Abstract
The interaction of the human immunodeficiency virus type 1 (HIV-1) Pr55Gag molecule with the plasma membrane of an infected cell is an essential step of the viral life cycle. Myristic acid and positively charged residues within the N-terminal portion of MA constitute the membrane-binding domain of Pr55Gag. A separate assembly domain, termed the interaction (I) domain, is located nearer the C-terminal end of the molecule. The I domain is required for production of dense retroviral particles, but has not previously been described to influence the efficiency of membrane binding or the subcellular distribution of Gag. This study used a series of Gag-green fluorescent protein fusion constructs to define a region outside of MA which determines efficient plasma membrane interaction. This function was mapped to the nucleocapsid (NC) region of Gag. The minimal region in a series of C-terminally truncated Gag proteins conferring plasma membrane fluorescence was identified as the N-terminal 14 amino acids of NC. This same region was sufficient to create a density shift in released retrovirus-like particles from 1.13 to 1.17 g/ml. The functional assembly domain previously termed the I domain is thus required for the efficient plasma membrane binding of Gag, in addition to its role in determining the density of released particles. We propose a model in which the I domain facilitates the interaction of the N-terminal membrane-binding domain of Pr55Gag with the plasma membrane.
The assembly of human immunodeficiency virus (HIV) particles occurs on the cytoplasmic face of the plasma membrane of infected cells. Interactions between HIV type 1 (HIV-1) Pr55Gag and cellular membranes are mediated through the N-terminal membrane-binding domain of Gag. This domain consists of myristic acid as well as regions of the matrix protein (MA) downstream from the myristylation site (40, 47). Ionic interactions between negatively charged membrane phospholipids and positively charged residues within the first 31 amino acids of MA contribute to the stability of membrane binding. The MA region of Gag is also essential for plasma membrane targeting of Gag, as demonstrated by studies using mutagenesis of MA which redirected the site of particle assembly from the plasma membrane to intracellular locations (16, 18, 40, 46). However, results of membrane-binding studies performed with mammalian cells and with cell-free systems have demonstrated that MA interacts with membranes less efficiently than the intact Pr55Gag molecule (39, 48). Moreover, Pr55Gag was found by immunofluorescence localization at discrete plasma membrane sites, while MA was predominantly intracellular and cytoplasmic in distribution (39).
The Pr55Gag molecule is cleaved in the maturing particle by the viral protease to produce the major cleavage products MA, capsid (CA), nucleocapsid (NC), and p6 (22, 42). Small peptide sequences, termed spacer peptides 1 and 2 (SP1 and SP2), are also present between CA and NC (SP1) and between NC and p6 (SP2) (26, 29, 35). The structure of NC as determined by molecular modeling based on nuclear magnetic resonance spectroscopy data consists of a central globular domain flanked by flexible N- and C-terminal domains (11, 31). The central globular region is made up of two zinc finger domains separated by the highly basic linker RAPRKKG. NC plays multiple roles in the viral life cycle, including an essential role in genomic RNA encapsidation and dimerization (4, 5, 9, 10, 24, 33, 37), annealing of the tRNA to its binding site on the viral RNA (2, 12, 38), and strand transfer during reverse transcription (1, 34). The NC portion of Pr55Gag also plays a role in particle assembly. Gag molecules lacking p6 and most of NC remain competent for particle formation, but truncation at the level of the CA-NC junction eliminates particle formation (27). An assembly domain termed the interaction (I) domain which is required for the formation of dense retroviral particles has been identified within NC. Studies with Rous sarcoma virus (RSV) Pr76Gag indicate that mutants lacking this assembly domain release particles of abnormally light density and that domains within the HIV-1 NC can substitute for the RSV I domain in allowing the formation of particles of authentic retroviral density (3, 44). Two separate subdomains of HIV-1 NC, each containing a zinc finger motif, are capable of conferring dense particle formation upon RSV Gag (3). The mechanism underlying the dense particle formation mediated by the I domain is unknown, although it has been postulated that this region itself may be involved in the tight packing of Gag molecules in the developing particle (3).
The existence of an assembly domain within Pr55Gag but outside of MA which increases the efficiency of membrane binding has been suggested by previous work (39). The present study was undertaken to determine the location of this proposed domain within Pr55Gag, to define its relationship, if any, to the previously described I domain, and to determine if this same domain influences the intracellular versus plasma membrane distribution of Gag. A small region in the N-terminal one-third of NC was determined to be sufficient to confer plasma membrane distribution of Gag in the context of the myristylated N-terminal sequences. Membrane-binding efficiency was significantly increased by this same domain. The appearance of Gag protein on the plasma membrane by fluorescence microscopic studies correlated precisely with the shift in released retroviruslike particles from low- to high-density particles which defines the I domain. The formation of dense particles mediated by the I domain did not require the zinc finger motifs or the basic linker region of NC, although inclusion of these regions contributed to overall membrane-binding efficiency. We conclude that the I domain of HIV-1 contributes significantly to the efficiency of plasma membrane binding and to the subcellular appearance of Gag at the plasma membrane in addition to allowing packing of Gag molecules to form retroviruslike particles of native density.
MATERIALS AND METHODS
Construction of plasmids expressing Gag-GFP fusion proteins.
