Abstract
Hepatitis G virus (HGV or GB-C virus) is a newly described virus that is closely related to hepatitis C virus (HCV). Based on sequence analysis and by evaluation of translational initiation codon preferences utilized during in vitro translation, HGV appears to have a truncated or absent core protein at the amino terminus of the HGV polyprotein. Consequently, the biophysical properties of HGV may be very different from those of HCV. To characterize HGV particle types, we evaluated plasma from chronically infected individuals with and without concomitant HCV infection by using sucrose gradient centrifugation, isopycnic banding in cesium chloride, and saline density flotation centrifugation. Similar to HCV, HGV particles included an extremely-low-density virion particle (1.07 to 1.09 g/ml) and a nucleocapsid of ∼1.18 g/ml. One major difference between the particle types was that HGV was consistently more stable in cesium chloride than HCV. Plasma samples from chronically HGV-infected individuals and controls were assessed by a synthetic peptide-based immunoassay to determine if they contained HGV antibody specific for a conserved region in the coding region upstream of the E1 protein. Chronically HGV-infected individuals contained antibody to the HGV core protein peptide, whereas no binding to a hepatitis A virus peptide control was observed. Competitive inhibition of binding to the HGV peptide confirmed the specificity of the assay. These data indicate that HGV has a nucleocapsid and that at least part of the putative core region of HGV is expressed in vivo.
Hepatitis G virus (HGV [also called GB-C virus]) is a newly described virus that has a genome organization similar to that of hepatitis C virus (HCV) (14, 19, 27). Although it was initially thought to be associated with acute, posttransfusion hepatitis in humans and tamarins (14) and has been reported in some individuals with fulminant non-A, non-B, non-C hepatitis (5, 9, 14, 35), subsequent clinical studies suggest that HGV does not cause chronic liver disease in humans (1, 2, 6, 11–13, 16, 17, 21, 33). Nevertheless, chronic viremia occurs in some infected individuals, and viral RNA has been detected in multiple plasma samples obtained from infected people over a 16-year period (16). Among highly transfused individuals who have been exposed to HGV, approximately 20% are viremic. This suggests that up to 80% of people with HGV infection are able to clear their viremia (20).
Like HCV, HGV contains a positive-sense, single-stranded RNA genome approximately 9.4 kb in length that encodes a single, long open reading frame (ORF) (14, 19, 27). Based on amino acid sequence homology between the two viruses, the predicted HGV polyprotein contains two putative envelope proteins (E1 and E2), an RNA helicase, a trypsin-like serine protease, and an RNA-dependent RNA polymerase (14, 19). One difference between HGV and HCV is the limited homology within the amino terminus of the HGV polyprotein and the HCV core protein. The putative HGV core protein appears truncated or even absent in different isolates (14, 19, 26). Also, there is a great deal of variability in the 5′ nontranslated region of HGV compared with that of HCV. Comparison of five different HGV isolates revealed that nucleotide substitutions are nearly equally distributed among the first, second, and third codon positions (19). This impartial distribution suggests that this region is unlikely to contain a gene that encodes a core protein. In addition, careful analysis of protein products translated in vitro indicated that translation was only initiated at the AUG codon located immediately upstream of the signal sequence of the putative E1 glycoprotein (6, 26).
These data have led to speculations that the biophysical structure of HGV may be very different from HCV or other flaviviruses, perhaps producing particles without a nucleocapsid (26). However, there is no precedent for this among currently identified RNA viruses. Alternatively, HGV could utilize a cellular protein or a protein encoded by another coexisting virus such as HCV, instead of encoding its own capsid protein. This would be analogous to delta hepatitis virus using hepatitis B virus surface antigen for its capsid (reviewed in references 6 and 31), but would be unique among flavi- and pestiviruses. Taken together, these findings suggest that the biophysical structure of HGV may be quite different from that of HCV and may be unique among animal RNA viruses.
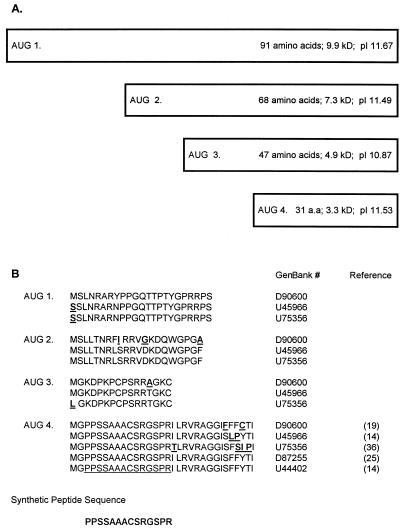
There are up to eight AUGs found upstream of the putative E1 protein in different HGV isolates, with as many as four of these in frame with the HGV polyprotein (1, 14, 19, 25, 36). Therefore, translation could potentially initiate at several sites upstream of the HGV E1 protein (Fig. 1). In this study, we characterized the sedimentation profiles of HGV and compared them with those of HCV. In addition, we evaluated patients with chronic HGV infection to determine if their serum contained antibody to a peptide representing a conserved region of the HGV ORF, 29 amino acids upstream of the putative E1 protein (Fig. 1B). These studies demonstrated that HGV particles shared many similarities with HCV particles, suggesting that a nucleocapsid was present. In addition, HGV-infected individuals produced antibodies that reacted with a peptide representing the putative core region, indicating that this protein is expressed in some humans with chronic HGV infection.
FIG. 1.
