Abstract
With the emergence of drug-resistant strains, the treatment of tuberculosis (TB) is becoming more difficult and there is an urgent need to find new anti-TB drugs. Mycobacterium marinum, as a model organism of Mycobacterium tuberculosis, can be used for the rapid and efficient screening of bioactive compounds. The 14-membered resorcylic acid lactones (RALs) have a wide range of bioactivities such as antibacterial, antifouling and antimalarial activity. In order to further study their bioactivities, we initially constructed a 14-membered RALs library, which contains 16 new derivatives. The anti-M. marinum activity was evaluated in vitro. Derivatives 12, 19, 20 and 22 exhibited promising activity with MIC90 values of 80, 90, 80 and 80 μM, respectively. The preliminary structure–activity relationships showed that the presence of a chlorine atom at C-5 was a key factor to improve activity. Further studies showed that 12 markedly inhibited the survival of M. marinum and significantly reduced the dosage of positive drugs isoniazid and rifampicin when combined with them. These results suggest that 12 is a bioactive compound capable of enhancing the potency of existing positive drugs, and its effective properties make it a very useful leads for future drug development in combating TB resistance.
Keywords: 14-membered resorcylic acid lactones, Mycobacterium marinum, Mycobacterium tuberculosis, marine natural products, anti-tuberculosis
1. Introduction
Tuberculosis (TB) is caused by Mycobacterium tuberculosis and is one of the deadliest infectious diseases worldwide [1,2]. By 2022, TB became the second leading cause of death globally from a single infectious source after COVID-19, with almost twice as many deaths as HIV/AIDS [3]. Multidrug-resistant tuberculosis (MDR-TB) is a public health crisis and health security threat, and cases of drug-resistant tuberculosis are increasing gradually due to the emergence of multidrug-resistant strains [4,5,6,7]. However, due to the difficulty of anti-tuberculosis drug development and other factors, only bedaquiline, delamanid and pretomanid have been approved for clinical treatment in the past decade, and the first-line drugs are still mainly isoniazid and rifampicin, which were discovered in the 1950s [8,9,10]. Therefore, the development of new anti-TB drugs is crucial.
M. tuberculosis is highly infectious and pathogenic, and its culture must be carried out in biosafety level-3 (BSL-3) laboratories [11,12]. Specifically, M. tuberculosis grows slowly under in vitro culture conditions, taking about 15 to 20 h to proliferate one generation and 12 days to culture, resulting in a long study cycle. These factors limit the in-depth research of anti-TB drugs [13,14]. In contrast, Mycobacterium marinum, from the same genus as M. tuberculosis, which grows rapidly with a growth time of about 4 h, is less pathogenic and can be operated in BSL-2 laboratories. More importantly, M. marinum and M. tuberculosis have high genetic and protein sequence homology. The former’s genome size is 6.5 Mb, which is 2.1 Mb longer than the genes of M. tuberculosis [15]. It shares more than 85% of its genome with M. tuberculosis and shares major virulence factors. Additionally, M. marinum infection in humans usually occurs when broken skin comes into direct contact with infected fish or water sources. After infection, patients exhibit pathological features of TB, such as granuloma. Clinically, M. marinum can be effectively treated with anti-TB drugs such as rifampicin, ethambutol and quinolones. Based on the advantages of its good biosafety and ease of operation, M. marinum is also widely used as one of the model organisms to study the pathogenesis of M. tuberculosis [16,17,18].
Natural products are an important source of new drug development, and more than two-thirds of small molecules approved by the FDA between 1981 and 2019 were related to natural products [19]. The value of natural products of marine origin cannot be ignored. The unique marine environment has created complex, novel and diverse natural products, and endow marine natural products with diversity and particularity in pharmacological activity [20,21]. In terms of anti-TB activity, marine compounds have shown important research value and are valuable resources for the development of anti-TB drugs [22].
The search for new bioactive natural products and derivatives from marine-derived fungi is an ongoing focus of our laboratory. One of our research subjects focuses on resorcylic acid lactones (RALs), polyketide natural products with a 14-membered macrocyclic ring fused to a resorcylic acid residue, which have antibacterial, antifouling, antimalarial and other activities [23,24]. In our previous research, a series of bioactive natural 14-membered RALs (Cochliomycins A–G, 5-Bromozeaenol and 3,5-Dibromozeaenol) were isolated from the marine-derived fungus Cochliobolus lunatus (Figure 1) [23,25,26,27]. Especially, cochliomycin A, at a concentration of 1.2 μg/mL, showed significant antifouling activity against the barnacle Balanus Amphitrite [23]. In addition, a series of 14-membered RAL derivatives with antiplasmodial and antifouling activities have been discovered [25,26,27,28,29,30].
Figure 1.
Fourteen-membered lactones isolated from Cochliobolus lunatus [23,25,26,27].
In this study, we constructed a library consisting of zeaenol (1) and its derivatives, 2–97, aimed to enrich the structural diversity of 14-membered RALs and evaluate their structure–activity relationships. Among the synthesized compounds, 19 and 24–38 are new derivatives. In addition, six different bacteria and fungi were selected for the in vitro screening of activity. Derivatives 12, 19, 20 and 22 have selective activity against M. marinum. A further study showed that compound 12 significantly inhibited the survival of M. marinum, and the combination with positive drugs significantly reduced the dose of positive drugs isoniazid and rifampicin.
2. Results and Discussion
2.1. Chemistry
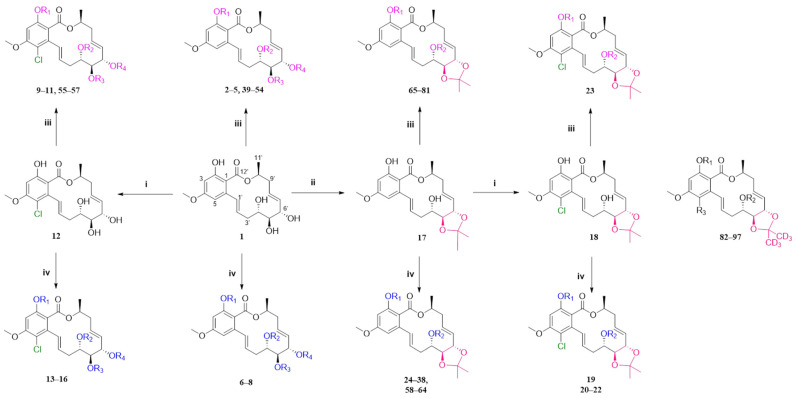
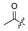
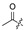
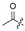
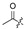
The fermentation condition of Cochliobolus lunatus (CHNSCLM-0009) was liquid fermentation. The crude extract obtained after fermentation was subjected to silica gel column chromatography (CC) and recrystallization, and a total of 7.54 g of zeaenol (1) was obtained [29,30]. 97 derivatives were semi-synthesized using zeaenol (1) as a starting material through one to three steps (Scheme 1). Synthetic schemes for some of the compounds (2–18, 20–23 and 39–97) can be seen in reference [30].
Scheme 1.
The synthetic route. i SO2Cl2, 0 °C; ii p-TsOH, acetone, room temperature, 5 h; iii ArCH2Br reagent, K2CO3, acetone, 50 °C, 24 h; iv anhydride, acyl chloride reagents or carboxylic acids, DMAP, EDCl, DCM, 45 °C, 3–4 h. Compounds 82–97 were synthesized in the same way as other compounds were, except acetone-d6 was used for acetal formation. The specific information of compounds 2–18, 20–23, 39–97 was shown in Ref. [27].
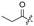
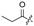
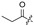
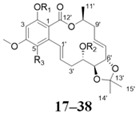
Among the 97 compounds of the library, 38 representative derivatives are shown in Table 1, including the new compounds 19 and 24–38. Compounds 39–97 are shown in Table S1.
Table 1.
Zeaenol (1) and its derivatives 2–38.