Gag protein expression constructs fused to a human codon-optimized form of green fluorescence protein (GFP) were used in this study. The Gag coding sequences for all constructs were derived from the full-length proviral DNA clone HXB2gpt. The GFP sequence originated from plasmids pEGFP-N2 or pEGFP-N3 (Clontech, Palo Alto, Calif.). Plasmids pTM1 and pTM3, which employ the T7 promoter and the untranslated region from encephalomyocarditis virus for cap-independent translation of mRNA, were used for all expression plasmid construction (32). For all Gag expression constructs except GAGP/GFP and GAGB/GFP, PCR cloning was used to generate gag gene fragments with an NcoI site at the 5′ ATG and a BamHI site at the 3′ fusion site. The resulting PCR product was ligated into pTM1, using the NcoI and BamHI sites of the polylinker region to generate intermediate cloning constructs. GFP fusion constructs were then generated from the appropriate enhanced GFP vector by digestion with BamHI and NotI and ligation of the BamHI-NotI GFP cassette into the pTM/Gag intermediate construct digested with BamHI and EagI. GAGP/GFP and GAGB/GFP were constructed from the previously described expression construct p55G1, which contains the HXB2 gag gene in a pTM3 backbone, through ligation of the GFP cassette from pEGFP-N3 or pEGFP-N2 into the PstI or BglII site, respectively, of the gag gene within p55G1 (40). The expression plasmid design allows production of full-length MA fused to GFP (MA/GFP), the MA and CA regions of Gag up the C-terminal CA proteolytic cleavage site (final Gag residue, Leu-363) fused to GFP (MACA/GFP), and full-length Gag fused to GFP (55GAG/GFP), as well as intermediate fusion sites within the N-terminal portion of CA (GAGP/GFP) and within SP2 which follows NC (GAGB/GFP). The amino acid number of the last intact gag codon for each fusion construct is indicated at the fusion site in Fig. 1A and shown directly in Fig. 1B, with the numbering beginning with the gag initiator ATG (Met-1). Use of the BamHI site from the pEGFP-N vectors placed a short amino acid sequence corresponding to codons of the polylinker region between the Gag protein and the GFP initiator methionine codon (Gly-Ile-His-Arg-Pro-Val-Ala-Thr for MA/GFP; Gly-Ser-Ile-Ala-Thr for 55GAG/GFP, MACA/GFP, GAG377/GFP, GAG391/GFP, GAG405/GFP, and GAG411/GFP; and slightly larger intervening sequences for GAGB/GFP and GAGP/GFP). A myristylation-deficient construct (myr− GAGB/GFP) was constructed by transfer of an NcoI-PstI fragment from plasmid p55A1, which contains a mutation of the N-terminal glycine codon to alanine, into the identical position within GAGB/GFP. The GFP expression plasmid was generated by ligation of the 724-bp NcoI-NotI fragment containing the gfp gene from pEGFP-N2 into pTM1 which had been digested with NcoI and EagI. All Gag-GFP fusions were completely sequenced through the Gag open reading frame and across the fusion boundary to confirm correct construction.
FIG. 1.
Construction of Gag-GFP chimeric constructs. (A) The initial panel of Gag-GFP constructs. Shaded regions represent the 27-kDa GFP protein fused to truncated or full-length HIV Gag protein (open bars). The number above the fusion site represents the C-terminal Gag residue expressed (numbered from the amino acid sequence of HXB2CG, with the initiator methionine as residue 1). Note that GAGB/GFP and GAGP/GFP utilize restriction sites as the sites of truncation of Gag and fusion to GFP; fusion sites for the other constructs were created at proteolytic cleavage sites (MA/GFP and MACA/GFP) or following the final residue of Gag (55GAG/GFP) by PCR cloning techniques. (B) Gag-GFP chimeric constructs subdividing HIV NC. Asterisks indicate the sites of Gag truncation and fusion with GFP; numbering is as in panel A. Cleavage sites used by HIV protease are indicated by arrows.
Oligonucleotides used in the above-described constructions were GAAGGAGAGCCATGGGTGCGAGAGCG (N-terminal MA oligonucleotide, all Gag-GFP fusion constructs except myr− GAGB/GFP), AGAGCCATGGCTGCGAGAGCGTCAGTA (N-terminal MA oligonucleotide, myr− GAGB/GFP), CGCGGATCCCGTAATTTTGGCTGACCTGATT (MA/GFP, C-terminal fusion), GGTGGATCCCAAAACTCTTGCCTTATG (MACA/GFP), CGGGATCCA TTATGGTAGCTGAATTTG (GAG377/GFP), GGTGGATCCCTTAACAA TCTTTCTTTG (GAG391/GFP), GGTGGATCCGCAATTTCTGGCTGTGTG (GAG405/GFP), GGTGGATCCCTTTTTCCTAGGGGCCCTG (GAG411/GFP), and TTGGATCCTTGTGACGAGGGGTC (55GAG/GFP).
Expression of Gag-GFP fusion proteins.
The vaccinia virus-T7 RNA polymerase hybrid system was used to express Gag-GFP fusion proteins in the African green monkey kidney cell line BSC-40 as previously described (39, 40). T7 RNA polymerase was provided by infection with 10 PFU of the recombinant vaccinia virus VTF 7-3 per cell. Cells were grown in 100-mm3 tissue culture dishes for subcellular fractionation experiments or on glass coverslips within 35-mm3 dishes for microscopic analysis. Cells were examined by microscopy or collected for analysis at 4 to 5 h posttransfection unless otherwise indicated.
Conventional epifluorescence and laser confocal fluorescence microscopy.
Living cells expressing Gag-GFP fusion proteins and grown on glass coverslips in 35-mm3 dishes were photographed without fixation by using a Nikon Diaphot inverted microscope equipped for epifluorescence photomicrography (Nikon, Tokyo, Japan). Confocal microscopy was performed in a similar manner, using a Zeiss LSM410 confocal laser scanning microscope (Carl Zeiss, Thornwood, N.Y.) equipped for simultaneous fluorescence and Nomarski differential interference contrast (DIC) imaging and optical sectioning with three-dimensional z-plane reconstruction. Images were reconstructed by using Zeiss LSM software, and levels were adjusted with Adobe Photoshop version 4.0 for Macintosh (Adobe Systems, Mountain View, Calif.). Photomicrographs were obtained from cells which were representative of the subcellular distribution observed in at least 80% of the fluorescing cells examined during three independent experiments with each construct.
Subcellular fractionation and quantitation of protein.
