Abstract
We previously reported the development of an in vitro adeno-associated virus (AAV) DNA replication system. The system required one of the p5 Rep proteins encoded by AAV (either Rep78 or Rep68) and a crude adenovirus (Ad)-infected HeLa cell cytoplasmic extract to catalyze origin of replication-dependent AAV DNA replication. However, in addition to fully permissive DNA replication, which occurs in the presence of Ad, AAV is also capable of partially permissive DNA replication in the absence of the helper virus in cells that have been treated with genotoxic agents. Limited DNA replication also occurs in the absence of Ad during the process of establishing a latent infection. In an attempt to isolate uninfected extracts that would support AAV DNA replication, we discovered that HeLa cell extracts grown to high density can occasionally display as much in vitro replication activity as Ad-infected extracts. This finding confirmed previous genetic analyses which suggested that no Ad-encoded proteins were absolutely essential for AAV DNA replication and that the uninfected extracts should be useful for studying the differences between helper-dependent and helper-independent AAV DNA replication. Using specific chemical inhibitors and monoclonal antibodies, as well as the fractionation of uninfected HeLa extracts, we identified several of the cellular enzymes involved in AAV DNA replication. They were the single-stranded DNA binding protein, replication protein A (RFA), the 3′ primer binding complex, replication factor C (RFC), and proliferating cell nuclear antigen (PCNA). Consistent with the current model for AAV DNA replication, which requires only leading-strand DNA synthesis, we found no requirement for DNA polymerase α-primase. AAV DNA replication could be reconstituted with purified Rep78, RPA, RFC, and PCNA and a phosphocellulose chromatography fraction (IIA) that contained DNA polymerase activity. As both RFC and PCNA are known to be accessory proteins for polymerase δ and ɛ, we attempted to reconstitute AAV DNA replication by substituting either purified polymerase δ or polymerase ɛ for fraction IIA. These attempts were unsuccessful and suggested that some novel cellular protein or modification was required for AAV DNA replication that had not been previously identified. Finally, we also further characterized the in vitro DNA replication assay and demonstrated by two-dimensional (2D) gel electrophoresis that all of the intermediates commonly seen in vivo are generated in the in vitro system. The only difference was an accumulation of single-stranded DNA in vivo that was not seen in vitro. The 2D data also suggested that although both Rep78 and Rep68 can generate dimeric intermediates in vitro, Rep68 is more efficient in processing dimers to monomer duplex DNA. Regardless of the Rep that was used in vitro, we found evidence of an interaction between the elongation complex and the terminal repeats. Nicking at the terminal repeats of a replicating molecule appeared to be inhibited until after elongation was complete.
The replication of adeno-associated virus (AAV) DNA is a unique and highly regulated event. The productive replication cycle of the virus requires coinfection with an unrelated helper virus (usually adenovirus [Ad] or herpes simplex virus) (1, 38). In the absence of the helper virus, the AAV genome undergoes only limited amplification, which results in the integration of a tandemly repeated provirus into a host chromosome (5, 23, 34). In addition, some transformed cell lines which have been treated with genotoxic agents (heat shock, hydroxyurea, UV light, or carcinogens) can become semipermissive for AAV DNA replication in the absence of helper virus (44, 66–68).
The molecular mechanism by which AAV DNA replication is regulated by an unrelated helper virus is not fully understood. In the case of Ad, five genetic regions, the E1A, E1B, E4, E2A, and VA regions (1, 38), are required for complete helper function. However, none of these genes appears to be directly involved in AAV DNA replication. Genetic analyses have suggested that the role of Ad coinfection is primarily to maximize the synthesis of AAV-encoded gene products and possibly cellular genes that are required for AAV DNA replication. Thus, the study of AAV DNA replication should provide a useful approach for studying mammalian DNA replication enzymes and their regulation.
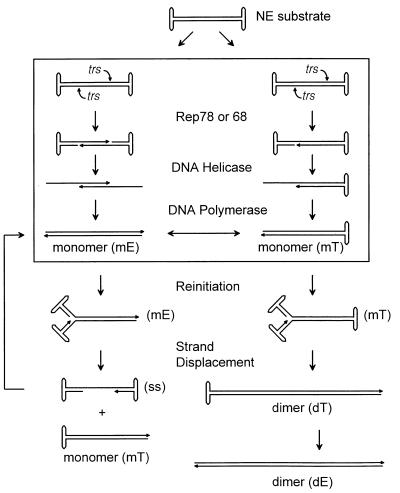
AAV is a single-stranded DNA (ssDNA) virus whose genome is approximately 4.7 kbp. Both ends of the genome contain identical palindromes of 125 nucleotides that can fold back into a T-shaped hairpin structure (Fig. 1). The genome replicates by a self-priming strand displacement mechanism (2, 16, 17, 28, 29, 43, 49). Replication is initiated from the 3′ hairpin primer of the single-stranded input genome to generate a linear duplex molecule in which one of the ends is covalently joined (Fig. 1, mT). To replicate the covalently closed terminal sequence, the termini are nicked on one of the two strands and the newly exposed 3′-OH primer that is generated by the nick is used to repair the terminal sequence. This process, called terminal resolution or hairpin transfer, is a unique mechanism used by the parvovirus family to maintain the integrity of the terminal sequences (Fig. 1, box). The linear duplex end is then unwound to reform a terminal hairpin, thus providing a 3′-OH primer for strand displacement synthesis (Fig. 1, reinitiation step). Elongation from the hairpin primer then generates a single-stranded genome (which is presumably packaged) and a new replicative-form (RF) molecule, which again can undergo terminal resolution.
FIG. 1.
Mechanism of AAV DNA replication. The diagram at the top illustrates a working model for the formation of replicative AAV DNA intermediates when NE DNA is used as a template for replication in vitro. Designations mE, mT, dE, dT, and ss stand for monomer extended, monomer turnaround, dimer extended, dimer turnaround, and single-stranded DNA, respectively. The series of reactions on the left depict steps involved in the generation of monomer duplex and single-stranded DNA species, and those on the right depict the steps involved in the formation of dimer duplex DNA. The boxed region illustrates the steps in terminal resolution on one or both ends of NE DNA. In contrast to the in vitro reaction with NE substrate, in a normal virus infection a single-stranded input DNA molecule (ss) is converted to a monomer turnaround (mT) form, which is then converted via terminal resolution to a monomer extended form (mE). See text for further details.
In previous work, we showed that either of the two virus-encoded proteins, Rep68 or Rep78, is capable of catalyzing the site-specific and strand-specific endonucleolytic cleavage event at the terminal resolution site (trs) within the terminal repeat (TR) (Fig. 1) (21, 48). Rep binds to a bipartite element that consists of a 22-bp Rep binding element (6, 7, 32, 33, 42, 47, 64) within the stem of the hairpinned TR and a smaller five-base motif within the cross arms of the T-shaped structure (42). It then cleaves a specific sequence at the trs and becomes covalently attached to the 5′ end of the cut site via a tyrosine phosphate linkage (46, 47). Cleavage at the trs is much more efficient on hairpinned substrates than on linear ones (6, 47). Unwinding of the TR, possibly by the Rep-associated DNA helicase activity (21) and repair of the TR with cellular DNA polymerases, completes the process of terminal resolution (48) (Fig. 1, box).
More recently, we have developed a cell-free assay for efficient AAV DNA replication (39) that mimics AAV DNA replication in vivo. The in vitro system required the presence of one of the large Rep proteins encoded by AAV (Rep68 or Rep78), the AAV TRs, which are the origins for DNA replication, and a cytoplasmic extract from Ad-infected HeLa cells. The substrate used in this assay was a linear AAV DNA molecule in which both ends were covalently joined, no-end (NE) DNA (Fig. 1). As expected, uninfected HeLa cell extracts were significantly less effective in supporting AAV DNA replication. The defect was found to be either at the reinitiation step or in elongation (Fig. 1). Ward and Berns (63), using linear AAV substrates, subsequently demonstrated that uninfected extracts were defective in the elongation step. Replication assays using circular plasmid substrates containing AAV genomes have also been explored (18, 19). These require both excision and replication of the AAV sequences.
The study of in vitro simian virus 40 (SV40) DNA replication (a circular double-stranded DNA virus) has led to the identification of several cellular replication proteins. SV40 DNA replication has been reconstituted with highly purified SV40-encoded T antigen and 10 cellular factors (see reference 60 and references therein). They are DNA polymerase α (pol α)-primase and DNA pol δ, replication protein A (RPA), replication factor C (RFC), proliferating cell nuclear antigen (PCNA), topoisomerase (topo) I and topo II, DNA ligase I, RNase H, and maturation factor I (MF-I). In view of the mechanism of AAV DNA replication, only a subset of the cellular factors needed for SV40 DNA replication, namely, those involved in leading-strand synthesis (PCNA, RPA, RFC, and pol δ), should be necessary for AAV DNA replication. As yet, however, nothing is known about the cellular factors required for AAV DNA replication. Additionally, it is not known why uninfected cellular extracts are incapable of supporting efficient AAV DNA synthesis either in vitro or in vivo.