(A) Schematic diagram of potential HGV core proteins. The boxes represent the potential amino termini of the HGV polyprotein. All AUGs shown are in frame with the putative E1 protein for one HGV isolate (GenBank accession no. D90600) (19). The predicted number of amino acids, molecular mass, and pI are noted for each protein. Predicted molecular mass and pI were calculated by the Network Scientific Toolkit (DNA and Protein Analysis Toolkit, provided by The Rockefeller University). (B) The translated amino acid sequence starting with each AUG until the next AUG for one HGV isolate is shown on the top line (19). This sequence is compared with those of four additional HGV isolates below. The GenBank accession number and reference are shown in the right column. Nucleotide insertions created frameshifts for isolates D87255 and U44402 between the third and fourth AUGs of D90600. If these frameshifts did not occur, there would be greater than 90% amino acid homology for the 91-amino-acid “core” protein. The synthetic peptide sequence utilized in the immunoassay is shown at the bottom of the figure.
(This work was presented in part at the 91st Meeting of the American Society of Microbiology, 6 May 1997, Miami Beach, Fla.)
MATERIALS AND METHODS
Patients and specimens.
Patients with known HCV infection and chronic liver disease or chronic liver disease without HCV infection from the University of Iowa Liver Clinic, the University of Iowa General Clinical Research Center, or the California Pacific Medical Center were studied. All patients were evaluated by HCV enzyme immunoassay (EIA) 2.0 antibody tests (Abbott Laboratories, North Chicago, Ill.) and for HCV RNA by our laboratory method of reverse transcription (RT)-PCR (22, 23). Plasma samples were prepared from anticoagulated blood samples by centrifugation at 600 × g for 10 min and stored at −70°C prior to use. Additional specimens from HGV RNA-positive individuals, and hypogammaglobulinemic and agammaglobulinemic patients with HCV viremia were kindly provided by Harvey Alter (National Institutes of Health, Bethesda, Md.) and Christopher Wilson and Hans D. Ochs (University of Washington, Seattle), respectively. This study was approved by the Institutional Review Board, and informed consent was obtained from all subjects.
RNA extraction and RT-PCR.
The one-step guanidinium isothiocyanate RNA extraction method of Chomczynski and Sacchi (4) was used to isolate HGV and HCV RNAs from patient plasma (200 μl) and gradient fractions (100 μl) as previously described (8, 22). Previously described oligonucleotide primers from the HCV 5′ nontranslated region were used for HCV PCR (22, 24). Nested RT-PCR was performed for HGV with the following primers from the 5′ nontranslated region (14): outer sense, AAGCCCCAGAAACCGACGCC; antisense, TGAAGGGCGACGTGGACCGT; and inner sense, CGGCCAAAAGGTGGTGGATG; antisense, GTAACGGGCTCGGTTTAACG. RT was performed at 41°C for 1 h, followed by 35 cycles of PCR. Two microliters of the first round was amplified by nested PCR. DNA products (HCV, 250 bp; HGV, 220 bp) were detected with ethidium bromide following electrophoresis on a 1.5% agarose gel.
Gradient centrifugation.
Individual plasma samples from patients with HCV-HGV coinfection or with isolated HGV infection were fractionated by sucrose density gradient centrifugation, isopycnic banding in cesium chloride (CsCl), and differential flotation in saline. For sucrose gradient equilibrium centrifugation, 500-μl diluted samples were layered onto 10.5 ml of a 20 to 60% sucrose-preformed gradient, and centrifugation was performed with a Beckman SW41 rotor at 156,000 × g for 16 h at 4°C. Fractions (750 μl each) were collected and evaluated for HCV or HGV RNA (10). For isopycnic banding in cesium, 10 μl of plasma was mixed with CsCl (final concentration, 1.35 g/ml) and centrifuged in a Beckman SW41 rotor (200,000 × g, 72 h, 6°C), and 12 fractions were collected (420 μl each) (28). Where noted, plasma samples were twice extracted by equal volumes of chloroform prior to application to sucrose or CsCl gradients (10, 29).
Saline differential density flotation was performed as described by Hijikata et al. (10). Briefly, 300 μl of undiluted plasma was mixed with 8 ml of NaCl solution (1.063 g/ml), and centrifugation was carried out in a Beckman SW41 rotor (139,500 × g, 22 h, 14°C) as previously described (8, 10). The top 1 ml, middle 6 ml, and bottom 1 ml were collected for analysis. One hundred microliters from each fraction was subsequently analyzed in sucrose or cesium chloride gradients as described above.
Detection of anti-HGV core peptide IgG.
