatment, secondary effects induced under these conditions are independent of TisB, highlighting that downstream consequences following bactericidal antibiotics are specific to certain classes of antibiotics.
A single molecular determinant for multiple OFX-induced downstream events
Since OFX induces the SOS system, the parsimonious hypothesis was that genes controlled by this response might be responsible for downstream events. We identified the SOS-regulated TisB protein as the unique molecular determinant of cytoplasmic condensation, PMF dissipation, and H2O2 accumulation following DNA gyrase corruption. Previous studies reported that TisB inserts in the E. coli inner membrane (28, 30, 31, 43) and provokes depolarization and superoxide generation (28, 30, 31, 43). In vitro data and molecular dynamics simulations proposed that TisB assembles in the inner membrane as a dimer or tetramer and might form a narrow anionic pore that allows the leakage of small molecules (28, 29, 56, 57). We observed that tisB induction by OFX leads to cytoplasmic condensation, which is characterized by an increase in intracellular concentration, most likely due to the loss of water from the cytoplasm to the periplasm. In cells undergoing cytoplasmic condensation, we detected concomitant loss of Δψ and ΔpH, resulting in immediate PMF collapse. Recent studies showed that ciprofloxacin, a related fluoroquinolone, causes loss of intracellular potassium ions simultaneously with DiBAC4(3)staining (10, 58). Potassium ions are involved in both establishing the Δψ component of PMF and generating turgor pressure. It is therefore tempting to speculate that TisB is responsible for a controlled loss of potassium. In the case of plasmolysis induced by an osmotic shock, which is unrelated to tisB expression, cells respond rapidly to equilibrate the internal turgor pressure by importing ions such as potassium and synthesizing solutes, allowing them to return to homeostasis (59). In contrast, TisB-dependent cytoplasmic condensation remained stable for the entire duration of the treatment. This is probably due to the concomitant loss of Δψ and ΔpH, most likely preventing any membrane-related metabolic activity that would allow a return to homeostasis. While cytoplasmic condensation and PMF collapse occur simultaneously, we observed that H2O2 accumulates at the later time of OFX treatment in a TisB-dependent manner. It was proposed that bactericidal antibiotics, notably the fluoroquinolone norfloxacin, increase respiration and thereby NADH consumption and electron flux through the electron transport chain which ultimately generate lethal doses of hydroxyl radicals via Fenton chemistry (5). However, studies from the 1970s reported that ultraviolet irradiation (which induces DNA damage and the SOS response) causes respiration cessation within an hour after irradiation (60, 61), and subsequently, there have been several reports indicating that O2 consumption decreases with time during treatment with fluoroquinolones, indicative of respiration reduction (58, 62). The antibiotic-mediated accumulation of ROS has also been questioned because of findings showing that the expression of scavenger enzymes was not induced by bactericidal antibiotics and survival of the corresponding mutants was not affected under the same conditions (62, 63). Using the HyPer-3 reporter, we were able to detect TisB-dependent H2O2 accumulation subsequently to cytoplasmic condensation and PMF dissipation. We further confirmed these data using the H2DCFDA chemical dye that detects ROS-dependent oxidation (64). Moreover, as OFX rapidly leads to an arrest of transcription, cells are unable to mount an oxidative stress response at the time of ROS accumulation, potentially explaining the lack of oxidative scavenging agents following high fluoroquinolone treatment. The origins of TisB-dependent H2O2 remain unclear. O2− and H2O2 are thought to be produced primarily by accidental autooxidation of non-respiratory flavoproteins with a minor fraction originating from the respiratory chain (65, 66). The rate of endogenous H2O2 production in the cytoplasm in oxic conditions is estimated at ~10 to 15 μM/s, and accumulation is continuously dampened by scavenging enzymes (65). On the basis of our results, we speculate that the observed H2O2 accumulation in the wild-type strain is not the result of increased H2O2 production but seems rather due to problems in H2O2 scavenging possibly because cytoplasmic condensation could lead to an efflux of cofactors needed for basal H2O2 scavenging enzymes to maintain their activity.
The role of TisB in antibiotic cell death
Having identified the molecular determinant responsible for OFX-induced downstream effects, we were able to assess their contribution to antibiotic lethality. Our data showed that the survival of the ΔtisB-istR1 mutant at the high dose of OFX is similar to that of the wild-type strain, indicating that the metabolic perturbations resulting from DNA gyrase poisoning by OFX do not enhance antibiotic lethality. This is in agreement with the observation that cells with high-ROS and low-ROS levels generated during treatment at low doses of norfloxacin showed similar viability (64). Consequently, primary target poisoning appears to be the major contributor to OFX-mediated lethality. It is well known that quinolones and fluoroquinolones induce DNA fragmentation (52, 67, 68). DNA damage is extremely toxic and one non-repaired double-strand break per chromosome is sufficient to induce cell death (69). Therefore, we propose that cells are ultimately killed by OFX-induced DNA damage, without the intervention of ROS. Following OFX treatment, TisB-dependent cells with condensed cytoplasm were not stained by PI, suggesting that this phenotype is not a consequence of membrane damage. We cannot exclude that TisB-independent cytoplasmic condensation induced by Kan reflects some inner membrane defects, as aminoglycosides are known to promote amino acid misincorporation into nascent proteins resulting in misfolded proteins that can accumulate in the inner membrane. However, in our experiments, only 5 to 10% of cells treated with Kan showed cytoplasmic condensation, indicating that this phenotype is not likely to be a marker for cell death. Moreover, both wild-type and ΔtisB-istR1 cells accumulate H2O2 during treatment with Kan, showing that the pathways leading to ROS accumulation are specific to each antibiotic.
