Abstract
The double-stranded DNA genome of human hepatitis B virus (HBV) and related hepadnaviruses is reverse transcribed from a pregenomic RNA by a viral polymerase (Pol) harboring both priming and RNA- and DNA-dependent elongation activities. Although hepadnavirus replication occurs inside viral nucleocapsids, or cores, biochemical systems for analyzing this reaction are currently limited to unencapsidated Pols expressed in heterologous systems. Here, we describe cis and trans classes of replicative HBV cores, produced in the recombinant baculovirus system via coexpression of HBV core and Pol proteins from either a single RNA (i.e., in cis) or two distinct RNAs (in trans). Upon isolation from insect cells, cis and trans cores contained Pol-linked HBV minus-strand DNA with 5′ ends mapping to the authentic elongation origin DR1 and also plus-strand DNA species. Only trans cores, however, were highly active for the de novo priming and reverse transcription of authentic HBV minus strands in in vitro endogenous polymerase assays. This reaction strictly required HBV Pol but not the ɛ stem-loop element, although the presence of one ɛ, or better, two ɛs, enhanced minus-strand synthesis up to 10-fold. Compared to unencapsidated Pol enzymes, encapsidated Pol appeared to be (i) highly processive, able to extend minus-strand DNAs of 400 nucleotides from DR1 in vitro, and (ii) more active for HBV plus-strand synthesis. These observations suggest possible contributions to the replication process from the HBV core protein. These novel core reagents should facilitate the analysis of HBV replication in its natural environment, the interior of the capsid, and also fuel the development of new anti-HBV drug screens.
Human hepatitis B virus (HBV), the prototype member of a small family of hepadnaviruses, is a major cause of liver disease ranging in severity from chronic infection of hepatocytes to liver cirrhosis and hepatocellular carcinoma (5). HBV utilizes a reverse transcription (RT) step to convert a greater-than-genome-length RNA intermediate known as pregenomic RNA (pgRNA) into its non-covalently closed, partially double-stranded, circular DNA genome (12, 35, 41). HBV replication occurs inside the newly synthesized nucleocapsid, or core particle (23), and is catalyzed by the multifunctional viral polymerase (Pol) which is involved in all phases of the replication reaction. Specifically, Pol (i) mediates selective packaging of the pgRNA template into the nucleocapsid (2, 16); (ii) serves as the primer for initiation of minus-strand DNA synthesis (3, 50); (iii) possesses both RNA- and DNA-dependent polymerase activities; and (iv) carries an RNase H activity that degrades pgRNA during RT (29). In addition to Pol, genetic studies suggest that the hepadnavirus core (C) protein, which harbors RNA and DNA binding activities (14), plays an essential role in hepadnavirus replication and hence propagation (14, 25, 33, 52). The nature of this role remains to be elucidated.
The biochemical dissection of hepadnavirus genome replication has long been hampered by failure to liberate enzymatically active Pol from nucleocapsids (30) or to express functional, capsid-free Pol in heterologous systems. The latter obstacle has recently been overcome, first for the duck hepatitis B virus (DHBV) and soon after for HBV. Studies of in vitro-translated (50) and yeast retrotransposon-derived DHBV Pol (45) challenged the established hepadnavirus replication model by revealing that a novel priming reaction precedes the major phase of viral first-strand DNA synthesis. This priming step yields a discrete 3- to 4-deoxyribonucleotide oligomer which is covalently linked to Pol, the result of a unique priming mechanism (49) wherein deoxyribonucleoside triphosphates (dNTPs) are polymerized directly onto a tyrosine acceptor residue located in the N-terminal priming (TP) domain of Pol. Priming is templated by a bulge sequence within a stem-loop structure, called epsilon or ɛ, located near the 5′ end of the pgRNA. The Pol-primer adduct subsequently translocates to the 3′ end of the pgRNA and binds to a complementary sequence in an element called direct repeat 1 (DR1) where the elongation of minus-strand DNA is initiated (47, 49), making 3′ DR1 the apparent origin for minus-strand DNA synthesis.
While genetic studies suggest that HBV adheres to the revised DHBV replication scheme, HBV Pol has proved more refractory to biochemical analysis than its avian counterpart; thus, neither of the above systems has yielded active HBV Pol. An in vitro assay for HBV Pol based on Xenopus laevis oocytes injected with an HBV Pol mRNA was developed previously (37). This system is probably too complex to allow a precise dissection of HBV replication steps but has nevertheless led to some interesting findings; for example, our HBV Pol appears to be less strictly dependent on DR1 and ɛ for in vitro activity than is DHBV Pol, and further, HBV-specific replication is retained in the absence of these elements. Fortunately, authentic priming and minus-strand elongation have now been demonstrated for HBV Pol purified from insect cells infected with recombinant baculovirus (19, 20). This work identified Tyr 63 as the specific priming acceptor site and revealed that the priming and polymerase domains of HBV Pol can be separated without loss of function. As was seen in the oocyte system, HBV Pol activity did not depend strictly on ɛ.
In this report, we set out to develop recombinant HBV nucleocapsids carrying HBV Pol and viral RNAs as a second-generation reagent for studying HBV replication. By using the recombinant baculovirus expression system, HBV core and Pol were supplied either in cis from a single baculovirus construct designed to express a pregenome-like RNA or else by transcomplementation of separate core- and Pol-expressing baculoviruses. Surprisingly, though both classes of cores were competent for HBV replication in vivo, only the transcomplementation-derived cores were highly active for HBV replication in vitro. The reaction occurring in trans nucleocapsids reflects primarily the early phases of HBV replication, such as priming and RT, and retains hallmark characteristics of authentic HBV replication.
MATERIALS AND METHODS
Constructs.
Numbering of the HBV sequence (subtype ayw) follows the designation of Galibert et al. (11). HBV genes were excised from plasmid pTHBV-1 or pHBV-1, generously supplied by G. Acs (Mt. Sinai Medical Center, New York, N.Y.), and introduced into the baculovirus transfer vector pVL1393 (Invitrogen, San Diego, Calif.) by standard molecular cloning techniques. The constructs used in transcomplementation experiments carry the following HBV inserts (Fig. 1): pBV-EC contains 5′ ɛ plus the C-protein open reading frame (ORF) (nucleotides [nt] 1847 to 2459); pBV-C encodes just ORF C (nt 1884 to 2459); pBV-PE consists of the Pol (P) ORF followed by 3′ ɛ and 3′ DR1 (nt 2309 to 1986); pBV-P carries the P ORF only (nt 2309 to 1804). In addition, a full-length pregenome construct, pBV-ECPE (nt 1847 to 3182 and 1 to 1986), as well as 5′- and/or 3′-truncated derivatives was created (pBV-CPE, nt 1884 to 3182 and 1 to 1986; pBV-ECP, nt 1847 to 3182 and 1 to 1804; and pBV-CP, nt 1884 to 3182 and 1 to 1804). All constructs yield RNAs bearing a baculovirus leader sequence of ∼120 nt. To eliminate any risk of generating mature HBV virions, four amber stop codons were placed in the surface protein ORF of pBV-ECPE via site-directed mutagenesis (17). These mutations were silent in the P ORF. A Pol mutant (pBV-PE-YMHA) bearing YMHA in place of the catalytic site YMDD motif and pBV-PE-Y63F, in which phenylalanine replaced the tyrosine 63 priming residue, were prepared by site-directed mutagenesis with a Morph kit (5′→3′ Inc., Boulder, Colo.).
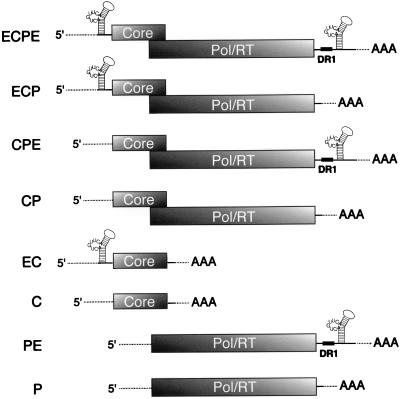
FIG. 1.
Structures of the baculovirus-expressed HBV transcripts used to generate nucleocapsids. The core (C) and Pol (P) ORFs are indicated (shaded boxes). ECPE carries a full-length pregenome-like HBV insert flanked by HBV regulatory sequences harboring the ɛ stem-loop structure (E); ECP, CPE, and CP are ECPE derivatives lacking, respectively, the 3′ DR1 and ɛ element (ECP), 5′ ɛ (CPE), or both (CP). Note that DR1 is included with the 3′ but not the 5′ ɛ element in these RNAs. EC and C encode the core protein only and are similar except only the former harbors the 5′ ɛ; likewise, PE and P are both monocistronic Pol mRNAs which differ by the inclusion of the 3′ DR1 and ɛ element in PE. Shown for all the RNAs are the polyadenylated 3′ ends (AAA) and the short (120-bp) 5′ baculovirus-derived leader sequence (dotted line at the 5′ end).
Cells and infections.
