Abstract
While pancreatic ductal adenocarcinomas (PDACs) are addicted to KRAS-activating mutations, inhibitors of downstream KRAS effectors, such as the MEK1/2 kinase inhibitor trametinib, are devoid of therapeutic effects. However, the extensive rewiring of regulatory circuits driven by the attenuation of the KRAS pathway may induce vulnerabilities of therapeutic relevance. An in-depth molecular analysis of the transcriptional and epigenomic alterations occurring in PDAC cells in the initial hours after MEK1/2 inhibition by trametinib unveiled the induction of endogenous retroviruses (ERVs) escaping epigenetic silencing, leading to the production of double-stranded RNAs and the increased expression of interferon (IFN) genes. We tracked ERV activation to the early induction of the transcription factor ELF3, which extensively bound and activated nonsilenced retroelements and synergized with IRF1 (interferon regulatory factor 1) in the activation of IFNs and IFN-stimulated genes. Trametinib-induced viral mimicry in PDAC may be exploited in the rational design of combination therapies in immuno-oncology.
MEK1/2 inhibition activates endogenous retroviruses in pancreatic cancer cells, thus inducing an interferon response.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is the most lethal common solid malignancy, with a median survival time after diagnosis of 8 to 12 months and a 5-year survival rate lower than 10% considering all disease stages (1). As PDAC is predicted to become the second leading cause of cancer-related deaths by 2040 (2), there is the pressing need to increase our understanding of this cancer to develop rational approaches to therapy.
Although extensive molecular profiling studies carried out during the past two decades allowed identifying the genetic and molecular bases of PDAC (3, 4), they did not translate into measurable changes in the overall survival, which has significantly improved only for the small fraction of patients who could undergo surgical resection (5).
KRAS-activating mutations (KRAS G12D, G12V, and G12R) are detected in 94% of PDACs (6, 7), and their role as essential founders and drivers in this cancer has been extensively validated (8). The early selection of KRAS mutations in PDAC precursor lesions, the pancreatic intraepithelial neoplasia (PanINs) (9), is explained by their ability to increase cellular fitness via various mechanisms (10), notably the rapid inactivation of acinar gene expression in response to damage, which prevents acinar enzyme release and the ensuing tissue injury, thus eventually preserving organ integrity (11).
Although PDAC is addicted to KRAS (12–14), blocking the constitutively active KRAS–RAF–MEK [mitogen-activated protein kinase (MAPK) kinase]–ERK (extracellular signal–regulated kinase) pathway by drugs targeting downstream KRAS effectors, such the clinically approved MEK1/2 kinase inhibitor trametinib, is devoid of significant effects in patients, and in vitro, it only reduces cell proliferation without causing cell death, in part due to the induction of adaptive mechanisms during sustained treatment (15, 16).
While inhibition of KRAS effectors has limited, if any, clinical impact in PDAC, disabling the KRAS-ERK axis may expose vulnerabilities of possible therapeutic relevance. For instance, increased expression of autophagy genes following KRAS-MEK1/2 inactivation suggested the possible synergy between trametinib and autophagy inhibitors, which in some cases resulted in clinical effects (13, 17, 18).
With these notions in mind, we set out to molecularly dissect how transcriptional regulatory networks driven by constitutively active KRAS in PDAC cells are rewired following acute inhibition of MEK1/2 by trametinib. The most striking and unexpected finding was the identification of a transcriptional circuit rapidly activated upon MEK1/2 inhibition that directly caused the strong induction of endogenous retroviruses (ERVs) that escaped epigenetic silencing.
ERVs account for 9% of the human genome and include about half a million sequences divided into 100 families (19). About 90% of ERVs exist as single or “solo” long terminal repeats (LTRs) generated by recombination of the 5′ and 3′ LTRs of an integrated provirus. Owing to the high density of transcription factor binding sites in LTRs (20) and their ability to drive transcription initiation, they were extensively coopted as enhancers and promoters controlling various transcriptional networks such as those involved in placentation (21), immune response (22), and oncogenesis (23–25).
Because of such a high regulatory potential, ERVs are subjected to stringent control and are extensively repressed. In somatic cells, ERV silencing is mainly enforced by DNA methylation and repressive histone marks. In particular, ERVs are repressed by the local deposition of H3K9me3 by the methyltransferase SETDB1 (26–29), which is recruited upon DNA sequence–specific recognition of ERVs by KRAB domain–containing zinc finger proteins (KZFPs) (30–32) associated with the KAP1 (TRIM28) corepressor (33, 34).
Nevertheless, despite actively enforced and efficient silencing, ERVs can elude repression. Because most of the binding sites for KZFP transcription factors are in internal ERV regions that are lost upon recombination of the 5′ and the 3′ LTRs (24), solo LTRs commonly escape H3K9me3-mediated repression. Silencing escape can also occur in the context of highly regulated developmental transitions, thus allowing ERVs to acquire chromatin profiles characteristic of active enhancers (35) and to contribute to endogenous gene regulation (21, 22).
Derepression of ERVs leading to increased expression of ERV-derived double-stranded RNAs (dsRNAs) from both intact and solo ERVs (36, 37) can be induced by treatment with inhibitors of both DNA and histone methyltransferases (38–41). In these conditions, detection of ERV-derived dsRNAs by cytoplasmic viral sensors is interpreted as a sign of viral infection, resulting in a state of viral mimicry causing the induction of type I and III interferons (IFNs) (38, 39). The therapeutic relevance of viral mimicry for antitumor therapy (42, 43) relates to the ability of IFNs to stimulate immune responses both by inducing the expression of genes encoding components of the antigen presentation machinery and by promoting the recruitment of innate and adaptive immune cells to the tumor microenvironment (40). In addition, the production of immunogenic ERV-derived peptides can determine antitumor adaptive responses, as shown in renal cancer (44).
Here, we described an unreported effect of trametinib, which involves the induction of ERV expression in PDAC. We found that one of the earliest events caused by MEK1/2 inhibition by trametinib in all PDAC cell lines tested was the up-regulation of the ETS family transcription factor ELF3, which directly bound and activated a large number of retroelements escaping H3K9me3-mediated repression. Increased expression of ERV-derived dsRNAs caused a robust IFN response that represented one of the strongest components of the trametinib-induced transcriptional reprogramming in multiple PDAC cell lines. The viral mimicry caused by trametinib in PDAC may have practical relevance in the design of combination therapies in immuno-oncology, as indicated by the effects of the combined treatment with trametinib and an anti-PD1 immune checkpoint inhibitor in an immunocompetent PDAC mouse model.
RESULTS
Rapid induction of the IFN response by trametinib
Treatment of PDAC cells with trametinib at a concentration corresponding to the Cmax, the maximal concentration that can be reached in patients’ blood (https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/217513s000lbl.pdf), did not affect viability over 3 days (fig. S1A), while it caused the sustained down-regulation of ERK1/2 phosphorylation (fig. S1B).
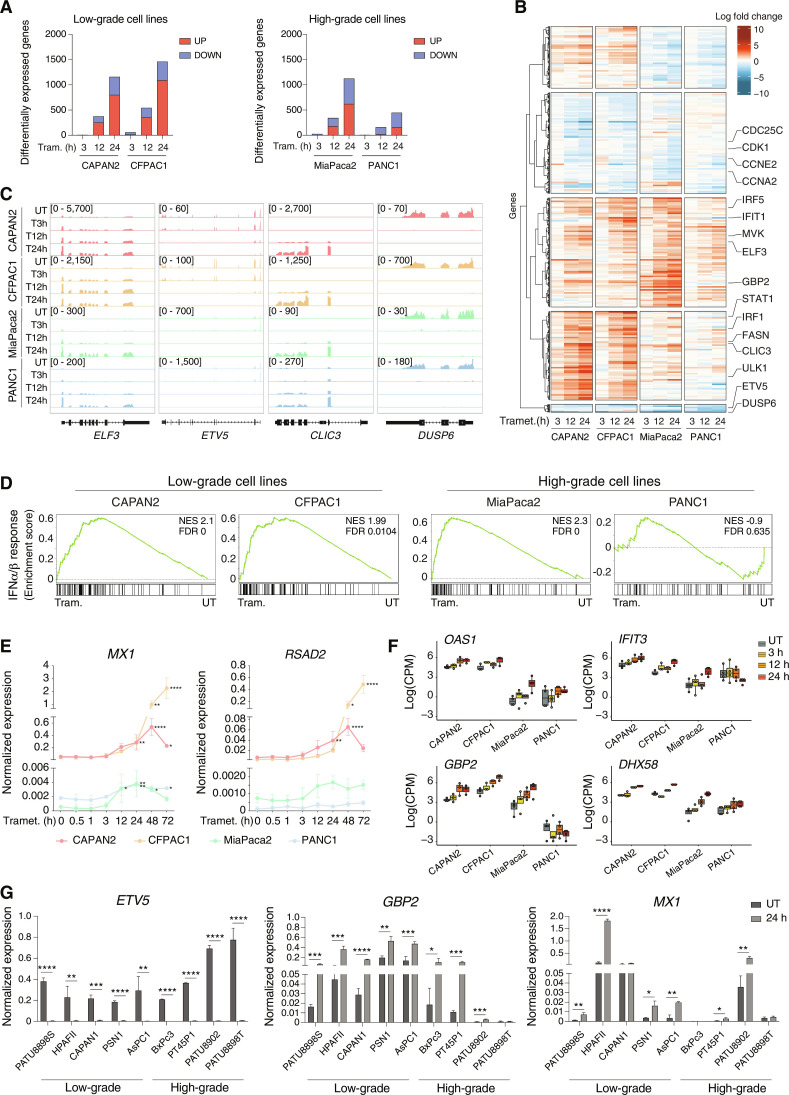
We generated RNA-sequencing (RNA-seq) datasets in two low-grade (CAPAN2 and CFPAC1) and two high-grade (MiaPaca2 and PANC1) PDAC cell lines that were sampled at multiple time points after trametinib treatment. Data clustered by cell line and time, with the 12- and 24-hour time points clustering together and separately from the 3-hour time point and untreated cells (fig. S1, C to F). While only a handful of genes were differentially expressed at 3 hours [log2 fold change (log2FC) ≥ 1 and false discovery rate (FDR) ≤ 0.001], the response was greatly amplified over time, with hundreds of differentially expressed genes (DEGs) (Fig. 1, A and B; fig. S1G; and table S1) that included a core set of genes showing shared profiles either in cells of the same grade or across all cell lines tested (fig. S1H and table S2). Among them, the down-regulation of the ERK phosphatase DUSP6 and the ERK-regulated transcription factor ETV5 was consistent with the efficient MEK1/2 inactivation (Fig. 1C and tables S1 and S2). Core up-regulated genes included the transcription factor ELF3 and the chloride intracellular channel CLIC3 (Fig. 1C and tables S1 and S2).
Fig. 1. Activation of the IFN response upon trametinib treatment.
(A) Bar plots showing the number of DEGs (up-regulated in red and down-regulated in blue) in low-grade (left panel) and high-grade (right panel) PDAC cell lines after trametinib treatment with respect to untreated samples. (B) DEGs in low-grade and high-grade PDAC cell lines after trametinib treatment. RNA-seq normalized counts are shown as row z scores. The number of DEGs in the five clusters shown is as follows (from top to bottom): 441, 721, 779, 633, and 57. (C) Genome browser snapshots showing RNA-seq profiles for selected up- or down-regulated genes in low- and high-grade PDAC cell lines after trametinib treatment. (D) GSEA plots showing the differential enrichment of the type I IFN signature in low- and high-grade PDAC cell lines treated with trametinib for 24 hours versus untreated (UT) cells. (E) Expression of two IFN-stimulated genes, MX1 and RSAD2, was measured by RT-qPCR during a trametinib treatment time course. Values represent 2−ΔCq relative to the reference gene (C1ORF43). n = 3 independent biological replicates. Means ± SD are shown. Significance was assessed using one-way ANOVA and indicated as follows: *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001. (F) RNA-seq data counts showing the expression of selected IFN-regulated genes in low- and high-grade PDAC cell lines. Data are reported as the log of count per million (CPM) after TMM normalization with EdgeR. (G) RT-qPCR analysis of the ETV5, GBP2, and MX1 genes in a panel of human PDAC cell lines after treatment with trametinib or vehicle for 24 hours. Values represent 2−ΔCq relative to C1ORF43. n = 3 independent biological replicates. Means ± SD are shown. Significance was assessed using t test and indicated as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
In addition to the expected down-regulation of ERK-dependent gene expression programs, a Gene Set Enrichment Analysis (GSEA) (table S3) showed a strong induction across multiple cell lines of two main groups of functional signatures related to cholesterol synthesis (see below) and to the IFN response (Fig. 1D), which was validated by a time-resolved quantitative reverse transcription polymerase chain reaction (RT-qPCR) on two IFN-stimulated genes (MX1 and RSAD2) (Fig. 1E). The IFN response was significantly induced in both low-grade cell lines and high-grade MiaPaca2 cell line, but the intensity of the response was of lower magnitude in the latter (Fig. 1E), as also indicated by the level of induction of representative IFN-stimulated genes in the RNA-seq datasets (Fig. 1F). A more extensive analysis confirmed the induction of IFN-stimulated genes in response to trametinib stimulation in a broad panel of PDAC cell lines, with an imperfect trend toward higher responses in low-grade than high-grade cells (Fig. 1G).
Requirement for the dsRNA sensing pathway for IFN induction by trametinib
Induction of IFN-stimulated genes was associated with the induction of signal transducer and activator of transcription 1 (STAT1) phosphorylation at Tyr701, which occurs in response to Janus kinase 1 (JAK1)/TYK2 kinase activation upon IFNα/β receptor (IFNAR) triggering (Fig. 2A). Consistent with an autocrine and paracrine activation of the IFN response, the IFNB1 gene was strongly induced in response to trametinib in the low-grade CFPAC1 and CAPAN2 cell lines (Fig. 2B). Instead, MiaPaca2 cells contain a large deletion of the INK4/ARF locus that extends to the type I IFN gene cluster (45). However, in these cells, the type III IFN gene IFNL3 was induced in response to trametinib (Fig. 2B), with its comparatively lower activation correlating with the mild induction of STAT1 Tyr701 phosphorylation. Treatment with baricitinib, a JAK family kinase inhibitor, abrogated the induction of STAT1 phosphorylation by trametinib (Fig. 2C), thus confirming the involvement of the IFNAR pathway.
Fig. 2. MAVS requirement for the induction of the IFN response by trametinib.