In this report, we demonstrate that uninfected cell extracts that are capable of sustaining in vitro AAV DNA replication at levels similar to those seen with Ad-infected extracts can be prepared. Such extracts may prove useful in analyzing the differences between uninfected and Ad-infected cells that control the level of AAV DNA synthesis. We also use the in vitro replication assay to fractionate uninfected HeLa cell extracts and to identify some of the cellular replication proteins involved in AAV DNA synthesis. As mentioned earlier, we expected that all of the enzymes identified in the SV40 DNA replication system which were involved in the generation or processing of lagging-strand Okazaki fragments (pol α, RNase H, ligase I, and MF-I) would be unnecessary for AAV DNA replication. Indeed, our results confirmed that the purified cellular factors PCNA, RPA, and RFC and a partially purified fraction containing cellular DNA polymerases are necessary for reconstituting AAV DNA replication. However, we are unable to reconstitute activity in assays using any combination of pol α, δ, and ɛ, suggesting that an additional cellular factor, not been previously identified in the SV40 system, is necessary for AAV DNA synthesis. Finally, using two-dimensional (2D) gels, we show that the products of the in vitro assay are consistent with the current model for AAV DNA replication, in which AAV DNA undergoes only leading-strand DNA synthesis and no RNA priming is involved.
MATERIALS AND METHODS
Substrates, antibodies, and chromatography materials.
NE DNA substrate was prepared from psub201 plasmid DNA as previously described (48). Polyclonal antibody against PCNA (40) and monoclonal antibodies against DNA pol α (SJK132-20) (53), RPA (recognizing the 34-kDa subunit), and RFC (monoclonal antibody 19, recognizing the 140-kDa subunit) (3) have been previously described. All antibodies were purified by protein A-Sepharose chromatography prior to use. Monoclonal antibody against large T antigen (pAb419) (15) was a generous gift from Peter Tegtmeyer (State University of New York at Stony Brook). Aphidicolin, N2-(p-n-butylphenyl)-2′-deoxyguanosine 5′-triphosphate (BuPdGTP), and 2-(p-n-butylanilino)-2′-deoxyadenosine 5′-triphosphate (BuAdATP) were generous gifts from George E. Wright. Ribo- and deoxyribonucleoside triphosphates were purchased from Pharmacia or Sigma. They were dissolved in water and neutralized with NaOH. Radioactive nucleotides were purchased from ICN. Creatine phosphate, creatine phosphokinase, and dithiothreitol (DTT) were purchased from Sigma and dissolved in water. Restriction and DNA-modifying enzymes and λ bacteriophage DNA were from New England Biolabs. DEAE-cellulose and phosphocellulose were purchased from Whatman; Mono-S and Mono-Q FPLC columns were purchased from Pharmacia, and ssDNA-agarose was purchased from Life Technologies.
Preparation of Ad-infected and uninfected HeLa cell crude extracts.
Ad-infected extracts were prepared as previously described (39). Low-density uninfected HeLa cell [HeLa(S)] extracts and Ad-infected HeLa cell [Ad(S)] extracts were prepared as previously described from cells that had been grown to a density of 5 × 105 cell per ml. High-density uninfected HeLa cell suspension [HeLa(H)] extracts were grown to a density of 9 × 105 to 10 × 105 cells per ml, diluted 1:1 with fresh medium, and incubated for another 16 to 20 h. Cells were then counted to determine if the cell density was again above 9 × 105 cells per ml and harvested by low-speed centrifugation. Cells were broken and extracted with 0.2 M NaCl as described previously (39), and the extracts were dialyzed against 20 mM Tris (pH 7.5)–5 mM NaCl–10% glycerol–0.1 mM EDTA–1 mM DTT. The final protein concentration of the extracts was approximately 30 mg/ml, and the extracts were stored at −80°C. Protein was measured with the Bradford reagent (Bio-Rad), using pooled bovine gamma globulin as the standard.
Rep protein.
Baculovirus expression vectors and the preparation of crude baculovirus extracts containing Rep78 or Rep68 have been described previously (39). The crude extracts contained approximately 104 U of replication activity per mg of protein (see below for unit definition) and 6 to 7 mg of protein per ml. Homogeneously pure Rep68 (Mono-Q fraction) was prepared as described previously (39). It had a specific activity of 106 U/mg and was approximately 100-fold purified relative to the crude preparation. Partially purified Rep78 was prepared from baculovirus-infected SF9 crude extracts as follows. Four milliliters of the crude extract was diluted with buffer A (25 mM Tris-HCl [pH 7.5], 0.1 mM EDTA, 0.05% Nonidet P-40, 10% glycerol, 1 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, 0.5 μg of leupeptin per ml, 0.7 μg of pepstatin per ml), adjusted to a conductivity of 0.2 M NaCl, and then applied to a phosphocellulose (P-cell) column that had been equilibrated with buffer A containing 0.2 M NaCl. The column was washed with 5 column volumes of 0.2 M NaCl buffer A and eluted with 10 column volumes of an ascending linear gradient (0.2 to 0.8 M NaCl) in buffer A. The active P-cell fractions were identified by using the in vitro AAV DNA replication assay. Only one major peak was found and pooled, and the specific activity of the P-cell fraction was 9.3 × 104 U/mg. The pooled Rep78 fractions were diluted with buffer A to adjust the conductivity to the equivalent of 0.1 M NaCl in buffer A and then applied to an ssDNA-agarose column that had been equilibrated with the loading buffer. The column was washed with loading buffer and eluted with 10 column volumes of a linear gradient (0.1 to 0.8 M NaCl) in buffer A containing 2 mM MgCl2 and 20% glycerol. Active fractions were identified, pooled, and dialyzed against buffer A containing 2 mM MgCl2 and 20% glycerol. Rep78 was enriched fivefold after phosphocellulose column and another twofold after ssDNA-agarose chromatography. The final specific activity in the DNA replication assay was approximately 2 × 105 U/ml at a total protein concentration of 275 μg/ml. The ssDNA fraction was stable at −80°C for at least 6 months.
2D agarose gel electrophoresis.
In vitro synthesized AAV DNA replication products were treated with proteinase K and run in the first dimension on a 0.8% neutral agarose gel. The gel was then soaked for 1 h in alkaline running buffer (30 mM NaOH, 1 mM EDTA), turned 90°, and run in the second dimension in alkaline running buffer as described (31). The gel then was dried and exposed to X-ray film −70°C with an intensifying screen.
Fractionation of uninfected HeLa cell extracts.
The preparation of phosphocellulose fractions I, II, IIA, IIB, IIC, and IID from HeLa(H) extracts was performed as described previously (54, 56, 57). Briefly, the crude uninfected HeLa cell S100 extract was adjusted to the conductivity of buffer B (25 mM Tris HCl [pH 7.5], 1 mM EDTA, 0.01% Nonidet P-40, 10% glycerol, 0.1 mM phenylmethylsulfonyl chloride) containing 0.2 M NaCl and loaded onto a phosphocellulose column that had been equilibrated with buffer A containing 0.2 M NaCl. The column was washed with the same buffer, and the protein pool that came off the column was collected (fraction I). The column was then eluted with buffer A containing 1 M NaCl (fraction II). Alternatively, the column was eluted discontinuously with buffer A containing 0.33, 0.4, 0.66, and 1 M NaCl to produce fractions IIA, IIB, IIC, and IID, respectively. Each fraction was dialyzed against buffer A containing 25 mM NaCl and stored at −80°C.
Mammalian replication enzymes.
Pol δ (6.4 U/ml) was purified from calf thymus through five steps as described previously (24, 56) and assayed using by poly(dA-dT) substrate as described by Syvaoja et al. (51). Purified human pol α (3.1 U/ml) was prepared by antibody affinity chromatography as described (37, 58) and assayed by using activated salmon sperm DNA as a template. Purified pol ɛ (9.01 U/ml) was a kind gift from Stuart Linn. It was purified and assayed as described elsewhere (50, 51), using poly(dA)4000 primed with dT16 (both purchased from Midland Certified Reagent Co.). One unit of DNA polymerase catalyzes the incorporation of 1 nmol of nucleotide per h. Purified RPA (0.54 mg/ml) was prepared as described by Fairman and Stillman (12). Purified RFC (1.8 mg/ml) was the second phosphocellulose fraction (fraction IV) described previously (58). PCNA (0.8 mg/ml) was purified from a bacterial expression vector as described elsewhere (13). Topo I (27) and topo II (45) were purified as described elsewhere.
AAV in vitro DNA replication assay.