The Spodoptera frugiperda cell line Sf9 was maintained and infected as described previously (28). Briefly, 200-ml suspension cultures (106 cells per ml) were kept in serum-free medium (SF-900 SFM; GIBCO BRL, Gaithersburg, Md.) at 27°C. Recombinant baculoviruses were generated by employing standard techniques. Single infections were initiated by adding the appropriate recombinant baculovirus at a multiplicity of infection (MOI) of 2.5 to 5. In transcomplementation experiments, cells were typically coinfected with core- and Pol-expressing baculoviruses (at MOIs of 2 to 2.5 and 8 to 10, respectively). The infected cells were harvested approximately 3.5 days postinfection by low-speed centrifugation (700 × g, 10 min, 4°C). After one rinse with phosphate-buffered saline, the dry cell pellet was stored at −80°C. To block DNA synthesis in nucleocapsids in vivo, insect cells were fed with 1.5 mM sodium phosphonoformate (PFA) (Sigma, St. Louis, Mo.) starting circa 1 h postinfection and then refed at 24-h intervals during the course of infection.
Preparation of lysates and immunoprecipitation (IP) of nucleocapsids.
Crude lysate from insect cells was prepared by resuspending the thawed cell pellet in 1/10 of the original volume of lysis buffer (50 mM Tris [pH 7.4]–150 mM NaCl–10 mM EDTA–0.75% Nonidet P-40). After 15 min on ice, the lysate was clarified by low-speed centrifugation (2,000 × g, 4°C, 15 min). Aliquots (0.5 to 1 ml) were immunoprecipitated under native conditions with a rabbit anti-HBe/c antibody (Dako, Carpinteria, Calif.) and protein A-Sepharose CL-4B (Pharmacia, Piscataway, N.J.) overnight at 4°C. Thereafter, the immunobeads were rinsed twice with endogenous polymerase assay (EPA) wash buffer (50 mM Tris [pH 7.4]–75 mM NH4Cl–1 mM EDTA) as described by Bruss and Ganem (7).
EPA.
For standard EPA reactions, immunocomplexed capsids were resuspended in 50 μl of 50 mM Tris (pH 7.4)–75 mM NH4Cl–1 mM EDTA–20 mM MgCl2–0.1 mM β-mercaptoethanol–0.5% Tween 20–50 μM unlabeled dNTPs (dATP, dCTP, and dTTP)–10 μCi of [α-32P]dGTP (3,000 Ci/mmol; NEN-DuPont, Boston, Mass.). The reaction mixtures were typically incubated for 6 h at 37°C unless noted otherwise. For primer extension analyses, EPAs were performed in the presence of all four unlabeled dNTPs at 200 μM.
Product analyses.
For the analysis of in vivo-generated products, cores were partially purified via limited nuclease-protease treatment and standard ultracentrifugation procedures. Nucleic acids were deproteinized by incubation in 100 mM Tris (pH 7.6)–150 mM NaCl–12.5 mM EDTA–1% sodium dodecyl sulfate (SDS)–1 mg of proteinase K (Boehringer Mannheim, Indianapolis, Ind.) per ml for 1 h at 37°C followed by several extractions with phenol-chloroform and ethanol precipitation in the presence of 30 μg of carrier RNA (Boehringer Mannheim). For the analysis of non-protein-linked DNA, protease digestion was omitted; instead, capsids were disrupted by being boiled for 8 min in the presence of 2% SDS and 10 mM dithiothreitol (DTT). Protein-free nucleic acids were then recovered by three rounds of phenol extractions followed by chloroform extraction and ethanol precipitation as described above. Nucleic acid was dissolved in 30 mM NaOH–1 mM EDTA, electrophoresed through 1% alkaline agarose in 30 mM NaOH–1 mM EDTA for 4 h at 45 V, transferred to a positively charged nylon membrane (Qiagen, Santa Clarita, Calif.) by capillary transfer in 0.4 M NaOH, and hybridized for 2.5 h at 70°C with strand-specific HBV riboprobes in Rapid-hyb buffer (Amersham, Arlington Heights, Ill.). The riboprobes were generated by in vitro transcription (T7-Maxiscript; Ambion, Austin, Tex.) in the presence of [α-32P]UTP (800 Ci/mmol; NEN-DuPont), yielding sense and antisense ∼600-bp transcripts complementary to HBV nt 1401 to 2016.
For the analysis of in vitro-generated EPA products on agarose gel, capsids were first freed of exogenous nucleic acid contamination by limited micrococcal nuclease treatment in the presence of 5 mM CaCl2. Encapsidated nucleic acid was then deproteinized as described above, dissolved in TE (10 mM Tris [pH 8.0]–1 mM EDTA), electrophoresed through 1% agarose in TBE (90 mM Tris [pH 8.0]–90 mM borate–0.1 mM EDTA), and visualized by autoradiography of the dried gel as described previously (37).
For analysis of Pol-linked RT products by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), nucleocapsids were immunoprecipitated under native conditions, subjected to EPA, and then washed twice with IP wash buffer (50 mM Tris [pH 7.4]–150 mM NaCl–5 mM EDTA–1% Triton X-100–0.02% SDS) to remove the majority of unincorporated dNTPs. To release the radiolabeled Pol-DNA adducts, the nucleocapsids were disrupted by being boiled for 7 min in TNE (10 mM Tris [pH 7.4]–150 mM NaCl–1 mM EDTA) containing 1% SDS and 10 mM DTT and then diluted 10-fold into buffer (50 mM Tris [pH 7.4]–190 mM NaCl–6 mM EDTA–1.25% Triton X-100) supplemented with 1 mM benzamidine-HCl and 1 mM Pefabloc (Boehringer Mannheim). The liberated Pol-DNA complexes were immunoprecipitated by being rocked overnight at 4°C with rabbit anti-Pol antiserum 1-832 (19) bound to protein A-Sepharose CL-4B. After being rinsed twice with IP wash buffer as described above, the beads were heated to 100°C for 5 min in Laemmli sample buffer containing 5% DTT. The liberated Pol adducts were resolved by SDS-PAGE and visualized by autoradiography following fixing and drying of the minigels. In some experiments, 35S-labeled, in vitro-translated HBV Pol was run as a reference.
Biochemical analysis of native nucleocapsids.
Cores were enriched from crude EC- plus PE-expressing (EC+PE) cell lysates by standard ultracentrifugation procedures essentially as described elsewhere (53). Aliquots (∼2 μg of core protein) were subjected to EPA reactions as described above but without prior IP. The accessibility of the nascent DNA products to different antibodies was assessed by IP performed under native or denaturing conditions essentially as described above. The antibodies used were polyclonal rabbit anti-Pol- and anti-HBe/c-specific antisera (Dako Corp.) as well as two monoclonal antibodies (MAbs) that specifically recognize either intact hepatitis B core antigen-carrying capsids (MAb 3120) (43) or nonparticulate hepatitis B e antigen-expressing core proteins (MAb 2221) (22). Both antibodies were a kind gift of M. Mayumi (Jichi Medical School, Tochigi-ken, Japan). The IP samples were examined for the presence or absence of radiolabeled RT products by SDS-PAGE as described above.
For native agarose gel analysis, cores were first freed of unincorporated isotope by being pelleted through a cushion of 20% (wt/vol) sucrose in TNE at 550,000 × g for 1 h at 4°C with a tabletop ultracentrifuge (Beckman Instruments, Fullerton, Calif.). After resuspension in TNE, the nucleocapsids were electrophoresed through 1% agarose in 40 mM Tris-acetate (pH 7.8)–2 mM EDTA (TAE) prior to capillary transfer onto a nitrocellulose membrane (Schleicher & Schuell, Keene, N.H.). Membrane-immobilized nucleocapsids were visualized by immunostaining with anti-HBe/c antibody (Dako) and alkaline phosphatase-coupled anti-rabbit immunoglobulin G (Bio-Rad Laboratories, Richmond, Calif.) followed by autoradiography of the dried membrane.
Primer extension.
Covalently linked minus-strand DNA was recovered from disrupted capsids via IP with anti-Pol-coupled protein A-Sepharose and then deproteinized and precipitated as described above. The 5′ ends of minus-strand DNA were mapped by reiterated primer extension using Vent (exonuclease-deficient) polymerase (New England BioLabs, Beverly, Mass.) and a thermocycler essentially as described previously (21). The primers (Genosys, The Woodlands, Tex.) were complementary to minus-strand DNA nt 1764 to 1783 (5′-AGGTCTTTGTACTAGGAGGC-3′) and 1382 to 1401 (5′-GCTAGGCTGTGCTGCCAACT-3′) of the HBV ayw sequence; they were 5′ end labeled with [γ-32P]ATP (6,000 Ci/mmol; NEN-DuPont) and T4 polynucleotide kinase (New England Biolabs). Primer extension reaction cocktails containing 5.7 μl of DNA in H2O, 0.6 pmol of primer, 1.0 μl of 10× Vent polymerase buffer (New England Biolabs), 0.3 μl of 100 mM MgSO4, 0.2 mM dNTPs, and 1 U of Vent (exo-minus) polymerase in a total volume of 10 μl were subjected to PCR and then analyzed on 6 or 8% sequencing gels along with sequencing ladders generated with the corresponding primers. According to the manufacturer’s technical instructions, the Vent (exo-minus) polymerase extension products are typically 0.5 to 1.5 nt longer than the corresponding sequencing reaction species; this discrepancy reflects (i) the phosphate present on the extension but not the sequencing primer and (ii) and a nontemplated nucleotide frequently left on the extension product due to the lack of the 3′-5′ exonuclease activity.
RESULTS
Expression of replicative HBV nucleocapsids.