Several studies highlighted positive effects associated with TisB-dependent downstream metabolic perturbations and it was recently suggested that induction of TisB could stabilize the persister state over long-term fluoroquinolone treatment (70). However, under the conditions tested here, we were unable to detect any difference in survival between the wild-type and the tisB-istR1 strains over a 24-hour period of treatment. In another study, the group of Matic showed that TisB-dependent membrane depolarization induced by a trimethoprim treatment is detrimental as cells could not be rescued by the pyrimidine salvage pathway. Yet, this condition proved to be beneficial by preventing the import of toxic compounds, such as aminoglycosides and colicin M that require active PMF for import. The wild-type strain outcompeted the ΔtisB-istR1 mutant during intestine colonization in mice, potentially limiting the uptake of toxic components produced by competing gut microbes (35). Last, several antibiotics belonging to different classes were shown to cause a decrease in intracellular pH (71–73). Here, we showed that OFX does not lead per se to cytoplasm acidification but rather to the loss of control of ΔpH as proposed for the quinolone nalidixic acid (71). Intracellular pH has been shown to modulate the SOS response by modifying the activity of the SOS master regulators LexA and RecA (11, 74–76). Depending on the environmental conditions, TisB-dependent loss of ΔpH might influence the dynamics of the SOS response. Moreover, intracellular pH acidification was shown to occur during thymineless death before ROS accumulation and contribute to cell death (77). Whether this phenotype is TisB-dependent remains to be tested.
Together, our data indicate that TisB “freezes” any membrane-related metabolic activity needed to maintain homeostasis. As a consequence, downstream metabolic modifications including loss of PMF, intracellular pH acidification, and H2O2 accumulation are observed. However, in the conditions tested here, corruption of DNA gyrase leading to inhibition of replication and transcription is sufficient to cause lethality.
Limitations of this study
The TisB protein appears to have been acquired recently in evolution as it is mostly limited to the recent Enterobacter-Escherichia clade of the Enterobacterales (35). Therefore, it is plausible that functional analogs might play similar roles in bacterial species devoid of tisB. As an example, the small DinQ protein shares common features with TisB: Its expression is SOS-regulated and its activity modulates membrane-related processes (43, 78).
Our experiments were performed with OFX at a high dose in controlled conditions. We cannot exclude that the TisB-dependent downstream effects confer a selective advantage at lower doses of OFX. For instance, ROS induced by a low dose of ciprofloxacin were shown to generate mutable “gambler” subpopulations which may promote adaptability (79, 80). Also, whether other bactericidal antibiotics targeting different cellular processes exhibit similar downstream effects in vitro and how they contribute to lethality remains to be investigated. Last, in an infectious context, pathogenic bacteria encounter a variety of niches that might alter their physiology. Testing bacterial responses to antibiotics in an in vivo context might inform on potential means to boost the efficiency of our current antibiotic arsenal.
MATERIALS AND METHODS
Bacterial strains and plasmids
The strains used in experiments are all derivatives of the E. coli MG1655 laboratory strain (81). Strains, plasmids, and primers used in this study are listed in tables S1, S2, and S3, respectively. Chromosomal gene loci were transferred by phage P1 transduction to generate the final strains. When multiple insertions of modified genes were required, the antibiotic resistance gene markers were removed using site-specific recombination induced by expression of the FLP recombinase from pCP20 (82).
Deletion of the tisB-istR1 locus
The tisB-istR1 locus was deleted and replaced by the Kan resistance gene in MG1655 using the pKD4 plasmid as template DNA for amplification of the Kan resistance cassette using the P1 and P2 primers (table S3) as previously described (82). The deletion was transduced into the final strain by P1 phage transduction and confirmed by polymerase chain reaction (PCR) with primers P3 and P4 (table S3). The Kan resistance cassette was removed using the pCP20 thermosensitive plasmid encoding the FLP recombinase as described (82).
Construction of the plasmids expressing the tisB-istR1 locus or the tisB gene
The tisB-istR1 locus or the tisB gene were PCR-amplified using the P11 and P12 and P10 and P11 primers (table S3), respectively, using the genomic DNA of MG1655 as a template. The single-copy pNF02-EV plasmid was constructed by PCR amplification using P5 and P6 to remove the proDp-mScarlet-I fragment. The PCR product was phosphorylated by T4 polynucleotide kinase circularized by T4 ligase, and DpnI endonuclease was used to remove the template DNA. The final construct was confirmed by sequencing. The pNF02-EV was PCR-amplified using the P5 and P6 primers. The Gibson method was used to assemble the fragments containing the tisB-istR1 locus or the tisB gene using P8 and P9 and P7 and P9 primers, respectively (table S3). Final constructs were confirmed by sequencing using P13 and P14 primers (table S3).
