Abstract
Herpes simplex virus (HSV) type 1 DNA synthesis and packaging occur within the nuclei of infected cells; however, the extent to which the two processes are coupled remains unclear. Correct packaging is thought to be dependent upon DNA debranching or other repair processes, and such events commonly involve new DNA synthesis. Furthermore, the HSV UL15 gene product, essential for packaging, nevertheless localizes to sites of active DNA replication and may link the two events. It has previously been difficult to determine whether packaging requires concomitant DNA synthesis due to the complexity of these processes and of the viral life cycle; however, we have recently described a model system which simplifies the study of HSV assembly. Cells infected with HSV strain tsProt.A accumulate unpackaged capsids at the nonpermissive temperature of 39°C. Following release of the temperature block, these capsids proceed to package viral DNA in a single, synchronous wave. Here we report that, when DNA replication was inhibited prior to release of the temperature block, DNA packaging and later events in viral assembly nevertheless occurred at near-normal levels. We conclude that, under our conditions, HSV DNA packaging does not require detectable levels of DNA synthesis.
Herpes simplex virus (HSV) type 1 (HSV-1) is a complex double-stranded DNA virus which replicates its genome in the nuclei of infected cells (21, 29, 35). Seven viral genes are known to be essential for HSV DNA replication (4, 5, 8, 13, 21, 25, 35), and replication is thought to occur by a rolling circle mechanism, generating head-to-tail concatamers of the 152-kb viral chromosome (4, 29). An additional, inherently recombinogenic replication mechanism may also exist and give rise to branched forms of viral DNA (4, 17, 30).
Packaging, which also occurs in the nucleus, involves cleavage of the concatameric viral DNA at the flanking “a” sequences and engulfment of a single full-length copy of the viral chromosome by the maturing viral capsids (19, 29, 31–33). Cleavage at the a sequence appears to be inseparable from packaging and can be readily monitored by Southern blotting since it results in cleavage of the viral SQ BamHI fragment to discrete S and Q fragments. The molecular apparatus responsible for packaging is poorly understood, but one key component is the product of the HSV UL15 gene. This protein has possible homology with the large subunit of the terminase complex responsible for cleavage and packaging of the phage T4 genome (3, 7, 9), and HSV-1 mutants defective in UL15 fail to cleave and package viral DNA (1, 2, 23, 37). At least five other HSV gene products, UL6, UL25, UL28, UL32, and UL33, are also required for the packaging process (references 1, 29, and 37 and references therein).
Replication of viral DNA takes place in large globular regions termed replication compartments (8, 25, 28) which initially form adjacent to nuclear subdomains known as ND10 (12, 18). Replication compartments contain all seven virally encoded proteins essential for DNA replication (5, 8, 11, 14–16, 22) but surprisingly also contain the packaging factor UL15 (34). UL15 colocalizes with the DNA synthesis machinery even late in infection, when further levels of nuclear compartmentation become apparent. At these late times, when capsid assembly is thought to be maximal, at least some HSV strains generate dense structures termed assemblons at the periphery of their nuclei (34). Assemblons are recognized by antibodies which react with the major viral capsid protein VP5 and the capsid scaffold protein ICP35 (34). It has been hypothesized that they may represent sites of capsid assembly and maturation, positioned so as to gain access to the pool of actively replicating viral DNA in replication compartments. It has further been proposed that UL15 could actually be an accessory component of the DNA replication machinery (34) and may couple the process of DNA replication to that of genome cleavage and packaging.
One of a number of issues which remain unaddressed concerning replication and packaging in HSV-1 is the degree of interdependence between the two events. Does packaging require ongoing DNA synthesis, or can the UL15-dependent packaging machinery capture and process DNA from a previously replicated pool? Similarly, the resolution of branched HSV DNA may be important for production of packaged nucleocapsids capable of further maturation (17, 30). Such events and other kinds of DNA repair generally require DNA synthesis, at least in uninfected cells (36).
Determination of the dependence of packaging upon DNA replication would help advance our understanding of the molecular nature of the packaging process. Unfortunately, the inherent complexity of the assembly pathway of HSV and the fact that packaging and replication occur simultaneously and continuously from a point very early in infection make it extremely difficult to dissect the relationship between these two processes. We have recently described a model system to facilitate the study of late events in HSV assembly: the virus strain tsProt.A carries a reversible temperature-sensitive mutation in its maturational protease, which results in accumulation of unpackaged capsids at the nonpermissive temperature of 39°C (10, 24). Following downshift to the permissive temperature of 31°C, these capsids mature, package DNA, and give rise to infectious particles in a single, synchronized wave (6). In the present study, we made use of this system to test the requirement for DNA synthesis in packaging. When DNA replication was inhibited by addition of acyclovir (ACV), we found that packaging and PFU production could subsequently take place at near-normal levels. We conclude that, although DNA packaging and detectable levels of DNA synthesis occur simultaneously during a normal infection, they are not coupled together in an obligate fashion.