BSC-40 cells grown in 100-mm3 plates and expressing Gag-GFP fusion proteins were harvested 4 to 5 h posttransfection and processed for differential sedimentation centrifugation as previously described (40). Briefly, cells were subjected to Dounce homogenization in 1 ml of hypotonic buffer with protease inhibitors and then centrifuged at 1,000 × g for 10 min to remove unbroken cells and nuclei. Supernatants containing cytosolic components and cellular membranes were subjected to centrifugation at 100,000 × g for 30 min in a Sorvall RCM120EX microultracentrifuge. Soluble and membrane pellet fractions were collected and analyzed by Western blotting or by fluorescence spectrophotometry. Samples for Western blotting were immunoprecipitated with pooled HIV patient sera, separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and detected with monoclonal antibodies directed against HIV MA. Supernatants and pellets for quantitation were adjusted to 0.5% Triton X-100 in NTE buffer (100 mM NaCl, 10 mM Tris [pH 8.0], 1 mM EDTA); vigorous vortexing of the membrane/cytoskeletal pellet was required for generation of a uniform suspension. Standards of recombinant EGFP (Clontech) were prepared in the same solution. Four serial 10-fold dilutions were prepared beginning from an initial concentration of 1 μg/ml, and fluorescence was measured to generate a standard curve. Fluorescence intensity of Gag-GFP samples and of standards was determined with a Perkin-Elmer (Foster City, Calif.) model 650-40 fluorescence spectrophotometer with an excitation filter of 450 nm and emission filter of 510 nm. The percentage of membrane-bound protein in each experiment was calculated as protein in membrane pellet/protein in pellet plus protein in soluble fraction.
Equilibrium density measurements of Gag-GFP pseudovirions.
Gag-GFP fusion proteins were produced in BSC-40 cells as described above. HIV-1 strain H9/IIIB (III B) cells were obtained from Robert Gallo through the NIH/NIAID AIDS Research and Reference Reagent Program and maintained in RPMI medium supplemented with 10% fetal bovine sera and antibiotics. Culture medium was removed 3 to 4 h posttransfection and replaced with Dulbecco Modified Eagle medium deficient in cysteine and methionine and supplemented with 200 μCi of [35S]cysteine-methionine per ml. Following an overnight incubation, supernatants were collected, layered on top of a 20% sucrose cushion in NTE, and subjected to centrifugation at 100,000 × g for 60 min. The media and cushion were removed; the pelleted material was resuspended in NTE and layered on the top of a linear 20 to 60% (wt/vol) sucrose gradient. The gradient and pelleted material were subjected to centrifugation at 100,000 × g for 16 h, after which equal fractions were collected. Ninety percent of each fraction was used for immunoprecipitation with pooled HIV patient sera, separation via SDS-PAGE, and autoradiographic analysis. The remaining 10% was saved and used for meticulous determination of sucrose density with a refractometer. Analysis of autoradiograms was accompanied in some cases by scanning of the film on a flatbed scanner with a transparency adapter and quantitation of the relative density of the bands by using NIH Image software, version 1.61.
RESULTS
Design of Gag-GFP fusion constructs.
To define the differences in subcellular distribution exhibited by HIV-1 Pr55Gag and HIV-1 MA, we adopted a Gag-GFP fusion approach. This approach was taken to allow simultaneous and rapid subcellular localization by fluorescence microscopy and quantitation of protein in subcellular fractions generated by differential sedimentation centrifugation. Fusion proteins were constructed with Gag proteins fused to the N terminus of GFP so that the myristylated N terminus of Gag was preserved. An initial series of Gag-GFP fusions was generated to identify the region(s) responsible for the enhanced membrane binding and plasma membrane localization of Pr55Gag compared to MA (Fig. 1A). Four additional constructs were designed to further refine the region between MACA/GFP and GAGB/GFP following initial results (Fig. 1B). We used the vaccinia virus-T7 expression system because of its proven utility in subcellular fractionation studies of retroviral Gag proteins and for its ability to readily produce Gag retroviruslike particles which are released into the cellular supernatant (13, 39, 40).
Differential membrane binding of Gag proteins.
Previous work has demonstrated that HIV-1 Pr55Gag binds more efficiently to cellular or phospholipid vesicle membranes than MA (39, 40, 48). To validate the Gag-GFP fusion strategy for identification of the domains outside of MA which contribute this membrane-binding efficiency, the cytosolic versus membrane-associated distribution of a full-length Pr55Gag molecule fused to GFP (55GAG/GFP) was compared to that of MA fused to GFP (MA/GFP), using differential sedimentation centrifugation. Western blot analysis demonstrated that most of 55GAG/GFP sedimented with the membrane pellet, consistent with previous results (Fig. 2A). The distribution of MA/GFP was markedly different, with more protein present in the soluble fraction. To obtain more accurate quantitation of this result, the quantity of fusion protein present in the soluble and pellet fractions from identically transfected plates harvested at three separate timepoints was measured by fluorometric quantitation methods (Fig. 2B). The percentage of 55GAG/GFP in the pelleted fraction varied from 74 to 84%, while the percentage of MA/GFP in the membrane pellet ranged from 14 to 18%. Remarkably, the difference in percentage of membrane-bound protein between the full-length Gag construct and the MA construct varied very little over time (Fig. 2B). The total amount of Gag protein present at the 5- and 30-h time points differed dramatically in this experiment, from 27 ng/100-mm3 plate at 5 h to 221 ng/100-mm3 plate at 30 h for MA/GFP and from 6 ng/100-mm3 plate at 5 h to 121 ng/100-mm3 plate at 30 h for 55GAG/GFP. Thus, the difference in efficiency of membrane binding between the full-length Gag and MA constructs was consistent over a wide range of protein concentration and did not vary significantly with time.
FIG. 2.
