Abstract
Background:
Orthodontic tooth movement (OTM) relies on bone remodeling and controlled aseptic inflammation. Autophagy, a conserved homeostatic pathway, has been shown to play a role in bone turnover. We hypothesize that autophagy participates in regulating bone remodeling during OTM in a force-dependent and cell-type specific manner.
Materials & Methods:
A split mouth design was used to load molars with one of three force levels (15g, 30g, or 45g) in mice carrying a GFP-LC3 transgene to detect cellular autophagy. Fluorescent microscopy and qPCR analyses were used to evaluate autophagy activation and how it correlates with force level. Cell type-specific antibodies were utilized to identify cells with GFP positive puncta (autophagosomes) in periodontal tissues.
Results:
Autophagic activity increased shortly after loading with moderate force and was associated with expression of bone turnover, inflammatory and autophagy markers. Different load levels resulted in altered degrees of autophagic activation, gene expression and osteoclast recruitment. Autophagy was specifically induced by loading in macrophages and osteoclasts found in the periodontal ligament and alveolar bone. Data suggest autophagy participates in regulating bone turnover during OTM.
Conclusions:
Autophagy is induced in macrophage-lineage cells by orthodontic loading in a force-dependent manner and plays a role during OTM, possibly through modulation of osteoclast bone resorption. Exploring roles of autophagy in OTM is medically relevant given that autophagy is associated with oral and systemic inflammatory conditions.
Keywords: autophagy, orthodontic tooth movement, force loading, bone turnover, bone remodeling, frontal resorption, undermining resorption, osteoclasts, osteoblasts, macrophages, inflammation
Introduction
Well-controlled bone turnover is critical for efficient and safe orthodontic care; to this end, researchers have studied optimal force for orthodontic tooth movement (OTM).1,2 Ample data indicate excessive load slows OTM with undermining resorption and increases the risk of inflammatory root resorption.2-5 It behooves the clinician to apply an appropriate load to maximize rate of tooth movement and minimize risk of adverse outcomes like root resorption.2 With an optimal load, OTM is achieved through coordinated bone adaptation and reversible periodontal injury, involving osteoclast frontal resorption under compression and osteoblast bone formation with tension. This process is orchestrated by an aseptic inflammatory response with release of mediators, such as prostaglandins and cytokines. In healthy patients, load-induced inflammation is well-controlled, with periodontal ligament (PDL) dimensions remaining fairly constant.2 Inflammation resolves and homeostasis returns, yet it is unknown how inflammation is downregulated during OTM. Additionally, force-titration studies have characterized clinical and histological effects of differential load on OTM, with less attention paid to force-dependent molecular sequelae.2-5
To explore possible regulatory mechanisms, we investigated the role of autophagy, a conserved intracellular pathway in homeostasis.6,7 Macroautophagy, referred to throughout as autophagy, is a catabolic pathway induced by stress associated with starvation, hypoxia, toxins, or damaged organelles.7-10 Under normal conditions, autophagy helps to resolve inflammation through sequestration, breakdown and recycling of damaged cytosolic components in the autophagosome, a double membraned autophagy organelle.7 Autophagy can inhibit inflammasome activation and proinflammatory cytokine production including Interleukin-1β (IL-1β) and Interleukin-18 (IL-18).7,11,12 Th1 proinflammatory cytokines IL-1β and Tissue Necrosis Factor-α (TNF-α) induce autophagy as a negative feedback mechanism.13 Not surprisingly, aberrant autophagy contributes to chronic inflammatory conditions observed with rheumatoid arthritis, psoriasis, inflammatory bowel disease, multiple sclerosis, and systemic lupus erythematosus.6-9,14 Autophagy is also active in the developing and adult dentition under normal conditions, yet knowledge of its roles is limited.15-18
Autophagy has also been linked to bone remodeling in a variety of contexts. TNF-α induces autophagy in osteoclasts, while activation of autophagy through Beclin-1 (BECN1) overexpression promotes osteoclastogenesis and bone resorption.19 With microgravity, autophagy is induced in osteoclast precursors, prompting differentiation, and under oxidative stress, autophagy is induced in osteoblasts during mineralization.20,21 Osteoblasts with aberrant autophagy secrete Receptor Activator of Nuclear Factor Kappa-β Ligand (RANKL), a protein which activates RANK intracellular signaling, resulting in osteoclast differentiation, recruitment and bone resorption in a Rheumatoid Arthritis model.21 Mice selectively lacking autophagy in osteoblasts lost half of their trabecular bone mass while conditional knockouts, where autophagy gene expression was shut off early in life, failed to ever develop normal bone mass.21,22 These phenotypes are attributed to reduced osteoblasts, bone mineralization and Osteoprotegerin (OPG) secretion paired with increased osteoclasts and RANKL.22 These data highlight the importance of autophagy in normal bone turnover.
We previously discovered novel autophagy activation during OTM.23 In this study, we aimed to investigate the role of autophagy in bone turnover during OTM and to determine if varied force level influences autophagy activation, gene expression and osteoclast recruitment. We hypothesized that optimal loading induces autophagy in specific cell-types of the PDL and alveolar bone to regulate force-induced bone turnover. Using a transgenic mouse model with a fluorescently tagged autophagy protein LC3, we demonstrate autophagy activation in response to mechanical loading in a force-dependent and cell-specific manner.
Materials and Methods
Mouse model for studying OTM
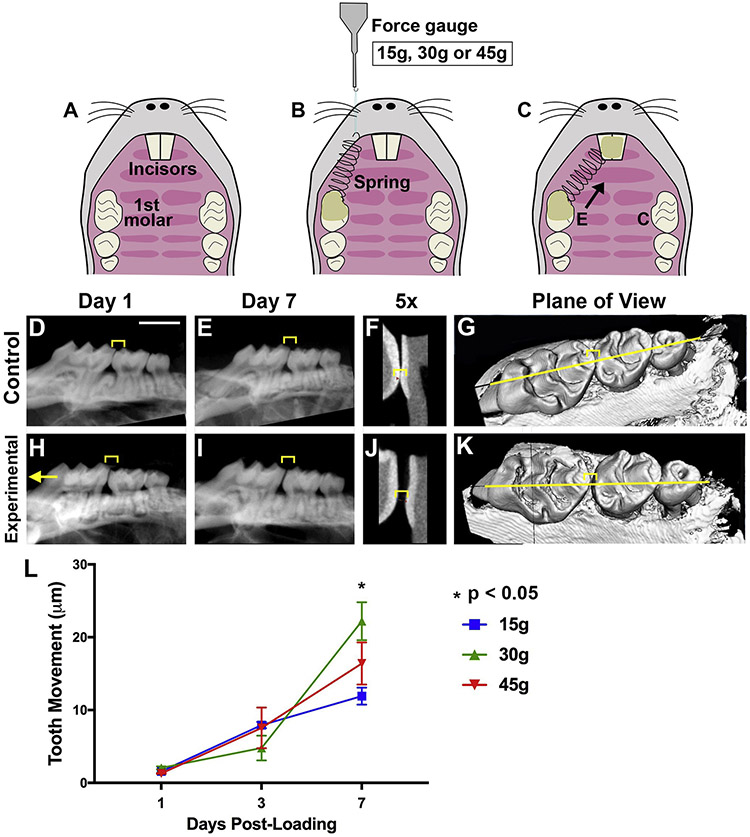
Orthodontic force application in a murine model has been characterized in studies where optimal conditions were established and recapitulated by Sambandam et al20 and Li.23 Mice were anesthetized by intraperitoneal injection of xylazine (10 mg/kg) and ketamine (100 mg/kg) solution. A split mouth design was employed, where fifteen, thirty or forty-five grams (=0.15 N, 0.3 N or 0.45 N) of force was delivered to the maxillary right first molar in the mesial direction, by bonding a nickel-titanium (NiTi) closed coil spring (American Orthodontics, Cat# 855-181, Sheboygan, WI, USA, length adapted to each mouse’s mouth) between the maxillary right first molar and incisors with light-cured resin (Transbond Supreme LV, 3M Unitek, Morovia, CA, USA) (Fig 1A-C). Lighter forces were used in the preliminary stages of this study, but little tooth movement was observed. These force levels were chosen because they fall within ranges used in previous studies and resulted in significant tooth movement.2-5, 24 No spring reactivation was performed after bonding. Animal procedures followed ethical regulations defined by Institutional Animal Care and Use Committee (IACUC) of the University of North Carolina at Chapel Hill (#17-139.0).