A synthetic peptide was synthesized which represented a highly conserved 14-amino-acid region beginning 29 amino acids upstream of the predicted start of the HGV E1 protein and in frame with the HGV ORF (Macromolecular Synthesis Facility, Michigan State University, East Lansing, Mich.). This peptide (PPSSAACSRGSPR) was selected on the basis of its hydrophilicity and conservation among reported HGV isolates (14, 19, 25, 36). The location of this sequence is depicted in Fig. 1B. Following conjugation to bovine serum albumin (BSA), a range of concentrations of peptide were applied to 96-well enzyme-linked immunosorbent assay (ELISA) microtiter plates (Costar, Cambridge, Mass.) in 0.025 M carbonate buffer (pH 9.6) for 4 h at 37°C (30). A peptide representing hepatitis A virus (HAV) VP3 (1C) protein (also conjugated to BSA) was applied to separate wells to serve as the control peptide (28). Following washing, HGV RNA-positive and HGV RNA-negative patient sera (diluted 1:5 to 1:500 in phosphate-buffered saline) were applied to the wells and the mixture was incubated overnight at 4°C. In addition, sera from normal healthy HCV antibody-negative individuals without HCV or HGV RNA detected in their plasma served as control sera. The wells were washed, and alkaline phosphatase-conjugated goat anti-human immunoglobulin G (IgG) (Sigma, St. Louis, Mo.) was added for 1 h at 37°C. The wells were washed again, and substrate was added (1 mg of p-nitrophenyl phosphate per ml in diethanolamine buffer [Sigma Chemicals]). The reaction was stopped with 3.5 M HCl after 15 to 30 min, and the A405 was measured by an ELISA reader (model EL-309; Microtek, Winooski, Vt.). Results were expressed as the S/N ratio (mean sample absorbance minus the background absorbance [S] divided by the background absorbance [N]). The background was determined by measuring the absorbance in wells not coated with peptide to which each serum sample was added. To further confirm the specificity of this peptide-based immunoassay, competition with either the HGV peptide or the HAV peptide (both conjugated to BSA) was evaluated. Briefly, wells were coated with the HGV-BSA conjugate (1 μg/well) as described above. Serum samples were diluted in buffer containing 0.001, 0.01, or 0.1 μg of either the HAV peptide or the HGV peptide and subsequently were applied to the HGV peptide-coated wells. Following overnight incubation, wells were washed, and human IgG was detected as described above.
RESULTS
Although there has been a great deal of information published regarding HGV genome sequence and organization (1, 14, 19), biophysical characterization of the particle types found in the plasma of HGV-infected individuals has not been previously described. Because HGV infection frequently coexists with HCV infection, direct comparison of the particle types of these two agents was possible by evaluation of plasma obtained from HCV-HGV-coinfected individuals. In addition, HGV RNA-positive samples were evaluated from individuals without HCV infection (both HCV RNA and antibody negative) to ensure that the presence of HCV infection did not alter HGV buoyant density. Three samples from patients with HGV infection without HCV infection were analyzed by sucrose, CsCl, and saline gradients, and five samples from HGV-HCV-coinfected patients were similarly analyzed. A total of 30 gradients were evaluated. There were no differences between the HGV buoyant densities detected in individuals with isolated HGV infection and HGV-HCV coinfection (data not shown). To allow direct comparison between HGV and HCV particles, all of the data presented below represent HGV and HCV particle types found in HGV-HCV coinfection.
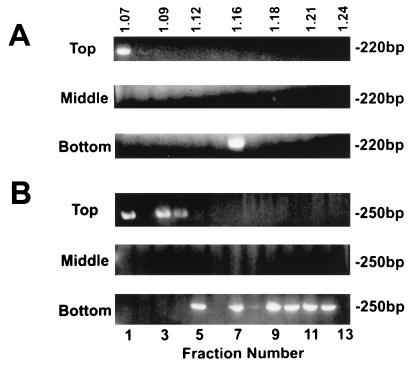
Plasma from HCV-HGV-coinfected individuals was fractionated by equilibrium centrifugation on sucrose gradients (Fig. 2). RNA was extracted from each fraction, and HGV or HCV RNA was detected by RT-PCR from the same gradient fractions. Two HGV particle types were identified in this experiment with densities of 1.07 and 1.17 g/ml (Fig. 2A). In different experiments, the density of the low-density particles ranged from 1.07 to 1.09 g/ml, and among samples tested, there were always more HGV RNA-containing fractions in the more rapidly sedimenting viral peak. Similarly, two HCV particle types were found with buoyant densities of 1.09 and 1.17 to 1.21 g/ml (Fig. 2B). The interexperiment range of densities for the low-density particles was 1.07 to 1.10 g/ml. The extremely-low-density and intermediate-density particle types have previously been described for HCV (3, 18), although not for HGV. Previous studies have shown that the very-low-density HCV particle is associated with infectivity (3, 10), and this particle is thought to represent the complete virion. The more dense species has been termed both a nucleocapsid (18, 32) and a virus-immune complex (HCV-IgG) (10). The range of particles seen in HCV has also been attributed to HCV binding lipoproteins and Igs (34).
FIG. 2.
Plasma from an HGV (A)- and HCV (B)-coinfected individual was fractionated on a 20 to 60% (wt/wt) sucrose equilibrium gradient. The sucrose density of each fraction is shown on top in grams per milliliter. RNA was extracted from each fraction, and viral RNA was detected by RT-PCR. HGV-specific products (250 bp) and HCV-specific products (220 bp) demonstrated that both HGV and HCV consisted of two particle types, an extremely-low-density peak (1.07 and 1.09 g/ml, respectively) and an intermediate-density peak. Following two extractions of plasma with an equal volume of chloroform, a shift to a higher density was demonstrated for both HGV and HCV.
Chloroform (CHCl3) was previously shown to cause HCV particles to shift to a higher buoyant density (10). Furthermore, CHCl3 extraction abolished HCV infectivity (7), presumably by removing the HCV lipid envelope. Following CHCl3 extraction, HGV RNA-containing fractions were detected with buoyant densities of 1.22 to 1.24 g/ml, which presumably represented nucleocapsids (18, 32) or free RNA (Fig. 2A). Similarly, HCV particle detection shifted to 1.19 and 1.22 to 1.24 g/ml following CHCl3 treatment, consistent with extraction of the virus envelope (Fig. 2B). Incubation of the fractions with RNase (0.5 mg/ml for 1 h) prior to RNA extraction revealed that only the RNA associated with the 1.22- to 1.24-g/ml density fractions was RNase sensitive (Table 1). In addition to shifting the buoyant density of HGV and HCV particles, CHCl3 treatment reproducibly decreased the number of fractions containing HCV RNA, perhaps by increasing the susceptibility of these particles to RNase.