MATERIALS AND METHODS
Cells and viruses.
All cells were grown in Dulbecco modified Eagle’s medium supplemented with 1% penicillin-streptomycin (GIBCO Laboratories) and 10% newborn calf serum (Vero cells) or 10% fetal calf serum (the UL15-complementing cell line M-3). During routine passage, M-3 cells were cultured in the presence of 250 μg of G418 (Sigma Chemical Co.) per ml. HSV strains tsProt.A and SC16 were grown as previously described (6). The UL15 null mutant hr81-2 was grown by low-multiplicity infection of M-3 cells, and its titer was determined by plaque assay on preformed M-3 monolayers.
Preparation of total infected cell DNA and packaged viral DNA.
Cell extracts were prepared as previously described (6). Packaged DNA was isolated by a modification of the method of Shao et al. (30): extracts were incubated with 70 U of DNase I (Sigma) per ml in the presence of 2 mM Mg2+ for 2 h at 37°C and then adjusted to 10 mM EDTA–0.3% sodium dodecyl sulfate–50 μg of proteinase K per ml, and incubation was continued for a further 2 h. Total cell DNA was prepared in a similar way but with omission of the incubation with DNase. Finally, samples were subjected to exhaustive extraction with phenol and chloroform prior to ethanol precipitation. When DNA was to be Southern blotted, it was first digested to completion with BamHI and then blotted and probed with the SQ junction fragment and/or a loading control fragment derived from the UL22 gene, as previously described (6).
Trichloroacetic acid (TCA) precipitation assay to measure HSV DNA packaging.
Vero or M-3 cells were infected with HSV at a multiplicity of infection of 10 and then incubated in Dulbecco modified Eagle’s medium containing 1% dialyzed newborn calf serum (to deplete intracellular pools of thymidine). After 2 h, [3H]thymidine (New England Nuclear) was added to a final concentration of 25 μCi/ml and incubations were continued as required. Total DNA and packaged DNA were prepared as described above, except that samples were neither phenol-chloroform extracted nor ethanol precipitated. Instead, aliquots were spotted onto 24-mm-diameter glass fiber filters (Whatman GF/C) and incubated in ice-cold TP buffer (5% TCA, 20 mM sodium pyrophosphate) for 5 min. After incubation in a second sample of TP buffer at 65°C for 5 min, filters were rinsed in 70% ethanol at room temperature for 2 min and then dried, and bound counts per minute of tritium were determined by liquid scintillation counting.
Scoring levels of infectious progeny virus.
The yield of progeny virions from infected Vero cells was determined as described previously (6). Briefly, following infection cells were rinsed in a pH 3.0 buffer to inactivate virus that had not penetrated, overlaid with prewarmed medium, and then incubated. At the appropriate time, cells were frozen, thawed, collected by scraping, and sonicated, and the resulting extract was titrated onto preformed Vero cell monolayers.
RESULTS
Establishing conditions suitable for inhibition of DNA replication during synchronized assembly of the tsProt.A virus.
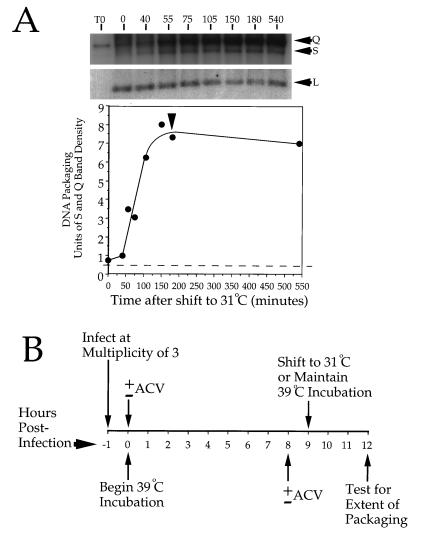
To establish conditions for these studies, we first wished to determine the time required for accumulated capsids to package viral DNA following downshift of tsProt.A-infected cells from 39 to 31°C. In earlier studies, which made use of Southern blotting to detect BamHI SQ fragment cleavage, we could not properly define the kinetics of DNA packaging due to a relatively high background of cleaved fragments from input viral DNA (6). We modified our earlier conditions by reducing the multiplicity of infection to 3 and increasing the length of the 39°C incubation to 9 h. This substantially improved the quality of our DNA packaging data (Fig. 1A), and these conditions were adopted for our current study. Figure 1A demonstrates the kinetics of DNA packaging, as determined by Southern blot analysis of the cleavage of the SQ junction BamHI fragment to S and Q fragments. Following the shift to 31°C, there was no detectable packaging for about 50 min. There was then a burst of DNA packaging, which appeared to be complete within 3 h of the temperature downshift. We therefore designed our experiments as depicted in Fig. 1B, to test how much DNA packaging could occur by the 3-h point when DNA synthesis had been inhibited by addition of the replication inhibitor ACV (26, 27). ACV was added 1 h prior to shifting cells to 31°C, in order to ensure maximum opportunity for the drug to inhibit replication before packaging could begin.