(A) pY701-STAT1 and total STAT1 abundance in trametinib-treated PDAC cell lines. Vinculin: loading control. Representative of n = 3 independent experiments. (B) IFNB1 and IFNL3 mRNA abundance in PDAC cell lines after trametinib treatment. Values represent 2−ΔCq relative to the C1ORF43 reference gene. n = 3 biological replicates. Means ± SD are shown. Significance was assessed using one-way ANOVA (*P ≤ 0.05). (C) pY701-STAT1 abundance in low-grade PDAC cell lines after treatment with trametinib and/or baricitinib (72 hours). Representative of n = 3 independent experiments. (D) Left: MX1 gene expression in CFPAC1 cells after treatment with trametinib and/or RU.521 (72 hours). Values represent 2−ΔCq relative to C1ORF43. n = 3. Means ± SD are shown. One-way ANOVA (ns P > 0.05, **P ≤ 0.0). Right: IFNB1 mRNA abundance in CFPAC1 cells after transfection with ssDNA and treatment with RU.521 (6 hours). Values represent 2−ΔCq relative to C1ORF43. (E) Effects of MAVS gene inactivation on trametinib-induced pY701-STAT1. Left: pY701-STAT1 and total STAT1 abundance in CFPAC1 cells transduced with nontargeting or MAVS-targeting single-guide RNAs (sgRNAs) and treated with trametinib or vehicle (72 hours). Right: MAVS abundance in CFPAC1 infected with lenti-V2 expressing a nontargeting (NT) or a MAVS-targeting sgRNA (n = 3). (F) RT-qPCR analysis of IFNB1 and selected IFN-stimulated genes in CFPAC1 transduced with nontargeting (NT) or MAVS-specific sgRNAs (n = 3) after treatment with trametinib or vehicle (72 hours). Values represent 2−ΔCq relative to C1ORF43. Means ± SD are shown. Two-way ANOVA (ns P > 0.05, *P ≤ 0.05, **P ≤ 0.01). (G) ETV5 and IFNB1 mRNA abundance in primary PDAC cell lines in vitro upon trametinib treatment. Values represent 2−ΔCq to C1ORF43. n = 3. Means ± SD are shown. Two-way ANOVA (*P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001). (H) ETV5 and IFNB1 mRNA abundance in mouse xenografts of primary human PDAC cell lines before or after treatment with trametinib. Values represent 2−ΔCq versus C1ORF43. n = 3. Means ± SD are shown. Two-way ANOVA (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
Induction of type I and type III IFN genes occurs in response to the detection of abnormal nucleic acids by specific sensors (46). Inhibition of the single-stranded DNA (ssDNA) sensor cGAS by RU-521 (47) was devoid of effects on the induction of IFN-stimulated genes by trametinib, while it efficiently inhibited IFNB1 induction by transfected ssDNA (Fig. 2D). Conversely, inactivation of the MAVS gene, which encodes a downstream effector of dsRNA sensors (46), abrogated trametinib-induced STAT1 phosphorylation and the associated induction of STAT1 protein levels (Fig. 2E), as well as induction of IFNB1 and the IFN-stimulated genes MX1 and RSAD2, without instead affecting expression of the IRF1 gene (Fig. 2F).
Trametinib treatment of three different primary human PDAC cell lines in vitro (Fig. 2G) and in mouse xenografts (Fig. 2H) similarly resulted in the induction of IFNB1 gene expression concurrently with the down-regulation of the KRAS-MEK effector ETV5, indicating that an increased IFN response is a common consequence of MEK1/2 inhibition in PDAC.
Trametinib treatment also induced the expression of a subset of IFN-stimulated genes involved in antigen presentation such as those encoding selected class II major histocompatibility complex (MHC) molecules and two of the genes (TAP1 and TAPBP) encoding components of the peptide loading complex required for antigen loading onto class I MHC molecules (fig. S2A). However, it did not increase surface expression of class II MHC molecules (fig. S2B), a finding that may relate to the reported high level of autophagy of MHC molecules in PDAC (48).
Independence of the trametinib-stimulated IFN program from the cholesterol pathway
Previous reports linking altered cholesterol synthesis to the induction of IFNB1 and IFN-stimulated gene expression (49) prompted us to investigate the relationship between the cholesterol homeostasis signatures up-regulated in three of the four analyzed trametinib-treated cell lines (fig. S3A) and the activation of the IFN response.
The increased cholesterol homeostasis signature was accounted for by the increased expression of all genes encoding components of the cholesterol synthesis pathway generating cholesterol from acetyl–coenzyme A, while genes encoding enzymes that generate geranyl or farnesyl isoprenoid groups from pathway intermediates, or that metabolize cholesterol into various hydroxylated derivatives, were not affected (fig. S3, B to D).
The main stimulus inducing the activation of cholesterol synthesis is the reduced intracellular content of cholesterol, which is sensed at the endoplasmic reticulum membrane and results in cleavage-mediated activation of the transcription factor SREBP-2, the master regulator of cholesterol synthesis genes (50). Analysis of cholesterol content in CFPAC1 and CAPAN2 cells showed a significant decrease at 12 hours after trametinib, with a subsequent trend to restoration of prestimulation levels (fig. S3E). Addition of water-soluble cholesterol [cholesterol–methyl-β-cyclodextrin (Cholesterol-MBCD)] dampened trametinib-induced activation of the LSS gene, encoding the lanosterol synthase and even reduced it below basal levels (fig. S3F), suggesting that reduced cholesterol abundance after trametinib is a driver of cholesterol synthesis in this system.
Reduced cholesterol abundance correlated with reduced expression of the low-density lipoprotein receptor (LDLR) at the RNA level and even more so at the level of the protein loaded on the surface of cells exposed to trametinib (fig. S3, G and H).
Notably, addition of Cholesterol-MBCD or depletion of SREBF-2 did not affect the induction of the IFN-stimulated marker gene RSAD2 (fig. S3, I and J), and reciprocally, inactivation of the JAK kinases by baricitinib did not affect activation of the cholesterol synthesis gene LSS (fig. S3K), indicating that regulation of cholesterol synthesis and induction of the IFN gene expression program occur independently in trametinib-treated PDAC cells.
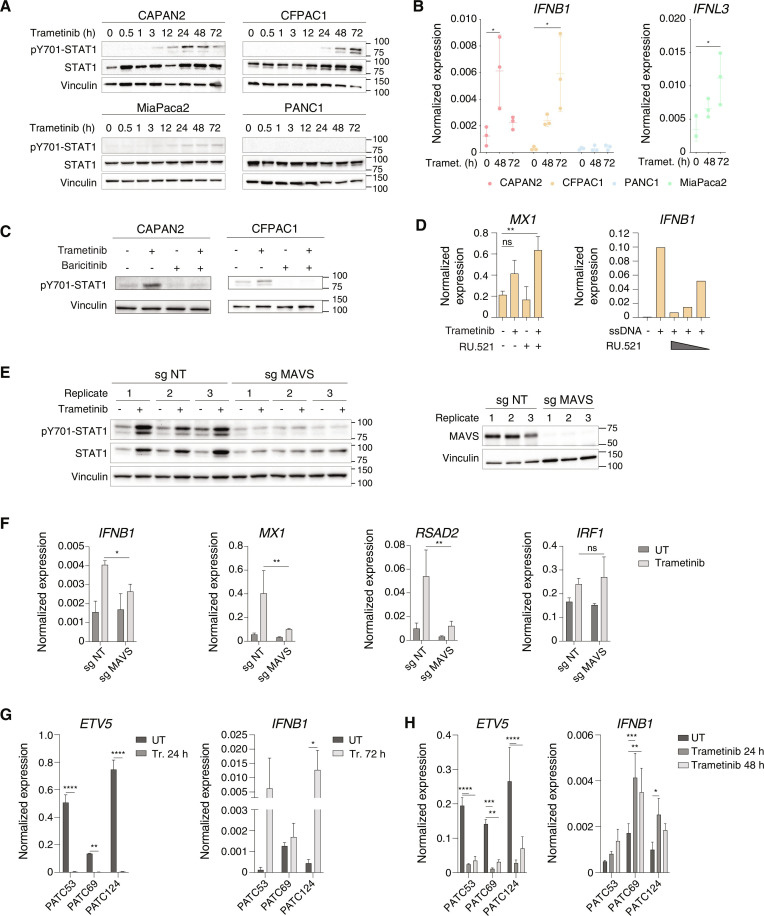
Induction of ERV-derived dsRNAs upon trametinib treatment
Since MAVS is a downstream effector of dsRNA sensors, its requirement for the induction of the IFN program hinted at the increased production of dsRNAs in response to trametinib. The overall abundance of dsRNAs measured by staining of trametinib-treated cells with a dsRNA-specific antibody showed a progressive increase over time (Fig. 3A).
Fig. 3. Induction of ERVs and other repetitive elements by trametinib.
(A) Immunofluorescence analysis of trametinib-treated CFPAC1 cells using the anti-dsRNA J2 antibody. The box plot shows the quantification of dsRNA levels in the immunofluorescence calculated as mean cell intensity. n = 3 biological replicates. Means ± SD are shown. ***P ≤ 0.001, ****P ≤ 0.0001. (B) Expression levels of differentially expressed subfamilies of repetitive elements in RNA-seq data from low-grade and high-grade PDAC cell lines after trametinib treatment. Values are reported as row Z score of mean-centered CPM expression. (C) Expression of selected subfamilies of ERVs in CAPAN2, CFPAC1, and MiaPaca2 cells. (D) Expression of selected ERVs was measured by RT-qPCR in low- and high-grade PDAC cell lines after treatment with trametinib or vehicle (24 hours). Values represent 2−ΔCq relative to C1ORF43. n = 3 biological replicates. Means ± SD are shown. Significance was assessed using two-way ANOVA (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). Reverse transcriptase (RT)–minus controls are shown to exclude amplification of repeats in genomic DNA. (E and F) RT-qPCR analyses of selected ERVs in primary human PDAC cell lines in vitro (E) and in vivo (F) before or after treatment with trametinib or vehicle (24 hours). Values represent 2−ΔCq versus C1ORF43. n = 3 biological replicates. Means ± SD are shown. Significance was assessed using two-way ANOVA (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). RT-minus controls are shown. (G) Observed versus expected enrichment ratio of retroelement subfamilies in trametinib–up-regulated H3K27ac ChIP-seq peaks. We estimated the enrichment as the ratio between the amount of overlap with up-regulated H3K27ac peaks with the overlap obtained using all peaks. Enrichment was tested through Fisher’s exact test considering two significance thresholds (see Materials and Methods). (H) Intersection between the enrichment of the indicated retroelements at trametinib–up-regulated H3K27ac peaks (y axis) and their trametinib-induced expression in RNA-seq datasets (x axis).
Consistent with these data, the expression of polyadenylated transcripts derived from transposable elements and, in particular, from many families of ERVs (including ERV1, ERVK, and ERVL) and, to a lesser extent, LINE1 retrotransposons was strongly induced in the RNA-seq datasets from all tested cell lines except PANC1 (Fig. 3, B and C, and table S4), in which the IFN signature was not up-regulated after trametinib treatment (Fig. 1D). Among the dsRNAs induced by trametinib are those generated by the MLT1C49 and MER57B1 ERVs (Fig. 3D), retroviral sequences embedded in antisense orientation in the 3′ untranslated regions of some IFN-stimulated genes, and thus able to generate dsRNAs and further amplify the IFN response (51). We also tested the induction of LTR family repeats in primary human PDAC cell lines. Both in vitro and in mouse xenografts, treatment with trametinib for 24 or 48 hours resulted in the induction of LTR family repeats (Fig. 3, E and F). The analysis of a broad panel of PDAC cell lines confirmed that the induction of ERVs by trametinib is a widespread occurrence (fig. S4).
We next analyzed H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) datasets generated at the same time points (from 3 to 24 hours) after trametinib treatment to determine the enrichment of retrotransposon sequences in differentially acetylated genomic regulatory regions. These analyses showed that the abundance of several ERV families and LINE1 retrotransposons in regions where H3K27ac increased in response to trametinib was significantly higher than randomly expected, indicating increased activity (Fig. 3G). In keeping with these findings, increased H3K27ac positively correlated with increased expression in RNA-seq data (Fig. 3H).
Overall, these data show that trametinib treatment caused a strong induction of retroelements and in particular ERVs, leading to the increased production and abundance of dsRNAs.
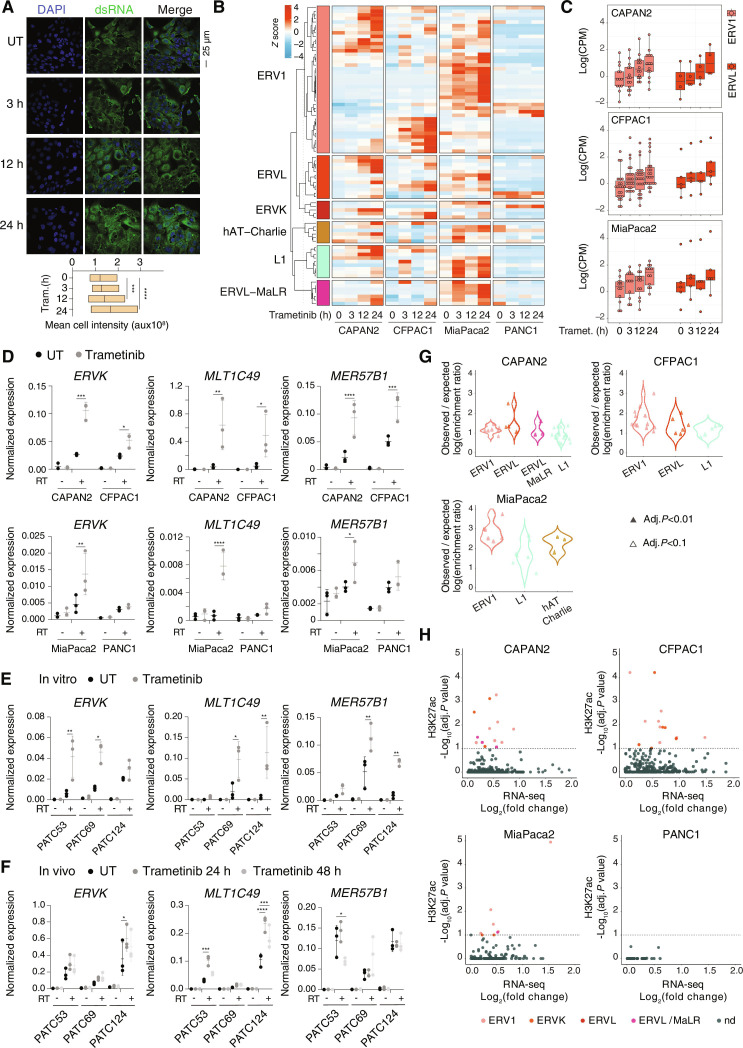
Escape of trametinib-induced ERVs from H3K9me3-mediated silencing
We set out to determine the mechanism accounting for ERV activation induced by trametinib. The expression of all the tested components of the KRAB-KAP1-SETDB1 pathway, which is responsible for the deposition of H3K9me3 at ERVs and their ensuing silencing, was steady over an extended treatment kinetics and thus substantially unaffected by trametinib (Fig. 4A).
Fig. 4. Trametinib-induced retroelements escape H3K9me3-mediated silencing.