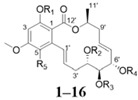
| |||||||||||
| No. | R1 | R2 | R3 | R4 | R5 | No. | R1 | R2 | R3 | R4 | R5 |
| 1 | H | H | H | H | H | 9 |

|
H | H | H | Cl |
| 2 |
|
H | H | H | H | 10 |
|
H | H | H | Cl |
| 3 |
|
H | H | H | H | 11 |
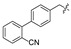
|
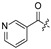
H | H | H | Cl |
| 4 |
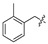
|
H | H | H | H | 12 | H | H | H | H | Cl |
| 5 |
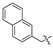
|
H | H | H | H | 13 |
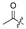
|
|
|
|
Cl |
| 6 |
|
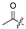
|
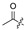
|
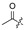
|
H | 14 |
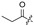
|
|
|
|
Cl |
| 7 |
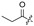
|

|
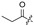
|
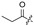
|
H | 15 |

|

|
H |
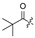
|
Cl |
| 8 |
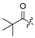
|
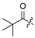
|
H |
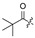
|
H | 16 |
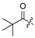
|
H | H | H | Cl |
| |||||||||||
| No. | R1 | R2 | R3 | No. | R1 | R2 | R3 | ||||
| 17 | H | H | H | 28 |
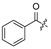
|
H | H | ||||
| 18 | H | H | Cl | 29 |
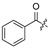
|
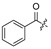
|
H | ||||
| 19 |
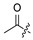
|
H | Cl | 30 |
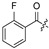
|
H | H | ||||
| 20 |

|
H | Cl | 31 |
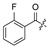
|
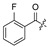
|
H | ||||
| 21 |
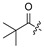
|
H | Cl | 32 | H |
|
H | ||||
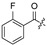
| 22 |
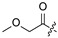
|
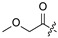
|
Cl | 33 |
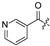
|
H | H | ||||
| 23 |
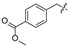
|
H | Cl | 34 |
|
|
H | ||||
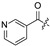
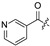
| 24 |
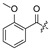
|
H | H | 35 | H |
|
H | ||||
| 25 |

|
H | H | 36 |
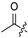
|
H | H | ||||
| 26 |

|
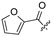
|
H | 37 |
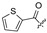
|
H | H | ||||
| 27 | H |
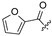
|
H | 38 |
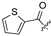
|
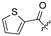
|
H | ||||
2.2. Evaluation of Biological Activity
2.2.1. Anti-M. marinum and Other Antimicrobial Activity
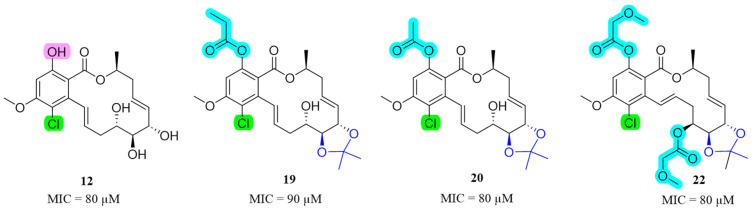
The cases of drug-resistant tuberculosis are increasing gradually due to the emergence of multidrug-resistant strains [31,32]. As a model organism of M. tuberculosis, M. marinum can quickly and efficiently screen bioactive compounds. The antibacterial and antifungal activity of the 97 RAL derivatives were evaluated. We discovered several compounds exhibiting potent antibacterial activity, as evidenced by the data presented in Table 2. The activities of the remaining derivatives (MIC90 > 200 µM) are not shown in Table 2. Derivatives 12, 19, 20 and 22 exhibited promising anti-M. marinum activity with MIC90 values of 80, 90, 80 and 80 μM, respectively (Figure 2). Comparatively, the MIC90 of isoniazid was 40 μM, which indicated the presence of derivatives 12, 19, 20 and 22, and these exhibit good activity. It is worth noting that they also have antibacterial activity selectivity. Compound 1 was the raw material for all derivatives and did not exhibit antimicrobial activity. Derivatives 12, 19, 20 and 22 showed substantially improved anti-M. marinum activity compared with compound 1. In order to determine the safety of these compounds, we selected the active derivatives 12, 19, 20 and 22 to evaluate non-small cell lung cancer (A549), and the results showed that the IC50 values of the above compounds were between 100 and 600 µM, which fully verified that the compounds had own antibacterial effects rather than toxicity.
Table 2.
Antimicrobial activity of representative compounds in the RAL library 1.
| Compound | MIC90 (µM) | |||||
|---|---|---|---|---|---|---|
| M. marinum | S. aureus | E. coli | P. aeruginosa | C. albicans | A. fumigatus | |
| 1 | >200 | >100 | >100 | >100 | >100 | >100 |
| 4 | >200 | 12.5 | >100 | >100 | >100 | >100 |
| 11 | 100 | >100 | >100 | >100 | >100 | >100 |
| 12 | 80 | >100 | >100 | >100 | >100 | >100 |
| 18 | >200 | >100 | >100 | >100 | >100 | >100 |
| 19 | 90 | >100 | >100 | >100 | >100 | >100 |
| 20 | 80 | >100 | >100 | >100 | >100 | >100 |
| 22 | 80 | >100 | >100 | >100 | >100 | >100 |
| Isoniazid | 40 | nt | nt | nt | nt | nt |
| Rifampicin | 10 | nt | nt | nt | nt | nt |
| Ciprofloxacin | nt | 3.13 | 0.10 | 1.56 | nt | nt |
| Amphotericin B | nt | nt | nt | nt | 0.84 | 0.07 |
1 Results are the average of three independent experiments, each performed in duplicate. Standard deviations were less than ±10%. nt = not tested.
Figure 2.
Chemical structures of derivatives 12, 19, 20 and 22, and their anti-M. marinum activity.
An overview of the MICs of this 14-membered RAL library in combination with their structures gives preliminary insights of SARs: (1) the anti-M. marinum activity of 14-membered RALs can be significantly improved via the introduction of the chlorine atom at the C-5 position, as seen by comparing the MICs of 1/12; (2) bearing an acetonide group at position 5’6’ reduces activity, as seen by comparing the MICs of 12/18; (3) the acetyl- and propanoyl-substituted phenolic hydroxy group enhanced activity when both the chlorine atom, and an acetonide group at 5’6’ were also present (19, 20 and 22); (4) a comparison of compound 12 with compounds 9–11 indicates that when the phenolic hydroxy group is transformed into an ether, the activity decreases.
2.2.2. Time−Growth Curves of M. marinum Strain Treated with Different Concentrations of Compound 12
According to the above results, derivatives 12, 19, 20 and 22 have selective anti-M. marinum activity. Initially, we tested the solubility of the active compounds. In the aqueous solution of 1% DMSO, the solubility of 12 was 0.30 mg/mL, and the solubility of derivatives 19, 20 and 22 was less than 0.10 mg/mL. Due to the high solubility and strong activity of 12, it was selected for further study.
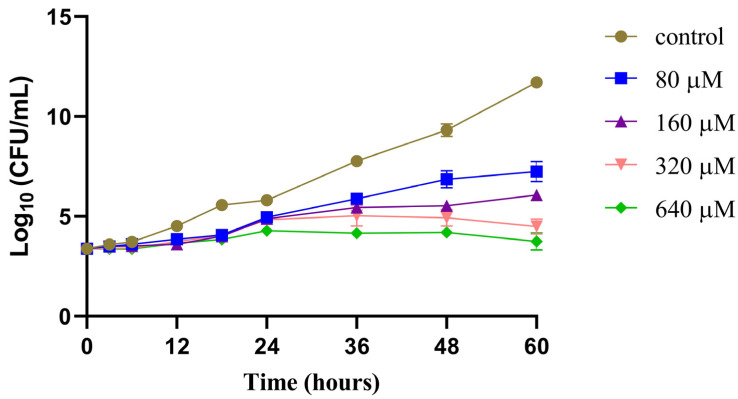
To investigate its effect on the survival of the M. marinum strain, we treated the M. marinum strain with different concentrations of compound 12 and plotted a time–growth curve (Figure 3). The results showed that the higher the concentration of compound 12, the stronger its inhibitory effect on the survival of M. marinum, showing a certain concentration dependence. The anti-M. marinum effect of 12 began to play a role in 24 h. From that point onwards, the inhibition rate of 12 significantly exceeded the growth rate of the bacteria, resulting in a gradually widening gap between the number of bacteria in the treated group and the control group. By 48 h, a noticeable difference between the groups emerged; however, neither completely eradicated the bacteria.
Figure 3.
Time−growth curves of M. marinum strain treated with different concentrations of compound 12. Results are expressed as mean ± SEM, n = 6.