Optimization and construction of the HyPer-3, HyPer-3 C208A, and pHluorin2 reporters
For HyPer-3, the original HyPer-3 sequence (48) was codon-optimized for E. coli (synthetic fragment MluI-34b-6His-HyPer-3-N1-ApaL1; table S4). The synthetic fragment was cloned in a pBeloBAC11 plasmid derivative (NEB, GenBank Accession # U51113). In the final construct, the optimized Hyper-3 sequence was cloned downstream of a control region including the strong proDp synthetic promoter (83) and the ribosome binding site sequence BBa_0034 (synthetic fragment MluI-proDp-RBS0034b-NheI; table S4). For HyPer-3 C208A, the mutated HyPer-3 version containing a cysteine-208 to alanine substitution (HyPer-3 C208A) was constructed by PCR using the HyPer-3 plasmid as a template, and the P15 and P16 primers (table S3). After amplification, the PCR product was phosphorylated by T4 polynucleotide kinase and circularized by T4 ligase, and DpnI endonuclease was used to remove the template DNA. For pHluorin2, the original pHluorin2 sequence was codon-optimized for E. coli (synthetic fragment XbaI-34b-pHluorin2-N3-NotI; table S4) and subcloned in the pET28 plasmid (Novagen). The XbaI-34b-pHluorin2-N3-NotI fragment was amplified by PCR using primers P17 and P18 (table S3) and cloned in a pBeloBAC11 plasmid derivative. In the final construct, the optimized pHluorin2 sequence was cloned downstream of a control region including the proDp synthetic promoter and the ribosome binding site sequence BBa_0034. All final constructs were confirmed by sequencing.
Medium and growth conditions
For molecular biology experiments, E. coli strains were grown at 37°C under agitation in Luria Bertani (LB) medium [tryptone (10 g/liter), yeast extract (5 g/liter), and NaCl (10 g/liter)]. M9 medium was used for nutrient downshift experiments (27 mM KH2PO4, 8.6 mM NaCl, 9 mM Na2HPO4, 18.8 mM NH4Cl, 2 mM MgSO4, and 0.1 mM CaCl2). For all the other experiments, cells were grown overnight (16 to 18 h) in MOPS medium supplemented with 0.4% glucose (84). Overnight cultures were diluted to an OD600nm of 0.01 in MOPS 0.4% glucose and grown to an OD600nm of 0.3, except if otherwise indicated. When appropriate, culture media were supplemented with chloramphenicol (Cm; 25 μg/ml), Kan (50 μg/ml), ampicillin (100 μg/ml), OFX (5 μg/ml), MMC (20 μg/ml), tetracycline (10 μg/ml), and IPTG (isopropyl-β-d-thiogalactopyranoside; 250 μM).
Growth curve
Overnight cultures grown in MOPS 0.4% glucose were diluted in 2 ml of fresh medium as described above and placed in clear bottom 24-well plates. Plates were inserted into an automated microplate reader and the OD600nm was measured every 15 min for 24 hours while being shaken at 140 rpm at 37°C.
Minimal inhibitory concentration
The MIC of OFX and MMC was determined using the agar dilution method as described (85). Briefly, overnight cultures grown in LB were adjusted to a cell density of 107 colony-forming units/ml in sterile phosphate-buffered saline (PBS). Two microliters was spotted on an LB agar medium containing increasing concentrations of OFX or MMC. The MIC was determined as the lowest dose at which colony growth was inhibited.
Killing assays
Killing assays were performed as previously described (84). Briefly, cultures were grown as described above and treated with OFX (5 μg/ml) at an OD600nm of 0.3. Samples were withdrawn at indicated times, diluted in 10 mM MgSO4, and plated on LB agar plates. Survival frequency was calculated as the ratio of the number of colony-forming units per milliliter at a given time to the number of colony-forming units per milliliter at the treatment time.
Excitation spectra and calibration of pHluorin2 and Hyper-3 reporters
See the Supplementary Materials.
