Abstract
The 2′,5′-oligoadenylate (2-5A) system is an RNA degradation pathway which plays an important role in the antipicornavirus effects of interferon (IFN). RNase L, the terminal component of the 2-5A system, is thought to mediate this antiviral activity through the degradation of viral RNA; however, the capacity of RNase L to selectively target viral RNA has not been carefully examined in intact cells. Therefore, the mechanism of RNase L-mediated antiviral activity was investigated following encephalomyocarditis virus (EMCV) infection of cell lines in which expression of transfected RNase L was induced or endogenous RNase L activity was inhibited. RNase L induction markedly enhanced the anti-EMCV activity of IFN via a reduction in EMCV RNA. Inhibition of endogenous RNase L activity inhibited this reduction in viral RNA. RNase L had no effect on IFN-mediated protection from vesicular stomatitis virus. RNase L induction reduced the rate of EMCV RNA synthesis, suggesting that RNase L may target viral RNAs involved in replication early in the virus life cycle. The RNase L-mediated reduction in viral RNA occurred in the absence of detectable effects on specific cellular mRNAs and without any global alteration in the cellular RNA profile. Extensive rRNA cleavage, indicative of high levels of 2-5A, was not observed in RNase L-induced, EMCV-infected cells; however, transfection of 2-5A into cells resulted in widespread degradation of cellular RNAs. These findings provide the first demonstration of the selective capacity of RNase L in intact cells and link this selective activity to cellular levels of 2-5A.
The interferons (IFNs) are a family of cytokines which were originally discovered as a result of their abilities to interfere with virus infection and replication (24) but which are now known to effect a diverse set of activities important to higher organisms, including control of cell proliferation and differentiation and regulation of the immune response (reviewed in references 26 and 45). The effects of IFN on cells are initiated by the binding of IFN to specific cellular receptors. The resultant signaling cascade, known as the JAK-STAT pathway, culminates in the transcriptional induction of IFN-stimulated genes (ISGs) and has recently been described in considerable detail (44). Although more than 30 ISGs have been identified to date (26, 52), very little is known about the mechanisms by which they mediate the biological activities of IFN, including its antiviral effects.
Among the ISG-encoded products known to function in the antiviral activity of IFN, the Mx family of proteins (40) and two double-stranded RNA (dsRNA)-dependent enzymes, a 68-kDa protein kinase (PKR) (26, 45) and a family of 2′,5′-oligoadenylate (2-5A) synthetases (9, 43) whose enzymatic products regulate 2-5A-dependent RNase (RNase L) (46), have been the most extensively characterized. The 2-5A system is an IFN-regulated RNA degradation pathway (27; reviewed in references 26 and 45). 2-5A synthetase, the first enzyme of the 2-5A system, produces a series of 5′-phosphorylated, 2′,5′-linked oligoadenylates from ATP when activated by dsRNA (9, 23). The only well-established function of 2-5A is the activation of RNase L (46). 2-5A binding results in the dimerization of RNase L, which constitutes its active form (14). Activated RNase L mediates the biological activity of the 2-5A system through the cleavage of single-stranded RNA 3′ of UpN residues, with a preference for UU and UA sequences (16, 55). Free 2-5A is unstable in cells, being degraded by cellular phosphatases and a 2′-phosphodiesterase (25).
A number of studies have established that the 2-5A system functions in the antiviral effects of IFN. For example, elevated levels of 2-5A and the appearance of specific rRNA cleavage products characteristic of RNase L activation are correlated with IFN-mediated inhibition of encephalomyocarditis virus (EMCV) (47, 53), vaccinia virus (13), and reovirus infections (39). Introduction of 2-5A into cells has been shown to reduce the cytopathic effects of several viruses, including vesicular stomatitis virus (VSV), EMCV, poliovirus, and Semliki Forest virus (1), whereas a 2-5A analog inhibitor of RNase L has reduced the anti-EMCV activity of IFN in intact cells (51). Further, in cell lines in which 2-5A pathway activity is defective, EMCV replication is resistant to the inhibitory effects of IFN (31). The cloning of cDNAs encoding 2-5A synthetase (35, 43) and RNase L (56) has provided a means of definitively addressing the antiviral function of the 2-5A system in cells. Constitutive expression of the 40-kDa form of 2-5A synthetase conferred resistance to EMCV and mengovirus (8, 41). Similarly, expression of a dominant negative mutant of RNase L inhibited the anti-EMCV activity of IFN, confirming its role in IFN’s antiviral activity (21). In these experiments, the antiviral activity of the 2-5A system in cells was restricted to picornaviruses. Accordingly, modulation of 2-5A pathway activity in cells had no effect on IFN-mediated anti-VSV activity (8, 21), suggesting that a restricted range of viruses is sensitive to inhibition by the 2-5A system.
Although the role of the 2-5A system in IFN’s antiviral effects has been well documented, the mechanism by which RNase L activation results in inhibition of virus replication remains unclear. Studies of cell-free systems have suggested that 2-5A pathway-mediated antiviral activity occurs through the direct action of RNase L on viral RNA (2, 37); however, the effect of RNase L on viral RNAs has not been carefully studied in intact cells. Further, the recent finding that RNase L functions in apoptosis (6) suggests that its antiviral activity may occur through the degradation of cellular RNAs, triggering apoptosis to inhibit viral spread. Indeed, specific substrates of RNase L have not been identified. rRNA is the only RNA which has been demonstrated to be cleaved by RNase L in intact cells, and its degradation to discrete cleavage products (47, 48, 54) is detected only in the presence of high levels of 2-5A (≥10 nM) (29, 54). However, RNase L has been shown to mediate the antiviral and antiproliferative effects of IFN in the absence of high levels of 2-5A and detectable rRNA cleavage (21, 39). Thus, rRNA may not represent a physiologically relevant substrate but rather may serve as an indicator of widespread RNase L activity under conditions of high levels of 2-5A. Indeed, RNase L has the capacity to degrade both viral and cellular RNAs, as indiscriminate cleavage has been reported under conditions that produce high levels of 2-5A, e.g., infection with a high multiplicity of infection (MOI) of reovirus (39) or treatment with exogenous dsRNA (38). The high levels of 2-5A produced are thought to result in widespread activation of RNase L, leading to a global degradation of RNA. However, the finding that RNase L can exert its biological activities in the absence of high levels of 2-5A and rRNA cleavage supports earlier studies which suggested that RNase L can function in a selective fashion under conditions of limiting 2-5A (2, 39). Specifically, viral replicative complexes or mRNAs possessing a contiguous double-stranded structure were preferentially degraded by RNase L in vitro. This observation led to the localized activation model of RNase L selectivity, in which dsRNA in viral replicative intermediates activates 2-5A synthetase, resulting in localized production of 2-5A, activation of RNase L, and cleavage of viral RNA (37). Consistent with this model, physical complexes containing EMCV dsRNA and 2-5A synthetase which produce authentic 2-5A have been isolated from virus-infected cells (19). Taken together, these studies suggest that RNase L can function in both selective and nonselective manners depending on cellular 2-5A levels. However, the capacity of RNase L to selectively target specific RNAs in intact cells has not been demonstrated to date.