Differential sedimentation analysis of Gag-GFP fusion constructs. (A) Western blot of soluble (S) and pellet (P) fractions from cells expressing 55GAG/GFP and MA/GFP. The positions of molecular mass markers are indicated in kilodaltons at the left. (B) Quantitation of Gag-GFP membrane binding by fluorescence spectrophotometry. Results of differential sedimentation for identically transfected cells collected at three different time points are shown. Percent membrane-bound protein was calculated as protein in membrane-enriched pellet/protein in pellet plus protein in supernatant.
Differential subcellular localization of Gag proteins.
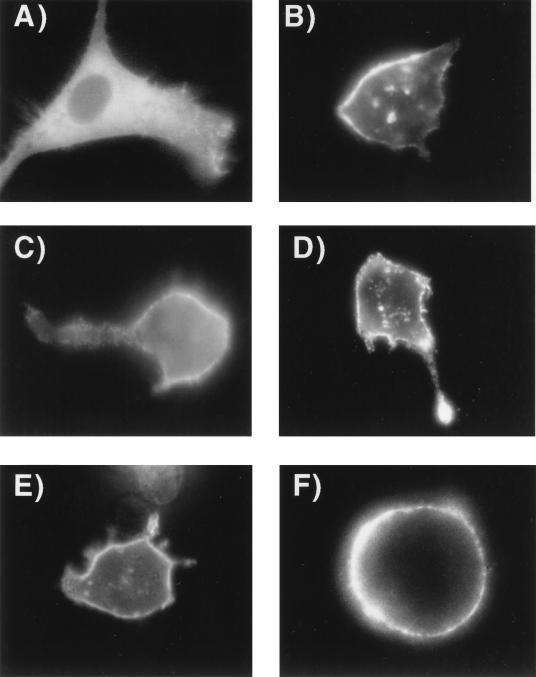
The MA region of Pr55Gag has been demonstrated to be essential for plasma membrane targeting of the precursor polyprotein and for efficient particle assembly (18, 39, 40, 46). Myristic acid plays a key role in this process, along with additional domain(s) within MA. We recently observed, however, that the specific plasma membrane localization of Pr55Gag was not demonstrated when the Gag cleavage product MA was produced within mammalian cells (39). To determine the relationship of the subcellular localization of Gag to the membrane-binding studies illustrated above, we created a panel of Gag-GFP fusion constructs spanning the region from the end of MA (MA/GFP) to full-length Pr55Gag (55GAG/GFP). The subcellular distribution of the fusion constructs within living cells was examined by epifluorescence microscopy (Fig. 3). A control plasmid expressing GFP alone revealed bright cytoplasmic fluorescence (Fig. 3A). Fusion constructs expressing MA, MA and the N-terminal segment of CA, and all of MA and CA also demonstrated a diffuse intracellular fluorescence pattern (Fig. 3B to D). A marked difference in subcellular localization was noted with GAGB/GFP, which exhibited bright, punctate membrane fluorescence (Fig. 3E). Similarly, 55GAG/GFP was found in a plasma membrane distribution, with little intracellular fluorescence (Fig. 3F).
FIG. 3.
Epifluorescence microscopy of BSC-40 cells expressing the Gag-GFP fusion constructs. Photographs were obtained 5 h after transfection with the indicated Gag-GFP expression plasmid. Photographs were taken of unfixed cells attached to a glass coverslip and are representative of more than 80% of Gag-GFP-expressing cells viewed for each construct. (A) GFP control; (B) MA/GFP; (C) GAGP/GFP; (D) MACA/GFP; (E) GAGB/GFP; (F) 55GAG/GFP.
Laser confocal microscopy was next performed to verify the results of the epifluorescence studies and to obtain localization data in three dimensions. Serial 1- to 2- μm two-dimensional images through cells expressing each Gag-GFP construct were obtained. The data were then used to construct a view of Gag protein intracellular distribution in the third dimension. A representative image from optical sectioning of a cell expressing MACA/GFP is shown in Fig. 4A. The diffuse cytoplasmic fluorescence pattern previously observed by conventional wide-field epifluorescence microscopy was confirmed, as represented by the bottom image (the x-y plane). Analysis of a reconstructed optical slice in the third dimension (the z plane) through this cell is presented at the top of Fig. 4A. This technique also revealed diffuse cytoplasmic fluorescence and lack of peripheral signal. In contrast, 55GAG/GFP analyzed in a similar manner demonstrated a peripheral pattern in all three dimensions (Fig. 4B). To further demonstrate the differential distribution of these constructs, we used a combination of laser confocal sectioning and Nomarski DIC microscopy. The appearance of GAGB/GFP is shown with this technique in Fig. 4C as bright peripheral signal. MA/GFP was again demonstrated to be cytoplasmic in distribution (Fig. 4D). Inclusion of the region of Gag between MACA/GFP and GAGB/GFP (Fig. 1B) thus resulted in a marked change in subcellular distribution of GFP fluorescence. To determine whether the plasma membrane distribution influenced by this region was dependent on myristylation of the N terminus of Gag, a similar analysis was carried out with a myristylation-deficient mutant construct (myr− GAGB/GFP). Elimination of the myristylation of Gag resulted in the complete loss of the membrane fluorescence pattern (Fig. 4E). The punctate nature of the peripheral staining pattern of 55GAG/GFP was confirmed by obtaining 1-μm sections through the attachment site of the cell to the coverglass. As shown in Fig. 4F, the Gag-GFP fusion proteins localize to focal regions along this attachment plane.
FIG. 4.
Laser scanning confocal microscopy of BSC-40 cells expressing Gag-GFP constructs. Digital images of living cells were acquired with a Zeiss LSM410 laser confocal microscope; reconstructed views were created by using Zeiss LSM software, version 3.84. (A) Confocal reconstructed view of a series of optical sections of MACA/GFP (bottom). The line indicates plane used for reconstructing the z-plane section (top). (B) Confocal section of cell expressing 55GAG/GFP (bottom). The line indicates plane of optical z section displayed on top of the figure. (C) Confocal and transmission DIC image, GAGB/GFP represented by bright peripheral signal. (D) Confocal and transmission DIC image, MA/GFP. (E) Confocal and transmission DIC image, myr− GAGB/GFP. (F) Confocal section from a series of 1-μm optical sections corresponding to the plane of the coverslip, 55GAG/GFP.