2.2.3. Anti-M. marinum Effects of Compound 12 in Combination with Positive Drugs
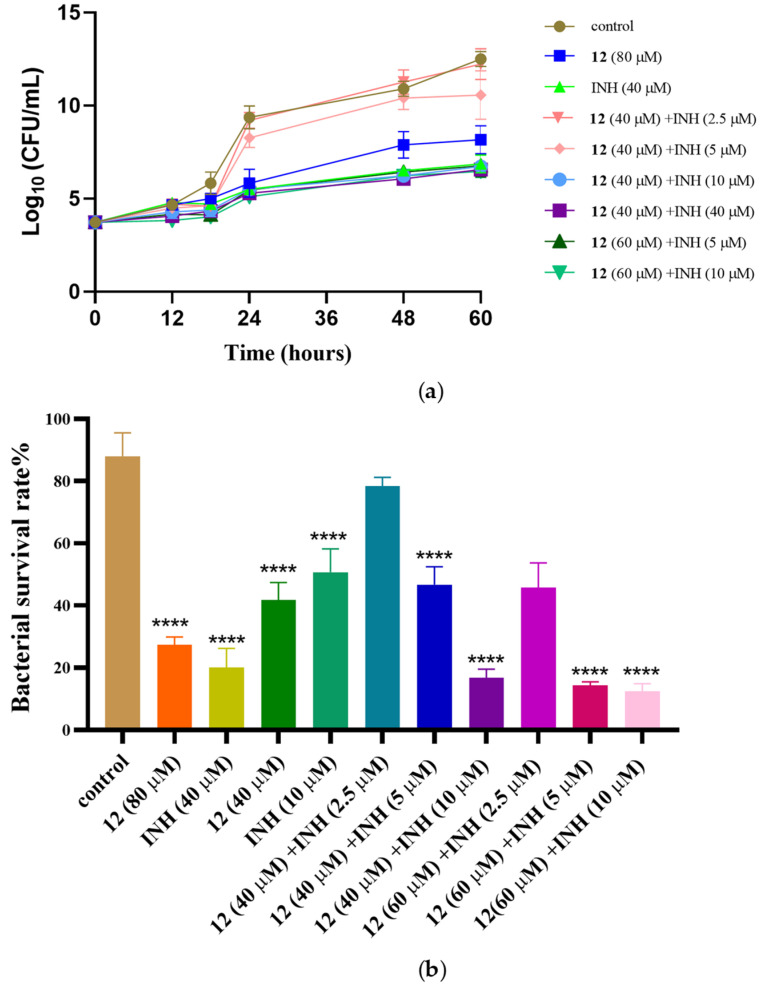
Current TB treatment regimens are extremely challenging, with about 20% of TB deaths caused by drug-resistant M. tuberculosis. MDR-TB is at least resistant to isoniazid and rifampicin, the two most important first-line drugs with which to treat TB. It is necessary to reduce the use of both drugs. We used the checkerboard method [33] to combine 12 with positive drugs for a drug sensitivity test, and the MIC90 values for combined medication are shown in Table 3. The results showed that 12 combined with isoniazid and rifampicin had an obvious additive effect. We observed that 12 can significantly reduce the dosage of the positive drugs isoniazid and rifampicin. Compound 12 at 40 μM made M. marinum four-fold more sensitive to isoniazid and rifampicin, while 20 μM isoniazid or 2.5 μM rifampicin did not inhibit bacterial growth on average. In particular, when the concentration of 12 was adjusted to 60 μM, M. marinum was eight-fold more sensitive to isoniazid and six-fold more sensitive to rifampicin. We selected 12 in combination with isoniazid for bacterial count statistics (Figure 4). It is worth mentioning that 12 did not significantly inhibit bacterial growth in the concentration range of 40–60 μM. Therefore, 12 is considered to be an active compound that can improve the sensitivity of positive drugs.
Table 3.
Anti-M. marinum effects of compound 12 in combination with positive drugs.
| Positive Drugs MIC90 (µM) | Compound 12 MIC90 (µM) | FICI 1 | Mode of Action | |||
|---|---|---|---|---|---|---|
| Alone | Combined | Alone | Combined | |||
| Isoniazid | 40 | 10 | 80 | 40 | 0.75 | additive |
| Rifampicin | 10 | 2.5 | 80 | 40 | 0.75 | additive |
1 The mode of action was determined using the fractional inhibitory concentration index (FICI): (1) FICI ≤ 0.5, synergistic effect; (2) 0.5 < FICI ≤ 1, additive effect; (3) 1 < FICI ≤ 2, irrelevant; (4) FICI > 2, antagonistic effect.
Figure 4.
Compound 12 in combination with positive drugs isoniazid (INH). (a) Time–growth curves of M. marinum strain treated with different concentrations of compound 12 and INH; (b) different concentrations of 12 and INH inhibit the growth of M. marinum. The number of bacterial cells was measured at 48 h. Data are presented as the mean of three experiments ± SD. **** p < 0.0001 compared with the control group.
3. Materials and Methods
3.1. General Experimental Procedures
Reagents and solvents were purchased from commercial suppliers and used without further purification. Column chromatography (CC) was performed on silica gel (Qingdao Haiyang Chemical Group Co., Qingdao, China; 200–300 mesh) and Sephadex LH-20 (Amersham Biosciences, Amersham, UK). TLC silica gel plates (Yantai Zifu Chemical Group) were used for thin-layer chromatography. Semi-preparative HPLC was performed on a Waters 1525 system using a C18 column (Amsterdam, The Netherland; Kromasil, 5 μm, 10 × 250 mm) equipped with a Waters 2996 photodiode array detector, and the flow rate was 2.0 mL/min. NMR spectra were recorded on Bruker Advance NEO 400. Chemical shifts, δ, were measured in ppm, the internal standard was TMS and coupling constants (J) were measured in Hz.
3.2. Fungal Material
The fungal strain Cochliobolus lunatus (CHNSCLM-0009) was isolated from a piece of tissue from the inner part of the freshly collected gorgonian coral Dichotella gemmacea (GX-WZ-20080034), which was collected from the Weizhou coral reef in the South China Sea in September 2008. The fungus was identified as C. lunatus via 16S rRNA gene analysis, and code ZJ2008002 was obtained. The strain was stored at the Key Laboratory of Marine Drugs, the Ministry of Education of China, School of Medicine and Pharmacy, Ocean University of China, Qingdao, China.
3.3. Fermentation, Extraction and Isolation
The fermentation condition of Cochliobolus lunatus (CHNSCLM-0009) was liquid fermentation. This fungus was placed in a 500 mL flask with 200 mL of liquid medium (soluble starch 10 g/L, NaNO3 5 g/L, NaOAc 1 g/L, 1% salinity), and fermented on a rotating shaker at 120 r/min at 28 °C for 10 days [29,30].
The fermentation liquid was extracted with an equal volume of EtOAc 3–4 times, and concentrated to obtain 25 g of crude extract. The total crude extract was separated via silica gel column chromatography (CC). Ethyl acetate and petroleum ether were selected as eluents. Zeaenol (1) was obtained in a 60% ethyl acetate/petroleum ether composition. After repeated recrystallization of the crude product (ethyl acetate/petroleum ether/methanol), a total of 7.54 g of zeaenol (1) was obtained.
3.4. General Synthetic Methods for Compounds 2–97
The synthesis of reported compounds (2–18, 20–23 and 39–97) is not described in this article. The synthesis and detailed data of these compounds can be found in reference [30]. Here, we describe in detail the synthetic steps of compound 19 and 24–38.
3.4.1. General Procedure for the Synthesis of 19
Compound 18 (438.14 µmol, 1 equiv), acetyl chloride (77.99 µmol, 2 equiv), DMAP (753.56 µmol, 3 equiv) and EDCl (753.56 µmol, 3 equiv) in dry CH2Cl2 (15 mL) were stirred at 45 °C for 2 h, and the reaction process was monitored via TLC. After the reaction was completed, it was quenched with saturated NaHCO3 (30 mL) aqueous solution and extracted with CH2Cl2 (30 mL), and the organic layer was evaporated to dryness, leaving the crude product. The crude product was purified via silica gel CC column chromatography (EtOAc/petroleum ether, 1:5, v/v) to give derivative 19.
3.4.2. General Procedure for the Synthesis of 24–38
Compounds 17 (404.18 µmol, 1 equiv), anhydride, acyl chloride or carboxylic acid reagents (5 equiv, Supplementary Table S2), DMAP (753.56 µmol, 3 equiv) and EDCl (753.56 µmol, 3 equiv) in dry CH2Cl2 (15 mL) were stirred at 45 °C for 4 h, and the reaction process was monitored via TLC. After the reaction was completed, it was quenched with saturated NaHCO3 (30 mL) aqueous solution and extracted with CH2Cl2 (30 mL), and the organic layer was evaporated to dryness, leaving the crude product. Derivatives 24–38 were purified via silica gel column chromatography.
3.4.3. Characterization Data of Compounds 19, and 24–38
The structures of all compounds were identified using NMR data and the HR-ESI-MS spectrum. Compounds 19 and 24–38 were new derivatives, the details of which are in the Supplementary Materials. Detailed structural information on other known compounds is not indicated.
Compound 19, white, solid; yield 70.7%; –14.6° (c 0.05, MeOH); 1H NMR (400 MHz, CDCl3): δ ppm 6.61 (1H, s), 6.50 (1H, dd, J = 16.0, 1.9 Hz), 5.95 (1H, dt, J = 16.0, 4.6 Hz), 5.78 (1H, ddd, J = 16.0, 10.0, 3.8 Hz), 5.67–5.48 (2H, overlapped), 4.51 (1H, t, J = 8.4 Hz), 4.12 (1H, m), 3.89 (3H, s), 3.80 (1H, dd, J = 8.4, 2.2 Hz), 2.77 (1H, ddd, J = 14.6, 6.5, 3.9 Hz), 2.47–2.39 (2H, overlapped), 2.32 (1H, m), 2.28 (3H, s), 1.42 (3H, s), 1.38 (3H, s), 1.30 (3H, d, J = 6.3 Hz). 13C NMR (100 MHz, CDCl3): δ 169.0 (C), 165.0 (C), 156.9 (C), 147.7 (C), 136.8 (C), 133.2 (CH), 131.4 (CH), 128.9 (CH), 127.1 (CH), 119.7 (C), 119.4 (CH), 108.5 (CH), 105.4 (CH), 81.2 (CH), 75.5 (CH), 69.2 (CH), 67.9 (CH), 56.5(CH2), 37.0 (CH3), 35.5 (CH2), 26.9(CH3 × 2), 26.9(CH3 × 2), 20.8 (CH3), 20.7 (CH3). HRESIMS m/z 479.1480 [M − H]− (calcd for C24H28O8Cl−, 479.1467).