TABLE 1.
RNase sensitivity of HCV and HGV particles
| Gradient | Virus | Chloroform treatment | Fraction characteristica
|
||
|---|---|---|---|---|---|
| Light | Intermediate | Heavy | |||
| Sucrose | HGV | No | R | R | NA |
| Yes | NA | NA | S | ||
| HCV | No | R | R | NA | |
| Yes | NA | NA | S | ||
| Cesium chloride | HGV | No | R | R | S |
| Yes | NA | NA | S | ||
| HGV RNAb | No | NA | NA | S | |
| HCV | No | NA | R | NA | |
| Yes | NA | NA | S | ||
| HCV RNAb | No | NA | NA | S | |
R, RNase resistant; S, RNase sensitive; NA, not applicable (no viral peak identified).
HGV or HCV RNA applied to cesium gradient instead of plasma.
Plasma samples containing both HCV and HGV were also analyzed by isopycnic banding in cesium chloride (CsCl). As described above, HGV or HCV RNA was detected by RT-PCR. Two peaks of HGV RNA were detected with buoyant densities of 1.07 g/ml (fraction 1) and 1.12 to 1.16 g/ml (Fig. 3A). In different experiments, the range of these intermediate particles was from 1.12 to 1.20 g/ml (data not shown). The same fractions contained only a single HCV particle with a buoyant density of 1.20 g/ml (Fig. 3B). Thus, it appeared that HGV virions were more stable in CsCl than HCV.
FIG. 3.
Distribution of HGV (A) and HCV (B) after isopycnic banding in cesium chloride with and without chloroform extraction. To determine if the higher-density peak detected following CHCl3 extraction represented nucleocapsids or free RNA, plasma RNA was prepared and banded in parallel gradients. The density is shown at the top in grams per milliliter.
CHCl3 treatment of plasma prior to CsCl centrifugation resulted in a shift of both HGV and HCV particles to 1.32 to 1.34 g/ml (Fig. 3). To determine if this higher-density RNA was associated with a nucleocapsid or represented free RNA, GITC-extracted, phenol-chloroform-purified HCV RNA and HGV RNA were individually mixed with CsCl and isopycnically banded. Tubes were treated with RNasin, and care was taken to avoid RNase. The density of HCV and HGV RNA were identical to the RNA detected in chloroform-treated plasma (1.34 g/ml [Fig. 3]), thus the combined effects of CHCl3 extraction and CsCl appeared to extract both envelopes and nucleocapsids. RNA and the “heavy” particles were only found in the bottom, or next to the bottom gradient fraction. Since fractions were collected from the top of the gradient, it is likely that there was some mixing of these two fractions, and the density of the RNA was greater than that measured. As with the sucrose gradient particles, only the RNA associated with the chloroform-extracted, high-density material was RNase sensitive (Table 1).
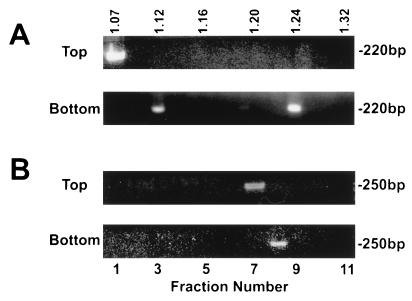
Because CsCl is harsh and can disrupt the viral envelope, HGV and HCV were fractionated by saline flotation sedimentation (1.063 g of NaCl/ml) as described by Hijikata et al. (8, 10). For HCV, the top 1 ml of the gradient was thought to represent HCV virions (putative antibody-free virus), and this fraction correlated with viral infectivity (10). The bottom 1 ml was thought to represent HCV-IgG immune complexes (10) because it was associated with lower infectivity, and HCV in this fraction was immunoprecipitated with anti-human IgG, IgA, and IgM (10). HGV RNA was detected in both the top and bottom fractions (but not the middle) of the saline gradients (data not shown). To characterize these fractions further, 100-μl aliquots of the top and bottom of the saline gradients were applied to 20 to 60% sucrose gradients, and their buoyant densities were determined. HGV particles present in the top of the saline gradients had a buoyant density of 1.07 g/ml (fraction 1, Fig. 4A). No viral RNA was detected from the middle of the saline gradient, and HGV RNA from the bottom fraction was detected at a buoyant density of 1.16 g/ml. HCV RNA was also detected in the top and bottom fractions of the saline gradients, but not the middle fraction (data not shown). When these fractions were applied to sucrose gradients, HCV RNA was detected in fractions with densities of 1.07 and 1.09 to 1.11 g/ml (fractions 1 and 3 and 4, respectively, Fig. 4B). The HCV particles in the bottom fraction of the saline gradient separated at a variety of densities between 1.12 and 1.22 g/ml (Fig. 4B).
FIG. 4.
Plasma from an HGV (A)- and HCV (B)-coinfected individual was fractionated by saline flotation density gradients as previously described (8, 10). The top, middle, and bottom fractions were subsequently fractionated on 20 to 60% sucrose gradients. HGV separated into two distinct peaks (A), whereas HCV demonstrated a mixture of particle types (B). The density is shown at the top in grams per milliliter.
The top and bottom fractions produced by saline flotation density gradient centrifugation were also evaluated by isopycnic banding in CsCl. The top fraction of the saline gradients contained HGV particles with a buoyant density of 1.07 g/ml in CsCl (fraction 1, Fig. 5A), whereas the bottom of the saline gradient had two particle types with buoyant densities of 1.12 and 1.24 g/ml, respectively (Fig. 5A). HCV particles present in the top buoyant densities of 1.12 and 1.24 g/ml, respectively (Fig. 5A). HCV particles present in the top fraction of the saline gradient had a buoyant density of 1.20 to 1.22 g/ml (Fig. 5B), and the bottom fraction contained HCV with a buoyant density of 1.24 g/ml (Fig. 5B).