FIG. 1.
Time course of DNA packaging and choice of incubation conditions. (A) Vero cells were infected with HSV tsProt.A and incubated at 39°C for 9 h. Following cycloheximide addition (to inhibit new protein synthesis and to ensure a single wave of packaging) and downshift to 31°C, total infected cell DNA was prepared at particular times, digested with BamHI, separated on a 1.0% agarose gel, blotted onto a nylon membrane, and hybridized to a mixed 32P-labeled probe corresponding to the SQ cleavage junction and to a region within the UL22 gene. The latter probe hybridizes to a BamHI fragment which should not change in abundance during packaging and which provides a convenient loading control. The top of the figure shows different exposures of the region of the filter containing the S and Q fragments or the UL22 fragment (L) as indicated. Numbers above lanes indicate the time of incubation at 31°C in minutes. Lane T0, sample recovered immediately postinfection. The lower part of the panel represents a densitometric analysis of the data. The broken line indicates background levels of S and Q fragments contributed by residual input virus (calculated from the T0 lane). The vertical arrowhead indicates 3 h after downshift. (B) Time course and conditions used for many of the experiments in this study. Numbers correspond to the times after completion of viral infection, in hours. ACV was added either immediately postinfection (0 h) to confirm the efficacy of the drug or after 8 h at 39°C or omitted. After 9 h at 39°C, samples were incubated for a further 3 h at 31 or 39°C.
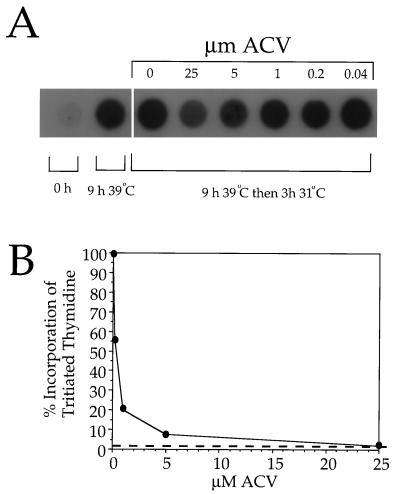
We next determined what concentration of ACV was required to completely inhibit measurable DNA replication under these unusual conditions. Varying concentrations of ACV were added to infected cells, and incubations were performed as described for Fig. 1B. At the end of the 31°C incubation, cells were collected and used to prepare total infected cell DNA. This was dot blotted onto a filter and incubated with a radiolabeled SQ junction probe. Figure 2A demonstrates that either 5 or 25 μM ACV appeared to prevent any increase in the levels of viral DNA during the 31°C incubation. Indeed, in cells treated with these concentrations of ACV, levels of viral DNA at the end of the 3-h incubation were lower than those in non-drug-treated cells harvested at the start of the 3-h incubation. This implies that the ACV had shut down viral DNA synthesis before the end of the final hour of incubation at 39°C or that DNA was undergoing turnover in ACV-inhibited cells. As expected, the probe was specific for viral DNA since only low levels of hybridization to DNA prepared immediately postinfection (0-h sample) occurred.
FIG. 2.
Effect of ACV on DNA synthesis by tsProt.A. (A) Dot blot of total infected cell DNA. Vero cells were infected with tsProt.A and then incubated for 9 h at 39°C and 3 h at 31°C. ACV was added after 8 h at 39°C to the concentrations indicated. Total infected cell DNA was prepared immediately postinfection (0 h), at the time of shift to 31°C (9 h 39°C), or 3 h later and then dot blotted and hybridized to a radiolabeled probe prepared from the HSV-1 SQ junction fragment. (B) The same procedure as for panel A was followed except that 25 μCi of [3H]thymidine per ml was added at the time of shift to 31°C. Following purification, infected cell DNA was counted in a liquid scintillation counter. The vertical axis indicates the degree of radiolabeled thymidine incorporation into DNA as a percentage of that in the absence of ACV. The horizontal axis indicates the concentration of ACV added at 8 h of infection. The broken line represents background incorporation, determined from a sample which received no ACV but which was shifted to 4°C for 3 h at the time of addition of [3H]thymidine.
In an effort to confirm the efficacy of the drug in an alternative and more quantitative way, cells were infected and incubated as shown in Fig. 1B except that, at the time of downshift to 31°C, [3H]thymidine was added to the samples. At the end of the incubation, total infected cell DNA was purified and levels of incorporated radioactivity were determined with a liquid scintillation counter. Figure 2B demonstrates that 25 μM ACV was sufficient to reduce the incorporation of radiolabeled thymidine to background levels under these conditions.
Viral DNA becomes packaged and cleaved despite inhibition of DNA replication.