Compound 24, White, solid; yield 83.2%; –23.6° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 8.05 (1H, dd, J = 7.8, 1.8 Hz), 7.53 (1H, ddd, J = 8.4, 7.4, 1.8 Hz), 7.10–6.99 (3H, overlapped), 6.81 (1H, d, J = 2.5 Hz), 6.70 (1H, d, J = 2.5 Hz), 5.98 (1H, dt, J = 15.8, 5.1 Hz), 5.83 (1H, ddd, J = 15.1, 10.8, 3.9 Hz), 5.57–5.37 (2H, overlapped), 4.54 (1H, t, J = 8.4 Hz), 4.16(1H, m), 3.94–3.86 (4H, overlapped), 3.83 (3H, s), 2.67 (1H, dtd, J = 14.0, 4.1, 2.2 Hz), 2.57–2.41 (3H, overlapped), 2.29 (1H, ddd, J = 13.9, 12.0, 10.6 Hz), 1.43 (3H, s), 1.34 (6H, m).13C NMR (100 MHz, CDCl3) δ 165.6 (C), 164.1 (C), 161.9 (C), 160.5 (C), 151.6 (C), 139.9 (C), 134.8 (CH), 133.3 (CH), 133.0 (CH), 131.8 (CH), 130.7 (CH), 128.4 (CH), 120.7 (C), 119.1 (CH), 117.9 (C), 112.6 (CH), 110.7 (C), 108.9 (CH), 108.5 (CH), 81.5 (CH), 76.2 (CH), 69.8 (CH), 67.0 (CH), 56.5 (CH2), 56.1 (CH3), 37.5 (CH3), 36.4 (CH2), 27.4 (CH3 × 2), 20.5 (CH3). HRESIMS m/z 539.2273 [M + H]+ (calcd for C30H35O9+, 539.2276).
Compound 25, White, solid; yield 70.3%; –21.4° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 7.66 (1H, dd, J = 1.7, 0.8 Hz), 7.37 (1H, dd, J = 3.5, 0.9 Hz), 7.05 (1H, dd, J = 15.6, 2.0 Hz), 6.82 (1H, d, J = 2.5 Hz), 6.68 (1H, d, J = 2.5 Hz), 6.58 (1H, dd, J = 3.5, 1.7 Hz), 5.98 (1H, dt, J = 15.7, 5.2 Hz), 5.84 (1H, ddd, J = 15.1, 10.8, 3.9 Hz), 5.49 (1H, ddt, J = 15.7, 8.9, 1.8 Hz), 5.39 (1H, pd, J = 6.4, 3.8 Hz), 4.54 (1H, t, J = 8.4 Hz), 4.16 (1H, ddd, J = 12.2, 4.3, 2.3 Hz), 3.88 (1H, dd, J = 8.0, 2.3 Hz), 3.83 (3H, s), 2.67 (1H, dtd, J = 14.0, 4.3, 2.3 Hz), 2.59–2.46 (3H, overlapped), 2.29 (1H, ddd, J = 14.0, 12.0, 10.6 Hz), 1.42 (3H, s), 1.36–1.31 (6H, overlapped).13C NMR (100 MHz, CDCl3) δ 165.1 (C), 161.7 (C), 156.7 (C), 150.5 (C), 147.4 (CH), 143.9 (C), 139.9 (C), 132.8 (CH), 131.4 (CH), 130.6 (CH), 128.4 (CH), 119.9 (C), 117.3 (CH), 112.4 (C), 110.6 (CH), 108.6 (CH), 108.0 (CH), 81.2 (CH), 75.9 (CH), 69.7 (CH), 68.6 (CH), 55.8 (CH2), 37.1 (CH3), 36.1 (CH2), 27.1 (CH3), 27.0 (CH3), 20.1 (CH3). HRESIMS m/z 499.1968 [M + H]+ (calcd for C27H31O9+, 499.1963).
Compound 26, White, solid; yield 60.6%; –17.2° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 7.67 (1H, dd, J = 1.8, 0.8 Hz), 7.60 (1H, dd, J = 1.8, 0.8 Hz), 7.38 (1H, dd, J = 3.5, 0.9 Hz), 7.23 (1H, dd, J = 3.5, 0.9 Hz), 7.12 (1H, dd, J = 15.5, 1.9 Hz), 6.84 (1H, d, J = 2.5 Hz), 6.70 (1H, d, J = 2.5 Hz), 6.59 (1H, dd, J = 3.5, 1.7 Hz), 6.53 (1H, dd, J = 3.5, 1.7 Hz), 6.06 (1H, dt, J = 15.8, 5.2 Hz), 5.93 (1H, ddd, J = 15.1, 10.7, 3.9 Hz), 5.65–5.49 (2H, overlapped), 5.41 (1H, m), 4.73 (1H, t, J = 8.2 Hz), 4.06 (1H, dd, J = 7.8, 2.3 Hz), 3.85 (3H, s), 2.77 (1H, dtd, J = 13.8, 4.3, 2.2 Hz), 2.55–2.40 (3H, overlapped), 1.34 (6H, overlapped), 1.26 (3H, s).13C NMR (100 MHz, CDCl3) δ 165.0 (C), 161.8 (C), 158.1 (C), 156.7 (C), 150.6 (C), 147.4 (CH), 146.6 (CH), 144.7 (CH), 143.9 (CH), 139.6 (C), 133.4 (CH), 131.3 (CH), 130.6 (CH), 127.3 (CH), 119.9 (C), 118.3 (CH), 117.3 (CH), 112.4 (C), 112.0 (CH), 110.5 (CH), 109.3 (CH), 108.3 (CH), 79.8 (CH), 70.8 (CH), 69.8 (CH), 55.8 (CH), 37.3 (CH2), 35.2 (CH3), 29.8 (CH2), 27.3 (CH3), 26.7 (CH3), 20.1 (CH3). HRESIMS m/z 593.2027 [M + H]+ (calcd for C32H33O11+, 593.2017).
Compound 27, White, solid; yield 73.4%; –19.7° (c 0.1, MeCN); 1H NMR (400 MHz, CDCl3) δ 11.51 (1H, s), 7.61 (1H, dd, J = 1.8, 0.8 Hz), 7.24 (1H, d, J = 2.5 Hz), 6.54 (1H, dd, J = 3.5, 1.8 Hz), 6.49 (1H, d, J = 2.6 Hz), 6.41 (1H, d, J = 2.6 Hz), 6.08 (1H, ddd, J = 15.3, 7.5, 5.5 Hz), 5.81 (1H, ddd, t, J = 15.4, 10.6, 3.1 Hz), 5.71–5.54 (2H, overlapped), 5.45 (1H, m), 4.76 (1H, t, J = 8.1 Hz), 4.06 (1H, dd, J = 7.8, 2.3 Hz), 3.83 (3H, s), 2.84 (1H, ddt, J = 14.4, 5.4, 2.9 Hz), 2.61–2.39 (3H, overlapped), 1.46 (3H, d, J = 6.3 Hz), 1.35 (3H, s), 1.26 (3H, s).13C NMR (100 MHz, CDCl3) δ 170.9 (C), 164.9 (C), 164.2 (C), 158.1 (C), 146.7 (CH), 144.7 (CH), 141.9 (C), 134.7 (CH), 132.5 (CH), 129.6 (CH), 125.8 (CH), 118.4 (C), 112.0 (CH), 109.3 (CH), 107.4 (C), 104.5 (CH), 100.6 (CH), 80.3 (CH), 76.2 (CH), 71.0 (CH), 70.9 (CH), 55.6 (CH2), 38.1 (CH3), 35.1 (CH2), 27.4 (CH3), 26.7 (CH3), 19.6 (CH3). HRESIMS m/z 499.1962 [M + H]+ (calcd for C27H31O9+, 499.1963).