Mapping the domain within NC essential for plasma membrane localization.
Four additional Gag-GFP constructs were generated in order to define the region between MACA/GFP and GAGB/GFP which contributed the difference in plasma membrane fluorescence pattern described above. The additional Gag-GFP fusion constructs were designed to test whether inclusion of SP1 was sufficient to confer plasma membrane localization (GAG377/GFP), if the N-terminal portion of NC was sufficient to confer plasma membrane localization (GAG391/GFP), or if the first zinc finger (GAG405/GFP), first zinc finger and basic linker region (GAG411/GFP), or both zinc fingers and the C-terminal region (GAGB/GFP) of NC were required. Representative epifluorescence microscopy findings are presented in Fig. 5. SP1 did not confer plasma membrane localization, as no plasma membrane staining was observed with GAG377/GFP (Fig. 5A). A marked difference was noted with GAG391/GFP, demonstrating a bright peripheral signal (Fig. 5B). Distinctive plasma membrane fluorescence was evident also for GAG405/GFP, GAG411/GFP, and GAGB/GFP (Fig. 5C, D, and E, respectively). It should be noted that although the peripheral pattern of fluorescence with all three of the constructs containing a fusion site downstream of GAG377/GFP was striking, there was noticeably more intracellular cytoplasmic fluorescence with these intermediate constructs than with GAGB/GFP. The pattern of subcellular distribution varied from entirely cytoplasmic fluorescence (GAG377/GFP) to plasma membrane and cytoplasmic signal (GAG391/GFP, GAG405/GFP, and GAG411/GFP) to almost entirely plasma membrane fluorescence (GAGB/GFP). This finding is best illustrated by comparing Fig. 5C (GAG405/GFP) to Fig. 5E or F (GAGB/GFP). Figure 5F illustrates the marked peripheral fluorescence pattern of cells expressing GAGB/GFP which had detached from the glass coverslip. No apparent quenching of GFP fluorescence at membrane surfaces was noted in these experiments, although some alteration of the relative signal intensity in a particular subcellular compartment cannot be excluded. Quantitation of the transition from cytoplasmic to plasma membrane fluorescence with the addition of portions of NC was not attempted from the microscopic images but was instead carried out with differential sedimentation experiments described below.
FIG. 5.
Epifluorescence microscopy of Gag-GFP fusion constructs from Fig. 1B. Acquisition of images was as for Fig. 3. (A) GAG377/GFP; (B) GAG391/GFP; (C) GAG405/GFP; (D) GAG411/GFP; (E) GAGB/GFP; (F) cell expressing GAGB/GFP which separated from the coverglass and assumed a round conformation.
Defining the contribution of domains within NC to efficient membrane binding.
Our experiments using fluorescence microscopy revealed that focal plasma membrane staining was absent in constructs containing MA or MA and CA alone but present in constructs containing the N-terminal third of NC. We next used the panel of Gag-GFP constructs to determine whether the domain conferring plasma membrane distribution by microscopy was also responsible for the efficiency of membrane binding as measured by differential sedimentation centrifugation. Membrane- and cytoskeleton-enriched fractions were separated from soluble cytosolic components by differential sedimentation centrifugation, and the amount of Gag-GFP fusion protein in the fractions quantified by fluorescence spectrophotometry. The markedly higher percentage of membrane-bound 55GAG/GFP compared to MA/GFP was again demonstrated (Fig. 6). No significant increase in membrane-bound protein was found with inclusion of the N-terminal portion of CA (GAGP/GFP). Extension of the Gag sequence to include all of MA and CA (MACA/GFP) or MA, CA, and SP1 (GAG377/GFP) resulted in only a slight increase in the percentage of membrane-bound protein. In contrast, the addition of the N-terminal 14 amino acids of NC (GAG391/GFP) resulted in a significant increase in membrane-bound Gag protein, from a mean of 25% membrane-bound Gag protein (GAG377/GFP) to 45%. A second significant increase in the percentage of membrane-bound Gag-GFP protein occurred with the addition of the six-amino-acid linker region RAPRKK, represented by GAG411/GFP. Addition of this small linker led to an increase in membrane-bound Gag protein from 55 to 78% (mean values). It must be noted that the optimal membrane-binding construct in this study measured in this manner was GAGB/GFP, which includes the entire NC region but lacks p6. However, as is evident in Fig. 7A and B, 55GAG/GFP was more efficiently released from cells than GAGB/GFP, which may have altered the total amount of membrane-bound protein measured. Thus, the efficient membrane binding of myristylated Pr55Gag was mapped to the NC region. Although the N-terminal portion and basic linker regions of NC contributed the greatest increases in membrane binding, optimal membrane binding required the presence of the entire NC sequence.
FIG. 6.
Quantitation of membrane binding of Gag-GFP fusion constructs. The protein content of membrane-enriched pellet and soluble fractions from differential sedimentation experiments was determined by fluorescence spectrophotometry using sample measurements compared to a standard curve generated from recombinant GFP. The percentage of protein present in the membrane-enriched pellet was calculated as protein in pellet/protein in soluble fraction plus protein in pellet. Results are means of three independent experiments for each construct, with error bars representing 1 standard deviation from the mean.
FIG. 7.
Determination of buoyant density of Gag-GFP particles. Following overnight labeling with [35S]cysteine-methionine, supernatants were collected and subjected to centrifugation through 20% sucrose pellets. Pelleted material was analyzed via equilibrium centrifugation on sucrose gradients as described in Materials and Methods. Gradient fractions were analyzed by SDS-PAGE and autoradiography following immunoprecipitation with pooled HIV patient sera. The data represent the bottom 20 fractions of a total of 30 collected fractions, with the bottom of the gradient to the left. The precise density of each gradient fraction was determined; densities of peak fractions are indicated above the autoradiogram. (A) 55GAG/GFP; (B) GAGB/GFP; (C) GAG377/GFP; (D) MACA/GFP; (E) GAGP/GFP. The positions of molecular mass markers are indicated at the left in kilodaltons.