(A) Western blot analysis of the abundance of the indicated components of the KRAB-KAP-SETDB1 ERV silencing pathway in CFPAC1 cells after trametinib treatment time course. Vinculin is shown as loading control. Molecular weight markers are indicated on the right. The experiment is representative of n = 3 independent experiments. (B) Heatmap showing L/H ratios normalized on the average ratio at the 0-hour time point for the indicated modified histone tail peptides as measured by mass spectrometry in CFPAC1 lysates after treatment with trametinib or vehicle. (C) Normalized H3K9me3 counts in CFPAC1 cells across time points. For each time point, the signal was divided into quintiles of increasing signal intensity in the untreated (UT) group. (D) Correlation between the H3K9me3 signals at each trametinib treatment time point relative to untreated (UT) CFPAC1 cells. (E) Metaplots showing H3K27ac and H3K9me3 normalized coverage in the indicated groups of ERVs. Metaplots were centered on the first annotated nucleotide of the repetitive element.
We next measured the global and locus-specific changes in H3K9me3 levels after trametinib stimulation. In keeping with the inactivation of the MEK1/2-ERK pathway and the ensuing loss of its stimulatory effects on gene activation, quantitative mass spectrometry–based analysis of histone posttranslational modifications showed a significant reduction of acetylation at 3 hours after treatment, with a more marked effect on multiply acetylated peptides, such as diacetylated H3K9-K14 as well as tri- and tetra-acetylated histone H4 tail peptides (Fig. 4B). On the contrary, H3K9 methylated peptides as well as peptides bearing other repressive methylations (such as H3K27me3) were substantially unmodified during treatment (Fig. 4B).
To determine the genomic distribution of H3K9me3 during trametinib treatment, we generated ChIP-seq datasets in CFPAC1 cells. Even with relaxed thresholds, the analysis of differential H3K9me3 regions in all comparisons between trametinib-treated and untreated cells returned only 3 down-regulated peaks at 48 hours and 1 to 81 up-regulated peaks depending on the time point, suggesting that the genomic distribution of H3K9me3 during trametinib treatment was extremely stable. As an additional analysis independent of the thresholds used for differential peak calling, we divided H3K9me3 peaks into quintiles of increasing signal strength in untreated cells and then determined the impact of trametinib treatment on each quintile. This analysis showed only minimal, if any, reduction of H3K9me3 across quintiles and, on the contrary, even a small increase in H3K9me3 in the first quintile (Fig. 4C). A time-resolved scatterplot analysis comparing signals in untreated versus trametinib-treated cells similarly showed no obvious differences induced by trametinib (Fig. 4D).
Although global and locus-specific levels of H3K9me3 were unaffected, we investigated the abundance of this repressive modification at trametinib-induced ERVs and other retrotransposons. This analysis revealed that repetitive elements at which H3K27ac was increased by trametinib, were not associated with detectable H3K9me3 levels; conversely, H3K9me3-positive ERVs did not show any basal H3K27ac and did not undergo any increase in acetylation upon treatment (Fig. 4E). Therefore, induction of ERVs and other retrotransposons by trametinib selectively occurred at repetitive elements escaping constitutive H3K9me3 repression.
Activation of ERVs and IFN gene expression by the trametinib-induced ELF3 transcription factor
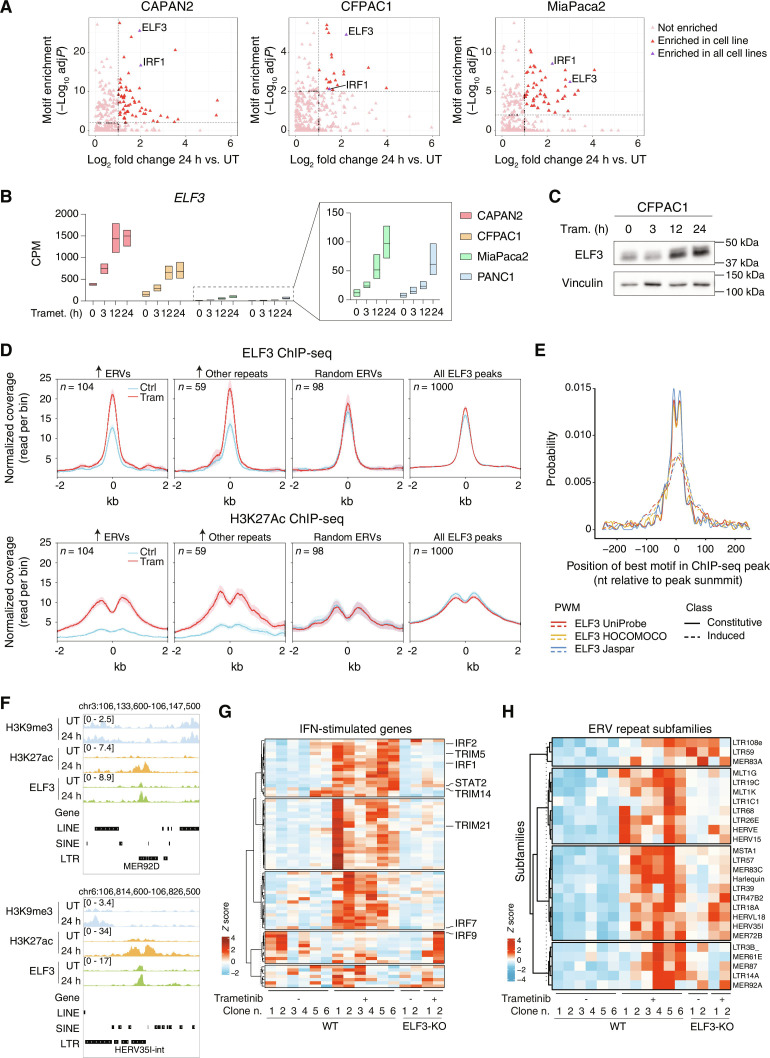
We next set out to identify the transcription factors responsible for trametinib-induced activation of H3K9me3-escaped retrotransposons. To this aim, we first identified transcription factor DNA binding motifs that were statistically overrepresented in regions that gained H3K27ac and contained trametinib-induced repetitive elements; then, we intersected overrepresented motifs with RNA-seq data to determine expression changes of the transcription factors recognizing the overrepresented motifs. Using this analysis, the only hits that were detected in common in all the three cell lines in which ERVs were induced by trametinib were IRF1 and ELF3, which belongs to a subgroup of the ETS transcription factor family with a distinct DNA binding specificity (Fig. 5A) (52). ELF3 expression was strongly induced by trametinib at the transcriptional and protein level in all PDAC cell lines (including PANC1, in which retroelements were not activated by trametinib), with pre- and poststimulation levels being higher in the low-grade relative to the high-grade cells (Fig. 5, B and C). ELF3 induction represented one of the earliest events induced by trametinib as it was clearly detectable at 3 hours after stimulation, reaching plateau levels at 12 hours (Fig. 5B).
Fig. 5. ELF3 induction drives ERV activation and viral mimicry in trametinib-treated cells.
(A) Intersection between the statistical enrichment of TF DNA binding motifs in trametinib-induced H3K27ac peaks associated with retroelements and the expression of the corresponding TFs in RNA-seq data. (B) RNA-seq data counts showing the expression of ELF3 in low- and high-grade PDAC cell lines after trametinib treatment. Data are reported as the log of CPM after TMM normalization with EdgeR. Data for MiaPaca2 and PANC1 are shown magnified in the boxed panel on the right. (C) Western blot analysis of ELF3 expression in CFPAC1 cells after trametinib treatment. Vinculin: loading control. Data are representative of n = 3 independent experiments. (D) Binding of ELF3 to trametinib-activated retroelements. Top: Metaplots representing ELF3 normalized coverage in different groups of ERVs centered on the ELF3 peak summit. Bottom: Metaplots representing H3K27ac normalized coverage in different groups of ERVs centered on the ELF3 peak summit. (E) Estimated affinity of ELF3 motifs in constitutive (continuous lines) and trametinib-induced (dotted lines) ELF3 ChIP-seq peaks. The probability distribution of the best ELF3 motif match is reported on the y axis as a function of the relative distance [nucleotides (nt)] from the peak summit (x axis). Distributions obtained with three different ELF3 Position Weight Matrices (PWMs) (from the UniProbe, HOCOMOCO, and Jaspar databases) are shown. (F) Representative genomic snapshots showing trametinib-induced ELF3 recruitment and histone acetylation at LTR retroelements. (G) Expression levels of IFN-stimulated genes included in the MSigDB signature “Hallmark response to IFN alpha” in CFPAC1 WT or ELF3-null clones treated with trametinib or vehicle for 24 hours. Values are reported as row Z score of mean-centered CPM (count per million) expression. (H) Differentially expressed ERVs in CFPAC1 WT or ELF3-null clones treated with trametinib or vehicle for 24 hours.
To determine whether ELF3 binds ERVs and other retroelements induced by trametinib, we generated ChIP-seq datasets in untreated and trametinib-treated (24 hours) CFPAC1 cells. ELF3 bound 65,540 and 77,904 sites before and after trametinib, respectively, and its overall genomic distribution was only minimally affected by treatment (Fig. 5D, “All ELF3 peaks”). However, when considering trametinib-induced retroelements, a strong induction of ELF3 binding was observed that paralleled a similarly strong increase in H3K27ac (Fig. 5D), indicating that ELF3 binding was associated with the activation of these repeats. Notably, the affinity of ELF3 motifs within trametinib-induced ChIP-seq peaks was significantly lower than that of motifs in constitutive peaks (Fig. 5E). This supports the dependence of de novo ELF3 binding on its increased expression following MEK1/2 inhibition. Two representative snapshots are reported in Fig. 5F.
To determine the role of ELF3 induction by trametinib in the transcriptional activation of retroelements and in the activation of the IFN response, we used ELF3-null CFPAC1 cells previously generated by genome editing in our laboratory (45, 53). The lack of ELF3 globally and strongly attenuated induction of IFN-stimulated genes in response to trametinib (Fig. 5G) and similarly reduced, yet not completely abrogated, the expression of trametinib-induced retroelements (Fig. 5H). Although a subset of retroelements was still expressed in ELF3-null cells, the abundance of the repeat-derived RNAs was significantly lower than that measured in wild-type (WT) cells (fig. S5), thus likely explaining the impaired activation of the IFN response. Overall, these data indicate that the early induction of ELF3 instructs ERV activation and the induction of a viral mimicry response.
Enforcement of trametinib-induced viral mimicry by an ELF3-IRF1 feed-forward loop
Among the transcription factors induced by trametinib and with cognate DNA binding motifs overrepresented in regions that gained H3K27ac and contained trametinib-induced repetitive elements, we also retrieved IRF1.
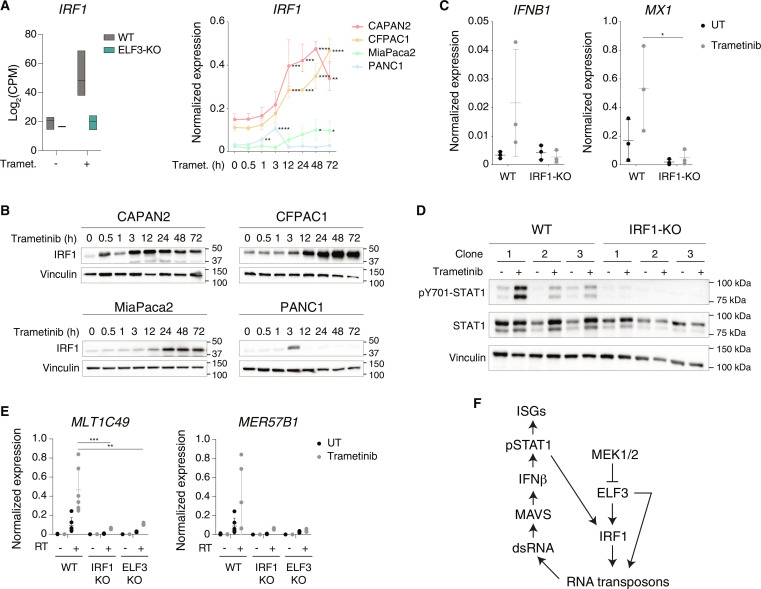
Previous data showed that constitutively high ELF3 expression in low-grade PDAC cells contribute to maintain basal expression of IRF1 (45, 53). IRF1 was strongly induced by trametinib both at the RNA and the protein level (Fig. 6, A and B), with its expression increasing over time in all cell lines except PANC1, in which a small expression spike at 3 hours (Fig. 6A) was paralleled by a transient increase in protein abundance (Fig. 6B). As shown in Fig. 2F, IRF1 expression was not affected by the deletion of MAVS, indicating another mechanism of transcriptional induction of this gene in response to trametinib. Notably, IRF1 induction by trametinib was virtually entirely dependent on ELF3 (Fig. 6A), which is explained by the direct binding of ELF3 to the IRF1 promoter (45). Hence, increased IRF1 abundance represents a consequence of increased ELF3 expression.
Fig. 6. An ELF3-IRF1 feed-forward loop in trametinib-induced viral mimicry.
(A) Induction of IRF1 expression by trametinib. Left panel: RNA-seq data counts showing the expression of IRF1 in WT and ELF3-null CFPAC1 cells after treatment with trametinib for 24 hours. Data are reported as the log of CPM after TMM normalization with EdgeR. Right panel: IRF1 expression was measured by RT-qPCR in low- and high-grade PDAC cell lines treated with trametinib. Values represent 2−ΔCq relative to C1ORF43. n = 3 biological replicates. Means ± SD are shown. Significance was assessed using one-way ANOVA and indicated as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. (B) IRF1 abundance in low- and high-grade PDAC cell lines after trametinib treatment. Vinculin is shown as loading control. The panel is representative of n = 3 independent experiments. (C) Expression of the IFNB1 and MX1 genes was measured by RT-qPCR in WT or IRF1-null CFPAC1 clones (n = 3) treated with trametinib or vehicle for 72 and 24 hours, respectively. Values represent 2−ΔCq relative to C1ORF43. Means ± SD are shown. Significance was assessed using two-way ANOVA and indicated as follows: *P ≤ 0.05. (D) pY701-STAT1 and total STAT1 abundance in WT and IRF1-null CFPAC1 clones (n = 3) after treatment with trametinib or vehicle for 72 hours. Vinculin is shown as loading control. Molecular weight markers are indicated on the right. (E) Expression of selected ERVs was measured by RT-qPCR in WT (n = 6), IRF1-null, and ELF3-null CFPAC1 clones (n = 3) after treatment with trametinib or vehicle for 24 hours. Values represent 2−ΔCq relative to C1ORF43. Means ± SD are shown. Significance was assessed using two-way ANOVA and indicated as follows: **P ≤ 0.01, ***P ≤ 0.001. RT-minus controls are shown to exclude amplification of repeats in genomic DNA. (F) Schematic model of the regulatory network controlling induction of retroelements in trametinib-treated PDAC cells.
To determine if IRF1 is required for the transcriptional response to trametinib, we analyzed previously generated IRF1-null CFPAC1 cells (45). Notably, induction by trametinib of IFNB1 and the IFN-stimulated gene MX1 were completely abrogated in IRF1-null cells (Fig. 6C). Consistent with these data, trametinib-induced STAT1 Tyr701 phosphorylation (Fig. 6D) and the induction of the MLT1C49 and MER57B1 repeat elements (Fig. 6E) were similarly abolished in IRF1- and ELF3-null cells.