Figure 1. Orthodontic tooth movement (OTM) is reduced with insufficient and excessive force.
[A-C] Schematic of orthodontic force application in mice with a split-mouth design. Occlusal view of the maxilla before [A] and after [C] placement of a NiTi coil spring on the experimental (E) side. The control (C) side has no spring placed and the molar remains unloaded. [B] 15g, 30g or 45g of force measured by a force gauge prior to spring cementation. [D-K] CT radiographs of control [D-G] and experimental [H-K] molars. 2D CT radiographs [D-F, H-J], zoomed in view of molar-premolar contact point [F, J] and 3D CT [G, K] with a yellow line indicating the plane of view shown in the 2D images. Left: mesial, compression side. Right: distal, tension side. Scale = 2 mm. Yellow Bracket: Molar-premolar contact point where OTM measurements were made. [L] OTM quantification at days post-loading in mice with 15g, 30g, or 45g of force. Graph displays the mean and SEM. Convention: * p<0.05.
For OTM distance measurements, detection of autophagic activity and TRAP staining, we utilized a Green Fluorescent Protein-LC3 (GFP-LC3) reporter mouse line, as recently described 23. Briefly, GFP-tagged LC3 is expressed under the LC3 promoter and is inserted into the autophagosome membrane with autophagy induction, yielding green fluorescent puncta throughout the cytoplasm.25,26 Orthodontic force application in mice using a split-mouth design is a well-accepted model (Fig 1A-C).23, 26 Fifty-four GFP-LC3 adult mice (8-9 weeks old, in C57BL/6 background) were subdivided into 9 groups (n=6 for each) for sacrifice at 3 different time points (days 1, 3, and 7) after spring loading (15g, 30g, and 45g; contralateral first molar was the 0g control). For molecular analyses, i.e. mRNA/qRT-PCR, forty-five wild-type adult mice (C57BL/6, 8-9 weeks old, obtained from Jackson Laboratory, Bar Harbor, ME, USA) were subdivided into 9 groups (n=5 for each), to have 3 different force levels applied (15g, 30g and 45g; contralateral first molar was the 0g control) and to be sacrificed at 3 different time points (days 1, 3, and 7).
OTM distance and PDL measurements
The occlusal view of the maxilla was imaged using a stereomicroscope (SMZ18, Nikon Instruments, Melville, NY, USA) with an adapted digital camera (Nikon Instruments, Melville, NY, USA).23 NIS-Elements Basic Research imaging software was used for distance measurements, as described (n=5 for each time point).23 The OTM distance measurement of the upper right first molar was calculated by subtracting the measurement of the Control (C) unloaded side from the Experimental (E) loaded side (OTM distance = E side – C side) (Fig 1). To confirm measurements taken on the stereomicroscope, microCT radiographs were taken at days 1, 3, and 7 post-loading on fixed mice maxillae using a Skyscan 1275 MicroCT, as described.23 2D images from the bucco-lingual midpoint of the first and second molar were extracted from the 3D volumetric file. MicroCT 2D images and measurements were taken at the height of contour between the molars (Fig 1).
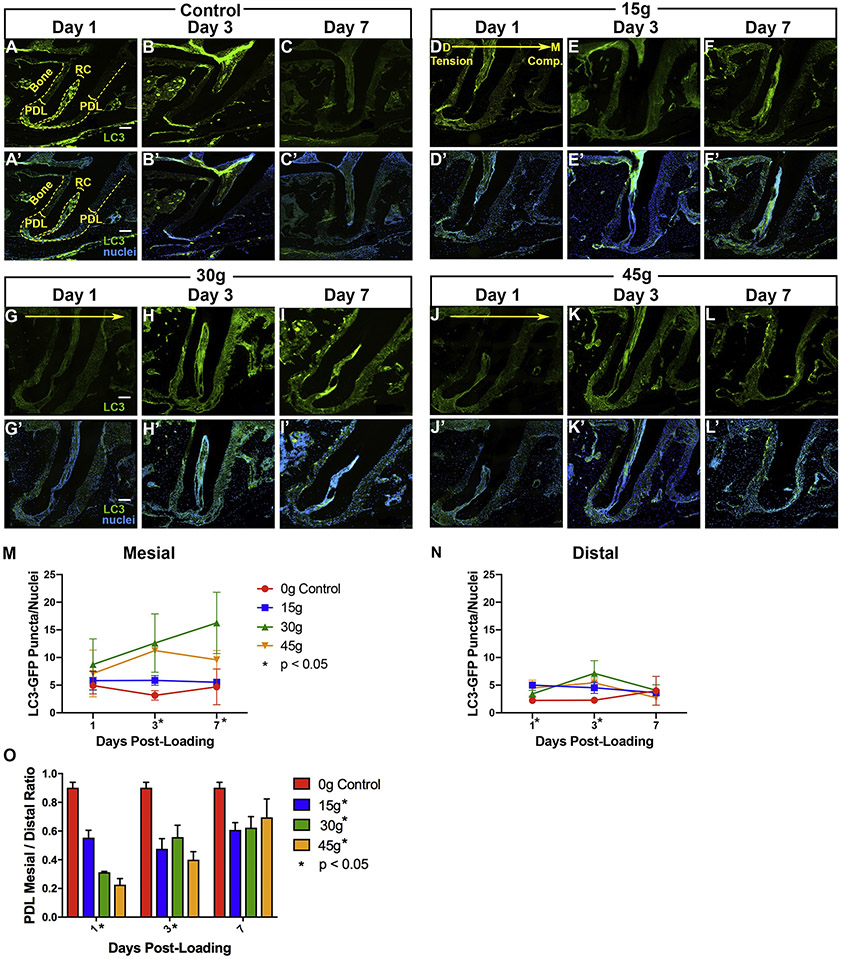
For PDL width, measurements were taken using NIS-Element Basic Research program at 20X magnification on fluorescent and brightfield images of the distal molar root (Fig 2O). Ten measurements each were taken on the mesial and distal aspects of the mid-root (20 measurements total), from the outer surface of the root to the innermost surface of the alveolar bone, along the shortest perpendicular path. Measurements were averaged and then divided to give the ratio of mesial (compression) to distal (tension) mean PDL width (n=51 total with 5-6 animals per time point [t=1, 3, and 7 days post-loading] at each force level [0g control, 15g, 30g, or 45g]).