To investigate the mechanism of RNase L-mediated antiviral activity, we examined viral and cellular RNAs following EMCV infection of cell lines in which expression of transfected RNase L is induced or endogenous RNase L activity is inhibited. Increased expression of RNase L enhanced the anti-EMCV activity of IFN through a reduction in EMCV RNA. IFN treatment and RNase L induction reduced EMCV RNA synthesis, suggesting that RNAs involved in virus replication are degraded by RNase L at early times in the virus life cycle, resulting in an inhibition of viral RNA synthesis. At late times postinfection (PI), RNase L induction had no effect on the decay rate of viral RNA, consistent with its activity early in infection. The RNase L-mediated reduction in viral RNA occurred in the absence of detectable effects on steady-state levels of specific cellular mRNAs and without any global alteration in the cellular RNA profile. Furthermore, extensive rRNA cleavage, indicative of high levels of 2-5A, was not observed in RNase L-induced, EMCV-infected cells, whereas transfection of 2-5A into cells resulted in widespread degradation of cellular RNAs. These findings are the first demonstration of the selective capacity of RNase L in intact cells and link this selective activity to cellular levels of 2-5A.
MATERIALS AND METHODS
Cell culture and transfections.
All cells were maintained in a humidified atmosphere of 5% CO2–95% balanced air at 37°C. Cells were cultured in growth medium consisting of Dulbecco modified Eagle medium, 10% fetal calf serum (Atlanta Biologicals), and 1× antibiotic-antimycotic (Life Technologies); G418 (500 μg/ml) and hygromycin (250 μg/ml) were added in stably transfected cell lines. NIH 3T3 cells were transfected with the Lac repressor expression plasmid (p3′SS; Stratagene) by calcium phosphate coprecipitation, by standard methods (42). Stable transfectants were selected by growth in hygromycin and were clonally isolated; cell lines expressing high levels of Lac repressor, as determined by Western blot analysis, were chosen for further study. Lac repressor-expressing cells were transfected with an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible RNase L expression plasmid in which the human RNase L cDNA had been cloned into the NotI site of pOP13 (Stratagene) in the sense orientation. Stable transfectants were selected by growth in hygromycin and G418 and were clonally isolated. RNase L was induced by treatment of cells with 3mM IPTG. SVT2/ZB1 and SVT2/pSVL cells were cultured as previously described (21). Interferon treatment was at 1,000 U/ml, unless otherwise indicated, with murine IFN-α/β (Lee Biomolecular). Cells were harvested by trypsinization, stained with trypan blue, and counted by hemocytometer.
Antiviral assays and in vivo labeling.
For antiviral assays, cells were seeded in 96-well plates at 104 cells/well and allowed to attach overnight; cells were treated with IFN for 5 h, and then IPTG (3 mM) was added to induce RNase L and cells were incubated for 24 h prior to virus infection. Cells were infected with virus at an MOI of 1.0. Infection was for 1 h in low-serum (2% fetal calf serum) medium, and then the virus was removed and cells were washed twice in phosphate-buffered saline, refed with growth medium, and incubated for 18 h. Viable cells were then stained with a modification of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Cell Titer-96; Promega) (49), as described by the supplier. Percent viable cells protected by IFN from virus-induced killing was calculated for each data point with the following formula: sample value − virus control (no IFN)/cell control (no virus) − virus control (no IFN).
For labeling studies, cells were seeded in 24-well plates (5 × 104 cells/well) and infected with virus as described above. After 1 h of infection, virus was removed and cells were refed with growth medium; 2 h later, the cells were treated with 5 μg of actinomycin D per ml for 30 min, and then [3H]uridine (Amersham) at 5 μCi/ml was added to the medium. At various times PI, cells were harvested by brief trypsinization and then lysed in ice-cold 5% trichloroacetic acid (TCA) (Sigma). Incorporated radioactivity was determined by filtration through glass fiber filters followed by consecutive washes with 5% TCA, ethanol, ethanol-acetone (1:1), and acetone; dried filters were then counted in 5 ml of RediSafe scintillation fluid (Beckman). In pulse-chase experiments, [3H]uridine was removed at 8 h PI and cells were refed with growth medium containing unlabeled uridine (10 mM) and cytidine (5 mM). TCA-insoluble radioactivity was measured at various postchase time points.
2-5A transfection.
2-5A trimer–triphosphate, at a concentration of 1 μM, was transfected into cells by calcium phosphate coprecipitation for 75 min (42). The cells were then washed with phosphate-buffered saline, refed with growth medium, and incubated for 2.5 h; total RNA was then harvested for analysis with Trizol reagent (Life Technologies).
Analyses of gene expression. (i) Protein.
Transfected RNase L protein was measured in postmitochondrial supernatants by Western blot analysis with a monoclonal antibody specific for the human enzyme (kindly provided by Robert H. Silverman, The Cleveland Clinic Foundation) (14); RNase L was visualized by reaction of blots with ECL (Amersham) and exposure on X-Omat AR film (Kodak). Lac repressor expression was measured by reacting Western blots to a monoclonal antibody specific for the Lac repressor (Stratagene) and visualized by ECL.
(ii) RNA.
Total RNA was analyzed on glyoxal-agarose gels by ethidium staining or Northern blot hybridization. Hybridization probes were labeled with [α-32P]dCTP by random priming (Pharmacia). A plasmid containing EMCV cDNA, pEC9 (20), was generously provided by Ann C. Palmenberg, University of Wisconsin. cDNA hybridization probes for ISG15 (4), c-myc (34), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (17), and 18S rRNA (21) have been previously described. A PhosphorImager (Molecular Dynamics) was used for quantitation of the data given in Fig. 3A. Autoradiographic images shown in the figures were reproduced by the Eagle Eye (Stratagene) photodocumentation system with real-time imaging without enhancement.
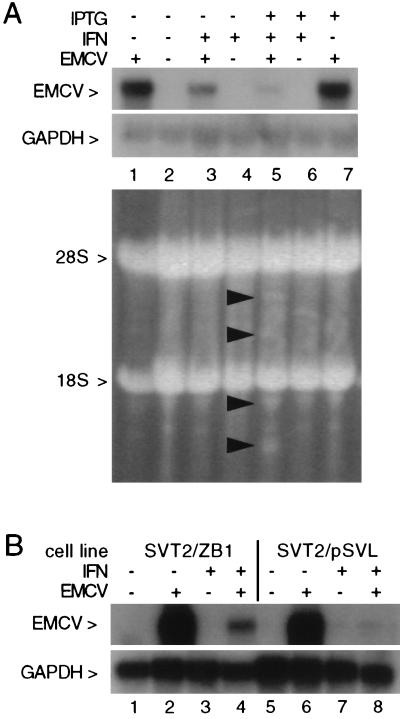
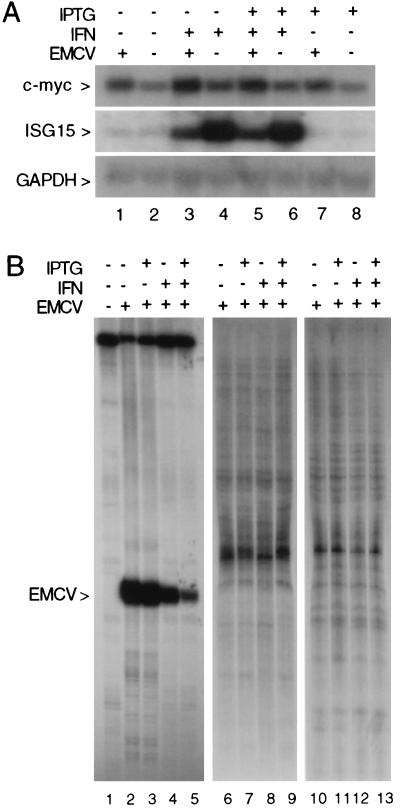
FIG. 3.