The standard AAV DNA replication assay (39) contained, in 30 μl, 30 mM HEPES (pH 7.5), 7 mM MgCl2, 0.5 mM DTT, 100 μM each dATP, dGTP, dCTP, and dTTP, 25 μCi of [α-32P]dATP (3 μCi/pmol), 4 mM ATP, 40 mM creatine phosphate, 1 μg of creatine phosphokinase, 255 μg of Ad-infected or uninfected HeLa S100 extract, 0.1 μg of NE substrate DNA (0.032 pmol of AAV DNA or 300 pmol of nucleotide), and 1 to 80 U of Rep78 or Rep68 baculovirus crude extract, the Mono-Q or ssDNA Rep68 fraction, or the ssDNA Rep78 fraction. A unit of Rep activity was arbitrarily defined as an amount of Rep protein that catalyzed the incorporation of 1 pmol of radioactive deoxynucleoside monophosphate into DpnI-resistant monomer or dimer duplex AAV DNA in 2 h at 37°C. Following incubation, the reaction mixture was adjusted to 70 μl containing 0.3% sodium dodecyl sulfate, 0.7 mg of proteinase K per ml, and 17 mM EDTA. Proteinase K digestion was at 37°C for 1 h. The products were then extracted with phenol and chloroform and precipitated with ethanol. The ethanol precipitate was dissolved in 18 μl of water and where indicated digested with DpnI for 2 h at 37°C. The products (or a portion of them) were separated on either 0.8 or 1% agarose gels by electrophoresis for 4 h at 6 V/cm. The radioactivity in monomer and dimer RF products was counted in dried gels by a scanning gas flow (AMBIS) counter. X-ray film was exposed for 5 min to 16 h without a screen at room temperature. Additionally, the DE-81 filter assay was used to measure total incorporation into acid-insoluble product as described previously (39).
RESULTS
Replication with uninfected and Ad-infected HeLa cell extracts.
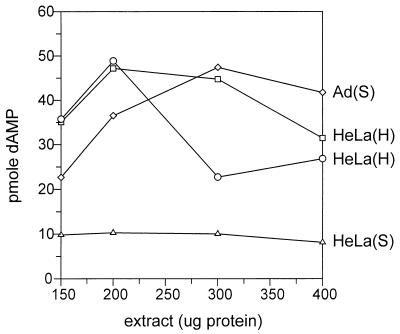
We previously reported that Ad-infected HeLa cell extracts were more efficient in supporting AAV DNA replication than uninfected extracts. As mentioned earlier, this appeared to be due to a problem either in the reinitiation step (Fig. 1) or in strand displacement and elongation (39). More recently, Ward and Berns (63) demonstrated that the defect in uninfected extracts was at the level of elongation. However, genetic analyses by a number of laboratories had suggested that no Ad-encoded proteins were directly involved in AAV DNA synthesis (38). Ad coinfection is known to be essential for the expression of the AAV-encoded Rep proteins (which are required for AAV DNA synthesis), but the fact that addition of exogenous Rep protein to uninfected extracts was not sufficient to support AAV DNA synthesis suggested that Ad was either inducing or inhibiting the expression of some critical cellular or Ad-encoded factor. In this respect, several reports have demonstrated that transformed tissue culture cells that have been subjected to stress (for example, UV irradiation) became semipermissive for AAV DNA replication (44, 66–68). As a first step toward defining the biochemical differences between permissive and semipermissive conditions for AAV replication, we tested several alternative methods of preparing HeLa cell extracts to see if a permissive HeLa cell extract could be made in the absence of Ad infection. As shown in Fig. 2, we found that HeLa(H) extracts were significantly better than HeLa(S) extracts and almost as efficient as Ad(S) extracts. Although as yet we do not understand the mechanism of this observation, we have used the modified HeLa(H) extracts for many of the experiments described in this report. Elsewhere, we will compare these extracts with Ad(S) extracts to define factors necessary for permissive AAV DNA replication (33a).
FIG. 2.
Abilities of Ad(S), HeLa(S), and HeLa(H) extracts to support AAV DNA replication in the standard in vitro AAV DNA replication assay containing partially purified Rep78 (ssDNA fraction; 0.25 μg). Two different high-density extracts are shown. The amount of incorporation into DNA product was measured by DE-81 filter binding assay.
Inhibition of AAV DNA synthesis by monoclonal antibodies to specific cellular replication enzymes.
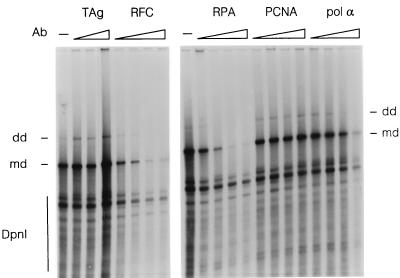
To identify some of the cellular factors in the uninfected extract that are necessary for AAV DNA replication, we used monoclonal antibodies that had been shown to inhibit the activity of RPA, RFC, or pol α-primase. These antibodies were added to the standard in vitro AAV DNA replication assay which contained, in addition to the linear AAV NE DNA substrate, the optimal amount of crude uninfected HeLa cell extract and crude baculovirus extract containing Rep78. The intensities of the monomer and dimer duplex molecules that were resistant to DpnI digestion were used to determine the level of AAV DNA synthesis (Fig. 3). In the reactions that were incubated with antibodies against either RFC or RPA, significant inhibition of AAV DNA replication was observed. This implied that both RFC and RPA are cellular factors that are essential for AAV DNA replication in vitro. As expected, a monoclonal antibody against SV40 T antigen did not inhibit AAV DNA synthesis. In addition, a polyclonal antibody against PCNA, which was known not to inhibit SV40 DNA replication in vitro, did not inhibit AAV DNA synthesis either. Finally, the monoclonal antibody against pol α seemed to have an inhibitory effect only at the highest level of antibody tested. This effect could not be reproduced in two additional antibody titrations (data not shown), and we concluded that pol α was not necessary for AAV DNA synthesis. We also note that inhibition by any of the antibodies tested does not conclusively demonstrate that the target enzyme is directly involved in AAV DNA synthesis. Conceivably, inhibition might be due to the presence of another protein that is present in a complex with the target protein.
FIG. 3.
Effects of neutralizing monoclonal antibodies on AAV DNA synthesis in vitro. Standard DNA replication reactions mixtures containing uninfected HeLa cell crude extract (255 μg) and crude baculovirus-expressed Rep78 (8 μg) were incubated with monoclonal antibodies against T antigen (TAg; lanes 2 to 4; 0.4, 0.8, and 2.4 μg), RFC (lanes 5 to 8; 0.5, 1.0, 2.0, and 3.0 μg), RPA (lanes 10 to 13; 0.75, 1.5, 3.0, and 4.5 μg), PCNA (lanes 14 to 17; 0.4, 0.8, 1.6, and 2.4 μg), or DNA pol α (lanes 18 to 21; 0.4, 0.8, 1.6, and 2.4 μg) for 2 h at 37°C. DNA products were digested with DpnI and analyzed on a 0.8% agarose gel. md and dd indicate monomer duplex and dimer duplex DNA species that are resistant to DpnI digestion, respectively. DNA products that are sensitive to DpnI digestion are marked with a line on the left. Lanes 1 and 9 represent reaction mixtures incubated without antibody.
Inhibition of AAV DNA synthesis by specific chemical inhibitors.
A variety of chemical inhibitors have been shown to have differential effects on the known cellular DNA polymerases. These were tested in the standard in vitro AAV DNA replication assay to determine if one or another of the DNA polymerases was more likely to be involved in AAV DNA synthesis (Table 1). Aphidicolin, which is a potent inhibitor of pol α, pol δ, and pol ɛ (14, 20, 26) but is without effect on pol β and pol γ (20), completely abolished AAV DNA synthesis at a concentration of 10 μg/ml (Table 1). Dimethyl sulfoxide, the solvent used to dissolve aphidicolin, had no effect on the reaction at the same concentration (0.1%). This result suggested that one or more of the three cellular DNA polymerases α, δ, and ɛ, were primarily responsible for AAV DNA replication in vitro. The possibility of the involvement of DNA pol β and γ without the presence of either DNA polymerase α, δ, or ɛ was thus ruled out. The base analog 2′,3′-dideoxythymidine 5′-triphosphate, a compound that inhibits pol β and pol γ with at least an order of magnitude greater potency than it does pol α or pol δ (11, 61, 62), inhibited AAV DNA synthesis in vitro with a potency reflecting that which it displays for pol α and pol δ, rather than pol β and pol γ. This finding also suggested that pol β and pol γ were not required for AAV DNA synthesis.
TABLE 1.
Effects of specific inhibitors on AAV DNA replication in vitro
| Inhibitor | Concn | Relative activity (%) |
|---|---|---|
| None | 100 | |
| Aphidicolina | 10 μg/ml | 0 |
| ddTTPa | 500 μM | 42 |
| BuPdGTPb | 40 nM | 100 |
| 25 μM | 50 | |
| BuAdATPb | 20 nM | 100 |
| 120 μM | 50 | |
| Dimethyl sulfoxidea | 0.1% | 79 |
| 10% | 0 |
The level of AAV DNA synthesis was measured by scanning the intensity of monomer and dimer duplex DNA that were resistant to DpnI digestion by a gas flow counter to calculate the amount of incorporation of [α-32P]dAMP into NE DNA substrate in the standard in vitro AAV DNA replication assay using uninfected crude HeLa cell extract (255 μg/ml), crude baculovirus Rep78 (8 μg), and the indicated inhibitors in a 2-h reaction. The percent activity remaining was then calculated by comparison to the reaction that contained no inhibitor.