Compound 28, White, solid; yield 63.7%; –38.1° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 8.20–8.13 (2H, overlapped), 7.63 (1H, m), 7.50 (2H, overlapped), 7.06 (1H, dd, J = 17.2, 2.0 Hz), 6.82 (1H, d, J = 2.5 Hz), 6.69 (1H, d, J = 2.5 Hz), 5.98 (1H, dt, J = 15.8, 5.1 Hz), 5.84 (1H, ddd, J = 15.2, 10.8, 3.9 Hz), 5.50 (1H, ddt, J = 15.7, 8.8, 1.8 Hz), 5.39 (1H, m), 4.54 (1H, t, J = 8.4 Hz), 4.17–4.09 (1H, overlapped), 3.89 (1H, dd, J = 8.0, 2.3 Hz), 3.84 (3H, s), 2.67 (1H, dtd, J = 14.0, 4.1, 2.2 Hz), 2.58–2.44 (3H, overlapped), 2.30 (1H, ddd, J = 14.0, 12.0, 10.6 Hz), 1.42 (3H, s), 1.35 (3H, s), 1.30 (3H, d, J = 6.3 Hz).13C NMR (100 MHz, CDCl3) δ 165.2 (C), 165.1 (C), 161.7 (C), 151.3 (C), 139.8 (C), 133.8 (CH), 132.7 (CH), 131.4 (CH), 130.4 (CH), 129.4 (C), 128.7 (CH), 128.3 (CH), 117.4 (CH), 110.4 (CH), 108.6 (CH), 108.0 (C), 81.2 (C), 75.9 (CH), 69.5 (CH), 68.6 (CH), 60.5 (CH), 55.8 (CH), 37.1 (CH), 36.1 (CH2), 27.1 (CH3), 27.1 (CH2), 21.2 (CH3), 20.1 (CH3), 14.3 (CH3). HRESIMS m/z 509.2176 [M + H]+ (calcd for C29H33O8+, 509.2170).
Compound 29, White, solid; yield 77.8%; –8.1° (c 0.1, MeCN); 1H NMR (400 MHz, CDCl3) δ 8.21–8.15 (2H, overlapped), 8.12–8.06 (2H, overlapped), 7.61 (2H, m), 7.49 (4H, overlapped), 7.13 (1H, dd, J = 15.5, 1.9 Hz), 6.85 (1H, d, J = 2.5 Hz), 6.72 (1H, d, J = 2.5 Hz), 6.07 (1H, dt, J = 15.7, 5.2 Hz), 5.97 (1H, ddd, J = 15.1, 10.7, 3.9 Hz), 5.66 (1H, ddd, J = 12.3, 4.6, 2.2 Hz), 5.58 (1H, dd, J = 15.8, 8.6 Hz), 5.41 (1H, q, J = 5.9 Hz), 4.78 (1H, t, J = 8.2 Hz), 4.11 (1H, dt, J = 7.8, 2.4 Hz), 3.86 (3H, s), 2.78 (1H, m), 2.56–2.43 (3H, overlapped), 1.32–1.31 (6H, overlapped), 1.23 (3H, s).13C NMR (100 MHz, CDCl3) δ 165.9 (C), 165.2 (C), 161.8 (C), 151.4 (C), 139.5 (C), 133.8 (C), 133.4 (CH), 133.3 (CH), 131.2 (CH), 130.7 (C), 130.4 (C), 130.4 (CH), 129.8 (CH), 129.4 (CH), 128.8 (CH×2), 128.6 (CH×2), 127.5 (CH), 117.5 (CH), 110.4 (C), 109.3 (C), 108.3 (CH), 79.9 (CH), 70.8 (CH), 69.7 (CH), 55.8 (CH), 37.3 (CH), 35.3 (CH2), 29.9 (CH3), 27.2 (CH2), 27.0 (CH3 × 2), 20.2 (CH3). HRESIMS m/z 613.2433 [M + H]+ (calcd for C36H37O9+, 613.2433).
Compound 30, White, solid; yield 83.5%; –19.3° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 8.10 (1H, td, J = 7.5, 1.9 Hz), 7.59(m, 1H), 7.28 (1H, dd, J = 7.7, 1.1Hz), 7.19 (1H, ddd, J = 10.8, 8.3, 1.1 Hz), 7.07 (1H, dd, J = 15.5, 2.0 Hz), 6.83 (1H, d, J = 2.5 Hz), 6.70 (1H, d, J = 2.5 Hz), 5.98 (1H, dt, J = 15.8, 5.1 Hz), 5.84 (1H, ddd, J = 15.2, 10.8, 3.9 Hz), 5.50 (1H, ddt, J = 15.6, 8.8, 1.8 Hz), 5.41 (1H, q, J = 5.9 Hz), 4.54 (1H, t, J = 8.4 Hz), 4.16 (1H, ddd, J = 12.3, 4.4, 2.2 Hz), 3.89 (1H, dd, J = 8.0, 2.3 Hz), 3.84 (3H, s), 2.68 (1H, dtd, J = 14.0, 4.2, 2.3 Hz), 2.56–2.46 (3H, overlapped), 2.30 (1H, ddd, J = 14.0, 12.0, 10.6 Hz), 1.42 (3H, s), 1.37–1.31 (6H, overlapped).13C NMR (100 MHz, CDCl3) δ 165.2 (C), 163.8 (C), 162.6 (C), 161.8 (C), 151.1 (C), 139.9 (C), 135.4 (CH), 135.4 (CH), 132.9 (CH), 132.9 (CH), 131.5 (CH), 130.4 (CH), 128.4 (CH), 124.3 (C), 124.3 (C), 117.9 (CH), 117.2 (C), 110.7 (CH), 108.6 (CH), 108.0 (CH), 81.2 (CH), 75.9 (CH), 69.6 (CH), 68.6 (CH2), 55.8 (CH3), 37.2 (CH2), 36.1 (CH3), 27.1 (CH3), 20.1 (CH3). HRESIMS m/z 527.2080 [M + H]+ (calcd for C29H32O8F+, 527.2076).
Compound 31, White, solid; yield 87.2%; –28.4° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 8.12 (1H, td, J = 7.5, 1.9 Hz), 8.03 (1H, m), 7.65–7.49 (2H, overlapped), 7.32–7.11 (5H, overlapped), 6.85 (1H, d, J = 2.6 Hz), 6.72 (1H, d, J = 2.6 Hz), 6.06 (1H, dt, J = 15.8, 5.1 Hz), 5.94 (1H, ddd, J = 15.1, 10.8, 3.8 Hz), 5.69 (1H, ddd, J = 12.4, 4.8, 2.3 Hz), 5.56 (1H, ddt, J = 15.6, 8.5, 1.8 Hz), 5.44 (1H, td, J = 6.5, 4.5 Hz), 4.76 (1H, t, J = 8.3 Hz), 4.08 (1H, dd, J = 7.9, 2.3 Hz), 3.86 (3H, s), 2.80 (1H, ddq, J = 13.8, 4.3, 2.3 Hz), 2.60–2.43 (3H, overlapped), 1.38–1.31 (6H, overlapped), 1.26 (3H, s).13C NMR (100 MHz, CDCl3) δ 169.2 (C), 165.2 (C), 163.4 (C), 162.6 (C), 161.8 (C), 161.4 (C), 151.1 (C), 139.6 (C), 135.7 (CH), 135.5 (CH), 134.9 (CH), 133.5 (CH), 132.9 (CH), 132.9 (CH), 132.4 (CH), 131.3 (CH), 130.3 (CH), 127.5 (CH), 124.3 (C), 117.4 (C), 117.3 (C), 117.2 (CH), 117.1 (CH), 110.6 (C), 109.1 (CH), 108.2 (CH), 79.7 (CH), 77.0 (CH), 71.2 (CH), 69.7 (CH), 55.8 (CH2), 37.3 (CH3), 35.2 (CH2), 27.2 (CH3), 26.7 (CH3), 20.2 (CH3). HRESIMS m/z 649.2242 [M + H]+ (calcd for C36H35O9F2+, 649.2244).
Compound 32, White, solid; yield 73.9%; –31.5° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 11.53 (1H, s), 8.01 (1H, td, J = 7.5, 1.9 Hz), 7.55 (1H, m), 7.23 (1H, m), 7.17 (1H, ddd, J = 10.9, 8.4, 1.1 Hz), 6.49 (1H, d, J = 2.6 Hz), 6.42 (1H, d, J = 2.6 Hz), 6.06 (1H, ddd, J = 15.4, 7.2, 5.6 Hz), 5.82 (1H, ddd, J = 15.4, 10.6, 3.1 Hz), 5.72 (1H, ddd, J = 12.4, 5.2, 2.2 Hz), 5.61 (1H, ddt, J = 15.5, 8.5, 1.5 Hz), 5.44 (1H, qd, J = 6.4, 3.6 Hz), 4.77 (1H, t, J = 8.1 Hz), 4.07 (1H, dd, J = 7.8, 2.2 Hz), 3.83 (3H, s), 2.87 (1H, ddt, J = 14.4, 5.4, 2.8 Hz), 2.50 (3H, m), 2.59–2.42 (4H, overlapped), 1.46 (3H, d, J = 6.3 Hz), 1.35 (3H, s), 1.27 (3H, s).13C NMR (100 MHz, CDCl3) δ 171.0 (C), 165.0 (C), 164.2 (C), 142.0 (C), 134.9 (C), 134.8 (C), 134.7 (CH), 132.5 (CH), 132.4 (CH), 129.6 (CH), 126.0 (CH), 124.2 (CH), 118.7 (CH), 117.4 (C), 117.1 (C), 109.1 (CH), 107.4 (C), 104.5 (CH), 100.5 (CH), 80.2 (CH), 76.3 (CH), 71.4 (CH), 71.0 (CH), 55.6 (CH2), 38.2 (CH3), 35.2 (CH2), 27.3 (CH3), 26.8 (CH3), 19.7 (CH3). HRESIMS m/z 527.2077 [M + H]+ (calcd for C29H32O8F+, 527.2076).