FIG. 5.
HGV (A) and HCV (B) particles present in the top and bottom fractions of saline flotation density gradients were isopycnically banded in cesium chloride. As in Fig. 3, HGV appeared more stable in cesium chloride than HCV. The density is shown at the top in grams per milliliter.
Because the intermediate particle type (1.18 g/ml) found in HCV-infected patient plasma has been called both a nucleocapsid (18, 32) and an immune complex (10), we evaluated plasma from four HCV-infected patients who have either common variable immunodeficiency or X-linked agammaglobulinemia. Figure 6 demonstrates the particle types present in one of the agammaglobulinemic patients, who was representative of the four patients. Prior to Ig therapy, this patient lacked IgG, IgM, and IgA antibodies. Because the patient receives regular intravenous IgG therapy, the plasma sample studied did have IgG detected (769 g/dl); however, no IgM or IgA antibody was detected. It is noteworthy that since 1991, intravenous IgG preparations are screened for HCV antibodies, and thus this patient has not received anti-HCV antibody (against antigens tested for by the commercial EIA 2.0 assay). Consequently, this patient has no HCV-specific IgG or rheumatoid factor in his plasma and should be incapable of producing HCV-IgG immune complexes. This plasma was fractionated on a 20 to 60% sucrose gradient, and two particle types were identified with buoyant densities of 1.09 to 1.11 g/ml (fractions 5 and 6) and 1.17 g/ml (fraction 12) (Fig. 6).
FIG. 6.
Evaluation of HCV particles present in the plasma of an HCV-infected agammaglobulinemia patient. Two distinct particle types (1.09 to 1.11 g/ml and 1.17 g/ml) were identified. The density is shown at the top in grams per milliliter.
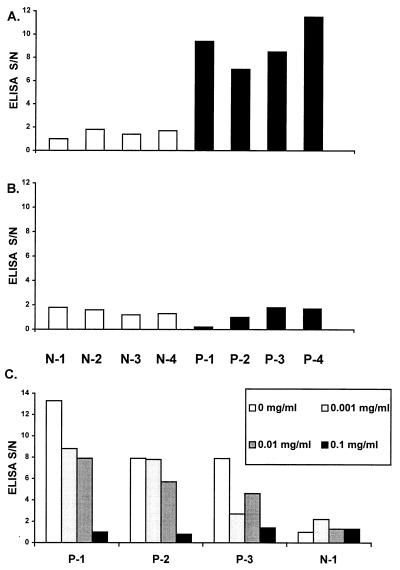
Although the studies described above suggest that HGV particles are very similar to HCV and that they have an extremely-low-density particle type which is quite different from those of most other viruses, the studies do not indicate the composition of the nucleocapsid. Although the putative HGV core protein described by Linnen et al. is much smaller than the HCV core protein, there are several similarities between this HGV coding region and flavivirus core proteins (15). The predicted protein is very basic (pI 11.67 [Fig. 1A]), and the predicted hydrophobicity profile shares structural patterns seen in several flaviviruses (15). To determine if the amino acids located upstream of the envelope region in the HGV ORF are expressed in vivo, plasma were evaluated for evidence of antibody directed against a conserved peptide coding region within the HGV ORF (Fig. 1B). This peptide (conjugated to BSA) was applied to wells of an ELISA plate as described in Materials and Methods. An unrelated HAV peptide conjugated to BSA served as the negative control (28). Four HGV RNA-negative serum samples from healthy donors with no history of blood transfusion or hepatitis (N-1 to N-4, negative control plasma), and four plasma samples from HGV RNA-positive patients with chronic liver disease (P-1 to P-4) were evaluated. Testing a range of antigen concentrations as the capture assay demonstrated that 1 μg per well was optimal for coating wells, and this concentration was subsequently used for both the HGV peptide and HAV peptide (data not shown). Figure 7A demonstrates that the negative control sera did not bind to the HGV peptide at a 1:100 dilution, whereas the HGV RNA-positive sera did. Figure 7B demonstrates that all eight serum samples failed to bind the HAV peptide at a 1:100 dilution. All four positive serum samples remained positive in the HGV peptide immunoassay at the 1:500 dilution.
FIG. 7.
Plasma samples (1:100 dilution) from four individuals without liver disease, who were negative for HGV RNA (N-1 to N-4), and four patients who had HGV RNA in their plasma and chronic liver disease (but who were HCV RNA and HCV antibody negative) were applied to wells of a 96-well ELISA plate coated with 1 μg of an HGV peptide representing the putative core protein (Fig. 1B) per well (A) or 1 μg of an HAV peptide (VP3 or 1C protein) per well (B) (28). Results are expressed as S/N (absorbance of sample − background absorbance [S]/background absorbance [N]). (C) Competitive inhibition of binding for three HGV RNA-positive samples (P-1, P-2, and P-3) and one negative (N-1) sample is shown. As the concentration of competing peptide was increased from 0 to 0.1 mg/ml, the S/N ratio decreased to background levels. The HAV 1C peptide did not demonstrate any competition with these plasma samples (data not shown).