Our goal was to reconstitute in vitro replication-competent HBV nucleocapsids as a reagent for studying the polymerase in its authentic environment, the core particle interior. To this end, we created two types of recombinant baculovirus vectors harboring the C and Pol (P) genes. The structures of the resultant HBV RNAs are illustrated in Fig. 1; all contain ∼120 nt of baculovirus 5′ leader sequence. The first vector type yields an RNA that resembles pgRNA and is equipped to coexpress the C and Pol proteins in cis from overlapping ORFs within the same RNA, resulting in cores that are referred to here as cis cores. The prototype vector of this class, pBV-ECPE, produces ECPE RNA, in which the C and P ORFs are flanked by the 5′ ɛ and 3′-DR1 and ɛ elements (designated E in Fig. 1). pBV-ECP, pBV-CPE, and pBV-CP are pBV-ECPE derivatives lacking the 3′ and/or 5′ ɛ element. The remaining vectors yield monocistronic RNAs harboring a single HBV protein: in this case, the C and Pol proteins were provided in trans (i.e., via transcomplementation with two separate recombinant baculoviruses), yielding trans cores. The C and EC RNAs are similar except the latter includes a 5′-terminal extension bearing ɛ. Likewise, for the pBV-PE and pBV-P Pol constructs, only PE RNA includes 3′-DR1 and ɛ elements.
Expression of the C and Pol proteins from the above constructs in infected Sf9 cells was verified by immunoblotting (data not shown). Irrespective of their precise structure, all the core-containing constructs expressed C protein at high levels (2 to 6 mg per liter of culture) and generated core particles. Pol expression levels were lower but respectable (∼130 to 260 μg per liter), with the monocistronic P constructs giving greater expression.
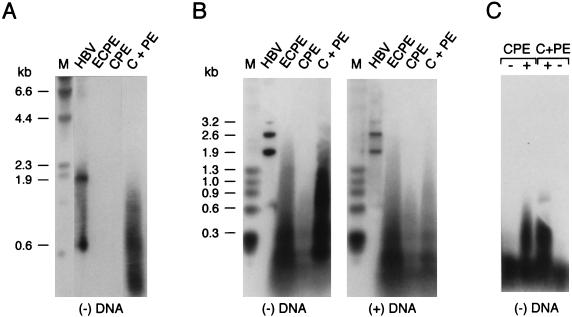
Since hepadnavirus polymerases initiate HBV DNA synthesis in vivo when expressed in yeast or insect cell systems (19, 45), we first examined whether cores expressed in baculovirus-infected insect cells contain HBV DNA (Fig. 2). Lysates were prepared from cells 90 h postinfection, and the cores were substantially enriched by mild nuclease-protease treatment followed by centrifugation through sucrose. Encapsidated nucleic acids were deproteinized, electrophoresed through denaturing alkaline agarose gels, and detected by Southern blotting using 32P-labeled strand-specific riboprobes. An early experiment (Fig. 2A) compared the minus-strand DNA contents of similar amounts of baculovirus-derived cores created by coexpression of C and Pol in cis (ECPE and CPE) versus in trans (C+PE). trans cores gave a strong smear of HBV minus strands, indicating extensive in vivo HBV replication, whereas ECPE cis cores contained much less minus-strand DNA.
FIG. 2.
Southern analysis of the HBV DNA present in cis and trans cores. (A) HBV DNA extracted from equal amounts of purified cis (ECPE and CPE) or trans (C+PE) cores was resolved on alkaline agarose gels. After alkaline capillary transfer to a nylon membrane, minus-strand DNA [(−) DNA] was disclosed by hybridization with a radiolabeled plus-strand riboprobe. Lane M, labeled HindIII λ DNA markers (in kilobase pairs); lane HBV, 1.9- and 0.6-kb HBV DNA markers. (B) DNA extracted from purified ECPE, CPE, and C+PE cores was resolved on alkaline agarose gels and then membrane immobilized. Note that a threefold excess of cis cores was used for this experiment. HBV DNA species of minus (left) and plus (right) polarities were detected by hybridization with riboprobes of the opposite polarities. Migration of radiolabeled λ HindIII markers (lanes M) and HBV-specific 3.2-, 2.6-, 1.9-, and 0.6-kb DNA fragments (lanes HBV) is indicated on the left. (C) Analysis of phenol-extractable HBV minus-strand DNA was performed essentially as described for panel B, with a plus-strand riboprobe. Aliquots of HBV DNA extracted from CPE-derived cis cores and C+PE-derived trans cores were phenol extracted with (+) or without (−) prior digestion with proteinase K. The latter samples were boiled in the presence of SDS and DTT to disrupt the capsids prior to extraction.
A follow-up experiment (Fig. 2B) employing significantly more cis cores to compensate for their low HBV DNA content revealed that cis (ECPE and CPE lanes) and trans (C+PE) cores both contain HBV minus- and plus-strand DNA species but clearly differ with respect to the abundance and size distribution of these species. C+PE trans cores again contained predominantly minus-strand DNAs of diverse sizes, with the longest species being roughly the size expected for a full-length cDNA copy of PE RNA (∼2.8 kb). The corresponding plus-strand products appeared to be fewer and shorter overall. This product profile generally accords with a discontinuous HBV replication reaction in which minus-strand synthesis precedes plus-strand production.
In contrast to trans cores, cis cores contain broadly comparable amounts of both minus- and plus-strand HBV DNAs. These species (particularly the ECPE species), however, are not so clearly related to the HBV replication scheme since (i) they are mostly short and (ii) the minus strands are only marginally longer than the plus strands. However, evidence presented below leaves no doubt that ECPE cores conduct authentic HBV replication: specifically, they harbor correctly initiated minus strands that originated in vivo and also generate full-genome-length products in in vitro assays. In light of this, we suspect that the anomalous HBV DNA species seen in cis cores in this experiment probably result from partial degradation occurring subsequent to replication, most likely during the mild nuclease treatment used for capsid isolation; ECPE cis cores in particular appear highly susceptible to such degradation (data not shown). To further verify that the encapsidated HBV DNAs seen in this experiment result from HBV replication, we examined whether they could be extracted into phenol (Fig. 2C): authentic hepadnavirus minus-strand DNA is rendered soluble in phenol by virtue of its covalent linkage to Pol (3, 19, 45, 49). The Southern blotting data revealed that most HBV DNA minus strands present in enriched CPE and EC+PE cores were removed by phenol extraction unless they were first deproteinized with proteinase K. This implies that these DNAs are protein linked and thus presumably derive from HBV replication; conversely, they are unlikely to represent contaminating DNA species from baculovirus vectors.
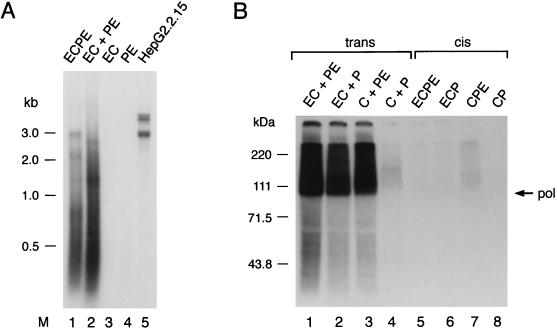
Next, we used a standard EPA to establish the in vitro replication competence of these HBV nucleocapsids. EPAs were conducted with cores immunoprecipitated with anticore antibody from equivalent amounts of baculovirus-infected Sf9 cell lysates prepared 90 h postinfection (Fig. 3A). The resultant 32P-labeled DNA products were deproteinized and resolved by native agarose electrophoresis. For comparison, this analysis included cores derived from 2.2.15 hepatoblastoma cells (39) 10 days postconfluence; these cores are largely mature with respect to HBV replication and thus gave mainly labeled double-stranded circular and linear HBV genomes (Fig. 3A, lane 5). In contrast, prolific disperse products were seen for trans (EC+PE) cores and, to a lesser extent, for cis (ECPE) cores; these products were mostly small but did include discrete species of the sizes expected for full-length copies of the ECPE and PE RNAs (∼3.2 and ∼2.8 kb, respectively). Controls expressing core (EC) or Pol (PE) protein alone gave no replication products, indicating that both proteins are required for the production of replicative cores.
FIG. 3.
Analysis of HBV DNA products generated in in vitro polymerase reactions. (A) Agarose gel analysis of endogenously radiolabeled DNA products from nucleocapsids. Sf9 cells were harvested 90 h postinfection with the recombinant baculoviruses ECPE, EC+PE, EC, and PE. 2.2.15 cells were collected after 10 days of culture. Core particles were immunoprecipitated from clarified cell extracts and subjected to EPAs as described in Materials and Methods. Samples of isolated nucleic acids were analyzed on a 1% agarose gel and then visualized by autoradiography. The approximate size range (in kilobase pairs) of the various products is indicated on the left. (B) SDS-PAGE analysis of Pol-DNA complexes after in vitro polymerase reactions. HBV nucleocapsids were generated either in trans (lanes 1 to 4) or in cis (lanes 5 to 8) by infection of Sf9 insect cells with the baculovirus constructs indicated above each lane. The immunoprecipitated capsids were subjected to EPA and then boiled. Following radioimmunoprecipitation, the covalently linked minus-strand DNA-Pol complexes were examined by SDS-PAGE and autoradiography as described in Materials and Methods. The Pol protein band is indicated on the right; marker proteins (sizes in kilodaltons) are indicated on the left.