The level of [α-32P]dAMP incorporation in a standard (2-h) in vitro replication assay using uninfected crude HeLa cell extract and crude baculovirus Rep78 as described above was measured by DE-81 filter assay in the presence of the following inhibitor concentrations: 0.04, 0.4, 1, 10, 100, and 200 μM for BuPdGTP and 0.02, 0.2, 2, 20, 70, 100, and 200 μM for BuAdATP. The percent activity remaining was then calculated by comparison to a reaction that contained no inhibitor, and the concentration of inhibitor required to achieve 50% inhibition was calculated. Also shown, is the percent remaining activity at the lowest concentration of inhibitor used.
To distinguish between a requirement for pol α, pol δ, or pol ɛ, we used the base analogs BuPdGTP (65) and BuAdATP (22) in the in vitro replication reaction (Table 1). Both BuPdGTP and BuAdATP are known to differentially inhibit pol α and pol δ (or ɛ), inhibiting pol α with several orders of magnitude greater potency than pol δ or ɛ (4, 9, 10, 22, 25, 65). A titration of BuPdGTP and BuAdATP was performed in the standard AAV DNA synthesis reaction, and the level of synthesis was measured by the DE-81 filter binding assay. Fifty percent inhibition of AAV DNA replication was seen with 25 μM BuPdGTP and 120 μM BuAdATP (Table 1). No inhibition was seen at concentrations of BuPdGTP (40 nM) and BuAdATP (20 nM) that were expected to inhibit DNA pol α. The effect of the inhibitors BuPdGTP and BuAdATP on the complete AAV DNA synthesis reaction was similar to the effect seen previously on the AAV terminal resolution reaction in which the termini of AAV are repaired following site-specific nicking by the Rep protein (46). This result indicated that the requirements for a DNA polymerase in the terminal resolution reaction and the subsequent strand displacement reaction were probably the same. In both cases, the inhibition data were consistent with either pol δ or pol ɛ being primarily responsible for AAV DNA synthesis in vitro.
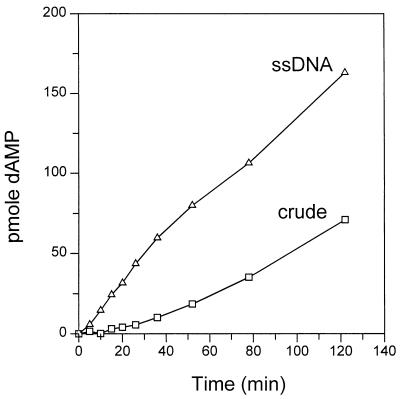
Time course of DNA synthesis.
We had previously reported that there was a lag in the onset of DNA synthesis in our in vitro reaction and suggested that the lag may be due to a requirement for assembling a replication complex (39). These studies had been done with a crude baculovirus extract in which Rep78 had been overexpressed. When homogeneously pure Rep68 and partially purified Rep78 became available, we repeated these experiments. As shown in Fig. 4, we found that the delay in onset of DNA synthesis was due to an inhibitor present in the baculovirus crude extract that had been the source of Rep protein. When the ssDNA-cellulose fraction of Rep78 (Fig. 4) or the homogeneously pure Rep68 (not shown) was used in a time course experiment, no lag in DNA synthesis was seen.
FIG. 4.
Time course of in vitro AAV DNA replication comparing purified and crude Rep78. The standard DNA replication assay contained 200 μg of Ad-infected HeLa cell extract and either crude baculovirus extract containing Rep78 (8 μg) or partially purified Rep78 (ssDNA fraction; 0.4 μg). Shown is the amount of dAMP incorporated per 30-μl standard reaction as determined from counting of the DpnI-resistant monomer and dimer RF species at each time point.
Reconstitution of AAV DNA synthesis in vitro with uninfected HeLa cell fractions.
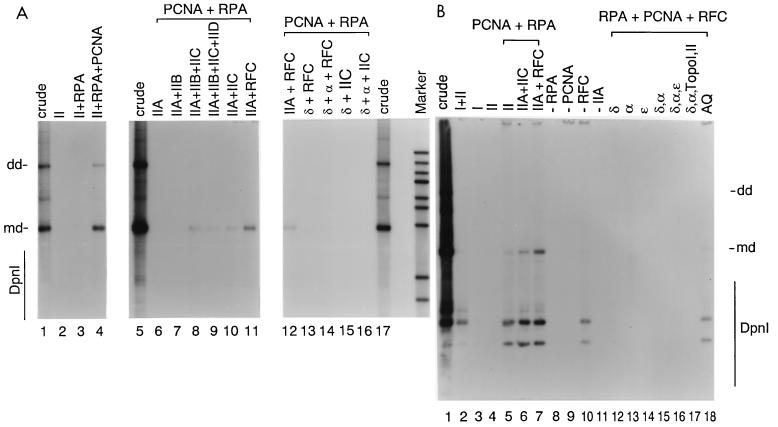
To further study the cellular factors that contribute to AAV in vitro DNA replication, the crude uninfected HeLa extract was fractionated by phosphocellulose chromatography essentially as described by Stillman and colleagues (54, 56, 57) and illustrated in Fig. 5. The phosphocellulose column was eluted discontinuously with two NaCl concentrations to produce a 0.2 M fraction (fraction I) and a 1.0 M fraction (fraction II). Fraction I had previously been shown to contain the cellular factors RPA and PCNA (12, 41). When fraction II alone or fraction I alone was used in the standard DNA replication assay, no AAV DNA synthesis could be detected (Fig. 6A, lane 2; Fig. 6B, lane 3 and 4). When fraction II was supplemented with purified RPA and PCNA, the level of DpnI-resistant monomer and dimer product synthesized was up to 40% of that seen with the starting crude extract (Fig. 6A, lane 4; Fig. 6B, lane 5). Omission of PCNA abolished DNA replication (Fig. 6A, lane 3; Fig. 6B, lane 9), suggesting that AAV DNA synthesis is PCNA dependent. Omission of RPA also eliminated synthesis (Fig. 6B, lane 8).
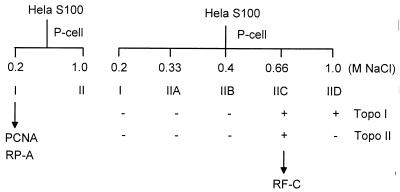
FIG. 5.
Scheme for fractionation of crude uninfected HeLa cell extracts by phosphocellulose chromatography. The presence or absence of previously characterized replication factors in each fraction is indicated (57).
FIG. 6.
(A) Reconstitution of AAV DNA replication in vitro using fractionated HeLa cell extracts. Standard DNA replication reaction mixtures (15 μl) contained the following concentrations of the indicated P-cell fractions and purified DNA replication factors: 50 μg of human RPA per ml, 8 μg of PCNA per ml, 72 μg of RFC (fraction IV per ml, 5 mg of fraction II per ml, 0.62 mg of fraction IIA per ml, 0.19 mg of fraction IIB per ml, 0.31 mg of fraction IIC per ml, and 0.11 mg of fraction IID per ml. Where indicated, the reaction mixtures contained, in 15 μl, 0.003 U of pol α, 0.02 U of pol δ, and 0.014 U of pol ɛ. Reaction products were processed and fractionated on 0.8% agarose gels as described in Materials and Methods. md and dd indicate monomer duplex and dimer duplex DNA species that are resistant to DpnI digestion, respectively. DNA products sensitive to DpnI digestion are denoted with a line at the left. Molecular weight markers are λ DNA molecules digested with BstEII. (B) Same as Fig. 6A except that fraction I was used at a final concentration of 5.3 mg/ml, topo I was used at 4 μg/ml, and topo II was used at 1.8 μg/ml. AQ, mono-Q fraction derived from P-cell fraction IIA.
To further investigate the nature of the protein factors present in fraction II, subfractions were prepared by elution of a phosphocellulose column with 0.33, 0.4, 0.66, and 1.0 M NaCl to produce fractions IIA, IIB, IIC, and IID, respectively (Fig. 5). Each subfraction was added to the standard AAV DNA replication reaction to see which combination of fractions could reconstitute activity in the presence of purified PCNA and RPA. Fraction IIA alone, which contained approximately 70% of the DNA polymerase activity as judged by incorporation into a poly(dA)-oligo(dT) template (data not shown), produced no detectable DNA synthesis (Fig. 6A, lane 6). When fraction IIA was supplemented with fraction IIC, approximately 10% of the full-length, DpnI-resistant, monomer or dimer DNA product that was seen with crude extract was obtained (Fig. 6A, lanes 8 to 10; Fig. 6B, lane 6). Omission of either fraction IIA (Fig. 6B, lane 11) or fraction IIC (Fig. 6A, lanes 6 and 7) eliminated DNA replication. Addition of fraction IIB alone (Fig. 6A, lane 8), fraction IID alone (data not shown), or both (Fig. 6A, lane 9) did not have a significant effect on replication. This result suggested that all the essential components that were present in fraction II were retained in fractions IIA and IIC.