Live-cell microscopy experiments
Time-course experiments
Cultures were grown as described above and treated with OFX (5 μg/ml), MMC (20 μg/ml), or Kan (50 μg/ml) at an OD600nm of 0.3. Ten microliters of cultures were collected at indicated time points, spotted on 1% agarose MOPS 0.4% glucose pads, and imaged by fluorescence microscopy. For PI staining, cells were collected at indicated time points, washed in NaCl solution (0.85%), and stained for 15 min using the LIVE/DEAD BacLightTM Bacterial Viability Kit (Thermo Fisher Scientific). Cells were then washed with the saline solution and transferred on a 1% agarose MOPS 0.4% glucose pad. For membrane potential assay, DiBAC4(3) (Thermo Fisher Scientific) was dissolved in 100% dimethyl sulfoxide (DMSO). At indicated time points, 1 ml of culture was collected and 1 µl of DiBAC4(3) (10 μg/ml) was added to the sample, followed by incubation for 15 min in the dark. Cells were then washed and transferred on 1% agarose MOPS 0.4% glucose pads. Negative control corresponds to untreated cells and positive control to heat-shocked cells at 72°C for 1 hour. For H2O2 detection after OFX treatment using H2DCFDA (Sigma-Aldrich), H2DCFDA was dissolved in 100% DMSO and, at indicated time points, 1 ml of cell culture was collected and incubated for 40 min with H2DCFDA (10 μg/ml). Cells were then washed and transferred on 1% agarose MOPS 0.4% glucose pads.
Time-lapse experiment on agarose pad
Cultures were grown as described above, spotted on 1% agarose MOPS 0.4% glucose pads at OD600nm of 0.3, and imaged using fluorescence microscopy at indicated time intervals (in minutes per frame). For time-lapse experiments during OFX treatment, OFX (5 μg/ml) was directly added to the 1% agarose Mops 0.4% glucose pads. For time-lapse experiments with the SSDsbA-sfGFP fusion, membrane potential assays, and osmotic shock, 1% agarose MOPS 0.4% glucose pads were supplemented with 250 μM IPTG and OFX (5 μg/ml), 1 µl of DiBAC4(3) (10 μg/ml), and OFX (5 μg/ml), or 20% sucrose, respectively.
Time-lapse in a microfluidic chamber
Time-lapse in microfluidic chambers was performed as described previously (16, 86). Briefly, cells were grown as described above to an OD600 nm of 0.3. Cells were then diluted in MOPS 0.4% glucose to an OD600nm of 0.05 and loaded into a B04A microfluidic chamber (ONIX, CellASIC) preheated at 37°C. Nutrient supply was maintained at 6.9 kPa for the duration of time-lapse imaging. For time-lapse in the presence of OFX, cells were supplied for 1 hour with MOPS 0.4% glucose injected at 6.9 kPa into the microfluidic chamber followed by the injection of MOPS 0.4% glucose containing OFX (5 μg/ml) at 6.9 kPa for 6 hours. For nutrient downshift experiments, cells were grown in LB medium at 37°C while shaking to an OD600nm of 0.3. Cells were diluted to an OD600nm of 0.01 in LB, loaded into the microfluidic chamber, and perfused for 2 hours with LB. Nutrient downshift was achieved by perfusing cells in M9 medium for 2 hours followed by perfusion with LB medium.
Image acquisition
Conventional wide-field fluorescence microscopy imaging was carried out on an AxioObserver inverted microscope (Carl Zeiss, Germany), equipped with 100×/1.45 oil Plan Apo Lambda phase objective, Orca FLash4 V2 CMOS camera (Hamamatsu) and a Colibri 7 LED light source (Carl Zeiss, Germany). Acquisition was performed using ZEN Blue 3.2 (Carl Zeiss, Germany) with different filters (Carl Zeiss, Germany) and parameters, depending on the fluorescent reporter or chemical dye (table S5).
Image analysis
Opening and visualization of original microscopy images were performed using the open-source ImageJ/Fiji [https://fiji.sc/; (87)] software. For quantitative image analysis, the open-source ImageJ/Fiji software was used together with the free MicrobeJ plugin [www.microbej.com; (88)]. Analysis of cell size and cell surface during time-course experiments was performed by automated detection of cells based on the segmentation of phase contrast images. Cell outlines were corrected, if necessary, by using the manual editing interface of the Fiji MicrobeJ plugin. Demographs and kymographs of HU-mCherry and periplasmic SSDsbA-sfGFP fluorescence distributions were generated automatically using MicrobeJ. Quantification of cell area and length and fluorescence intensity was performed using MicrobeJ and plotted with GraphPad Prism.
Calibration and H2O2 detection by the katG reporter
Calibration
MG1655 containing the pkatG-gfpmut2 plasmid (44) cultures were grown to an OD600nm of 0.3 and diluted 10-fold in MOPS 0.4% glucose containing 0, 5, 10, 20, 50, 100, or 150 μM H2O2. After 30-min incubation, fluorescence was measured by flow cytometry. Samples were diluted in PBS to an OD600nm of 0.01 and processed by Attune NXT flow cytometer (Thermo Fisher Scientific) at a flow rate of 12.5 μl/min. In each experiment, 50,000 events were analyzed with a blue laser (488 nm) for forward and side-scatter (FSC-H and SSC-H, respectively) and a 530/30 emission filter for GFP. Doublets were gated out on the basis of the area-to-height ratio of their side-scatter pulses. Photomultiplier tube gain was adjusted to obtain background median values of ~103 arbitrary units (A.U.).
katG reporter during OFX treatment
MG1655 containing the pkatG-gfpmut2 plasmid (44) was grown as described above to an OD600nm of 0.3 and treated with OFX (5 μg/ml). Thirty minutes before the indicated times, samples were collected and GFP fluorescence was measured via flow cytometry as described above.