Increased expression of both ELF3 and IRF1 upon treatment with trametinib was detected by RT-qPCR in a broad panel of PDAC cell lines (fig. S6A), suggesting that their induction is a common response to trametinib. Moreover, the reanalysis of publicly available datasets obtained in several PDAC cell lines treated with the ERK1/2 inhibitor SCH772984 (17) showed that the induction of the ELF3-IRF1-ERV pathway and the ensuing activation of the IFN response is a common outcome of MEK-ERK pathway inhibition in pancreatic cancer cells (fig. S6B).
We also tested whether MEK1/2 inhibition brings about the activation of this pathway in other tumors with a hyperactive RAS-RAF-MEK axis. We found that colon cancer cell lines treated with trametinib (54–56) showed the consistent induction of ERVs, ELF3 and IRF1, as well as the activation of an IFN response (fig. S6, C and D). A schematic representation of the transcriptional circuit instigated by exposure of PDAC cells to trametinib is reported in Fig. 6F.
Therapeutic effects of combined treatment with trametinib and anti-PD1
A possible implication of these data is that by inducing viral mimicry in tumor cells, trametinib may enhance their immunogenicity. At the same time, however, MEK1/2 inhibition may negatively affect T cell activation because of the role of the ERK pathway in normal T lymphocyte functions (57, 58). Nevertheless, such effects can be counteracted by treatments with various T cell agonists (59), thus suggesting that immune checkpoint inhibitors may neutralize the negative consequences of disabling the ERK pathway in T cells. Moreover, MEK inhibitors were reported to induce a stem cell memory phenotype in naïve CD8+ T cells, thus enhancing recall responses and antitumor activity (60).
Hence, while the possibility that trametinib-induced viral mimicry may augment the antitumor activity of immune checkpoint inhibitors exists, data available insofar are complex and not univocal, thus urging direct experimental investigation.
We first addressed the effects of trametinib on the activation of primary human memory CD4+ and CD8+ T cells induced by anti-CD3/CD28 treatment (fig. S7A). When using freshly isolated T cells, trametinib attenuated but did not abrogate T cell activation, exerting comparatively stronger effects on CD8+ than on CD4+ T lymphocytes (fig. S7B). When T cells were restimulated 10 days later, trametinib was virtually devoid of effects on CD4+ T cells, while still reducing the activation of memory CD8+ T cells. Hence, repeated activation may attenuate, while not completely abolishing, the negative consequences of MEK1/2 inhibition on T cell activation (fig. S7B). No significant induction of ERVs was detected in activated T lymphocytes, either with or without trametinib (fig. S7C). Conversely, trametinib increased anti-CD3/CD28–driven induction of IRF1, particularly in CD4+ T cells (fig. S7C).
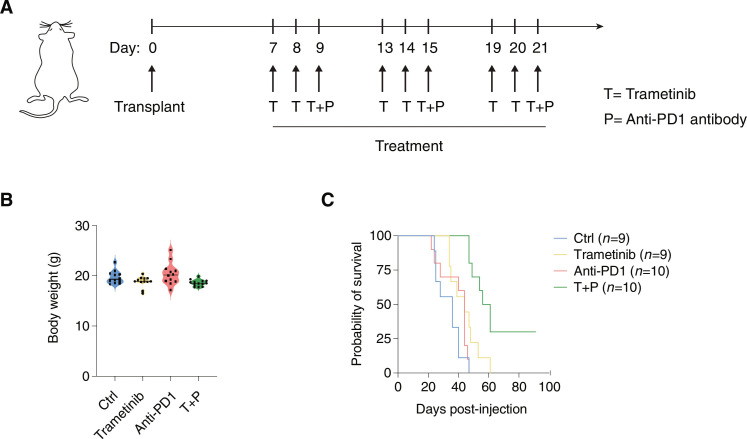
To determine the impact of trametinib on the response to immune checkpoint inhibitors, we analyzed the effects of individual or combined treatments on the survival of immunocompetent mice transplanted with syngeneic PDAC cells generated from KPC mice, in which PDAC is driven by the targeted pancreatic expression of the KrasG12D allele and mutant p53 (Trp53R172H) (61).
After orthotopic transplantation of PDAC cells, mice were subjected to three treatment cycles, each consisting of three consecutive doses of trametinib given by oral gavage, followed by one intraperitoneal injection of the anti-PD1 antibody (Fig. 7A). In the conditions used, mice showed no obvious signs of toxicity based on the measurement of their body weight (Fig. 7B). Treatment with trametinib or anti-PD1 alone had overall marginal effects on the survival of transplanted mice (Fig. 7C). Conversely, the combined treatment resulted in strong and significant increase of survival (from a median of 36 days to 58.5 days, P < 0.0001) and 3 of 10 mice subjected to combined treatment were still alive at 91 days (Fig. 7C).
Fig. 7. Combined treatment with trametinib and anti-PD1 in immunocompetent mice transplanted with syngeneic PDACs.
(A) Schematic representation of the experimental workflow. The transplanted PDAC cell line was obtained from KPC mice (KRasG12D/+, Trp53172H/+, p48-Cre). Trametinib treatment was administered by oral gavage, while the anti-mouse PD1 antibody was administered intraperitoneally. (B) Body weight in mice assigned to the four experimental groups was determined at the end of the treatment. (C) Survival curves of transplanted C57/BL6 mice in the different treatment groups. Significance was assessed using log-rank (Mantel-Cox) test and indicated as follows: ****P < 0.0001. n = 3 mice in the combined treatment group were still alive at the time of the analysis.
DISCUSSION
Here, we report a transcriptional regulatory circuit triggered by MEK1/2 inhibition that culminates in the induction of ERVs and a viral mimicry response with the strong induction of IFN-stimulated genes. In principle, the activation of this mechanism may create a window of opportunity during which the immunostimulatory effects of the trametinib-induced IFN response may synergize with immune checkpoint inhibitors in promoting recruitment and activation of immune cells in the tumor.
The notion that derepression of ERVs may boost the effects of immune checkpoint inhibitors in antitumor treatments has recently raised great interest (42, 43), particularly in the light of the evidence that disabling epigenetic mechanisms involved in the silencing of retroelements, such as DNA methylation (38, 39, 62) and H3K9 trimethylation (40, 63, 64), can increase the intrinsic immunogenicity of tumors.
What we show here, however, is an entirely different mechanism of retroelement activation and induction of viral mimicry in PDAC. Specifically, we report a regulatory circuit that does not involve any loss of epigenetic silencing and that instead entails the activation of repeats that escaped constitutive repression, yet were only marginally active in basal conditions. In response to MEK1/2 inhibition, the rapid and strong induction of the ELF3 transcription factor resulted in retroelement binding and activation, and the ensuing increase of ERV-derived dsRNAs triggered a MAVS-dependent IFN response. Therefore, the activation of this mechanism requires the presence of nonsilenced retroelements that, because of the lack of repressive chromatin modifications, are accessible and available for transcription factor binding. Notably, faulty repression of transposable elements that are otherwise efficiently silenced in normal cells is a common event in tumors (43, 65, 66) and specifically in PDAC (67, 68), and our data indicate the possible exploitation of such intrinsic epigenetic silencing failure to increase tumor immunogenic potential.
The viral mimicry response triggered by trametinib in principle provides the molecular bases for the synergistic effects of MEK1/2 inhibition and anti-PD1 antibody administration that we observed on the survival of PDAC-bearing mice. The results reported here are in line with many previously reported and mechanistically unexplained observations linking MEK1/2 inhibitors (alone or in combination with other kinase inhibitors) to tumor sensitization to immune checkpoint inhibitors in preclinical or clinical settings, both in PDAC (69, 70) and in other tumors with a hyperactive RAS-RAF-MEK pathway such as melanomas (71, 72). On a cautionary note, however, it is important to stress that the trametinib dose used (and tolerated) in mice (3 mg/kg per day) (73), which is also the one used in our study, corresponds to a human equivalent of ca. 14 to 16 mg/day, which is several-fold higher than the dose typically used in humans (ca. 2 mg/day). Hence, the possible translation to humans of our findings will require additional careful evaluations.
The detailed mechanistic analysis of the effects of trametinib on ERV expression and viral mimicry we report here has a number of implications. First, the effects we observed are particularly strong in well-differentiated PDAC cell lines, possibly due to the higher basal and trametinib-induced expression of the transcription factor ELF3 compared to high-grade cell lines, a finding in line with the distribution of ELF3 in PDAC tissues (53). On the other hand, the less differentiated and quasi-mesenchymal tumor component, which is a quantitatively less abundant constituent of most PDACs, shows extensive DNA demethylation and constitutive activation of some repeat elements (68). Therefore, trametinib treatment may enhance the activation of the IFN response in the tumor component in which this program is poorly active in basal conditions. Second, it is important to notice that ERV activation and the viral mimicry response are early events occurring hours after trametinib treatment, which may have practical implications for the administration schedule of combined treatments with immune checkpoint inhibitors. Finally, as the activation of ERVs can also result in the production of immunogenic neoantigens (44), the ability of trametinib to induce the activation of nonsilenced ERVs may be exploited in the design of anti-PDAC vaccination strategies that have very recently shown promising clinical results (74).
MATERIALS AND METHODS
Cell lines and culture
The following human PDAC cell lines were used: CAPAN2 [KRAS G12V; American Type Culture Collection (ATCC), HTB-80], CFPAC1 (KRAS G12V; ATCC, CRL-1918), MiaPaca2 (KRAS G12C; ATCC, CRL-1420), PANC1 (KRAS G12D; ATCC, CRL-1469), PATU8988S [KRAS G12V; Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ)], HPAFII (KRAS G12D; ATCC), CAPAN1 (KRAS G12V; DSMZ), PSN1 [KRAS G12R; Interlab Cell Line Collection (ICLC)], AsPC1 [KRAS G12D; European Collection of Cell Cultures (ECACC)], BxPc3 (ATCC), PT45P1 (KRAS G13D; G3 from primary tumor, obtained from P. Allavena, Humanitas, Milan), PATU8902 (KRAS G12V; DSMZ), and PATU8988T (KRAS G12V; DSMZ). In addition, human embryonic kidney (HEK) 293T (ATCC, CRL-3216) were also used in this study. The following human primary PDAC cell lines were gifted by M. Kim [University of Texas MD Anderson Cancer Center (UTMDACC): PATC53, PATC69, and PATC124. Cells were maintained in Iscove’s modified Dulbecco’s medium (IMDM) + 10% fetal bovine serum (FBS) (CFPAC1), RPM1 + 10% FBS (PSN1, AsPC1, BxPc3, and PT45P1), RPMI + 15% FBS (CAPAN2), RPMI + 20% FBS (CAPAN1), Dulbecco’s modified Eagle’s medium + 5% FBS + 5% horse serum (PATU8988S and PATU8988T), Dulbecco’s modified Eagle’s medium (DMEM) + 10% FBS (MiaPaca2, PANC1, PATU8988S, HEK293T, PATC53, PATC69, and PATC124), and Eagle’s minimum essential medium + 10% FBS + 1 mM sodium pyruvate + 0.1 mM nonessential amino acids (HPAFII). Media were all supplemented with 2 mM l-glutamine. All cell lines were authenticated by the Tissue Culture Facility of IEO using the GenePrint10 System (Promega) for the amplification of 10 short tandem repeat–containing loci, followed by Sanger sequencing. MAVS-, ELF3-, and IRF1-null CFPAC1 cells obtained by genome editing were described previously (45, 53).
Primary human T cell isolation, culture, and activation
Peripheral blood was collected from healthy donors at the Swiss Blood Donation Center of Lugano (Switzerland), with informed consent (authorization number CE 3428 from the Ethical Committee of the Canton Ticino). Peripheral blood mononuclear cells (PBMCs) were isolated through gradient centrifugation (GE Healthcare, Ficoll-Paque Plus) and further enriched for CD4+ or CD8+ T lymphocytes by positive selection using magnetic beads (Miltenyi Biotec), following the manufacturer’s protocols. Memory T lymphocyte subsets were then sorted on a SORP FACSymphony S6 (BD Biosciences) based on the expression of the following surface markers: CD4+ (or CD8+) CD25– CD45RA– CCR7+/−. Sorted cells were cultured in RPMI 1640 medium supplemented with 5% human serum, 1% nonessential amino acids, 1% sodium pyruvate, 1% l-glutamine, penicillin, streptomycin, and 50 μM β-mercaptoethanol. T cell activation was performed as previously described (75, 76) using plate-bound anti-CD3 antibody (0.7 μg/ml; clone TR66, recombinant, made in-house) (77) and anti-CD28 antibody (1 μg/ml, BD Pharmingen) in 96-well NUNC plates (Thermo Fisher Scientific). After 48 hours, cells were removed from the stimuli and further expanded for up to 10 days in culture. Recombinant interleukin-2 (IL-2) (50 U/ml, made in-house) was added to the cultures starting from day 5.
Chemicals and treatments
Trametinib (Selleckchem, S2673) was diluted in dimethyl sulfoxide (DMSO) and used at a concentration of 20 nM for all the experiments, with the exception of cumulative growth curves in which a 2 nM concentration was used. The cGAS inhibitor RU.521 (InvivoGen, inh-ru521) was diluted in DMSO and used at a concentration of 15 μg/ml. Baricitinib (Selleckchem, S2851) was diluted in DMSO and used at a concentration of 300 nM. Cholesterol-MBCD (Sigma-Aldrich, C4951) was diluted in H2O and used at a concentration of 0.25 mg/ml. MBCD (Sigma-Aldrich, C4555) was diluted in H2O and used at a concentration of 0.2375 mg/ml, which corresponds to the same concentration contained in Cholesterol-MBCD. Cells were plated in 96-well, 24-well, 6-well, 6-cm, or 10-cm plates 24 to 48 hours before treatment and treated for the indicated time at the reported concentration of drugs directly diluted in their own medium and then collected or fixed for the following analyses. In case of combined treatments, drugs were used simultaneously.
Cell viability assays
Cells (5 to 10,000 per well) were seeded in 96-well plates. One day after, cells were treated with trametinib and viability after 72 hours of treatment was measured using CellTiter-Glo (Promega, G9242) and GloMax (Promega). Each population was normalized for its untreated counterpart.
Western blots
Preparation of cell lysates and Western blots was carried out as described (45). Chemiluminescent horseradish peroxidase (HRP) signal was revealed using ECL Clarity Western Substrate or Clarity Max Western ECL Substate (Bio-Rad). For Western blot analyses, the following primary antibodies were used: phospho-p44/42 MAPK (Thr202/Tyr204) (Cell Signaling Technology, #9101), p44/42 MAPK (Cell Signaling Technology, #9102), phospho-Tyr701 STAT1 (Cell Signaling Technology, #7649), STAT1 (Cell Signaling Technology, #9172), MAVS (Abcam, ab264147), ESET (Cell Signaling Technology, #93212), KAP1 (Abcam, ab10483), HP1α (Abcam, ab109028), HP1β (Santa Cruz Biotechnology, sc-20699), HP1γ (Abcam, ab217999), ELF3 (Sigma-Aldrich, HPA003479), IRF-1 (Cell Signaling Technology, #8478), vinculin (Sigma-Aldrich, V9131), ELF3 (Sigma-Aldrich, HPA003479), and β-actin (Abcam, ab8227). Images were acquired using a ChemiDoc imaging system (Bio-Rad). Band quantification was performed using Image Lab Software (Bio-Rad).