RFC is known to be present primarily in fraction IIC (57). When fraction IIC was replaced by purified RFC, AAV DNA synthesis was stimulated (Fig. 6A, lane 11; Fig. 6B, lane 7), indicating that the major component of fraction IIC that is necessary for AAV DNA replication is RFC. We noted, however, that the level of DNA synthesis obtained with fraction IIA and purified RFC, RPA, and PCNA was only a portion of the activity seen with the starting crude extract. This suggested either that we had not achieved optimal concentrations of the components required for DNA synthesis or that in addition to the purified components RPA, RFC, and PCNA, there might be other factors in fractions I, IIB, IIC, and IID that were necessary for maximum DNA synthesis. It also is worth noting that although we compared only DpnI-resistant products generated in our reactions, we often saw a significant amount of DpnI-sensitive (i.e., incompletely replicated) product synthesis. The level of DpnI-sensitive product was a function of the particular extract used. Figures 6A and B illustrate two extracts that generated relatively low and high amounts of DpnI-sensitive products, respectively. When seen, the DpnI-sensitive products were absolutely dependent on the presence of Rep (39), RPA (Fig. 6B, lane 8), and PCNA (Fig. 6B, lane 9) but were not dependent on the presence of RFC (Fig. 6B, lane 10).
Because fraction IIA contained most of the DNA polymerase activity, we tried to replace fraction IIA with combinations of purified DNA pol α, δ, or ɛ in the presence of purified RPA, RFC, and PCNA. DNA pol δ alone (Fig. 6A, lane 13) or DNA pol α or ɛ alone (Fig. 6B, lanes 13 and 14) was unable to reconstitute DNA synthesis. Addition of pol α and pol δ together (Fig. 6A, lane 14; Fig. 6B, lane 15), as well as addition of all three DNA polymerases (α, δ, and ɛ) (Fig. 6B, lane 16) was also not sufficient to reconstitute activity. Finally, the addition of purified calf thymus topo I and II, which are known to be in fractions IIC and IID (Fig. 5), also did not reconstitute DNA synthesis (Fig. 6B, lane 17). All of the purified DNA polymerases used in these experiments, as well RPA, RFC, and PCNA, were active in a reconstituted SV40 DNA synthesis assay (pol α and pol δ) (36) or in a standard DNA polymerase assay using a poly(dA)-oligo(dT) substrate (pol ɛ) (data not shown). This result suggested that in addition to a DNA polymerase activity, some other factor(s) in fraction IIA was required for AAV DNA synthesis in vitro. The requirement for such a factor had not previously been seen in the studies of SV40 DNA replication.
Analysis of replication intermediates by 2D agarose gel electrophoresis.
To see if the products of the in vitro DNA replication assay were consistent with the intermediates predicted by the model for AAV DNA replication, we analyzed the products of the in vitro reaction by 2D gel electrophoresis. We would predict that the terminal sequence of AAV is either in the extended (or resolved) form (Fig. 1, mE or dE), in which the TR is linear, or in a turnaround (or unresolved) form (Fig. 1, mT or dT), in which the terminal sequence is covalently closed in a T-shaped hairpin structure. The extended and turnaround forms of different replication intermediates can be efficiently separated by 2D agarose gel electrophoresis (59) in which the first dimension is run under neutral conditions and the second dimension is run under alkaline conditions (Fig. 7 and 8). AAV DNA was synthesized by the incubation of NE DNA in the presence of uninfected HeLa cell crude extract and either baculovirus-expressed Rep78 (ssDNA-cellulose fraction) or homogeneously pure Rep68 in the standard reaction containing α-32P-deoxynucleoside triphosphates. The products of the reaction were treated with proteinase K, and then a portion of each reaction was fractionated in the first dimension on a 0.8% agarose gel under neutral conditions and in the second dimension under alkaline conditions (Fig. 7). We also examined the replicative intermediates generated in vivo in 293 cells following mixed infection with wild-type AAV and Ad (Fig. 8).
FIG. 7.
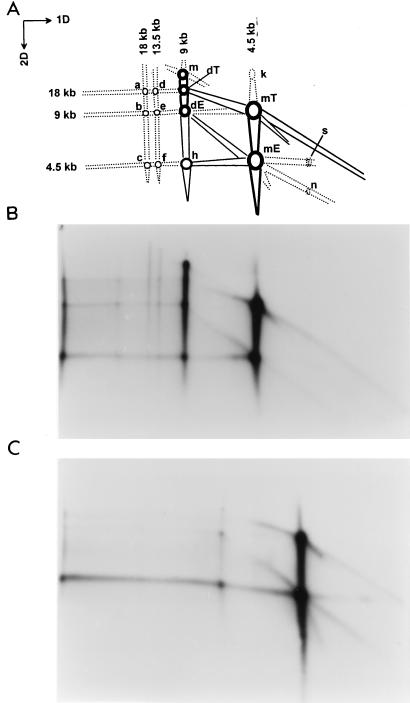
2D agarose gel analysis of in vitro-synthesized AAV DNA. Crude uninfected HeLa cell extracts were incubated with NE DNA and Rep78 (B; ssDNA-cellulose fraction) or Rep68 (C; ssDNA-cellulose fraction) in 30-μl reactions under standard DNA replication conditions. DpnI-digested DNA products (1/10 of each reaction) were fractionated on a 0.8% agarose gel under neutral conditions in the first dimension (1D) and alkaline conditions in the second dimension (2D). Panel A is a diagram of the RF species generated by Rep78, shown in panel B, and indicates relevant replicative DNA species (see the legend to Fig. 1 legend and text for a description). The relative molecular weights of each DNA species shown were derived from the migration pattern of BstEII-digested lambda DNA (not shown).
FIG. 8.
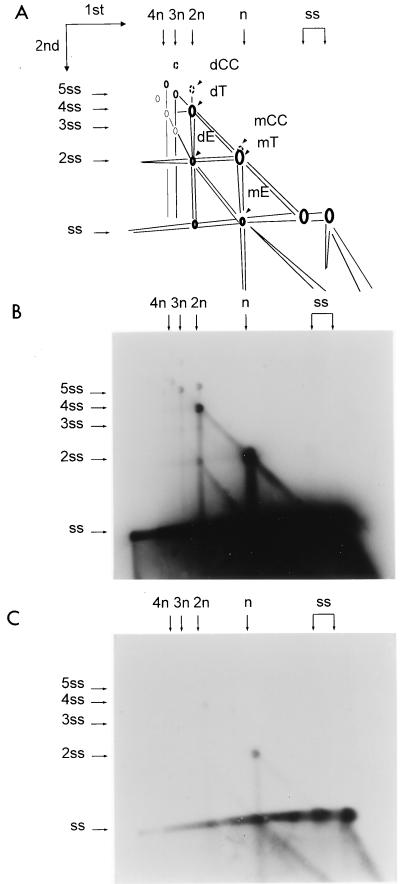
2D agarose gel analysis of DNA isolated from cells coinfected with Ad5 and AAV. 293 cells were infected with Ad5 and wild-type AAV at multiplicities of infection of 5 and 10, respectively. At 48 h postinfection, cultures were harvested and low-molecular-weight DNA was isolated by Hirt extraction. AAV DNA replicative forms were fractionated on a 0.8% agarose under neutral conditions in the first dimension and alkaline conditions in the second dimension. Panels B and C are 30-min and 2-day exposures of the same blot, respectively. Panel A is a schematic representation of the data. mE and dE represent linear monomer and dimer duplex DNA products with extended or open ends, respectively. mT and dT represent linear monomer and dimer duplex DNA products with a single covalently closed end or turn, respectively. cc denotes linear DNA products that have both ends covalently closed, and ss indicates single-stranded genomic DNA. The relative molecular weights of each DNA species shown were derived from the migration pattern of HindIII-digested lambda DNA (not shown). n indicates monomer size duplex DNA; ss indicates monomer size ssDNA.
(i) The double-stranded replication intermediates were the same in vitro and in vivo.
Both in vivo and in vitro, the replication intermediates separated into two dominant species in the first dimension. The sizes of these species were the expected sizes of monomer and dimer duplex linear DNA (Fig. 7 and 8, 4.5 kb [or n] and 9 kb [or 2n]). Two higher-molecular-weight species were also present in minor amounts, and their sizes were approximately those of trimer and tetramer duplex DNA. Under alkaline conditions, the monomer duplex DNA RF species was separated into two major forms: a 4.5-kb single-stranded monomer-size molecule which arose from monomer duplex molecules in which both ends of AAV were in the extended configuration (Fig. 7 and 8, mE), and a 9-kb ssDNA molecule which arose from monomer duplex DNA in which one of the termini was in the turnaround form (Fig. 7 and 8, mT). Similarly, dimer duplex DNA was separated in the second dimension into a 9-kb (or 2ss) extended dimer (dE, both ends of the dimer in the extended form) and an 18-kb turnaround dimer (dT, one end of the dimer in the covalently closed or turnaround form). A portion of dimer duplex DNA comigrated in the alkaline dimension with the 4.5-kb mE single-stranded monomer DNA molecules (Fig. 7A, h). These probably were derived from dimer duplex DNA that was nicked near the middle of molecule at the trs sites in one or both strands of the TR bridge. Another portion of dimer DNA (designated m in Fig. 7 and dCC in Fig. 8) migrated at a molecular weight higher than the 18-kb dT form and was present in significant amount. This was probably a circular tetrameric ssDNA molecule which arose from linear dimer duplex molecules in which both ends were covalently closed. A similar monomer species (form k in Fig. 7A and mCC in Fig. 8), whose structure should be identical to that NE DNA (Fig. 1), was also found. These structures could be formed if replication stalls at the TR and the nick is sealed by DNA ligase.