Figure 2. Orthodontic loading activates autophagy but varies by force level.
[A-L’] Nikon fluorescent microscope imaged sections of first molar distal roots from GFP-LC3 mice post-loading. [A-L] Green: GFP-LC3, Scale = 100 μm; [A’- L’] Green: GFP-LC3; Blue: DAPI nuclei, Scale = 100 μm. [D-L’] Experimental: Mesial, compression- right. Distal, tension- left. [D, G, J] Large yellow arrow indicates direction of force application with compression/mesial on the right and tension/distal on the left. PDL: Periodontal ligament. RC: root canal. Bone: alveolar bone. Yellow dashed lines- the outer lines the edge of the alveolar bone and the inner lines the root canal. [M-N] Quantification of autophagosome puncta / DAPI nuclei versus days post-loading in PDL of control (0g) and loaded (15g, 30g, 45g) molars on the mesial (M) and distal (N) sides of the root. Fluorescent puncta were quantified in a uniform (300l x 300 ul) area. Statistical significance (p< 0.05) is indicated by an asterisk next to the timepoint, when there were significant differences between all groups at that time point, and by an asterisk next to the force level, when there were significant trends over time within a force group. Graphs display the mean and SEM. Convention: * p<0.05. [O] Quantification of PDL dimension mesial / distal ratio versus days post-loading of control (0g) and loaded (15g, 30g, 45g) molars.
Histology and Imaging
Adult mice were sacrificed with CO2 asphyxiation and surgical dislocation. Tissue preparation, cryosectioning, staining (DAPI, tartrate-resistant acid phosphatase (TRAP) or Mayer/Harris Hematoxylin) and slide loading were conducted as described (Fig 2, 4).9,23,24 Images of sectioned molar roots were collected on a Nikon Eclipse Ti-U inverted microscope, with a 20x objective, and on a Zeiss LSM 710 Laser Scanning Confocal microscope retrofitted with an oil-immersion 40x PlanApo objective, with uniform imaging conditions (Figs 2, 4-6, Sup Fig 1).23 Images were analyzed using NIS-Element Basic Research Imaging software (Nikon Instruments, Melville, NY, USA), ImageJ (NIH, Bethesda, MD, USA), and Imaris (Bitplane, Windsor, CT, USA). A trained technician reviewed each image at a uniform magnification and quantified intracellular puncta and nuclei in a fixed area, as described.23,27
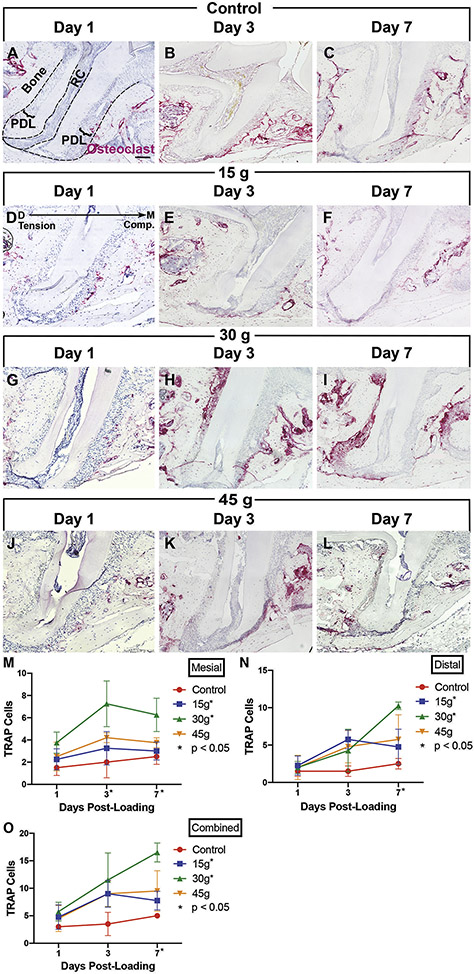
Figure 4. Osteoclasts increase after orthodontic loading, but numbers are suppressed with insufficient or excessive force.
[A-L] TRAP-stained sections. [A-C] Control: no loading. [D-L] Experimental: loaded at time 0, sacrificed and imaged at Days 1, 3 and 7, 15g [D-F], 30g [G-I], 45g [J-L]. PDL: Periodontal ligament. RC: root canal. Bone: alveolar bone. Black dashed lines- the outer lines the edge of the alveolar bone and the inner lines the root canal. Scale = 100 μm. Mesial, compression: right. Distal, tension: left. [M-O] Quantification of TRAP+ cells versus days post-loading on the mesial aspect (M), distal aspect (N), and combined mesial and distal regions (O). Statistical significance (p< 0.05) is indicated by an asterisk next to the timepoint, when there were significant differences between all groups at that time point, and by an asterisk next to the force level, when there were significant trends over time within a force group. Graphs display the mean and SEM. Convention: * p<0.05.