To confirm the specificity of this binding, sera were incubated with three concentrations of the HGV or HAV peptide prior to application to the peptide-coated well, and the immunoassay was performed. Concentration-dependent inhibition of binding to the HGV peptide was demonstrated (Fig. 7C); however, when the same concentrations of the HAV-BSA peptide conjugate were added to the sera, no changes in the S/N ratio were seen (data not shown).
DISCUSSION
Our data are the first to demonstrate that HGV particles separate similarly to HCV particles in both 20- to -60% sucrose gradients and in saline flotation gradients, supporting the hypothesis that HGV contains both a very-low-density virion and a putative nucleocapsid. Although there were slight differences in the HGV low-density particles compared with the HCV particles (Fig. 2), both HGV and HCV have extremely-low-buoyant densities relative to other enveloped flaviviruses (3, 10, 18). It was previously demonstrated that HCV infectivity in chimpanzees correlated with the low-density fraction (3, 10) and the top fraction of saline flotation gradients (10).
The differences between HGV and HCV observed in CsCl gradients suggest that the very-light-density particles (putative HGV virion) and intermediate-density particles are relatively more stable in CsCl than HCV (Fig. 3 and 5). The fact that the buoyant densities of HGV particles did not change significantly in CsCl (compared with the sucrose density equilibrium gradients), indicates that these particles exclude CsCl, whereas the HCV particles did not. In addition, no light particle was observed for HCV, suggesting that the HCV envelope was not stable in 1.35 g of CsCl per ml.
Our data provide new information regarding the intermediate-density HCV particles, which have been called both nucleocapsids (18, 32) and immune complexes (10). The range of particle densities seen in HCV has also been attributed to HCV binding lipoproteins and Igs (34). CHCl3 extraction caused a shift in density for HCV as previously described (10), and a similar shift was observed for HGV (Fig. 2 and 3). However, when purified RNA was simultaneously run in parallel CsCl gradients, it was clear that the more dense particles generated by CHCl3 extraction had the same density as RNA. Thus, both the virion and intermediate particles are sensitive to CHCl3 extraction. The resistance of these fractions to RNase treatment further supports this thesis. In addition, fractionation of HCV particles from patients with agammaglobulinemia clearly demonstrated the light- and intermediate-density particles (Fig. 6). Since there was no IgM detected (thus no cryoglobulins or rheumatoid factor) and there was no detectable anti-HCV antibody in these samples, immune complexes without nucleocapsids could not account for these intermediate-density particles. Nevertheless, the wide distribution of particle densities of HCV in the sucrose gradients may indicate that there is a combination of nucleocapsids, immune complexes, viral lipoprotein, or viral aggregates (Fig. 2). Consistent with this hypothesis, we were unable to resolve HCV found in the bottom fraction of saline gradients into a single fraction, although this was not the case for HGV (Fig. 4).
Takahashi et al. (32) demonstrated that the core protein, located at the amino terminus of the HCV polyprotein, was present in HCV nucleocapsids. These nucleocapsids migrated with a buoyant density of 1.12 g/ml in potassium bromide gradients (32). The origin of the protein for the HGV nucleocapsid remains unknown. The potential core protein coding regions at the amino terminus of the HGV polyprotein are much smaller than those of HCV, with the largest possible HGV core protein coding region (in frame with the polyprotein) having a predicted molecular mass of 9.9 kDa (Fig. 1), compared with 22 kDa for HCV. Core proteins among different flaviviruses are not typically conserved and average only 11 kDa in size (15), and flavivirus core proteins are very basic and have similar hydrophilicity profiles (15). As demonstrated in Fig. 1B, all potential HGV core proteins are very basic, and the 91-amino-acid core protein of one isolate (19) shares many similarities with the hydrophilicity profile of flaviviruses (data not shown). Although previous studies have noted equal distributions of nucleotide mutations in the three codon positions within the putative HGV core region (19), evaluation of the predicted amino acid sequences resulting from using all three reading frames of the 5′ nontranslated region and comparing the most divergent isolates reveals that a single nucleotide insertion 43 amino acids into the putative core protein changes the observed reading frame (for the 91-amino-acid core [Fig. 1A]) (14, 19, 25). If this insertion were not present, 85 of the 91 amino acids would be identical, and thus the “core” protein in this otherwise divergent isolate would also be highly conserved.
The sequence of the gene encoding the synthetic peptide representing the putative HGV core protein used in the immunoassays is highly conserved among HGV isolates. The fact that HGV RNA-positive individuals contain antibodies that bind this peptide and do not bind a control peptide and the fact that the binding was competitively inhibited indicate that this peptide sequence is expressed in vivo (Fig. 7). Thus, the RNA sequence upstream of the E1 protein appears to be translated in vivo.
To our knowledge, these data represent the first characterization of HGV particle types and are the first evidence that the HGV ORF upstream of the putative E1 protein is expressed. The biophysical data indicate that HGV has a nucleocapsid, and the peptide immunoassay suggests that the amino terminus of the HGV polyprotein is expressed and therefore may represent the HGV core protein.
ACKNOWLEDGMENTS
We thank Harvey Alter (National Institutes of Health, Bethesda, Md.) for supplying HGV RNA-positive sera to serve as a positive control for our assay, Christopher Wilson and Hans D. Ochs (University of Washington, Seattle) for supplying HCV RNA-positive, HCV antibody-negative plasma from individuals with X-linked agammaglobulinemia and common variable immunodeficiency syndrome, Robert Cook for performing serum protein electrophoresis, Mary Jeane Perino-Phillips and Bobby Cheng for assistance in obtaining clinical specimens, and Naomi Erickson for assistance with manuscript preparation.