In vivo replication, transcription, and translation assays
Cells were grown as described above and treated with OFX (5 μg/ml) at an OD600nm of 0.3. One milliliter of culture samples was collected at indicated time points and incubated with [3H]-thymidine (1 μCi/ml), [3H]-uridine (1 μCi/ml), or [35S]-methionine (3 μCi/ml; PerkinElmer) for 5 min. Untreated samples were collected before the addition of OFX. Samples were precipitated overnight at 4°C with 5 ml of cold 10% trichloroacetic acid (TCA). The next day, samples were filtered on 0.45-μm nitrocellulose and washed with 15 ml of 10% TCA. Filter-retained material was counted in 10 ml of scintillation liquid (Ecoscint H, National Diagnostics, LS-275) in a liquid scintillation counter (Beckman LS 6500). The integration time per read was 2 min. Background signals from cells without radioactivity were subtracted. The incorporation rate is the ratio of the CPM counted at each time point to the corresponding zero time. Controls for DNA, RNA, and protein synthesis arrest consist of cultures treated with MMC (10 μg/ml), rifampicin (100 μg/ml), and Cm (30 μg/ml), respectively.
Acknowledgments
We are grateful to I. Matic and M. Deghelt for constructive discussions and comments on the manuscript. We thank M. Maurizi, S. Gottesman, F. Goormaghtigh, D. Jurenas, G. Laloux, D. Mazel, and E. Cascales for helpful comments. We thank P. Goffin for implementing the pHluorin2 and Hyper-3 biosensors; C. Rousseau for constructing the ΔistR1-tisB strain; L. Dubuisson and R. Antheme for technical assistance; and T. Bernhardt, I. Matic, and D. Jurenas for providing strains, plasmids, and valuable reagents. We thank C. Jauniaux and B. André for help with the radioactive incorporation experiments.
Funding: This work was supported by the Wallonia Region (projet Pôles WAGRALIM-BIOWIN, Single Cells), ARC-Advanced (Actions 2018–2023), the Fonds National de la Recherche Scientifique (FNRS CDR J.0182.21F), and the International Brachet Stiftung (IBS). J.C. is supported by a FNRS Chargé de Recherches fellowship (2019–2022), T.O. (2022–2026), and T.S. (2018–2022) by a FNRS-FRIA fellowship.
Author contributions: Conceptualization: J.C., T.O., T.S., and L.V.M. Methodology: J.C., T.O., T.S., and L.V.M. Investigation: J.C., T.O., T.S., and S.Z. Supervision: L.V.M. Writing—original draft: J.C., T.O., and L.V.M. Writing—review and editing: J.C., T.O., and L.V.M. Funding acquisition: L.V.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Legends for movies S1 to S3
Tables S1 to S5
References
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S3
REFERENCES AND NOTES
- 1.Gutierrez A., Stokes J. M., Matic I., Our evolving understanding of the mechanism of quinolones. Antibiotics (Basel) 7, 32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohanski M. A., Dwyer D. J., Collins J. J., How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 8, 423–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darby E. M., Trampari E., Siasat P., Gaya M. S., Alav I., Webber M. A., Blair J. M. A., Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 21, 280–295 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J., A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Dwyer D. J., Kohanski M. A., Hayete B., Collins J. J., Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3, 91 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lobritz M. A., Belenky P., Porter C. B. M., Gutierrez A., Yang J. H., Schwarz E. G., Dwyer D. J., Khalil A. S., Collins J. J., Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. U.S.A. 112, 8173–8180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belenky P., Ye J. D., Porter C. B. M., Cohen N. R., Lobritz M. A., Ferrante T., Jain S., Korry B. J., Schwarz E. G., Walker G. C., Collins J. J., Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 13, 968–980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong Y., Li Q., Gao Q., Xie J., Huang H., Drlica K., Zhao X., Reactive oxygen species play a dominant role in all pathways of rapid quinolone-mediated killing. J. Antimicrob. Chemother. 75, 576–585 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Wang X., Zhao X., Malik M., Drlica K., Contribution of reactive oxygen species to pathways of quinolone-mediated bacterial cell death. J. Antimicrob. Chemother. 65, 520–524 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong F., Stokes J. M., Cervantes B., Penkov S., Friedrichs J., Renner L. D., Collins J. J., Cytoplasmic condensation induced by membrane damage is associated with antibiotic lethality. Nat. Commun. 12, 2321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmons L. A., Foti J. J., Cohen S. E., Walker G. C., The SOS regulatory network. EcoSal Plus. 3, 10.1128/ecosalplus.5.4.3 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Erill I., Campoy S., Barbé J., Aeons of distress: An evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31, 637–656 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Baharoglu Z., Mazel D., SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol. Rev. 38, 1126–1145 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Matic I., Taddei F., Radman M., Survival versus maintenance of genetic stability: A conflict of priorities during stress. Res. Microbiol. 155, 337–341 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Dörr T., Lewis K., Vulić M., SOS response induces persistence to fluoroquinolones in Escherichia coli. PLOS Genet. 5, e1000760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goormaghtigh F., Van Melderen L., Single-cell imaging and characterization of Escherichia coli persister cells to ofloxacin in exponential cultures. Sci. Adv. 5, eaav9462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Völzing K. G., Brynildsen M. P., Stationary-phase persisters to ofloxacin sustain DNA damage and require repair systems only during recovery. MBio 6, e00731–e00715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diver J. M., Wise R., Morphological and biochemical changes in Escherichia coli after exposure to ciprofloxacin. J. Antimicrob. Chemother. 18, 31–41 (1986). [DOI] [PubMed] [Google Scholar]