Real-time quantitative PCR
RNA was extracted using the Zymo Quick RNA Miniprep Kit (Zymo Research, R1055), TRIzol Reagent (Invitrogen, 15596026) for in vivo frozen samples, or Direct-zol RNA Microprep for T lymphocytes (Zymo Research, R2062), and 100 to 500 ng were reverse-transcribed using ImProm-II Reverse Transcriptase kit (Promega, A3802). For the amplification of repetitive elements, a reaction control was assembled, which did not contain the reverse transcriptase enzyme. The PCR was assembled using the Fast SYBR Green Master Mix (Applied Biosystems, 4385612) and run on the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). C1ORF43 was used as reference gene based on the analysis of data from the Human BodyMap 2.0 Project. For T lymphocyte–specific analyses, UBE2D2 was used as reference gene.
The following primers were designed using Primer3:
C1ORF43_FW: 5′-GGATGAAAGCTCTGGATGCC-3′
C1ORF43_REV: 5′-GCTTTGCGTACACCCTTGAA-3′
MX1_FW: 5′-CACCGTGACGGATATGGTCC-3′
MX1_REV: 5′-GCACCCCTGTATACCTGGTC-3′
RSAD2_FW: 5′-TGGGTGCTTACACCTGCTG-3′
RSAD2_REV: 5′-GAAGTGATAGTTGACGCTGGTT-3′
ETV5_FW: 5′-GCGGCCTGTGATTGACAGA-3′
ETV5_REV: 5′-GGAACTTGTGCTTCAGCTAACCA-3′
GBP2_FW: 5′-TTGGAAGCAAGGCGAGATGA-3′
GBP2_REV: 5′-CCTCTTTGGCCTGTATCCCC-3′
IFNB1_FW: 5′-ATGACCAACAAGTGTCTCCTCC-3′
IFNB1_REV: 5′-GGAATCCAAGCAAGTTGTAGCTC-3′
IFNL3_FW: 5′-CTGTGTGTCTGACCCTTCCG-3′
IFNL3_REV: 5′-ACATAAATAGCGACTGGGTGACA-3′
IRF1_FW: 5′-CAACATGCCCATCACTCGGA-3′
IRF1_REV: 5′-TGCTTTGTATCGGCCTGTGT-3′
LSS_FW: 5′-CAGGAGAAGTGTCCCCATGT-3′
LSS_REV: 5′-CCCTCCATCTGGATTTCTCA-3′
LDLR_FW: 5′-CTCGCTGGTGACTGAAAACA-3′
LDLR_REV: 5′-TCGATGCTTGAGATGGAGTG-3′
SREBF-2_FW: 5′-AGAAGGAGAAAGGCGGACAA-3′
SREBF-2_REV: 5′-TTCCTCAGAACGCCAGACTT-3′
UBE2D2_FW: 5′-GATCACAGTGGTCTCCAGCA-3′
UBE2D2_REV: 5′-CGAGCAATCTCAGGCACTAA-3′
ELF3_FW: 5′-TCTTCCCCAGCGATGGTTTTC-3′
ELF3_REV: 5′-TCCCGGATGAACTCCCACA-3′
The following primers (51, 78) were used to amplify selected ERVs:
ERVK_FW: 5′-AGAGTCTAAACCACGAGGCACAA-3′
ERVK_REV: 5′-TTCTTTAACCTGCTTTTGAGGTTGT-3′
MLT1C49_FW: 5′-TATTGCCGTACTGTGGGCTG-3′
MLT1C49_REV: 5′-TGGAACAGAGCCCTTCCTTG-3′
MER57B1_FW: 5′-CCTCCTGAGCCAGAGTAGGT-3′
MER57B1_REV: 5′-ACCAGTCTGGCTGTTTCTGT-3′
Lipid extraction and cholesterol quantification
Cells (150 to 350,000 per well) were seeded in six-well plates. At the indicated time of treatment, cells were scraped and one-fifth were lysed in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors for protein extraction and quantification. The remaining four-fifths were subjected to lipid extraction with methanol-acetonitrile (1:1). Total cholesterol quantification was performed using the Cholesterol Quantitation Kit (Sigma-Aldrich, MAK043) with esterase option. The relative fluorescence intensity was measured at GloMax (Promega).
FACS measurements
For MHC-II, 250,000 cells were seeded in 60-mm dishes. At the indicated time of treatment, cells were trypsinized and counted. Cells (150,000) were used for each labeling [not stained, immunoglobulin G (IgG)–Alexa Fluor 488 and pan–MHC-II–fluorescein isothiocyanate (FITC)], blocked, and stained with IgG1 Isotype Control-Alexa Fluor 488 (eBioscience, 53-4714-42) or FITC anti-human HLA-DR, DP, DQ Antibody (BioLegend, 361706). Fluorescence-activated cell sorting (FACS) analysis was performed on single-cell suspensions in phosphate-buffered saline (PBS). Samples were acquired on a FACSCelesta Cell Analyzer (BD Biosciences), and data were analyzed using FlowJo software.
For LDLR, 1 × 106 cells were seeded in 10-cm plates. At the indicated time of treatment, cells were trypsinized and counted. Cells (500,000) were used for each labeling [not stained, IgG-phycoerythrin (PE) and LDLR-PE], blocked, and stained with IgG1 Isotype Control-PE (eBioscience, 12-4714-82) or anti-human LDLR-PE (BD Pharmingen, 565653). FACS analysis was performed on single-cell suspensions in PBS. Samples were acquired on a FACSCelesta Cell Analyzer (BD Biosciences), and data were analyzed using FlowJo software.
For T lymphocytes, sorted CD4+ and CD8+ T cell subsets were treated with trametinib or DMSO at days 0 and 10 after isolation. Cells were pretreated with trametinib for 1 hour, followed by stimulation for 24 hours with plate-bound anti-CD3 and anti-CD28 antibodies as described above. Surface expression of activation markers was analyzed by flow cytometry using FACSymphony A5 (BD Biosciences) and directly conjugated antibodies anti-CD69 (BioLegend, clone FN50) and anti-OX40 (BioLegend, clone Ber-ACT35). Cell viability was determined using a Fixable Blue Stain UV live/dead staining (Thermo Fisher Scientific). Flow cytometry data were analyzed using the FlowJo software.
shRNA-mediated depletion
The following short hairpin RNA (shRNA) sequences were used: 5′-CCTCAGATCATCAAGACAGAT and 5′-CCTCAGATCATCAAGACAGAT targeting SREBF-2 and 5′-ACGCTGAGTACTTCGAAATGT specific for luciferase gene as a nontargeting control. The sequences were cloned into the pLKO.1 vector (Addgene #10878) using Eco RI and Age I cloning sites and verified by Sanger sequencing. Lentiviral particles were produced using calcium phosphate transfection of HEK293T cells with pLKO.1 and the packaging vectors pMD2.G (Addgene #12259) and psPAX2 (Addgene #12260). To obtain an SREBF-2–depleted polyclonal line, CFPAC1 cells were plated to near confluency and transduced twice with the lentiviral particles carrying pLKO.1-shSREBF-2 1 and 2 or the nontargeting shRNA vector. Cells were selected by puromycin and grown as bulk population for downstream analyses.
Immunofluorescence analysis
Two-color immunofluorescence analysis was performed on CFPAC1 cells grown onto glass coverslips. Briefly, paraformaldehyde-fixed cells were permeabilized with 0.5% Triton X-100, blocked, and incubated with anti-dsRNA (J2) antibody (Nordic-MUbio, 10010200). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI), and samples were mounted with glycerol mounting medium. CFPAC1 cells were imaged on a Yokogawa spinning disk field scanning confocal system (CSU-W1Nikon Europe B.V., Stroombaan 14, 1181 VX Amstelveen, The Netherlands) with a 60×/1.25 NA (numerical aperture) oil immersion objective lens. Sixty-four fields of view (FOVs) per sample were acquired at the nuclear middle plane in z, because of an autofocus function on the DAPI channel. The J2 (Alexa Fluor 488) and the DAPI signals were acquired with a multi-band dichroic mirror and single-band emission filters on a Photometrics Prime BSI sCMOS camera. Images were made of 1024 × 1024 pixels with a pixel size of 110 × 110 nm. The J2 signal was quantified because of a custom ImageJ/Fiji macro. The DAPI channel images were pre-processed (Gaussian blur filter with sigma set to 30 pixels and background subtraction with rolling ball radius set to 100 pixels), and then the nuclei in each FOV were counted because of the Find Maxima function. The J2 channel images were background-corrected (rolling ball radius set to 100 pixels), and the total intensity in each FOV was quantified. The J2–Alexa Fluor 488 approximate mean cell intensity was then calculated as the total FOV intensity divided by the number of nuclei in each FOV for each replica.
Histone posttranslational modification mass spectrometry analysis
CFPAC1 cells (1.5 × 106 per plate) were seeded in 10-cm plates. At the indicated time of treatment, cells were scraped and collected. Nuclei were purified, and histone proteins were isolated as described (79). Approximately 4 μg of histone octamer was mixed with an equal amount of heavy isotope–labeled histones, which were used as an internal standard (80), and separated on a 17% SDS–polyacrylamide gel electrophoresis (PAGE) gel. Histone bands were excised, chemically acylated with propionic anhydride, and in-gel–digested with trypsin, followed by peptide N-terminal derivatization with phenyl isocyanate (PIC) (81). Peptide mixtures were separated by reversed-phase chromatography on an EASY-Spray column (Thermo Fisher Scientific), 25 cm long (inner diameter 75 μm, PepMap C18, 2-μm particles), which was connected online to a Q Exactive Plus instrument (Thermo Fisher Scientific) through an EASY-Spray Ion Source (Thermo Fisher Scientific), as described (81). The acquired RAW data were analyzed using EpiProfile 2.0 (82), followed by manual validation. For each histone modified peptide, a % relative abundance (%RA) value for the sample [light channel (L)] or the internal standard [heavy channel (H)] was estimated by dividing the area under the curve of each peptide for the sum of the areas corresponding to all the observed forms of that peptide and multiplying by 100. Light/heavy (L/H) ratios of %RA were then calculated and used for data display. L/H ratios are reported in table S5. The mass spectrometry data have been deposited to the ProteomeXchange Consortium (83) via the PRIDE partner repository with the dataset identifier PXD043559.
RNA-seq sample preparation and sequencing
Total RNA from was extracted using the Quick-RNA Miniprep Kit (Zymo Research, R1055). RNA-seq was carried out using the SMART-seq2 protocol (84) with minor modifications, as described (45). Briefly, 10 ng of total RNA was reverse-transcribed using oligo(dT) primers and a locked nucleic acid–containing template-switching oligo. The resulting complementary DNA (cDNA) was pre-amplified, purified, and tagmented using in-house made Tn5 transposase (84). cDNA fragments generated after tagmentation were gap-repaired, enriched by PCR, and purified to create the final cDNA libraries. Single-read [51 base pairs (bp)] (for CAPAN2, CFPAC1, MiaPaca2, and PANC1 untreated and treated with trametinib for 3–12–24 hours) and paired-end (for CFPAC1 WT and ELF3-KO (knockout) clones untreated and treated with trametinib for 24 hours) sequencing were performed on Illumina NextSeq 500 and NovaSeq6000 platforms.
ChIP-seq sample preparation and sequencing
ChIP-seq was carried out as previously described (85) using 1 × 106 cells for the H3K27ac and H3K9me3 ChIP and 20 × 106 cells for the ELF3 ChIP. The following antibodies were used: anti-H3K27ac (Abcam, ab4729), anti-H3K9me3 (Abcam, ab8898), and anti-ELF3 antibody (rabbit polyclonal antibody generated in-house using a peptide corresponding to amino acids 1 to 150 of the human ELF3 reference protein sequence). DNA libraries were prepared as described (86) and sequenced (51 bp single-read) on Illumina NextSeq 500 and NovaSeq6000 platforms.
In vivo studies
Patient-derived PDAC samples were obtained from consented patients under the Institutional Review Board (IRB)–approved protocol LAB00-0396 chaired by M. Kim (UTMDACC). NSG and C57BL/6J female mice were purchased from the Experimental Radiation Oncology at MD Anderson Cancer Center and The Jackson Laboratory (strain #000664), respectively. All animal studies and procedures were approved by the UTMDACC Institutional Animal Care and Use Committee. All experiments conformed to the relevant regulatory standards and were overseen by the IRB. For subcutaneous transplantation of human PDAC cells and trametinib treatment, tumor single-cell suspensions were resuspended in DMEM without phenol red (Sigma-Aldrich) and Matrigel (BD Biosciences, 356231) (1:1 dilution) at a density of 1.5 × 106 cells/100 μl for early-passage patient-derived pancreatic cancer cells (PATC53, PATC69, and PATC124). Cell suspensions were injected subcutaneously into the flank of 6-week-old NSG mice. Mice were monitored every 7 days, and tumor measurements were calculated according to the following formula: tumor volume (mm3) = (W2 × L)/2, where W is width and L is length. Trametinib was purchased from Selleckchem (S2673) and resuspended in corn oil (MilliporeSigma, C8267) according to the manufacturer’s protocol. Once tumors reached approximately 400 to 500 mm3, mice were randomized into two treatment groups. On the basis of trametinib half-life (73), the first group received two consecutive oral administrations of trametinib (3 mg/kg) at 0 and 24 hours, and tumors were collected at 28 hours for molecular analysis. The second group received three consecutive oral administrations of trametinib at 0, 24, and 48 hours, and tumors were collected at 52 hours for molecular analysis. Mice were euthanized by exposure to CO2 followed by cervical dislocation. A necropsy form was filled in with mouse information, tumor size, and weight. Each primary tumor was flash-frozen in dry ice and kept at −80°C.