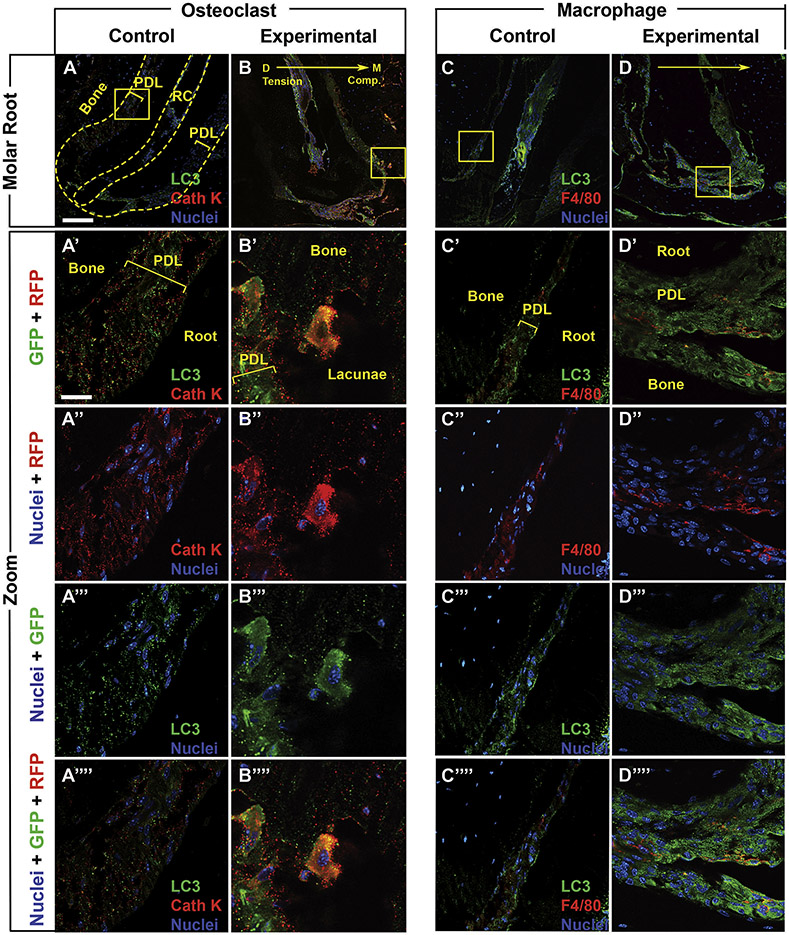
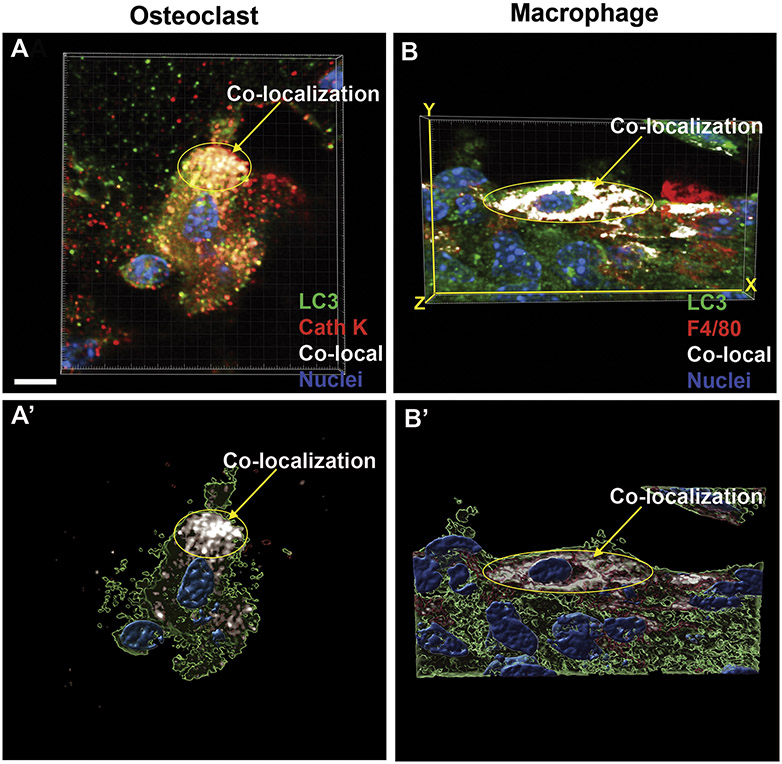
Figure 6: 3D reconstruction demonstrates co-localization of autophagy, macrophage and osteoclasts markers.
3D reconstructions of confocal Z-stacks of peri-dental cells from the first molar distal root. GFP [A-B’], Cathepsin K [A, A’] and F4/80 [B, B’] immunolabeling were used to visualize LC3-GFP puncta, osteoclasts and macrophages, respectively. Experimental molars of LC3-GFP mice were loaded at time 0 with 30g of force and sacrificed at day 3, n=3 mice. LC3-GFP: green. Cathepsin K (Cath K) and F4/80: red with nuclear DAPI: blue. [A, A’] 3D reconstruction of an osteoclast demonstrating co-localization of red fluorescent signal (Cathepsin K of osteoclasts) and LC3-GFP puncta (autophagic activity). [A’] Confocal Z-stack illustrative projection for visualization of percent volume of fluorescence cellular co-localization. White regions demonstrate colocalization of Cathepsin K with LC3-GFP puncta. [B, B’] 3D reconstruction of a macrophage demonstrating co-localization of red fluorescence (F4/80 of macrophages) and LC3-GFP puncta (autophagic activity). [B’] Confocal Z-stack illustrative projection for visualization of percent volume of green co-localized with red fluorescence. White regions demonstrate colocalization of F4/80 with LC3-GFP puncta. Scale = 5 μm.
For cell-specific labeling, 10 μm frozen sections (from day 3, 30g-loaded experimental molars and 0g contralateral control molars, n=4 animals each) were permeabilized with 0.3% Triton-X-100 for one hour and blocked for an additional hour with 10% normal goat serum at room temperature.28 Sections were then incubated overnight at 4°C with a chicken anti-GFP antibody (1:500, Aves Lab, #GFP-1020, Tigard, OR, USA) along with one other primary antibody including: rabbit anti-Osterix (OSX) for osteoblasts (1:250, Abcam, ab209484, Cambridge, MA, USA), rat anti-F4/80 for macrophages (1:100, BioRad, MCA497, Hercules, CA, USA), and rabbit anti-Cathepsin K for osteoclasts (1:100, Abcam, ab19027) (Fig 5). Slides were then incubated for two hours at room temperature with AlexaFluor® 488 goat anti-chicken IgG (1:500, Lifetech #A11039, Waltham, MA) along with one other secondary antibody as follows: AlexaFluor® 594 goat anti-rabbit IgG(1:500, Lifetech #A11037) or AlexaFluor® 594 goat anti-rat IgG (1:500, Lifetech #A11007). Slides were coverslipped using Fluoro-Gel II with DAPI (a fluorescent stain that labels DNA for nuclear labeling, EMS, 17985-50, Hatfield, PA, USA) and imaged on a Zeiss LSM 710 Laser Scanning Confocal microscope. Image deconvolution was performed with AutoQuantX3 while Imaris (, USA) was used for image analysis and 3D cellular reconstruction according to the Herranz et al.29 Photoshop (Adobe) was used for figure assembly. Co-occurrence of elevated intracellular GFP puncta (autophagosomes) and cell-specific antibody label (RFP) were noted for identification of cell types exhibiting autophagic activity. Cellular 3D reconstructions displayed co-localization in white.
Figure 5: Cell type-specific autophagy activation is found in macrophages and osteoclasts after orthodontic loading.
Sections of the first molar distal root with GFP [A-D””], Cathepsin K [A-B””] and F4/80 [C-D””] immunolabeling, to visualize LC3-GFP, osteoclasts and macrophages, respectively. Anti-GFP: green. Anti-Cathepsin K (Cath K) and F4/80: red with nuclear DAPI: blue. PDL: Periodontal ligament. RC: root canal. Bone: alveolar bone. Yellow dashed lines- the outer lines the edge of the alveolar bone and the inner lines the root canal. [A, C] Control: no loading, n= 3 mice with 4-8 sections each. [B, D] Experimental: loaded at time 0 with 30g, sacrificed at day 3 and imaged, n=3 mice with 4-8 sections each. Mesial, compression: right. Distal, tension: left. Yellow arrow indicates direction of force application. Yellow box indicates area that is enlarged below in the zoomed in images to visualize individual cells. Scale = 100 μm. [A’-A””, C’-C””] Control: no loading. Pairs of fluorescent channels (A’-A’”, C’-C’”) followed by all fluorescent channels (A””, C””). [B’-B””, D’-D’”'] Experimental: loaded with 30g. Pairs of fluorescent channels (B’-B’”, D’-D’”) followed by all fluorescent channels (B””, D””), demonstrating co-localization of osteoclast and macrophage red fluorescence with green LC3 fluorescence. Scale =10 μm.