RNase L-dependent reduction in EMCV RNA in IFN-treated cells. (A) Total RNA isolated from LS1 cells following RNase L induction, IFN treatment, and EMCV infection, as indicated, was analyzed on a glyoxal-agarose gel (5 μg of RNA/lane); IFN and IPTG treatments were as described in Materials and Methods, and EMCV infection was for 6 h. The ethidium-stained gel was photographed (lower panel) or transferred to a nylon membrane and hybridized to EMCV and GAPDH cDNA probes; an autoradiograph of these blots is shown in the upper panel. (B) Total RNA isolated from mutant RNase L (SVT2/ZB1) or vector control (SVT2/pSVL) cells following IFN treatment or EMCV infection, as indicated, was analyzed on a glyoxal-agarose gel (7 μg of RNA/lane). A Northern blot of this gel was hybridized to EMCV (upper panel) and GAPDH (lower panel) cDNA probes; an autoradiograph of this blot is shown.
Differential display analysis.
Total cellular RNA (0.2 μg/reaction) was reverse transcribed (SuperScript II; Life Technologies) as described by the supplier with oligo(dT)11A used as primer, and 4 μl of the 20-μl reaction mixture was PCR amplified (Amplitaq; Perkin-Elmer) in the presence of [α-33P]dATP and arbitrary sequence upstream primers, as indicated in the figure legends. Amplification conditions were as described by the supplier (RNAmap; GenHunter). Reaction products were analyzed on 6% acrylamide denaturing sequencing gels and autoradiographed.
RESULTS
Inducible expression of enzymatically active RNase L.
To examine the role and mechanism of action of RNase L in the antiviral activity of IFN, we sought to express transfected RNase L in cells. Constitutive expression of transfected RNase L was previously demonstrated to be incompatible with cell viability (20a); therefore, an inducible RNase L expression system (LacSwitch; Stratagene) was employed. The human and murine proteins are highly homologous and exhibit essentially identical enzymatic properties in vitro (15, 56). The human form of RNase L was thus chosen for transfection, since monoclonal antibodies specific for the human enzyme were available (14) and permit direct measurement of transgene expression without contribution from the endogenous murine enzyme. NIH 3T3 cells were transfected with an expression vector driving constitutive expression of the Lac repressor and an RNase L expression vector under the control of a Lac operator. Constitutive synthesis of the Lac repressor inhibits expression of RNase L; treatment of cells with IPTG inactivated the Lac repressor and resulted in a 2- to 10-fold induction in RNase L in different clonal cell lines (compare control and IPTG-induced lanes in Fig. 1A). Subsequent experiments were performed with the LS1 cell line, as it displayed a high level of induction and a low level of leaky expression in uninduced cells. These inducible cells thus permitted study of the biological effects of ectopic RNase L expression without the compromised growth properties and viability associated with constitutive expression of the transfected enzyme.
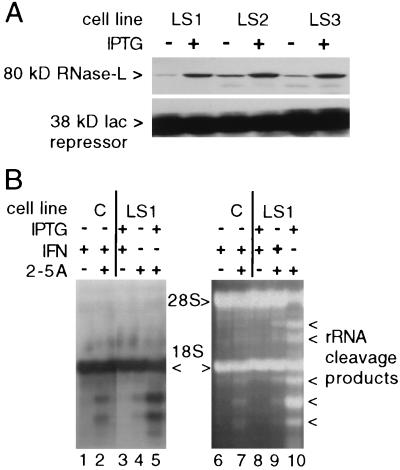
FIG. 1.
Inducible expression and enzymatic activation of transfected RNase L. (A) RNase L in 100 μg of postmitochondrial supernatant protein from independent transfected cell lines (LS1, LS2, and LS3) was analyzed on Western blots before and 24 h after treatment with 3 mM IPTG. Blots were reacted with a monoclonal antibody specific for transfected human RNase L and with a polyclonal antibody to the constitutively expressed Lac repressor (Stratagene). A film exposed to an ECL (Amersham)-reacted blot is shown. (B) Total RNA from control (C) and RNase L-inducible LS1 cells which had been transfected with 1 μM 2-5A trimer and treated with 1,000 U of murine IFN-α/β (Lee Biomolecular), as indicated, was analyzed on a glyoxal-agarose gel (15 μg of RNA/lane). A Northern blot of this gel was hybridized with an 18S rRNA probe; an autoradiograph of the blot is shown in the lefthand panel. A photograph of the ethidium-stained gel is shown in the righthand panel. Arrows indicate specific rRNA cleavage products.
To determine if the transfected RNase L was enzymatically active, cellular RNA was analyzed for the presence of RNase L-specific rRNA cleavage products following 2-5A transfection. RNase L-mediated rRNA cleavage occurs in the presence of high levels of 2-5A (i.e., ≥10nM [29, 54]), which can be achieved by 2-5A transfection, providing a convenient assay for RNase L activity in intact cells (48). Following transfection with 2-5A trimer–triphosphate, characteristic 18S rRNA cleavage products were observed in RNA from IFN-treated vector control or uninduced LS1 cells (Fig. 1B, lanes 2, 4, 7, and 9). rRNA cleavage was dramatically increased following 2-5A transfection in RNase L-induced LS1 cells (compare lanes 4 and 9 to lanes 5 and 10 in Fig. 1B). No rRNA degradation was observed following RNase L induction and IFN treatment in the absence of 2-5A transfection (Fig. 1B, lanes 3 and 8), indicating that RNase L induction alone does not lead to widespread RNA decay (also see below). Vector control and uninduced LS1 cells exhibited identical RNase L activities upon 2-5A transfection (compare lanes 2 and 4 in Fig. 1B); therefore, uninduced LS1 cells were used as controls in subsequent experiments. Both 18S and 28S rRNA cleavage products were detected following activation of the transfected human enzyme, whereas activation of the endogenous murine enzyme resulted in only 18S degradation products (compare lanes 7 and 10 in Fig. 1B). These distinct cleavage activities provide a qualitative measurement of enzyme activity from the transfected RNase L and demonstrate that the endogenous enzyme is functional in these cells. Importantly, the presence of human RNase L-specific 28S cleavage products indicates that the increased enzymatic activity is a direct effect of the transfected human enzyme rather than an enhanced activity of the endogenous murine enzyme due to titration of a reported RNase L inhibitor (5). The ability to exogenously regulate expression of a functional RNase L enzyme in these cell lines establishes a system to study the specific biological activities and antiviral mechanisms of this enzyme.
Increased expression of RNase L enhances the anti-EMCV, but not the anti-VSV, activity of IFN.