- 19.Dougherty T. J., Saukkonen J. J., Membrane permeability changes associated with DNA gyrase inhibitors in Escherichia coli. Antimicrob. Agents Chemother. 28, 200–206 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher J. K., Bourniquel A., Witz G., Weiner B., Prentiss M., Kleckner N., Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell 153, 882–895 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinh T., Bernhardt T. G., Using superfolder green fluorescent protein for periplasmic protein localization studies. J. Bacteriol. 193, 4984–4987 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odsbu I., Skarstad K., DNA compaction in the early part of the SOS response is dependent on RecN and RecA. Microbiology (Reading). 160, 872–882 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Lesterlin C., Ball G., Schermelleh L., Sherratt D. J., RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature 506, 249–253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vickridge E., Planchenault C., Cockram C., Junceda I. G., Espéli O., Management of E. coli sister chromatid cohesion in response to genotoxic stress. Nat. Commun. 8, 14618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Little J. W., Harper J. E., Identification of the lexA gene product of Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 76, 6147–6151 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin L. L., Little J. W., Isolation and characterization of noncleavable (Ind-) mutants of the LexA repressor of Escherichia coli K-12. J. Bacteriol. 170, 2163–2173 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unoson C., Wagner E. G. H., A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 70, 258–270 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Gurnev P. A., Ortenberg R., Dörr T., Lewis K., Bezrukov S. M., Persister-promoting bacterial toxin TisB produces anion-selective pores in planar lipid bilayers. FEBS Lett. 586, 2529–2534 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinbrecher T., Prock S., Reichert J., Wadhwani P., Zimpfer B., Bürck J., Berditsch M., Elstner M., Ulrich A. S., Peptide-lipid interactions of the stress-response peptide TisB that induces bacterial persistence. Biophys. J. 103, 1460–1469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berghoff B. A., Hoekzema M., Aulbach L., Wagner E. G. H., Two regulatory RNA elements affect TisB-dependent depolarization and persister formation. Mol. Microbiol. 103, 1020–1033 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Vogel J., Argaman L., Wagner E. G. H., Altuvia S., The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 14, 2271–2276 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Darfeuille F., Unoson C., Vogel J., Wagner E. G. H., An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell 26, 381–392 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Wagner E. G. H., Unoson C., The toxin-antitoxin systemtisB-istR1. RNA Biol. 9, 1513–1519 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Romilly C., Lippegaus A., Wagner E. G. H., An RNA pseudoknot is essential for standby-mediated translation of the tisB toxin mRNA in Escherichia coli. Nucleic Acids Res. 48, 12336–12347 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su W.-L., Bredèche M.-F., Dion S., Dauverd J., Condamine B., Gutierrez A., Denamur E., Matic I., TisB protein protects Escherichia coli cells suffering massive DNA damage from environmental toxic compounds. MBio 13, e0038522 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulder E., Woldringh C. L., Plasmolysis bays in Escherichia coli: Are they related to development and positioning of division sites? J. Bacteriol. 175, 2241–2247 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi H., Westfall C. S., Kao J., Odermatt P. D., Anderson S. E., Cesar S., Sievert M., Moore J., Gonzalez C. G., Zhang L., Elias J. E., Chang F., Huang K. C., Levin P. A., Starvation induces shrinkage of the bacterial cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 118, e2104686118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kogoma T., Torrey T. A., Connaughton M. J., Induction of UV-resistant DNA replication in Escherichia coli: Induced stable DNA replication as an SOS function. Mol. Gen. Genet. 176, 1–9 (1979). [DOI] [PubMed] [Google Scholar]