Dense retroviruslike particle formation conferred by the I domain correlates with plasma membrane fluorescence pattern.
The I domain is an assembly domain present in retroviral Gag proteins which is distinct from N-terminal membrane-binding domains (3, 44). The I domain is required for the formation of retroviruslike particles of normal density (1.16 to 1.18 g/ml). To determine the relationship of the subcellular localization domain outlined above to the I domain, the density of retroviruslike particles formed by the Gag-GFP constructs was assessed. [35S]cysteine-methionine-labeled Gag-GFP particles were collected from the cellular supernatant, pelleted through a sucrose cushion, and subjected to equilibrium density centrifugation on sucrose gradients. Labeled products of the predicted molecular mass were found to pellet through sucrose cushions and into the 20 to 60% sucrose gradients for all constructs shown in Fig. 1 except the GFP control (data not shown). 55GAG/GFP attained an equilibrium density of 1.175 g/ml; GAGB/GFP attained a similar density but had a broader (less uniform) peak (Fig. 7A and B). MA/GFP was released, pelleted through sucrose, and attained an equilibrium density of 1.13 g/ml (data not shown). GAG377/GFP, MACA/GFP, and GAGP/GFP were demonstrated to have similar equilibrium densities (1.12 to 1.13 g/ml) well below that of retroviral particles (Fig. 7C to E).
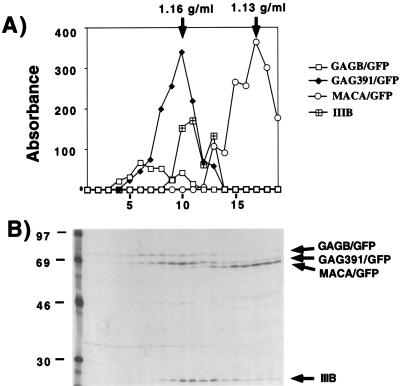
To further demonstrate the transition between light (1.12 to 1.13 g/ml) and dense (1.16 to 1.18 g/ml) Gag-GFP particles, pelleted retroviruslike particles from three constructs (GAGB/GFP, GAG391/GFP, and MACA/GFP) were analyzed on a single linear sucrose gradient together with HIV-1 isolate IIIB as a marker for normal retroviral particle density (Fig. 8). A striking distinction was demonstrated in this experiment between MACA, with an equilibrium density of 1.13 g/ml, and the denser particles. HIV-1 IIIB virions were demonstrated with a peak density of 1.16 g/ml. GAGB/GFP particles were noted to be slightly more dense than HIV-1 IIIB virions (1.17 g/ml). GAG391/GFP expression produced particles of normal retroviral particle density, as indicated by the measured density (1.16 g/ml) which overlapped the IIIB peak. The transition between light retroviruslike particles, represented by MACA/GFP (Fig. 8) or GAG377/GFP (Fig. 7D), and dense retroviruslike particles (GAG391/GFP) thus correlated precisely with the appearance of Gag protein in a plasma membrane distribution as indicated by fluorescence microscopy.
FIG. 8.
Comparison of buoyant densities of Gag-GFP constructs and HIV IIIB. Particles were prepared and labeled as described in Materials and Methods. Labeled supernatants from cells expressing GAGB/GFP, GAG391/GFP, MACA/GFP, and HIV-1 IIIB were separately centrifuged through a 20% cushion. The resuspended pellets were loaded onto the top of a single 20 to 60% sucrose gradient and centrifuged to equilibrium. Thirty fractions were collected; data for the bottom 20 fractions are presented. Gag proteins and Gag-GFP proteins in each fraction were immunoprecipitated with HIV patient sera and further analyzed by SDS-PAGE and autoradiography. Relative absorbance values were obtained by scanning the resulting autoradiogram on a flatbed scanner equipped for transparencies, and quantitation was performed with NIH Image software. (A) Plot of absorbance (y axis, relative values) for immunoprecipitated bands on the autoradiogram shown below. The x axis indicates fraction numbers, with the bottom of the gradient to the left. (B) Autoradiogram corresponding to the graphed results. Arrows point to the individual Gag-GFP fusion proteins. IIIB indicates the position of HIV-1 IIIB virions as demonstrated by radiolabeled and immunoprecipitated p24 (CA). The positions of molecular mass markers are indicated at the left in kilodaltons.
DISCUSSION
Retroviral Gag proteins direct the particle assembly process; they require no other viral gene products in order to make their way to the plasma membrane, interact with each other, generate a budding particle, and separate from the plasma membrane as a retroviral pseudovirion. The means by which these complex polyproteins travel to the plasma membrane and the specific interactions with cellular components which may occur at the assembly site remain largely unknown. An essential function of the Gag polyprotein is to interact with components of the plasma membrane. This interaction is mediated by the myristylated N-terminal region of the molecule. However, the efficiency with which myristylated MA binds to membranes is much lower than that of the intact Pr55Gag molecule (39, 48). Furthermore, MA expressed in mammalian cells is found in a predominant cytoplasmic distribution which differs markedly from the plasma membrane pattern seen with Pr55Gag (39). The experiments described in this report were designed to identify the region outside of MA which contributes to the efficiency of plasma membrane binding and to examine the relationship of this proposed domain to the previously described I domain responsible for dense particle formation.
The I domain is required for plasma membrane localization or retention of Gag.