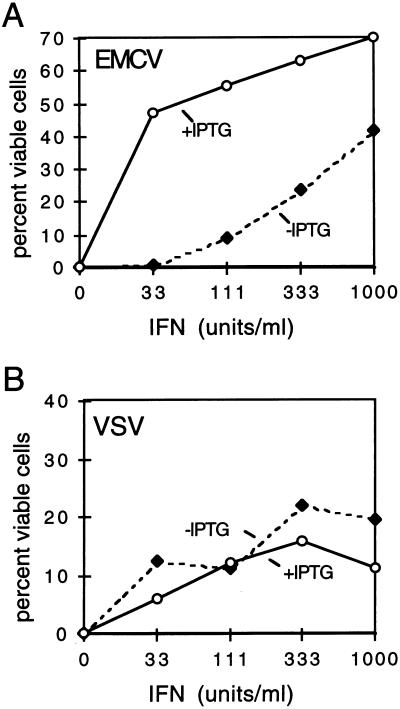
We have previously demonstrated that RNase L activity is required for the anti-EMCV effects of IFN by using a dominant negative RNase L mutant (21). This antiviral effect is thought to be due to the enzymatic activation of RNase L by 2-5A rather than an increase in levels of RNase L protein; indeed, the transcriptional induction of RNase L by IFN is modest (two- to fivefold [56]) compared to other ISGs (20- to 100-fold [26, 52]). We sought to determine directly if the increased expression of transfected human RNase L protein would confer enhanced antiviral activity in mouse NIH 3T3 cells. Cells were treated with IFN in the presence or absence of IPTG for 24 h (at which time induced levels of RNase L were maximal [data not shown]) prior to infection of cells with EMCV or VSV at an MOI of 1.0. At 18 h PI, viable cells were measured by MTT assay. Induction of RNase L in LS1 cells resulted in a dramatic enhancement of IFN-induced protection from EMCV cell killing. Nearly 50% protection was achieved with treatment of 33 U of IFN per ml in RNase L-induced cells, whereas control cells required more than 1,000 U/ml to obtain a similar level of protection (Fig. 2A). In contrast to the enhanced RNase L-dependent antiviral activity observed in EMCV-treated cells, IPTG induction of RNase L had little effect on IFN-mediated protection from VSV infection (Fig. 2B). This result is consistent with several previous studies which have demonstrated that VSV is not sensitive to inhibition by the 2-5A pathway (8, 13, 21). The increased levels of RNase L in the induced cells thus cannot override the restricted targeting of EMCV and not VSV by the 2-5A pathway. In the absence of IFN, induction of RNase L had no detectable antiviral activity in NIH 3T3 cells. This IFN dependence likely reflects a requirement for elevated, IFN-induced levels of 2-5A synthetase to produce 2-5A and activate RNase L in these cells. Alternatively, IFN may function to block the activity of an IFN-sensitive RNase L inhibitor which has been reported to be present in EMCV-infected cells (7, 47; also see Discussion).
FIG. 2.
RNase L enhances the anti-EMCV, but not the anti-VSV, activity of IFN. LS1 cells were treated with the indicated concentrations of murine IFN-α/β for 24 h, in the presence and absence of IPTG (3 mM), to induce RNase L. Cells were then infected with EMCV (A) or VSV (B) at an MOI of 1.0, as described in Materials and Methods. At 18 h PI, viable cells were stained. Data points on the graphs represent means of quadruplicate samples; for each point, the coefficient of variability is ≤3%.
RNase L-dependent reduction of EMCV RNA in IFN-treated cells.
The antiviral activity of RNase L is thought to occur through the degradation of viral RNA; however, the effect of directly manipulating RNase L expression on viral and cellular RNAs has not been examined in virus-infected cells. To determine if the enhanced protection from EMCV observed following IPTG induction of RNase L was mediated through a reduction in EMCV RNA by RNase L, RNA was isolated from control or IPTG-treated LS1 cells which had been pretreated with IFN and then infected with EMCV. A Northern blot of the RNA samples was hybridized with an EMCV cDNA probe (20). A dramatic RNase L-dependent downregulation of the 7.7-kb EMCV RNA was observed in the RNA from RNase L-induced IFN-treated cells compared to cells treated with IFN in the absence of RNase L induction (compare lanes 3 and 5 in Fig. 3A). PhosphorImager analysis revealed a sixfold decrease in EMCV RNA following RNase L induction. Ethidium staining of the RNA gel and hybridization with a GAPDH cDNA probe further confirmed that comparable amounts of rRNA were present in each lane (Fig. 3A, lower panels). Interestingly, despite the elevated levels of 2-5A synthetase (not shown) and RNase L in the IFN-treated, IPTG-induced LS1 cells, rRNA cleavage products were barely detectable following EMCV infection (Fig. 3A, lane 5). This observation indicates that the levels of 2-5A produced at the low MOI used in these experiments were insufficient to induce extensive rRNA cleavage. The RNase L-dependent reduction in EMCV RNA thus occurred in the absence of widespread rRNA cleavage, indicating that low levels of 2-5A are sufficient to effect a dramatic biological response.
To confirm our results with transfected RNase L in LS1 cells and demonstrate that endogenous RNase L also exerts its antiviral activity through a reduction in EMCV RNA, we used a cell line which stably expresses a dominant negative mutant of RNase L. We have previously reported that a truncated clone of RNase L lacking the 89 carboxy-terminal amino acids specifically and efficiently inhibits endogenous RNase L activity when expressed in murine SVT2 cells and results in reduced sensitivity to the anti-EMCV activity of IFN (21). RNA from vector control (SVT2/pSVL) and mutant RNase L (SVT2/ZB1) cells which had been IFN pretreated and EMCV infected, as described above, was analyzed by Northern blotting. Consistent with our results in RNase L-induced LS1 cells, inhibition of endogenous RNase L activity inhibited the IFN-induced reduction in EMCV RNA (Fig. 3B, lanes 4 and 8). Inhibition of RNase L activity in the mutant-expressing cell line did not restore EMCV RNA to levels observed in the absence of IFN, indicating either that endogenous RNase L activity is not completely inhibited or that other IFN-induced activities contribute to its anti-EMCV effects. Taken together, these results indicate that the anti-EMCV activity of the 2-5A system is mediated through an RNase L-dependent reduction in EMCV RNA.
Induction of RNase L inhibits viral RNA synthesis.
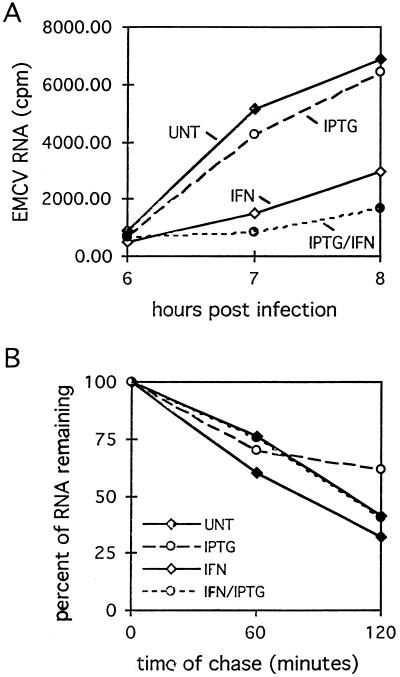
The EMCV genome is a positive-strand, single-stranded RNA. Since RNase L degrades single-stranded RNA, RNase L activation can potentially affect both the synthesis and decay of viral RNA through the degradation of positive- and negative-strand template RNAs and mRNAs, respectively. To determine whether the RNase L-mediated reduction in the steady-state levels of EMCV RNA observed in Northern blot analyses occurred through changes in the accumulation or decay of viral RNA, pulse-chase RNA labeling experiments were conducted. Control and RNase L-induced LS1 cells in the presence and absence of IFN pretreatment were labeled with [3H]uridine beginning at 2 h PI. Significant incorporation of label into TCA-precipitable counts began at 6 h PI and was subsequently measured every hour through 8 h PI. Cells were treated with actinomycin D during the labeling period to inhibit cellular RNA synthesis; therefore, incorporated label represents viral RNA only. As observed in the Northern blot analyses (Fig. 3A), IPTG induction of RNase L in the absence of IFN had no detectable effect on viral RNA synthesis (Fig. 4A). IFN treatment reduced both the absolute levels of [3H]uridine incorporation and the rate of EMCV RNA synthesis. RNase L induction resulted in a significant (P < 0.01 [Student’s t test]) further reduction in viral RNA and the synthesis rate. Specifically, RNase L induction in IFN-treated cells reduced the rate of viral RNA synthesis to 42% of that observed in cells treated with IFN alone and to 17% of that observed in control, uninduced cells (Fig. 4A). To examine the decay of viral RNA in control and RNase L-induced cells, viral RNA was labeled from 2 to 8 h PI, after which the [3H]uridine was removed and chase medium was added; EMCV RNA accumulation prior to this time of chase was insufficient for decay analysis. Interestingly, IFN treatment and RNase L induction had no detectable effects on the stability of viral RNA compared to untreated cells at late times in the virus life cycle (Fig. 4B). The decay rate of viral RNA, as indicated by the slopes of the lines in Fig. 4B, was nearly identical under all treatment conditions at 8 to 10 h PI, exhibiting a half-life of approximately 90 min. These kinetic analyses suggest that the antiviral activity of RNase L occurs at early times PI, possibly through the degradation of RNAs involved in active replication of viral RNA, to inhibit viral RNA synthesis.