- 39.Edelmann D., Leinberger F. H., Schmid N. E., Oberpaul M., Schäberle T. F., Berghoff B. A., Elevated expression of toxin TisB protects persister cells against ciprofloxacin but enhances susceptibility to mitomycin C. Microorganisms. 9, 943 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams D. S., Levin M., Measuring resting membrane potential using the fluorescent voltage reporters DiBAC4(3) and CC2-DMPE. Cold Spring Harb. Protoc. 2012, 459–464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krulwich T. A., Sachs G., Padan E., Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahon M. J., pHluorin2: An enhanced, ratiometric, pH-sensitive green florescent protein. ABB. 02, 132–137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edelmann D., Berghoff B. A., Type I toxin-dependent generation of superoxide affects the persister life cycle of Escherichia coli. Sci. Rep. 9, 14256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silander O. K., Nikolic N., Zaslaver A., Bren A., Kikoin I., Alon U., Ackermann M., A genome-wide analysis of promoter-mediated phenotypic noise in Escherichia coli. PLOS Genet. 8, e1002443 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loewen P. C., Switala J., Purification and characterization of catalase HPII from Escherichia coli K12. Biochem. Cell Biol. 64, 638–646 (1986). [DOI] [PubMed] [Google Scholar]
- 46.Sen A., Imlay J. A., How microbes defend themselves from incoming hydrogen peroxide. Front. Immunol. 12, 667343 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willmott C. J., Critchlow S. E., Eperon I. C., Maxwell A., The complex of DNA gyrase and quinolone drugs with DNA forms a barrier to transcription by RNA polymerase. J. Mol. Biol. 242, 351–363 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Bilan D. S., Pase L., Joosen L., Gorokhovatsky A. Y., Ermakova Y. G., Gadella T. W. J., Grabher C., Schultz C., Lukyanov S., Belousov V. V., HyPer-3: A genetically encoded H(2)O(2) probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem. Biol. 8, 535–542 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Dwyer D. J., Belenky P. A., Yang J. H., MacDonald I. C., Martell J. D., Takahashi N., Chan C. T. Y., Lobritz M. A., Braff D., Schwarz E. G., Ye J. D., Pati M., Vercruysse M., Ralifo P. S., Allison K. R., Khalil A. S., Ting A. Y., Walker G. C., Collins J. J., Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U.S.A. 111, E2100–E2109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martínez S. R., Durantini A. M., Becerra M. C., Cosa G., Real-time single-cell imaging reveals accelerating lipid peroxyl radical formation in Escherichia coli triggered by a fluoroquinolone antibiotic. ACS Infect Dis. 6, 2468–2477 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Goss W. A., Deitz W. H., Cook T. M., Mechanism of action of nalidixic acid on Escherichia coli II. Inhibition of deoxyribonucleic acid synthesis. J. Bacteriol. 89, 1068–1074 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pohlhaus J. R., Kreuzer K. N., Norfloxacin-induced DNA gyrase cleavage complexes block Escherichia coli replication forks, causing double-stranded breaks in vivo. Mol. Microbiol. 56, 1416–1429 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maxwell A., The molecular basis of quinolone action. J. Antimicrob. Chemother. 30, 409–414 (1992). [DOI] [PubMed] [Google Scholar]
- 54.Howard M. T., Neece S. H., Matson S. W., Kreuzer K. N., Disruption of a topoisomerase-DNA cleavage complex by a DNA helicase. Proc. Natl. Acad. Sci. U.S.A. 91, 12031–12035 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wentzell L. M., Maxwell A., The complex of DNA gyrase and quinolone drugs on DNA forms a barrier to the T7 DNA polymerase replication complex. J. Mol. Biol. 304, 779–791 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Schneider V., Wadhwani P., Reichert J., Bürck J., Elstner M., Ulrich A. S., Kubař T., Tetrameric charge-zipper assembly of the TisB peptide in membranes-computer simulation and experiment. J. Phys. Chem. B 123, 1770–1779 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Grage S. L., Afonin S., Kara S., Buth G., Ulrich A. S., Membrane thinning and thickening induced by membrane-active amphipathic peptides. Front. Cell Dev. Biol. 4, 65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smirnova G. V., Tyulenev A. V., Muzyka N. G., Peters M. A., Oktyabrsky O. N., Ciprofloxacin provokes SOS-dependent changes in respiration and membrane potential and causes alterations in the redox status of Escherichia coli. Res. Microbiol. 168, 64–73 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Brauer A. M., Shi H., Levin P. A., Huang K. C., Physiological and regulatory convergence between osmotic and nutrient stress responses in microbes. Curr. Opin. Cell Biol. 81, 102170 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swenson P. A., Schenley R. L., Evidence for the control of respiration by DNA in ultraviolet-irradiated Escherichia coli B-r cells. Mutat. Res. 9, 443–453 (1970). [DOI] [PubMed] [Google Scholar]
- 61.Swenson P. A., Schenley R. L., Evidence relating cessation of respiration, cell envelope changes, and death in ultraviolet-irradiated Escherichia coli B/r cells. J. Bacteriol. 117, 551–559 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y., Imlay J. A., Cell death from antibiotics without the involvement of reactive oxygen species. Science 339, 1210–1213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ezraty B., Vergnes A., Banzhaf M., Duverger Y., Huguenot A., Brochado A. R., Su S.-Y., Espinosa L., Loiseau L., Py B., Typas A., Barras F., Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 340, 1583–1587 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Keren I., Wu Y., Inocencio J., Mulcahy L. R., Lewis K., Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339, 1213–1216 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Seaver L. C., Imlay J. A., Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J. Biol. Chem. 279, 48742–48750 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Korshunov S., Imlay J. A., Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol. Microbiol. 75, 1389–1401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Snyder M., Drlica K., DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J. Mol. Biol. 131, 287–302 (1979). [DOI] [PubMed] [Google Scholar]