We then examined (Fig. 3B), for a variety of trans (lanes 1 to 4) and cis (lanes 5 to 8) core preparations, whether the in vitro EPA reaction yields DNA that is covalently linked to Pol. Following immunoprecipitation and EPA (see above), the cores were disrupted by being boiled with SDS and DTT. The liberated DNA-Pol adducts were recovered with anti-Pol antibody (19) and examined by SDS-PAGE. Only trans cores generated abundant smears of radiolabeled material extending from ∼93 kDa in mass to the top of the gel, as expected for variable lengths of [32P]DNA linked to full-length Pol (∼93 kDa). Quantitation of these data revealed that the cis cores gave, on average, ∼30-fold less Pol-linked material than EC+PE cores. Interestingly, product synthesis was highest in trans cores when both RNAs harbored ɛ (EC+PE), falling by 30 to 40% when only one ɛ was present (EC+P or C+PE) and by ∼10-fold in the absence of ɛ (C+P).
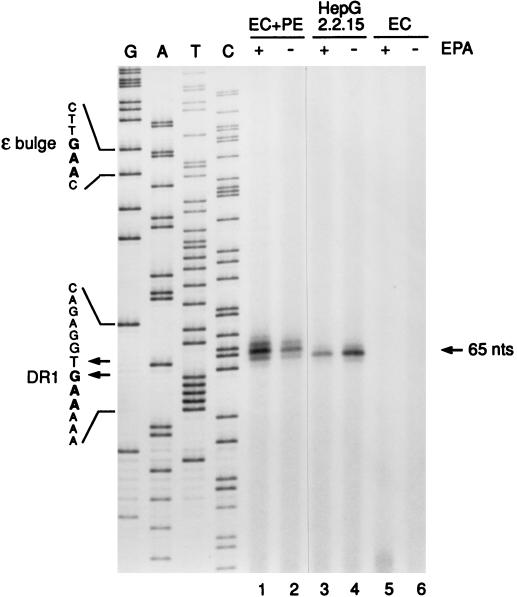
In authentic HBV DNA replication, the 5′ ends of nascent minus strands map by primer extension to the sequence (3′-CACUU-5′) within the 3′ DR1 element, although their true origin lies within ɛ. To establish if the 5′ ends of HBV minus-strand DNAs isolated from immunoprecipitated EC+PE cores map to DR1, we performed a PCR primer extension analysis (Fig. 4) using an end-labeled plus-strand primer which anneals to HBV nt 1764 to 1783, ∼65 nt upstream of 3′ DR1. For positive and negative controls, we also analyzed cores from 2.2.15 cells, which closely resemble cores from infected patients (1, 39), as well as Pol-minus EC capsids. In an effort to distinguish between HBV DNA strands that arise in vivo (i.e., in intact insect cells) versus in the in vitro EPA reaction, the EC+PE and 2.2.15 core samples were divided in two; one half was analyzed with no further treatment, while the other was first subjected to an EPA to allow in vitro DNA synthesis.
FIG. 4.
Primer extension analysis of Pol-associated DNA isolated from recombinant HBV nucleocapsids reveals DR1 as the origin of minus-strand DNA elongation. Following in vitro polymerase reactions in the presence (+ EPA) or absence (− EPA) of deoxynucleotides, covalently linked minus-strand DNA-Pol adducts were recovered from immunoprecipitated HBV capsids from EC+PE (lanes 1 and 2)- or EC (lanes 5 and 6)-expressing Sf9 cell lysates as described in Materials and Methods. Intracellular cores from 2.2.15 cells (lanes 3 and 4) served as a positive control. Purified DNA was annealed with a 5′-end-labeled plus-polarity oligonucleotide spanning nt 1764 to 1783 and then extended with a thermostable (exo-minus) polymerase and PCR cycling. Extension products were resolved on an 8% polyacrylamide sequencing gel along with a plus-strand dideoxy sequencing ladder (lanes G, A, T, and C) generated with the same primer and recombinant HBV DNA. The 65-nt extension product mapping to DR1 is indicated on the right. Shown on the left are minus-strand DNA sequences derived from the indicated portions of the plus-strand sequence ladder; these sequences include parts of the priming site in the bulge of 3′ ɛ and the elongation site in DR1. Minus-strand DNA 5′ ends mapping to nt 1829 (T residue) and 1828 (G residue) within DR1 are also indicated (arrows). Note that the primer extension products migrated approximately 0.5 to 1.5 nt more slowly than the corresponding position indicated by the sequencing ladder (see text).
To map the 5′ ends to the nucleotide level, the primer extension products were resolved on a sequencing gel alongside a plus-strand dideoxy sequencing ladder generated from the same primer and HBV plasmid DNA. The sequence alignment was corrected (46) for the primer extension products being retarded by 0.5 to 1.5 nt due to (i) the extra phosphate on the oligonucleotide primer and (ii) the addition of a nontemplated nucleotide by the exonuclease-deficient polymerase used for primer extension.
The autoradiogram revealed that the minus-strand 5′ ends indeed arise by copying the sequence 3′-CACUU-5′ within DR1. The primary 5′ ends mapped to minus-strand residues T1829 for EC+PE (Fig. 4, lanes 1 and 2) and G1828 for 2.2.15 (lanes 3 and 4) cores. Lesser start sites were G1828 and G1830 (trans cores) and T1829 (2.2.15 cores); the latter was more prominent in some experiments (data not shown). As expected, EC negative controls (Fig. 4, lanes 5 and 6) gave no signal. Authentic 5′ ends were seen in the absence of EPA, confirming the finding (Fig. 2) that HBV replication had occurred in vivo. For EC+PE capsids, the minus-strand signal appeared to increase somewhat with EPA, hinting at the production of additional HBV minus strands in vitro; a more explicit proof of this is found later in this paper.
In summary, the preceding experiments (and others below) demonstrate conclusively that both cis and trans baculovirus-expressed cores (i) contain a functional HBV Pol; (ii) undergo HBV replication in insect cells, yielding both minus- and plus-strand DNAs with a profile reminiscent of hepdnavirus replicative intermediates seen in immature cores from infected livers (41); (iii) generate minus strands with the correct 5′ ends; and (iv) remain competent for further replication in an in vitro setting.
In terms of in vitro Pol activity, trans cores appeared significantly more active than cis cores in the in vitro EPA reaction and were fully 30-fold more active in the production of Pol-linked minus strands; the reasons for the different activities of the two classes of cores remain uncertain but presumably reflect, at least in part, their intrinsically different HBV DNA contents prior to the EPA (see above). In any case, since we were interested primarily in reconstituting the earliest phases of HBV replication, the remaining experiments focus mainly on trans cores and, unless otherwise noted, employ EC+PE cores due to their consistently high activity.
DNA synthesis occurs within core particles.
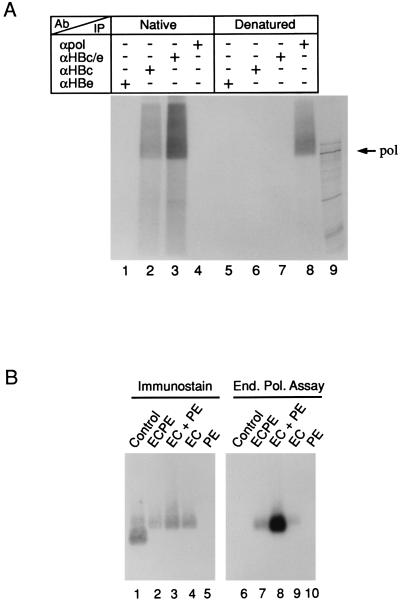
To provide formal proof that in vitro HBV DNA synthesis occurs inside intact cores, we used two experimental approaches (Fig. 5). First, enriched EC+PE cores were subjected to an EPA, and then the accessibility of the radiolabeled Pol-DNA adducts to a panel of antibodies (see Materials and Methods) was determined by IP (Fig. 5A) under either native (lanes 1 to 4) or denaturing (lanes 5 to 8) conditions. SDS-PAGE revealed that only under native conditions were radiolabeled Pol-DNA adducts recovered by antibodies directed against antigens present on intact cores, such as MAb 3120 (43), which recognizes HBc/β (lane 2), or a rabbit polyclonal antiserum (lane 3). Conversely, only an anti-Pol antiserum recovered adducts following capsid disruption (Fig. 5A, lane 8). MAb 2221 (Fig. 5A, lanes 1 and 5), which is specific for the HBe/b epitope and recognizes only nonparticulate core proteins (42), provided a negative control.
FIG. 5.
Pol activity is confined to the interior of the core particle. (A) Core particles from EC+PE-infected insect cells were purified by ultracentrifugation and then subjected to in vitro EPAs as described in Materials and Methods. For SDS-PAGE analysis of the covalently linked Pol-DNA complexes, reaction products were radioimmunoprecipitated under native (lanes 1 to 4) or denaturing (lanes 5 to 8) conditions using the following antibodies: HBe-specific MAb 2221 (lanes 1 and 5), HBc-specific MAb 3120 (lanes 2 and 6), rabbit anti-HBe/c-specific antiserum (lanes 3 and 7), and rabbit anti-Pol antiserum (lanes 4 and 8). See the text for further details. For visualization, the gel was dried and exposed to X-ray film. The migration of the ∼93-kDa in vitro-translated HBV Pol protein (lane 9) is indicated on the right. (B) Colocalization of radiolabeled in vitro Pol products and immunostained HBV nucleocapsids. Core particles were harvested from insect cell lysates ∼90 h postinfection with the ECPE (lanes 2 and 6), EC+PE (lanes 3 and 8), EC (lanes 4 and 9), and PE (lanes 5 and 10) recombinant baculoviruses as indicated above and then partially purified by ultracentrifugation. Aliquots were subjected to an in vitro EPA (End. Pol. Assay) and then resolved on a native 1% agarose gel. Following transfer to a nitrocellulose membrane, core proteins were immunostained with anti-HBe/c antiserum (lanes 1 to 5) and then exposed to X-ray film to visualize the 32P-labeled products (lanes 6 to 10). Core particles expressed in Escherichia coli (lanes 1 and 6) served as a reference.