FIG. 4.
RNase L inhibits EMCV RNA synthesis. (A) LS1 cells were treated with IFN and IPTG, as indicated, and then infected with EMCV in the presence of [3H]uridine, as described in Materials and Methods. Cells were harvested at the indicated times, and [3H]uridine incorporation was determined by TCA precipitation. (B) At 8 h PI, [3H]uridine was removed from the medium and chased with unlabeled UTP and CTP for an additional 2 h; percent RNA remaining was calculated based on TCA-precipitable counts at the beginning of the chase for each treatment. Data points on both graphs are means of six samples at each time point. UNT, untreated (control) cells.
RNase L selectively targets EMCV RNA without altering the cellular RNA profile.
To examine if the RNase L-mediated reduction in EMCV RNA was selective for viral RNA, the steady-state levels of selected cellular mRNAs of distinct stability and abundance classes were analyzed in the RNA isolated from EMCV-infected cells. Specifically, Northern blots were hybridized with a GAPDH cDNA probe representing high-abundance stable mRNA (17), a c-myc cDNA probe representing a medium-abundance labile mRNA (18), and an ISG15 cDNA probe representing an IFN-inducible mRNA (4). The GAPDH and c-myc mRNAs were present at comparable levels in all samples (Fig. 5A), indicating that RNase L induction and IFN treatment had no effect on the steady-state levels of these mRNAs. Similarly, the kinetics and magnitude of ISG15 induction by IFN were unchanged in control and RNase L-induced cells (Fig. 5A). Interestingly, IFN-induced levels of ISG15 mRNA were consistently reduced in EMCV-infected cells independent of RNase L induction, suggesting a virus-mediated inhibition of ISG15 expression. Thus, while RNase L induction and IFN treatment resulted in a dramatic reduction in EMCV RNA, this activity had no detectable effect on the steady-state levels of specific cellular mRNAs.
FIG. 5.
RNase L induction does not alter the cellular RNA profile. (A) The Northern blot shown in Fig. 3A was rehybridized with c-myc (top panel), ISG15 (middle panel), and GAPDH (bottom panel) cDNA hybridization probes; an autoradiograph of the blot is shown. (B) Total RNA from the EMCV-infected cells shown in Fig. 3 (200 ng/reaction) was analyzed by differential display, as described in Materials and Methods; the reverse primer was oligo(dT)11CA, and the forward primers were EMCV specific (5′GAGTCTGTTCTGG3′ [lanes 1 to 5]) or were arbitrary sequence decamers (5′CTGATCCATG3′ [lanes 6 to 9]; 5′CGTAGATCGT3′ [lanes 10 to 13]). An autoradiograph of the gel is shown.
To further demonstrate the selective action of RNase L on EMCV RNA, differential display analysis (33) was used to examine the effects of ectopic RNase L induction on a larger portion of the cellular mRNA population. In this procedure, a subset of cellular mRNAs are amplified by reverse transcription-PCR so that their expression in cells following RNase L induction, IFN treatment, or EMCV infection can be simultaneously compared. The majority of bands detected in the differential display analysis did not change following RNase L induction (compare lanes 6 to 9 and 10 to 13 in Fig. 5B), indicating that the reduction in EMCV RNA occurs in the absence of any global changes in the cellular RNA population. The primer sets used in lanes 6 to 13 of Fig. 5B were not homologous to EMCV; therefore, bands corresponding to changes in EMCV RNA were not detected. As a positive control for the differential display analysis, we chose a forward primer from the EMCV RNA sequence so that EMCV RNA would be represented in the set of amplified RNAs. Consistent with our Northern blot analyses, an EMCV-specific PCR product was observed in the samples from virus-infected cells and reduced in samples from IFN-treated, RNase L-induced cells (Fig. 5B, lanes 2 to 5). Importantly, cellular RNAs amplified with the EMCV primer do not change with RNase L induction or IFN treatment (compare lanes 1 and 5 in Fig. 5B). These results demonstrate the capacity of RNase L to selectively target viral RNA within a milieu of nontarget RNAs in intact cells.
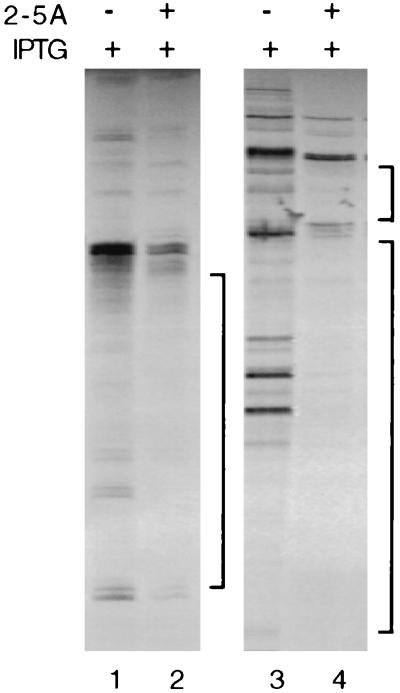
Previous studies have demonstrated that enhanced levels of 2-5A are required for RNase L-mediated rRNA cleavage and are correlated with indiscriminate degradation of cellular RNA (38, 39). Consistent with these reports, rRNA cleavage products were barely detectable in RNase L-induced, IFN-treated, EMCV-infected cells in which EMCV RNA was selectively reduced (Fig. 3A). However, rRNA cleavage was readily detectable in 2-5A-transfected, RNase L-induced cells (Fig. 1B). To further examine the relationship between 2-5A levels and the selective action of RNase L, cellular RNA was analyzed by differential display following 2-5A transfection into control and RNase L-induced cells. In contrast to EMCV-infected cells in which RNase L induction had no detectable effects on cellular RNA (Fig. 5B), 2-5A transfection resulted in the disappearance or reduction of a large portion of bands in the RNase L-induced cells (Fig. 6, lanes 2 and 4). Maintenance of low levels of 2-5A in IFN-treated, EMCV-infected cells was thus critical for the selective action of RNase L on EMCV RNA, as transfection of excess 2-5A into cells led to widespread RNA degradation.