The only major difference seen between DNA synthesized in vivo and in vitro was the substantially higher level of ssDNA (Fig. 8, ss) seen in vivo. Most of the DNA in the cell was single stranded and migrated to the right of monomer duplex in the neutral dimension. This may reflect the accumulation of ssDNA in a packaged form in vivo, whereas in vitro packaging was not possible due to the absence of capsid protein.
(ii) Processing of dimer bridges may be coordinated with strand elongation.
Replication that initiates from one end of an mE molecule would generate DNA that migrates slower than monomeric DNA in the neutral dimension. In the alkaline dimension, such molecules would dissociate into a 4.5-kb single-stranded monomer molecule (the displaced strand, visible as a line of radioactivity between mE and h in Fig. 7A) and a growing strand which would have sizes between 4.5 and 9 kb (the line between mE and dE) (see also Fig. 9). Similarly, replication initiated from the extended end of mT molecules would generate replicative intermediates migrating between 9 and 18 kb in the alkaline dimension. If no terminal resolution occurred during strand displacement synthesis, the migration pattern of the intermediates would correspond to the line between mT and dT in Fig. 7A. The end product of such intermediates would be a dimer molecule that is covalently closed at the end where strand displacement synthesis originated. The other end would be in the extended configuration, and the former hairpinned palindrome of mT would be in the middle of the dimer molecule (the dimer bridge) (see also Fig. 9). We note that although the line of radioactivity between mT and dT is consistent with strand displacement synthesis that originates on mT molecules, it could also be the result of random, nonspecific nicking of dT molecules during or after their synthesis; there is no way to distinguish the relative contribution of these two mechanisms. The same is true for the line of radioactivity between mE and dE.
FIG. 9.
Diagram of potential Rep cuts at trs sites that might occur during strand displacement synthesis on monomer extended (mE) and monomer turnaround (mT) replicative forms. See text for discussion.
If a single nick occurred on the template strand after replication on an mT template went through the dimer bridge region (Fig. 9, cut i), it would generate a 4.5-kb strand and a strand between 9 and 13.5 kb, to form a line of radioactivity migrating between mT and a position between dT and dE. If a nick occurred on the growing strand (Fig. 9, cut ii), it would generate a 13.5-kb strand and a strand between 0 and 4.5 kb (i.e., a line of radioactivity below and between h and mE). However, none of these products were detected in vitro or in vivo, suggesting that a single cut in the dimer bridge is not a feature of AAV DNA replication until after dT synthesis is complete. An alternative possibility is that molecules that are singly nicked in the dimer bridge are so unstable that a second nick on the opposite strand is immediately generated, but this possibility would produce a 4.5-kb strand, a 9-kb strand, and a 0- to 4.5-kb strand. Although a 4.5-kb band (line between h and mE) and a faint 9-kb band (line between mT and dE) were both seen, nothing corresponding to a 0- to 4.5-kb band was seen between mE and h. Finally, if terminal resolution occurred on the covalently closed hairpin end formed shortly after replication of an mT molecule was initiated (Fig. 9, cut iii), such intermediates would dissociate into 0- to 9-kb strands and 9-kb strands. However, the 0- to 9-kb strands would produce a line between dE and a position well below mE, and no such line was seen. By a similar argument, there appears to have been no terminal resolution until replication from mE molecules had been completed (Fig. 9, mE to dE); otherwise there would have been a line of radioactivity between mE and h from 0 to 4.5 kb in the alkaline dimension. Taken together, these data suggested that resolution of a dimer bridge or a hairpinned end occurred predominantly after replication of the entire DNA molecule had been completed. How the coordination of terminal resolution and strand elongation is accomplished is not clear.
(iii) Rep68 appears to be more efficient in nicking the dimer bridge.
In previous experiments, a difference between Rep78 and Rep68 was observed in the in vitro DNA replication assay (39). Rep78 made significantly higher levels of dimer RF species in vitro than are normally seen in vivo, and Rep68 made hardly any dimer intermediate. This was confirmed by the data in Fig. 8, where it is clear that Rep68 generated significantly less dimer RF species than Rep78. Similar observations have been made in vivo by Samulski et al. (43a). There are at least two possible explanations for the difference between Rep78 and Rep68. The first is that Rep68 is incapable of synthesizing dimers; the other is that it synthesizes dimers but processes the dimer bridge as well as hairpinned ends more rapidly than Rep78, so that dimers are short lived. Inspection of the 2D patterns obtained with Rep68 and Rep78 supports the latter explanation (Fig. 7). The fact that both Rep78 and Rep68 can process dimer bridges at some level was demonstrated clearly by the spot marked h, which represents a monomer single-stranded 4.5-kb species that is generated in alkali from dimer RF molecules. However, the lines of intermediates between mE and dE and between mT and dT were of similar intensity with the two Rep proteins. This result indicated that Rep68 was able to initiate dimer duplex DNA synthesis via the same pathway as Rep78 and to approximately the same extent but was more efficient in processing dimers to monomers and in resolving normal hairpinned ends. Inspection of the single-stranded species present in the dimer duplex pool supported this conclusion. First, h molecules, which are single-stranded 4.5-kb DNA molecules present in the dimer duplex RF population, were a much higher proportion of the DNA in the dimer duplex pool when Rep68 was used. Second, the total amount of dimer duplex RF species was less in the Rep68 reaction, suggesting that monomer species on the way to becoming dimers were likely to be resolved just prior to or shortly after completion of dimer synthesis.
(iv) No evidence of discontinuous synthesis.
Finally, we note that we could detect no evidence of discontinuous synthesis by 2D gel analysis. If de novo priming by pol α-primase and the synthesis of Okazaki fragments accounted for a significant portion of AAV DNA synthesis, then we would have expected to see ssDNA fragments shorter than 2 kb. In fact, very little radioactivity was detected in this region (Fig. 7). This implied that pol α was not likely to account for much of the synthesis in the in vitro AAV DNA replication reaction.
DISCUSSION
Uninfected HeLa cell cytoplasmic extracts.
The AAV in vitro replication assay that we previously described (39) is dependent on the addition of one of two larger Rep proteins encoded by AAV and proteins provided by Ad-infected HeLa cell extracts. Little if any AAV-specific DNA replication occurred when uninfected extracts were used (39). Here we report that the difference between uninfected and Ad-infected extracts could be diminished if HeLa cells are harvested after continuous high-density growth. The extracts made from this preparation of HeLa cells were essentially as active as Ad-infected HeLa cell extracts for supporting AAV DNA replication in vitro. The mechanism of this effect is currently unknown.
As mentioned earlier, it is generally believed that none of the Ad genes is directly involved in AAV DNA replication (1, 38). Coinfection with Ad seems to activate or induce a cellular factor(s) that is missing or inactive in a normal cell cycle as well as increasing the expression of the AAV rep gene. However, other experiments suggest that this is possible without Ad coinfection. It has been shown that some cells can become partially permissive for AAV DNA replication in vivo if the cells are transformed with either a viral or a cellular oncogene and further treated with reagents that transiently arrest cellular DNA synthesis (hydroxyurea, carcinogens, heat shock, or UV light) (44, 66–68). Presumably, the use of high cell densities described here activates similar pathways required for AAV DNA replication. Our hope is that the identification of the cellular factors in these kinds of activated extracts and their comparison with extracts prepared from Ad-infected cells will lead to the identification of the key differences in viral DNA replication under helper-dependent and helper-independent conditions. These differences are expected to be important for understanding how AAV chooses whether to establish a persistent latent infection or to undergo productive viral replication.
Identification of cellular factors from uninfected cells necessary for AAV DNA replication.
Our primary approach to identifying the cellular factors that are necessary for AAV DNA replication has been to fractionate the crude HeLa cell extract and to reconstitute AAV DNA synthesis in vitro, using purified viral and cellular proteins. Since much of what we know about cellular replication enzymes has come from the in vitro studies of SV40 DNA replication, we adopted for our initial studies the fractionation scheme of Stillman and colleagues (12, 54, 57) (Fig. 5). Fractionation of the crude extract by phosphocellulose chromatography showed that a 0.2 to 1.0 M NaCl fraction (fraction II) could successfully reconstitute AAV DNA synthesis to levels comparable to those seen with the crude extract provided that it was supplemented with purified RPA and PCNA (as well as purified Rep78 or Rep68). The requirement for RPA was also supported by antibody inhibition studies which demonstrated that monoclonal antibodies to RPA could inhibit AAV DNA synthesis up to 90%. These experiments clearly demonstrated that both RPA and PCNA were essential components for AAV DNA replication.
Two lines of evidence indicated that AAV DNA replication also required the cellular factor RFC. First, fractionation by phosphocellulose chromatography allowed us to reconstitute AAV DNA synthesis with a 0.2 to 0.33 M NaCl eluate (fraction IIA) when it was supplemented with purified RPA, PCNA, and RFC (or with fraction IIC which contains RFC). Second, monoclonal antibody inhibition experiments demonstrated that a monoclonal antibody directed against RFC inhibits AAV DNA synthesis.