In this study, we found that the membrane-targeting/binding function within MA by itself was insufficient to direct GFP to the plasma membrane in a manner sufficient to produce visible fluorescence at this location. The extension of the Gag region of a Gag-GFP fusion protein to include CA and SP1 did not significantly alter the observed intracellular distribution of the protein. However, when all or an N-terminal portion of NC was included, a dramatic redistribution of fluorescence to the plasma membrane was noted. The region required for the punctate plasma membrane localization was thus mapped to NC, with the minimal requirement for plasma membrane localization in a series of C-terminally truncated Gag molecules lying within the 14-amino-acid sequence between GAG377/GFP and GAG391/GFP. Analysis of the density of particles released by cells expressing the Gag-GFP fusion constructs revealed that the region responsible for the appearance of focal plasma membrane fluorescence precisely matched the region necessary for the production of dense (1.16 to 1.18 g/ml) retroviruslike particles. The influence of the I domain on Gag protein subcellular distribution could reflect either an effect on Gag protein transport to the plasma membrane or increased retention of Gag molecules on the plasma membrane. The latter explanation is more consistent with multiple previous studies indicating that the major transport function within Gag lies within the MA region (16, 18, 39–41, 43, 46, 47). The plasma membrane interaction of HIV-1 Gag requires myristylation of the N terminus of Gag, as indicated by the purely cytoplasmic signal seen with a myristylation-deficient mutant which included the I domain. Thus, the I domain contributes to or facilitates the function of the more N-terminal membrane-binding domain of Pr55Gag but is not sufficient to direct the plasma membrane interaction of Gag in the absence of the myristylated membrane-binding domain.
Contribution of multiple subdomains of NC to membrane-binding efficiency.
The NC region of Gag was found in this study to be essential for the efficient membrane binding of Gag. In contrast to the discrete mapping of the location of the minimal N-terminal portion of the I domain required for dense particle formation, the membrane binding of Gag was not optimal with the addition of this region of NC alone (represented by GAG391/GFP) but improved sequentially with the inclusion of the basic linker region between the two zinc fingers and with the addition of the second zinc finger and C-terminal portion of NC. This result confirms previous findings from our laboratory and from others that suggested that a domain or domains outside of MA played an important role in membrane binding and is in agreement with several previous studies which have localized an assembly defect to deletions or site-specific mutations within NC (14, 27, 28, 37). It is important to note that the appearance of Gag protein on the plasma membrane as judged from conventional epifluorescence and confocal fluorescence microscopy correlated well with the membrane-binding quantitation obtained by differential sedimentation. The constructs which efficiently bound membranes (GAGB/GFP) were seen almost exclusively at the plasma membrane, while those that bound with intermediate efficiency (GAG391/GFP) exhibited both cytoplasmic and plasma membrane fluorescence, and those which bound least efficiently (MA/GFP) demonstrated a complete lack of plasma membrane fluorescence. We conclude from this that the membrane binding measured in this study by differential sedimentation represents primarily plasma membrane binding rather than interaction with intracellular membranes.
We propose the following model to explain how these two widely separated domains contribute to plasma membrane interaction. Gag molecules interact with components of the plasma membrane through the myristylated N terminus of the molecule. However, the binding energy of a single membrane-binding domain to the lipid bilayer may be insufficient to support a stable membrane interaction, leading to dissociation from the membrane. In the presence of the I domain, Gag-Gag or Gag-RNA interactions occur which allow Gag molecules to coalesce in an orientation that brings their respective membrane-binding domains together for membrane interaction. Thus, the binding energy of the developing bud becomes sufficient to maintain stable plasma membrane interaction in a cooperative manner. An important aspect of this model is its application to the understanding of the role of MA in postentry events of the HIV life cycle. MA plays an essential role in events following viral entry (45), and has been shown to direct nuclear targeting of the viral preintegration complex (6, 19–21). To perform this role, MA must be released from the lipid bilayer of the entering virion. According to the model proposed, proteolytic cleavage during virion maturation would separate the I domain from the N-terminal membrane-binding region of Gag. Although MA remains largely associated with the lipid bilayer in mature virions (23), release of MA from the constraints imposed by the I domain may facilitate membrane dissociation of a fraction of MA molecules and thus allow their participation in postentry events such as the nuclear transport of the preintegration complex.
Location of the I domain within HIV-1 NC.
In this study, we used serially truncated Gag molecules fused to GFP to map the I domain to the N-terminal portion of NC. Previous studies have also pointed to an essential assembly function located in this region. Jowett et al. described a critical assembly domain located between Gag residues 372 and 379 (27). In their study, Gag constructs which were truncated at the C terminus at a point proximal to the SP1-NC cleavage site were incapable of forming and releasing particles. This region is also part of the region described as required for Gag-Gag multimerization, although the precise location of the multimerization domain has not yet been defined (17). Previous studies have established that deletion of SP1 alone does not alter the density of released particles, although it does decrease particle release and virion infectivity (29, 35). The role of a C-terminal Gag domain in enhancing plasma membrane interactions has also been reported in a study using a cell-free membrane-binding assay (36). This study reported that a domain within NC was required for Gag protein membrane binding. The data from this study suggested that an important determinant of membrane binding is located within the basic linker region between the two zinc fingers of NC, which is consistent with our finding that this region contributes to overall membrane binding efficiency (Fig. 8). However, as discussed below, the basic linker region is not required for the production of dense retroviral particles.
Our results defining the minimal I domain will be useful in future experiments designed to elucidate the mechanism through which the I domain functions to promote membrane interaction and dense particle formation. However, in light of the finding that dense particle formation correlates precisely with the appearance of Gag molecules in focal collections beneath the plasma membrane, and that multiple subdomains within NC contribute to the efficiency of Gag protein plasma membrane binding, the I domain may actually represent an assembly domain present throughout NC. This would be consistent with the finding of Bennett et al. that the I domain is present in two copies within HIV NC (3). Using a truncated RSV Gag construct, these investigators demonstrated that addition of either an N- or a C-terminal segment of HIV-1 NC changed the character of released particles to that of dense retroviruslike particles. The minimal domain that we have mapped represents further mapping of the N-terminal I domain segment described by these investigators and represented in the RSV-HIV Gag chimera RHA (3).