For orthotopic transplantation of mouse PDAC cells in syngeneic models, cell lines from the KPC PDAC model were used. Briefly, p48-Cre mice (11), LSL-KrasG12D (The Jackson Laboratory, #008179), and Trp53R172H (provided by G. Lozano, UTMDACC) were crossed. Spontaneous pancreatic tumors were harvested and digested, and KPC cells were derived. The entire tumors were minced into small pieces with sterile blades and incubated at 37°C for 45 min with collagenase IV (Gibco)–dispase II (Roche), 2 mg/ml for enzymatic digestion. Cells were then centrifuged and further digested with 0.25% trypsin (Gibco) for 5 min at 37°C to obtain a single-cell suspension. After expansion in DMEM (Gibco) supplemented with 10% FBS (Gibco) and penicillin/streptomycin (100 μg/ml) (Gibco), tumor cells were enriched by FACS using an anti-EpCAM antibody. For orthotopic transplantation, 8-week-old C57BL/6J female mice were anesthetized using isoflurane (Henry Schein Animal Health). Analgesia was achieved with buprenorphine ER (0.1 mg/kg, twice daily) (Par Pharmaceutical) via subcutaneous injection, and skin was disinfected with chlorhexidine and 70% ethanol. A 0.5-cm incision was performed on the left flank through the skin/subcutaneous and muscular/peritoneal layers. Pancreas was exposed, and a 20-μl cell suspension [2.5 × 104 KPC cells per mouse suspended in 1:1 DMEM/Matrigel (BD Biosciences, #356231)] was injected using a 27-gauge Hamilton syringe. The pancreas was carefully repositioned into the abdominal cavity, and muscular/peritoneal planes were sutured individually by absorbable sutures (Ethicon). The skin/subcutaneous planes were closed using metal clips (Fine Science Tools). Mice were monitored daily for the first 3 days for signs of illness and surgical wounds. Thereafter, orthotopic tumor formation has been evaluated twice/week by transabdominal palpation or magnetic resonance imaging (MRI) when needed. At day 7 after injection, mice were randomized into four treatment groups: vehicle as control, trametinib, anti-mouse PD1, and trametinib + anti-PD1. The anti-mouse PD1 antibody (Bio X Cell, #BP0146) was resuspended in 1× PBS and administered by intraperitoneal injection at the dose of 8 mg/kg on days 9, 15, and 21 after transplantation. Trametinib was resuspended in corn oil (MilliporeSigma, #C8267) and administered by oral gavage at the dose of 3 mg/kg on days 7, 8, 9, 13, 14, 15, 19, 20, and 21. At the end of treatment (day 22 after injection), at least n = 9 mice per group were monitored for survival studies.
Statistical analysis
All statistical analysis present in Results were performed using GraphPad Prism V.9. One-way analysis of variance (ANOVA), two-way ANOVA, and t test were used.
Computational methods
RNA-seq data analysis
Fastq raw reads were processed with FASTQC tool v0.11.9 for quality base check and to exclude any technical bias in the data. Sequences were then checked for the presence of the adaptors using trimmomatic_v0.39 (87) with the following settings: TRAILING:3, LEADING:3, ILLUMINACLIP: adapters.fasta:2:30:10, SLIDING-WINDOW:4:15, MILEN:30. Trimmed single-rend reads were then mapped onto the human genome GRCh38 assembly version using STAR-v.2.7.8a (88) with the following parameters: --limitSjdbInsertnsJ = 500,000, --alignIntronMax = 1,000,000, --alignIntronMin = 20. After uniquely mapped reads were retained, gene counts were retrieved using featureCounts on Gencode GRCh38 annotation (Ensembl 104 version). Next, protein-coding genes were selected and processed with the edgeR_3.39 R package (89) for gene filtering, normalization, and differential testing. Specifically, low-expression filtering was performed using the default setting of at least 1 CPM (count per million) in n samples, where n corresponds to the smallest group size. TMM (trimmed mean of M value) normalization was applied to correct for both sequencing depth and RNA composition bias across samples. TMM-normalized CPM was used as expression unit. Analyses were separately performed for low-grade cell line data and high-grade cell line data. Using generalized linear models and likelihood ratio testing, we identified DEGs as those genes having absolute log fold change > 1 and FDR < 0.001. Hierarchical clustering of DEGs was represented using the ComplexHeatmap R package v2.13.3 (90). Venn diagrams were generated using the eulerr (v7.0) and the VennDiagram (v1.7.0) R package. Gene set enrichment analyses were done with the GSEA tool (91) and carried out in the pre-ranked mode, using the decreasing log(fold change) in each comparison as a ranking metric.
ChIP-seq data analysis
Single-end reads were processed using Trimmomatic (v0.39) with the following settings: TRAILING:3, LEADING:3, ILLUMINACLIP: adaters.fasta:2:30:10, SLIDING-WINDOW:4:15, MILEN:30. Next, high-quality trimmed reads were aligned onto the human genome build GRCh38/hg38 using Shortstack v3.8.5 (92), an aligner tool to recover best-matched alignments from multi-mappers. In summary, Shortstack uses bowtie (93) to perform a first round of mapping to distribute uniquely and multi-mapped reads according to the maximum number of matches allowed. In a second step, each ambiguous read is assigned to a location by estimating a probability that depends on the frequency of uniquely mapped reads in that region. Finally, the alignment with the highest probability is considered as the best candidate mapping for that read. Using Shortstack, we generally observed an appreciable increase in the amount of multi-mapping reads that could be recovered as “unique” in the final BAM, with H3K27ac ChIP and ELF3 ChIP samples having 5 to 8% more reads compared to standard mapping, while the multi-match read fractions in H3K9me3 data were even higher (10 to 15%). Thus, we ran Shortstack with the following settings: --mismatches 2 --bowtie_m 100 --align_only --mmap u. Mapped reads were filtered to remove randomly placed multi-mappers and unmapped reads using samtools v1.16.9 (94).
Any read alignment matching hg38-blacklist regions (https://github.com/Boyle-Lab/Blacklist/blob/master/lists/hg38-blacklist.v2.bed.gz) was discarded using bedtools_v.2.30.0 (95), and duplicate reads were removed with the samtools “markdup –r” option. For the H3K27ac ChIP data, peaks were called using MACS2 v.2.2.7.1 (96) with the following parameters: --nomodel –extsize 250 –mfold 5 50 –broad –broad-cutoff 0.01. For the ELF3 ChIP data, we run MACS in “narrow-peak” mode with parameters –nomodel –extsize 200 –mfold 5 50 -q 0.01.
H3K9me3 islands were called using Sicer2 v1.0.3 (97) with the following settings: --window_size = 200 –fragment_size = 300 -fdr = 0.01 –gap_size = 600. A corresponding input DNA from the same cell line was used as a control in peak detection in each set.
For each ChIP-seq sample, fragment pile-up in “read per million” was generated using MACS2 callpeak with parameters –format BAM –nomodel –B –SPMR, with the –extsize parameter set as the same used in the calling step. The resulting BedGraph files from the ChIP were compared to that of the input to generate a signal profile in BigWig format for browser visualization.
All differential peak analyses were carried out using DiffBind R package v3.4.11 (98). Before differential testing, peaks/islands in each sample were merged and compared to build a consensus set of peaks taking regions with a minimum overlap of two samples in each dataset, and differentially enriched regions were detected for each time point versus control using the DESeq2 method (99). Differentially acetylated regions were defined as those with absolute log(fold change) ≥ 1 and FDR ≤ 0.1 at 3 hours and absolute log(fold change) ≥ 1.5 and FDR ≤ 0.01 for later points. The identification of differential H3K9me3 islands was carried out using a relaxed log(fold change) threshold of 0.5 with FDR < 0.01. For ELF3 differential binding, we consider peaks with absolute log(fold change) ≥ 1 and FDR ≤ 0.01.
Repetitive element analysis
Detection and quantification of repetitive elements were achieved using TEtranscript tool v.2.2.1 (100). According to the classification of RepBase (101), repetitive loci (“insertions”) are organized in Transposable Elements (TE) subfamilies of loci that are highly related at the sequence level but relatively distinct from other families. By taking advantage of the repeat sequence similarities in the same TEs, TEtranscripts distributes reads among closely related TE sequences to infer the expression at the TE subfamily level. The read counting can be run in the “unique” mode or in the “multi” mode. In the first approach, only uniquely mapped reads are used, while in the second one, both uniquely and multi-mapped reads are counted. In doing so, each ambiguous alignment is weighted with a value of 1/n, where n is the number of multiple matches for a given read. An expectation/maximization (EM) step is also performed to define the maximum likelihood estimates for all TE insertions from multi-alignments. The multi-mapper estimate is then integrated with the unique read count to calculate the relative expression count for that insertion. TE subfamily abundances are then computed by summing up the estimated counts from all insertions belonging to the same subfamily.
Single-end RNA-seq data were re-aligned with STAR-v.2.7.8a (88) on the hg38 genome with only few changes in parameter settings: --outFilterMultimapNmax 500 –winAnchorMultimapNmax 500, while other STAR parameters remained unchanged. The resulting BAM files were then processed by TEtranscript in the “multi” modality, using the same gene annotation (GTF file) integrated in primary RNA-seq analysis and a UCSC-RepeatMasker (v-4.1.1) GTF file for TE annotation. Subfamily-level counts were then processed with the edgeR_v3.39 (89) R package for data normalization and differential testing, with the same statistical workflow as described before (see the “RNA-seq data analysis” section). Differentially expressed subfamilies in each cell line were defined setting an absolute log(fold change) threshold of 1 and FDR ≤ 0.05.
ELF3-KO data analysis
The ELF3-KO dataset was processed using the same RNA-seq workflow described in the previous section with some minor modifications. Specifically, sequencing reads were aligned with STAR-v.2.7.8a (88) on the hg38 genome using --outFilterMultimapNmax 500 –winAnchorMultimapNmax 500 to recover multi-mapper alignments for TE quantification. Properly mapped pairs were then selected and processed by TEtranscript in the “multi” mode, using the same gene annotation and TE annotation GTF files as described previously. Subfamily-level counts were then processed and normalized with edgeR_v3.39 (89), and differentially expressed subfamilies were defined imposing absolute log(fold change) ≥ 1 and FDR ≤ 0.05.
Enrichment of H3K27ac and H3K9me3 ChIP-seq signals in repeat subfamilies
To identify subfamilies significantly enriched in a given set of H3K27ac peaks (e.g., hyper-acetylated peaks at 24 hours of trametinib treatment), we assessed the overlap between peaks and RepeatMasker-v.4.1.1 annotations, considering a match if the peak was located no more far than 1 kb from the repeat boundaries. A Fisher’s exact test was then performed by assessing the overlap of each subfamily for a specific set of peaks compared to all peaks. Fisher’s exact test P values were corrected for multiple testing using Benjamini-Hochberg method. For the H3K9me3 data, the total list of 5233 islands was divided into quintile groups based on the mean signal intensity (count per million reads) of each region in the untreated groups. According to the signal distribution in Fig. 3C, islands from the fourth and fifth quintile groups were tested for their overlap with different repeat subfamilies, as previously mentioned. Any subfamily with an adjusted Fisher’s exact test P value of less than 0.1 was considered statistically significant.
Transcription factor motif analysis
Statistically overrepresented TF motifs were identified using PSCAN (102) considering a motif score adjusted P value (Benjamini-Hochberg correction) of 0.01 or smaller. For the TF-motif database, we used a previously published collection of 4304 position weight matrices (53). Motifs were scanned in a window of 1 kb around the summit of hyper-acetylated peaks at 24 hours of trametinib treatment, focusing on those peaks overlapping one or more repeats in subfamilies associated with hyper-acetylation at 24 hours. The same amount of repeat-overlapping H3K27ac peaks that were not differential across time points were included as a background for the enrichment test.
Meta-plot data and visualization
All meta-plots were generated using DeepTools_v3.5.1 (103). More specifically, BigWig files (reported in reads per bin, scaled to 1× coverage) were generated using bamCoverage with the following settings: --binSize 10 –normalizeUsing RPGC –effectiveGenomeSize 2913022398 –extendReads 250. The “reads per bin” value was then averaged across replicates using Wiggle-tools (104) to obtain a mean normalized value in each time point. The resulting Wig files were reconverted in BigWig and processed in DeepTools for plot signal visualization.
Affinity of ELF3 binding
Probability distributions of the best match for three different ELF3 motifs were generated using MEME-CentriMo v5.5.4 (105) to also assess the local enrichment in the top 500 constitutive (unchanged) peaks compared to the same number of top-induced peaks.
Gene set enrichment score
The IFN response signature (https://www.gsea-msigdb.org/gsea/msigdb/human/geneset/HALLMARK_INTERFERON_ALPHA_RESPONSE.html) was used to calculate an enrichment score per sample using GSVA R package with the “ssGSEA” option (https://bioconductor.org/packages/release/bioc/html/GSVA.html).
Hierarchical clustering and heatmaps
Hierarchical clustering analysis was performed using the ComplexHeatmap_v2.13.3 (90) R package using complete linkage method and Pearson correlation as distance metric.
Principal components analysis
Principal components analyses were generated in R using the “prcomp” function on the CPM-normalized data.
Genome browser tracks
Tracks for visualization in the IGV browser were generated using bedGraphTo-BigWig tool (106) and rescaled in reads per million (RPM).
Genomic datasets
Datasets are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE238200. Additional RNA-seq datasets were obtained from European Nucleotide Archive (ENA) database (http://www.ebi.ac.uk/ena/) under the accession number PRJEB25806 (17) and from GEO database (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE218404, GSE118548, and GSE78519 (54–56).
The complete list of datasets generated or used in this study is reported in table S6.
Acknowledgments
We thank M. Kim at the MD Anderson Cancer Center for providing human primary PDAC cell lines. We thank C. Ghirardi for technical support.
Funding: This work was supported by AIRC, the Italian Association for Research on Cancer (AIRC Investigator Grant 20251 to G.N. and AIRC 5x1000 Grant ISM). This work was also partially supported by the Italian Ministry of Health with “Ricerca Corrente” and 5x1000 funds to the IEO IRCCS and by the Ministry of University and Research (MUR) with the Progetto Eccellenza (2023–2027) to the Department of Pharmacological and Biomolecular Sciences, Università degli Studi di Milano (NM) and grant GR-2016-02361721 to G.R.D. F.A. is supported by a long-term fellowship from AIRC. T.B.’s work was supported by AIRC Investigator Grant #2018-15741. E.V. has been supported by fellowships from the Fondazione Umberto Veronesi (FUV). E.-Y.Y. is supported by a CPRIT Training Award (RP210028). A.V. is supported by NIH/NCI SPORE in gastrointestinal cancer grant P50CA221707 and NIH grant NIH/NCI R01CA258917.