Compound 33, White, solid; yield 67.7%; –24.4° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 9.35 (1H, dd, J = 2.2, 0.9 Hz), 8.85 (1H, dd, J = 4.9, 1.8 Hz), 8.43 (1H, dt, J = 7.9, 2.0 Hz), 7.46 (1H, ddd, J = 7.9, 4.9, 0.9 Hz), 7.08 (1H, dd, J = 15.5, 2.0 Hz), 6.85 (1H, d, J = 2.5 Hz), 6.70 (1H, d, J = 2.5 Hz), 5.98 (1H, dt, J = 15.7, 5.2 Hz), 5.85 (1H, ddd, J = 15.1, 10.8, 3.8 Hz), 5.48 (1H, ddt, J = 15.7, 8.9, 1.8 Hz), 5.35 (1H, m), 4.55 (1H, t, J = 8.4 Hz), 4.17 (1H, dt, J = 12.3, 3.5 Hz), 3.91–3.84 (4H, overlapped), 2.69 (1H, dtd, J = 14.0, 4.1, 2.3 Hz), 2.56 (1H, d, J = 1.5 Hz), 2.49 (2H, overlapped), 2.30 (1H, m), 1.42 (3H, s), 1.35 (3H, s), 1.31 (3H, d, J = 6.4 Hz).13C NMR (100 MHz, CDCl3) δ 165.1 (C), 164.0 (C), 161.9 (C), 154.1 (C), 151.6 (CH), 151.1 (CH), 140.2(C), 137.8 (CH), 132.8 (CH), 131.4 (CH), 130.5 (CH), 128.5 (C), 125.6 (CH), 123.7 (CH), 117.0(C), 110.7 (C), 108.6 (CH), 108.0 (CH), 81.2 (CH), 75.9 (CH), 69.8 (CH), 68.6 (CH), 55.8 (CH2), 37.1 (CH3), 36.2 (CH2), 27.1 (CH3 × 2), 20.1 (CH3). HRESIMS m/z 510.2127 [M + H]+ (calcd for C28H32O8N+, 510.2122).
Compound 34, White, solid; yield 85.6%; –10.3° (c 0.05, MeOH); 1H NMR (400 MHz, CDCl3) δ 9.36 (1H, dd, J = 2.2, 0.9 Hz), 9.29 (1H, dd, J = 2.2, 0.9 Hz), 8.85 (1H, dd, J = 4.9, 1.8 Hz), 8.81 (1H, dd, J = 4.9, 1.8 Hz), 8.44 (1H, dt, J = 8.0, 2.0 Hz), 8.35 (1H, dt, J = 8.0, 2.0 Hz), 7.45 (2H, dddd, J = 16.4, 8.0, 4.9, 0.9 Hz), 7.17 (1H, dd, J = 15.5, 1.9 Hz), 6.88 (1H, d, J = 2.5 Hz), 6.73 (1H, d, J = 2.5 Hz), 6.07 (1H, dt, J = 15.7, 5.2 Hz), 5.96 (1H, ddd, J = 15.1, 10.7, 3.8 Hz), 5.70 (1H, ddd, J = 12.5, 4.7, 2.3 Hz), 5.55 (1H, ddt, J = 15.7, 8.7, 1.7 Hz), 5.39 (1H, td, J = 6.5, 4.4 Hz), 4.74 (1H, t, J = 8.2 Hz), 4.10 (1H, dd, J = 7.8, 2.3 Hz), 3.87 (3H, s), 2.80 (1H, dtd, J = 13.8, 4.3, 2.3 Hz), 2.59–2.45 (3H, overlapped), 1.33 (6H, overlapped), 1.22 (3H, s).13C NMR (100 MHz, CDCl3) δ 165.1 (C), 164.6 (C), 164.0 (C), 162.0 (C), 154.2 (C), 153.8 (CH), 151.6 (CH), 151.2 (CH), 151.0 (CH), 139.7 (C), 137.9 (CH), 137.3 (CH), 133.5 (CH), 131.3 (CH), 130.6 (CH), 127.3 (C), 126.2 (C), 125.6 (CH), 123.7 (CH), 123.6 (CH), 117.0 (C), 110.7 (C), 109.3 (CH), 108.4 (CH), 79.7 (CH), 71.3 (CH), 69.9 (CH), 55.9 (CH), 37.2 (CH2), 35.2 (CH3), 27.2 (CH2), 27.0 (CH3 × 2), 20.2 (CH3). HRESIMS m/z 615.2318 [M + H]+ (calcd for C34H35O9N2+, 615.2337).
Compound 35, White, solid; yield 72.4%; –17.0° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 11.52 (1H, s), 9.29 (1H, m), 8.81 (1H, d, J = 4.0 Hz), 8.35 (1H, dt, J = 7.9, 1.9 Hz), 7.44 (1H, dd, J = 7.9, 4.8 Hz), 7.25 (1H, d, J = 15.4, 2.2Hz), 6.50 (1H, d, J = 2.6 Hz), 6.42 (1H, d, J = 2.6 Hz), 6.09 (1H, ddd, J = 15.3, 7.6, 5.5 Hz), 5.83 (1H, ddd, J = 15.4, 10.5, 3.1 Hz), 5.74 (1H, m), 4.77 (1H, t, J = 8.0 Hz), 4.11 (1H, dd, J = 7.7, 2.3 Hz), 3.83 (3H, s), 2.86 (1H, ddt, J = 14.4, 5.5, 2.9 Hz), 2.60–2.44 (3H, overlapped), 1.47 (3H, d, J = 6.3 Hz), 1.35 (3H, s), 1.23 (3H, s).13C NMR (100 MHz, CDCl3) δ 170.9 (C), 165.0 (C), 164.6 (C), 164.2 (C), 153.8 (CH), 151.0 (CH), 141.8 (C), 137.3 (CH), 134.8 (CH), 132.6 (CH), 129.7 (CH), 125.6 (C), 123.6 (CH), 109.3 (CH), 107.5 (C), 104.5 (C), 100.6 (CH), 80.2 (CH), 77.4 (CH), 76.4 (CH), 71.5 (CH), 70.9 (CH), 55.6 (CH2), 38.1 (CH3), 35.2 (CH2), 27.2 (CH3), 27.0 (CH3), 19.6 (CH3). HRESIMS m/z 510.2126 [M + H]+ (calcd for C28H32O8N+, 510.2122).
Compound 36, White, solid; yield 63.7%; –24.6° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 7.05 (1H, dd, J = 15.6, 2.1 Hz), 6.79 (1H, d, J = 2.5 Hz), 6.55 (1H, d, J = 2.5 Hz), 5.99 (1H, dt, J = 15.7, 5.3 Hz), 5.81 (1H, ddd, J = 15.5, 10.8, 3.8 Hz), 5.56–5.34 (2H, overlapped), 4.55 (1H, t, J = 8.4 Hz), 4.16 (1H, ddd, J = 12.3, 4.5, 2.4 Hz), 3.93–3.79 (4H, overlapped), 2.67 (1H, dtd, J = 14.0, 4.1, 2.3 Hz), 2.57–2.44 (3H, overlapped), 2.29 (4H, s), 1.39 (9H, m).13C NMR (100 MHz, CDCl3) δ 169.7 (C), 165.2 (C), 161.8 (C), 151.4 (C), 140.0 (C), 132.9 (CH), 131.3 (CH), 130.7 (CH), 128.2 (CH), 117.0 (C), 110.4 (C), 108.6 (CH), 108.0 (CH), 81.2 (CH), 75.9 (CH), 69.7 (CH), 68.7 (CH), 55.7 (CH2), 37.2 (CH3), 36.1 (CH2), 27.1 (CH3 × 2), 21.1 (CH3), 20.0 (CH3). HRESIMS m/z 447.2011 [M + H]+ (calcd for C24H31O8+, 447.2013).