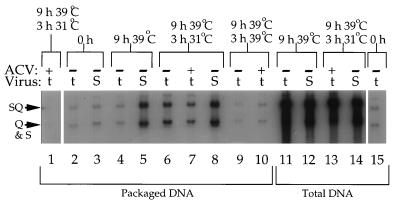
Having confirmed the efficacy of 25 μM ACV addition under these conditions, we tested whether packaging of the viral genome could occur after inhibition of DNA replication. Figure 3 demonstrates packaging of the viral genome, as measured by protection from DNase I digestion and cleavage of the BamHI SQ junction fragment, under a variety of incubation conditions for strain tsProt.A and the wild-type virus SC16. When ACV was added to tsProt.A-infected cells immediately postinfection and cells were subsequently taken through the complete time course of incubation, levels of packaged viral DNA actually fell below that detectable in samples harvested immediately postinfection (compare lane 1 with lanes 2, 3, and 15), which presumably derives from input virus. This suggests that some level of turnover of viral DNA proceeds during incubation under these conditions. After 9 h of incubation at 39°C, SC16 and tsProt.A had synthesized comparable amounts of DNA (lanes 11 and 12) and DNA packaging was easily visible for SC16 (lane 5). A small amount of DNA packaging was measurable for tsProt.A (lane 4), suggesting some leakage through the temperature block in this experiment; however, no additional leakage through the block was observed if incubations were continued for 3 further h at 39°C (compare lanes 4 and 9). In contrast, following downshift to 31°C for 3 h a substantial amount of tsProt.A packaging occurred (lane 6), as expected from the data in Fig. 1A. Wild-type virus, which does not accumulate a synchronized population of immature capsids at 39°C, showed only a modest increase in packaging over the same time period (compare lanes 5 and 8). Levels of DNA replication in the wild-type virus were also quite low over this 3-h period, perhaps because of the temperature of incubation (compare total DNA levels at the beginning and the end of the 31°C incubation [lanes 12 and 14, respectively]). When ACV was added to the tsProt.A infection, it prevented this small amount of viral DNA synthesis (compare lanes 11 and 13).
FIG. 3.
Packaging of the HSV genome in the absence of measurable DNA replication. Vero cells were infected with tsProt.A or with SC16 (t or S, respectively, as indicated). ACV was added at a concentration of 25 μM either immediately postinfection (lane 1) or 8 h postinfection (lanes 7, 10, and 13) or omitted (all other lanes) as indicated by + or −. Samples were harvested immediately postinfection (lanes 2, 3, and 15), at the end of 9 h at 39°C (lanes 4, 5, 11, and 12), or after a further 3 h at 31°C (lanes 1, 6, 7, 8, 13, and 14) or 39°C (lanes 9 and 10), as indicated at the top of the figure. Packaged DNA (lanes 1 to 10) or total infected cell DNA (lanes 11 to 15) was then prepared, digested with BamHI, Southern blotted, and probed exactly as described in the legend to Fig. 1A. The positions of the SQ junction fragment and the S and Q cleavage products are indicated at the left of the figure.
In the presence of 25 μM ACV, sufficient to reduce DNA replication to undetectable levels (lane 1 and data presented above), there was still a substantial degree of DNA cleavage and packaging (lane 7). Although levels were lower than those in the absence of drug (lane 6), they were above background (lanes 4 and 10).
To attempt to more carefully quantitate the amount of packaging which could occur in the presence and absence of 25 μM ACV, we performed an experiment similar to the one presented in Fig. 3 but probed the filter with a radiolabeled UL22-specific probe. Following exposure to film, the regions of the filter which had hybridized to the probe were excised, and the amount of bound probe was determined by Cerenkov counting. The effects of ACV upon total levels of viral DNA for two independent Southern blots are reported in Table 1. Comparison of the amounts of total viral DNA at the beginning and the end of the 3-h, 31°C incubation revealed a 50 to 71% increase in the absence of drug, but a 21 to 50% reduction in the presence of drug, consistent with our earlier findings (Fig. 2A). From Table 1, it is also clear that when 25 μM ACV was added at the beginning of the 39°C incubation it was able to prevent any measurable level of new viral DNA synthesis. The amounts of total and protected DNA were used to calculate the percentage of packaged DNA under each set of conditions, and the results from two independent Southern blots are listed in Table 2. A complication of these studies is that total DNA levels continued to increase during the 3-h, 31°C incubation in the absence, but not the presence, of ACV. Thus, even packaging of equal numbers of HSV genomes would lead to an apparent higher percentage of packaged DNA in the presence of drug. For this reason, the percentage of packaged DNA was calculated by comparison with the total amount of DNA present at the beginning of the 31°C incubation (at which time levels were similar between samples) rather than at the end. From Table 2, it is apparent that packaging occurs in the presence of ACV, although at somewhat lower levels than in non-drug-treated controls. Possible reasons for this will be considered in the discussion.
TABLE 1.