- 68.Malik M., Zhao X., Drlica K., Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol. Microbiol. 61, 810–825 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Pennington J. M., Rosenberg S. M., Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 39, 797–802 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edelmann D., Berghoff B. A., A shift in perspective: A role for the type I Toxin TisB as persistence-stabilizing factor. Front. Microbiol. 13, 871699 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Booth J. A., Špírek M., Lobie T. A., Skarstad K., Krejci L., Bjørås M., Antibiotic-induced DNA damage results in a controlled loss of pH homeostasis and genome instability. Sci. Rep. 10, 19422 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reyes-Fernández E. Z., Schuldiner S., Acidification of cytoplasm in Escherichia coli provides a strategy to cope with stress and facilitates development of antibiotic resistance. Sci. Rep. 10, 9954 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van den Bergh B., Schramke H., Michiels J. E., Kimkes T. E. P., Radzikowski J. L., Schimpf J., Vedelaar S. R., Burschel S., Dewachter L., Lončar N., Schmidt A., Meijer T., Fauvart M., Friedrich T., Michiels J., Heinemann M., Mutations in respiratory complex I promote antibiotic persistence through alterations in intracellular acidity and protein synthesis. Nat. Commun. 13, 546 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Padan E., Schuldiner S., [27] Intracellular pH regulation in bacterial cells. Methods Enzymol. 125, 337–352 (1986). [DOI] [PubMed] [Google Scholar]
- 75.Dri A. M., Moreau P. L., Control of the LexA regulon by pH: Evidence for a reversible inactivation of the LexA repressor during the growth cycle of Escherichia coli. Mol. Microbiol. 12, 621–629 (1994). [DOI] [PubMed] [Google Scholar]
- 76.Sousa F. J. R., Lima L. M. T. R., Pacheco A. B. F., Oliveira C. L. P., Torriani I., Almeida D. F., Foguel D., Silva J. L., Mohana-Borges R., Tetramerization of the LexA repressor in solution: Implications for gene regulation of the E.coli SOS system at acidic pH. J. Mol. Biol. 359, 1059–1074 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Ketcham A., Freddolino P. L., Tavazoie S., Intracellular acidification is a hallmark of thymineless death in E. coli. PLOS Genet. 18, e1010456 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weel-Sneve R., Kristiansen K. I., Odsbu I., Dalhus B., Booth J., Rognes T., Skarstad K., Bjørås M., Single transmembrane peptide DinQ modulates membrane-dependent activities. PLOS Genet. 9, e1003260 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pribis J. P., García-Villada L., Zhai Y., Lewin-Epstein O., Wang A. Z., Liu J., Xia J., Mei Q., Fitzgerald D. M., Bos J., Austin R. H., Herman C., Bates D., Hadany L., Hastings P. J., Rosenberg S. M., Gamblers: An antibiotic-induced evolvable cell subpopulation differentiated by reactive-oxygen-induced general stress response. Mol. Cell 74, 785–800.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pribis J. P., Zhai Y., Hastings P. J., Rosenberg S. M., Stress-induced mutagenesis, gambler cells, and stealth targeting antibiotic-induced evolution. MBio 13, e0107422 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blattner F. R., Plunkett G., Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Glasner J. D., Rode C. K., Mayhew G. F., Gregor J., Davis N. W., Kirkpatrick H. A., Goeden M. A., Rose D. J., Mau B., Shao Y., The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997). [DOI] [PubMed] [Google Scholar]
- 82.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davis J. H., Rubin A. J., Sauer R. T., Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 39, 1131–1141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goormaghtigh F., Van Melderen L., Optimized method for measuring persistence in escherichia coli with improved reproducibility. Methods Mol. Biol. 1333, 43–52 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Wiegand I., Hilpert K., Hancock R. E. W., Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008). [DOI] [PubMed] [Google Scholar]
- 86.Oms T., Schlechtweg T., Cayron J., Van Melderen L., Population and single-cell analysis of antibiotic persistence in Escherichia coli. J. Vis. Exp., 10.3791/64550 (2023). [DOI] [PubMed] [Google Scholar]
- 87.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ducret A., Quardokus E. M., Brun Y. V., MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cayron J., Dedieu-Berne A., Lesterlin C., Bacterial filaments recover by successive and accelerated asymmetric divisions that allow rapid post-stress cell proliferation. Mol. Microbiol. 119, 237–251 (2023). [DOI] [PubMed] [Google Scholar]
- 90.Goormaghtigh F., Fraikin N., Putrinš M., Hallaert T., Hauryliuk V., Garcia-Pino A., Sjödin A., Kasvandik S., Udekwu K., Tenson T., Kaldalu N., Van L., Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type ii persister cells. mBio 9, e00640-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Legends for movies S1 to S3
Tables S1 to S5
References
Movies S1 to S3