Quantitative Real-Time PCR (qRT-PCR)
RNA extraction and qRT-PCR were performed for each animal’s tissues individually, as described (n=5 at all time points and force levels).23 Primers were designed using Primer Express (Applied Biosystems, Foster City, CA, USA) and synthesized by Invitrogen (Table 1). Genes assayed include inflammatory cytokines (Il-1b, and Nuclear Factor of Activated T Cells-1 (NFATC1)), bone turnover markers (Osteocalcin (Ocn), Osterix (Osx), Osteoprotegerin (Opg), Receptor Activator of Nuclear Factor Kappa-B Ligand (RankL), Runt-related transcription factor (Runx2), and Matrix Metallopeptidase 9 (Mmp9)) and autophagy pathway genes (Becn1, LC3 and Autophagy Related 5 and 7 (Atg5 and Atg7)). Relative differences in gene expression between groups were determined from cycle time (Ct) values. The values were normalized to beta-2-microglobulin (B2M, an internal positive control) in the same sample (ΔCt) and expressed as fold-change over 0 g control (2−ΔΔCt) which is expressed as a fold change of 1 in all graphs.25
Table I:
Primers
| Pathway | Gene | Forward (5’ to 3’) | Reverse (5’ to 3’) |
|---|---|---|---|
| Autophagy | Atg5 | TGTGCTTCGAGATGTGTGGTT | GTCAAATAGCTGACTCTTGGCAA |
| Atg7 | GTTCGCCCCCTTTAATAGTGC | TGAACTCCAACGTCAAGCGG | |
| Becn1 | ATGGAGGGGTCTAAGGCGTC | TCCTCTCCTGAGTTAGCCTCT | |
| Lc3 | GACCGCTGTAAGGAGGTGC | CTTGACCAACTCGCTCATGTTA | |
| Il-1β | TTCAGGCAGGCAGTATCACTC | GAAGGTCCACGGGAAAGACAC | |
| Inflammation | Nfatc1 | GACCCGGAGTTCGACTTCG | TGACACTAGGGGACACATAACTG |
| Ocn | CCGGGAGCAGTGTGAGCTTA | AGGCGGTCTTCAAGCCATACT | |
| Bone Turnover | Opg | ACCCAGAAACTGGTCATCAGC | CTGCAATACACACACTCATCACT |
| Osx | CCTCTCGACCCGACTGCAGATC | AGCTGCAAGCTCTCTGTAACCATGAC | |
| Mmp9 | AATCTCTTCTAGAGACTGGGAAGGAG | AGCTGATTGACTAAAGTAGCTGGA | |
| RankL | CAGCATCGCTCTGTTCCTGTA | CTGCGTTTTCATGGAGTCTCA | |
| Runx2 | CCGCACGACAACCGCACCAT | CGCTCCGGCCCACAATCTC | |
| Control | B2m | TTCTGGTGCTTGTCTCACTGA | CAGTATGTTCGGCTTCCCATTC |
Statistical analysis
Data for each group was expressed as the mean ± standard error of the mean (SEM). Data were normally distributed, so comparison among different groups from different time points were analyzed by two-way Analysis of Variance (ANOVA), followed by effect and contrast test (p=0.05). For GFP+ puncta and TRAP osteoclast data (Figs 2, 4), repeated measures ANOVA was used to test whether the means of the experimental outcomes were different than the control outcome and if changes between time points were significant. The analysis was adjusted for repeated measurements from the same mouse using random effects to account for dependence between outcomes in the same mouse. For qPCR data (Fig 3), repeated measures ANOVA was used to test whether the mean of experimental outcome is different than 1 (control reference value) and if changes between time points were significant. Tukey post-hoc tests were used to evaluate pairwise differences.
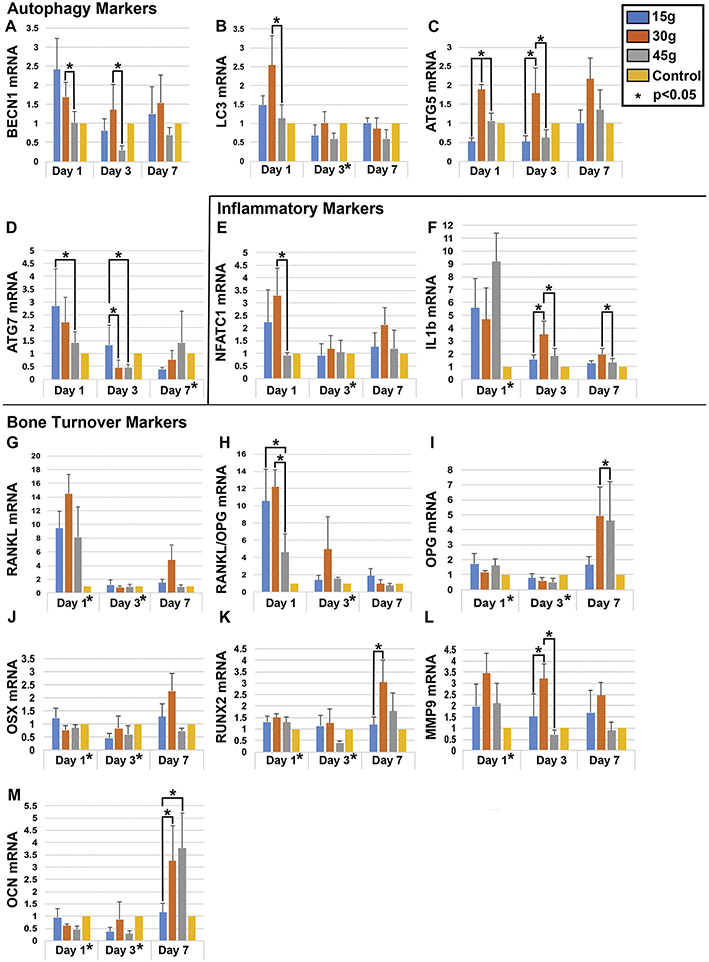
Figure 3. Expression qPCR analysis of autophagy, inflammatory and bone turnover markers.
Expression of autophagy [A-D; BECN1, LC3, ATG5, ATG7], inflammatory [E,F; NFATC1, IL-1β], and bone turnover markers [G-M; RANKL, RANKL/OPG, OPG, OSX, RUNX2, MMP9, OCN] increases in peri-dental tissues after orthodontic loading at time points 1, 3, and 7 days after loading but fold change varies with force level applied (n=3 each at each time point and force level, 0g control, 15g, 30g and 45g). Y axis displays fold change in mRNA level. Statistical significance (p< 0.05) is indicated by an asterisk next to the timepoint, when there were significant differences between all groups at that time point. Significant pairwise comparisons (p< 0.05) are noted with an asterisk and a bracket. Convention: * p<0.05.
Results
Autophagy is activated under orthodontic compression within a specific force-range
To track autophagic activity following orthodontic loading, we utilized a GFP-LC3 reporter mouse line and an established device placement system.23,30 Adult GFP-LC3 mice had a Nickel-Titanium (NiTi) spring bonded to their central incisors and first molars, applying 15g, 30g or 45g of force to orthodontically move teeth (Fig 1A-C). Molar displacement increased from days 1 to 7, with maximal displacement observed for 30g of loading (Fig 1H-L). The least movement was seen for 15g, suggesting it offers insufficient force, while 45g caused less displacement than 30g, suggesting it provides excessive force with slowed OTM. With a split mouth design, the contralateral control molar was not loaded (0g, no spring) and displayed no tooth movement (Fig 1A-C, D-G). OTM was seen at an intermediate force level of 30g.