Levels of viral DNA synthesis in the absence and the presence of ACV
| Exptl condition and no. | % Increase in HSV DNA after:
|
|
|---|---|---|
| 3 h at 31°Ca | 9 h at 39°C and 3 h at 31°Cb | |
| No ACV | ||
| Expt 1 | +71 | +2,133 |
| Expt 2 | +50 | +1,718 |
| With ACV | ||
| Expt 1 | −21 | −69 |
| Expt 2 | −50 | −72 |
When ACV was present, it was added after 8 h at 39°C.
When ACV was present, it was added at the beginning of the 39°C incubation.
TABLE 2.
Percentage of viral DNA packaged under various growth conditions
| Growth conditions | % Packaged
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| 9 h at 39°C, no ACV | 1.2 | 1.7 |
| 9 h at 39°C, with ACV | 1.2 | 1.2 |
| 9 h at 39°C, 3 h at 31°C, no ACV | 3.6 | 3.9 |
| 9 h at 39°C, 3 h at 31°C, with ACV | 3.4 | 2.7 |
| 12 h at 39°C, no ACV | 1.1 | 0.9 |
Nucleocapsids formed without attendant DNA synthesis can mature to infectious particles.
Although genomic cleavage and protection from DNase I occurred in the presence of ACV, the inhibition of DNA replication could nevertheless have resulted in abnormal packaging not detectable by these assays. There is a precedent for such a phenomenon; HSV mutants lacking the gene encoding alkaline nuclease are able to cleave and encapsidate DNA at near-wild-type levels; however, yields of infectious progeny are drastically reduced (17, 30), suggesting some defect in the structure of the viral genome or the matured nucleocapsid. To test whether capsids packaged in the absence of DNA replication could efficiently mature to infectious virions, we performed a drug addition and temperature shift experiment similar to that described for Fig. 3, but this time we scored PFU yields. Figure 4 demonstrates that when accumulated, immature tsProt.A virions were shifted from 39 to 31°C, a 2.7-log increase in PFU occurred (samples 3 and 5), compared with a 0.5-log increase for wild-type virus (samples 4 and 6). This increase was dependent upon return to 31°C (sample 8). As we demonstrated previously (6), the final yield of infectious tsProt.A virions following release of the temperature block is similar to the yield resulting from gradual accumulation of virus in a wild-type infection (samples 5 and 6). When DNA replication was inhibited by addition of ACV at 8 h, yields of progeny virus were diminished by about 0.2 logs (samples 5 and 7). Addition of 25 μM ACV immediately postinfection (sample 9) resulted in PFU yields similar to those obtained when infected cells were harvested immediately postinfection (samples 1 and 2), providing an additional internal control for the efficacy of the drug.
FIG. 4.
Production of infectious progeny virions in the absence of measurable DNA replication. An experiment identical to that for Fig. 3 was performed, except that the resulting cell extracts were titrated for PFU production. Plotted values indicate the means and standard deviations from the means of single samples titrated in duplicate (samples 1, 2, 8, and 9) or duplicate samples, each titrated in duplicate (samples 3, 4, 5, 6, and 7). Hatched bars represent data from tsProt.A-infected cells, and open bars represent data from SC16-infected cells.
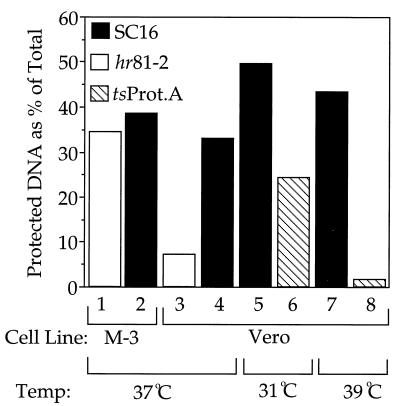
Measurement of DNA packaging by TCA precipitation.
We sought to test DNA packaging by independent means and established conditions in which packaged, radiolabeled DNA could be TCA precipitated, washed free of low-molecular-weight radioactive contaminants, and then quantitated by scintillation counting. Figure 5 summarizes two different experiments to test the validity of this approach. When wild-type virus or the UL15 null HSV strain hr81-2 (37) was grown in M-3 cells (stably transformed to express UL15 [37]) for 14 h, approximately 35% of the total high-molecular-weight DNA became protected (samples 1 and 2), similar to that of wild-type SC16 virus growing in a noncomplementing cell line (sample 4). However, the apparent level of packaging fell to 7% when the UL15 null virus was grown on a noncomplementing cell line (sample 3). In a separate experiment, wild-type or tsProt.A virus was grown for 14 h at 31 or 39°C. Whereas tsProt.A packaged about half as much DNA as SC16 at 31°C (samples 5 and 6), it packaged only about 2% as much at 39°C (samples 7 and 8).
FIG. 5.
Characterization of the TCA precipitation assay. Vero cells or the Vero cell-derived UL15-complementing cell line M-3 was infected with tsProt.A (hatched bars), SC16 (black bars), or the UL15 null virus hr81-2 (open bars) and incubated at 31, 37, or 39°C in the presence of tritiated thymidine. At 14 h postinfection, cell extracts were prepared and processed as described in Materials and Methods. The vertical axis represents DNase I-resistant (packaged) DNA as a percentage of the total DNA in the sample. Samples 1 to 4 and 5 to 8 represent data from separate experiments.