In a second approach, we performed in vitro EPAs on enriched ECPE, EC+PE, EC, and PE cores and then examined whether the EPA products comigrate with the core proteins upon native agarose gel electrophoresis (Fig. 5B) under conditions under which cores remain intact (6, 14). After the EPA, the cores were pelleted through 20% (wt/vol) sucrose to remove unincorporated label and then resolved on a native agarose gel and transferred to nitrocellulose. Core proteins were disclosed by immunostaining (Fig. 5B, lanes 1 to 5), while the polymerase reaction products were detected by autoradiography of the dried membrane (lanes 6 to 10). Immunostaining verified the expected presence of core proteins in a reference sample of purified, recombinant bacterial cores (Fig. 5B, lane 1) as well as in the ECPE, EC+PE, and EC lanes (lanes 2 to 4) but not in the free polymerase (PE) sample (lane 5); the faster migration of bacterial cores is presumably due to a short leader sequence carried by the bacterial core protein (48). In accord with our earlier results, a strong EPA product signal comigrated with EC+PE cores (Fig. 5B, lane 8) and a weaker signal comigrated with ECPE cores (lane 7), while the free polymerase (PE) control lane is empty; the faint signal seen with empty (i.e., Pol-minus) EC capsids (lane 9) may reflect a spillover from the EC+PE lane. These results again suggest that DNA synthesis occurs inside capsids; consistent with this interpretation, the products migrated significantly ahead of the core proteins if the cores were disrupted with SDS prior to electrophoresis (data not shown).
Characterization of the HBV Pol activity.
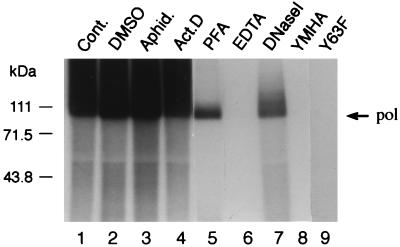
To confirm that the reverse transcriptase activity of HBV Pol mediates the synthesis of Pol-linked products in vitro, we conducted additional control EPA reactions on equal amounts of immunoprecipitated EC+PE cores (Fig. 6). The readout was the covalent linkage SDS-PAGE assay described above (see Materials and Methods). Consistent with the known properties of HBV Pol (19, 50), the standard in vitro reaction (Fig. 6, lane 1) was eliminated by EDTA (lane 6) but was not affected by the cellular DNA polymerase inhibitor aphidicolin (lane 3) or by actinomycin D (lane 4), which inhibits DNA-dependent DNA synthesis but not RNA-templated RT. Dimethyl sulfoxide, the solvent for some of the inhibitors, did not affect the reaction (Fig. 6, lane 2).
FIG. 6.
HBV Pol is responsible for in vitro DNA synthesis. Immunoprecipitated cores from EC+PE-infected Sf9 cells (lanes 1 to 7) were subjected to an in vitro EPA reaction in the presence of inhibitors as follows: lane 1, control (Cont.) (standard reaction); lane 2, 0.4% dimethyl sulfoxide (DMSO); lane 3, 30 μM aphidicolin (Aphid.); lane 4, 80 μM actinomycin D (Act. D); lane 5, 1 mM PFA; lane 6, 40 mM EDTA; and lane 7, DNase I posttreatment with 4 U for 30 min at 37°C. Also included are EC+PE coinfections carried out with mutant Pol baculoviruses bearing either catalytic site (YMHA [lane 8]) or priming site (Y63F [lane 9]) mutations. These were analyzed under standard in vitro EPA conditions as described in Materials and Methods and examined by SDS-PAGE and autoradiography. Pol is indicated on the right, and the positions of marker proteins (in kilodaltons) are indicated on the left.
In accord with previous reports (19, 50), the drug PFA had a highly characteristic impact (Fig. 6, lane 5), leaving the priming reaction intact but blocking all further minus-strand elongation, resulting in a discrete, labeled Pol band. Posttreatment of the standard reaction product (Fig. 6, lane 1) with DNase I (lane 7) verified that the products are of DNA origin; the short Pol-DNA products resisting DNase digestion are presumably sterically protected by Pol.
Finally, and most importantly, we confirmed the role of both the polymerase and the priming functions of HBV Pol in the in vitro reaction via the inactivity of two previously reported mutants (20, 37) bearing changes in either the YMDD catalytic site motif (YMHA [Fig. 6, lane 8]) or the Tyr 63 priming acceptor site, which was replaced with Phe (Y63F [lane 9]).
In vitro priming.
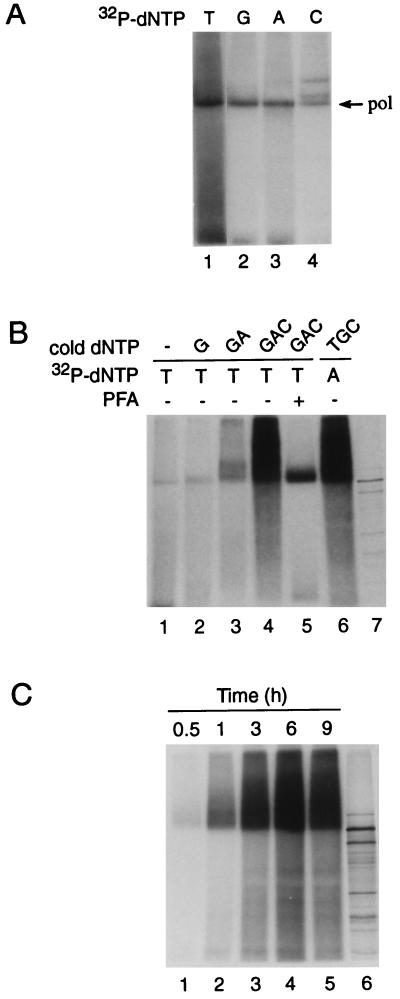
The effect of PFA on the in vitro EPA for EC+PE cores (see above) strongly suggested the occurrence of a discrete in vitro priming reaction, the initial event in HBV replication. The HBV priming reaction initiates at the ɛ bulge, the true origin of hepadnavirus minus strands (26, 47, 49), and is templated by the sequence 3′-ACUU-5′. It results in a Pol-DNA adduct with 3 or 4 nt of DNA which are added in a specific order, with the first deoxynucleotide being preferably dG, as reported for the DHBV Pol priming reaction (50), or T, which appears to be the first choice for free HBV Pol expressed in insect cells (19). To verify the occurrence of priming in immunoprecipitated EC+PE cores and examine the preference for the first nucleotide added, we conducted EPAs in which only a single radiolabeled dNTP was included; all four radiolabeled dNTPs (with similar specific activities) were tested in this manner. The SDS-PAGE results show that HBV Pol can incorporate any of the four dNTPs (Fig. 7A) but, in accord with the findings of Lanford et al. (19), preferred TTP, followed by dGTP, dATP, and finally dCTP, which resulted in two additional bands of unknown origin; the high background seen in Fig. 7A, lane 1, presumably reflects impurities in the batch of [32P]TTP (cf. Fig. 7B).
FIG. 7.
Characterization of in vitro priming and elongation reactions. In vitro Pol reactions (EPAs) were performed with immunoprecipitated nucleocapsids from EC+PE-coinfected insect cell lysates. (A) Nucleotide preference for priming. EPAs were conducted in the presence of a single radiolabeled deoxyribonucleotide as specified above the lanes. The Pol band is indicated on the right. (B) Sequential addition of dNTPs. EPAs were performed with [32P]TTP (lanes 1 to 5) or [32P]dATP (lane 6) followed by the specified unlabeled dNTPs. The reaction products shown in lanes 4 and 5 were generated analogously except that elongation of the Pol-primer complex was inhibited by PFA in the latter. Lane 7 shows the migration of in vitro-translated 35S-Pol protein. (C) Time course of in vitro priming-RT. Standard EPA mixtures containing [32P]dATP and unlabeled dGTP, dCTP, and TTP were incubated at 37°C for 0.5, 1, 3, 6, or 9 h. 35S-labeled in vitro-translated HBV Pol is shown in lane 6. All reaction products were analyzed by SDS–8% PAGE and autoradiography to visualize covalently Pol-linked products as described in Materials and Methods.
This nucleotide incorporation preference (T > dG > dA > dC) seemed fully consistent with the possibility of authentic priming directed by ɛ. If this is correct, we reasoned that we should be able to “grow” the Pol-linked oligonucleotide primer via the sequential addition of dNTPs, as predicted by the ɛ bulge sequence (ACUU). To test this (Fig. 7B), immunoprecipitated EC+PE cores were subjected to EPA reactions in the presence of only [32P]TTP, resulting in a Pol-[32P]T adduct (lane 1) which comigrated upon SDS-PAGE with an in vitro-translated HBV Pol marker (lane 7). Immediately after the EPA, this adduct was chased by addition of unlabeled dNTPs in the order indicated above Fig. 7B. The Pol-T band was clearly shifted to a slightly higher molecular mass by addition of dGTP (Fig. 7B, lane 2) and further still by dGTP plus dATP (lane 3). For comparison, control lanes show the disperse smear of labeled products generated in the presence of all four dNTPs with either [32P]TTP (Fig. 7B, lane 4) or [32P]dATP (lane 6) used as the label and the strong priming band resulting from the use of PFA (lane 5). These data are consistent with the idea that EC+PE cores are competent for authentic de novo priming in vitro, resulting in the synthesis of a Pol-T-G-A or Pol-T-G-A-A adduct.