Changes in the periodontal ligament (PDL) dimensions were tracked over time (Fig 2O). On day 1, load levels correlate directly with the amount of mesial compression relative to distal expansion under tension, with more force causing a smaller mesial PDL, a larger distal PDL and a smaller overall ratio. At day 3, however, the 30g load offered the greatest relative mesial to distal ratio, possibly with the onset of frontal resorption under compression.2 By day 7, 15g had the smallest ratio and 45g had the largest, suggesting that undermining resorption due to heavy force may have caused a delayed but rapid change in mesial PDL dimension.2
Autophagic activity was monitored. After loading, LC3-GFP puncta were visible by day 1 and increased over time, on the compression (mesial) side with 30g of force (Fig 2G-I’, M-N). However, there was less change in puncta on the tension (distal) side of 30g loaded molars and on both sides of the control (0g), 15g and 45g loaded molars (Fig 2A-L’, M-N). Results indicate that with a moderate force range, autophagy is induced after mechanical loading with tissue compression.
Autophagic activity correlates with bone turnover markers
To evaluate correlation between autophagy, inflammation and bone turnover, we examined mRNA expression of autophagy pathway genes (Atg5, Atg7, Becn1, LC3), inflammatory cytokines (Il-1β and NFATC1), and bone turnover markers (Ocn, Osx, Opg, Rankl, Runx2, and Mmp9) in peri-dental tissues after force loading by qRT-PCR (Fig 3). Expression of autophagy markers Becn1, LC3 and Atg7 peaked early with 30g of force on day 1 (Fig 3A-D). Atg5 peaked later on day 7, with little variation between days (Fig 3C). Moderate force (30g) was associated with significantly higher levels of autophagy marker expression (days 1 & 3) compared to other force levels (Fig 3A-D). Inflammatory markers NFATC1 and IL-1β peaked early at day 1 and then decreased with all load levels (Fig 3E-F). Moderate 30g force was associated with an early peak followed by more sustained expression, particularly for Il-1β. This is notable as release of cytokine IL-1β induces osteoclast differentiation, survival and resorption.31-33 Bone resorption genes Rankl and Mmp9 were upregulated after force loading, though Rankl had a more precipitous drop off on days 3 and 7 than Mmp9 which demonstrated continued expression with 30g of force (Fig 3G-H, L). Ocn, Osx, Opg, and Runx2 were all upregulated by day 7, most markedly with moderate 30g force (Fig 3I-K, M). Increased expression of these genes is suggestive of bone formation. Osteoblast-secreted OCN and OPG are associated with bone deposition; OPG binds RANKL, inhibiting osteoclast differentiation and bone resorption while favoring apposition.32,34 OSX and RUNX2 stimulate osteoblast differentiation and bone formation.35
The time course of expression indicates that autophagy activation, inflammatory and bone resorptive genes are upregulated shortly after loading by day 1, while a week later, bone formation genes are upregulated. There appears to be an ordered sequence of events where shortly after force application, autophagy activation, inflammation and bone breakdown predominate, and by day 7, bone formation takes over with inflammation and autophagic activity subsiding. This sequence appears to function to the greatest degree at a moderate force level of 30g.
Bone turnover and osteoclast recruitment: Maximal osteoclasts after moderate loading
To visualize how cellular changes correlate with force level, orthodontically loaded molars were TRAP stained for detection of osteoclasts (Fig 4). Osteoclasts appear as early as day 3 on the compression (mesial) side, with the highest overall levels detected at day 7 with moderate 30g loading (distal and combined, Fig 4G-I, M-O). The highest force (45g) was also associated with osteoclast recruitment, but osteoclast numbers never reached the level of 30g and ingress was delayed, consistent with tissue necrosis and undermining resorption (Fig 4J-L, M-O). The lightest force (15g) also had reduced TRAP positive cells compared to 30g (Fig 4D-F, M-O). Minimal TRAP staining was found in controls (Fig 4A-C, M-O). Sufficient but appropriate force must be applied for efficient recruitment of osteoclasts for OTM.
Orthodontic loading activates autophagy in osteoclasts and macrophages
To determine which cell types exhibit autophagic activity in peri-dental tissues, sections of control and orthodontically-loaded molars from LC3-GFP mice were antibody labeled for cell-specific markers including macrophages (F4/80), osteoclasts (Cathepsin K), and osteoblasts (OSX), (Fig 5, 6, Supp Fig 1). Confocal imaging allowed for evaluation of colocalization of red fluorescent cell markers with elevated GFP puncta indicative of autophagy induction. We observed autophagy activity in osteoclasts and macrophages in peri-dental tissues of orthodontically-loaded molars (Figs 5B-B””, D-D””, 6, Sup Fig 1). Osteoclasts with autophagic puncta were primarily located on the mesial/compression side of the PDL and in boney lacunae adjacent to the mesial root, likely at sites of resorption (Fig 5B-B””, Sup Fig 1); these locations are consistent with the areas displaying TRAP staining (Fig 4G-I, M) and autophagy active cells (Fig 2G-I,M). Macrophages were concentrated around the root apex (Fig 5D-D””, Sup Fig 1).
To further confirm cellular colocalization of LC3 puncta (GFP) and cell-specific labels (RFP), we assembled three-dimensional reconstructions of osteoclasts (Fig 6A-A’) and macrophages (Fig 6B-B’). White regions represent areas of overlap between the cell-type (in red) and LC3 green fluorescent autophagic puncta, confirming macrophages and osteoclasts activate autophagy. No autophagy induction was found in osteoblasts (data not shown). Autophagy active osteoclasts and macrophages were missing from the peri-dental tissues of control molars, suggesting that force application is required for autophagy induction (Fig 5A-A””, C-C””, Sup Fig 1).
Discussion
Orthodontic loading is associated with activation of autophagy in a force-dependent and cell-specific manner. Orthodontic tooth movement (OTM) and histological changes observed in our assays are consistent with the literature, with molar displacement increasing over time.2,23,36 Loaded molars demonstrate significantly more GFP-LC3 punctate cells than controls, consistent with an increase in autophagic activity and supported by expression analysis of autophagy genes (Fig 3). GFP puncta enriched and TRAP positive osteoclasts are primarily on the mesial side of the root, suggesting autophagy may play more of a role in bone resorption with compression than deposition under tension (Fig 2, 3, 5). The discovery that macrophages and osteoclasts activate autophagy after loading is consistent with autophagy acting upstream of bone resorption (Fig 5, 6).37,38
Expression data (Fig 3) demonstrate a correlated increase in autophagy, inflammatory and bone resorption markers secondary to orthodontic loading and a later rise in markers of bone apposition. Due to the technical limitations of working with a miniscule mouse molar, we are unable to separately analyze compression and tension tissues to say how expression correlates with location around the root. However, the temporal sequence after loading begins with autophagy activation, inflammation and bone breakdown, while by day 7, bone formation takes over with inflammation and autophagic activity subsiding. This expression sequence was most apparent at a moderate force of 30g, with reduced gene expression associated with light (15g) and heavy (45g) loads. Our light force may be insufficient to elicit a response, and 45g may cause tissue damage altering the normal course of gene expression and autophagy’s regulation of inflammation and bone turnover. Autophagy protein levels were not evaluated in this investigation and could be a valuable next step.