FIG. 6.
2-5A transfection induces a widespread reduction in cellular RNA. Total RNA from the control and 2-5A-transfected LS1 cells shown in Fig. 1 (200 ng/reaction) was analyzed by differential display, as described in Materials and Methods; the reverse primer was oligo(dT)12A, and the forward primers were 5′TTTTGGCTCC3′ (lanes 1 and 2) or 5′CTTTCTACCC3′ (lanes 3 and 4). An autoradiograph of the gel is shown.
DISCUSSION
An accumulation of evidence has established the role of the 2-5A system in mediating the antipicornavirus effects of IFN. Isolation of physical complexes containing EMCV dsRNA has demonstrated the means by which 2-5A synthetase is activated and 2-5A is produced in virus-infected cells (19). The cloning of cDNAs encoding RNase L (56) has provided the tools for functional modulation of RNase L in cells and direct testing of the hypothesis that RNase L-mediated antiviral activity occurs through the direct and selective degradation of viral RNA by RNase L (2, 37). The aim of this study was thus to investigate how RNase L affects the fate of viral and cellular RNAs in EMCV-infected cells and how this relates to the mechanism of RNase L-mediated antiviral activity.
RNase L is present at basal levels in most, if not all, mammalian cell types (46). Although RNase L levels are increased in growth-arrested cells (30) and following IFN treatment (56), its biological activity is thought to be controlled at the level of enzymatic activation rather than through regulation of its transcription and translation. Indeed, transfection of exogenous 2-5A into cells results in increased RNase L activity (48), supporting the view that intracellular levels of 2-5A are rate limiting in the activation of RNase-L and that cellular levels of RNase L are sufficient for maximal biological activity. Ectopic expression of RNase L in a vaccinia vector system, however, resulted in an enhanced protection from vaccinia infection (13). Our results demonstrate an RNase L-dependent increase in anti-EMCV activity, providing further evidence that increased expression of the ribonuclease component alone can result in enhanced biological activity.
While increased levels of RNase L enhanced the anti-EMCV activity of IFN, no anti-EMCV activity was observed in the absence of IFN. In contrast, ectopic RNase L expression was reported in a previous study to confer resistance to vaccinia virus independent of IFN treatment. This difference may reflect the higher basal level of 2-5A synthetase in cell lines used in the latter study (13). Alternatively, the different virus types and MOI employed in the vaccinia system may have resulted in higher levels of viral dsRNA, leading to activation of 2-5A synthetase in the absence of IFN. In the case of EMCV infection, IFN may serve a function in addition to the induction of 2-5A synthetase to facilitate 2-5A pathway-mediated antiviral activity. Previous studies have reported an EMCV-induced RNase L inhibitor which resulted in nearly complete inhibition of 2-5A binding activity by 6 h PI (7, 47). IFN pretreatment abolished this inhibition of RNase L, in agreement with the requirement for IFN observed in our studies. In VSV-infected cells, RNase L induction did not potentiate IFN-mediated antiviral activity. Similarly, inhibition of endogenous RNase L (21) or expression of exogenous RNase L with a vaccinia vector system (13) did not affect IFN-mediated protection from VSV. The increased levels of RNase L obtained in IPTG-induced LS1 cells did not extend the range of viruses sensitive to 2-5A pathway antiviral activity to include VSV, confirming previous studies which demonstrated that multiple mechanisms are responsible for the full range of IFN’s antiviral effects (31, 36).
IPTG induction of RNase L clearly reduced viral RNA to levels below that observed in cells treated with IFN alone. In cells expressing a dominant negative RNase L mutant, the IFN-induced reduction in EMCV RNA was inhibited, thus establishing that the antiviral activity of the endogenous enzyme functions through a reduction in viral RNA. Distinct viral RNA populations function as mRNA, replication templates, and genomes for progeny virions in the course of EMCV infection; therefore, we investigated if the RNase L-mediated reduction in EMCV RNA affected viral RNAs at a specific stage of infection. IPTG induction of RNase L inhibited EMCV RNA synthesis, indicating that RNase L activation occurred at an early point in the virus life cycle. Consistent with this observation, an increase in 2-5A has been reported to parallel the formation of EMCV replicative intermediates at early times in the virus life cycle (19, 47, 53). Inhibition of EMCV RNA synthesis by RNase L suggested that viral RNAs associated with active replication complexes are targeted by RNase L. Studies in a cell-free system which indicated that viral replicative intermediates were preferentially degraded by RNase L (37) support this interpretation.
Interestingly, although IFN treatment and RNase L induction dramatically reduced the absolute levels and synthetic rate of viral RNA synthesis at early times PI, neither treatment had a detectable effect on the decay rate of viral transcripts late in EMCV infection. This observation is consistent with RNase L activation at early steps of virus replication; however, the low levels of viral RNAs at this stage of infection precluded direct measurement of their decay rates. The uniform stability of viral RNAs under all treatment conditions at late times of infection suggests that once viral RNAs dissociate from the replication complex, they behave like the majority of cellular RNAs, being refractory to degradation by RNase L (also see below). Indeed, distinct viral RNA populations have been shown to be differentially localized (50) and to exhibit different kinetic profiles during the picornavirus life cycle (3).
A central issue in elucidating the mechanism by which RNase L elicits its antiviral activity lies in ascertaining its capacity to target specific RNAs for degradation; indeed, this has been a pivotal question since the discovery of the 2-5A system. Previous studies have reported indiscriminate degradation of viral and cellular RNAs by RNase L in the presence of high levels of 2-5A (38, 39); however, high levels of 2-5A are not required for the antiviral and antiproliferative activities of RNase L in cells (21, 39). Studies using a cell-free system have demonstrated 2-5A-dependent, preferential degradation of nascent reovirus mRNA over nonviral mRNAs, further supporting the view that RNase L can function in a selective fashion (2). Our data support a selective mechanism of action for RNase L, as the RNase L-mediated reduction in EMCV RNA was specific for viral RNA and occurred in the absence of detectable changes in specific cellular mRNAs or in the global RNA profile. In addition, significant rRNA cleavage was not observed under conditions in which EMCV RNA was selectively targeted (i.e., in RNase L-induced, IFN-treated cells), indicating that cellular levels of 2-5A were low. Increasing the 2-5A concentration via transfection of exogenous 2-5A into RNase L-induced LS1 cells resulted in rRNA cleavage and widespread degradation of cellular mRNA. These results extend previous correlative and in vitro data, providing the first demonstration of the selective capacity of RNase L in intact cells; our data further link this selective activity to low levels of cellular 2-5A.
Mammalian cells employ diverse strategies in the defense against viral infection depending on the type of virus and conditions of infection. In the classical scenario of IFN-mediated antiviral activity, IFN produced in virus-infected cells protects surrounding cells from subsequent rounds of infection through the collective action of ISG-encoded products. An alternate antiviral strategy is the induction of apoptosis in the host cell to prevent the production and spread of progeny virions (10, 11). The recent findings that the IFN-regulated enzymes PKR (12, 28, 32) and RNase L (6) can function to promote apoptosis suggested that apoptotic and nonapoptotic antiviral pathways are mediated through common mechanisms. In the case of RNase L, recent studies have demonstrated that transfection of cells with 2-5A or constitutive expression of transfected RNase L can induce apoptosis and that inhibition of endogenous RNase L can protect from apoptosis (6). In addition, rRNA cleavage products identical to those generated by RNase L were observed in apoptotic cells (22), suggesting that the role of RNase L in apoptosis may involve widespread degradation of RNA. Conversely, RNase L-dependent anti-EMCV activity occurred through selective reduction in viral RNA in the absence of high levels of 2-5A and rRNA cleavage through a nonapoptotic mechanism, as apoptosis was not observed under the conditions of transient RNase L induction and EMCV infection employed in this study (20a). The ability to regulate the extent of RNase L activation directly through the levels of cellular 2-5A, or indirectly through the levels of dsRNA, thus provides a potential mechanism of directing its biological activity to an apoptotic or nonapoptotic pathway.