We next examined the effect of reaction time (Fig. 7C) on the overall quantity and size of the labeled Pol-DNA adducts generated by EC+PE cores in a standard EPA at 37°C. The EPA was sampled at intervals of 0.5, 1, 3, 6, and 9 h (Fig. 7C, lanes 1 to 5, respectively) and analyzed as before by SDS-PAGE along with a 35S-labeled Pol marker (lane 6). The autoradiogram reveals the labeled products increasing progressively with time in both amount and average size (molecular weight), plateauing at ∼6 h as judged by phosphorimaging. Thus, the encapsidated polymerase seems both stable and quite processive in vitro, as is further evidenced below.
In vitro processivity of HBV Pol.
The production of HBV DNAs with sizes approaching those of the input Pol RNA or pgRNA templates (see above) indirectly implies that cis and trans cores harbor a highly processive HBV Pol enzyme. For a more direct test of processivity and, in particular, of in vitro processivity, we designed a primer extension experiment (Fig. 8) to establish if Pol could extend HBV DNA minus strands from the vicinity of DR1 to a point ∼400 bp downstream in a pure in vitro elongation reaction. This experiment used enriched ECPE cis cores which were generated either by the standard procedure or by a modified procedure in which the insect cells were exposed to 1.5 mM PFA immediately postinfection (see Materials and Methods). This trick effectively blocks HBV DNA elongation within insect cells (see below).
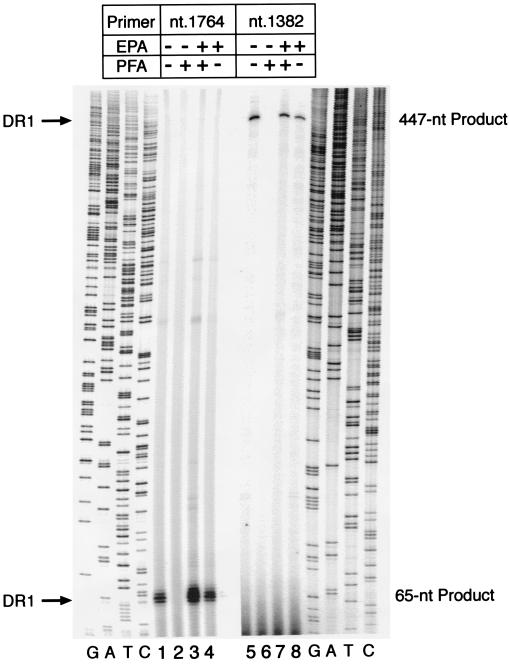
FIG. 8.
In vitro processivity of HBV Pol. HBV nucleocapsids were expressed in insect cells via infection with a recombinant baculovirus carrying a full-length HBV pregenome (ECPE) in the absence (−PFA) or presence (+PFA) of 1.5 mM PFA. Purified capsids were incubated in vitro without (−EPA) or with (+EPA) deoxynucleotides as described in Materials and Methods. Aliquots of the extracted DNA were annealed to 32P-end-labeled oligomers spanning nt 1764 to 1783 and 1382 to 1401 of the HBV sequence and extended as described above. Primer extension products were analyzed on a 6% polyacrylamide sequencing gel adjacent to sequencing ladders (lanes G, A, T, and C) generated with the analogous primers and HBV plasmid DNA. The positions of the 65- and the 447-nt extension products mapping to DR1 are indicated.
The ECPE cores were subjected to either a standard EPA or a mock EPA with no dNTPs as a control for any HBV DNA losses associated with the reaction. Recovered HBV DNA samples were annealed to one of two 32P-labeled HBV primers: one (nt 1764 to 1783) which gives an ∼65-nt extension product (see above) and an upstream primer (nt 1382 to 1401) yielding a band of ∼447 nt. The primers were extended and analyzed alongside the corresponding sequencing ladders as described above.
The results show that unreacted ECPE cores from standard infections gave products of around 65 (Fig. 8, lane 1) and 447 (lane 5) nt which were not enhanced by EPA (lanes 4 and 8), indicating that these HBV minus strands predated the EPA. In contrast, cores from PFA-treated cells gave no such products without EPA (Fig. 8, lanes 2 and 6), showing that PFA had effectively suppressed minus-strand DNA production in vivo. The full amount of products reappeared following EPA (Fig. 8, lanes 3 and 7).
From these results, we draw two conclusions. Most importantly, the encapsidated HBV Pol is able to reverse transcribe some 400 nt in vitro. Moreover, the in vitro-derived products seem comparable in amount to those generated in vivo (Fig. 8, cf. lanes 1 and 5 versus lanes 3 and 7), suggesting that the PFA-inhibited cores retain their full HBV DNA production capacity. We also note that this autoradiogram shows faint products (most noticeable in lane 3) mapping to ɛ, as well as to an unrelated site at ∼ nt 1930 as reported by Lanford et al. (19).
DISCUSSION
Persistent infection by HBV ultimately relies on continuous active viral replication. Thus, there is a need to develop in vitro systems suitable for (i) reconstituting HBV replication; (ii) providing a detailed biochemical characterization of HBV Pol, the key enzyme in HBV replication; and (iii) identifying novel inhibitors of HBV replication. This need has been partially met by the recent discovery of unencapsidated DHBV and HBV Pol enzymes with in vitro replicative activity (19, 37, 45, 50). While these Pol reagents have advanced our knowledge of hepadnavirus replication, they are still limited: they lack the core protein, which plays a role in viral replication (14, 25, 33, 52); they have not been shown to synthesize viral plus strands; and they may exhibit suboptimal specific activity and processivity. Finally, it is not clear that results obtained with DHBV Pol extrapolate to the less well characterized and distantly related HBV Pol; the two enzymes may differ functionally, e.g., with respect to their dependence on ɛ (20, 37, 38, 46, 50).
Since authentic HBV replication in infected cells occurs inside nucleocapsids, we have been interested in developing recombinant HBV nucleocapsids as a second-generation reagent for studying HBV replication in vitro. In principle, nucleocapsid formation requires just C protein, Pol, and pgRNA. However, despite considerable effort and the use of a wide variety of constructs, we repeatedly failed to demonstrate productive interactions between these three components or to generate replicative cores in Xenopus oocytes (36). These failures, which could be for trivial or profound reasons, serve to illustrate that there may be as-yet-unknown requirements for the production of replicative HBV cores.
For the present study, we turned to the baculovirus expression system in light of its ability to drive high expression and also to produce active HBV Pol (19). We generated cis and trans classes of nucleocapsids, based on whether C protein and Pol were supplied in cis from a pgRNA-like RNA or in trans from two monocistronic viral RNAs. Both types of cores could be generated in quantity (up to 6 mg/liter), were reasonably easy to purify, conducted HBV replication in a reaction which occurs strictly on the interior of the core particle (Fig. 5), and could be stored for up to 2 years without noticeable loss of Pol activity (36). Both classes of cores were active for HBV replication in in vivo and in vitro settings, as was also the case for unencapsidated forms of HBV Pol in the baculovirus system and DHBV Pol in the yeast retrotransposon system (19, 45).
HBV replication in vivo was assessed by Southern analyses of DNA from cores newly isolated from insect cells (Fig. 2). Both cis and trans cores harbored HBV DNA products of minus and plus polarities and of varied lengths. The minus-strand products were phenol extractable and had authentic 5′ ends (see below); the longest of them appeared close in size to unit-length copies of the cognate pgRNA or Pol RNA. trans cores typically contained more HBV DNA than an equal amount of cis cores. This DNA was asymmetric in character, with a molar excess of minus strands over shorter plus strands, a profile suggestive of the replicative intermediates seen in immature cores from infected cells. These findings imply that our recombinant cores execute a substantial portion of the HBV replication cycle while still within intact insect cells. The smaller amount of DNA present in cis cores typically comprised more-equal parts of minus and plus strands which appeared to be somewhat degraded, possibly due to the procedure used to enrich the cores.
In terms of in vitro HBV replication activity, cis cores were weakly active overall, and particularly so for the production of Pol-linked minus strands. trans cores showed more endogenous polymerase activity, were up to 30-fold more active (for EC+PE cores) for the production of Pol-linked minus strands, and, moreover, retained the capacity to execute the earliest stages of HBV replication in vitro (see below). Thus, although trans cores support HBV replication in vivo, some fraction evidently remains naive or “frozen” for minus-strand production until the in vitro reaction. These replication differences between cis and trans cores could reflect mechanistic distinctions between the replication of pgRNA versus subviral RNA templates, but they could alternatively be trivial in nature; factors specific to the trans setting, such as the greater expression of Pol, the higher DNA content, and the predominance of minus strands, could simply sway the efficiency of the in vitro replication reaction. Whatever the reasons, fortunately we can study early HBV replication events in both settings by maintaining the insect cells in PFA, starting soon after the infection. This simple trick blocks essentially all in vivo HBV replication and significantly increases the in vitro replicative activities of both cis and trans cores, although the trans cores remain more active (36). cis and trans cores offer potentially distinct experimental advantages for studying HBV replication; a specific advantage of the latter is the greater ease of mutagenizing key regulatory elements.