The applied force range between high and low is somewhat narrow (15g- 45g), with all load values being relatively conservative. As a result, it is notable to see significant differences in amount of tooth movement (Fig 1), PDL dimensions (Fig 2O), autophagy activation (Fig 2), gene expression (Fig 3) and osteoclast recruitment (Fig 4). Excessive force causes the tooth to fully compress its PDL, occluding blood flow and causing sterile necrosis, undermining resorption, and slowed OTM, consistent with our observations from 45g samples.2,22 A lighter load allows reduced but continued blood flow with frontal resorption and efficient OTM, as seen with 30g. However, an insufficient load is not effective, as demonstrated by 15g data. The importance of sufficient but appropriate force translates directly to clinical practice, where insufficient and excessive force slows OTM with reduced osteoclast recruitment, autophagy induction and altered gene expression. It behooves the orthodontist to understand these biological mechanisms and therefore apply an appropriate load to maximize rate of OTM and minimize risk of adverse outcomes (like root resorption). 2
Optimal force varies with the species and type of tooth movement. Even within the root of a single tooth, the stress experienced by the tissues varies with anatomy and type of force applied. Published ranges in humans fall between 110-130g for bodily translation and between 28-33g for tipping.24 Consistent with previous reports, ideal force in mice is 30-35g for translation.24 Force levels tested here were similar to those applied during rat OTM assays (force range: 10-50g), where odontoclast-mediated orthodontic root resorption and apoptosis occurred following days of heavy force.39,40 RANKL and TRAP positive cells increased along the root, consistent with our data.39,40 Because autophagy increases the stress threshold needed to induce cell death, (thereby reducing apoptosis,) and apoptosis can in turn inhibit autophagy, a future direction would include looking at autophagy induction, apoptosis and osteoclast/odontoclast recruitment in wild-type and autophagy loss-of-function models, to further elucidate mechanisms of orthodontic root resorption.41
Furthermore, findings suggest that autophagy activation in osteoclasts and macrophages may be a force-dependent step towards bone resorption and OTM; unloaded molars displayed no increase in autophagy activation (Fig 5, 6, Sup Fig 1). Macrophages and osteoclasts are both derived from the macrophage/monocyte hematopoietic cell lineage; a common precursor differentiates into macrophages in the presence of Macrophage-Colony Stimulating Factor (M-CSF) while osteoclasts form in the presence of M-CSF and RANKL.42 Conversely, osteoblasts, which arise from mesenchymal stem cells (MSCs), showed no autophagic activity in our OTM model, unlike in the Nollet et al. study.21,42 This suggests that the monocyte lineage may be particularly tied to induction of autophagy and bone turnover in an orthodontic context. Apart from osteoclasts and macrophages, it is possible that other cell types in the PDL are activating autophagy after loading; performing a more extensive screen of cell-specific labels could be a fruitful follow-up inquiry.
Autophagy activity in the macrophage/monocyte lineage of the mouth is consistent with published literature (Fig 5, 6). Macrophages and osteoclasts demonstrate autophagy in other settings, as they are digestive, phagocytic cells. Macrophages induce autophagy as innate immune cells, where they engulf foreign debris and pathogens for protection and antigen-presentation.37,43,44 Essential autophagy proteins Atg5, Atg7, and LC3, are important for generating the osteoclast ruffled border and bone resorption in vivo and are also upregulated with orthodontic loading in our study (Fig 3A-D).44
Macrophages and osteoclasts are notable players in autoimmune conditions, making data from this oral context more generalizable. Macrophages function in both induction and suppression of autoimmune responses, with pathology developing from dysregulation when macrophages fail to support repair and homeostatic restoration.37 Uncontrolled inflammation is linked to reduced bone production by osteoblasts and cartilage and bone degradation by osteoclasts, with net loss of bone mass in Rheumatoid arthritis, Type 1 Diabetes mellitus, autoimmune thyroid diseases, and Systemic Lupus Erythematosus (SLE).38 Similarly, chronic inflammation leads to loss of attachment and supporting bone in periodontal disease. All of these conditions have been linked to aberrant autophagy.14,45,46 One hypothesis is that autophagy functions in peri-dental macrophages and osteoclasts to regulate bone resorption and timely resolution of orthodontically-induced inflammation. Failure of these mechanisms could underlie orthodontically-induced apical root resorption. Future analyses are needed to study mechanisms of autophagy’s role in OTM, regulation of inflammation and bone turnover, including loss of function, gain of function, cell-specific and molecular inquiries. Identifying autophagy’s roles during OTM holds potential for improved understanding of bone remodeling and oral inflammatory disease.47,48
Conclusions
Orthodontic loading activates autophagy in a force-dependent and cell type-specific manner.
Autophagy is induced in peri-dental osteoclasts and macrophages by orthodontic loading.
Autophagic activity, gene expression and osteoclast recruitment were correlated with load.
Autophagy may play a role in regulating bone turnover needed for orthodontic tooth movement.
Supplementary Material
Highlights.
Orthodontic loading activates autophagy in a force-dependent manner.
Autophagy is induced in peri-dental osteoclasts and macrophages.
Autophagic activity, gene expression and osteoclast recruitment were correlated with load.
Autophagy may play a role in regulating bone turnover of orthodontic tooth movement.
Acknowledgements
We thank Dr. Pablo Ariel, Ms. Victoria Madden and the excellent staff of the UNC Microscopy Services Laboratory for their guidance and instruction; J. Ashley Ezzell of the UNC Histology Research Core Facility in the Department of Cell Biology and Physiology for histology services and expertise; and we acknowledge John Whitley, Peng Xue and other members of the Ko lab for their support and assistance.
Funding
This work was supported by the National Institute of Dental and Craniofacial Research (NIDCR) at the National Institutes of Health [grant #R01DE022816 to C.K.] and the UNC Hale Professorship fund [to C.K.]. This work was also supported by the NIEHS [#1ZIAES10328601 to J.M.] and the American Association of Orthodontics Foundation (AAOF) Martin ‘Bud’ Schulman Postdoctoral Fellowship [to L.J.] Y.L. and L.J. were supported by the Graduate School Masters Merit Assistantship for study in Dentistry awarded by the UNC Graduate School, the Southern Association of Orthodontics (SAO) research awards, and the Masters Research Support Grants awarded by the Office of the Associate Dean for Research at UNC School of Dentistry and the Dental Foundation of North Carolina.