The level of DNA synthesis achieved in the reconstituted reactions containing RFC, RPA, and PCNA were significantly lower than those seen with the starting crude extracts. The reason for this was not clear. It is possible that the levels of cellular factors used in these studies were not optimal. Alternatively, we may have purified away a factor that is not present in fraction IIA and that stimulates AAV DNA synthesis (see below). Recently, Christensen et al. (8) identified a cellular factor called parvovirus initiation factor which was present in fraction I and required for nicking of the dimer bridge and initiation of DNA synthesis from the 3′ origin in the related parvovirus minute virus of mice. Conceivably, this or a similar factor is needed for AAV as well. Christensen et al. (8) also demonstrated that RPA and PCNA were required for minute virus of mice DNA synthesis.
The requirement for RFC and PCNA is not surprising in light of previous studies of the SV40 DNA replication system. RFC has been shown to be an ATP-dependent primer recognition complex which assembles a PCNA complex at a 3′-OH end of a primer-template (55). PCNA in turn is an accessory protein which stimulates pol δ by making it more processive (52). Together, RPA, PCNA, RFC, and pol δ are the essential proteins for leading-strand synthesis during SV40 DNA replication (60). This finding, coupled with the fact that AAV DNA replication occurs entirely by leading-strand synthesis, suggests that pol δ is most likely the DNA polymerase required for AAV DNA replication.
Fraction IIA contains all three of the known eukaryotic DNA polymerases (α, δ, and ɛ) involved in chromosome DNA replication. Our chemical inhibition experiments suggest that either pol δ or pol ɛ is responsible for AAV DNA replication. DNA synthesis is not affected by concentrations of the inhibitors BuPdGTP and BuAdATP, to which pol α is sensitive. Furthermore, the in vitro reaction was insensitive to inhibition by monoclonal antibodies directed against pol α. Finally, the fact that we were unable to find any evidence for discontinuous synthesis in the in vitro reaction by 2D gel electrophoresis also suggests that pol α is not required for AAV DNA synthesis. The chemical inhibition experiments also suggested that two other DNA polymerases that might have been present in our cell extracts, specifically pol β and pol γ, are probably not involved. Taken together, our inhibition experiments suggest that either pol δ or pol ɛ is the DNA polymerase responsible for AAV DNA replication.
Pol ɛ is similar to pol δ in that it has a tightly associated 3′-5′ exonuclease activity and has a chemical inhibition spectrum similar to that of pol δ. However, the two DNA polymerases are structurally and functionally different, and both appear to be required for cellular DNA replication (51). The most striking difference between them is that pol δ is highly processive only in the presence PCNA, whereas DNA pol ɛ is intrinsically processive. Nevertheless, it has been shown recently that pol ɛ can also be stimulated by the presence of PCNA (30). Therefore, we cannot distinguish between pol δ and pol ɛ as the primary DNA polymerase required for AAV DNA replication. We note that HeLa pol ɛ has been shown to be resistant to 500 μM ddTTP whereas pol δ from HeLa cells is relatively sensitive (51). The relative sensitivity of AAV DNA replication to 500 μM ddTTP is thus consistent with the requirement of DNA pol δ.
Additional unidentified factors are required for AAV DNA synthesis.
An interesting finding from our in vitro reconstitution studies was that the addition of the three DNA polymerases (α, δ, and ɛ), either alone or in various combinations, could not substitute for fraction IIA. Studies of SV40 DNA replication have shown that fraction IIA provides pol α and pol δ to the reaction (36). This suggested that an additional unknown protein(s) was present in fraction IIA that was required for AAV DNA replication but was not needed for SV40 DNA replication. We note that a similar observation has recently been made for papillomavirus in vitro DNA replication (35).
The possible involvement of an unknown factor for AAV DNA replication is not entirely surprising. If the combination of Rep, RPA, PCNA, and DNA pol δ were all that were needed for AAV DNA replication, it would be hard to explain why productive AAV DNA replication needs the coinfection of a helper virus. We will present evidence elsewhere (33a) that at least one of the additional factors that is present in Ad-infected cells is the Ad-encoded DNA binding protein. Reconstitution experiments done with fractionated Ad-infected extracts suggest that the requirement for RPA is replaced by this protein. However, even when this is done, there is still a need for additional, as yet uncharacterized factors present in fraction IIA.
Rep78 versus Rep68.
Finally, we previously reported that Rep68 generates significantly lower levels of dimer RF than Rep78 (39). The results of the 2D gel analysis reported here suggest that this is due to Rep68 being more efficient in nicking the dimer bridge. The 2D analysis also suggested a peculiar property of AAV DNA replication both in vivo and in vitro. Resolution of termini that were undergoing elongation seemed to occur only after elongation had been completed. This implied some type of coordinate interaction between the replication complex and the TR that had been used to prime elongation. The mechanism by which this is accomplished is not clear, but this may be important if packaging of newly displaced strands and elongation are biochemically linked.
ACKNOWLEDGMENTS
This work was supported by grant RO1 GM35723 from the National Institute of General Medical Sciences (N.M.) and by grant CA 13106 from the National Cancer Institute (B.S.).
REFERENCES
- 1.Berns K I. Parvovirus replication. Microbiol Rev. 1990;54:316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berns K I, Hauswirth W W, Fife K H, Lusby E. Adeno-associated virus DNA replication. Cold Spring Harbor Symp Quant Biol. 1979;43(part 2):781–787. doi: 10.1101/sqb.1979.043.01.085. [DOI] [PubMed] [Google Scholar]
- 3.Bunz F, Kobayashi R, Stillman B. cDNAs encoding the large subunit of human replication factor C. Proc Natl Acad Sci USA. 1993;90:11014–11018. doi: 10.1073/pnas.90.23.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrnes J J. Differential inhibitors of DNA polymerases alpha and delta. Biochem Biophys Res Commun. 1985;132:628–634. doi: 10.1016/0006-291x(85)91179-9. [DOI] [PubMed] [Google Scholar]
- 5.Cheung A K, Hoggan M D, Hauswirth W W, Berns K I. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J Virol. 1980;33:739–748. doi: 10.1128/jvi.33.2.739-748.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiorini J A, Wiener S M, Owens R A, Kyostio S R, Kotin R M, Safer B. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J Virol. 1994;68:7448–7457. doi: 10.1128/jvi.68.11.7448-7457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiorini J A, Yang L, Safer B, Kotin R M. Determination of adeno-associated virus Rep68 and Rep78 binding sites by random sequence oligonucleotide selection. J Virol. 1995;69:7334–7338. doi: 10.1128/jvi.69.11.7334-7338.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen J, Cotmore S F, Tattersall P. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dresler S L, Frattini M G. Analysis of butylphenyl-guanine, butylphenyl-deoxyguanosine, and butylphenyl-deoxyguanosine triphosphate inhibition of DNA replication and ultraviolet-induced DNA repair synthesis using permeable human fibroblasts. Biochem Pharmacol. 1988;37:1033–1037. doi: 10.1016/0006-2952(88)90506-0. [DOI] [PubMed] [Google Scholar]
- 10.Dresler S L, Frattini M G. DNA replication and UV-induced DNA repair synthesis in human fibroblasts are much less sensitive than DNA polymerase alpha to inhibition by butylphenyl-deoxyguanosine triphosphate. Nucleic Acids Res. 1986;14:7093–7102. doi: 10.1093/nar/14.17.7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edenberg H J, Anderson S, DePamphilis M L. Involvement of DNA polymerase alpha in simian virus 40 DNA replication. J Biol Chem. 1978;253:3273–3280. [PubMed] [Google Scholar]
- 12.Fairman M P, Stillman B. Cellular factors required for multiple stages of SV40 replication in vitro. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol Cell Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goscin L P, Byrnes J J. DNA polymerase delta: one polypeptide, two activities. Biochemistry. 1982;21:2513–2518. doi: 10.1021/bi00539a034. [DOI] [PubMed] [Google Scholar]
- 15.Harlow E, Crawford L V, Pim D C, Williamson N M. Monoclonal antibodies specific for the simian virus 40 tumor antigens. J Virol. 1981;78:488–499. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauswirth W W, Berns K I. Adeno-associated virus DNA replication: nonunit-length molecules. Virology. 1979;93:57–68. doi: 10.1016/0042-6822(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 17.Hauswirth W W, Berns K I. Origin and termination of adeno-associated virus DNA replication. Virology. 1977;78:488–499. doi: 10.1016/0042-6822(77)90125-8. [DOI] [PubMed] [Google Scholar]
- 18.Hong G, Ward P, Berns K I. In vitro replication of adeno-associated virus DNA. Proc Natl Acad Sci USA. 1992;89:4673–4677. doi: 10.1073/pnas.89.10.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong G, Ward P, Berns K I. Intermediates of adeno-associated virus DNA replication in vitro. J Virol. 1994;68:2011–2015. doi: 10.1128/jvi.68.3.2011-2015.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikegami S, Taguchi T, Ohashi M, Oguro M, Nagano H, Mano Y. Aphidocolin prevents mitotic cell division by interfering with the activity of DNA polymerase α. Nature. 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- 21.Im D S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 22.Khan N N, Wright G E, Dudycz L W, Brown N C. Elucidation of the mechanism of selective inhibition of mammalian DNA polymerase alpha by 2-butylanilinopurines: development and characterization of 2-(p-n-butylanilino)adenine and its deoxyribonucleotides. Nucleic Acids Res. 1985;13:7093–7102. doi: 10.1093/nar/13.17.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laughlin C A, Cardellichio C B, Coon H C. Latent infection of KB cells with adeno-associated virus type 2. J Virol. 1986;60:515–524. doi: 10.1128/jvi.60.2.515-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee M Y, Tan C K, Downey K M, So A G. Further studies on calf thymus DNA polymerase delta purified to homogeneity by a new procedure. Biochemistry. 1984;23:1906–1913. doi: 10.1021/bi00304a003. [DOI] [PubMed] [Google Scholar]
- 25.Lee M Y, Toomey N L, Wright G E. Differential inhibition of human placental DNA polymerases delta and alpha by BuPdGTP and BuAdATP. Nucleic Acids Res. 1985;13:8623–8630. doi: 10.1093/nar/13.23.8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M Y W T, Tan C-K, Downery K M, So A G. Structural and functional properties of calf thymus DNA polymerase δ. Prog Nucleic Acids Res Mol Biol. 1981;26:83–96. doi: 10.1016/s0079-6603(08)60396-7. [DOI] [PubMed] [Google Scholar]
- 27.Liu L F, Miller K G. Eukaryotic DNA topoisomerases: two forms of type I DNA topoisomerases from HeLa cell nuclei. Proc Natl Acad Sci USA. 1981;78:3487–3491. doi: 10.1073/pnas.78.6.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lusby E, Bohenzky R, Berns K I. Inverted terminal repetition in adeno-associated virus DNA: independence of the orientation at either end of the genome. J Virol. 1981;37:1083–1086. doi: 10.1128/jvi.37.3.1083-1086.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lusby E, Fife K H, Berns K I. Nucleotide sequence of the inverted terminal repetition in adeno-associated virus DNA. J Virol. 1980;34:402–409. doi: 10.1128/jvi.34.2.402-409.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maga G, Hubscher U. DNA polymerase epsilon interacts with proliferating cell nuclear antigen in primer recognition and elongation. Biochemistry. 1995;34:891–901. doi: 10.1021/bi00003a023. [DOI] [PubMed] [Google Scholar]
- 31.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 32.McCarty D M, Pereira D J, Zolotukhin I, Zhou X, Ryan J H, Muzyczka N. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J Virol. 1994;68:4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarty D M, Ryan J H, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.McDonald, W. F., and N. Muzyczka. Submitted for publication.