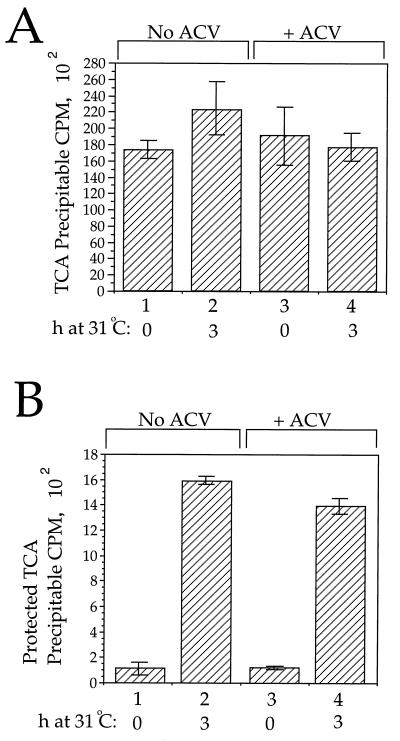
Figure 6 demonstrates the effect of ACV addition upon DNA replication and packaging as measured by this method. As can be seen in Fig. 6A, when ACV was absent the mean amount of tritium incorporated into TCA-precipitable material increased by 28% over the course of the 3-h incubation (samples 1 and 2). However, when ACV was added prior to the start of the 31°C incubation the mean level of TCA-precipitable tritium fell by 7% over the 3-h period (samples 3 and 4). In other experiments, TCA-precipitable tritium levels fell by as much as 30% in the presence of ACV (data not shown).
FIG. 6.
Quantitation of replication and packaging by a TCA precipitation assay. Vero cells were infected with tsProt.A and incubated in the presence of tritiated thymidine, and then extracts were prepared after 9 h at 39°C (samples 1 and 3) or after a further 3 h at 31°C (samples 2 and 4). Samples 3 and 4 received ACV 1 h before the end of the 39°C incubation. (A) Amounts of total DNA present in each sample, determined by TCA precipitation as described in Materials and Methods. Plotted values and error bars indicate the means and standard deviations from the means for cell extracts TCA precipitated in two separate experiments, each in triplicate. (B) Amounts of DNase I-resistant (packaged) DNA present, determined as described in Materials and Methods. TCA precipitations were performed in two separate experiments, each in triplicate.
We next tested whether packaging occurred despite inhibition of incorporation of tritiated thymidine into DNA. The cell extracts used to prepare the data in Fig. 6A were assayed for their level of DNase I-resistant DNA. The data in Fig. 6B indicates that comparable amounts of DNA packaging occurred in the presence or absence of detectable levels of DNA replication. We have not attempted to represent packaging as a percentage of total DNA since total TCA-precipitable counts may include not only viral DNA but also host cell DNA synthesis at this relatively early time in infection. Nevertheless, comparison of tritium incorporation in infected cells with that in uninfected cells suggests that less than 30% of the total TCA-precipitable counts are contributed by host cell DNA synthesis under these conditions (data not shown).
DISCUSSION
A number of basic questions in herpesvirus biology remain unresolved. Some of the most intractable problems concern late events in virus assembly. Despite the central importance of DNA packaging to the viral life cycle, and to potential therapeutic intervention, very little is known about the molecular details of the process. At least six gene products are known to be required for packaging and cleavage; however, their role, and that of other viral and cellular proteins, remains cryptic.
Studies with double-stranded DNA bacteriophage have revealed that, in some cases, the structure of the viral DNA profoundly affects the rate and extent of packaging (3). Phage T4 packaging is inhibited in the absence of a functional topoisomerase II (40) or following inactivation of gene 49, which encodes endonuclease VII (3). In the latter case, it is thought that packaging arrests as a result of the accumulation of DNA branches generated by recombination, and a similar mechanism has been proposed to explain why packaging in alkaline nuclease-defective HSV mutants gives rise to defective nucleocapsids (17, 30). T4 packaging also requires a DNA ligase activity (38) and can be slowed by UV irradiation (39), suggesting that repair and replication functions may be required for encapsidation (3).
We have addressed the question of whether the HSV packaging machinery requires a functional DNA replication apparatus or actively replicating DNA as a substrate. Such a requirement could result from the need to carry out DNA debranching or other repair processes (17, 30, 36), although the role of DNA synthesis in HSV DNA repair remains unclear. The localization of the DNA packaging factor UL15 in a compartment enriched for replicating DNA (34) also suggests the possibility that these two phenomena might be coupled during viral assembly.