Our data indicate that the zinc finger and basic linker regions of HIV Gag are not required for dense particle production. This finding is consistent with studies in which site-directed mutagenesis of conserved residues within the zinc finger domains of RSV and Moloney murine leukemia virus did not result in defects in particle assembly or release (15, 25, 30). One of these studies further demonstrated that a mutation within the Moloney murine leukemia virus zinc finger which disrupted viral RNA packaging did not alter the density of released particles (25). Thus, the I domain does not require specific viral RNA interaction in order to allow dense particle formation. Nonspecific RNA interactions have been reported to play an important role in Gag-Gag interactions in an in vitro particle assembly system (7, 8). Mutant Gag proteins which have lost specific viral RNA-binding ability but preserved the minimal I domain region may retain the ability to bind cellular RNAs and therefore remain competent to produce dense retroviruslike particles. Further investigation is required to elucidate the role that Gag-RNA interactions may play in influencing retroviral particle density.
Specificity of plasma membrane interaction.
The punctate appearance of Gag protein on the plasma membrane in this study and as previously reported (39) suggests that there is specificity to the location of Gag protein targeting. If the I domain functions to bring Gag molecules together and to allow multiple N-terminal membrane-binding domains to be correctly oriented for membrane interaction, this would explain increased membrane-binding efficiency but not the specificity of binding to the plasma membrane. Focal collections of Gag beneath the plasma membrane suggest the existence of plasma membrane or cytoskeletal compartments which serve as particle assembly sites. It is possible that Gag proteins interact with cellular components at these focal sites. Ongoing studies in our laboratory are directed toward further defining Gag assembly domains and identifying the factors which determine the specificity and focality of the plasma membrane particle assembly site.
ACKNOWLEDGMENTS
P.S. was supported through PHS awards AI40338-01A1 and N01-AI-45210. V.V. was supported by PHS award N01-AI-45210. Experiments and analyses were performed in part through the use of the VUMC Cell Imaging Resource Center (supported by CA68485 and DK20593).
We thank Jonathan Sheehan for technical assistance with confocal microscopy and Chris Aiken and Terence Dermody for discussions and critical reviews of the manuscript.
REFERENCES
- 1.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barat C, Schatz O, Le Grice S, Darlix J L. Analysis of the interactions of HIV 1 replication primer tRNA(Lys,3) with nucleocapsid protein and reverse transcriptase. J Mol Biol. 1993;231:185–190. doi: 10.1006/jmbi.1993.1273. [DOI] [PubMed] [Google Scholar]
- 3.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 11.Demene H, Dong C Z, Ottmann M, Rouyez M C, Jullian N, Morellet N, Mely Y, Darlix J L, Fournie-Zaluski M C, Saragosti S, et al. 1H NMR structure and biological studies of the His23→Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critical for virus infectivity. Biochemistry. 1994;33:11707–11716. doi: 10.1021/bi00205a006. [DOI] [PubMed] [Google Scholar]
- 12.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski M C, Roques B, Darlix J L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong J, Hunter E. Analysis of retroviral assembly using a vaccinia/T7-polymerase complementation system. Virology. 1993;194:192–199. doi: 10.1006/viro.1993.1249. [DOI] [PubMed] [Google Scholar]
- 14.Dorfman T, Luban J, Goff S P, Haseltine W A, Gottlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupraz P, Oertle S, Meric C, Damay P, Spahr P F. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J Virol. 1990;64:4978–4987. doi: 10.1128/jvi.64.10.4978-4987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Facke M, Janetzko A, Shoeman R L, Krausslich H G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franke E K, Yuan H E, Bossolt K L, Goff S P, Luban J. Specificity and sequence requirements for interactions between various retroviral Gag proteins. J Virol. 1994;68:5300–5305. doi: 10.1128/jvi.68.8.5300-5305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 21.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 22.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–637. [PubMed] [Google Scholar]
- 23.Gelderblom H R, Hausmann E H, Ozel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 24.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelick R J, Henderson L E, Hanser J P, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson L E, Bowers M A, Sowder R C d, Serabyn S A, Johnson D G, Bess J W, Jr, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jowett J B, Hockley D J, Nermut M V, Jones I M. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle. J Gen Virol. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. [DOI] [PubMed] [Google Scholar]
- 28.Kaye J F, Lever A M. trans-acting proteins involved in RNA encapsidation and viral assembly in human immunodeficiency virus type 1. J Virol. 1996;70:880–886. doi: 10.1128/jvi.70.2.880-886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krausslich H G, Facke M, Heuser A M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meric C, Goff S P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morellet N, de Rocquigny H, Mely Y, Jullian N, Demene H, Ottmann M, Gerard D, Darlix J L, Fournie-Zaluski M C, Roques B P. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J Mol Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- 32.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. Product review. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 33.Ottmann M, Gabus C, Darlix J L. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peliska J A, Balasubramanian S, Giedroc D P, Benkovic S J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 35.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platt E J, Haffar O K. Characterization of human immunodeficiency virus type 1 Pr55gag membrane association in a cell-free system: requirement for a C-terminal domain. Proc Natl Acad Sci USA. 1994;91:4594–4598. doi: 10.1073/pnas.91.10.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poon D T, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prats A C, Sarih L, Gabus C, Litvak S, Keith G, Darlix J L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988;7:1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spearman P, Wang J J, Vander Heyden N, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verderame M F, Nelle T D, Wills J W. The membrane-binding domain of the Rous sarcoma virus Gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wills J W, Craven R C. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 43.Wills J W, Craven R C, Weldon R A, Jr, Nelle T D, Erdie C R. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu X, Yu Q C, Lee T H, Essex M. The C terminus of human immunodeficiency virus type 1 matrix protein is involved in early steps of the virus life cycle. J Virol. 1992;66:5667–5670. doi: 10.1128/jvi.66.9.5667-5670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan X, Yu X, Lee T H, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]