Author contributions: Conceptualization: G.N. and A.C. Investigation: A.C. carried out most of the experiments with help from F.A., E.V., S.P., R.N., S.A., F.C., I.H., R.S., E.-Y.Y., M.C.S., and S.Ron. Formal analysis: F.Ga. carried out most of the analyses with help from P.D.C. and F.Gu. A.C., F.Ga., and G.N. wrote the manuscript with contributions from all the authors. Supervision: G.N., S.Rod., N.M., T.B., S.M., S.G., A.V., and G.R.D. Writing—original draft: A.C. and G.N. Writing—review and editing: G.N. Funding acquisition: G.N., N.M., T.B., and A.V.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Datasets are available in the GEO database (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE238200. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Legends for tables S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S6
REFERENCES AND NOTES
- 1.Aslan M., Shahbazi R., Ulubayram K., Ozpolat B., Targeted therapies for pancreatic cancer and hurdles ahead. Anticancer Res 38, 6591–6606 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Rahib L., Wehner M. R., Matrisian L. M., Nead K. T., Estimated projection of US cancer incidence and death to 2040. JAMA Netw. Open 4, e214708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collisson E. A., Bailey P., Chang D. K., Biankin A. V., Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Hayashi A., Hong J., Iacobuzio-Donahue C. A., The pancreatic cancer genome revisited. Nat. Rev. Gastroenterol. Hepatol. 18, 469–481 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Bengtsson A., Andersson R., Ansari D., The actual 5-year survivors of pancreatic ductal adenocarcinoma based on real-world data. Sci. Rep. 10, 16425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones S., Zhang X., Parsons D. W., Lin J. C.-H., Leary R. J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., Hong S.-M., Fu B., Lin M.-T., Calhoun E. S., Kamiyama M., Walter K., Nikolskaya T., Nikolsky Y., Hartigan J., Smith D. R., Hidalgo M., Leach S. D., Klein A. P., Jaffee E. M., Goggins M., Maitra A., Iacobuzio-Donahue C., Eshleman J. R., Kern S. E., Hruban R. H., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E., Kinzler K. W., Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biankin A. V., Waddell N., Kassahn K. S., Gingras M.-C., Muthuswamy L. B., Johns A. L., Miller D. K., Wilson P. J., Patch A.-M., Wu J., Chang D. K., Cowley M. J., Gardiner B. B., Song S., Harliwong I., Idrisoglu S., Nourse C., Nourbakhsh E., Manning S., Wani S., Gongora M., Pajic M., Scarlett C. J., Gill A. J., Pinho A. V., Rooman I., Anderson M., Holmes O., Leonard C., Taylor D., Wood S., Xu Q., Nones K., Fink J. L., Christ A., Bruxner T., Cloonan N., Kolle G., Newell F., Pinese M., Mead R. S., Humphris J. L., Kaplan W., Jones M. D., Colvin E. K., Nagrial A. M., Humphrey E. S., Chou A., Chin V. T., Chantrill L. A., Mawson A., Samra J. S., Kench J. G., Lovell J. A., Daly R. J., Merrett N. D., Toon C., Epari K., Nguyen N. Q., Barbour A., Zeps N., Initiative A. P. C. G., Kakkar N., Zhao F., Wu Y. Q., Wang M., Muzny D. M., Fisher W. E., Brunicardi F. C., Hodges S. E., Reid J. G., Drummond J., Chang K., Han Y., Lewis L. R., Dinh H., Buhay C. J., Beck T., Timms L., Sam M., Begley K., Brown A., Pai D., Panchal A., Buchner N., De Borja R., Denroche R. E., Yung C. K., Serra S., Onetto N., Mukhopadhyay D., Tsao M.-S., Shaw P. A., Petersen G. M., Gallinger S., Hruban R. H., Maitra A., Iacobuzio-Donahue C. A., Schulick R. D., Wolfgang C. L., Morgan R. A., Lawlor R. T., Capelli P., Corbo V., Scardoni M., Tortora G., Tempero M. A., Mann K. M., Jenkins N. A., Perez-Mancera P. A., Adams D. J., Largaespada D. A., Wessels L. F. A., Rust A. G., Stein L. D., Tuveson D. A., Copeland N. G., Musgrove E. A., Scarpa A., Eshleman J. R., Hudson T. J., Sutherland R. L., Wheeler D. A., Pearson J. V., Pherson J. D. M., Gibbs R. A., Grimmond S. M., Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waters A. M., Der C. J., KRAS: The critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb. Perspect. Med. 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanda M., Matthaei H., Wu J., Hong S.-M., Yu J., Borges M., Hruban R. H., Maitra A., Kinzler K., Vogelstein B., Goggins M., Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 142, 730–733.e9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Commisso C., Davidson S. M., Soydaner-Azeloglu R. G., Parker S. J., Kamphorst J. J., Hackett S., Grabocka E., Nofal M., Drebin J. A., Thompson C. B., Rabinowitz J. D., Metallo C. M., Vander Heiden M. G., Bar-Sagi D., Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Poggetto E., Ho I.-L., Balestrieri C., Yen E.-Y., Zhang S., Citron F., Shah R., Corti D., Diaferia G. R., Li C.-Y., Loponte S., Carbone F., Hayakawa Y., Valenti G., Jiang S., Sapio L., Jiang H., Dey P., Gao S., Deem A. K., Rose-John S., Yao W., Ying H., Rhim A. D., Genovese G., Heffernan T. P., Maitra A., Wang T. C., Wang L., Draetta G. F., Carugo A., Natoli G., Viale A., Epithelial memory of inflammation limits tissue damage while promoting pancreatic tumorigenesis. Science 373, eabj0486 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ying H., Kimmelman A. C., Lyssiotis C. A., Hua S., Chu G. C., Fletcher-Sananikone E., Locasale J. W., Son J., Zhang H., Coloff J. L., Yan H., Wang W., Chen S., Viale A., Zheng H., Paik J. H., Lim C., Guimaraes A. R., Martin E. S., Chang J., Hezel A. F., Perry S. R., Hu J., Gan B., Xiao Y., Asara J. M., Weissleder R., Wang Y. A., Chin L., Cantley L. C., DePinho R. A., Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viale A., Pettazzoni P., Lyssiotis C. A., Ying H., Sánchez N., Marchesini M., Carugo A., Green T., Seth S., Giuliani V., Kost-Alimova M., Muller F., Colla S., Nezi L., Genovese G., Deem A. K., Kapoor A., Yao W., Brunetto E., Kang Y., Yuan M., Asara J. M., Wang Y. A., Heffernan T. P., Kimmelman A. C., Wang H., Fleming J. B., Cantley L. C., DePinho R. A., Draetta G. F., Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapoor A., Yao W., Ying H., Hua S., Liewen A., Wang Q., Zhong Y., Wu C. J., Sadanandam A., Hu B., Chang Q., Chu G. C., al-Khalil R., Jiang S., Xia H., Fletcher-Sananikone E., Lim C., Horwitz G. I., Viale A., Pettazzoni P., Sanchez N., Wang H., Protopopov A., Zhang J., Heffernan T., Johnson R. L., Chin L., Wang Y. A., Draetta G., DePinho R. A., Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158, 185–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettazzoni P., Viale A., Shah P., Carugo A., Ying H., Wang H., Genovese G., Seth S., Minelli R., Green T., Huang-Hobbs E., Corti D., Sanchez N., Nezi L., Marchesini M., Kapoor A., Yao W., Francesco M. E. D., Petrocchi A., Deem A. K., Scott K., Colla S., Mills G. B., Fleming J. B., Heffernan T. P., Jones P., Toniatti C., DePinho R. A., Draetta G. F., Genetic events that limit the efficacy of MEK and RTK inhibitor therapies in a mouse model of KRAS-driven pancreatic cancer. Cancer Res. 75, 1091–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayes T. K., Neel N. F., Hu C., Gautam P., Chenard M., Long B., Aziz M., Kassner M., Bryant K. L., Pierobon M., Marayati R., Kher S., George S. D., Xu M., Wang-Gillam A., Samatar A. A., Maitra A., Wennerberg K., Petricoin E. F. III, Yin H. H., Nelkin B., Cox A. D., Yeh J. J., der C. J., long-term erk inhibition in kras-mutant pancreatic cancer is associated with MYC degradation and senescence-like growth suppression. Cancer Cell 29, 75–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryant K. L., Stalnecker C. A., Zeitouni D., Klomp J. E., Peng S., Tikunov A. P., Gunda V., Pierobon M., Waters A. M., George S. D., Tomar G., Papke B., Hobbs G. A., Yan L., Hayes T. K., Diehl J. N., Goode G. D., Chaika N. V., Wang Y., Zhang G. F., Witkiewicz A. K., Knudsen E. S., Petricoin E. F. III, Singh P. K., Macdonald J. M., Tran N. L., Lyssiotis C. A., Ying H., Kimmelman A. C., Cox A. D., der C. J., Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 25, 628–640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinsey C. G., Camolotto S. A., Boespflug A. M., Guillen K. P., Foth M., Truong A., Schuman S. S., Shea J. E., Seipp M. T., Yap J. T., Burrell L. D., Lum D. H., Whisenant J. R., Gilcrease G. W. III, Cavalieri C. C., Rehbein K. M., Cutler S. L., Affolter K. E., Welm A. L., Welm B. E., Scaife C. L., Snyder E. L., McMahon M., Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 25, 620–627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., Hugh W. F., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., Le Vine R., Ewan P. M., Kernan K. M., Meldrim J., Mesirov J. P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann Y., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., Murray A. M., Matthews L., Mercer S., Milne S., Mullikin J. C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R. H., Wilson R. K., Hillier L. W., Pherson J. D. M., Marra M. A., Mardis E. R., Fulton L. A., Chinwalla A. T., Pepin K. H., Gish W. R., Chissoe S. L., Wendl M. C., Delehaunty K. D., Miner T. L., Delehaunty A., Kramer J. B., Cook L. L., Fulton R. S., Johnson D. L., Minx P. J., Clifton S. W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J. F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R. A., Muzny D. M., Scherer S. E., Bouck J. B., Sodergren E. J., Worley K. C., Rives C. M., Gorrell J. H., Metzker M. L., Naylor S. L., Kucherlapati R. S., Nelson D. L., Weinstock G. M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D. R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H. M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R. W., Federspiel N. A., Abola A. P., Proctor M. J., Myers R. M., Schmutz J., Dickson M., Grimwood J., Cox D. R., Olson M. V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G. A., Athanasiou M., Schultz R., Roe B. A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., Combie W. R. M., de la Bastide M., Dedhia N., Blöcker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J. A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D. G., Burge C. B., Cerutti L., Chen H. C., Church D., Clamp M., Copley R. R., Doerks T., Eddy S. R., Eichler E. E., Furey T. S., Galagan J., Gilbert J. G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L. S., Jones T. A., Kasif S., Kaspryzk A., Kennedy S., Kent W. J., Kitts P., Koonin E. V., Korf I., Kulp D., Lancet D., Lowe T. M., Lysaght A. M., Mikkelsen T., Moran J. V., Mulder N., Pollara V. J., Ponting C. P., Schuler G., Schultz J., Slater G., Smit A. F., Stupka E., Szustakowki J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y. I., Wolfe K. H., Yang S. P., Yeh R. F., Collins F., Guyer M. S., Peterson J., Felsenfeld A., Wetterstrand K. A., Patrinos A., Morgan M. J., de Jong P., Catanese J. J., Osoegawa K., Shizuya H., Choi S., Chen Y. J., Szustakowki J., Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Bourque G., Leong B., Vega V. B., Chen X., Lee Y. L., Srinivasan K. G., Chew J. L., Ruan Y., Wei C. L., Ng H. H., Liu E. T., Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 18, 1752–1762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuong E. B., Rumi M. A., Soares M. J., Baker J. C., Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 45, 325–329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuong E. B., Elde N. C., Feschotte C., Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deniz O., Ahmed M., Todd C. D., Rio-Machin A., Dawson M. A., Branco M. R., Endogenous retroviruses are a source of enhancers with oncogenic potential in acute myeloid leukaemia. Nat. Commun. 11, 3506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson P. J., Macfarlan T. S., Lorincz M. C., Long terminal repeats: From parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol. Cell 62, 766–776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babaian A., Mager D. L., Endogenous retroviral promoter exaptation in human cancer. Mob. DNA 7, 24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., Rauscher F. J. III, SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919–932 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Xia L., Wu D. Y., Wang H., Chansky H. A., Schubach W. H., Hickstein D. D., Zhang Y., Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene 21, 148–152 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Collins P. L., Kyle K. E., Egawa T., Shinkai Y., Oltz E. M., The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 112, 8367–8372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato M., Takemoto K., Shinkai Y., A somatic role for the histone methyltransferase Setdb1 in endogenous retrovirus silencing. Nat. Commun. 9, 1683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf D., Goff S. P., Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458, 1201–1204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobs F. M., Greenberg D., Nguyen N., Haeussler M., Ewing A. D., Katzman S., Paten B., Salama S. R., Haussler D., An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516, 242–245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imbeault M., Helleboid P. Y., Trono D., KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Matsui T., Leung D., Miyashita H., Maksakova I. A., Miyachi H., Kimura H., Tachibana M., Lorincz M. C., Shinkai Y., Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Rowe H. M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P. V., Layard-Liesching H., Verp S., Marquis J., Spitz F., Constam D. B., Trono D., KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Pehrsson E. C., Choudhary M. N. K., Sundaram V., Wang T., The epigenomic landscape of transposable elements across normal human development and anatomy. Nat. Commun. 10, 5640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domansky A. N., Kopantzev E. P., Snezhkov E. V., Lebedev Y. B., Leib-Mosch C., Sverdlov E. D., Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 472, 191–195 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Chen Y. G., Hur S., Cellular origins of dsRNA, their recognition and consequences. Nat. Rev. Mol. Cell Biol. 23, 286–301 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roulois D., Loo Yau H., Singhania R., Wang Y., Danesh A., Shen S. Y., Han H., Liang G., Jones P. A., Pugh T. J., O’Brien C., de Carvalho D. D., DNA-Demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiappinelli K. B., Strissel P. L., Desrichard A., Li H., Henke C., Akman B., Hein A., Rote N. S., Cope L. M., Snyder A., Makarov V., Budhu S., Slamon D. J., Wolchok J. D., Pardoll D. M., Beckmann M. W., Zahnow C. A., Merghoub T., Chan T. A., Baylin S. B., Strick R., Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 164, 1073 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Sheng W., LaFleur M. W., Nguyen T. H., Chen S., Chakravarthy A., Conway J. R., Li Y., Chen H., Yang H., Hsu P. H., van Allen E. M., Freeman G. J., de Carvalho D. D., He H. H., Sharpe A. H., Shi Y., LSD1 Ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M., Thomas S. L., DeWitt A. K., Zhou W., Madaj Z. B., Ohtani H., Baylin S. B., Liang G., Jones P. A., Dual inhibition of DNA and histone methyltransferases increases viral mimicry in ovarian cancer cells. Cancer Res. 78, 5754–5766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones P. A., Ohtani H., Chakravarthy A., De Carvalho D. D., Epigenetic therapy in immune-oncology. Nat. Rev. Cancer 19, 151–161 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Jansz N., Faulkner G. J., Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 22, 147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith C. C., Beckermann K. E., Bortone D. S., de Cubas A. A., Bixby L. M., Lee S. J., Panda A., Ganesan S., Bhanot G., Wallen E. M., Milowsky M. I., Kim W. Y., Rathmell W. K., Swanstrom R., Parker J. S., Serody J. S., Selitsky S. R., Vincent B. G., Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J. Clin. Invest. 128, 4804–4820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alfarano G., Audano M., di Chiaro P., Balestrieri C., Milan M., Polletti S., Spaggiari P., Zerbi A., Diaferia G. R., Mitro N., Natoli G., Interferon regulatory factor 1 (IRF1) controls the metabolic programmes of low-grade pancreatic cancer cells. Gut 72, 109–128 (2023). [DOI] [PubMed] [Google Scholar]