Using two different approaches, we found that, when DNA synthesis fell below detectable levels due to ACV addition, packaging nevertheless could continue. Furthermore, the resulting nucleocapsids were able to mature to infectivity. Although we cannot exclude the possibility that some undetectable level of DNA synthesis persists under our conditions (see below), we were able to inhibit the apparent level of DNA replication to less than 6% of normal (compare the 5 and 25 μM points in Fig. 2B) but still retain an efficiency of packaging within about 80% of non-drug-treated controls (Table 2 and Fig. 6). This data implies that normal levels of ongoing DNA replication are not a prerequisite for proper packaging under our conditions. Our findings are consistent with earlier work which demonstrated that a plasmid incapable of replication but containing a sequences could be specifically cleaved during the course of an HSV infection (20), although packaging was not directly demonstrated in that study.
The ACV-dependent decrease in the extent of packaging (Fig. 3, Table 2, and Fig. 6) and in infectious virus production (Fig. 4) may reflect a genuine reduction in the efficiency of packaging of prereplicated rather than replicating DNA or could suggest that DNA synthesis does play some role in the proofreading or repair of molecules immediately prior to their encapsidation. Alternatively, it may be a consequence of our experimental design. Infected cells which did not receive ACV were free to continue replicating their DNA for 4 h longer than drug-treated samples (1 h at 39°C and then 3 h at 31°C). It is conceivable that a higher concentration of packageable DNA, or of gene products expressed from it, could explain the differences observed in the presence and the absence of drug. Consistent with this latter possibility, levels of protein synthesis over the 3-h, 31°C incubation were 20% lower in ACV-treated cells than in untreated cells, as determined by the extent of incorporation of [35S]cysteine and [35S]methionine into a TCA-precipitable fraction (data not shown).
The two different methods used to measure packaging, Southern blotting and TCA precipitation, may indicate the fates of different populations of viral DNA molecules. As noted by Roizman and Sears (29), changes in intracellular levels of deoxynucleotides during the course of an infection mean that incorporation of labeled nucleotides is most rapid at early time points. Our TCA precipitation study therefore may tend to reflect the fate of older viral genomes, whereas Southern blotting reveals the behavior of the entire population. Furthermore, it is important to emphasize that this study was conducted under extremely unusual incubation conditions. Artificially separating early and late events in HSV assembly by use of the tsProt.A temperature-sensitive mutant could lead to conclusions not relevant to a normal viral infection, and it is certainly possible that under normal conditions the processes of replication and packaging are mechanistically coupled. Nevertheless, our growth conditions do generate packaged, infectious virus particles at an efficiency comparable to that of similarly grown wild-type virus. A further concern is that DNA appears to turn over in ACV-treated cells (Fig. 2A and 3; Table 1); we therefore cannot rule out the possibility that this DNA degradation could be masking a low level of new DNA synthesis. Nevertheless, we suggest that, although replication and packaging could still be linked under normal circumstances, our data implies that under our conditions normal levels of the former are not an absolute prerequisite for the latter.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI38265 and DK41918 to D.W.W. and by NIH training grant T32 GM07491 to G.A.C. Core support was provided by NIH Cancer Center grant P30-CA13330.
HSV strain hr81-2 and the cell line M-3 were generous gifts from Sandra Weller. We thank Lily Huang for excellent technical assistance and Sandra Weller and Carol Harley for helpful discussions.
REFERENCES
- 1.Baines J D, Cunningham C, Nalwanga D, Davison A. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Poon A P, Rovnak J, Roizman B. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J Virol. 1994;68:8118–8124. doi: 10.1128/jvi.68.12.8118-8124.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black L W. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 4.Boehmer P E, Lehman I R. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 5.Bush M, Yager D R, Gao M, Weisshart K, Marcy A I, Coen D M, Knipe D M. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J Virol. 1991;65:1082–1089. doi: 10.1128/jvi.65.3.1082-1089.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church G A, Wilson D W. Study of herpes simplex virus maturation during a synchronous wave of assembly. J Virol. 1997;71:3603–3612. doi: 10.1128/jvi.71.5.3603-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 8.de Bruyn Kops A, Knipe D M. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 9.Dolan A, Arbuckle M, McGeoch D J. Sequence analysis of the splice junction in the transcript of herpes simplex virus type 1 gene UL15. Virus Res. 1991;20:97–104. doi: 10.1016/0168-1702(91)90064-3. [DOI] [PubMed] [Google Scholar]