Although the full spectrum of biological activities attributable to the 2-5A system are not yet known, it is clear that it serves functions beyond the antipicornavirus effects of IFN. For example, we have previously demonstrated that RNase L is required for the growth-inhibitory response to IFN (21), and it has recently been shown to play a role in apoptosis independent of IFN (6). Our current findings that RNase L can act in a selective manner suggest that RNase L may elicit its growth-inhibitory effects via the degradation of specific cellular mRNAs. Indeed, RNase L-dependent growth inhibition in IFN-treated cells occurs in the absence of detectable rRNA cleavage (21), consistent with low levels of 2-5A and a selective mode of action. The present study has established the capacity of RNase L to selectively reduce viral RNA in intact cells. This provides a biologically relevant substrate which can be used to identify the cis- and trans-acting factors required for RNase L-mediated activity. An understanding of the molecular basis of selective RNA decay by RNase L may provide insights into the mechanisms which limit the range of its antiviral activity and into the nature of its cellular RNA substrates in the absence of viral infection.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant A1369608 from NIAID to B.A.H.
We are grateful to Robert H. Silverman and BeiHua Dong, The Cleveland Clinic Foundation, for providing monoclonal antibody to human RNase L and to Ann C. Palmenberg, University of Wisconsin, for helpful suggestions and for providing an EMCV cDNA. We thank E. C. Borden, J. A. Hewitt, B. Joshi, and T. Sharp, from Baltimore, and J. Castelli, from NIH, for critical readings of the manuscript.
REFERENCES
- 1.Alarcon B, Bugany H, Carrasco L. pppA2′p5A′ blocks vesicular stomatitis virus replication in intact cells. J Virol. 1984;52:183–187. doi: 10.1128/jvi.52.1.183-187.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baglioni C, DeBenedetti A, Williams G J. Cleavage of nascent reovirus mRNA by localized activation of the 2′-5′-oligoadenylate-dependent endoribonuclease. J Virol. 1984;52:865–871. doi: 10.1128/jvi.52.3.865-871.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltimore D, Girard M, Darnell J E. Aspects of the synthesis of poliovirus RNA and the formation of virus particles. Virology. 1966;29:179–189. doi: 10.1016/0042-6822(66)90024-9. [DOI] [PubMed] [Google Scholar]
- 4.Blostrom D C, Fahey S, Kutny R, Korand B D, Knight E., Jr Molecular characterization of the interferon-induced 15-kDa protein. J Biol Chem. 1986;261:8811–8816. [PubMed] [Google Scholar]
- 5.Brisbal C, Martinand C, Silhol M, LeBleu B, Salehzada T. Cloning and characterization of an RNase-L inhibitor: a new component of the interferon-regulated 2-5A pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- 6.Castelli J C, Hassel B A, Wood K A, Li X-L, Amemiya K, Dalakas M C, Torrence P F, Youle R J. A study of the interferon antiviral mechanism: apoptosis activation by the 2-5 a system. J Exp Med. 1997;186:1–6. doi: 10.1084/jem.186.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cayley P J, Knight M, Kerr I M. Virus-mediated inhibition of the ppp(A2′p)nA system and its prevention by interferon. Biochem Biophys Res Commun. 1982;104:376–382. doi: 10.1016/0006-291x(82)90647-7. [DOI] [PubMed] [Google Scholar]
- 8.Chebath J, Benech P, Revel M, Vigneron M. Constitutive expression of (2′-5′)oligo A synthetase confers resistance to picornavirus infection. Nature. 1987;330:587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- 9.Clemmens M J, Williams B R G. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell. 1978;13:565–572. doi: 10.1016/0092-8674(78)90329-x. [DOI] [PubMed] [Google Scholar]
- 10.Clouston W M, Kerr J F R. Apoptosis, lymphotoxicity and the containment of virus infections. Med Hypotheses. 1985;18:399–404. doi: 10.1016/0306-9877(85)90107-0. [DOI] [PubMed] [Google Scholar]
- 11.Collins M. Potential roles of apoptosis in viral pathogenesis. Am J Crit Care Med. 1995;152:S20–S24. doi: 10.1164/ajrccm/152.4_Pt_2.S20. [DOI] [PubMed] [Google Scholar]
- 12.Der S D, Yang Y L, Weissmann C, Williams B R G. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz-Guerra M, Rivas C, Esteban M. Inducible expression of the 2-5A synthetase/RNase-L system results in inhibition of vaccinia virus replication. Virology. 1997;227:220–228. doi: 10.1006/viro.1996.8294. [DOI] [PubMed] [Google Scholar]
- 14.Dong B, Silverman R H. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 15.Dong B, Xu L, Zhou A, Hassel B A, Lee X, Torrence P F, Silverman R H. Intrinsic molecular activities of the interferon-induced 2-5A-dependent RNase. J Biol Chem. 1994;269:14153–14158. [PubMed] [Google Scholar]
- 16.Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate-dependent endonuclease. Science. 1981;212:1020–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 17.Fort P M, Marty L, Piechaczyk M, el Sabrouty S, Dani C, Janteur P, Blanchard M. Various rat adult tissues express only one major species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg M E, Belasco J G. Control of the decay of labile protooncogene and cytokine mRNAs. In: Belasco J G, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press; 1993. pp. 199–218. [Google Scholar]
- 19.Gribaudo G, Lembo D, Cavallo G, Landolfo S, Lengyel P. Interferon action: binding of viral RNA to the 40-kilodalton 2′-5′-oligoadenylate synthetase in interferon-treated HeLa cells infected with encephalomyocarditis virus. J Virol. 1991;65:1748–1757. doi: 10.1128/jvi.65.4.1748-1757.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn H, Palmenberg A C. Encephalomyocarditis viruses with short poly(C) tracts are more virulent than their mengovirus counterparts. J Virol. 1995;69:2697–2699. doi: 10.1128/jvi.69.4.2697-2699.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Hassel, B. A. Unpublished results.