Compound 37, White, solid; yield 85.2%; –28.3° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 7.97 (1H, dd, J = 3.8, 1.3 Hz), 7.66 (1H, dd, J = 5.0, 1.3 Hz), 7.17 (1H, dd, J = 5.0, 3.8 Hz), 7.06 (1H, dd, J = 15.5, 2.0 Hz), 6.82 (1H, d, J = 2.5 Hz), 6.71 (1H, d, J = 2.5 Hz), 5.98 (1H, dt, J = 15.7, 5.1 Hz), 5.84 (1H, ddd, J = 15.2, 10.8, 3.9 Hz), 5.55–5.36 (2H, overlapped), 4.54 (1H, t, J = 8.5 Hz), 4.15 (1H, ddd, J = 12.2, 4.3, 2.3 Hz), 3.88 (1H, dd, J = 8.0, 2.3 Hz), 3.84 (3H, s), 2.67 (1H, dtd, J = 13.9, 4.1, 2.2 Hz), 2.49 (3H, overlapped), 2.29 (1H, ddd, J = 14.1, 12.0, 10.6 Hz), 1.42 (3H, s), 1.40–1.30 (6H, overlapped).13C NMR (100 MHz, CDCl3) δ 165.2 (C), 161.7 (C), 160.4 (C), 150.8 (C), 139.8 (C), 135.1 (C), 133.8 (CH), 132.9 (CH), 132.6 (CH), 131.5 (CH), 130.3 (CH), 128.4 (CH), 128.2 (CH), 117.4 (C), 110.6 (C), 108.6 (CH), 108.0 (CH), 81.2 (CH), 69.6 (CH), 68.6 (CH), 55.8 (CH), 37.2 (CH2), 36.1 (CH3), 29.8 (CH2), 27.1 (CH3), 27.1 (CH3), 20.2 (CH3). HRESIMS m/z 515.1730 [M + H]+ (calcd for C27H31O8S+, 515.1734).
Compound 38, White, solid; yield 87.7%; –26.7° (c 0.1, MeOH); 1H NMR (400 MHz, CDCl3) δ 7.98 (1H, dd, J = 3.8, 1.3 Hz), 7.86 (1H, dd, J = 3.8, 1.3 Hz), 7.66 (1H, dd, J = 5.0, 1.3 Hz), 7.58 (1H, dd, J = 5.0, 1.3 Hz), 7.17 (1H, dd, J = 5.0, 3.8 Hz), 7.12 (2H, m), 6.83 (1H, d, J = 2.5 Hz), 6.72 (1H, d, J = 2.5 Hz), 6.06 (1H, dt, J = 15.8, 5.0 Hz), 5.93 (1H, ddd, J = 15.1, 10.7, 3.9 Hz), 5.63–5.51 (2H, overlapped), 5.44 (1H, td, J = 6.5, 4.7 Hz), 4.72 (1H, t, J = 8.2 Hz), 4.05 (1H, dd, J = 7.9, 2.3 Hz), 3.85 (3H, s), 2.78 (1H, dtd, J = 13.6, 4.3, 2.2 Hz), 2.55–2.41 (3H, overlapped), 1.34 (6H, overlapped), 1.27 (3H, s).13C NMR (100 MHz, CDCl3) δ 165.1 (C), 161.8 (C), 161.5 (C), 160.4 (C), 150.9 (C), 139.5 (C), 135.1 (C), 133.9 (C), 133.7 (CH×2), 133.4 (CH), 132.7 (CH), 132.6 (CH), 131.4 (CH), 130.3 (CH), 128.2 (CH), 128.0 (CH), 127.5 (CH), 117.4 (C), 110.5 (C), 109.2 (CH), 108.3 (CH), 79.8 (CH), 71.0 (CH), 69.7 (CH), 55.8 (CH), 37.3 (CH2), 35.2 (CH3), 27.3 (CH2), 27.0 (CH3 × 2), 20.3 (CH3). HRESIMS m/z 625.1545 [M + H]+ (calcd for C32H33O9S2+, 625.1561).
3.5. Antimicrobial Activity
The methods described by Fromtling et al. were used to evaluate the derivatives’ antibacterial activity [34]. Isoniazid and rifampicin were used as a positive control anti-M. marinum, ciprofloxacin as a positive control anti-bacteria, and amphotericin B as a positive control anti-fungi. The strains were cultured in the corresponding medium at 32 °C for 8 h and diluted to 105 CFU/mL using 96-well plates with 2 μL of sample and 198 μL of bacterial solution. Incubation was carried out at 32 °C for 24 h or 48 h, and DMSO was used as a negative control.
3.6. Time–Growth Curve Assay
The time–growth curve was determined using the method of Li et al. [35]. Initially, the concentration of M. marinum was set at 105 CFU/mL, and 5 groups were chosen, each group tested on 6 times. The drug concentration of the dosing group was set at 640, 320, 160 and 80 μM. An equal amount of DMSO was added to the blank group, and the suspension of M. marinum was incubated at 32 °C (100 rpm) by oscillating it. The colony count was determined and counted using the OD600 at the planned time points (0, 3, 6, 12, 18, 24, 36, 48 and 60 h).
3.7. In Vitro Synergic Anti-M. marinum Activity Assay
An in vitro synergistic antibacterial assay was performed as described by Li et al., and synergistic activity was evaluated on 96-well plates [33]. Based on the MIC90 data of the compound and each positive drug as the design basis, the chessboard dilution method was used to combine 4 × MIC90, 2 × MIC90, MIC90, 1/2 × MIC90, 1/4 × MIC90, 1/8 × MIC90 and 1/16 × MIC90 in the 96-well plate with bacterial solution added. The measurements were repeated three times per well. The 96-well plates were placed in a constant-temperature incubator at 32 °C, the results were observed and recorded 48 h later, and an optical density of 600 nm (OD600) was measured.
3.8. Statistical Analysis
All statistical analyses were performed using GraphPad Prism 8.3 software. Data are presented as the mean of three experiments. For two-group comparison, the p value was derived from a one-way Student t test to determine the difference between groups with normally distributed data. For all comparisons, p < 0.05 was considered statistically significant. * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001.
4. Conclusions
In summary, 16 new derivatives were successfully synthesized through two to three steps, which enriched the diversity of 14-membered RALs. Through the activity evaluation of the derivative library, four derivatives showed promising anti-M. marinum activity. The preliminary structure–activity relationships showed that the anti-M. marinum activity of 14-membered RALs can be significantly improved via the introduction of a chlorine atom at the C-5 position. The substitution of positions 5’6’ of dihydroxy with an acetonide group reduced the activity. The etherification modification of the phenolic hydroxy group did not significantly improve the activity. Further studies showed that compound 12 enhanced the effects of positive drugs isoniazid and rifampicin on M. marinum. These results suggest that 12 is an active compound capable of enhancing the potency of existing positive drugs, and its effective properties make it a very useful lead for future drug development in combating TB resistance.
Acknowledgments
We thank Syngenta for the fellowship to Qun Zhang. We also thank Xiu-Li Zhang and Cong Wang at the School of Medicine and Pharmacy, Ocean University of China, for the NMR test.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/md22030135/s1; Table S1: All compounds that not appear in the text in the derivative library. Table S2: Anhydride, acyl chloride or carboxylic acid reagents used to generate compounds 24–38. Figures S1–S48: 1H NMR, 13C NMR and HRESIMS of compounds 19 and 24–38.
Author Contributions
Q.-Q.J. contributed to preparation of all compounds; writing—original draft; writing—review and editing. J.-N.Y. contributed to related work on bioactivity; writing—original draft; writing—review and editing, Y.-J.C., Q.Z., X.-Z.C. and W.-F.X. contributed to providing the compounds and their derivatives. C.-L.S. and M.-Y.W. were the project leaders, organizing and guiding the experiments and manuscript writing. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data are contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was supported by the Special Funds of Shandong Province for Qingdao National Laboratory of Marine Science and Technology (No. 2022QNLM030003), the State Key Laboratory for Chemistry and Molecular Engineering of Medicinal Resources, Guangxi Normal University (No. CMEMR2023-B16), Shandong Province Special Fund “Frontier Technology and Free Exploration” from Laoshan Laboratory (No. 8-01), the National Key Research and Development Program of China (No. 2022YFC2601305) and the Innovation Center for Academicians of Hainan Province.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Daniel T.M. The History of Tuberculosis. Respir. Med. 2006;100:1862–1870. doi: 10.1016/j.rmed.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Public Health Online. [(accessed on 13 October 2021)]. Available online: https://www.publichealthonline.org/worst-global-pandemics-in-history.