- 10.Gao M, Matusick-Kumar L, Hurlburt W, DiTusa S F, Newcomb W W, Brown J C, McCann P J, Deckman I, Colonno R J. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J Virol. 1994;68:3702–3712. doi: 10.1128/jvi.68.6.3702-3712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodrich L D, Schaffer P A, Dorsky D I, Crumpacker C S, Parris D S. Localization of the herpes simplex virus type 1 65-kilodalton DNA-binding protein and DNA polymerase in the presence and absence of viral DNA synthesis. J Virol. 1990;64:5738–5749. doi: 10.1128/jvi.64.12.5738-5749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knipe D M. The role of viral and cellular nuclear proteins in herpes simplex virus replication. Adv Virus Res. 1989;37:85–123. doi: 10.1016/s0065-3527(08)60833-7. [DOI] [PubMed] [Google Scholar]
- 14.Lukonis C J, Weller S K. Characterization of nuclear structures in cells infected with herpes simplex virus type 1 in the absence of viral DNA replication. J Virol. 1996;70:1751–1758. doi: 10.1128/jvi.70.3.1751-1758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukonis C J, Weller S K. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J Virol. 1997;71:2390–2399. doi: 10.1128/jvi.71.3.2390-2399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik A K, Shao L, Shanley J D, Weller S K. Intracellular localization of the herpes simplex virus type-1 origin binding protein, UL9. Virology. 1996;224:380–389. doi: 10.1006/viro.1996.0545. [DOI] [PubMed] [Google Scholar]
- 17.Martinez R, Sarisky R T, Weber P C, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 19.Mocarski E S, Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982;31:89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- 20.Nasseri M, Mocarski E S. The cleavage recognition signal is contained within sequences surrounding an a-a junction in herpes simplex virus DNA. Virology. 1988;167:25–30. doi: 10.1016/0042-6822(88)90050-5. [DOI] [PubMed] [Google Scholar]
- 21.Olivo P D, Challberg M D. Functional analysis of the herpes simplex virus gene products involved in DNA replication. In: Wagner E, editor. Herpesvirus transcription and its regulation. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 137–150. [Google Scholar]
- 22.Olivo P D, Nelson N J, Challberg M D. Herpes simplex virus type 1 gene products required for DNA replication: identification and overexpression. J Virol. 1989;63:196–204. doi: 10.1128/jvi.63.1.196-204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poon A P, Roizman B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preston V G, Coates J A, Rixon F J. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J Virol. 1983;45:1056–1064. doi: 10.1128/jvi.45.3.1056-1064.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan M P, Chen L B, Knipe D M. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 26.Reardon J E. Herpes simplex virus type 1 and human DNA polymerase interactions with 2′-deoxyguanosine 5′-triphosphate analogues. Kinetics of incorporation into DNA and induction of inhibition. J Biol Chem. 1989;264:19039–19044. [PubMed] [Google Scholar]
- 27.Reardon J E, Spector T. Herpes simplex virus type 1 DNA polymerase. Mechanism of inhibition by acyclovir triphosphate. J Biol Chem. 1989;264:7405–7411. [PubMed] [Google Scholar]
- 28.Rixon F J, Atkinson M A, Hay J. Intranuclear distribution of herpes simplex virus type 2 DNA synthesis: examination by light and electron microscopy. J Gen Virol. 1983;64:2087–2092. doi: 10.1099/0022-1317-64-9-2087. [DOI] [PubMed] [Google Scholar]
- 29.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press, Ltd.; 1993. pp. 11–68. [Google Scholar]
- 30.Shao L, Rapp L M, Weller S K. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology. 1993;196:146–162. doi: 10.1006/viro.1993.1463. [DOI] [PubMed] [Google Scholar]
- 31.Sherman G, Bachenheimer S L. DNA processing in temperature-sensitive morphogenic mutants of HSV-1. Virology. 1987;158:427–430. doi: 10.1016/0042-6822(87)90214-5. [DOI] [PubMed] [Google Scholar]
- 32.Stow N D, McMonagle E C, Davison A J. Fragments from both termini of the herpes simplex virus type 1 genome contain signals required for the encapsidation of viral DNA. Nucleic Acids Res. 1983;11:8205–8220. doi: 10.1093/nar/11.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vlazny D A, Kwong A, Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci USA. 1982;79:1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward P L, Ogle W O, Roizman B. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol. 1996;70:4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weller S K. Genetic analysis of HSV genes required for genome replication. In: Wagner E, editor. Herpesvirus transcription and its regulation. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 105–135. [Google Scholar]
- 36.Wood R D, Shivji M K. Which DNA polymerases are used for DNA-repair in eukaryotes? Carcinogenesis. 1997;18:605–610. doi: 10.1093/carcin/18.4.605. [DOI] [PubMed] [Google Scholar]
- 37.Yu D, Sheaffer A K, Tenney D J, Weller S K. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zachary A, Black L W. DNA ligase is required for encapsidation of bacteriophage T4 DNA. J Mol Biol. 1981;149:641–658. doi: 10.1016/0022-2836(81)90351-x. [DOI] [PubMed] [Google Scholar]
- 39.Zachary A, Black L W. UV irradiation impairs in vivo encapsidation of bacteriophage T4 DNA. J Virol. 1984;50:293–300. doi: 10.1128/jvi.50.2.293-300.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zachary A, Black L W. Topoisomerase II and other DNA-delay and DNA-arrest mutations impair bacteriophage T4 DNA packaging in vivo and in vitro. J Virol. 1986;60:97–104. doi: 10.1128/jvi.60.1.97-104.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]