In retrospect, it is arguably surprising that our transcomplementation system produces replicative HBV cores, given that subviral RNAs such as PE appear to be a poor substitute for an authentic pgRNA molecule, the basis for most successful prior transcomplementation studies (2, 8, 16, 34). Unencapsidated Pols, however, can replicate RNAs comparable to PE (19, 50), and our findings suggest that this precedent extends to cores. Recombination between the EC and PE baculoviruses to give an ECPE-like pgRNA molecule is unlikely to explain our results, given the distinctive properties of cis and trans cores (Fig. 2 and 3).
Several lines of evidence confirm that the robust in vitro HBV replication reaction seen in EC+PE trans cores includes the priming (see below) and early RT phases of minus-strand HBV DNA synthesis. In support of the latter point, the nascent DNA products are refractory to actinomycin D, confirming their RNA-templated synthesis by a reverse transcriptase; display the characteristic covalent linkage to full-length HBV Pol (13, 19, 24, 50); and are strictly dependent on HBV Pol, as proven by the inactivity of mutant Pols carrying either a YMDD-to-YMHA change in the reverse transcriptase domain active site or a Y63F substitution that ablates the Tyr acceptor residue in the TP priming domain (20). Finally, primer extension analyses confirm the authenticity of the minus-strand 5′ ends, which map primarily to T1829 at the third position of the 3′ DR1 element on PE RNA; minor species map to the two surrounding G residues (G1828 and G1830). G1828 was the major HBV minus-strand initiation site in 2.2.15 liver cells, although the use of the T1829 site was seen here and also by Rieger and Nassal (31).
With respect to in vitro priming, our data affirm earlier work with unencapsidated HBV Pol (19, 20). PFA, a specific inhibitor of hepadnavirus Pol elongation but not priming, characteristically intensified and isolated a discrete priming reaction; moreover, primer-sized Pol adducts (presumably representing Pol protein bearing no more than one to three dNTPs) were apparent among the shortest Pol-linked species deriving from the EPA. The results of incorporation assays employing single labeled dNTPs, or dNTPs added in a specific sequence, were consistent with priming templated by the ACUU sequence within the ɛ bulge. In accord with the primer extension results described above and the work of Lanford et al. (19), T was the preferred initial dNTP for incorporation, and the Pol-T adduct could be extended by sequential addition of dGTP and dATP. Finally, HBV minus-strand production strictly required the Tyr 63 acceptor residue (20). It may be dangerous to so infer authentic HBV priming and the use of ɛ as the true replication origin, given that HBV Pol (i) is less dependent than DHBV Pol on ɛ (37, 38, 46, 50) and (ii) even gives a discrete priming reaction in the absence of ɛ (20). However, in support of this idea, trace amounts of HBV minus strands mapped directly to ɛ (Fig. 8). Moreover, as will be reported elsewhere, mutation of ɛ has a profound effect on minus-strand DNA production (36). Taken together, these findings indicate that recombinant trans cores are competent for priming at ɛ, for the translocation of the Pol-primer adduct to the 3′ DR1 on PE RNA, and also for the subsequent RT of minus strands from DR1.
The 32P-labeled DNA-Pol adducts present in our cores implicate full-length HBV Pol (93 kDa) as the enzyme responsible for HBV negative-strand DNA synthesis. This finding accords with published work using animal cells (4), as well as with results obtained with nucleocapsid-free hepadnavirus Pols (19, 45, 50). While this one Pol molecule could execute the entire HBV replication reaction, it remains formally possible that the active form of HBV Pol is a dimer or a heterodimer, as is the case for human immunodeficiency virus RT (32). Enzymatically active subfragments of Pol reported for other systems (27, 51) would escape detection in our Pol assays if they lack covalently linked DNA or if they harbor other activities; indeed, preliminary evidence suggests the presence of RNase H activity in our recombinant cores (data not shown), but the relevance of this to the HBV life cycle, if any, remains to be established.
Although it is difficult to compare the properties of Pol molecules generated by different groups with different systems, we have unequivocally demonstrated that PFA-treated ECPE cores can elongate DNA chains of at least 400 nt in a typical in vitro reaction, with no sign that DNA synthesis is diminishing at this point. This processivity appears significantly superior to that of the nucleocapsid-free HBV (19) or in vitro-translated DHBV (9) Pol proteins, which were estimated to polymerize only ∼60 and 80 nt of DNA, respectively. We also believe that our recombinant cores harbor an HBV Pol with a specific activity higher than that of the corresponding unencapsidated enzyme from insect cells, where only 1 in 1,000 Pol molecules was estimated to be enzymatically active and 0.1 to 0.2 μg of purified Pol was required to obtain reasonable signals in vitro (19). In contrast, our assays give broadly comparable signals with ∼2 μg of core particles bearing at most ∼33 ng of Pol, assuming 1 Pol molecule per capsid of 240 core proteins. In reality, further analyses (data not shown) lead us to conclude that 2 μg of trans cores would contain <1 ng of Pol, thus making our encapsidated Pol at least 100-fold more active than its free counterpart. Along with the low Pol content, we have also confirmed earlier findings (15, 18) that baculovirus-expressed HBV cores contain significantly less RNA than cores from other systems (data not shown). The mechanistic basis of this phenomenon is unknown but is currently under investigation along with interesting issues it raises, most notably concerning the degree of selective packaging of Pol and HBV RNAs that occurs in this system.
Our findings with different trans cores shed interesting light on the influence of ɛ, which is reportedly essential for in vitro DNA synthesis by unencapsidated DHBV Pol (44, 50), although apparently not for HBV Pol (20, 37, 38). We have again confirmed, this time in the context of cores (C+P), the existence of an ɛ-independent mode of HBV replication; primer extension experiments suggest that in this case DNA initiates at cryptic sites on P mRNA (36). However, trans cores respond to the inclusion of ɛ and DR1 in two ways. First, one copy, or, better, two copies, of ɛ increase the in vitro polymerase activity of HBV cores up to 10-fold; strikingly, this is true whether ɛ is supplied by the 5′ end of C (EC+P) or by the 3′ end of Pol (C+PE); thus, ɛ and Pol do not need to be provided in cis. Second, ɛ, when present, appears to act as the dominant replication origin (see above). Together, these observations suggest that ɛ, though not essential for the reconstituted HBV replication reaction, is an important regulatory element when present. An intriguing issue which remains to be tested is whether the dependence of Pol on ɛ is modulated in any way by the C protein in trans cores.
Finally, we note that, unlike unencapsidated hepadnavirus Pol preparations (19, 45, 50), cis and trans recombinant cores clearly synthesize HBV plus strands, at least in vivo. By extrapolation, we suspect they may also make plus strands in the subsequent in vitro reaction, particularly in the case of cis cores, which, as judged by their comparable contents of HBV minus and plus strands, have already undergone significant plus-strand synthesis in vivo. cis cores may thus be analogous to mature cores from 2.2.15 cells which have largely completed DNA synthesis in vivo and conduct only limited plus-strand synthesis in in vitro assays (10). The idea that cis cores produce plus strands in vitro is supported by the observation that actinomycin D inhibits DNA synthesis in cis cores but not in trans cores (36). However, the modest DNA synthesis activity of cis cores may limit their utility for studying plus-strand biogenesis in vitro; moreover, the plus strands presumably arise by aberrant in situ priming (40), and our system does not necessarily allow de novo initiation of plus strands in vitro. The fact that our cores, unlike unencapsidated Pols, support plus-strand synthesis could imply a role for the core protein in this process; alternatively, however, plus-strand synthesis could be a simple consequence of the high processivity of encapsidated Pol.
In conclusion, novel baculovirus-generated cis and trans HBV nucleocapsids should provide valuable reagents for analysis of various aspects of HBV replication in vitro and in vivo, in the context of the authentic capsid environment. trans cores appear particularly well suited for study of the early priming and RT stages of HBV replication. These reagents should promote studies of the role of the HBV core protein in this process; our findings hint that core protein might influence such diverse functions as plus-strand synthesis, the use of ɛ, and overall Pol processivity. Finally, these cores will aid in the development of screens designed to identify new classes of inhibitors directed against Pol, such as the nonnucleoside reverse transcriptase inhibitors, which have therapeutic value in the HIV arena.
ACKNOWLEDGMENTS
We thank G. Acs (Mt. Sinai Medical Center) for plasmids pTHBV-1 and pHBV-1; R. Lanford (Southwest Foundation for Biomedical Research, San Antonio, Tex.) and M. Mayumi (Jichi Medical School) for valuable antibodies; and M.-A. Selby and P. Valenzuela (Chiron Corporation, Emeryville, Calif.) for pure bacterial core particles. We thank R. J. Colonno for his support and encouragement throughout this work, Don Ganem for insightful comments on the manuscript, and Steven F. Innaimo for 2.2.15-derived core particles, helpful discussions, and excellent support.
M.S. was supported in part by a fellowship from the Deutsche Forschungsgemeinschaft (SE 563/3-1).