This work was supported by a Merit Review grant from the Veterans Administration (J.T.S.) and a grant from the National Blood Foundation (J.T.S.). W.N.S. is supported by NIH grant 1K08-AI01460. Patient care was provided in part by the GCRC Program in NCRR, NIH grant RR0059.
REFERENCES
- 1.Alter H J, Nakatsuji Y, Melpolder J, Wages J, Wesley R, Shih J W K, Kim J P. The incidence of transfusion-associated hepatitis G virus infection and its relation to liver disease. N Engl J Med. 1997;336:747–754. doi: 10.1056/NEJM199703133361102. [DOI] [PubMed] [Google Scholar]
- 2.Alter M J, Gallagher M, Morris T T, Moyer L A, Meeks E L, Krawczynski K, Kim J P, Margolis H S. Acute non-A-E hepatitis in the United States and the role of hepatitis G virus infection. N Engl J Med. 1997;336:741–746. doi: 10.1056/NEJM199703133361101. [DOI] [PubMed] [Google Scholar]
- 3.Bradley D, McCaustland K, Krawcyznski K, Spelbring J, Humphrey C, Cook E H. Buoyant density of the factor CIII-derived isolate in sucrose. J Med Virol. 1991;34:206–208. doi: 10.1002/jmv.1890340315. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Colombatto P, Randone A, Civitico G, Gorin J M, Dolci L, Medaina N, Olivieri F, Verme G, Marchiaro G, Pagni R, Karayiannis P, Thomas H C, Hess G, Bonino F, Brunetto M R. Hepatitis G virus RNA in the serum of patients with elevated gamma glutamyl transpeptidase and alkaline phosphatase: a specific liver disease. J Viral Hepatol. 1996;3:301–306. doi: 10.1111/j.1365-2893.1996.tb00102.x. [DOI] [PubMed] [Google Scholar]
- 6.Dickens T, Lemon S M. GB virus C, hepatitis G virus, or human orphan flavivirus? Hepatology. 1997;25:1285–1286. doi: 10.1002/hep.510250541. [DOI] [PubMed] [Google Scholar]
- 7.Feinstone S M, Mihalik K B, Kamimura T, Alter H J, London W T, Purcell R H. Inactivation of hepatitis B virus and non-A, non-B hepatitis by chloroform. Infect Immun. 1983;41:816–821. doi: 10.1128/iai.41.2.816-821.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han, J.-Q., W. N. Schmidt, P. Wu, P. Loh, G. Neil, D. R. LaBrecque, and J. T. Stapleton. Specific binding of hepatitis C virus to the Fc fragment of immunoglobulin molecules. In M. Rizzeto (ed.), Viral hepatitis and liver disease, in press.
- 9.Heringlake S, Osterkamp S, Trautwein C, Tillmann H L, Boker K, Muerhoff S, Mushahwar I K, Hunsmann G, Manns M P. Association between fulminant hepatic failure and a strain of GBV virus C. Lancet. 1996;348:1626–1629. doi: 10.1016/S0140-6736(96)04413-3. [DOI] [PubMed] [Google Scholar]
- 10.Hijikata M, Shimizu Y K, Kato H, Iwamoto A, Shih J W, Alter H J, Purcell R H, Yoshikura H. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J Virol. 1993;67:1953–1958. doi: 10.1128/jvi.67.4.1953-1958.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanda T, Yokosuka O, Ehata T, Maru Y, Imazeki F, Saisho H, Shiratori Y, Omata M. Detection of GBV-C RNA in patients with non-A-E fulminant hepatitis by reverse-transcription polymerase chain reaction. Hepatology. 1997;25:1261–1265. doi: 10.1002/hep.510250534. [DOI] [PubMed] [Google Scholar]
- 12.Kao K, Chen P, Chen D. GBV-C in the aetiology of fulminant hepatitis. Lancet. 1996;347:120–121. doi: 10.1016/s0140-6736(96)90244-5. [DOI] [PubMed] [Google Scholar]
- 13.Kuroki T, Nishiguchi S, Tanaka M, Enomoto M, Kobayashi K. Does GBV-C cause fulminant hepatitis in Japan? Lancet. 1996;347:908. doi: 10.1016/s0140-6736(96)91395-1. [DOI] [PubMed] [Google Scholar]
- 14.Linnen J, Wages J, Zhang-Keck Z-Y, Fry K E, Krawczynski K Z, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih J W-K, Piatak M, Hoover C, Fernandez J, Chen S, Zou J-C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 15.Mandl C W, Heinz F X, Kunz C. Sequence of the structural proteins of tick-borne encephalitis virus (Western subtype) and comparative analysis with other flaviviruses. Virology. 1988;166:197–205. doi: 10.1016/0042-6822(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 16.Masuko K, Mitsui T, Iwano K, Yamazaki C, Okuda K, Meguro T, Murayama N, Inoue T, Tsuda F, Okamoto H, Miyakawa Y, Mayumi M. High prevalence of GB virus C in patients on maintenance hemodialysis. N Engl J Med. 1996;334:1485–1490. doi: 10.1056/NEJM199606063342301. [DOI] [PubMed] [Google Scholar]
- 17.Miyakawa Y, Mayumi M. Hepatitis G virus—a true hepatitis virus or an accidental tourist? N Engl J Med. 1997;336:795–796. doi: 10.1056/NEJM199703133361109. [DOI] [PubMed] [Google Scholar]
- 18.Miyamoto H, Okamoto H, Sato K, Tanaka T, Mishiro S. Extraordinarily low density of hepatitis C virus estimated by sucrose density gradient centrifugation and the polymerase chain reaction. J Gen Virol. 1992;73:715–718. doi: 10.1099/0022-1317-73-3-715. [DOI] [PubMed] [Google Scholar]