- 34.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melendy T, Sedman J, Stenlund A. Cellular factors required for papillomavirus DNA replication. J Virol. 1995;69:7857–7867. doi: 10.1128/jvi.69.12.7857-7867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melendy T, Stillman B. Purification of DNA polymerase delta as an essential simian virus 40 DNA replication factor. J Biol Chem. 1991;266:1942–1949. [PubMed] [Google Scholar]
- 37.Murakami Y, Wobbe C R, Weissbach L, Dean F B, Hurwitz J. Role of DNA polymerase alpha and DNA primase in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1986;83:2869–2873. doi: 10.1073/pnas.83.9.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 39.Ni T H, Zhou X, McCarty D M, Zolotukhin I, Muzyczka N. In vitro replication of adeno-associated virus DNA. J Virol. 1994;68:1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogata K, Kurki P, Celis J E, Nakamura R M, Tan E M. Monoclonal antibodies to a nuclear protein (PCNA/cyclin) associated with DNA replication. Exp Cell Res. 1987;168:475–486. doi: 10.1016/0014-4827(87)90020-6. [DOI] [PubMed] [Google Scholar]
- 41.Prelich G, Stillman B. Coordinated leading and lagging strand synthesis during SV40 DNA replication in vitro requires PCNA. Cell. 1988;53:117–126. doi: 10.1016/0092-8674(88)90493-x. [DOI] [PubMed] [Google Scholar]
- 42.Ryan J, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1995;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samulski R J, Berns K I, Tan M, Muzyczka N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci USA. 1982;79:2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Samulski, R. J., et al. Personal communication.
- 44.Schlehofer J R, Ehrbar M, zur Hausen H. Vaccinia virus, herpes simplex virus, and carcinogens induce DNA amplification in a human cell line and support replication of a helpervirus dependent parvovirus. Virology. 1986;152:110–117. doi: 10.1016/0042-6822(86)90376-4. [DOI] [PubMed] [Google Scholar]
- 45.Schomburg U, Grosse F. Purification and characterization of DNA topoisomerase II from calf thymus associated with polypeptides of 175 and 150 kDa. Eur J Biochem. 1986;160:451–457. doi: 10.1111/j.1432-1033.1986.tb10061.x. [DOI] [PubMed] [Google Scholar]
- 46.Snyder R O, Im D S, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) Rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snyder R O, Im D S, Ni T, Xiao X, Samulski R J, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snyder R O, Samulski R J, Muzyczka N. In vitro resolution of covalently joined AAV chromosome ends. Cell. 1990;60:105–113. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- 49.Straus S E, Sebring E D, Rose J A. Concatemers of alternating plus and minus strands are intermediates in adenovirus-associated virus DNA synthesis. Proc Natl Acad Sci USA. 1976;73:742–746. doi: 10.1073/pnas.73.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Syvaoja J, Linn S. Characterization of a large form of DNA polymerase delta from HeLa cells that is insensitive to proliferating cell nuclear antigen. J Biol Chem. 1989;264:2489–2497. [PubMed] [Google Scholar]
- 51.Syvaoja J, Suomensaari S, Nishida C, Goldsmith J S, Chui G S, Jain S, Linn S. DNA polymerases alpha, delta, and epsilon: three distinct enzymes from HeLa cells. Proc Natl Acad Sci USA. 1990;87:6664–6668. doi: 10.1073/pnas.87.17.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan C K, Castillo C, So A G, Downey K M. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. J Biol Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- 53.Tanaka S, Hu S-Z, Wang T S, Korn D. Preparation and preliminary characterization of monoclonal antibodies against human DNA polymerase alpha. J Biol Chem. 1982;257:8386–8390. [PubMed] [Google Scholar]
- 54.Tsurimoto T, Fairman M P, Stillman B. Simian virus 40 DNA replication in vitro: identification of multiple stages of initiation. Mol Cell Biol. 1989;9:3839–3849. doi: 10.1128/mcb.9.9.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsurimoto T, Stillman B. Functions of replication factor C and proliferating-cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc Natl Acad Sci USA. 1990;87:1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsurimoto T, Stillman B. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, alpha and delta. EMBO J. 1989;8:3883–3889. doi: 10.1002/j.1460-2075.1989.tb08567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsurimoto T, Stillman B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol Cell Biol. 1989;9:3839–3849. doi: 10.1128/mcb.9.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 59.Tullis G, Schoborg R V, Pintel D J. Characterization of the temporal accumulation of minute virus of mice replicative intermediates. J Gen Virol. 1994;75:1633–1646. doi: 10.1099/0022-1317-75-7-1633. [DOI] [PubMed] [Google Scholar]
- 60.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 61.Wahl A F, Crute J J, Sabatino R D, Bodner J B, Marraccino R L, Harwell L W, Lord E M, Bambara R A. Properties of two forms of DNA polymerase delta from calf thymus. Biochemistry. 1986;25:7821–7827. doi: 10.1021/bi00372a006. [DOI] [PubMed] [Google Scholar]
- 62.Waqar M A, Evans M J, Huberman J A. Effect of 2′,3′-dideoxythymidine-5′-triphosphate on HeLa cell in vitro DNA synthesis: evidence that DNA polymerase alpha is the only polymerase required for cellular DNA replication. Nucleic Acids Res. 1978;5:1933–1946. doi: 10.1093/nar/5.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward P, Berns K I. In vitro replication of adeno-associated virus DNA: enhancement by extracts from adenovirus-infected HeLa cells. J Virol. 1996;70:4495–4501. doi: 10.1128/jvi.70.7.4495-4501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weitzman M D, Kyostio S R, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wright G E, Dudycz L W. Synthesis and characterization of N2-(p-n-butylphenyl)-2′-deoxyguanosine and its 5′-triphosphate and their inhibition of HeLa DNA polymerase alpha. J Med Chem. 1984;27:175–181. doi: 10.1021/jm00368a012. [DOI] [PubMed] [Google Scholar]
- 66.Yakobson B, Hyrnko T A, Peak M J, Winocour E. Replication of adeno-associated virus in cells irradiated with UV light at 254 nm. J Virol. 1989;63:1023–1030. doi: 10.1128/jvi.63.3.1023-1030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yakobson B, Koch T, Winocour E. Replication of adeno-associated virus in synchronized cells without the addition of a helper virus. J Virol. 1987;61:972–981. doi: 10.1128/jvi.61.4.972-981.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yalkinoglu A O, Heilbronn R, Burkle A, Schlehofer J R, zur Hausen H. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. Cancer Res. 1988;48:3123–3129. [PubMed] [Google Scholar]