REFERENCES
- 1.Acs G, Sells M A, Purcell R H, Price P, Engle R, Shapiro M, Popper H. Hepatitis B virus produced by transfected Hep G2 cells causes hepatitis in chimpanzees. Proc Natl Acad Sci USA. 1987;84:4641–4644. doi: 10.1073/pnas.84.13.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Junker-Niepmann M, Schaller H. The P-gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Schaller H. The amino-terminal domain of the hepadnaviral P-gene encodes the terminal protein (genome-linked protein) believed to prime reverse transcription. EMBO J. 1988;7:4185–4192. doi: 10.1002/j.1460-2075.1988.tb03315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beasley R P, Hwang L Y. Overview on the epidemiology of hepatocellular carcinoma. In: Hollinger F B, Lemon S M, Margolis M, editors. Viral hepatitis and liver disease. Baltimore, Md: Williams and Willkins; 1991. pp. 532–535. [Google Scholar]
- 6.Birnbaum F, Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruss V, Ganem D. The role of the envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang L J, Pryciak P, Ganem D, Varmus H E. Biosynthesis of the reverse transcriptase of hepatitis B viruses involves de novo translational initiation, not ribosomal frameshifting. Nature (London) 1989;337:364–368. doi: 10.1038/337364a0. [DOI] [PubMed] [Google Scholar]
- 9.Dannaoui E, Trépo C, Zoulim F. Inhibitory effect of penciclovir-triphosphate on duck hepatitis B virus reverse transcription. Antiviral Chem Chemother. 1997;8:38–46. [Google Scholar]
- 10.Davis M G, Wilson J E, VanDraanen N A, Miller W H, Freeman G A, Daluge S M, Boyd F L, Aulabaugh A E, Painter G R, Boone L R. DNA polymerase activity of hepatitis B virus particles: differential inhibition by l-enantiomers of nucleotide analogs. Antiviral Res. 1996;30:133–145. doi: 10.1016/0166-3542(96)00938-2. [DOI] [PubMed] [Google Scholar]
- 11.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 12.Ganem D, Pollack J R, Tavis J. Hepatitis B virus reverse transcriptase and its many roles in hepadnavirus genomic replication. Infect Agents Dis. 1994;3:85–93. [PubMed] [Google Scholar]
- 13.Gerlich W, Robinson W S. Hepatitis B virus contains a protein attached to the 5′ terminus of its complete DNA strand. Cell. 1980;21:801–809. doi: 10.1016/0092-8674(80)90443-2. [DOI] [PubMed] [Google Scholar]
- 14.Hatton T, Zhou S, Standring D N. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J Virol. 1992;66:5232–5241. doi: 10.1128/jvi.66.9.5232-5241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilditch C M, Rogers L J, Bishop D H L. Physiochemical analysis of the hepatitis core antigen produced by a baculovirus expression vector. J Gen Virol. 1990;71:2755–2759. doi: 10.1099/0022-1317-71-11-2755. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch R C, Lavine J E, Chang L J, Varmus H E, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature (London) 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanford R E, Notvall L. Expression of hepatitis B virus core and precore antigens in insect cells and characterisation of a core-associated kinase activity. Virology. 1990;176:222–233. doi: 10.1016/0042-6822(90)90247-o. [DOI] [PubMed] [Google Scholar]
- 19.Lanford R E, Notvall L, Beames B. Nucleotide priming and reverse transcriptase activity of hepatitis B virus polymerase expressed in insect cells. J Virol. 1995;69:4431–4439. doi: 10.1128/jvi.69.7.4431-4439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanford R E, Notvall L, Lee H, Beames B. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J Virol. 1997;71:2996–3004. doi: 10.1128/jvi.71.4.2996-3004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loeb D D, Tian R. Transfer of the minus strand of DNA during hepadnavirus replication is not invariant but prefers a specific location. J Virol. 1995;69:6886–6891. doi: 10.1128/jvi.69.11.6886-6891.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machida A, Ohnuma H, Tsuda F, Yoshikawa A, Hoshi Y, Tanaka T, Kishimoto S, Akahane Y, Miyakawa Y, Mayumi M. Phosphorylation in the carboxyl-terminal domain of the capsid protein of hepatitis B virus: evaluation with a monoclonal antibody. J Virol. 1991;65:6024–6030. doi: 10.1128/jvi.65.11.6024-6030.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller R, Marion P, Robinson W. Hepatitis B viral DNA-RNA hybrid molecules in particles from infected liver are converted to viral DNA molecules during an endogenous DNA polymerase reaction. Virology. 1984;139:64–72. doi: 10.1016/0042-6822(84)90330-1. [DOI] [PubMed] [Google Scholar]
- 24.Molnar-Kimber K L, Summers J, Taylor J M, Mason W S. Protein covalently bound to minus-strand DNA intermediates of duck hepatitis B virus. J Virol. 1983;45:165–172. doi: 10.1128/jvi.45.1.165-172.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nassal M, Rieger A. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J Virol. 1996;70:2764–2773. doi: 10.1128/jvi.70.5.2764-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberhaus S M, Newbold J E. Detection of DNA polymerase activities associated with purified duck hepatitis B virus core particles by using an activity gel assay. J Virol. 1993;67:6558–6566. doi: 10.1128/jvi.67.11.6558-6566.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. New York, N.Y: Oxford University Press; 1994. [Google Scholar]
- 29.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radziwill G, Zentgraf H, Schaller H, Bosch V. The duck hepatitis B virus DNA polymerase is tightly associated with the viral core structure and unable to switch to an exogenous template. Virology. 1988;163:123–132. doi: 10.1016/0042-6822(88)90239-5. [DOI] [PubMed] [Google Scholar]
- 31.Rieger A, Nassal M. Specific hepatitis B virus minus-strand DNA synthesis requires only the 5′ encapsidation signal and the 3′-proximal direct repeat DR1*. J Virol. 1996;70:585–589. doi: 10.1128/jvi.70.1.585-589.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodgers D W, Gamblin S J, Harris B A, Ray S, Culp J S, Hellmig B, Woolf D J, Debouck C, Harrison S C. The structure of the unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlicht H J, Bartenschlager R, Schaller H. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J Virol. 1989;63:2995–3000. doi: 10.1128/jvi.63.7.2995-3000.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlicht H J, Radziwill G, Schaller H. Synthesis and encapsidation of duck hepatitis B virus reverse transcriptase do not require formation of core-polymerase fusion proteins. Cell. 1989;56:85–92. doi: 10.1016/0092-8674(89)90986-0. [DOI] [PubMed] [Google Scholar]
- 35.Seeger C, Summers J, Mason W S. Viral DNA synthesis. Curr Top Microbiol Immunol. 1991;168:41–59. doi: 10.1007/978-3-642-76015-0_3. [DOI] [PubMed] [Google Scholar]
- 36.Seifer, M., and D. N. Standring. Unpublished observations.
- 37.Seifer M, Standring D N. Recombinant human hepatitis B virus reverse transcriptase is active in the absence of the viral nucleocapsid or the replication origin, DR1. J Virol. 1993;67:4513–4520. doi: 10.1128/jvi.67.8.4513-4520.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seifer M, Standring D N. Ribonucleoprotein complex formation by the human hepatitis B virus polymerase. Intervirology. 1995;38:295–303. doi: 10.1159/000150454. [DOI] [PubMed] [Google Scholar]
- 39.Sells M A, Chen M-L, Acs G. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staprans S, Loeb D D, Ganem D. Mutations affecting hepadnaviral plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J Virol. 1991;65:1255–1262. doi: 10.1128/jvi.65.3.1255-1262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi K, Kishimoto S, Ohori K, Yoshizawa H, Machida A, Ohnuma H, Tsuda F, Munekata E, Miyakawa Y, Mayumi M. Molecular heterogeneity of e antigen polypeptides in sera from carriers of hepatitis B virus. J Immunol. 1991;147:3156–3160. [PubMed] [Google Scholar]
- 43.Takahashi K, Machida A, Funatsu G, Nomura M, Usuda S, Aoyagi S, Tachibana K, Miyakawa Y, Mayumi M. Immunochemical structure of hepatitis B antigen in the serum. J Immunol. 1983;130:2903–2907. [PubMed] [Google Scholar]
- 44.Tavis J E, Ganem D. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J Virol. 1996;70:5741–5750. doi: 10.1128/jvi.70.9.5741-5750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tavis J E, Ganem D. Expression of functional hepatitis B virus polymerase in yeast reveals it to be the sole viral protein required for correct initiation of reverse transcription. Proc Natl Acad Sci USA. 1993;90:4107–4111. doi: 10.1073/pnas.90.9.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tavis J E, Ganem D. RNA sequences controlling the initiation and transfer of duck hepatitis B virus minus-strand DNA. J Virol. 1995;69:4283–4291. doi: 10.1128/jvi.69.7.4283-4291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavis J E, Perri S, Ganem D E. Hepadnaviral reverse transcription initiates within a stem-loop of the RNA packaging signal and employs a novel strand transfer. J Virol. 1994;68:3536–3543. doi: 10.1128/jvi.68.6.3536-3543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valenzuela, P. Personal communication.
- 49.Wang G H, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang G H, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 51.Wu T-T, Condreay L D, Coates L, Aldrich C, Mason W. Evidence that less-than-full-length pol gene products are functional in hepadnavirus DNA synthesis. J Virol. 1991;65:2155–2163. doi: 10.1128/jvi.65.5.2155-2163.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu M, Summers J. A domain of the hepadnavirus capsid protein is specifically required for DNA maturation and virus assembly. J Virol. 1991;65:2511–2517. doi: 10.1128/jvi.65.5.2511-2517.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou S, Standring D N. Production of hepatitis B virus nucleocapsidlike core particles in Xenopus oocytes: assembly occurs mainly in the cytoplasm and does not require the nucleus. J Virol. 1991;65:5457–5464. doi: 10.1128/jvi.65.10.5457-5464.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]