- 3.World Health Organization; Geneva, Switzerland: 2023. [(accessed on 7 November 2023)]. Global Tuberculosis Report 2023. Licence: CC BY-NC-SA 3.0 IGOnization. Available online: https://www.who.int/publications/i/item/9789240083851. [Google Scholar]
- 4.Bloom B.R. A half-century of research on tuberculosis: Successes and challenges. J. Exp. Med. 2023;220:e20230859. doi: 10.1084/jem.20230859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maitre T., Aubry A., Jarlier V., Robert J., Veziris N., Bernard C., Sougakoff W., Brossier F., Cambau E., Mougari F., et al. Multidrug and Extensively Drug-Resistant Tuberculosis. Med. Mal. Infect. 2017;47:3–10. doi: 10.1016/j.medmal.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Lange C., Dheda K., Chesov D., Mandalakas A.M., Udwadia Z., Horsburgh C.R., Jr. Management of drug-resistant tuberculosis. Lancet. 2019;394:953–966. doi: 10.1016/S0140-6736(19)31882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elsevier Patient safety: Too little, but not too late. Lancet. 2019;394:895. doi: 10.1016/S0140-6736(19)32080-X. [DOI] [PubMed] [Google Scholar]
- 8.Kakkar A.K., Dahiya N. Bedaquiline for the treatment of resistant tuberculosis: Promises and pitfalls. Tuberculosis. 2014;94:357–362. doi: 10.1016/j.tube.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Ryan N.J., Lo J.H. Delamanid: First global approval. Drugs. 2014;74:1041–1045. doi: 10.1007/s40265-014-0241-5. [DOI] [PubMed] [Google Scholar]
- 10.Keam S.J. Pretomanid: First Approval. Drugs. 2019;79:1797–1803. doi: 10.1007/s40265-019-01207-9. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y., Wang G.Z., Xu M. Biohazard levels and biosafety protection for Mycobacterium tuberculosis strains with different virulence. Biosaf. Health. 2020;2:135–141. doi: 10.1016/j.bsheal.2020.04.001. [DOI] [Google Scholar]
- 12.van Soolingen D., Wisselink H.J., Lumb R., Anthony R., van der Zanden A., Gilpin C. Practical biosafety in the tuberculosis laboratory: Containment at the source is what truly counts. Int. J. Tuberc. Lung Dis. 2014;18:885–889. doi: 10.5588/ijtld.13.0629. [DOI] [PubMed] [Google Scholar]
- 13.Lambrecht R.S., Carriere J.F., Collins M.T. A model for analyzing growth kinetics of a slowly growing Mycobacterium sp. Appl. Environ. Microbiol. 1988;54:910–916. doi: 10.1128/aem.54.4.910-916.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao L.Y., Groger R., Cox J.S., Beverley S.M., Lawson E.H., Brown E.J. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect. Immun. 2003;71:922–929. doi: 10.1128/IAI.71.2.922-929.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cronin R.M., Ferrell M.J., Cahir C.W., Champion M.M., Champion P.A. Proteo-genetic analysis reveals clear hierarchy of ESX-1 secretion in Mycobacterium marinum. Proc. Natl. Acad. Sci. USA. 2022;119:e2123100119. doi: 10.1073/pnas.2123100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Seymortier P., Verellen K., De Jonge I. Mycobacterium marinum causing tenosynovitis. ‘Fish tank finger’. Acta. Orthop. Belg. 2004;70:279–282. [PubMed] [Google Scholar]
- 17.Stinear T.P., Seemann T., Harrison P.F., Jenkin G.A., Davies J.K., Johnson P.D., Abdellah Z., Arrowsmith C., Chillingworth T., Churcher C., et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008;18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Habjan E., Ho V.Q.T., Gallant J., van Stempvoort G., Jim K.K., Kuijl C., Geerke D.P., Bitter W., Speer A. An anti-tuberculosis compound screen using a zebrafish infection model identifies an aspartyl-tRNA synthetase inhibitor. Dis. Model. Mech. 2021;14:dmm049145. doi: 10.1242/dmm.049145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman D.J., Cragg G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 20.Hai Y., Cai Z.M., Li P.J., Wei M.Y., Wang C.Y., Gu Y.C., Shao C.L. Trends of antimalarial marine natural products: Progresses, challenges and opportunities. Nat. Prod. Rep. 2022;39:969–990. doi: 10.1039/D1NP00075F. [DOI] [PubMed] [Google Scholar]
- 21.Carroll A.R., Copp B.R., Davis R.A., Keyzers R.A., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2022;39:1122–1171. doi: 10.1039/D1NP00076D. [DOI] [PubMed] [Google Scholar]
- 22.Han J., Liu X., Zhang L., Quinn R.J., Feng Y. Anti-mycobacterial natural products and mechanisms of action. Nat. Prod. Rep. 2022;39:77–89. doi: 10.1039/D1NP00011J. [DOI] [PubMed] [Google Scholar]
- 23.Shao C.L., Wu H.X., Wang C.Y., Liu Q.A., Xu Y., Wei M.Y., Qian P.Y., Gu Y.C., Zheng C.J., She Z.G., et al. Potent antifouling resorcylic acid lactones from the gorgonian-derived fungus Cochliobolus lunatus. J. Nat. Prod. 2011;74:629–633. doi: 10.1021/np100641b. [DOI] [PubMed] [Google Scholar]
- 24.Jana N., Nanda S. Resorcylic acid lactones (RALs) and their structural congeners: Recent advances in their biosynthesis, chemical synthesis and biology. New J. Chem. 2018;42:17803–17873. doi: 10.1039/C8NJ02534G. [DOI] [Google Scholar]
- 25.Liu Q.A., Shao C.L., Gu Y.C., Blum M., Gan L.S., Wang K.L., Chen M., Wang C.Y. Antifouling and Fungicidal Resorcylic Acid Lactones from the Sea Anemone-Derived Fungus Cochliobolus lunatus. J. Agric. Food. Chem. 2014;62:3183–3191. doi: 10.1021/jf500248z. [DOI] [PubMed] [Google Scholar]
- 26.Xu W.F., Xue X.J., Qi Y.X., Wu N.N., Wang C.Y., Shao C.L. Cochliomycin G, a 14-membered resorcylic acid lactone from a marine-derived fungus Cochliobolus lunatus. Nat. Prod. Res. 2021;35:490–493. doi: 10.1080/14786419.2019.1633646. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W., Shao C.L., Chen M., Liu Q.A., Wang C.Y. Brominated resorcylic acid lactones from the marine-derived fungus Cochliobolus lunatus induced by histone deacetylase inhibitors. Tetrahedron Lett. 2014;55:4888–4891. doi: 10.1016/j.tetlet.2014.06.096. [DOI] [Google Scholar]
- 28.Wang K.L., Zhang G., Sun J., Xu Y., Han Z., Liu L.L., Shao C.L., Liu Q.A., Wang C.Y., Qian P.Y. Cochliomycin A inhibits the larval settlement of Amphibalanus amphitrite by activating the NO/cGMP pathway. Biofouling. 2016;32:35–44. doi: 10.1080/08927014.2015.1121245. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X.Q., Spadafora C., Pineda L.M., Ng M.G., Sun J.H., Wang W., Wang C.Y., Gu Y.C., Shao C.L. Discovery, Semisynthesis, Antiparasitic and Cytotoxic Evaluation of 14-Membered Resorcylic Acid Lactones and Their Derivatives. Sci. Rep. 2017;7:11822. doi: 10.1038/s41598-017-12336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu W.F., Wu N.N., Wu Y.W., Qi Y.X., Wei M.Y., Pineda L.M., Ng M.G., Spadafora C., Zheng J.Y., Lu L., et al. Structure modification, antialgal, antiplasmodial, and toxic evaluations of a series of new marine-derived 14-membered resorcylic acid lactone derivatives. Mar. Life Sci. Technol. 2022;4:88–97. doi: 10.1007/s42995-021-00103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koul A., Arnoult E., Lounis N., Guillemont J., Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 32.Smith T.C., 2nd, Aldridge B.B. Targeting drugs for tuberculosis. Science. 2019;364:1234–1235. doi: 10.1126/science.aay0211. [DOI] [PubMed] [Google Scholar]
- 33.Li Z., Huang Y., Tu J., Yang W., Liu N., Wang W., Sheng C. Discovery of BRD4-HDAC Dual Inhibitors with Improved Fungal Selectivity and Potent Synergistic Antifungal Activity against Fluconazole-Resistant Candida albicans. J. Med. Chem. 2023;66:5950–5964. doi: 10.1021/acs.jmedchem.3c00165. [DOI] [PubMed] [Google Scholar]
- 34.Fromtling R.A., Galgiani J.N., Pfaller M.A., Espinel-Ingroff A., Bartizal K.F., Bartlett M.S., Body B.A., Frey C., Hall G., Roberts G.D. Multicenter evaluation of a broth macrodilution antifungal susceptibility test for yeasts. Antimicrob. Agents Chemother. 1993;37:39–45. doi: 10.1128/AAC.37.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., Liu N., Tu J., Ji C., Han G., Sheng C. Discovery of simplified sampangine derivatives with potent antifung alactivities against cryptococcal meningitis. ACS Infect. Dis. 2019;5:1376–1384. doi: 10.1021/acsinfecdis.9b00086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are contained within the article or Supplementary Materials.