- 46.Tan X., Sun L., Chen J., Chen Z. J., Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol. 72, 447–478 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Vincent J., Adura C., Gao P., Luz A., Lama L., Asano Y., Okamoto R., Imaeda T., Aida J., Rothamel K., Gogakos T., Steinberg J., Reasoner S., Aso K., Tuschl T., Patel D. J., Glickman J. F., Ascano M., Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat. Commun. 8, 750 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto K., Venida A., Yano J., Biancur D. E., Kakiuchi M., Gupta S., Sohn A. S. W., Mukhopadhyay S., Lin E. Y., Parker S. J., Banh R. S., Paulo J. A., Wen K. W., Debnath J., Kim G. E., Mancias J. D., Fearon D. T., Perera R. M., Kimmelman A. C., Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581, 100–105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.York A. G., Williams K. J., Argus J. P., Zhou Q. D., Brar G., Vergnes L., Gray E. E., Zhen A., Wu N. C., Yamada D. H., Cunningham C. R., Tarling E. J., Wilks M. Q., Casero D., Gray D. H., Yu A. K., Wang E. S., Brooks D. G., Sun R., Kitchen S. G., Wu T. T., Reue K., Stetson D. B., Bensinger S. J., Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell 163, 1716–1729 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown M. S., Goldstein J. L., The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Canadas I., Thummalapalli R., Kim J. W., Kitajima S., Jenkins R. W., Christensen C. L., Campisi M., Kuang Y., Zhang Y., Gjini E., Zhang G., Tian T., Sen D. R., Miao D., Imamura Y., Thai T., Piel B., Terai H., Aref A. R., Hagan T., Koyama S., Watanabe M., Baba H., Adeni A. E., Lydon C. A., Tamayo P., Wei Z., Herlyn M., Barbie T. U., Uppaluri R., Sholl L. M., Sicinska E., Sands J., Rodig S., Wong K. K., Paweletz C. P., Watanabe H., Barbie D. A., Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med. 24, 1143–1150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei G. H., Badis G., Berger M. F., Kivioja T., Palin K., Enge M., Bonke M., Jolma A., Varjosalo M., Gehrke A. R., Yan J., Talukder S., Turunen M., Taipale M., Stunnenberg H. G., Ukkonen E., Hughes T. R., Bulyk M. L., Taipale J., Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29, 2147–2160 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diaferia G. R., Balestrieri C., Prosperini E., Nicoli P., Spaggiari P., Zerbi A., Natoli G., Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J. 35, 595–617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner S., Vlachogiannis G., de Haven Brandon A., Valenti M., Box G., Jenkins L., Mancusi C., Self A., Manodoro F., Assiotis I., Robinson P., Chauhan R., Rust A. G., Matthews N., Eason K., Khan K., Starling N., Cunningham D., Sadanandam A., Isacke C. M., Kirkin V., Valeri N., Whittaker S. R., Suppression of interferon gene expression overcomes resistance to MEK inhibition in KRAS-mutant colorectal cancer. Oncogene 38, 1717–1733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang B., Krall E. B., Aguirre A. J., Kim M., Widlund H. R., Doshi M. B., Sicinska E., Sulahian R., Goodale A., Cowley G. S., Piccioni F., Doench J. G., Root D. E., Hahn W. C., ATXN1L, CIC, and ETS transcription factors modulate sensitivity to MAPK pathway inhibition. Cell Rep. 18, 1543–1557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffmann T., Hörmann A., Corcokovic M., Zmajkovic J., Hinterndorfer M., Salkanovic J., Spreitzer F., Köferle A., Gitschtaler K., Popa A., Oberndorfer S., Andersch F., Schaefer M., Fellner M., Budano N., Ruppert J. G., Chetta P., Wurm M., Zuber J., Neumüller R. A., Precision RNAi using synthetic shRNAmir target sites. eLife 12, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharp L. L., Schwarz D. A., Bott C. M., Marshall C. J., Hedrick S. M., The influence of the MAPK pathway on T cell lineage commitment. Immunity 7, 609–618 (1997). [DOI] [PubMed] [Google Scholar]
- 58.D’Souza W. N., Chang C. F., Fischer A. M., Li M., Hedrick S. M., The Erk2 MAPK regulates CD8 T cell proliferation and survival. J. Immunol. 181, 7617–7629 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dushyanthen S., Teo Z. L., Caramia F., Savas P., Mintoff C. P., Virassamy B., Henderson M. A., Luen S. J., Mansour M., Kershaw M. H., Trapani J. A., Neeson P. J., Salgado R., McArthur G. A., Balko J. M., Beavis P. A., Darcy P. K., Loi S., Agonist immunotherapy restores T cell function following MEK inhibition improving efficacy in breast cancer. Nat. Commun. 8, 606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verma V., Jafarzadeh N., Boi S., Kundu S., Jiang Z., Fan Y., Lopez J., Nandre R., Zeng P., Alolaqi F., Ahmad S., Gaur P., Barry S. T., Valge-Archer V. E., Smith P. D., Banchereau J., Mkrtichyan M., Youngblood B., Rodriguez P. C., Gupta S., Khleif S. N., MEK inhibition reprograms CD8(+) T lymphocytes into memory stem cells with potent antitumor effects. Nat. Immunol. 22, 53–66 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hingorani S. R., Wang L., Multani A. S., Combs C., Deramaudt T. B., Hruban R. H., Rustgi A. K., Chang S., Tuveson D. A., Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469–483 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Mehdipour P., Marhon S. A., Ettayebi I., Chakravarthy A., Hosseini A., Wang Y., de Castro F. A., Loo Yau H., Ishak C., Abelson S., O’Brien C. A., de Carvalho D. D., Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 588, 169–173 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Cuellar T. L., Herzner A. M., Zhang X., Goyal Y., Watanabe C., Friedman B. A., Janakiraman V., Durinck S., Stinson J., Arnott D., Cheung T. K., Chaudhuri S., Modrusan Z., Doerr J. M., Classon M., Haley B., Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J. Cell Biol. 216, 3535–3549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Griffin G. K., Wu J., Iracheta-Vellve A., Patti J. C., Hsu J., Davis T., Dele-Oni D., du P. P., Halawi A. G., Ishizuka J. J., Kim S. Y., Klaeger S., Knudsen N. H., Miller B. C., Nguyen T. H., Olander K. E., Papanastasiou M., Rachimi S., Robitschek E. J., Schneider E. M., Yeary M. D., Zimmer M. D., Jaffe J. D., Carr S. A., Doench J. G., Haining W. N., Yates K. B., Manguso R. T., Bernstein B. E., Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature 595, 309–314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burns K. H., Transposable elements in cancer. Nat. Rev. Cancer 17, 415–424 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Grundy E. E., Diab N., Chiappinelli K. B., Transposable element regulation and expression in cancer. FEBS J. 289, 1160–1179 (2022). [DOI] [PubMed] [Google Scholar]
- 67.Ting D. T., Lipson D., Paul S., Brannigan B. W., Akhavanfard S., Coffman E. J., Contino G., Deshpande V., Iafrate A. J., Letovsky S., Rivera M. N., Bardeesy N., Maheswaran S., Haber D. A., Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Espinet E., Gu Z., Imbusch C. D., Giese N. A., Büscher M., Safavi M., Weisenburger S., Klein C., Vogel V., Falcone M., Insua-Rodríguez J., Reitberger M., Thiel V., Kossi S. O., Muckenhuber A., Sarai K., Lee A. Y. L., Backx E., Zarei S., Gaida M. M., Rodríguez-Paredes M., Donato E., Yen H. Y., Eils R., Schlesner M., Pfarr N., Hackert T., Plass C., Brors B., Steiger K., Weichenhan D., Arda H. E., Rooman I., Kopp J. L., Strobel O., Weichert W., Sprick M. R., Trumpp A., Aggressive PDACs show hypomethylation of repetitive elements and the execution of an intrinsic ifn program linked to a ductal cell of origin. Cancer Discov. 11, 638–659 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knudsen E. S., Kumarasamy V., Chung S., Ruiz A., Vail P., Tzetzo S., Wu J., Nambiar R., Sivinski J., Chauhan S. S., Seshadri M., Abrams S. I., Wang J., Witkiewicz A. K., Targeting dual signalling pathways in concert with immune checkpoints for the treatment of pancreatic cancer. Gut 70, 127–138 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Falcomata C., Bärthel S., Widholz S. A., Schneeweis C., Montero J. J., Toska A., Mir J., Kaltenbacher T., Heetmeyer J., Swietlik J. J., Cheng J.-Y., Teodorescu B., Reichert O., Schmitt C., Grabichler K., Coluccio A., Boniolo F., Veltkamp C., Zukowska M., Vargas A. A., Paik W. H., Jesinghaus M., Steiger K., Maresch R., Öllinger R., Ammon T., Baranov O., Robles M. S., Rechenberger J., Kuster B., Meissner F., Reichert M., Flossdorf M., Rad R., Schmidt-Supprian M., Schneider G., Saur D., Selective multi-kinase inhibition sensitizes mesenchymal pancreatic cancer to immune checkpoint blockade by remodeling the tumor microenvironment. Nat. Cancer 3, 318–336 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu L., Mayes P. A., Eastman S., Shi H., Yadavilli S., Zhang T., Yang J., Seestaller-Wehr L., Zhang S. Y., Hopson C., Tsvetkov L., Jing J., Zhang S., Smothers J., Hoos A., The BRAF and MEK inhibitors dabrafenib and trametinib: Effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin. Cancer Res. 21, 1639–1651 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Ribas A., Algazi A., Ascierto P. A., Butler M. O., Chandra S., Gordon M., Hernandez-Aya L., Lawrence D., Lutzky J., Miller W. H. Jr., Campbell K. M., Delafont B., Marshall S., Mueller N., Robert C., PD-L1 blockade in combination with inhibition of MAPK oncogenic signaling in patients with advanced melanoma. Nat. Commun. 11, 6262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilmartin A. G., Bleam M. R., Groy A., Moss K. G., Minthorn E. A., Kulkarni S. G., Rominger C. M., Erskine S., Fisher K. E., Yang J., Zappacosta F., Annan R., Sutton D., Laquerre S. G., GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin. Cancer Res. 17, 989–1000 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Rojas L. A., Sethna Z., Soares K. C., Olcese C., Pang N., Patterson E., Lihm J., Ceglia N., Guasp P., Chu A., Yu R., Chandra A. K., Waters T., Ruan J., Amisaki M., Zebboudj A., Odgerel Z., Payne G., Derhovanessian E., Müller F., Rhee I., Yadav M., Dobrin A., Sadelain M., Łuksza M., Cohen N., Tang L., Basturk O., Gönen M., Katz S., do R. K., Epstein A. S., Momtaz P., Park W., Sugarman R., Varghese A. M., Won E., Desai A., Wei A. C., D’Angelica M. I., Kingham T. P., Mellman I., Merghoub T., Wolchok J. D., Sahin U., Türeci Ö., Greenbaum B. D., Jarnagin W. R., Drebin J., O’Reilly E. M., Balachandran V. P., Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature 618, 144–150 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Emming S., Bianchi N., Polletti S., Balestrieri C., Leoni C., Montagner S., Chirichella M., Delaleu N., Natoli G., Monticelli S., A molecular network regulating the proinflammatory phenotype of human memory T lymphocytes. Nat. Immunol. 21, 388–399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chirichella M., Bianchi N., Džafo E., Foli E., Gualdrini F., Kenyon A., Natoli G., Monticelli S., RFX transcription factors control a miR-150/PDAP1 axis that restrains the proliferation of human T cells. PLOS Biol. 20, e3001538 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lanzavecchia A., Scheidegger D., The use of hybrid hybridomas to target human cytotoxic T lymphocytes. Eur. J. Immunol. 17, 105–111 (1987). [DOI] [PubMed] [Google Scholar]
- 78.Li M., Radvanyi L., Yin B., Rycaj K., Li J., Chivukula R., Lin K., Lu Y., Shen J. J., Chang D. Z., Li D., Johanning G. L., Wang-Johanning F., Correction: Downregulation of human endogenous retrovirus type K (HERV-K) viral env RNA in pancreatic cancer cells decreases cell proliferation and tumor growth. Clin. Cancer Res. 25, 2936 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Noberini R., Restellini C., Savoia E. O., Bonaldi T., Enrichment of histones from patient samples for mass spectrometry-based analysis of post-translational modifications. Methods 184, 19–28 (2020). [DOI] [PubMed] [Google Scholar]
- 80.Noberini R., Bonaldi T., A super-SILAC strategy for the accurate and multiplexed profiling of histone posttranslational modifications. Methods Enzymol. 586, 311–332 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Noberini R., Savoia E. O., Brandini S., Greco F., Marra F., Bertalot G., Pruneri G., McDonnell L. A., Bonaldi T., Spatial epi-proteomics enabled by histone post-translational modification analysis from low-abundance clinical samples. Clin. Epigenetics 13, 145 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuan Z. F., Sidoli S., Marchione D. M., Simithy J., Janssen K. A., Szurgot M. R., Garcia B. A., EpiProfile 2.0: A computational platform for processing epi-proteomics mass spectrometry data. J. Proteome Res. 17, 2533–2541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P.-A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H.-J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H., ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Picelli S., Faridani O. R., Björklund Å. K., Winberg G., Sagasser S., Sandberg R., Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Milan M., Balestrieri C., Alfarano G., Polletti S., Prosperini E., Nicoli P., Spaggiari P., Zerbi A., Cirulli V., Diaferia G. R., Natoli G., Pancreatic cancer cells require the transcription factor MYRF to maintain ER homeostasis. Dev. Cell 55, 398–412.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 86.Balestrieri C., Alfarano G., Milan M., Tosi V., Prosperini E., Nicoli P., Palamidessi A., Scita G., Diaferia G. R., Natoli G., Co-optation of tandem DNA repeats for the maintenance of mesenchymal identity. Cell 173, 1150–1164.e14 (2018). [DOI] [PubMed] [Google Scholar]
- 87.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 91.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Axtell M. J., ShortStack: Comprehensive annotation and quantification of small RNA genes. RNA 19, 740–751 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Langmead B., Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics Chapter 11, Unit 11.17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Danecek P., Bonfield J. K., Liddle J., Marshall J., Ohan V., Pollard M. O., Whitwham A., Keane T., McCarthy S. A., Davies R. M., Li H., Twelve years of SAMtools and BCFtools. Gigascience 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zang C., Schones D. E., Zeng C., Cui K., Zhao K., Peng W., A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ross-Innes C. S., Stark R., Teschendorff A. E., Holmes K. A., Ali H. R., Dunning M. J., Brown G. D., Gojis O., Ellis I. O., Green A. R., Ali S., Chin S. F., Palmieri C., Caldas C., Carroll J. S., Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jin Y., Tam O. H., Paniagua E., Hammell M., TEtranscripts: A package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31, 3593–3599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jurka J., Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005). [DOI] [PubMed] [Google Scholar]
- 102.Zambelli F., Pesole G., Pavesi G., Motif discovery and transcription factor binding sites before and after the next-generation sequencing era. Brief. Bioinform. 14, 225–237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ramirez F., Dundar F., Diehl S., Gruning B. A., Manke T., deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zerbino D. R., Johnson N., Juettemann T., Wilder S. P., Flicek P., WiggleTools: Parallel processing of large collections of genome-wide datasets for visualization and statistical analysis. Bioinformatics 30, 1008–1009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bailey T. L., Machanick P., Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 40, e128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kent W. J., Zweig A. S., Barber G., Hinrichs A. S., Karolchik D., BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for tables S1 to S6
Tables S1 to S6