- 21.Hassel B A, Zhou A, Sotomayor C, Maran A, Silverman R H. A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J. 1993;12:3297–3304. doi: 10.1002/j.1460-2075.1993.tb05999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houge G, Robaye B, Eikhom T S, Golstein J, Mellgren G, Gjertsen B T, Lanotte M, Doskeland S O. Fine mapping of 28S rRNA sites specifically cleaved in cells undergoing apoptosis. Mol Cell Biol. 1995;15:2051–2062. doi: 10.1128/mcb.15.4.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hovanessian A G, Brown R E, Kerr I M. Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature. 1977;268:537–539. doi: 10.1038/268537a0. [DOI] [PubMed] [Google Scholar]
- 24.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B. 1957;147:258–267. [PubMed] [Google Scholar]
- 25.Johnston M I, Hearl W G. Purification and characterization of a 2′-phosphodiesterase from bovine spleen. J Biol Chem. 1987;262:8377–8382. [PubMed] [Google Scholar]
- 26.Kalvakolanu D V, Borden E C. An overview of the interferon system: signal transduction and mechanisms of action. Cancer Investig. 1996;14:25–53. doi: 10.3109/07357909609018435. [DOI] [PubMed] [Google Scholar]
- 27.Kerr I M, Brown R E. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kibler K V, Shors T, Perkins K B, Zeaman C C, Banaszak M P, Biesterfeldt J, Langland J O, Jacobs B L. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. J Virol. 1997;71:1992–2003. doi: 10.1128/jvi.71.3.1992-2003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knight M, Cayley P J, Silverman R H, Wreschner D H, Gilbert C S, Brown R E, Kerr I M. Radioimmune, radiobinding and HPLC analysis of 2-5A and related oligonucleotides from intact cells. Nature. 1980;288:189–192. doi: 10.1038/288189a0. [DOI] [PubMed] [Google Scholar]
- 30.Krause D, Panet A, Arad G, Dieffenbach C W, Silverman R H. Independent regulation of ppp(A2′p)nA-dependent RNase in NIH3T3, clone 1 cells by growth arrest and interferon treatment. J Biol Chem. 1985;260:9501–9507. [PubMed] [Google Scholar]
- 31.Kumar R, Choubey D, Lengyel P, Sen G C. Studies on the role of the 2′-5′-oligoadenylate synthetase-RNase L pathway in beta interferon-mediated inhibition of encephalomyocarditis virus replication. J Virol. 1988;62:3175–3181. doi: 10.1128/jvi.62.9.3175-3181.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S B, Esteban M. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology. 1994;199:491–496. doi: 10.1006/viro.1994.1151. [DOI] [PubMed] [Google Scholar]
- 33.Liang P, Pardee A B. Differential display of eucaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 34.Marcu K B, Harris L J, Stanton L W, Erikson J, Watt R, Croce C M. Transcriptionally active c-myc oncogene is contained within NIARD, a DNA sequence associated with chromosome translocations in B-cell neoplasia. Proc Natl Acad Sci USA. 1983;80:519–523. doi: 10.1073/pnas.80.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merlin G, Chebath J, Benech P, Metz R, Revel M. Molecular cloning and sequence of partial cDNA for interferon-induced (2′-5′)oligo(A) synthetase mRNA from human cells. Proc Natl Acad Sci USA. 1983;80:4904–4908. doi: 10.1073/pnas.80.16.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meurs E F, Watanabe Y, Kadereit S, Barber G N, Katze M G, Chong K, Williams B R G, Hovanessian A G. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J Virol. 1992;66:5805–5814. doi: 10.1128/jvi.66.10.5805-5814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsen T W, Baglioni C. Mechanism for discrimination between viral and host mRNA in interferon-treated cells. Proc Natl Acad Sci USA. 1979;76:2600–2604. doi: 10.1073/pnas.76.6.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilsen T W, Maroney P A, Baglioni C. Double-stranded RNA causes synthesis of 2′,5′-oligo(A) and degradation of messenger RNA in interferon-treated cells. J Biol Chem. 1981;256:7806–7811. [PubMed] [Google Scholar]
- 39.Nilsen T W, Maroney P A, Baglioni C. Synthesis of (2′,5′)oligoadenylate and activation of an endoribonuclease in interferon-treated HeLa cells infected with reovirus. J Virol. 1982;42:1039–1045. doi: 10.1128/jvi.42.3.1039-1045.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavlovic J, Schroder A, Blank A, Pitossi F, Staeheli P. Mx proteins: GTPases involved in the interferon-induced antiviral state. Ciba Found Symp. 1993;176:233–243. doi: 10.1002/9780470514450.ch15. [DOI] [PubMed] [Google Scholar]
- 41.Rysiecki G, Gewert D R, Williams B R G. Constitutive expression of a 2′,5′-oligoadenylate synthetase cDNA results in increased antiviral activity and growth suppression. J Interferon Res. 1989;9:649–657. doi: 10.1089/jir.1989.9.649. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Saunders M E, Gewert D R, Tugwell M E, McMahon M, Williams B R G. Human 2-5A synthetase: characterization of a novel cDNA and corresponding gene structure. EMBO J. 1985;4:1761–1768. doi: 10.1002/j.1460-2075.1985.tb03848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindler C, Darnell J E. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 45.Sen G C, Lengyel P. The interferon system—a bird’s eye view of its biochemistry. J Biol Chem. 1992;267:5017–5020. [PubMed] [Google Scholar]
- 46.Silverman R H. 2-5A dependent RNase L: a regulated endoribonuclease in the interferon system. In: D’Alessio G 2nd, Riordan J F, editors. Ribonucleases: structure and function. New York, N.Y: Academic Press; 1997. pp. 515–551. [Google Scholar]
- 47.Silverman R H, Cayley P J, Knight M, Gilbert C S, Kerr I M. Control of the ppp(A2′p)nA system in HeLa cells. Eur J Biochem. 1982;124:131–138. doi: 10.1111/j.1432-1033.1982.tb05915.x. [DOI] [PubMed] [Google Scholar]
- 48.Silverman R H, Skehel J J, Tharappel J C, Wreschner D H, Kerr I M. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J Virol. 1983;46:1051–1055. doi: 10.1128/jvi.46.3.1051-1055.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tada H, Shiho O, Kuroshima K, Koyama M, Tsudamoto K. An improved colorimetric assay for interleukin 2. J Immunol Methods. 1986;93:157–165. doi: 10.1016/0022-1759(86)90183-3. [DOI] [PubMed] [Google Scholar]
- 50.Troxler M, Egger D, Pfister T, Bienz K. Intracellular localization of poliovirus RNA by in situ hybridization at the ultrastructural level using single-stranded riboprobes. Virology. 1992;191:687–697. doi: 10.1016/0042-6822(92)90244-j. [DOI] [PubMed] [Google Scholar]
- 51.Watling D, Serafinowska H T, Reese C B, Kerr I M. Analogue inhibitor of 2-5A action: effect on the interferon-mediated inhibition of encephalomyocarditis virus replication. EMBO J. 1985;4:431–436. doi: 10.1002/j.1460-2075.1985.tb03647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams B R G. Transcriptional regulation of interferon-stimulated genes. Eur J Biochem. 1991;200:1–11. doi: 10.1111/j.1432-1033.1991.tb21041.x. [DOI] [PubMed] [Google Scholar]
- 53.Williams B R G, Golgher R R, Brown R E, Gilbert C S, Kerr I M. Natural occurrence of 2-5A in interferon-treated EMC virus-infected L cells. Nature. 1979;282:582–586. doi: 10.1038/282582a0. [DOI] [PubMed] [Google Scholar]
- 54.Wreschner D H, James T C, Silverman R H, Kerr I M. Ribosomal RNA cleavage, nuclease activation and 2-5A (ppp(A2′p)nA) in interferon-treated cells. Nucleic Acids Res. 1981;9:1571–1581. doi: 10.1093/nar/9.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wreschner D H, McCauley J W, Skehel J J, Kerr I M. Interferon action: sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 56.Zhou A, Hassel B A, Silverman R H. Expression cloning of 2-5A dependent RNase: a uniquely regulated mediator of interferon action. Cell. 1993;72:1–20. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]