Footnotes
Declaration of Interests: The authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miles P. Accelerated orthodontic treatment - what’s the evidence? Aust. Dent. J 2017;62:63–70. [DOI] [PubMed] [Google Scholar]
- 2.Proffit WR, Fields HW, Larson B, Sarver DM. Contemporary Orthodontics. 6th ed. Elsevier Health Sciences; 2018. [Google Scholar]
- 3.Melsen B. Biological reaction of alveolar bone to orthodontic tooth movement. Angle Orthod. 1999. [DOI] [PubMed] [Google Scholar]
- 4.Ren Y, Maltha JC, Kuijpers-Jagtman AM. Optimum force magnitude for orthodontic tooth movement: A systematic literature review. Angle Orthod. 2003. [DOI] [PubMed] [Google Scholar]
- 5.Nakano T, Hotokezaka H, Hashimoto M, et al. Effects of different types of tooth movement and force magnitudes on the amount of tooth movement and root resorption in rats. Angle Orthod. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez J, Cunha LD, Park S, et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Yang Z, Goronzy JJ, Weyand CM. Autophagy in autoimmune disease. J. Mol. Med 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullon P, Cordero MD, Quiles JL, et al. Autophagy in periodontitis patients and gingival fibroblasts: Unraveling the link between chronic diseases and inflammation. BMC Med. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Netea-Maier RT, Plantinga TS, van de Veerdonk FL, Smit JW, Netea MG. Modulation of inflammation by autophagy: Consequences for human disease. Autophagy 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Yoshimori T, Ohsumi Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol 2011. [DOI] [PubMed] [Google Scholar]
- 11.Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 2008. [DOI] [PubMed] [Google Scholar]
- 12.Crișan TO, Plantinga TS, van de Veerdonk FL, et al. Inflammasome-independent modulation of cytokine response by autophagy in human cells. PLoS One 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris J. Autophagy and cytokines. Cytokine 2011. [DOI] [PubMed] [Google Scholar]
- 14.Memmert S, Nogueira AVB, Damanaki A, et al. Damage-regulated autophagy modulator 1 in oral inflammation and infection. Clin. Oral Investig 2018. [DOI] [PubMed] [Google Scholar]
- 15.Couve E, Osorio R, Schmachtenberg O. The amazing odontoblast: Activity, autophagy, and aging. J. Dent. Res 2013. [DOI] [PubMed] [Google Scholar]
- 16.Yang JW, Zhu LX, Yuan GH, et al. Autophagy appears during the development of the mouse lower first molar. Histochem. Cell Biol 2013. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Wan C, Nie S, et al. Localization of Beclin1 in mouse developing tooth germs: Possible implication of the interrelation between autophagy and apoptosis. J. Mol. Histol 2013. [DOI] [PubMed] [Google Scholar]
- 18.Zhuang H, Hu D, Singer D, et al. Local anesthetics induce autophagy in young permanent tooth pulp cells. Cell Death Discov. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin NY, Beyer C, Gießl A, et al. Autophagy regulates TNFα-mediated joint destruction in experimental arthritis. Ann. Rheum. Dis 2013. [DOI] [PubMed] [Google Scholar]
- 20.Sambandam Y, Townsend MT, Pierce JJ, et al. Microgravity control of autophagy modulates osteoclastogenesis. Bone 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nollet M, Santucci-Darmanin S, Breuil V, et al. Autophagy in osteoblasts is involved in mineralization and bone homeostasis. Autophagy 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Li D, Ma Z, et al. Defective autophagy in osteoblasts induces endoplasmic reticulum stress and causes remarkable bone loss. Autophagy 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y. Roles of Autophagy in Orthodontic Tooth Movement. Am. J. Orthod. Dentofac. Orthop 2021;(In pres). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de A Taddei SR, Moura AP, Andrade I, et al. Experimental model of tooth movement in mice: A standardized protocol for studying bone remodeling under compression and tensile strains. J. Biomech 2012. [DOI] [PubMed] [Google Scholar]
- 25.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In Vivo Analysis of Autophagy in Response to Nutrient Starvation Using Transgenic Mice Expressing a Fluorescent Autophagosome Marker. Mol. Biol. Cell 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizushima N. Chapter 2 Methods for Monitoring Autophagy Using GFP-LC3 Transgenic Mice. Methods Enzymol. 2009. [DOI] [PubMed] [Google Scholar]
- 27.García-Prat L, Martínez-Vicente M, Perdiguero E, et al. Autophagy maintains stemness by preventing senescence. Nature 2016. [DOI] [PubMed] [Google Scholar]
- 28.Jiang X, Kalajzic Z, Maye P, et al. Histological analysis of GFP expression in murine bone. J. Histochem. Cytochem 2005. [DOI] [PubMed] [Google Scholar]
- 29.Herranz M, Boyle MJ, Pardos F, Neves RC. Comparative myoanatomy of Echinoderes (Kinorhyncha): A comprehensive investigation by CLSM and 3D reconstruction. Front. Zool 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seifi M, Ezzati B, Saedi S, Hedayati M. The Effect of Ovariectomy and Orchiectomy on Orthodontic Tooth Movement and Root Resorption in Wistar Rats. J. Dent. (Shiraz, Iran) 2015. [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnan V, Davidovitch Z. Cellular, molecular, and tissue-level reactions to orthodontic force. Am. J. Orthod. Dentofac. Orthop 2006. [DOI] [PubMed] [Google Scholar]
- 32.Huang H, Williams RC, Kyrkanides S. Accelerated orthodontic tooth movement: Molecular mechanisms. Am. J. Orthod. Dentofac. Orthop 2014. [DOI] [PubMed] [Google Scholar]
- 33.Lee B. Force and Tooth Movement. Australas. Orthod. J 2007;23(3):155. [PubMed] [Google Scholar]
- 34.Li Y, Jacox LA, Little SH, Ko CC. Orthodontic tooth movement: The biology and clinical implications. Kaohsiung J. Med. Sci 2018. [DOI] [PubMed] [Google Scholar]
- 35.Nakashima K, De Crombrugghe B. Transcriptional mechanisms in osteoblast differentiation and bone formation. Trends Genet. 2003. [DOI] [PubMed] [Google Scholar]
- 36.Tyrovola JB, Odont XX. The “mechanostat Theory” of Frost and the OPG/RANKL/RANK System. J. Cell. Biochem 2015. [DOI] [PubMed] [Google Scholar]
- 37.Ushio A, Arakaki R, Yamada A, et al. Crucial roles of macrophages in the pathogenesis of autoimmune disease. World J. Immunol 2017. [Google Scholar]
- 38.Schett G, David JP. The multiple faces of autoimmune-mediated bone loss. Nat. Rev. Endocrinol 2010. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi M, Minato Y, Shimizu M, Kikuta J, Hikida T, Kasai K. Caspase-mediated Apoptosis by Compressive Force Induces RANKL in Cementoblasts . Int. J. Oral-Medical Sci 2018. [Google Scholar]
- 40.Minato Y, Yamaguchi M, Shimizu M, et al. Effect of caspases and RANKL induced by heavy force in orthodontic root resorption. Korean J. Orthod 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: The interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyamoto T, Suda T. Differentiation and function of monocyte/macrophage lineage cells and osteoblasts. Japanese J. Clin. Med 2005;63(9):1511–6. [PubMed] [Google Scholar]
- 43.Germic N, Frangez Z, Yousefi S, Simon HU. Regulation of the innate immune system by autophagy: monocytes, macrophages, dendritic cells and antigen presentation. Cell Death Differ. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeSelm CJ, Miller BC, Zou W, et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou XJ, Zhang H. Autophagy in immunity: Implications in etiology of autoimmune/autoinflammatory diseases. Autophagy 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng T, Xu C, Mao C, et al. Increased interleukin-23 in Hashimoto’s thyroiditis disease induces autophagy suppression and reactive oxygen species accumulation. Front. Immunol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan YQ, Zhang J, Zhou G. Autophagy and its implication in human oral diseases. Autophagy 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhuang H, Ali K, Ardu S, Tredwin C, Hu B. Autophagy in dental tissues: A double-edged sword. Cell Death Dis. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.