- 19.Okamoto H, Nakao H, Inoue T, Fukuda M, Kishimoto J, Iizuka H, Tsuda F, Miyakawa Y, Mayumi M. The entire nucleotide sequence of two GB virus C/hepatitis G virus isolates of distinct genotypes from Japan. J Gen Virol. 1997;78:737–745. doi: 10.1099/0022-1317-78-4-737. [DOI] [PubMed] [Google Scholar]
- 20.Pilot-Matias T J, Carrick R J, Coleman P F, Leary T P, Surowy T K, Simons J N, Muerhoff A S, Buijk S L, Chalmers M L, Dawson G J, Desai S M, Mushahwar I K. Expression of the GB virus C E2 glycoprotein using the Semliki Forest virus vector system and its utility as a serologic marker. Virology. 1996;225:282–292. doi: 10.1006/viro.1996.0602. [DOI] [PubMed] [Google Scholar]
- 21.Sallie R, Shaw J, Mutimer D. GBV-C virus and fulminant hepatic failure. Lancet. 1996;347:1552. doi: 10.1016/s0140-6736(96)90704-7. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt W N, Klinzman D, LaBrecque D, Macfarlane D E, Stapleton J T. Direct detection of hepatitis C virus (HCV) RNA from whole blood, and comparison with HCV RNA in plasma and peripheral blood mononuclear cells. J Med Virol. 1995;47:153–160. doi: 10.1002/jmv.1890470208. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt W N, Wu P, Cederna J, Mitros F A, LaBrecque D R, Stapleton J T. Surreptitious hepatitis C virus (HCV) infection detected in the majority of patients with cryptogenic chronic hepatitis and negative HCV antibody tests. J Infect Dis. 1997;176:27–33. doi: 10.1086/514033. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt W N, Wu P, Han J-Q, Perino M J, LaBrecque D R, Stapleton J T. Distribution of hepatitis C virus (HCV) RNA in whole blood and blood cell fractions: plasma HCV RNA analysis underestimates circulating virus load. J Infect Dis. 1997;176:20–26. doi: 10.1086/514024. [DOI] [PubMed] [Google Scholar]
- 25.Shao L, Shinzawa H, Ishikawa K, Zhang X, Ishibashi M, Misawa H, Yamada N, Togashi H, Takahashi T. Sequence of hepatitis G virus genome isolated from a Japanese patient with non-A-E-hepatitis: amplification and cloning by long reverse transcription-PCR. Biophys Res Commun. 1996;228:785–791. doi: 10.1006/bbrc.1996.1732. [DOI] [PubMed] [Google Scholar]
- 26.Simons J N, Desai S M, Schultz D E, Lemon S M, Mushahwar I K. Translation initiation in GB viruses A and C: evidence for internal ribosome entry and implications for genomic organization. J Virol. 1996;70:6126–6135. doi: 10.1128/jvi.70.9.6126-6135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoff A S, Schlauder G G, Desai S M, Mushahwar I K. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 28.Stapleton J T, Frederick J, Meyer B. Hepatitis A virus attachment to cultured cell lines. J Infect Dis. 1991;164:1098–1103. doi: 10.1093/infdis/164.6.1098. [DOI] [PubMed] [Google Scholar]
- 29.Stapleton J T, Jansen R, Lemon S M. Neutralizing antibody to hepatitis A virus in immune serum globulin and in the sera of human recipients of immune serum globulin. Gastroenterology. 1985;89:637–642. doi: 10.1016/0016-5085(85)90462-7. [DOI] [PubMed] [Google Scholar]
- 30.Stapleton J T, Lange D K, LeDuc J W, Binn L N, Jansen R W, Lemon S M. The role of secretory immunity in hepatitis A virus infection. J Infect Dis. 1991;163:7–11. doi: 10.1093/infdis/163.1.7. [DOI] [PubMed] [Google Scholar]
- 31.Stapleton J T, Lemon S M. Delta hepatitis: a recently recognized entity. Diagnosis. 1985;7:26–35. [Google Scholar]
- 32.Takahashi K, Kishimoto S, Yoshizawa H, Okamoto H, Yoshikawa A, Mishiro S. p26 protein and 33-nm particle associated with nucleocapsid of hepatitis C virus recovered from the circulation of infected hosts. Virology. 1992;191:431–434. doi: 10.1016/0042-6822(92)90204-3. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka E, Alter H J, Nakatsuji Y, Shih J W, Kim J P, Matsumoto A, Kobayashi M, Kiyosawa K. Effect of hepatitis G virus infection on chronic hepatitis C. Ann Intern Med. 1996;125:740–743. doi: 10.7326/0003-4819-125-9-199611010-00007. [DOI] [PubMed] [Google Scholar]
- 34.Thomssen R, Bonk S, Thiele A. Density heterogeneities of hepatitis C virus in human sera due to binding of beta-lipoproteins and immunoglobulins. Med Microbiol Immunol. 1993;182:329–334. doi: 10.1007/BF00191948. [DOI] [PubMed] [Google Scholar]
- 35.Yoshiba M, Okamoto H, Mishiro S. Detection of the GBV-C hepatitis virus genome in serum from patients with fulminant hepatitis of unknown aetiology. Lancet. 1995;346:1131–1132. doi: 10.1016/s0140-6736(95)91802-7. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Y S. cDNA cloning and sequencing of HGV genome from Chinese. Bull Acad Mil Med Sci. 1996;20:249–253. [Google Scholar]